Abstract
Background
In prostate cancer, mutated p53 alleles typically contain missense single-base substitution in codon 72 that resides within exons 5-8. Stable p53 proteins in tumor cell nuclei have been associated with malignancy. A role of p53 is the regulation of drug transporters like ABCC1 (MRP1) by an effect on promoter region.
Objectives
The objective of this study was to identify association of mutations of p53 at codon 72 and 282 and promoter region of ABCC1 with increased risks of prostate cancer.
Materials and Methods
Formalin fixed, paraffin-embedded malignant tissues of 45 patients and 45 control samples were evaluated. PCR-RFLP using BstUI for codon 72 and HpaII restriction enzyme for codon 282 p53 gene, and G-1666A promoter region of ABCC1 gene was performed. To assess the frequency of these mutations and to detect new mutations in cancerous samples, PCR-SSCP analysis was performed.
Results
The frequencies of CC, GC and GG genotypes of codon 72 of p53 were 33.33%, 46.67% and 20.00% in patients with cancer and 15.56%, 48.89% and 35.55% in controls, respectively. The relative allele frequencies of ABCC1 promoter polymorphism were 60.00% A and 40.00% G in patients as opposed to 37.78% for A and 62.22% for G in controls. Genotypic frequencies of p53 codon 72 and G1666A of ABCC1 in patients vs. Controls were statistically significant(p<0.05). The study of these samples with PCR-SSCP displayed some new banding patterns.
Conclusions
The present findings suggest that CC homozygosity in codon 72 of p53 gene and AA genotype in G-1666A of ABCC1 gene may play a role in combination in prostate cancer and increased susceptibility for this malignancy in the Iranian Kurdish population.
Keywords: ABCC1, Codon 72 p53, PCR-RFLP, PCR-SSCP, Polymorphism, Prostate cancer
1. Background
Prostate cancer ranks second after lung cancer and the sixth most common cause of cancer death among men in world (1) In Iran, prostate cancer is the third cause of death after coronary heart disease and car accidents. Many different risk factors such as the inherited genetic susceptibility to the development of prostate cancer have been identified (2).
Tumor suppressors use multiple mechanisms to suppress cancer cell growth. The most important of tumor suppressors, TP53, is associated with about 50% of human cancer cases (3). Human TP53 is a nuclear phosphoprotein with MW53 kDa and encoded by a 20-Kb gene containing 11 exons and 10 introns, on chromosome 17pl3 (4, 5). Single nucleotide polymorphisms (SNPs) are the most common form of tumor-associated mutations in p53. Among them, a G to C transversion in Codon 72 in exon 4 is a common polymorphism and is associated with an increased risk of various types of cancer (6). Moreover, p53 mutations are occurred throughout exons 5-8. Exons 7 and 8 are counting to be the highly conserved and most studied exons of this gene (7).
Two crucial aspects in cancer research are the roles of p53 in tumorigenesis and the establishment of chemoresistance or multidrug resistance (MDR) phenotype by the family of multidrug transporters (8). ABCC1 (MRP1) belongs to the ATP-binging cassette superfamily of cell-surface transport proteins. Human ABCC1as one of 13 members of human ABCC subfamily that has additional roles in drug resistance, is proposed, to have roles in cellular antioxidative defense system. SNPs in the coding region of ABCC1 gene have been shown to affect its function (9, 10). Recent studies have revealed that G-1666A polymorphism on promoter region of ABCC1 is a risk factor for cancer. It has been nearly 15years since the initial experiments of Chin et al. demonstrated transcriptional dependence of the MDR1 gene promoter by p53 (8, 10-11). To clarify the association of codon 72 and 282 polymorphisms of p53 gene and G-1666A promoter polymorphism of ABCC1 gene with the risk of prostate cancer, a case-control study of 45 controls and 45 paraffin embedded samples of patients with confirmed prostate cancer was performed. Additionally the possibility of the interaction between the two genes, with respect to the polymorphisms, was also investigated to identify if they participated either independently or jointly in disease pathogenesis.
