Abstract
Lakes at high altitude and latitude are typically unproductive ecosystems where external factors outweigh the relative importance of in‐lake processes, making them ideal sentinels of climate change. Climate change is inducing upward vegetation shifts at high altitude and latitude regions that translate into changes in the pools of soil organic matter. Upon mobilization, this allochthonous organic matter may rapidly alter the composition and function of lake bacterial communities. Here, we experimentally simulate this potential climate‐change effect by exposing bacterioplankton of two lakes located above the treeline, one in the Alps and one in the subarctic region, to soil organic matter from below and above the treeline. Changes in bacterial community composition, diversity and function were followed for 72 h. In the subarctic lake, soil organic matter from below the treeline reduced bulk and taxon‐specific phosphorus uptake, indicating that bacterial phosphorus limitation was alleviated compared to organic matter from above the treeline. These effects were less pronounced in the alpine lake, suggesting that soil properties (phosphorus and dissolved organic carbon availability) and water temperature further shaped the magnitude of response. The rapid bacterial succession observed in both lakes indicates that certain taxa directly benefited from soil sources. Accordingly, the substrate uptake profiles of initially rare bacteria (copiotrophs) indicated that they are one of the main actors cycling soil‐derived carbon and phosphorus. Our work suggests that climate‐induced changes in soil characteristics affect bacterioplankton community structure and function, and in turn, the cycling of carbon and phosphorus in high altitude and latitude aquatic ecosystems.
Keywords: allochthonous organic carbon, bacterial production, dissolved organic matter, diversity, heterotrophic, phosphorus limitation, terrestrial vegetation, treeline
Introduction
Climate change alters ecosystem properties and thereby threatens biodiversity and ecosystem functioning. Lake ecosystems integrate changes in the surrounding landscape and atmosphere, acting as sentinels of climate change (Sommaruga‐Wögrath et al., 1997; Carpenter et al., 2007; Pham et al., 2008; Williamson et al., 2008; Adrian et al., 2009). Several physical properties of lakes tend to respond directly to climatic forcing, such as water temperature and the duration of ice cover (Adrian et al., 2009; O'Reilly et al., 2015). Chemical lake characteristics often integrate catchment processes such as rock weathering and terrestrial primary productivity. Ecosystem properties, such as primary and bacterial secondary productivity, diversity of taxa, and taxa turnover integrate both physical and chemical alterations over time (Wagner & Adrian, 2009).
Lakes at high altitude and latitude are particularly vulnerable to climate change (Woodward et al., 2010). These remote ecosystems are typically small, oligotrophic, and ice‐covered for several months, amplifying the direct and indirect effects of climate change (Grimm et al., 2013). Compared to an expected global mean surface temperature increase of 1.0–3.7 °C until the end of the 21st century, surface temperatures in alpine and subarctic regions are expected to increase by ~3–6 °C (Beniston, 2005) and 2.2–8.3 °C (Collins et al., 2013), respectively. Moreover, snowpack duration is projected to decrease (Räisänen & Eklund, 2011; Steger et al., 2012), whereas the frequency and magnitude of extreme precipitation or drought events will increase (Fischer & Knutti, 2015). Finally, climate change induces range expansions of many plant and animal taxa at their altitudinal and latitudinal extremes (Grabherr et al., 1994; Lenoir et al., 2008; Harsch et al., 2009). Such shifts in local plant cover and plant community composition (Sturm et al., 2001; Parmesan & Yohe, 2003; Hinzman et al., 2005; Lenoir et al., 2008; Pearson et al., 2013), and their consequences on soil organic matter content and composition (Reuss et al., 2010; Classen et al., 2015), are well documented. However, observational and experimental knowledge on the effects of the treeline shift and the concomitant changes in soil chemical characteristics on lake ecosystems at high altitude and latitude regions is missing.
Heterotrophic bacteria play a central role in the cycling of terrestrial organic carbon and nutrients in inland waters (Jones, 1992; Tranvik, 1992; Bergström & Jansson, 2000). The quantity of carbon incorporated to bacterial biomass or released through respiration is determined by the bioavailability of organic matter and the ability of microorganisms to degrade this heterogeneous mixture (Del Giorgio & Cole, 1998; Guillemette & Del Giorgio, 2011). Extensive work on the quantity and quality of terrestrial organic matter in lake ecosystems revealed that the degree of afforestation or catchment composition controls the characteristics of the soil organic matter sources and thus, the rates of carbon processing (e.g., Bastidas Navarro et al. 2014; Berggren et al. 2010; Forsström et al. 2013; Judd et al. 2006; Kritzberg et al. 2004; Roiha et al. 2011). In turn, bacterial communities have been shown to respond quickly to variation in substrate availability and concentration, often favouring opportunistic bacteria such as members of Betaproteobacteria (Burkert et al., 2003; Šimek et al., 2005; Hornák et al., 2006; Posch et al., 2007) or Bacteroidetes (Cottrell & Kirchman, 2000; Battin et al., 2001; Eiler et al., 2003; Zeder et al., 2009). Besides the critical role of heterotrophic bacteria in processing soil‐derived organic matter in inland waters, knowledge on covarying factors, such as the release from nutrient limitation during pulses of soil run off in high altitude and latitude lakes remains limited (Pérez & Sommaruga, 2006).
Here, our aim was to assess how bacterial communities in high altitude and latitude lakes respond to alterations in soil organic matter composition as a response to an upward shift in vegetation. The experiments were conducted as pulse additions of soil‐derived organic matter from the local catchment (i.e., above treeline) compared to additions of soil extracts from a nearby catchment below the treeline. We hypothesized that lake bacteria will respond rapidly (within hours) to organic matter additions and that different taxa will prevail depending on the origin of the soil source. We expected lake communities to be readily adapted to inputs from local catchments, thus initially abundant taxa being able to utilize these soil sources. In contrast, addition of soil extracts from below the treeline may stimulate the growth of initially rare bacterial taxa, potentially causing shifts in community structure. Moreover, we hypothesized that soil extracts will relieve phosphorus limitation and may lead to a reduction in bacterial diversity with fast‐growing taxa dominating bacterial community composition.
Materials and methods
In summer 2014, an in situ experiment was done in one alpine and in one subarctic oligotrophic lake, located above and north of the current treeline, respectively. In both experiments, the in situ microbial community was incubated for 72 h and amended with two different soil extracts, namely, one from the lake own catchment above the treeline and one from a nearby site located below the treeline.
Study sites
The subarctic, oligotrophic lake Saanajärvi (SAA; Fig. S1a) is located at 679 m a.s.l. in north‐western Finland (69°05′N 20°87′E). This medium‐sized lake (70 ha) has a steep shore, a maximum depth of 24 m, and is ice‐covered for up to 9 months per year. The catchment (461 ha) mainly comprises steep mountain slopes and consists of bare rocks (limestone and calcareous dolomite) and a thin soil layer. Plant diversity is rich and dominated by dwarf shrubs, lichens and subarctic flowering plants. More information on SAA and the Kilpisjävi region can be found in Sorvari (2001). The alpine lake Gossenköllesee (GKS; Fig. S1b) is located at 2417 m a.s.l. in the Austrian Alps (47°13′N 11°01′E). Gossenköllesee is small (1.7 ha) with a maximum depth of 9.9 m and usually ice‐covered for up to 7 months per year. The catchment (30 ha) comprises crystalline bedrock, scarce vegetation and a poor soil layer.
Soil extract preparation
For each experiment, soil extracts from the local catchment (local soil) and from the catchment of a lake located below the treeline (hereafter, foreign soil) were prepared according to Kalbitz et al. (2003). Foreign soil extracts were obtained from the lake catchments of Bajit lvgujärvi, Norway (69°21′N 20°19′E) and Piburgersee, Austria (47°11′N 10°53′E) (see Table S1 for key physicochemical parameters). Soils were sampled from six sites in each catchment. The uppermost soil layer, stones, roots and large organic debris were removed, and in total, 4 kg of soil was soaked in 4 L Milli‐Q water for 18–19 h at 4 °C. Then, the water–soil mixture was passed through a cotton cloth and centrifuged at 2 500–10 000 g for 20–30 min. The supernatant was sequentially filtered through 3‐μm (Polycarbonate, Millipore, Vienna, Austria), 1.2‐μm (GF/C, Whatman, GE Healthcare, Vienna, Austria), 0.7‐μm (GF/F, Whatman, GE Healthcare), 0.5‐μm (GF‐5, Marchery Nagel, Düren, Germany), 0.45‐μm (Cellulose ester, Millipore) and 0.2‐μm (Cellulose acetate, Sartorius, Vienna, Austria) filters to obtain a bacteria‐free soil extract (cross‐checked by DAPI‐staining and epifluorescence microscopy). Nutrient and DOC concentrations in the soil extracts were analysed as described later on.
