Abstract
Despite their essential role in the process of chromosome segregation in most eukaryotes, centromeric histones show remarkable evolutionary lability. Not only have they been lost in multiple insect lineages, but they have also undergone gene duplication in multiple plant lineages. Based on detailed study of a handful of model organisms including Drosophila melanogaster, centromeric histone duplication is considered to be rare in animals. Using a detailed phylogenomic study, we find that Cid, the centromeric histone gene, has undergone at least four independent gene duplications during Drosophila evolution. We find duplicate Cid genes in D. eugracilis (Cid2), in the montium species subgroup (Cid3, Cid4) and in the entire Drosophila subgenus (Cid5). We show that Cid3, Cid4, and Cid5 all localize to centromeres in their respective species. Some Cid duplicates are primarily expressed in the male germline. With rare exceptions, Cid duplicates have been strictly retained after birth, suggesting that they perform nonredundant centromeric functions, independent from the ancestral Cid. Indeed, each duplicate encodes a distinct N-terminal tail, which may provide the basis for distinct protein–protein interactions. Finally, we show some Cid duplicates evolve under positive selection whereas others do not. Taken together, our results support the hypothesis that Drosophila Cid duplicates have subfunctionalized. Thus, these gene duplications provide an unprecedented opportunity to dissect the multiple roles of centromeric histones.
Keywords: positive selection, gene conversion, protein motifs, molecular evolution
Introduction
Centromeres are the chromosomal regions that link DNA to the spindle during cell division, thus ensuring faithful segregation of genetic material. Proper centromere function is critical for eukaryotic life. Centromeric defects can result in aneuploidy and cycles of chromosome breakage (McClintock 1939; Hassold and Hunt 2001) with catastrophic consequences for genome stability and fertility. Despite the fact that centromeres are essential for life, centromere architecture is remarkably diverse (Kursel and Malik 2016). Centromeric DNA sequences (Lohe and Brutlag 1987; Schueler et al. 2001; Lee et al. 2005) and centromeric proteins (Malik and Henikoff 2001; Talbert et al. 2004; Schueler et al. 2010) also evolve rapidly in diverse organisms. This diversity and rapid evolution make it nearly impossible to name a single defining feature of all centromeres. However, the hallmark of many centromeres is the presence of a specialized centromeric H3 variant called CenH3 [CENP-A in mammals (Earnshaw and Rothfield 1985; Palmer et al. 1991), Cid in Drosophila (Henikoff et al. 2000)]. Despite being essential for chromosome segregation in most eukaryotes (Stoler et al. 1995; Howman et al. 2000; Blower and Karpen 2001), CenH3 evolves rapidly (Malik and Henikoff 2001; Talbert et al. 2002) Thus, paradoxically, proteins and DNA that mediate chromosome segregation in eukaryotes are less conserved than one would expect given their participation in an essential process. This rapid evolution despite the expectation of constraint is referred to as the “centromere paradox” (Henikoff et al. 2001).
Genetic conflicts provide one potential explanation for the rapid evolution of centromeric DNA and proteins. In both animals and plants, the asymmetry of female meiosis provides an opportunity for centromere alleles to act selfishly to favor their own inclusion in the oocyte and subsequent passage into offspring rather than the polar body. In female meiosis, centromeric expansions (Fishman and Saunders 2008) and differential recruitment of centromeric proteins resulting in centromere strength variation between homologs (Chmatal et al. 2014) may provide the molecular basis of segregation distortion. In males, however, expanded centromeres and centromere strength variation are thought to result in reduced fertility (Daniel 2002; Fishman and Saunders 2008). This lower fertility is predicted to drive the evolution of genetic suppressors of centromere drive, including alleles of centromeric proteins with altered DNA-binding affinity. Under this model, centromeric proteins evolve rapidly in order to mitigate fitness costs associated with centromere drive (Henikoff et al. 2001).