2. Objectives
All prostate cancer samples were confirmed histologically at the Tohid Hospital of Sanandaj, Iran. The age of patients ranged between 22-90 years, with a mean of 56 years. These samples were fixed in formalin and embedded in paraffin.
As a non-malignant control group, blood of 45 randomly selected men with no previous history of cancer and no signs and symptoms of malignancy were used for DNA extraction from the same population that case patients were sampled.
3. Materials and Methods
3.1. Genomic DNA Extraction and PCR Amplification
Genomic DNA was extracted from paraffin embedded tissues. Tissue sections (2 pieces with 20 mm thickness each) were scraped into a 1.5 ml microcentrifuge tube. Tissue fragments were deparaffinized with xylene, ethanol and washed with PBS buffer ( pH 7.4; 1.7 mM KH2PO4, 5.2 mM Na2HPO4, 150 mM NaCl) two times. For digestion, lysis buffer (1 M Tris, 0.5M EDTA, SDS 10% w/v and proteinase K (20 mg.mL-1)) were added to each microtube and incubated 16 h at 55ºC and DNA was extracted by phenol-chloroform extraction method (12-14).
PCR amplification of exon 4 and exon 8 of p53 gene and G-1666A promoter region of ABCC1 was carried out using designed primers with Gene runner software (Hastings Software, Inc. http://www.generunner.net) (Table 1).
Table 1. Primer sequences of codons, 72 and 282, and ABCC1 .
| Gene | Sequence (→) | Product length |
|
Codon 72 exon 4 P53 Codon 282 exon 8 P53 ABCC1 |
Forward primer: GCTCTTTTCACCCATCTACAGTC Reverse primer: CGTAGCTGCCCTGGTAGGTTTTC Forward primer: GTGGTAATCTACTGGGACGGAAC Reverse primer: GTGTTGTTGGGCAGTGCTAGGA Forward primer: CAAGCAACAGCATAACTGGC Reverse primer: GTGTGTGCATTTGAGACCTC |
247bp 246bp 175bp |
3.2. RFLP and SSCP Analysis
PCR-RFLP was used for genotype identification. BstUI was used to digest the PCR product of codon 72, and HpaI was used to restrict both codon 282 of p53 and G-1666A of ABCC1. The digestion was carried out for 16 h at 37ºC. Restricted DNA fragments were separated on an 8% acrylamide gel for 3 h at constant power of 150 w and stained with silver nitrate (15). BstUI cleaves 274 bp PCR product of G allele at codon 72 p53 gene, generating 140 bp and 107 bp fragments (Figure 1A). HpaII cleaves 175 bp PCR product of G allele of ABCC1, generating 102 bp and 73 bp fragments (Figure 1C). All PCR products were screened for sequence variations via SSCP. PCR products (4 μL) were mixed with 4 μL of loading buffer (0.05% bromophenol blue, 0.05% xylene cyanol, 95% formamide, 20 mM EDTA) and 8 μL of ddH2O. The mixture was heat denatured at 95ºC for 10 min and immediately placed on ice for 5 min until loading on 12% w/v acrylamide at constant power of 200 w for 17-18h and stained with silver nitrate (Figure 2).
Figure 1 .