Experimental set‐up
Water was sampled from 1‐m depth from a boat anchored above the deepest point of the lakes and filtered through 1.0‐μm (Polycap 75 filter column, Whatman, GE Healthcare) using a peristaltic pump (Model Vampire, Bürkle, Bad Bellingen, Germany) to obtain the bacterioplankton fraction. The filtered water (10 L) was collected in HCl‐washed and Milli‐Q‐rinsed polycarbonate bottles for SAA, and in polyethylene bottles for GKS. Triplicates were prepared for both soil extract additions and the control (no soil extract addition). The bottles containing the filtrates were allowed to acclimatize for 24–40 h in the lakes prior to the soil extract additions. Between 213 and 438 mL of soil extracts were added to 10 L of filtered lake water to increase the DOC concentration by a factor of 3. The bottles were placed in wooden racks or steel mesh and incubated at the northern lake shore in SAA and at the eastern lake shore in GKS.
The bottles (n = 9) were sampled at the beginning of the experiment (t = 0), after 6, 24 and 72 h, thus reducing the end volume to ca. 3 L. The water samples were collected into HCl‐ and Milli‐Q‐rinsed polyethylene bottles and subsequently split to measure bulk uptake rates (33P‐orthophosphate, 33P‐ATP and 3H‐leucine), taxon‐specific uptake using the same substrates (MAR‐CARD‐FISH; only in SAA) and basic physicochemical parameters. Furthermore, 500 mL were filtered on 0.2‐μm filter (GPWP, Millipore) and stored at −80 °C for molecular analyses, and 10 mL subsamples were fixed with 3.7% formaldehyde (final concentration) and stored at 4 °C to assess bacterial abundance by flow cytometry.
DOC and nutrient analysis
For SAA, water chemical analyses were run by the METLA Institute in Rovaniemi, Finland. Briefly, samples for total dissolved phosphorus (TDP) and inorganic phosphate (Pi‐P) analyses were filtered through 0.4‐μm (Track‐Etched polycarbonate, Nuclepore) and analysed on a Lachat QuikChem 8 000 automated ion analyzer. Dissolved organic carbon (DOC) and total dissolved nitrogen (TDN) concentrations of the soil extracts and SAA lake water samples were determined on a Shimadzu TOC‐LCSH/CSN, while samples taken during the experiment in SAA and the GKS experiment were filtered through two precombusted (450 °C for 4 h) filters (GF/F, Whatman, GE Healthcare), adjusted to pH 2 (HCl) and analysed on a Shimadzu TOC‐VCPH series. Total dissolved phosphorus concentrations for the GKS experiment were estimated by the molybdenum blue method (Vogler, 1966). Dissolved organic matter (DOM) was characterized by means of absorbance and fluorescence spectroscopy, and details can be found in the supplementary information.
DNA extraction and 16S rRNA gene amplicon sequencing
DNA was extracted using a commercial kit (PowerWater DNA Isolation Kit, MOBIO, Carlsbad, CA, USA), and samples were shipped to LGC Genomics (Germany) for library preparation and 300‐bp paired‐end sequencing (Miseq V3, Illumina, San Diego, CA, USA). There, the V2–V3 region of the bacterial 16S rRNA gene was PCR‐amplified using barcoded (7 bp) primers (341F‐785R) (Klindworth et al., 2013). The PCR products were quality‐screened and equimolar‐mixed. A total of 20.1 million reads were obtained from a single MiSeq run and de‐multiplexed using bcl2fastq 1.8.4 software (Illumina). Reads were sorted allowing for no mismatch in barcode sequences, and primer and barcodes were clipped from the sequences. Forward and reverse reads were merged, and the sequence quality was controlled using mothur (Kozich et al., 2013). Sequences with more than two ambiguous bases, more than eight homopolymers and shorter than 400 bp were discarded. The sequences were aligned against a reference database (SILVA Release 123), and sequences with <4 different bases were merged using mothur's pre.cluster algorithm. Chimeric sequences were identified using mothur's UCHIME implementation. Finally, singletons and sequences not classified as bacteria by mothur's Naïve Bayesian classifier were also removed from the data set. In total, 1 864 746 sequences remained after quality screening. Sequences were clustered into operational taxonomic units (OTUs) based on the neighbour joining method of mothur's cluster algorithm using a 97% similarity cut‐off. As samples obtained in our experiments had different numbers of sequences, the data set was rarefied to 10 000 sequences to allow for comparability of diversity estimates between samples. Samples with <10 000 sequences and samples which appeared to be outliers (i.e. two samples derived from GKS resembled more communities found in SAA than all other samples from GKS) were removed. As two of three replicates of the foreign soil treatment in GKS at time point 72 h were compromised, we removed this data point from downstream analyses. Sequences have been deposited at the Sequence Read Archive under accession number: SRP077041.
Incubations for microautoradiography (MAR)
For microautoradiography, 10‐mL samples were incubated at in situ temperatures with one of the following substrates (Perkin Elmer): 33P‐Pi (specific activity 155.8 Ci mg−1; final concentration 40 pM), 33P‐ATP as model compound for dissolved organic phosphorus (specific activity 3000 Ci mmol−1; final concentration 100 pM) and 3H‐leucine as indicator for bacterial production (specific activity 53 Ci mmol−1; final concentration 20 nm). Samples for 3H‐leucine uptake were incubated for 2 h, while the uptake of 33P‐Pi and 33P‐ATP was stopped after 3 h by adding formaldehyde (2% final conc.). The fixed samples were kept at 4 °C overnight and then filtered onto white 0.22‐μm polycarbonate filters (GTTP, Millipore). Filters were rinsed with 10 mL sterile filtered double‐distilled water and stored at −20 °C until further processing. The most dominant bacterial taxa in the lakes were targeted by catalysed‐reporter‐deposition‐fluorescence‐in‐situ‐hybridization (CARD‐FISH; Alphaproteobacteria, Betaproteobacteria, Bacteroidetes, and the AcI lineage of Actinobacteria). We also targeted important members within the Betaproteobacteria such as the R‐BT cluster (lineage of genus Limnohabitans). Information on filter preparation and processing for MAR and CARD‐FISH can be found in the supplementary information.
Substrate bulk uptake rates
Incubations for 33P‐Pi, 33P‐ATP and 3H‐leucine bulk uptake rates were performed as described for MAR. Additionally, one formaldehyde‐killed blank was fixed 15 min before adding the radioactive substrate. Incubations lasted for 45 min in the case of 33P‐Pi and 33P‐ATP, and for 1 h for 3H‐leucine and were terminated by adding formaldehyde. Samples were filtered onto white 0.22‐μm polycarbonate filters (Poretics). The 33P incubations were filtered within 10–60 min to minimize isotope leakage, and the samples for leucine uptake were extracted with trichloroacetic acid (5%) for 5 min and rinsed with the same solution. Filters were placed in scintillation vials with 5 mL of scintillation cocktail (Ready‐safe, Beckman Coulter, Brea, CA, USA). The radioactivity of the filters was assessed after 16 h on a scintillation counter (LS 6 000IC, Beckman Coulter).
Bacterial abundance
Bacterial abundance was assessed by flow cytometry according to Del Giorgio et al. (1996). Briefly, formaldehyde‐fixed cells were stained with 2.5 μm SYTO13 which binds to DNA and RNA. The cells were identified in plots of fluorescence at 520 nm vs. side scatter of a 488‐nm laser on a MoFlo Astrios (Beckman Coulter). Bacterial abundance was calculated from the ratio of cells to 1‐μm fluorescent reference beads (Sigma Aldrich, Vienna, Austria), which were counted under an epifluorescence microscope and adjusted for dilution due to the addition of fixative and dye.
Statistics
Multivariate statistical analyses such as analysis of similarity (ANOSIM) were performed using r (R Development Core Team, 2015) package ‘vegan’ (Oksanen et al., 2011), and phylogenetic diversity was estimated using package ‘picante’ (Kembel et al., 2010). A one‐way analysis of variance (anova) or a several sample repeated measure test were run on Past (Hammer et al., 2001) to detect whether soil extract additions caused significant differences in the proportions of cells taking up the substrates or in the bulk substrate uptake rates. Sample means were compared between treatments and time. When significant differences (P < 0.05) were found, a post hoc test (Tukey) was applied. Normal distribution of data was visually checked with histograms, normal probability plots and Shapiro–Wilk test. When not normally distributed, data was log‐transformed.