Centromere drive and its suppression provide an explanation for the rapid evolution of both centromeric DNA and centromeric proteins. However, it invokes the relentless, rapid evolution of essential proteins such as CenH3, whose mutation could be highly deleterious (Stoler et al. 1995; Howman et al. 2000; Blower and Karpen 2001; Logsdon et al. 2015). A simpler way to allow for the rapid evolution of centromeric proteins without compromising their essential function would be via gene duplication. Duplication and specialization of centromeric proteins would allow one paralog to function as a drive suppressor in the male germline, while allowing the other to carry out its canonical centromeric role. Gene duplication as a way of separating functions with divergent fitness optima has been previously invoked to explain the high frequency of duplicate gene retention, including retention of testis-expressed gene duplicates that carry out mitochondrial functions (Gallach and Betran 2011). Even though both somatic and testis mitochondrial functions are similar, they have different fitness maxima, which may not be simultaneously achievable using the same set of genes. For example, the most important selective constraint shaping mitochondrial function in sperm may be the increased production of faster-swimming sperm even at the expense of a higher mutation rate. A high mitochondrial mutation rate in sperm is mitigated by the fact that sperm mitochondria are not transmitted to offspring; however such a high mutation rate would be deleterious for somatic tissues. Gene duplications allow organisms to achieve optimal mitochondrial function simultaneously in somatic tissues and testes. By the same reasoning, if a single-copy gene is incapable of achieving the multiple fitness optima that are required for multiple centromeric functions (e.g., mitosis versus meiosis), gene duplication could allow each duplicate to achieve optimality for different functions, thereby resolving intralocus conflict (Gallach and Betran 2011). The potential for functional interrogation of intralocus conflict within CenH3 makes the identification and study of CenH3 duplications intriguing.
At least five independent gene duplications of CenH3 have been described in plants (Kawabe et al. 2006; Moraes et al. 2011; Sanei et al. 2011; Neumann et al. 2012; Finseth et al. 2015; Ishii et al. 2015; Neumann et al. 2015). In most cases, both protein variants are widely expressed and co-localize at centromeres during cell divisions (Neumann et al. 2012, 2015). However, in barley, one CenH3 paralog is widely expressed whereas the other is only expressed in embryonic and reproductive tissues (Ishii et al. 2015). In cases that have been examined closely, CenH3 duplicates are subject to divergent selective pressures (i.e., one paralog evolves under positive selection but the other does not) (Finseth et al. 2015; Neumann et al. 2015). Indeed, CenH3 duplications in Mimulus guttatus have been hypothesized to result from centromere drive suppression (Finseth et al. 2015).
In animals, CenH3 is thought to have independently duplicated in the holocentric nematodes Caenorhabditis elegans and C. remanei (Monen et al. 2005, 2015). Detailed studies have only been performed on the CenH3 duplicate in C. elegans, and these have yet to elucidate a clear function (Monen et al. 2015). CenH3 duplications have also been described in Bovidae (including cows) where recent gene family expansion has resulted in ten copies of CenH3 (Li and Huang 2008). However, only two of the 10 cow CenH3 duplicates have retained open reading frames and all cow CenH3 duplicates remain poorly characterized (Li and Huang 2008) Furthermore, many systems in which CenH3 has been extensively studied (predominant mammalian systems, such as mice and humans, and model organisms like D. melanogaster) have only one copy of CenH3.
To comprehensively study the incidence of CenH3 duplication in a well-studied animal lineage, we took advantage of the recent sequencing of high-quality genomes from multiple Drosophila species. These genomes are at a close enough evolutionary distance to allow inferences of gains, losses and selective constraints. Despite there being only one copy of CenH3 in D. melanogaster, we were surprised to find that some Drosophila species had two or more copies of CenH3. This motivated our broader analysis of CenH3 duplication and evolution throughout Drosophila. In total, we find at least four independent Cid duplications over Drosophila evolution. Cytological analyses confirm that these Cid duplicates encode bona fide centromeric proteins, two of which are expressed primarily in the male germline. Based on their retention without loss over long periods of Drosophila evolution, and analysis of their selective constraints, we infer that these duplicates now perform nonredundant centromeric roles, possibly as a result of subfunctionalization. Overall, this suggests that Drosophila species encoding a single CenH3 gene may be in the minority. The sheer number of available Drosophila species and their experimental tractability make Drosophila an ideal system to study the evolution and functional specialization of duplicate Cid genes. Our results suggest the intriguing possibility that CenH3 duplications may allow Drosophila species to better achieve functional optimality of multiple centromeric functions (e.g., mitotic cell division in somatic cells and centromere drive suppression in the male germline) than species encoding a single CenH3 gene.