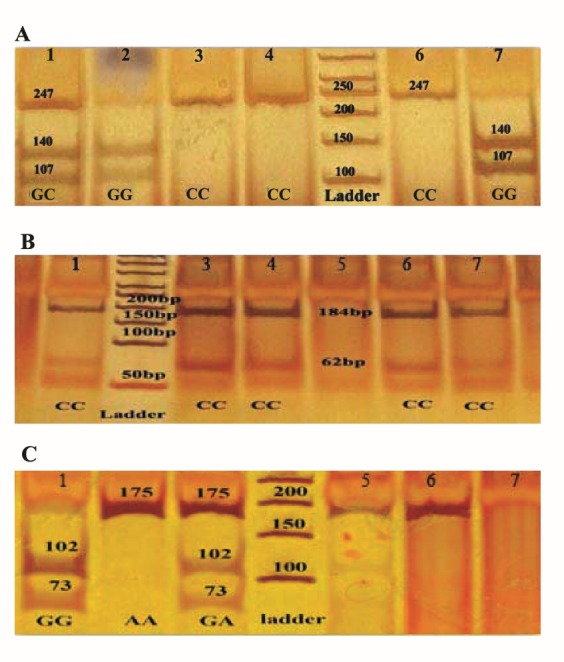
A: PCR-RFLP of codon 72 p53 gene. Lanes 3, 4 and 6 CC homozygous genotype (Pro/Pro, 247 bp); lane 1 GC heterozygous genotype (Pro/Arg, 247,140 and 107 bp); Lanes 2 and 7 GG homozygous genotype (Arg/Arg, 140 and 107 bp). B: PCR-RFLP of codon 282 p53 gene. All lanes were homozygous (184 and 62 bp). C: PCR-RFLP of G- 1666A ABCC1. Lanes 2, 5 and 6 AAhomozygous genotype (175 bp); lane 3 GA heterozygous genotype (175, 102 and 73 bp); lane 1 GG homozygous genotype (102 and 73 bp)
Figure 2 .
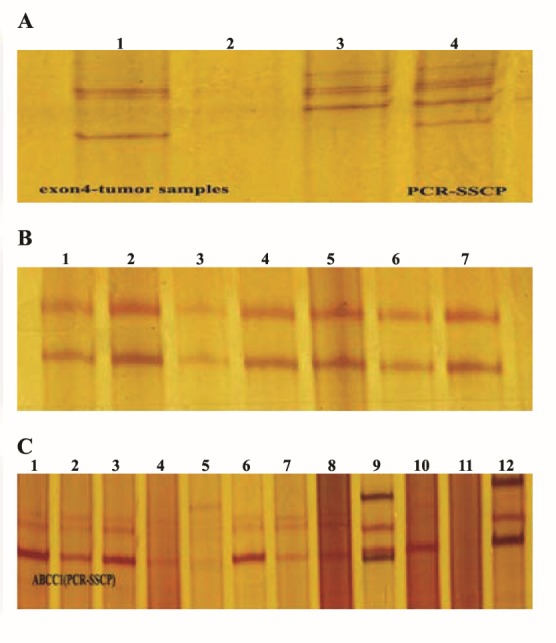
A: PCR-SSCP of codon 72 p53 gene. Lane 3 had pattern 1 with 5 bands that like control samples; lane 1had pattern 2 with 3 bands; lane 2 had pattern 3 with 4 bands. B: PCR-SSCP of codon 282 p53 gene. All lanes had one pattern with 2 bands. C: PCR-SSCP of G-1666A ABCC1. Lanes 1-4, 6-8 had pattern 1 with 3 bands; lane 5 had pattern 2 with 5 bands; lane 9 had pattern 4 with 4 bands and lane 12 had pattern 3 with 3 bands
3.3. Statistical Analysis
Population allelic frequencies were tested for Hardy-Weinberg equilibrium by χ2 test. The association between p53 and ABCC1 polymorphisms and the risk of development of prostate cancer was analyzed with χ2 test. P values below 0.05 were considered to be statistically significant. All analyses were performed by POPGENE V.1.31 (16).
4. Results
4.1. The Correlation between p53 Codons 72 and 282, and ABCC1G-1666A Polymorphic Variants and Risk of Prostate Cancer Occurrence
The allele and genotype frequencies of p53 codon 72 and ABCC1 promoter region of G-1666A genotypes are summarized in Table 2. The genotype frequencies in patients and controls did not deviate significantly from the Hardy-Weinberg equilibrium (p>0.05). Detection of p53 and ABCC1 polymorphisms were successfully conducted in all patient and control samples by PCR-RFLP (Figure 1). The cases had more CC (Pro/Pro) individuals and a higher C allele frequency in the codon 72 than that of the controls (56.66% vs. 40.00%). Moreover, they had more AA individuals and a higher A allele frequency for the G-1666A of ABCC1 gene than that of the controls (60.00% vs. 37.78%; Table 2).
