Abstract
Background
Gossypium thurberi is a wild diploid species that has been used to improve cultivated allotetraploid cotton.G. thurberi belongs to D genome, which is an important wild bio-source for the cotton breeding and genetic research. To a certain degree, chloroplast DNA sequence information are a versatile tool for species identification and phylogenetic implications in plants. Different chloroplast loci have been utilized for evaluating phylogenetic relationships at each classification level among plant species, including at the interspecies and intraspecies levels. Present study was conducted in order to analyse the sequence of its chloroplast.
Objectives
Present study was conducted to study and compare the complete chloroplast sequence of G. thurberi, analyses of its genome structure, gene content and organization, repeat sequence and codon usage and comparison with two cultivated allotetraploid sequenced cotton species.
Materials and Methods
The available sequence was assembled by DNAman (Version 8.1.2.378). Gene annotation was mainly performed by DOGMA. The map of genome structure and gene distribution were carried out using OGDRAW V1.1. Relative synonymous codon usage (RSCU) of different codons in each gene sample was calculated by codonW in Mobyle. To determine the repeat sequence and location, an online version of REPuter was used.
Results
The G. thurberi chloroplast (cp) genome is 160264 bp in length with conserved quadripartite structure. Single copy region of cp genome is separated by the two inverted regions. The large single copy region is 88,737 bp, and the small single copy region is 20,271 bp whereas the inverted repeat is 25,628 bp each. The plastidic genome has 113 single genes and 20 duplicated genes. The singletones encode 79 proteins, 4 ribosomal RNA genes and 30 transfer RNA genes.
Conclusions
Amongst all plastidic genes only 18 genes appeared to have 1-2 introns and when compared with cpDNA of two cultivated allotetraploid, rps18 was the only duplicated gene in G.thurberi. Despite the high level of conservation in cp genome SSRs ,these are useful in analysis of genetic diversity due to their greater efficiency as opposed to genomic SSRs. Low GC content is a significant feature of plastidic genomes, which is possibly formed after endosymbiosis by DNA replication and repair.
Keywords: Chloroplast genome, Complete sequence, Gossypium thurberi
1. Background
Most plastidic genomes have four regions, namely large single copy region (LSC, 80 Kb), small single copy region (SSC, 20 kb) and two inverted repeat regions (IR, 25 kb). The single copy region is separated by two IRs. This structural conservation however breaks in some plants sauch as Vicia faba (1) and Cryptomeria japonica (2, 3) by loss of an IR, and in Euglena gracilis that has three tandem repeats (4).
Variations among different species provide large information for the phylogenetic studies. Chloroplasts have low mutation rate with great deal of conservation in their genome size and structure, gene content and organization. Few differences have been reported in the same species, but significant differences could be detected between the different species in genome size and gene orientation (5). It has been reported that, chloroplast genes like 16S, 23S, ndhB, psbA, psbD, psaB, pasA, psbC, psbB and rbcL are appropriate to study the relationship among higher plants; ycf1, ycf2, accD, matK, rpoC2 and ndhF are more suitable to study the relationship of the close species (5).
Transplastomics have proved to be a powerful tool to improve the plant genetic architecture with high expression of the foreign protein, low risk of the pollen pollution (6) and no gene silencing. Therefore and In addition to phylogenetic analysis based on plastidic genomes, it is imperative to understand the chloroplast genome in order to logically design our next generation transplastomics. Accordingly, chloroplast genomes of many species have been sequenced (7-14).
2. Objectives
Gossypium includes 52 species that are divided to eight diploid genome A-G and K (2n=26), and one allotetraploid genome (AADD, 2n=52). G. barbadense and G. hirsutum are extensively cultivated in the world and their chloroplast genome sequences have been published. G. thurberi belongs to D genome, which is an important wild bio-source for the cotton breeding and genetic research. Present study was conducted to study and compare the complete chloroplast sequence of G. thurberi, analyses of its genome structure, gene content and organization, repeat sequence and codon usage. Meanwhile the comparison of the three sequenced cotton species was performed.
3. Materials and Methods
3.1. Chloroplast Sequence
Compleate chloroplast genome sequence of Gossypium thurberi with accession number NC_015204.1 downloaded from NCBI (http://www.ncbi.nlm.nih.gov/nuccore/?term=Gossypiumthurberi).
3.2. Genome Assembly and Gene Annotation
The available sequence was assembled by DNAman (Version 8.1.2.378). Gene annotation was mainly performed by DOGMA (Dual Organellar Geno Me Annotator, http://dogma.ccbb.utexas.edu/; Wyman, 2004). DOGMA uses BLAST against 11 plant chloroplast database (Adiantum capillus-veneris, Arabidopsis thaliana, Chlorella vulgaris, Lotus japonicus, Marchantia polymorpha, Mesostigma viride, Nephroselmis olivacea, Nicotiana tabacum, Oenothera elata, Oryza sativa, Pinus thunbergii, Psilotum nudum, Spinacia oleracea, Triticum aestivum, Zea mays). Identity cutoff for protein coding genes was set at 60%. Identity cutoff for RNAs was set at 80%. The map of genome structure and gene distribution were carried out using OGDRAW V1.1 (OrganellarGenomeDRAW, http://ogdraw.mpimp-golm.mpg.de/), which takes a Genbank file or a special accession number (15).