Results
Four Cid Duplications in the Drosophila Genus: Ancient Retention and Recent Recombination
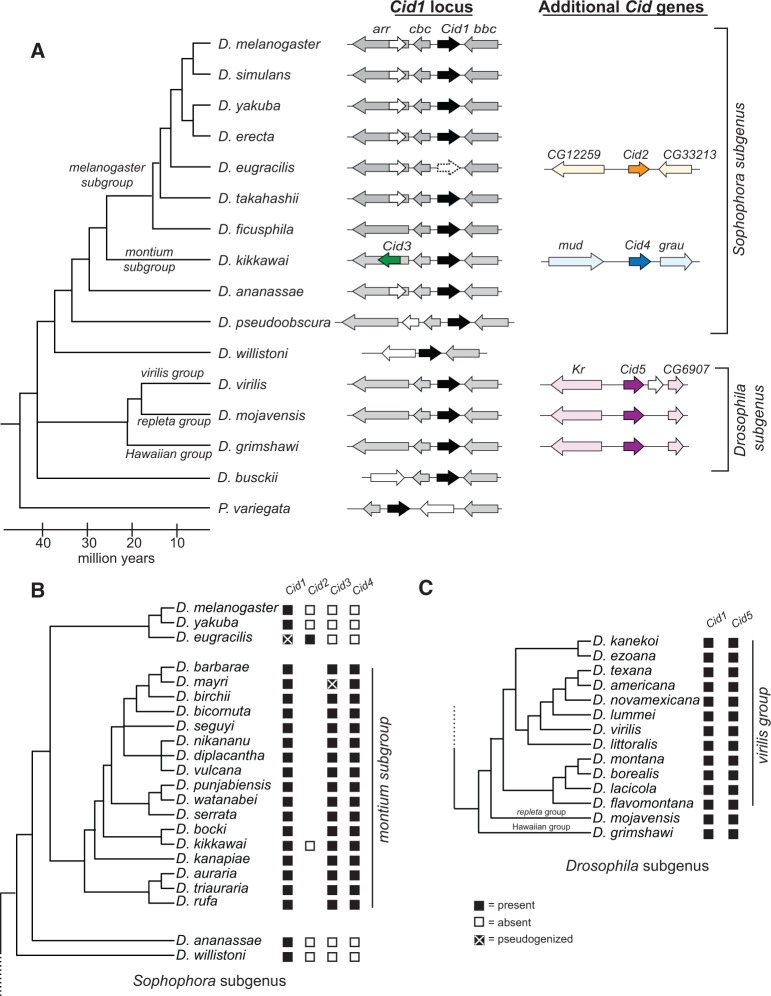
Although their N-terminal tails are highly divergent, CenH3 histone fold domains (HFD, ∼100 aa) are highly conserved and recognizably related to canonical H3 (Palmer et al. 1991; Malik and Henikoff 2003). Thus, sequence similarity searches based on either CenH3 or even canonical H3 HFDs are sufficient to identify putative CenH3 homologs in fully sequenced genomes; inability to find homologous genes can be indicative of true absence (Drinnenberg et al. 2014). To identify all CenH3 homologs in Drosophila, we performed a tBLASTn search using both the canonical H3 and the D. melanogaster CenH3 (Cid) HFD as a query against 22 sequenced Drosophila genomes, as well as genomes from two additional dipteran species. We recorded each Cid gene “hit” as well as its syntenic locus in each species (fig. 1A, supplementary table S1, Supplementary Material online). Consistent with previous studies, we found no additional Cid genes in the D. melanogaster genome or in closely related species of the melanogaster species subgroup (Henikoff et al. 2000; Malik et al. 2002). In addition, we found that orthologs of the Cid gene in D. melanogaster have been preserved in their shared syntenic location in each of the Drosophila species we examined, except in D. eugracilis where it has clearly pseudogenized (supplementary fig. S1, Supplementary Material online). We also found Cid orthologs in the shared syntenic context in a basal Drosophila species, D. busckii, as well as Phortica variegata, which belongs to an outgroup sister clade of Drosophila. Based on these findings, we conclude that an ortholog of D. melanogaster Cid1 was present in the common ancestor of Drosophila in the shared syntenic location. We denote this orthologous set of genes in this shared syntenic location as Cid1.