Table 2. Genotypic frequencies of p53 at codons 72 and 282 and ABCC1 in promoter region G-1666A among patients and controls .
| Genotype |
Patient
N (%) |
Control
N (%) |
|
|
Codon 72 exon 4 P53 Codon 282 exon 8 P53 G-1666A ABCC1 |
Pro/Pro(CC) Pro/Arg(GC) Arg/Arg(GG) C G Trp/Trp (TT) Arg/Trp (CT) Arg/Arg (CC) T C AA GA GG A G |
15 (33.33%) 21(46.67%) 9 (20.00%) 56.66% 43.34% 0 (0.00%) 0 (0.00%) 45 (100.00%) 0.00% 100.00% 14 (31.11%) 26 (57.78%) 5 (11.11%) 60.00% 40.00% |
7 (15.56%) 22 (48.89%) 16 (35.55%) 40.00% 60.00% (0.00%) (0.00%) 45 (100.00%) 0.00% 100.00% 5 (11.11%) 24 (53.44%) 16 (35.55%) 37.78% 62.22% |
The frequencies of homozygous and heterozygous genotypes among patients were significantly different from those among controls. Results of this study showed the p53 Pro/Pro genotype in codon 72 was associated with an increased risk of developing prostate cancer (33.33% in patients with cancer and 15.56% in controls p<0.05). Similarly the ABCC1 AA genotype of G-1666A was also correlated with prostate cancer risk (31.11% in patients and 11.11% in controls p<0.05). In p53 gene, polymorphism was also analyzed for codon 282. However, no polymorphism was observed for this codon among the cases and the controls and all had CC genotype (Table 2).
4.2. Nucleotide Changes and Mutation Analysis in p53 and ABCC1
Mobility shifts in banding pattern between samples during SSCP is indicative of sequence variation. The PCR-SSCP analyses of codons 72 and 282 of the p53 gene, and G-1666A mutation of ABCC1 gene was performed for all 90 patient and control samples. PCR-SSCP analysis of codon 72 of p53 showed three patterns (Figure 2A). Pattern 1 (lane3) with 5 bands was seen in all control samples and in 41 of 45 tumor samples. Pattern 2 (lane 1) with 3 bands was displayed in 1tumor sample. Two patient samples displayed pattern 3 (lane 2) with 4 bands. These 3 samples apparently had a different migration pattern. Further analysis need to confirm sequence variation. PCR-SSCP analysis of codon 282 of p53 showed no new banding pattern (Figure 2B), all with 2 bands.
The representative PCR-SSCP analysis of G-1666A mutation of ABCC1 showed four patterns (Figure 2C). Pattern 1 had 3 bands (lane 1-4, 6-8) that was detected in most tumor samples. Three tumor samples had 4 bands (pattern 4, lane 9) and another two samples 3 bands (pattern 3). These patterns (4 and 3) can be considered as new mutations in this cohort.
5. Discussion
TP53 as “the guardian” of the human genome is involved in DNA repair, the cell cycle, gene transcription and apoptosis (15, 16). Many of p53 mutations produce abnormal proteins without transcriptional regulatory activities (17).A brief search of reported mutations in EXAC browser beta (URL: http://exac.broadinstitute.org) showed that missense mutation in codon 72 (rs1042522) has high incidence in some populations and located in the third place of highly occurred mutations in this gene. One of the roles of p53 is regulation of the ABCC1 promoter. In one study, the researchers found a concomitant reduction in ABCC1 mRNA levels with wtp53 expression (8).