Results
Water chemistry and environmental parameters
Soil extract additions increased the dissolved organic carbon (DOC) concentration by a factor 2.5–3 in SAA, and by a factor 3 or 7 in GKS (Fig. 1). In the SAA experiment, DOC concentrations gradually decreased in all incubations, whereas in GKS, concentrations remained constant or slightly decreased (Fig. 1; foreign soil treatment). The soil extract additions increased the initial total dissolved phosphorus (TDP) concentrations (0.10 μm) to 0.41 ± 0.03 μm in SAA; and from 0.02 μm to 0.15 ± 0.03 μm in GKS (Fig. 1). In SAA, TDP concentrations increased slightly to 0.45 μm in the local soil treatment and decreased to 0.31 μm in the foreign soil treatment by the end of the experiment. In GKS, TDP concentrations remained rather constant during the experiment in all treatments (Fig. 1). Inorganic phosphate (Pi‐P) was only determined in the SAA experiment and decreased from 0.12 to 0.06 μm and 0.18 to 0.10 μm in the local and foreign soil treatments, respectively. Total dissolved nitrogen (TDN) concentrations were more than double in the soil treatments than in the control in SAA, and about one‐third higher in GKS. In all soil treatments, TDN concentrations decreased steadily until the end of the experiment (Fig. 1). The soil extracts changed the relative contribution of chromophoric DOM and of the terrestrial to microbial‐derived organic matter (Table S2). Lake water temperature increased from 13 to 18 °C during the experiment in SAA (Table S1), whereas water temperature remained low (i.e., between 10 and 11 °C) throughout the experiment in GKS.
Figure 1.
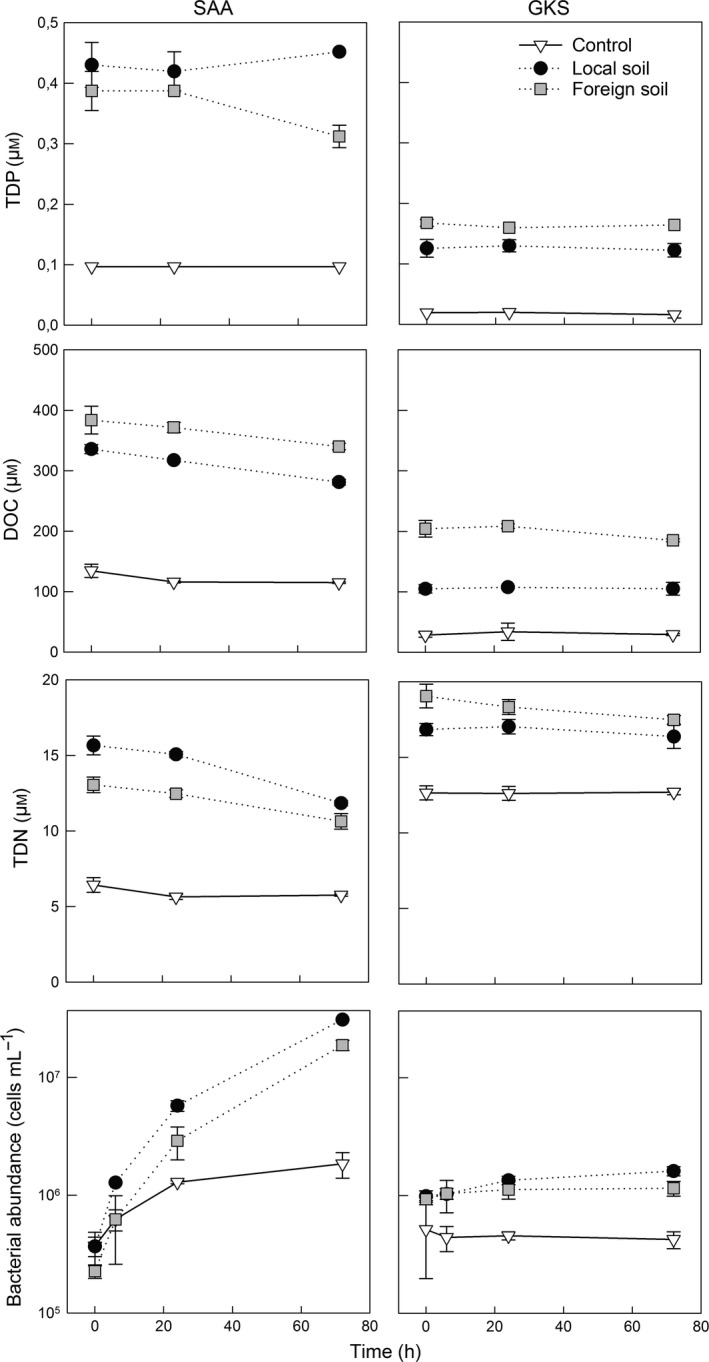
Temporal changes in chemical parameters and bacterial abundance. Different symbols indicate the control and the two treatments in lakes SAA and GKS for which total dissolved phosphorus (TDP), dissolved organic carbon (DOC), total dissolved nitrogen (TDN) and bacterial abundance were determined. Each data point represents the average of triplicate incubations. Error bars represent ± 1 SD, and in some cases are smaller than the symbol. Note that bacterial abundance is given on a logarithmic scale.
Bacterial community structure and succession
Bacterial abundance increased in the course of the experiment in SAA (Fig. 1), but this increase was more pronounced in soil treatments (85‐fold) compared with the control (5‐fold). Bacterial abundance in GKS remained rather constant in the control, but increased by a factor 1.6 in the local and foreign soil treatments (Fig. 1).
Bacterial community composition based on 16S rRNA gene amplicon sequencing differed significantly between treatments and times in GKS (ANOSIM, R = 0.23, P < 0.01), but not in SAA (ANOSIM, R = −0.02, P = 0.60). However, nonmetric multidimensional scaling plots (Fig. 2) showed that in both lakes, bacterial community composition overlapped in the controls and the local soil treatments, whereas the foreign soil treatments showed more separation.
Figure 2.
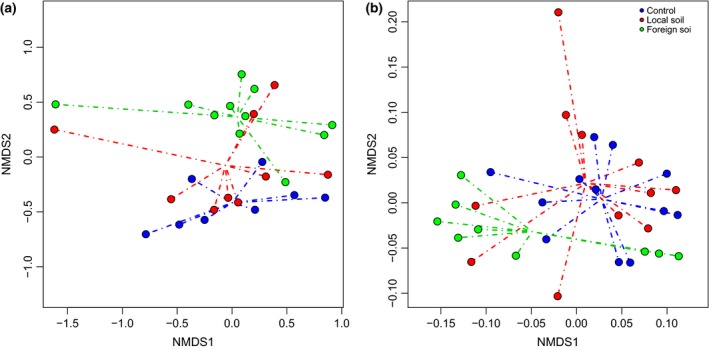
Nonmetric multidimensional scaling plot based on Bray–Curtis similarity in lakes (a) SAA and (b) GKS. Filled circles show communities in the control, the local soil and the foreign soil treatments. Lines connect the single communities to the centroids of each treatment.
Bacterial community composition in the two lakes was very different at the beginning of the experiments (Fig. 3). In SAA, bacterial sequences were dominated by different classes of Proteobacteria and in GKS, mainly by Actinobacteria. During the course of the experiments, a bacterial succession was observed in the controls and the soil treatments of both lakes (Fig. 3, Figs S2 and S4), where initially rare OTUs belonging to Bacteroidetes increased in relative abundance. In SAA, members of Sphingobacteriaceae and Flavobacteriales dominated (Fig. 3a), whereas in GKS mainly Flavobacteriales were responsible for the enrichment (Fig. 3b). At the end of the experiments (72 h), Bacteroidetes accounted for 30–60% of the bacterial assemblage in the soil treatments, but taxa related to Betaproteobacteria (Oxalobacteraceae) and Actinobacteria (Actinomycetales) also gained in relative abundance (Fig. 3, Figs S2 and S4).
Figure 3.