3.3. Chloroplast Genome Analysis
Relative synonymous codon usage (RSCU) of different codons in each gene sample was calculated by codonW in Mobyle (http://mobyle.pasteur.fr/cgi-bin/portal.py). To determine the repeat sequence and location, an online version of REPuter (http://bibiserv.techfak.uni-bielefeld.de/reputer/) was used (16). Searching condition was followed as Saski (3).
4. Results
4.1. Overall Structure
Chloroplast genome of G. thurberi (Figure 1) has a conserved quadripartite structure. Total genome is a circular DNA molecule of 160,264 bp, which is shorter than G. barbadnese (17) and G. hirsutum (18). The two single copy regions are separated by the two inverted repeats. The whole genome was analyzed (Table 1). The large single copy region is 88,737 bp, the small single copy is 20,271 bp and the two inverted repeats are 25, 628 bp each. The coding region is 91,485 bp in length, accounting for 57.08% of the whole plastidic genome, which is similar to Gossypium hirsutum by 56.46% (18), Bambusa oldhamii by 53.4% (13) and Dendrocalamus latiflorus by 53.4% (13), genus Megaleranthis 52.4% (5), genus Alsophila 53.2% (8). where as it is smaller than Glycine max (60%). G. thurberi plastidic genome codes for proteins (49.76%), tRNA genes (1.73%) and rRNA (5.60%), similar to Manihot esculenta (19), cucumber (20) and coffee (21). The non-coding region is 70,351 bp in length (43.90% of the genome). The proportions of intergenic spacers and intron are 31.15% and 12.75%, respectively.
Figure 1 .
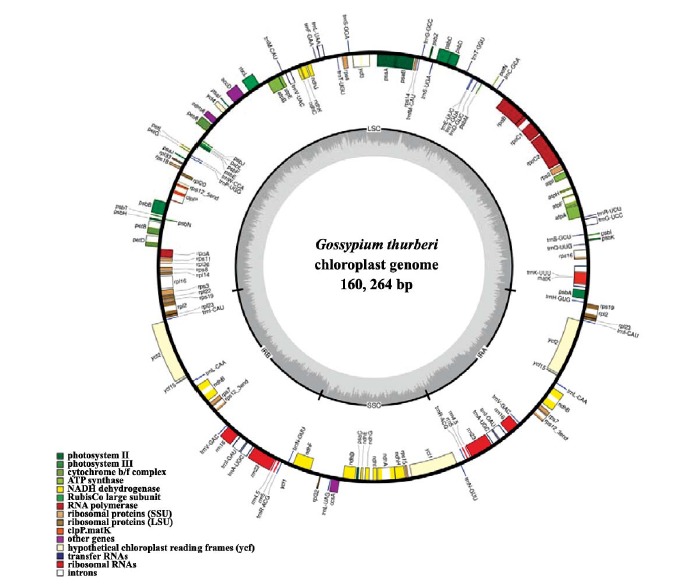
Gossypium thureri chloroplast genome structure and gene organization Note: genes shown outside of the circle transcribed anticlockwise and shown inside of the circle transcribed clockwise. tRNA genes are shown by 1 letter of the coded amino acid followed by anticodon (genome map was created by using OGDRAW V 1.1, Lohse, 2007)
Table 1. Repeat sequences detected in chloroplast genome of G. thurberi .