Previous reports suggested that a functional polymorphism in codon 72 of p53 gene, which encodes polyproline domain with a vital role in apoptosis induction, is in relation to the most cancers (18-23) and prevalence of exon 5-8 mutations, which encodes the DNA binding domain of the TP53 molecule are found in many of human cancers (5, 26). On the other hand, a report by Zhao et al., indicated that promoter polymorphism of ABCC1 gene too, is associated with cancer development (10). Here, polymorphisms of 72 and 282 codons of p53 gene and promoter of ABCC1 were investigated. Results showed that Pro/Pro genotype in codon 72 of p53 and AA genotype in ABCC1 in patients are higher than controls and are correlated with the risk of prostate cancer in our population. It was also found that the Pro allele in codon 72 of p53 and the A allele in ABCC1 were significantly associated with prostate cancer in the cohort. Overall, most of cancer samples had GCGA and CCAA genotypes (18 and 11 of 45, respectively), whereas most of controls had GCGA and GGGG genotypes (20 and 16 of 45, respectively). These results showed that CCAA genotype may be more susceptible to developing cancer than other possible genotypes. Our results need confirmation in larger sample size assays. Different human populations have different allele frequencies. Many factors can influence genotyping studies such as genetic background of populations, different life style, selection of control samples and inter-laboratory variation in the genotyping methods and samples conditions (20, 22). Moreover the size of fragment that must be amplified would be an important factor in working with severely degraded DNA specimens, especially DNA obtained from stored tissues like paraffin embedded tissues. Many studies suggested that shorter fragments are better (22, 27). The p53 pathway is composed of many genes with different responses in diverse environmental signals of various populations that may be other explanations for different results (28).
In codon 72 polymorphism of p53, transversion mutation of G to C, leading to substitution of Arg with Pro will result different biological properties. Arg allele as a wild type form of p53 is more efficient in apoptosis induction than Pro allele, which is resulted from increased localization potential of Arg variant in the mitochondria that regulates the release of cytochrome c into cytosol (24, 25). The function of p53 in transcription activation differs between the Arg and the Pro variants, with Pro variant demonstrating increased interaction with TFII (29, 30).In our population, higher frequencies were noted for Pro/Pro as opposed to Arg/Arg. Such differences of two alleles can demonstrate the probable correlation of Pro/Pro genotype with increased risk of prostate cancer.
ABCC1 promoter studies showed that N-myc regulates expression of ABCC1 gene through interaction with a putative E-box element and other cis-acting factors (31). Evidences indicated that SNPs within the ABCC1 (MRP1) are important in predicting the response to chemotherapy in different cancers. This gene can induce a characteristic mutation in p53 (10, 30). Previous studies have shown that p53 mutations are significantly associated with a poor prognosis for patients with cancer (32). SNPs in the promoter region of a gene can potentially alter the affinity of interactions between DNA and nuclear proteins and, so that, affect the efficiency of transcription. Allele G in this prompter region of ABCC1 has a stronger binding affinity for nuclear proteins like p53 than allele A (10). Therefore, TP53 can affect transcription of ABCC1 by binding to its promoter.
Higher frequency of AA genotype was noted in our study. Existence of allele A may be creating abnormal protein that cannot bear an accurate function in drug resistance, affecting nuclear proteins. TP53 cannot bind to abnormal allele A in promoter region of ABCC1 and regulate its transcription. Survey of gene expression of these two genes can confirm the results and hypothesis.
In this study, we also investigated probable new mutation in samples using PCR-based SSCP technique. Results showed new banding patterns only in cancerous patients’ samples than controls that can be novel polymorphic sites for these regions. These abnormal SSCP patterns can characterize single point mutation, gene deletions or rearrangements, which would result in amino acid substitution within the p53 protein. Sequencing of these samples with uncommon patterns may propose a hypothesis that some of these new polymorphisms may contribute insusceptibility to the development of some cancers.