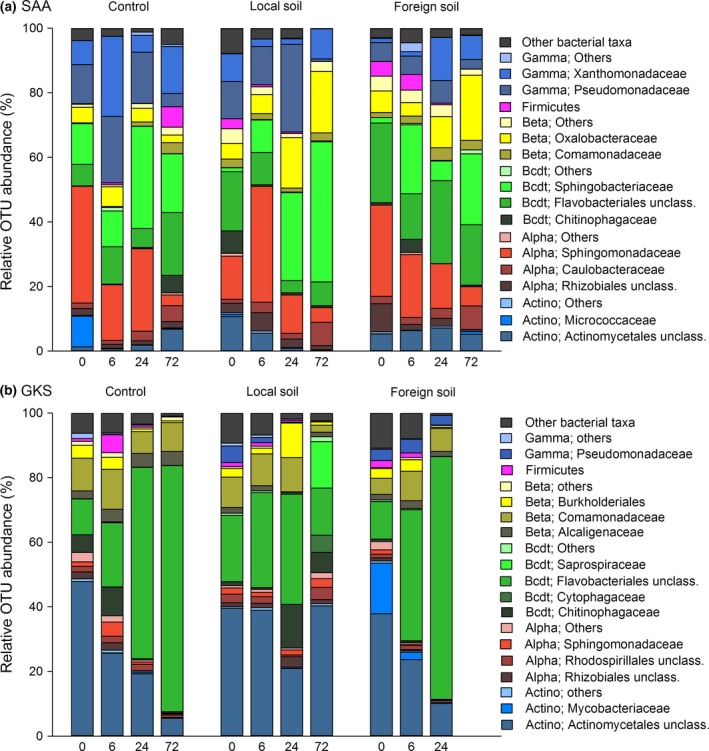
Community composition at the family level in (a) SAA and (b) GKS. Relative OTU abundance from left to right, in the control, the local and the foreign soil treatments determined at every sampling point (0, 6, 24, 72 h). Bacterial taxa that contributed most to the total number of sequences per sample were Gammaproteobacteria (Gamma), Firmicutes, Betaproteobacteria (Beta), Bacteroidetes (Bcdt), Alphaproteobacteria (Alpha) and Actinobacteria (Actino). Unclassified bacteria and other typical freshwater taxa that accounted for <3% of total sequence number were categorized as ‘Others’.
In total, 5 070 OTUs were detected throughout both experiments. In GKS, 297 ± 108 OTUs were detected at the beginning of the experiment, whereas 162 ± 69 OTUs were found in SAA. In all treatments and controls, diversity measured as the number of OTUs, evenness (Shannon H’) and phylogenetic diversity (Faith's pd) gradually decreased during the incubations (Fig. S3). However, when comparing final vs. initial OTU abundances, initially rare bacterial taxa dominated community structure in the soil treatments, whereas in the control, initial dominant taxa prevailed throughout the incubations (Fig. S4).
Bulk community phosphate, ATP and leucine uptake rates
Phosphate, ATP and leucine bulk uptake rates decreased initially in the soil treatments, but increased at the end of the incubations (Fig. 4). In SAA, both soil extract additions initially reduced the uptake of radiolabelled leucine to ~2 pmol L−1 h−1 (Fig. 4a). However, towards the end of the experiment, these rates increased significantly to 783 and 610 pmol L−1 h−1 in the local and foreign soil treatments (Tukey, P < 0.001), respectively. During the first 24 h, phosphate uptake rates were two magnitudes lower in the soil treatments (0.38 ± 0.16 pmol L−1 h−1) when compared to the control (30 ± 0.81 pmol L−1 h−1), whereas ATP bulk uptake rates ranged between 0.92 and 10.4 pmol L−1 h−1. At the end of the experiment, Pi and ATP uptake rates in the local soil treatment reached levels similar to the control (Fig. 4a), but these rates remained significantly lower in the foreign soil treatment (Pi: 2.12 pmol L−1 h−1, ATP: 3.48 pmol L−1 h−1; Tukey, P < 0.001).
Figure 4.
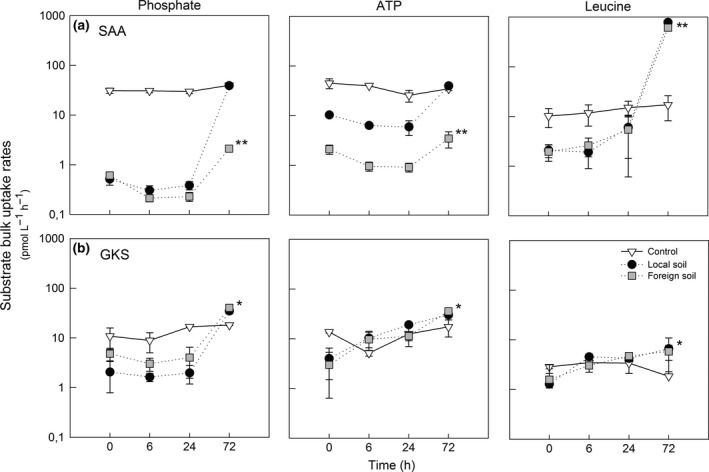
Phosphate, ATP and leucine bulk uptake rates assessed during the experiments. Different symbols indicate the control and the two treatments in lakes (a) SAA and (b) GKS. Data points represent the mean of triplicate incubations, and error bars represent ± 1 SD which are in some cases smaller than the symbol. Asterisks indicate significant difference (Tukey, *P < 0.01, **P < 0.001) in substrate bulk uptake rates between the control and the soil treatments at the end of the experiments. Note that the y‐axis is in logarithmic scale.
In GKS, leucine uptake rates remained low throughout the experiment in all incubations (<7 pmol L−1 h−1), but increased significantly in both soil treatments after 72 h (Fig. 4b; Tukey, P < 0.01). Phosphate uptake rates were significantly lower in the local (1.89 ± 0.23 pmol L−1 h−1) and foreign soil treatments (3.95 ± 0.91 pmol L−1 h−1) when compared to the control (12.16 ± 4.09 pmol L−1 h−1) during the first 24 h (Tukey, P < 0.05). At the end, Pi uptake rates in both soil treatments exceeded the control rates, reaching 37.55 ± 3.96 pmol L−1 h−1. By contrast, ATP bulk uptake rates in the soil treatments exceeded the control rates already after 6 h and were double as high as in the control after 72 h (foreign soil: 35.54 pmol L−1 h−1; local soil: 30.12 pmol L−1 h−1).
Taxon‐specific uptake of phosphate, ATP and leucine in Saanajärvi
In the control and throughout the experiment, the most common bacterial taxa examined using MAR‐CARD‐FISH (Betaproteobacteria, its R‐BT cluster, Bacteroidetes, Alphaproteobacteria) contributed substantially to Pi and ATP uptake (range: 35–88% of hybridized cells), whereas AcI Actinobacteria only contributed 14–33% to the uptake of these substrates (Fig. 5, Fig. S5). A large percentage of bacteria from all taxa examined (range: 59–100% of hybridized cells) took up leucine at the beginning of the experiment in the control, except for Bacteroidetes. Generally, the proportions of cells taking up a substrate declined throughout the experiment in the control (Fig. 5, Fig. S5).
Figure 5.
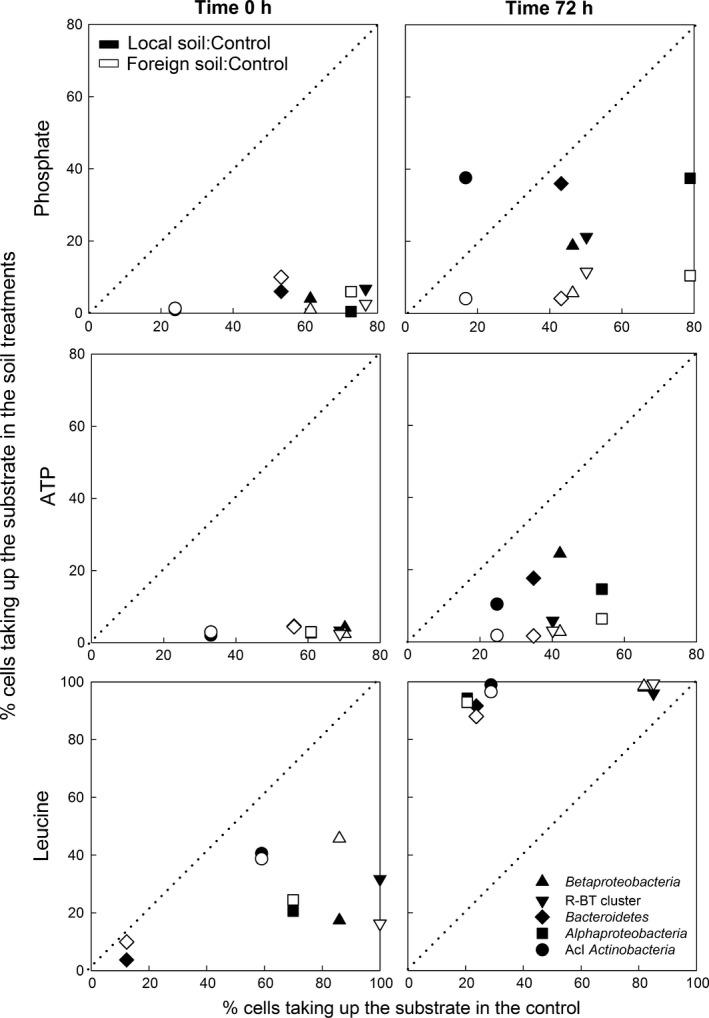
Taxon‐specific uptake of phosphate, ATP and leucine in the soil treatments vs. in the control. Results are given as the percentage of Betaproteobacteria, its R‐BT cluster, Bacteroidetes, Alphaproteobacteria, and AcI Actinobacteria taking up the substrates in the local and foreign soil treatment plotted against the control at the beginning (0 h) and the end of the experiment (72 h) in SAA.