| Number | Size(bp) | Location | Match direction |
| 1 | 30 | intron, IGS | C |
| 2 | 32 | IGS | C |
| 3 | 30 | IGS | F |
| 4 | 30 | IGS-trnS, | F |
| 5 | 30 | Itron, IGS | F |
| 6 | 30 | ycf2 | F |
| 7 | 31 | IGS | F |
| 8 | 31 | IGS | F |
| 9 | 31 | IGS | F |
| 10 | 32 | IGS | F |
| 11 | 33 | IGS | F |
| 12 | 34 | IGS | F |
| 13 | 34 | IGS | F |
| 14 | 34 | IGS | F |
| 15 | 34 | IGS | F |
| 16 | 34 | ycf2 | F |
| 17 | 34 | ycf2 | F |
| 18 | 35 | Intron | F |
| 19 | 36 | Intron, IGS | F |
| 20 | 38 | IGS, Intron | F |
| 21 | 38 | ycf2 | F |
| 22 | 38 | ycf2 | F |
| 23 | 38 | ycf2 | F |
| 24 | 41 | Intron | F |
| 25 | 43 | IGC | F |
| 26 | 47 | ycf2 | F |
| 27 | 52 | ycf2 | F |
| 28 | 64 | ycf2 | F |
| 29 | 64 | ycf2 | F |
| 30 | 72 | psaB, psaA | F |
| 31 | 30 | trnS | P |
| 32 | 31 | IGS | P |
| 33 | 31 | IGS | P |
| 34 | 31 | IGS | P |
| 35 | 34 | IGS | P |
| 36 | 34 | ycf2 | P |
| 37 | 34 | ycf2 | P |
| 38 | 34 | IGS | P |
| 39 | 34 | IGS | P |
| 40 | 34 | ycf2 | P |
| 41 | 34 | ycf2 | P |
| 42 | 36 | Intron, IGS | P |
| 43 | 38 | Intron, IGS | P |
| 44 | 38 | ycf2 | P |
| 45 | 38 | ycf2 | P |
| 46 | 41 | IGS | P |
| 47 | 43 | IGS | P |
| 48 | 43 | IGS | P |
| 49 | 48 | IGS | P |
| 50 | 52 | ycf2 | P |
| 51 | 52 | ycf2 | P |
| 52 | 64 | ycf2 | P |
| 53 | 64 | ycf2 | P |
| 54 | 30 | ycf1 | R |
| 55 | 30 | IGS | R |
| 56 | 30 | IGS | R |
| 57 | 30 | IGS | R |
| 58 | 30 | IGS | R |
| 59 | 31 | IGS | R |
| 60 | 32 | Intron | R |
| 61 | 32 | IGSS | R |
| 62 | 32 | IGS | R |
| 63 | 33 | IGS | R |
| 64 | 33 | IGS | R |
| 65 | 35 | IGS | R |
| 66 | 38 | IGS | R |
Note: IGS represents intergenic spacer sequence. F represents forward (direct) match, R represents reverse match, C represents complement match, P represents palindromic (invert) match
4.2. Repeat Sequence
Chloroplast genome structures are similar to prokaryotes, it has been considered uncommon to have large scale of repeat sequences in these genomes. Here, PEPuter was used to detect the repeat sequence of cp genome of G. thurberi. Four types of repeats were detected; forward (direct) match, reverse match, complements match and palindromic (inverted) match. Sixty six repeats having more than 30 bp in length are listed in (Table 1). There are 2 complementary repeats, 28 forward repeats, 23 inverted repeats and 13 reverse repeats. Most of the repeats are located at ycf2 and intergenic spacers (IGS), and few located at trnS and introns. The largest repeat is 72 bp, which is located at psaB and psaA, while the most of the repeats are 30-40 bp. In addition to the four types of repeats, there are few simple sequence repeats (SSRs).
Simple sequence repeats were screened in G. thurberi chloroplast genome and 67 cpSSRs (³10 bp) were obtained. Most of the SSRs are mononucleotide repeats, while 16 dinucleotide repeats and 3 trinucleotide repeats. The longest repeat is the repeat of “CT”, which is 14 bp, but the most of the repeats are C and T having 13 bp (Table 2).
Table 2. Simple sequence repeat (SSR) in G. thurberi chloroplast genome .
| Repeat | Repeat sequence | Number | Max (bp) |
|
mononucleotide dinucleotide trinucleotide total |
A C T AT CT TA TC TG AAT ATA TTA |
14 2 32 8 1 5 1 1 1 1 1 67 |
12 13 13 12 14 12 10 10 12 12 12 14 |
4.3. Gene Content and Codon Usage
Genes coded by the cp of G. thurberi are listed (Table 3). Among the total 79 protein coded genes, there are 4 rRNA genes, 30 tRNA genes and 113 single genes; out of which 20 genes are duplicated, locating at IR. According to the gene function, all genes can be classified as genes of the functional genetic system, the photosynthetic system, the biosynthesis and some with unknown function. In G. thurberi cp genome, five genes with unknown function (ycf gene) were detected and considered as to be essential in plants, which were highly conserved between species (22). Intrestingly, two genes, namely rps12 has an intron (5). rps12 was separated (by an intron) into two fragments with one exon locating at LSC (5'-end) and the other at 3'-end at IR. matK is 1.5 Kbp in length, and was found in the intron of trnK-UUU, which is the only gene located in an intron and encodes maturase K. This gene has both conserved and variable fragments (23). Thus, it is frequently used in phylogenetic studies (24, 25, 23, 14).