In conclusion, our study showed both p53 codon 72 polymorphism (Pro/Pro genotype) and ABCC1 promoter region G-1666A polymorphism (A allele) appear to have significant association with an increased risk of developing prostate cancer. The precision role of the two polymorphisms can be confirmed through future studies with larger number of samples. This is the first attempt to establish a correlation between p53 and ABCC1 polymorphisms, and cancer development. We believe that the presented data can be used as a basis for future studies to study the functional effects of such polymorphisms and other gene polymorphisms in p53 pathway.
Acknowledgments
We thank the staff of pathological lab of the Tohid Hospital of Sanandaj for their technical assistance.
References
- 1.Ahmedin J, Freddie B, Melissa M, Jacques F, Elizabeth W, David F. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Damin AP, Frazzon AP, Damin DC, Roehe A, Hermes V, Zettler C. et al. Evidence for an association of TP53 codon 72 polymorphism with breast cancer risk. Cancer Detect Prev. 2006;30:523–529. doi: 10.1016/j.cdp.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Sun W, Yang J. Functional mechanisms for human tumor suppressors. J Cancer. 2010;1:136–40. doi: 10.7150/jca.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 5.Bai L, Zhu WG. p53: Structure, function and therapeutic applications. http://www.mupnet.com J Cancer. 2006;2(4):141–153. [Google Scholar]
- 6.Van Heemst D, Mooijaart SP, Beekman M, Schreuder J, de Craen AJ, Brandt BW, Bijlsma R, van Heemst D, Heijmans BT, van Houwelingen JC, Knook DL, Meulenbelt I, de Meijer PHEM, Mooijaart SP, Pijpe J, Schoenmaker M, Slagboom PE, Westendorp RGJ, van de Zande LPWGM, Zwaan BJ. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 7. Schneuber SE, Regiting P, Petru E, Winter R. Breast metastasis 56 months before the diagnosis of primary ovarian cancer: a case study. Anticancer Res 2008;28:3047-3050. PMID: 19031954 [PubMed - indexed for MEDLINE]. [PubMed]
- 8.Bush JA, Li G. Cancer chemoresistance: the relationship between p53 and multidrug transporters. Int J Cancer. 2002;98:323–330. doi: 10.1002/ijc.10226. [DOI] [PubMed] [Google Scholar]
- 9.Yin JY, Huang Q, Yang Y, Zhang JT, Zhong MZ, Zhou HH, Liu ZQ. Characterization and analyses of multidrug resistance-associated protein 1 (MRP1/ABCC1) polymorphisms in Chinese population. Pharmacogenet Genomics. 2009;19:206–216. doi: 10.1097/FPC.0b013e328323f680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Yu BY, Wang DY, Yang JE. Promoter polymorphism of MRP1 associated with reduced survival in hepatocellular carcinoma. World J Gastroenterol. 2010;16:6104–6110. doi: 10.3748/wjg.v16.i48.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin KV, Ueda K, Pastan I, Gottesman MM. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science. 1992:255. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 12.Mohammadi A, Vaziri Gohar A, Shakibaie MR. Mutations in tumor suppressor TP53 Gene in formalin-fixed, paraffin embedded tissues of squamous cell carcinoma (SCC) of lung cancer. Am J Bioch Biotechnol. 2008;4(1):1–6. [Google Scholar]
- 13. Sato Y, Mukai K, Matsuno Y, Furuya S, Kagami Y, Miwa M, et al. The AMeX method: a multipurpose tissue-processing and paraffin-embedding method. II. Extraction of spooled DNA and its application to Southern blot hybridization analysis. Am J Pathol 1990;136:267-271. PMID: 2407122 [PubMed - indexed for MEDLINE]. [PMC free article] [PubMed]
- 14.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y. et al. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 15.Han YC, Teng CZ, Hu ZL, Song YC. An optimal method of DNA silver staining in polyacrylamide gels Electrophoresis. Electrophoresis. 2008;29(6):1355–1358. doi: 10.1002/elps.200700558. [DOI] [PubMed] [Google Scholar]
- 16. Yeh FC, RC Yang, J Mao, Z Ye, TJB Boyle. Pop Gene Version 1.31. Microsoft Window-Based Freeware for Population Genetic Analysis. University of Alberta, Edmonton. http://www.ualberta.ca/wfyeh/. 1999.