Soil extract additions caused a significant decrease in the proportions of cells taking up Pi, ATP and leucine (Fig. 5; anova, P < 0.05). These percentages dropped below 10% for Pi and ATP incorporation, whereas the percentage of cells taking up leucine comprised 4–41%. During the experiment, these proportions increased (Fig. S5), but the magnitude of increase depended on soil origin. For instance, cells positive for P uptake increased among most bacterial taxa in the local soil treatment reaching 30.2 ± 9.4% and 14.6 ± 7.1% of cells positive in Pi and ATP uptake, respectively. However, the percentage of Pi‐ and ATP‐labelled cells in the foreign soil treatment remained low (Fig. 5). By contrast, all bacterial taxa examined were significantly overrepresented (data points above the 1 : 1 line) in leucine uptake in both soil treatments when compared with the control at the end of the experiment (Fig. 5; anova, P < 0.01).
In the control, the contribution of a specific bacterial taxon to the uptake of Pi and ATP was generally proportional to its contribution to leucine incorporation (Fig. 6), only AcI Actinobacteria were underrepresented in P uptake. The contribution to substrate uptake declined during the experiment in the control for all taxa except Bacteroidetes, which increased their contribution to P and leucine uptake. In contrast to the control, we found that soil extract additions caused considerable shifts in the contribution of major taxa to substrate uptake (Fig. 6). For instance, Betaproteobacteria were overrepresented for Pi and ATP compared to leucine uptake in the local soil treatment, whereas they contributed equally to substrate uptake in the foreign soil treatment (Fig. 6). By the end of the experiment, this class dominated the uptake of the three substrates in both soil treatments and contributed to P uptake in relation to leucine incorporation. Similarly, Bacteroidetes were overrepresented in Pi and ATP uptake when compared to leucine uptake (2.5 ± 0.9%) in both soil treatments at the beginning of the experiment. Towards the end, this phylum increased its relative contribution to cells taking up leucine (34.8 ± 1.5%) and thus contributed to Pi and ATP uptake (38.5 ± 15.8%) proportionally to leucine incorporation. The contribution of Bacteroidetes in the local soil treatment was higher for Pi uptake than for leucine one. Although AcI Actinobacteria accounted for the majority of cells taking up leucine at the beginning in the soil treatments (Fig. 6), their relative contribution to the uptake of any substrate declined during the experiment and was negligible by the end of it.
Figure 6.
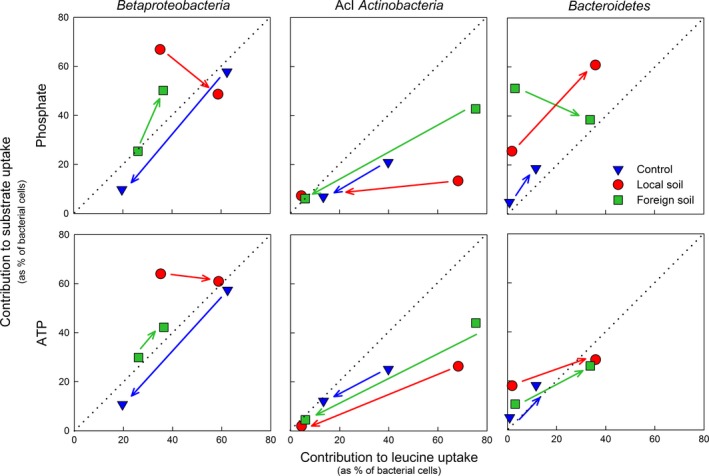
Relative contribution of the dominant bacterial taxa to phosphate and ATP uptake plotted against leucine uptake. Different symbols indicate the control and the two treatments in the SAA experiment. Shifts in contribution to substrate uptake from the beginning of the experiment (0 h) towards the end (72 h) are indicated by arrows. Values are mean of triplicate incubations and the dashed line indicates a 1 : 1 relationship.
Discussion
Soil extract additions cause rapid bacterial successions
Soil extract additions strongly affected the structure and abundance of the bacterioplankton community of lakes located above the treeline (Figs 1 and 2, Fig. S3). Shifts in lake bacterial community structure and cell abundance have been previously linked to the source of organic matter entering a lake (Crump et al., 2003; Roiha et al., 2011; Forsström et al., 2013), and our results indicate that changes in soil organic matter quality induce quick shifts in community composition (Figs 2 and 3, Fig. S2).
In the high‐altitude lake GKS, the initial bacterial community was dominated by the members of Actinobacteria and Betaproteobacteria which agrees with previous reports from mountain lakes in the central Alps (Warnecke et al., 2005; Salcher et al., 2010; Pérez & Sommaruga, 2011). The contribution of major freshwater phyla to community structure in the high‐latitude lake was rather evenly distributed and represented by classes of Proteobacteria, Actinobacteria and Bacteroidetes. Upon soil extract additions, bacterial community composition followed successional patterns, with rare taxa rapidly dominating over the initial community, and persistent taxa gaining importance towards the end (Fig. 3, Fig. S2). These patterns were consistent in both the local and the foreign soil treatments, but the increase of initially rare bacteria was more rapid in the foreign soil treatments at both locations. This was probably mediated by a different composition of the soil sources (e.g., low‐ vs. high‐molecular‐weight compounds) (Moran & Hodson, 1990; Berggren et al., 2010; Logue et al., 2016), which favoured community members having the required metabolic capacities (Judd et al., 2006; Logue et al., 2016). The foreign soil treatments showed a higher aromatic character, most likely linked to higher lignin‐derived compound concentrations from the forest vegetation, whereas in the local soil treatments, this property was less pronounced (Table S2). The stronger effects on bacterial structure observed in treatments with foreign than local soils were likely due to larger differences in DOM quality characteristics. However, in both soil treatments, the specific and rapid community change, concomitantly with the decrease in diversity (Fig. S3), indicated that soil extract additions selected for copiotrophic bacteria, rare under oligotrophic conditions, but preponderant during periods of high allochthonous loadings. This agrees with previous observations from field (Crump et al., 2003; Roiha et al., 2011) and laboratory experiments (Logue et al., 2016) showing that rapid shifts in community composition take place upon soil organic matter input. The loss and prevalence of the taxa determined in our study may be explained by their different functional adaptability, including differences in resource affinities (Cottrell & Kirchman, 2000; Salcher et al., 2011; Heinrich et al., 2013), physiological characteristics (Hahn & Pöckl, 2005; Šimek et al., 2006) and genetic composition (Bauer et al., 2006; Gómez‐Pereira et al., 2012; Teeling et al., 2012; Tveit et al., 2013). Further, changes in lake bacterial community composition have been attributed not only to the differential response of individual bacterial taxa to DOM inputs, but also to the introduction of soil bacteria with allochthonous sources or to incubation effects (Crump et al., 2003; Logue et al., 2016). In our experiments, however, the addition of soil‐borne bacteria with the soil extracts was not confirmed when using staining techniques and microscopy. Nevertheless, potential bottle effects, which can favour the growth of copiotrophic bacteria (Christian & Capone, 2002), cannot be discarded. In any case, it is evident that bacteria interact in a complex way, partly depending on competition for or facilitation of resource utilization (Fuhrman et al., 2015). It can be assumed that the effects of soil source on bacterial community composition and succession were direct and mediated through a combination of ecological lifestyles (copiotrophs vs. oligotrophs) and taxon‐specific substrate preferences (specialists vs. generalists) (Cottrell & Kirchman, 2000; Eiler et al., 2003; Salcher et al., 2013; Pérez et al., 2015), as well as through species interactions.
Effects of soil extract additions on bacterial P limitation and leucine uptake
The degree to which allochthonous sources support lake ecosystem productivity depends on the availability of limiting nutrients and the bioavailability of the carbon source (Del Giorgio & Cole, 1998; Stets & Cotner, 2008; Berggren et al., 2009). Thus, changes in vegetation cover of lake catchments could have severe consequences for the productivity of oligotrophic lakes, if P and DOC availability increases (Jansson et al., 2006).