Fig. 1.
Identification of Cid duplication events across Drosophila evolution. (A) A Drosophila species cladogram is presented with Phortica variegata as an outgroup. The genomic context of representative Cid paralogs identified by tBLASTn using previously published genome sequences is schematized to the right of each species. Within a species, each locus depicted is contained on a unique genomic scaffold (see supplementary table S1, Supplementary Material online for detailed scaffold information). Cid1 is the ancestral locus based on its presence in almost all species, including the outgroup species P. variegata (black arrow, see column labeled “Cid1 locus”). In total, we found four Cid duplication events resulting in the birth of the genes Cid2, Cid3, Cid4, and Cid5 (see “Cid1 locus” and “Additional Cid genes” columns, dark orange, dark green, dark blue, and dark purple arrows). We also found one Cid1 pseudogene (“Cid1 locus” column, empty arrow, dashed outline) in D. eugracilis. Arrows colored in a lighter version of the corresponding Cid gene color represent genes that define the shared syntenic locus of each paralog. White arrows represent genes that are present in a locus, but do not define the locus since they are present in fewer than 50% of the represented species. We do not provide gene names for these “white arrow” genes. Genes that define each syntenic locus are named based on the D. melanogaster gene name. (B) Summary of Cid paralog presence across the Sophophora subgenus with an expanded montium subgroup. The presence (black box) or absence (white box) of each Cid paralog as determined by PCR and Sanger sequencing is displayed next to each species. The lack of a box means that we did not attempt to amplify the locus. Cid1, Cid3, and Cid4 were preserved in almost all montium subgroup species with the exception of a Cid3 pseudogene in Drosophila mayri (black box with a white X). This analysis indicated that Cid3 and Cid4 were born 20–30 Ma. (C) Summary of Cid paralog presence across the Drosophila subgenus with an expanded virilis group. Cid1 and Cid5 were completely preserved in all virilis group species. We conclude that Cid5 was born 40–50 Ma in the common ancestor of the Drosophila subgenus.
Our analysis also identified four previously undescribed Cid duplications in Drosophila (fig. 1A). The first of these was in D. eugracilis, which has a pseudogene at the ancestral Cid1 shared syntenic location but also encodes a full-length Cid gene in a new syntenic location in a new genomic location (fig. 1A, supplementary fig. S1, Supplementary Material online). We refer to this gene as Cid2. We sequenced an additional 8 strains of D. eugracilis to see if there were any cases of dual retention of both Cid1 and Cid2 in this species (supplementary data S1, Supplementary Material online). In all cases, we found that Cid1 orthologs were pseudogenized; they all contained a two base pair deletion leading to a frame shift after the first nine amino acids and a stop codon after 12 amino acids. D. eugracilis represents a unique case wherein the ancestral Cid1 was lost and replaced by a recent duplicate, Cid2. Based on additional sequencing (below) it remains the only case of Cid1 loss described in Drosophila.
In addition to the Cid duplicate in D. eugracilis, we found two new Cid paralogs in D. kikkawai, which belongs to the montium subgroup of Drosophila. Thus, D. kikkawai encodes three CenH3 genes: the ancestral Cid1, as well as Cid3 and Cid4 (fig. 1A). Cid3 is located in close proximity to the original Cid1 gene in the same genomic vicinity, whereas Cid4 is present at a distinct genomic location. Cid1, Cid3, and Cid4 are quite different from one another at the sequence level. Their N-terminal tails only share ∼25% amino acid identity, whereas pairwise amino acid identity of their HFD ranges from 80% (Cid1 and Cid3) to 55% (Cid3 and Cid4) to 45% (Cid1 and Cid4). To study the age and evolutionary retention of these Cid paralogs, we sequenced these three syntenic loci from 16 additional species of the montium subgroup, for which no genomic sequences are publically available. We found that Cid1, Cid3, and Cid4 have been almost completely preserved in the montium subgroup (fig. 1B) with one exception: the Cid3 ortholog is pseudogenized in D. mayri (fig. 1B, supplementary fig. S2, Supplementary Material online). Due to the lack of a complete genome sequence, we cannot rule out the possibility that D. mayri encodes a Cid3-like gene elsewhere in its genome. Based on these findings, we conclude that Cid3 and Cid4 were born from duplication events in the common ancestor of the montium subgroup at least 15 Ma (Russo et al. 2013).