Table 3. Genes coded by G. thurberi chloroplast genome .
| Group | Gene name | |
|
protein gene RNA gene |
Subunit of Acetyl-CoA-carboxylase Large subunit of rubisco Subunits of NADH-dehydrogenase Subunits of ATP synthase Subunits of cytochrome b/f complex subunits of photosystem I and II DNA dependendt RNA polymerase Large subunit of ribosome Small subunit of ribosome Others Function unknown ribosomal RNA gene transfer RNA gene |
accD rbcL ndhA*, ndhB*§, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK atpA, atpB, atpE, atpF*, atpH, atpI petA, petB*, petD*, petG, petL, petN ccsA psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ rpoA, rpoB, rpoC1*, rpoC2 rpl14, rpl16*, rpl2*§, rpl20, rpl22, rpl23§, rpl32, rpl33, rpl36 rps11, rps12*§, rps14, rps15*, rps16*, rps18, rps19§, rps2, rps3, rps4, rps7§, rps8 cemA, clpP**, matK ycf1§, ycf15§, ycf2§, ycf3**, ycf4 rrn16§, rrn23§, rrn4.5§, rrn5§ trnA-UGC*§, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnGUCC*, trnG-GCC, trnH-GUG, trnI-CAU§, trnI-GAU*§, trnK-UUU*, trnLCAA§, trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU§, trnP-UGG, trnQ-UUG, trnR-ACG§, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnTUGU, trnV-GAC§, trnV-UAC*, trnW-CCA, trnY-GUA |
Note: § reflects gene located in IR; * reflects gene which has one intron; ** reflects gene which has two introns
The codon usage was analyzed (Table 4). ATG and TGG code for methionine as the start codon and tryptophane, respectively with RSCU value=1. RSCU values of the three terminal codons TAA, TGA and TAG are 1.76, 0.64 and 0.6, respectively. Accoding to RSCU value, G. thurberi prefers TAA as its stop codon. The RSCU values grater than 1 indicates greater codon frequency. Most of the codons prefer A or T at the third position. The analysis of the composition for the codons showed that A+T content at the third position was 72.6%, similar to what was reported for Alophila (8) and Panax schinseng Nees (1).
Table 4. Codon analysis of G. thurberi chloroplast genes that code for proteins .
| AA | Codon | Number | RSCU | AA | Codon | Number | RSCU |
| Phe | TTT | 908 | 1.33 | Ser | TCT | 498 | 1.71 |
| TTC | 453 | 0.67 | TCC | 265 | 0.91 | ||
| Leu | TTA | 824 | 1.97 | TCA | 355 | 1.22 | |
| TTG | 517 | 1.23 | TCG | 156 | 0.54 | ||
| CTT | 506 | 1.21 | Pro | CCT | 373 | 1.51 | |
| CTC | 153 | 0.37 | CCC | 191 | 0.77 | ||
| CTA | 350 | 0.83 | CCA | 272 | 1.1 | ||
| CTG | 165 | 0.39 | CCG | 150 | 0.61 | ||
| Ile | ATT | 1000 | 150. | Thr | ACT | 479 | 1.56 |
| ATC | 384 | 58. | ACC | 251 | 0.82 | ||
| ATA | 619 | 93. | ACA | 368 | 1.2 | ||
| Met | ATG | 542 | 1 | ACG | 129 | 0.42 | |
| Val | GTT | 459 | 142. | Ala | GCT | 626 | 1.77 |
| GTC | 166 | 51. | GCC | 242 | 0.68 | ||
| GTA | 479 | 148. | GCA | 359 | 1.02 | ||
| GTG | 192 | 0.59 | GCG | 187 | 0.53 | ||
| Tyr | TAT | 713 | 1.61 | Cys | TGT | 190 | 1.52 |
| TAC | 174 | 0.39 | TGC | 60 | 0.48 | ||
| Ter | TAA | 47 | 1.76 | Ter | TGA | 17 | 0.64 |
| TAG | 16 | 0.6 | Trp | TGG | 458 | 1 | |
| His | CAT | 464 | 1.48 | Arg | CGT | 307 | 1.32 |
| CAC | 162 | 0.52 | CGC | 116 | 0.5 | ||
| Gln | CAA | 663 | 1.53 | CGA | 326 | 1.4 | |
| CAG | 203 | 0.47 | CGG | 106 | 0.46 | ||
| Asn | AAT | 859 | 1.54 | Ser | AGT | 364 | 1.25 |
| AAC | 253 | 0.46 | AGC | 107 | 0.37 | ||
| Lys | AAA | 909 | 1.51 | Arg | AGA | 390 | 1.68 |
| AAG | 294 | 0.49 | AGG | 148 | 0.64 | ||
| Asp | GAT | 767 | 1.59 | Gly | GGT | 554 | 1.29 |
| GAC | 196 | 0.41 | GGC | 195 | 0.45 | ||
| Glu | GAA | 926 | 1.5 | GGA | 653 | 1.52 | |
| GAG | 307 | 0.5 | GGG | 313 | 0.73 |
Note: Codon shown in bold represents RSCU value >1
5. Discussion
5.1. SSR in cpDNA
Despite the high level of conservation in cp genome SSRs are evident as stated in previous reports (20) cpSSRs are useful in analysis of genetic diversity (26) due to their greater efficiency as opposed to genomic SSRs (26). Furthermore and due to the greater level of conservation, the information of the other species can be used to design specific primers for a species with unknown sequence data (27, 28, 14).