- 17.Lane DP. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 18.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cance. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 19.Saxena N. Study of mutations in p53 tumor suppressor gene in human sporadic breast cancers. Indian J Clin Biochem. 2009;24:223–228. doi: 10.1007/s12291-009-0042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang P, Liu J, Li W, Zeng X, Tang J. Role of p53 and p21 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim Biophys Sin (Shanghai) 2010;42:671–676. doi: 10.1093/abbs/gmq069. [DOI] [PubMed] [Google Scholar]
- 21.Peltonen JK, Helppi HM, Paakko P, Turpeenniemi-Hujanen T, Vahakangas KH. p53 in head and neck cancer: functional consequences and environmental implications of TP53 mutations. Head Neck Oncol. 2010;2:36. doi: 10.1186/1758-3284-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki K, Matsui H, Ohtake N, Nakata S, Takei T, Nakazato H, Okugi H, Koike H, Ono Y, Ito K, Kurokawa K, Yamanaka H. A p53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci. 2003;10:430–435. doi: 10.1159/000071162. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, Lin B, Lu Y, Xie Y. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–33. doi: 10.1158/1078-0432. [DOI] [PubMed] [Google Scholar]
- 24.Zhuo W, Zhang Y, Xiang Z, Cai L, Chen Z. Polymorphisms of TP53 codon 72 with breast carcinoma risk: evidence from 12226 cases and 10782 controls. J Exp Clin Cancer Res. 2009;28:115. doi: 10.1186/1756-9966-28-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marelette V, Craig H, Thomas C, Paul D. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the gene in a population at high risk for esophageal cancer in South Africa TP53. Cancer Genet Cytogenet. 2003;140:23–30. doi: 10.1016/S0165-4608(02)00638-6. [DOI] [PubMed] [Google Scholar]
- 26.Makni H, Kaiano J, Villa LL, Matlashewski G. p53 polymorphism in codon 72 and risk of human papilomavirus-associated cervical cancer: Effect of inter-laboratory variation. Int J cancer. 2000;87:528–533. doi: 10.1002/1097-0215(20000815). [DOI] [PubMed] [Google Scholar]
- 27.Mojtahedi Z, Hashemi SB, Khademi B, Karimi M, Haghshenas MR, Fattahi MJ, Ghaderi A. p53 codon 72 polymorphism association with head and neck squamous cell carcinoma. Braz J Otorhinolaryngol. 2010;76:316–20. doi: 10.1590/S1808-86942010000300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henner WD, Evans AJ, Hough KM, Harris EL, Lowe BA, Beer TM. Association of codon 72 polymorphism of p53 with lower prostate cancer risk. Prostate. 2001;49:263–6. doi: 10.1002/pros.10021. [DOI] [PubMed] [Google Scholar]
- 29. Thomas M, Labrecque S, Pim D, Banks L, Matla- shewski G. Twopolymorphic variantsof wild-type p53 differ biochemically and biologically. Mol Cell Biol 1999;19:1092-1100. PMID: 9891044 [PubMed - indexed for MEDLINE]. [DOI] [PMC free article] [PubMed]
- 30.Munoz M, Henderson M, Haber M, Norris M. Role of the MRP1/ABCC1 multidrug transporter protein in cancer. IUBMB Life. 2007;59:752–757. doi: 10.1080/15216540701736285. [DOI] [PubMed] [Google Scholar]
- 31.Staib F, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 32.Yano M, Eguchi H, Hirai Y, MacPhee DG, Sug- ino K. et al. Prognosis in patients with hepatocellular carcinoma correlates to mutations of p53 and/or hMSH2 genes. Eur J Cancer. 2007;43:1092–1100. doi: 10.1016/j.ejca.2007.01.032. [DOI] [PubMed] [Google Scholar]