The fourth Cid duplication was found in the three species of the Drosophila subgenus: D. virilis, D. mojavensis, and D. grimshawi (fig. 1A, “Additional Cid genes” column). Each of these species encodes Cid1 and Cid5, which have an average pairwise amino acid identity of 60% in the HFD but only 15% in the N-terminal tail. To investigate the age and evolutionary retention of Cid1 and Cid5, we sequenced both genes from an additional 11 species from the virilis species group. We found that both Cid1 and Cid5 have been completely preserved (fig. 1C). Thus, we conclude that Cid5 was born in the common ancestor of Drosophila subgenus at least 40 Ma (Russo et al. 2013).
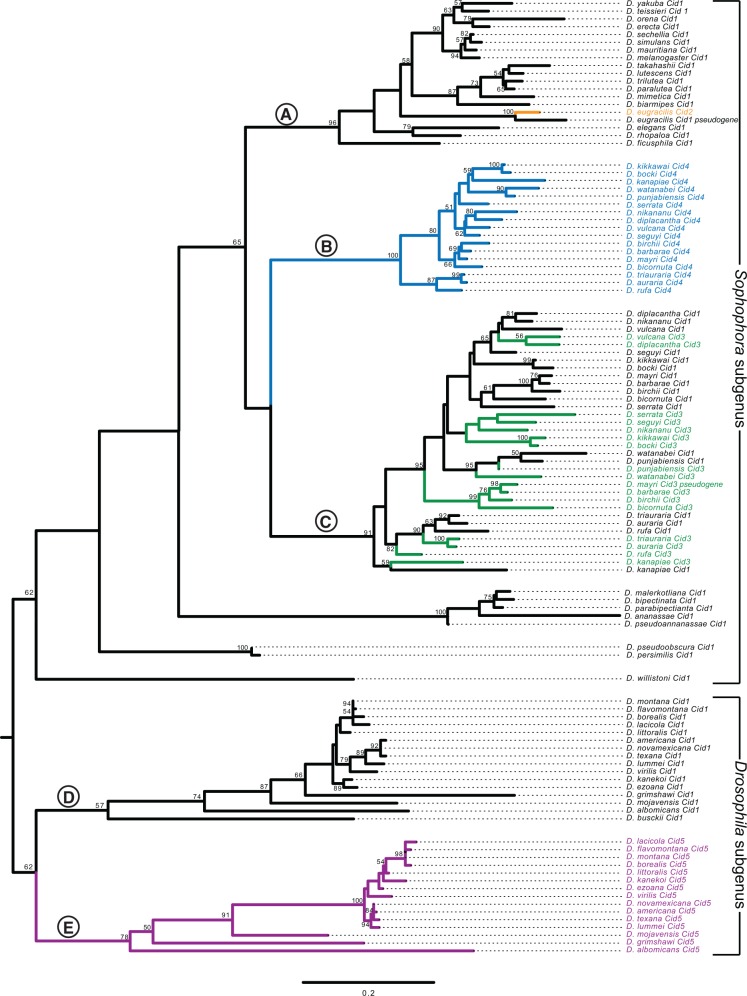
To more rigorously test the paralogy and age of the Cid duplicates, we performed phylogenetic analyses (fig. 2). The N-terminal tails of all the Cid proteins were too divergent to be aligned, so we built a codon-based DNA alignment of the HFD of all Drosophila Cid genes, including Cid1 orthologs sequenced in a previous survey (Malik et al. 2002) (for untrimmed sequences see supplementary data S2, for alignment see supplementary data S3, Supplementary Material online). We then used maximum likelihood (fig. 2) and neighbor-joining (supplementary fig. S3, Supplementary Material online) analyses to construct a phylogenetic tree based on this alignment. We were able to draw the same conclusions from both trees except for one major difference, which we discuss below. Both phylogenetic analyses were in agreement with expected branching topology of the Drosophila species (Russo et al. 2013) and concurred with our analyses of shared synteny (fig. 1A). For instance, D. eugracilis Cid2 (clade A, orange branch) grouped with Cid1 genes of the melanogaster group with high confidence. Its closest phylogenetic neighbor was the Cid1 pseudogene from D. eugracilis, supporting Cid2’s species-specific origin in a recent ancestor of D. eugracilis. We also found that the Cid1 and Cid5 genes of the Drosophila subgenus form monophyletic sister clades (clade D is sister to clade E, fig. 2 and supplementary fig. S3, Supplementary Material online). We found that D. busckii and D. albomicans encode Cid1 genes (clade E), based on phylogeny and shared synteny. However, whereas D. albomicans also encodes Cid5, D. busckii does not (clade D). The phylogenetic resolution between Cid1 and Cid5 clades is strong enough to suggest that the Cid5 duplication may have predated the split between D. busckii and other members of the Drosophila subgenus, but that Cid5 was subsequently lost in D. busckii.