5.2. Gene Loss in Chloroplast
During the course of evolution, loss and gain of genetic material have been noted for cpDNA. For instance ycf15, a non-functional gene in other plants (10, 22, 12) is also present in G. thurberi. The other example is infA, most mobile gene between chloroplast and nuclear genome, that codes for a translation initial factor 1 (29, 30, 31). In our study similar to cassava (19) G. hirsutum (18), the infA was absent. However, some others had the infA as a pseudogene (17, 22), while in others infA appeared as an intact gene (21). Similar to G. hirsutum (18) and G. barbadense (17) and angiosperms, trnP-GGG was absent in G. thurberi. However, trnP-GGG was reported in Cryptomeria japonica (2). Thus it can be suggested that the gene has lost before the divergence of angiosperms. The other gene that worth considering was rpl22 that codes for the large subunit of ribosomal protein 22. rpl22 is present in G. thurberi chloroplast genome similar to G. barbadense (17), but has been reported to be absent in G. hirsutum (18) and 3 legumes, namely Glycine, Lotus and Medicago (3). Therefore, its analysis may shed some light on the evolution of Gossypium.
5.3. Extent of IR
The border of the IR is usually different between species and the IR expansion and contraction are important as far as genome size is concerned. IR expansion often leads to larger sizes of genome. Usually the pseudogenes are residing at the junction of IR and LSC/ SSC. The differences of the junctions among G. thurberi and five other species were analyzed (Figure 2).
Figure 2 .
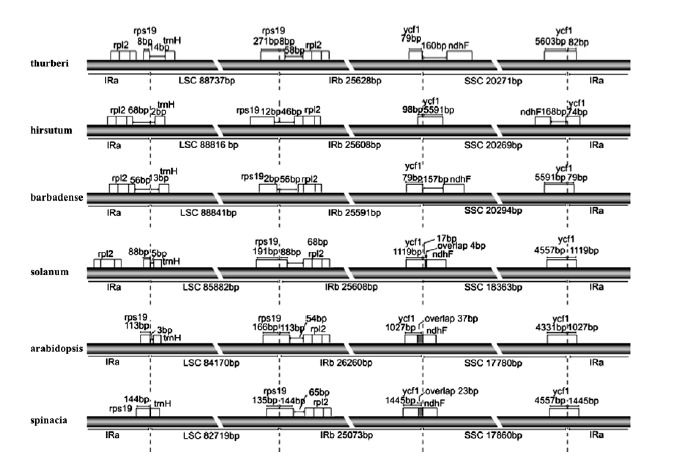
Comparison among LSC, IR and SSC border regions of three common reference species with studied genomes Note: all units of the sequence in the map is base pair
The IRb/LSC junction was found within rps19 in G. thurberi, Solanum lycopersicum, Arabidopsis thaliana and Spinacia oleracea, indicating that rps19 was duplicated at the junction at IRa and LSC. This duplication is very common in plants (20, 13). G. thurberi (8 bp) and Spinacia (144 bp) have the shortest and the longest duplications, respectively.
On the border of IRb and SSC, G. thurberi is similar with G. barbadense, having 79 bp of ycf1 fragment on the IRb border. Solanum, Arabidopsis and Spinacia have the same type of overlapping of ycf1 and ndhF at the junction. The longest overlap is in Arabidopsis with 37 bp and the shortest is in Solanum with 17 bp. The overlap is also found in Cucumis (20). The ycf1 is located at the junction of SSC/IRb. So ycf1 was duplicated in IRb at the border of IRb and SSC. In the Spinacia, ycf1 has the longest duplication with 1445 bp. In G. thurberi and G. barbadense, ycf1 has the shortest duplication with 79 bp. Gossypium hirsutum is in opposite direction of SSC as compared to other five speciesand therefore; ycf1 is located at the junction of IRb/SSC with 98 bp.
5.4. Intron
In the G. thurberi, 18 genes were found containing one or two introns, which is the same as in Panax schinseng Nees (1). In contrast to G. thurberi, introns were absent in rpoC1 and clpP in B. Oldhamii and D. Latiflorus (13). The number and location of the intron in chloroplast seems to be conserved. The comparison (Table 5) of introns among G. thurberi, G. hirsutum and G. barbadense shows that 18 genes have one or two introns in the cp genome; 6 of which are tRNA coding genes and the rest are protein-coding genes. The longest intron is located in trnK-UUU with 2542 bp in G. thurberi, which is the only intron in cotton with another gene, matK, inside. The smallest intron is located in rpl12-3end with 536 bp, which is situated in IR. Genes of ycf3 and clpP are located at LSC and are divided by two introns. Small variations among the introns were noted in three cotton species; intron lengths for ycf3-1, ycf3-2, rps12_3end, ndhB and ndhA are conserved, while the others have small variations. These intron sequences have a high identity, especially ycf3-2 with a 1 sequence identity among the three cotton species.