Soil organic matter is often dominated by high‐molecular‐weight (HMW) compounds and thus, an initial step of extracellular enzymatic activity is required to break down these compounds into low‐molecular‐weight (LMW) compounds prior to bacterial uptake (Arnosti, 2003; Cunha et al., 2010). In our experiments, soil organic matter additions led to the initial reduction of Pi, ATP and leucine uptake at the community (Fig. 4) and the single‐cell level (Fig. 5). These model compounds are readily accessible for bacteria and hence reflect the uptake of inorganic and organic P, as well as of labile DOC compounds. The initial low substrate uptake rates we observed (Fig. 4) could result from a dilution of the radiolabelled substrates with nonlabelled‐compounds, suggesting that LMW compounds were added with the soil extracts (De Haan & De Boer, 1978; Sweet & Perdue, 1982; Pérez & Sommaruga, 2007; Berggren et al., 2010). This trend reversed as the experiment progressed, and substrate uptake rates often reached levels similar to the control or exceeded the control's rates. Nevertheless, the degree of response clearly differed between locations (SAA vs. GKS) and soil treatments (local vs. foreign soil treatment in SAA). The patterns observed in substrate uptake rates were most likely the result of differences in initial bacterioplankton community composition (Logue et al., 2016), DOC lability (Kalbitz et al., 2003; Berggren et al., 2009) and nutrient (or rather P) availability (Moran & Hodson, 1990; Olsen et al., 2002). For instance, total dissolved phosphorus concentrations were three times lower in the local and in the foreign soil treatments in GKS than in SAA (Table S1). Furthermore, the higher leucine uptake rates concomitantly with increasing bacterial abundances observed in SAA may not only reflect the compositional differences of the soil amendments, but also the stimulatory effect of the increase in water temperature during the experiment (Table S1), which did not occur in GKS. For example, a 5 °C increase in water temperature in a mountain stream stimulates leucine uptake rates by four times (Tibbles, 1996).
The origin of the soil extract and its effect on bacterial functioning was also reflected by changes in the substrate utilization profiles of individual bacterial taxa in SAA (Figs 5 and 6) and was in agreement with results obtained in GKS previously (Pérez & Sommaruga, 2006, 2007). The substrate uptake patterns suggested that the dominant bacterial taxa contribute differently to the processing of soil‐derived resources in the treatments. For example, considering that the contribution to substrate uptake of some initially abundant taxa decreased (e.g., AcI Actinobacteria) suggests that their importance during soil run‐off events in general was low. By contrast, other taxa (e.g., Betaproteobacteria) were fundamental for C and P cycling irrespectively of the origin of the soil organic matter (Fig. 6). However, the high representation of rare taxa (e.g., Bacteroidetes) in Pi and ATP uptake when compared to leucine incorporation suggests that they accumulated excess P at the beginning of the experiments, probably to sustain fast growth. This agrees with the growth‐rate hypothesis which postulates that fast‐growing organisms have high P requirements (Elser et al. 2000) due to the increase of P‐rich cell structures required for growth (e.g., ribosomes; Franklin et al. 2011). These results are particularly relevant because the contribution of different taxa to the uptake of C and P substrates greatly affects the energy and nutrient supply to aquatic food webs (Šimek et al., 2005; Salcher et al., 2007). Our findings agree with a recent study showing that in grazer‐free incubations, and particularly during periods of high organic matter loadings, rare taxa dominated bacterial community composition (Neuenschwander et al., 2015). The authors suggested that the low abundance of these taxa at natural settings is controlled by bacterivorous grazing, implying that their biomass contributes substantially to the C channelled towards higher trophic levels. Our results suggest that copiotrophic bacteria do not only channel a substantial fraction of C, but also of P during soil run‐off events.
Given the pivotal role of inland waters in the global carbon cycle and the potentially large number of lakes affected by climate change (Raymond et al., 2013), understanding the effects of catchment alterations on the diversity and function of heterotrophic bacterial assemblages in high altitude and latitude lakes is crucial (Judd et al., 2006; Forsström et al., 2013; Solomon et al., 2015). Diverse assemblages of heterotrophic bacteria control the conversion of organic matter into biomass and regulate organic matter mineralization, determining carbon emissions to the atmosphere (Bass et al., 2010). Our experiments deal with the initial responses of lake bacterioplankton communities to soil organic matter amendments from above and below the treeline (Fig. S6), a scenario which gains realism given the increasing evidence for range expansion of many plant taxa at their altitudinal and latitudinal extremes (Sturm et al., 2001; Parmesan & Yohe, 2003; Hinzman et al., 2005; Lenoir et al., 2008; Pearson et al., 2013). Rare but fast‐growing taxa played a fundamental role in this initial response, but complex interactions formed by initially abundant and persistent bacteria may influence the long‐term effects. Although, the resilience of the communities to changes in soil‐derived inputs and to other climate‐change‐related factors need to be addressed in future experimental work, our results hint to potential major effects on the C and P cycling in lakes affected by the treeline shift (Fig. S6). Factors such as temperature rise or photochemical transformation of allochthonous organic matter (Vähätalo et al., 2003) will shape the functional response of bacteria under natural conditions. Further, specific lake characteristics, such as water residence time and ice‐cover duration (Bergström & Jansson, 2000; Adrian et al., 2009), and the chemical composition of atmospheric deposition (Kopáček et al., 2011) may be important in determining the relevance and magnitude of the response. Finally, the increasing frequency and magnitude of heavy‐precipitation events (Fischer & Knutti, 2015) might foster the input of terrestrial organic matter with strong consequences for lake metabolism and the dominance of heterotrophic processes (Sadro & Melack, 2012; Forsström et al., 2013).
Supporting information
Figure S1. The subarctic lake Saanajärvi (SAA) in Finland and the alpine lake Gossenköllesee (GKS) in Austria.
Figure S2. Relative abundance of specific bacterial taxa in the control and soil treatments in SAA and GKS.
Figure S3. Temporal dynamics of community diversity in SAA and GKS.
Figure S4. Initial vs. final sequence abundances in the control and soil treatments in SAA and GKS.
Figure S5. The proportions of probe‐specific bacterial taxa taking up phosphate, ATP and leucine in the SAA experiment.
Figure S6. Schematic depiction of the effects of climate‐induced change in soil run‐off composition on lake bacterial community composition and functioning.
Table S1. Summary of physicochemical and biological parameters determined in lakes SAA and GKS, and during the experiments.
Table S2. Optical characteristics of dissolved organic matter measured at the beginning of the experiments.
Acknowledgements
The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7/2007‐2013] under grant agreement no 262693 [INTERACT] to CR and the Austrian Science Fund (FWF) through project P‐24098‐B22 to MTP and RS. We thank Josef Franzoi, Gry Larsen and Salvador Morales‐Gomez for chemical analyses at the University of Innsbruck. Further, we would like to thank Antero Järvinen, Rauni Partanen, Oula Kalttopää and Pirjo Hakala from Kilpisjärvi Biological Station who helped with setting up and organizing the experiments in Finland and Laurent Moya and Jolien Scholten for help during the experiment in Austria. Special thanks to Jim Cotner for his feedback on this manuscript. NC hold a postdoctoral grant from the Wenner‐Gren Foundation (2014–2016) and a ‘Juan de la Cierva’ Postdoctoral Grant (FJCI‐2014‐23064). The work reported is part of the doctoral dissertation of CR. The authors declare no conflict of interest.
DNA Sequences have been deposited at the NCBI Sequence Read Archive under accession number: SRP077041.