Fig. 2.
Evolutionary relationship among all Drosophila Cid paralogs. We performed maximum likelihood phylogenetic analyses using PhyML with a nucleotide alignment of the histone fold domain of all Cid paralogs. We found that Drosophila subgenus Cid1 (clade E), Drosophila subgenus Cid5 (clade D) and montium subgroup Cid4 (clade B) all formed well-supported monophyletic clades suggesting a single origin for these Cid paralogs. In contrast, montium subgroup Cid1 and Cid3 grouped together (clade C), consistent with our finding that they may be undergoing recurrent recombination (fig. 3). Selected clades (labeled with letters A–E) are further discussed in the main text. Bootstrap values greater than 50 are shown. The tree is arbitrarily rooted to separate the Sophophora and Drosophila subgenera. Scale bar represents number of substitutions per site.
We also found that the Cid4 genes from the montium subgroup form a monophyletic clade (fig. 2, clade B) that forms sister clade to the montium subgroup Cid1 and Cid3 genes (clade C). The melanogaster subgroup Cid1 genes (clade A) formed an outgroup to montium subgroup genes Cid1, Cid3 and Cid4 (clade A is an outgroup to clade B and C). This was the only major difference in branching topology between the maximum likelihood and neighbor-joining analyses; the latter (supplementary fig. S3, Supplementary Material online) placed the Cid4 genes from the montium subgroup (clade B) as a sister lineage to the melanogaster subgroup Cid1 clade (clade A). Since Cid1 is expected to be the ancestral gene in both subgroups, we favor the tree topology suggested by the maximum likelihood analysis. Both analyses reveal an unexpected intermingling of the montium subgroup Cid1/Cid3 genes into a single clade (fig. 2, supplementary fig. S3, Supplementary Material online, clade C). This intermingled phylogenetic pattern could be the result of multiple, independent duplications of Cid3 from Cid1 in the montium subgroup. Alternatively, this pattern could reflect the effects of recurrent gene conversion, in which at least the HFD regions of Cid1 and Cid3 were homogenized by recombination.