Table 5. The comparison of introns among three cotton species; G. thurberi, G. hirsutum and G. barbadense .
| Intron | G. thurberi | G. hirsutum | G. barbadense | Sequence identity (%) |
| trnK-UUU | 2534 | 2542 | 2535 | 98.37 |
| rps16 | 868 | 871 | 870 | 99.24 |
| trnG-UCC | 770 | 771 | 763 | 98.29 |
| atpF | 790 | 804 | 805 | 98.97 |
| rpoC1 | 741 | 753 | 753 | 99.42 |
| ycf3-1 | 777 | 777 | 777 | 99.61 |
| ycf3-2 | 789 | 789 | 789 | 100 |
| trnL-UAA | 583 | 575 | 582 | 98.8 |
| trnV-UAC | 606 | 618 | 609 | 97.85 |
| rps12_3end | 536 | 536 | 536 | 99.94 |
| clpP1 | 891 | 891 | 890 | 99.52 |
| clpP2 | 682 | 683 | 679 | 98.83 |
| petB | 760 | 760 | 761 | 99.52 |
| petD | 757 | 757 | 754 | 99,87 |
| rpl16 | 1138 | 1140 | 1135 | 99.50 |
| rpl2 | 693 | 695 | 688 | 99,52 |
| ndhB | 683 | 683 | 683 | 99.95 |
| trnI-GAU | 954 | 954 | 959 | 99.48 |
| trnA-UGC | 797 | 797 | 795 | 99.79 |
| ndhA | 1076 | 1076 | 1076 | 99.78 |
5.5. GC Content
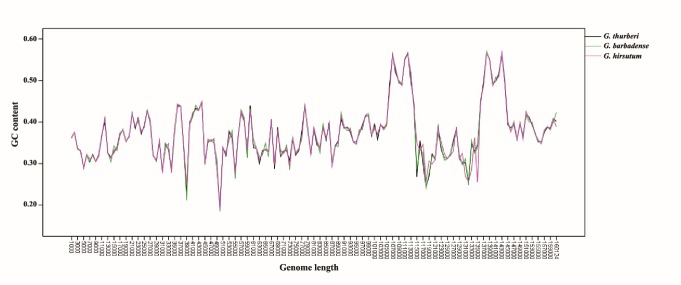
The GC content of G. thurberi cp genome is 37.22 %, similar to other plants, such as 37.86% in Solanum lycopersicum, 37.85% in Nicotiana tabacum, 37.56% in Atropa belladonna, 37.25% in G. hirsutum and 34% in Glycine max. Both coding and non-coding regions are low in GC content (32) with 40.35% and 33.08%, respectively in G. thurberi. Variation in GC content among four different regions in G. thurberi cpDNA was observed (Table 6) and IR was the richest (42.95%), similar to an earlier report (9). It is supposed that ribosomal genes (rrna4.5, rrna5, rrna16, rrna23) and coding regions (19, 8, 10, 5) are responsible for high GC content in IR. GC content distribution of the each region is similar with other species (20, 10, 5). According to Gao (8) GC content was uneven across cp genome in Alsophila. In this study, we cut genomes of the three Gossypium species into 1 kb-unit to compare uint to uint GC content across whole chloroplast genome. Our result showd that the distribution of GC is similar across the whole genome (Figure 3). At SSC region G. barbadense and G. thurberi are similar but different from G. hirsutum because of different direction of SSC. Across the whole genomes of the three cotton species, different fragments and even the adjacent fragments share different GC contents.
Table 6. GC content of G. thurberi chloroplast genome .