References
- Adrian R, O'Reilly CM, Zagarese H et al (2009) Lakes as sentinels of climate change. Limnology and Oceanography, 54, 2283–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti C (2003) Microbial extracellular enzymes and their role in dissolved organic matter cycling In: Aquatic Ecosystems: Interactivity of Dissolved Organic Matter (ed. Sinsabaugh RL.), pp. 315–342. Findlay SEG, Academic Press, San Diego. [Google Scholar]
- Bass AM, Waldron S, Preston T, Adams CE (2010) Net pelagic heterotrophy in mesotrophic and oligotrophic basins of a large, temperate lake. Hydrobiologia, 652, 363–375. [Google Scholar]
- Bastidas Navarro M, Balseiro E, Modenutti B (2014) Bacterial community structure in patagonian Andean Lakes above and below timberline: from community composition to community function. Microbial Ecology, 68, 528–541. [DOI] [PubMed] [Google Scholar]
- Battin TJ, Wille A, Sattler B, Psenner R (2001) Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Applied and Environmental Microbiology, 67, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Kube M, Teeling H et al (2006) Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environmental Microbiology, 8, 2201–2213. [DOI] [PubMed] [Google Scholar]
- Beniston M (2005) Mountain climates and climatic change: an overview of processes focusing on the European Alps. Pure and Applied Geophysics, 162, 1587–1606. [Google Scholar]
- Berggren M, Laudon H, Jansson M (2009) Aging of allochthonous organic carbon regulates bacterial production in unproductive boreal lakes. Limnology and Oceanography, 54, 1333–1342. [Google Scholar]
- Berggren M, Laudon H, Haei M, Strom L, Jansson M (2010) Efficient aquatic bacterial metabolism of dissolved low‐molecular‐weight compounds from terrestrial sources. The ISME Journal, 4, 408–416. [DOI] [PubMed] [Google Scholar]
- Bergström AK, Jansson M (2000) Bacterioplankton production in humic Lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microbial Ecology, 39, 101–115. [DOI] [PubMed] [Google Scholar]
- Burkert U, Warnecke F, Babenzien D, Zwirnmann E, Pernthaler J (2003) Members of a readily enriched β‐proteobacterial clade are common in surface waters of a humic lake. Applied and Environmental Microbioloy, 69, 6550–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter SR, Benson BJ, Biggs R et al (2007) Understanding regional change: a comparison of two lake districts. BioScience, 57, 323–335. [Google Scholar]
- Christian RR, Capone DG (2002) Overview of issues in aquatic microbial ecology In: Manual of Environmental Microbiology (eds Hurst CJ, Crawford RL, Knudsen GR, McInerney MJ, Stetzenbach LD.), pp. 323–328. ASM Press, Washington, USA. [Google Scholar]
- Classen AT, Sundqvist MK, Henning JA et al (2015) Direct and indirect effects of climate change on soil microbial and soil microbial‐plant interactions: what lies ahead? Ecosphere, 6, 130 doi: 10.1890/ES15‐00217.1. [Google Scholar]
- Collins M, Knutti R, Arblaster J et al (2013) Long‐term climate change: projections, commitments and irreversibility In: Climate Change 2013: The Physical Science Basis (eds Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM.), pp. 1055–1136. Cambridge University Press, Cambridge, UK and New York, NY, USA. [Google Scholar]
- Cottrell MT, Kirchman DL (2000) Natural assemblages of marine Proteobacteria and members of the Cytophaga‐Flavobacter cluster consuming low‐ and high‐molecular‐weight dissolved organic matter. Applied and Environmental Microbiology, 66, 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Kling GW, Bahr M, Hobbie JE (2003) Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Applied and Environmental Microbiology, 69, 2253–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha A, Almeida A, Coelho FJRC, Gomes NCM, Oliveira V, Santos AL (2010) Bacterial extracellular enzymatic activity in globally changing aquatic ecosystems In: Current Research, Technology and Education Topis in Applied Microbiology and Microbial Biotechnology (ed. Méndez‐Vilas A.), pp. 124–135. Formatex Research Center, Badajoz, Spain. [Google Scholar]
- De Haan H, De Boer T (1978) A study of the possible interactions between fulvic acids, amino acids and carbohydrates from Tjeukemeer, based on gel filtration at pH 7.0. Water Research, 12, 1035–1040. [Google Scholar]
- Del Giorgio PA, Cole JJ (1998) Bacterial growth efficiency in natural aquatic systems. Annual Review of Ecology and Systematics, 29, 503–541. [Google Scholar]
- Del Giorgio PA, Bird DF, Prairie YT, Planas D (1996) Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnology and Oceanography, 41, 783–789. [Google Scholar]
- Eiler A, Langenheder S, Bertilsson S, Tranvik LJ (2003) Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Applied and Environmental Microbiology, 69, 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Sterner RW, Gorokhova E, et al. (2000) Biological stoichiometry from genes to ecosystems. Ecology Letters, 3, 540–550. [Google Scholar]
- Fischer EM, Knutti R (2015) Anthropogenic contribution to global occurrence of heavy‐precipitation and high‐temperature extremes. Nature Climate Change, 5, 560–564. [Google Scholar]
- Forsström L, Roiha T, Rautio M (2013) Responses of microbial food web to increased allochthonous DOM in an oligotrophic subarctic lake. Aquatic Microbial Ecology, 68, 171–184. [Google Scholar]
- Franklin O, Hall EK, Kaiser C, Battin TJ, Richter A (2011) Optimization of Biomass Composition Explains Microbial Growth‐Stoichiometry Relationships. The American Naturalist, 177, 29–42. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM (2015) Marine microbial community dynamics and their ecological interpretation. Nature Reviews Microbiology, 13, 133–146. [DOI] [PubMed] [Google Scholar]
- Gómez‐Pereira PR, Schüler M, Fuchs BM et al (2012) Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environmental Microbiology, 14, 52–66. [DOI] [PubMed] [Google Scholar]
- Grabherr G, Gottfried M, Pauli H (1994) Climate effects on mountain plants. Nature, 369, 448. [DOI] [PubMed] [Google Scholar]
- Grimm NB, Chapin FS, Bierwagen B et al (2013) The impacts of climate change on ecosystem structure and function. Frontiers in Ecology and the Environment, 11, 474–482. [Google Scholar]
- Guillemette F, Del Giorgio PA (2011) Reconstructing the various facets of dissolved organic carbon bioavailability in freshwater ecosystems. Limnology and Oceanography, 56, 734–748. [Google Scholar]
- Hahn MW, Pöckl M (2005) Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Applied and Environmental Microbiology, 71, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper DT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4, 9. [Google Scholar]
- Harsch MA, Hulme PE, Mcglone MS, Duncan RP (2009) Are treelines advancing? A global meta‐analysis of treeline response to climate warming. Ecology Letters, 12, 1040–1049. [DOI] [PubMed] [Google Scholar]
- Heinrich F, Eiler A, Bertilsson S (2013) Seasonality and environmental control of freshwater SAR11 (LD12) in a temperate lake (Lake Erken, Sweden). Aquatic Microbial Ecology, 70, 33–44. [Google Scholar]
- Hinzman LD, Bettez ND, Bolton WR et al (2005) Evidence and implications of recent climate change in northern Alaska and other arctic regions. Climatic Change, 72, 251–298. [Google Scholar]
- Hornák K, Jezbera J, Nedoma J, Gasol JM, Šimek K (2006) Effects of resource availability and bacterivory on leucine incorporation in different groups of freshwater bacterioplankton, assessed using microautoradiography. Aquatic Microbial Ecology, 45, 277–289. [Google Scholar]
- Jansson M, Bergström AK, Lymer D, Vrede K, Karlsson J (2006) Bacterioplankton growth and nutrient use efficiencies under variable organic carbon and inorganic phosphorus ratios. Microbial Ecology, 52, 358–364. [DOI] [PubMed] [Google Scholar]
- Jones RI (1992) The influence of humic substances on lacustrine planktonic food‐chains. Hydrobiologia, 229, 73–91. [Google Scholar]
- Judd KE, Crump BC, Kling GW (2006) Variation in dissolved organic matter controls bacterial production and community composition. Ecology, 87, 2068–2079. [DOI] [PubMed] [Google Scholar]
- Kalbitz K, Schmerwitz J, Schwesig D, Matzner E (2003) Biodegradation of soil‐derived dissolved organic matter as related to its properties. Geoderma, 113, 273–291. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR et al (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463–1464. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Gloeckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Research, 41, e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopáček J, Hejzlar J, Vrba J, Stuchlík E (2011) Phosphorus loading of mountain lakes: terrestrial export and atmospheric deposition. Limnology and Oceanography, 56, 1343–1354. [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology, 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritzberg ES, Cole JJ, Pace ML, Granèli W, Bade DL (2004) Autochthonous vs. allochthonous carbon sources of bacteria: results from whole‐lake 13C addition experiments. Limnology and Oceanography, 49, 588–596. [Google Scholar]
- Lenoir J, Gegout JC, Marquet PA, De Ruffray P, Brisse H (2008) A significant upward shift in plant species optimum elevation during the 20th century. Science, 320, 1768–1771. [DOI] [PubMed] [Google Scholar]
- Logue JB, Stedmon CA, Kellerman AM et al (2016) Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. The ISME Journal, 10, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Hodson RE (1990) Bacterial production on humic and nonhumic components of dissolved organic carbon. Limnology and Oceanography, 35, 1744–1756. [Google Scholar]
- Neuenschwander SM, Pernthaler J, Posch T, Salcher MM (2015) Seasonal growth potential of rare lake water bacteria suggest their disproportional contribution to carbon fluxes. Environmental Microbiology, 17, 781–795. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al (2011) vegan: Community Ecology Package.