| coding region | non-coding region | ||||||||||
| protein | trna | rrna | total | IGS | intron | total | Complete genome | LSC | SSC | IR | |
| Length (bp) | 79740 | 2775 | 8970 | 91485 | 49915 | 20436 | 70351 | 160264 | 88737 | 20271 | 25628 |
| proportion% | 49.76 | 1.73 | 5.60 | 57.08 | 31.15 | 12.75 | 43.90 | 100.00 | 55.37 | 12.65 | 15.99 |
| T% | 31.05 | 23.14 | 22.32 | 30.34 | 34.32 | 32.28 | 33.73 | 31.83 | 33.15 | 34.46 | 28.52 |
| A% | 29.85 | 24.58 | 22.17 | 29.31 | 34.10 | 30.97 | 33.19 | 30.95 | 31.67 | 33.92 | 28.54 |
| C% | 19.28 | 26.13 | 27.79 | 20.56 | 15.99 | 19.12 | 16.90 | 18.99 | 18.11 | 16.54 | 20.66 |
| G% | 18.42 | 26.16 | 27.71 | 19.79 | 15.58 | 17.63 | 16.18 | 18.23 | 17.08 | 15.09 | 22.29 |
| A+T% | 60.90 | 47.71 | 44.49 | 59.65 | 68.42 | 63.25 | 66.92 | 62.78 | 64.81 | 68.38 | 57.05 |
| C+G% | 37.60 | 52.29 | 55.51 | 40.35 | 31.58 | 36.75 | 33.08 | 37.22 | 35.19 | 31.62 | 42.95 |
Note: matK and gene overlaps are analyzed twice. IGS represents inter gene space
Figure 3 .
GC content of three Gossypium species (G. thurberi, G. hirsutum and G. barbadense)
Gao (8) reported that GC contents in the chloroplast genomes are not the same between genes in different functional groups; rRNA (55.18%)>tRNA (54.55%)>photosynthetic (43.85%)>genetic system (40.80%)>NADH (39.54%). In G. thurberi, similar data was obtained. In the coding region, the rRNA genes have the highest GC content (55.51%) and the protein genes have the lowest (37.69%). In the non-coding region, GC content of IGS and intron is 31.58% and 36.75%, respectively. The non-coding regions experienced a fast evolution, thus the non-coding region is richer in GC than coding regions.
GC content is an important feature of a genome that is correlated to the number of microRNA binding sites (33), functional elements physical location (34), recombination rate and gene distribution (35), organelle RNA editing (36) and gene expression regulation (37). GC content varies in the 5'UTR and 3'UTR (34). GC content has a rare relationship with replication timing in human genome (38). GC content also have an effect on RNAi, because it is highly correlated to RNAi target site accessibility and negatively correlated with RNAi activity (39).
Low GC content is a significant feature of plastid genomes, which is possibly formed after endosymbiosis by DNA replication and repair (32). In viruses GC content is not dependent on genes constitution, but it is correlated with its location (40). Whether or not this exists in chloroplast genome needs more efferts on further studies.
References
- 1.Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11(4):247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 2.Hirao T, Watanabe A, Kurita M, Kondo T, Takata K. Complete nucleotide sequence of the Cryptomeria japonica D Don chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59(2):309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 4.Hallick RB, Hong L, Drager RG, Favreau MR, Monfort A, Orsat B, Spielmann A, Stutz E. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21(15):3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young-Kyu Kim CWP, Ki-Joong K. Complete Chloroplast DNA Sequence from a Korean Endemic Genus,Megaleranthis saniculifolia, and Its Evolutionary Implications Mol. Cells. 2009;27(3):365–381. doi: 10.1007/s10059-009-0047-6. [DOI] [PubMed] [Google Scholar]
- 6.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104(17):6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diekmann K, Hodkinson TR, Wolfe KH, van den Bekerom R, Dix PJ, Barth S. Complete chloroplast genome sequence of a major allogamous forage species, perennial ryegrass (Lolium perenne L) DNA Res. 2009;16(3):165–176. doi: 10.1093/dnares/dsp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao L, Yi X, Yang YX, Su YJ, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YK, Park CW, Kim KJ. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol Cells. 2009;27(3):365–381. doi: 10.1007/s10059-009-0047-6. [DOI] [PubMed] [Google Scholar]
- 10.Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, Kolganova TV, Skyabin KG. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 2008;66(6):555–564. doi: 10.1007/s00239-008-9091-7. [DOI] [PubMed] [Google Scholar]
- 11.Oliver MJ, Murdock AG, Mishler BD, Kuehl JV, Boore JL, Mandoli DF, Everett KD, Wolf PG, Duffy AM, Karol KG. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics. 2010;11:143–156. doi: 10.1186/1471-2164-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, Tragoonrung S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17(1):11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu FH, Kan DP, Lee SB, Daniell H, Lee YW, Lin CC, Lin NS, Lin CS. Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol. 2009;29(6):847–856. doi: 10.1093/treephys/tpp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Zhang X, Liu G, Yin Y, Chen K, Yun Q, Zhao D, Al-Mssallem IS, Yu J. The Complete Chloroplast Genome Sequence of Date Palm (Phoenix dactylifera L) PLoS One. 2010;5(9):143–152. doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52(5-6):267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahim RI, Azuma J, Sakamoto M. Complete nucleotide sequence of the cotton (Gossypium barbadense L) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet Syst. 2006;1(5):11–21. doi: 10.1266/ggs.81.311. [DOI] [PubMed] [Google Scholar]