- Olsen LM, Reinertsen H, Vadstein O (2002) Can phosphorus limitation inhibit dissolved organic carbon consumption in aquatic microbial food webs? A study of three food web structures in microcosms. Microbial Ecology, 43, 353–366. [DOI] [PubMed] [Google Scholar]
- O'Reilly CM, Sharma S, Gray DK et al (2015) Rapid and highly variable warming of lake surface waters around the globe. Geophysical Research Letters, 42, 10773–10781. [Google Scholar]
- Parmesan C, Yohe G (2003) A globally coherent fingerprint of climate change impacts across natural systems. Nature, 421, 37–42. [DOI] [PubMed] [Google Scholar]
- Pearson RG, Phillips SJ, Loranty MM, Beck PSA, Damoulas T, Knight SJ, Goetz SJ (2013) Shifts in Arctic vegetation and associated feedbacks under climate change. Nature Climate Change, 3, 673–677. [Google Scholar]
- Pérez MT, Sommaruga R (2006) Differential effect of algal‐ and soil‐derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnology and Oceanography, 51, 2527–2537. [Google Scholar]
- Pérez MT, Sommaruga R (2007) Interactive effects of solar radiation and dissolved organic matter on bacterial activity and community structure. Environmental Microbiology, 9, 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez MT, Sommaruga R (2011) Temporal changes in the dominance of major planktonic bacterial groups in an alpine lake: discrepancy with their contribution to bacterial production. Aquatic Microbial Ecology, 63, 161–170. [Google Scholar]
- Pérez MT, Rofner C, Sommaruga R (2015) Dissolved organic monomer partitioning among bacterial groups in two oligotrophic lakes. Environmental Microbiology Reports, 7, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham SV, Leavitt PR, McGowan S, Peres‐Neto P (2008) Spatial variability of climate and land‐use effects on lakes of the northern Great Plains. Limnology and Oceanography, 53, 728–742. [Google Scholar]
- Posch T, Mindl B, Hornák K et al (2007) Biomass reallocation within freshwater bacterioplankton induced by manipulating phosphorus availability and grazing. Aquatic Microbial Ecology, 49, 223–232. [Google Scholar]
- R Development Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/ (accessed 4 April 2016).
- Räisänen J, Eklund J (2011) 21st Century changes in snow climate in Northern Europe: a high‐resolution view from ENSEMBLES regional climate models. Climate Dynamics, 38, 2575–2591. [Google Scholar]
- Raymond PA, Hartmann J, Lauerwald R et al (2013) Global carbon dioxide emissions from inland waters. Nature, 503, 355–359. [DOI] [PubMed] [Google Scholar]
- Reuss NS, Hammarlund D, Rundgren M, Segerstrom U, Eriksson L, Rosen P (2010) Lake ecosystem responses to holocene climate change at the subarctic tree‐line in northern Sweden. Ecosystems, 13, 393–409. [Google Scholar]
- Roiha T, Tiirola M, Cazzanelli M, Rautio M (2011) Carbon quantity defines productivity while its quality defines community composition of bacterioplankton in subarctic ponds. Aquatic Sciences, 74, 513–525. [Google Scholar]
- Sadro S, Melack JM (2012) The effect of an extreme rain event on the biogeochemistry and ecosystem metabolism of an oligotrophic high‐elevation lake. Arctic, Antarctic, and Alpine Research, 44, 222–231. [Google Scholar]
- Salcher MM, Hofer J, Hornak K et al (2007) Modulation of microbial predator‐prey dynamics by phosphorus availability: growth patterns and survival strategies of bacterial phylogenetic clades. FEMS Microbiology Ecology, 60, 40–50. [DOI] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Posch T (2010) Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnology and Oceanography, 55, 846–856. [Google Scholar]
- Salcher MM, Pernthaler J, Posch T (2011) Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria ‘that rule the waves’ (LD12). The ISME Journal, 5, 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher MM, Posch T, Pernthaler J (2013) In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. The ISME Journal, 7, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Horňák K, Jezbera J, Mašín M, Nedoma J, Gasol JM, Schauer M (2005) Influence of top‐down and bottom‐up manipulations on the R‐BT065 subcluster of β‐Proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Applied and Environmental Microbiology, 71, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Horňak K, Jezbera J et al (2006) Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environmental Microbiology, 8, 1613–1624. [DOI] [PubMed] [Google Scholar]
- Solomon CT, Jones SE, Weidel BC et al (2015) Ecosystem consequences of changing inputs of terrestrial dissolved organic matter to lakes: current knowledge and future challenges. Ecosystems, 18, 376–389. [Google Scholar]
- Sommaruga‐Wögrath S, Koinig KA, Schmidt R, Sommaruga R, Tessadri R, Psenner R (1997) Temperature effects on the acidity of remote alpine lakes. Nature, 387, 64–67. [Google Scholar]
- Sorvari S (2001) Climate impacts on remote subarctic lakes in Finnish Lapland: Limnological and palaeolimnological assessment with a particular focus on diatoms and Lake Saanajärvi. PhD thesis. University of Helsinki, Finland.
- Steger C, Kotlarski S, Jonas T, Schär C (2012) Alpine snow cover in a changing climate: a regional climate model perspective. Climate Dynamics, 41, 735–754. [Google Scholar]
- Stets EG, Cotner JB (2008) The influence of dissolved organic carbon on bacterial phosphorus uptake and bacteria‐phytoplankton dynamics in two Minnesota lakes. Limnology and Oceanography, 53, 137–147. [Google Scholar]
- Sturm M, Racine C, Tape K (2001) Increasing shrub abundance in the Arctic. Nature, 411, 546–547. [DOI] [PubMed] [Google Scholar]
- Sweet SS, Perdue EM (1982) Concentration and speciation of dissolved sugars in river water. Environmental Science and Technology, 16, 692–698. [Google Scholar]
- Teeling H, Fuchs BM, Becher D et al (2012) Substrate‐controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science, 336, 608–611. [DOI] [PubMed] [Google Scholar]
- Tibbles BJ (1996) Effects of temperature on the incorporation of leucine and thymidine by bacterioplankton and bacterial isolates. Aquatic Microbial Ecology, 11, 239–250. [Google Scholar]
- Tranvik LJ (1992) Allochthonous dissolved organic matter as an energy source for pelagic bacteria and the concept of the microbial loop. Hydrobiologia, 229, 107–114. [Google Scholar]
- Tveit A, Schwacke R, Svenning MM, Urich T (2013) Organic carbon transformations in high‐Arctic peat soils: key functions and microorganisms. The ISME Journal, 7, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähätalo A, Salonen K, Münster U, Järvinen M, Wetzel R (2003) Photochemical transformation of allochthonous organic matter provides bioavailable nutrients in a humic lake. Archiv für Hydrobiologie, 156, 278–314. [Google Scholar]
- Vogler P (1966) Zur Analytik der Phosphorverbindungen in Gewässer. Limnologica (Berlin), 4, 437–444. [Google Scholar]
- Wagner C, Adrian R (2009) Exploring lake ecosystems: hierarchy responses to long‐term change? Global Change Biology, 15, 1104–1115. [Google Scholar]
- Warnecke F, Sommaruga R, Sekar R, Hofer JS, Pernthaler J (2005) Abundances, identity, and growth state of Actinobacteria in mountain lakes of different UV transparency. Applied and Environmental Microbiology, 71, 5551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CE, Dodds W, Kratz TK, Palmer MA (2008) Lakes and streams as sentinels of environmental change in terrestrial and atmospheric processes. Frontiers in Ecology and the Environment, 6, 247–254. [Google Scholar]
- Woodward G, Perkins DM, Brown LE (2010) Climate change and freshwater ecosystems: impacts across multiple levels of organization. Philosophical Transactions of the Royal Society Biological Sciences, 365, 2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder M, Peter S, Shabarova T, Pernthaler J (2009) A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environmental Microbiology, 11, 2676–2686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The subarctic lake Saanajärvi (SAA) in Finland and the alpine lake Gossenköllesee (GKS) in Austria.
Figure S2. Relative abundance of specific bacterial taxa in the control and soil treatments in SAA and GKS.
Figure S3. Temporal dynamics of community diversity in SAA and GKS.
Figure S4. Initial vs. final sequence abundances in the control and soil treatments in SAA and GKS.
Figure S5. The proportions of probe‐specific bacterial taxa taking up phosphate, ATP and leucine in the SAA experiment.
Figure S6. Schematic depiction of the effects of climate‐induced change in soil run‐off composition on lake bacterial community composition and functioning.
Table S1. Summary of physicochemical and biological parameters determined in lakes SAA and GKS, and during the experiments.
Table S2. Optical characteristics of dissolved organic matter measured at the beginning of the experiments.