- 18.Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniell H, Wurdack KG, Kanagaraj A, Lee SB, Saski C, Jansen RK. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor Appl Genet. 2008;116(5):723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR, Cho KY. Complete sequence and organization of the cucumber (Cucumis sativus L cv Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 2006;25(4):334–340. doi: 10.1007/s00299-005-0097-y. [DOI] [PubMed] [Google Scholar]
- 21.Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. The complete nucleotide sequence of the coffee (Coffea arabica L) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol J. 2007;5(2):339–353. doi: 10.1111/j.1467-7652.2007.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steane D A. Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae) DNA Res. 2005;12(3):215–220. doi: 10.1093/dnares/dsi006. [DOI] [PubMed] [Google Scholar]
- 23.Wilson C A. Phylogeny of Iris based on chloroplast matK gene and trnK intron sequence data. Mol Phylogenet Evol. 2004;33(2):402–412. doi: 10.1016/j.ympev.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ, Jermiin LS, Wolfe KH. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13(3):645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohsako T, Ohnishi O. Nucleotide sequence variation of the chloroplast trnK/matK region in two wild Fagopyrum (Polygonaceae) species, F leptopodum and F statice. Genes Genet Syst. 2001;76(1):39–46. doi: 10.1266/ggs.76.39. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro M M, Mariette S, Vendramin GG, Szmidt AE, Plomion C, Kremer A. Comparison of genetic diversity estimates within and among populations of maritime pine using chloroplast simple-sequence repeat and amplified fragment length polymorphism data. Mol Ecol. 2002;11(5):869–877. doi: 10.1046/j.1365-294X.2002.01490.x. [DOI] [PubMed] [Google Scholar]
- 27.Ishii T, Mori N, Takahashi C, Ikeda N, Kamijima O. Evaluation of allelic diversity at chloroplast microsatellite loci among common wheat and its ancestral species. Theor Appl Genet. 2001;103:9. doi: 10.1007/s001220100715. [DOI] [Google Scholar]
- 28.Weising K, Gardner RC. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome. 1999;42(1):9–19. doi: 10.1139/g98-104. [DOI] [PubMed] [Google Scholar]
- 29.Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 30.Miller JT, Bayer RJ. Molecular phylogenetics of Acacia (Fabaceae: Mimosoideae) based on the chloroplast MATK coding sequence and flanking TRNK intron spacer regions. Am J Bot. 2001;88(4):697–705. doi: 10.1071/SB01035. [DOI] [PubMed] [Google Scholar]
- 31.Sands JF, Cummings HS, Sacerdot C, Dondon L, Grunberg-Manago M, Hershey JW. Cloning and mapping of infA, the gene for protein synthesis initiation factor IF1. Nucleic Acids Res. 1987;15(13):5157–5168. doi: 10.1093/nar/15.13.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe CJ, Barbrook AC, Koumandou VL, Nisbet RE, Symington HA, Wightman TF. Evolution of the chloroplast genome. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):99–106. doi: 10.1098/rstb.2002.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis N, Biddlecom N, Hecht D, Fogel GB. On the relationship between GC content and the number of predicted microRNA binding sites by MicroInspector. Comput Biol Chem. 2008;32(3):222–226. doi: 10.1016/j.compbiolchem.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Kasif S, Cantor CR, Broude NE. GC/AT-content spikes as genomic punctuation marks. Proc Natl Acad Sci USA. 2004;101(48):16855–16860. doi: 10.1073/pnas.0407821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freudenberg J, Wang M, Yang Y, Li W. Partial correlation analysis indicates causal relationships between GC-content, exon density and recombination rate in the human genome. BMC Bioinformatics. 2009;10Suppl 1:S66. doi: 10.1186/1471-2105-10-S1-S66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith D R. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 2009;71(6):627–639. doi: 10.1007/s11103-9545-3. [DOI] [PubMed] [Google Scholar]
- 37.Paterson AH, Curt L, Wendel JF. A rapid method for extraction of cotton (Gossypium spp) genomic DNA suitable for RFLP or PCR analysis. Plant Molecular Biology Reporter. 1996;11:6–23. doi: 10.1007/BF02670470. [DOI] [Google Scholar]
- 38.Watanabe Y, Abe T, Ikemura T, Maekawa M. Relationships between replication timing and GC content of cancer-related genes on human chromosomes 11q and 21q. Gene. 2009;433(1-2):26–31. doi: 10.1016/j.gene.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Chan CY, Carmack CS, Long DD, Maliyekkel A, Shao Y, Roninson IB, Ding Y. A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinformatics. 2009;10Suppl 1:S33. doi: 10.1186/1471-2105-10-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khrustalev VV, Barkovsky EV. Mutational pressure is a cause of inter- and intragenomic differences in GC-content of simplex and varicello viruses. Comput Biol Chem. 2009;33(4):295–302. doi: 10.1016/j.compbiolchem.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]