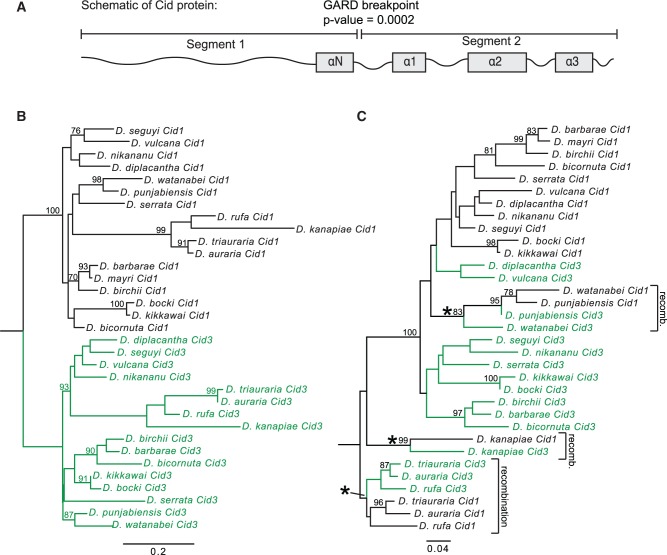
Gene conversion between Cid1 and Cid3 could be facilitated by the close proximity of their genomic locations (see fig. 1A, “Cid1 locus” column), since frequency of gene conversion is inversely proportional to the distance between recombining sequences (Schildkraut et al. 2005). We used GARD (Genetic Analysis for Recombination Detection) analyses (Kosakovsky Pond et al. 2006) to formally test for recombination between Cid1 and Cid3 from the montium subgroup. Consistent with our hypothesis of gene conversion, we found strong evidence for recombination between Cid1 and Cid3 (P = 0.0002) but not between Cid1 and Cid4. The predicted recombination breakpoint is at the transition between the N-terminal tail and HFD domains (fig. 3A). Indeed, when we made a maximum likelihood tree from segment 1 alone (consisting primarily of the N-terminal tail), Cid1 and Cid3 formed the expected monophyletic clades distinct from each other (fig. 3B). However, when we made a maximum likelihood tree of the HFD, we found evidence for at least three specific instances of gene conversion (fig. 3C, recombination highlighted by asterisks). The HFD is important for Cid’s interaction with other nucleosome proteins as well as for centromere targeting (Vermaak et al. 2002; Black et al. 2007; Tachiwana et al. 2011; Rosin and Mellone 2016, 2017). We speculate that such a recombination pattern allows Cid1 and Cid3 to perform distinct functions due to their divergent N-terminal tails whereas the homogenization of the HFD ensures that both proteins retain localization to the centromeric nucleosome. This pattern of ancient divergence followed by recurrent gene conversion may also partially explain the discrepant phylogenetic position of the Cid1/Cid3 clade from the montium subgroup relative to the Cid4 clade from the same subgroup (compare fig. 2 to supplementary fig. S3, Supplementary Material online).
Fig. 3.
Cid1 and Cid3 have undergone recurrent gene conversion in the montium subgroup. (A) We used the Genetic Algorithm for Recombination Detection (GARD; Kosakovsky Pond et al. 2006) to test for recombination in the montium subgroup Cid1 and Cid3. GARD identified one significant (P = 0.0002) breakpoint between the N-terminal tail and the histone fold domain. (B, C) Maximum likelihood phylogenetic trees from an alignment of GARD segment 1 (B) and GARD segment 2 (C) were subsequently generated using PhyML. Bootstrap values above 75 are displayed. Asterisks indicate branches along which gene conversion likely occurred. Scale bar represents nucleotide substitutions per site.
Drosophila Cid Paralogs Localize to Centromeres
There are three possible outcomes following a functional gene duplication event: subfunctionalization, neofunctionalization, and redundancy, which often leads to the loss of one paralog. Because we observe the co-retention of most Cid duplicates for millions of years (with the exception of Cid1 loss in D. eugracilis and Cid3 loss in D. mayri), it is unlikely that duplicate Cid genes have been retained for redundant functions. We therefore wanted to distinguish between the possibilities of subfunctionalization and neofunctionalization for duplicate Cid genes.
It is not unprecedented that a histone variant paralog might develop a new function. For example, in mammals, the H2B variant SubH2Bv acquired a non-nuclear role in acrosome development in sperm (Aul and Oko 2001). To assess the possibility that the Cid paralogs may have acquired a noncentromeric role (i.e., have become neofunctionalized), we turned to cell biological analyses to determine their localization. Previous studies showed that Cid1 orthologs (including those from D. bipectinata and D. virilis) can fail to localize to D. melanogaster centromeres, due to changes at the interface between Cid1 and its chaperone protein CAL1 (Rosin and Mellone 2016, 2017). We therefore decided to test the localization of selected Cid paralogs in tissue culture cells from the same species.
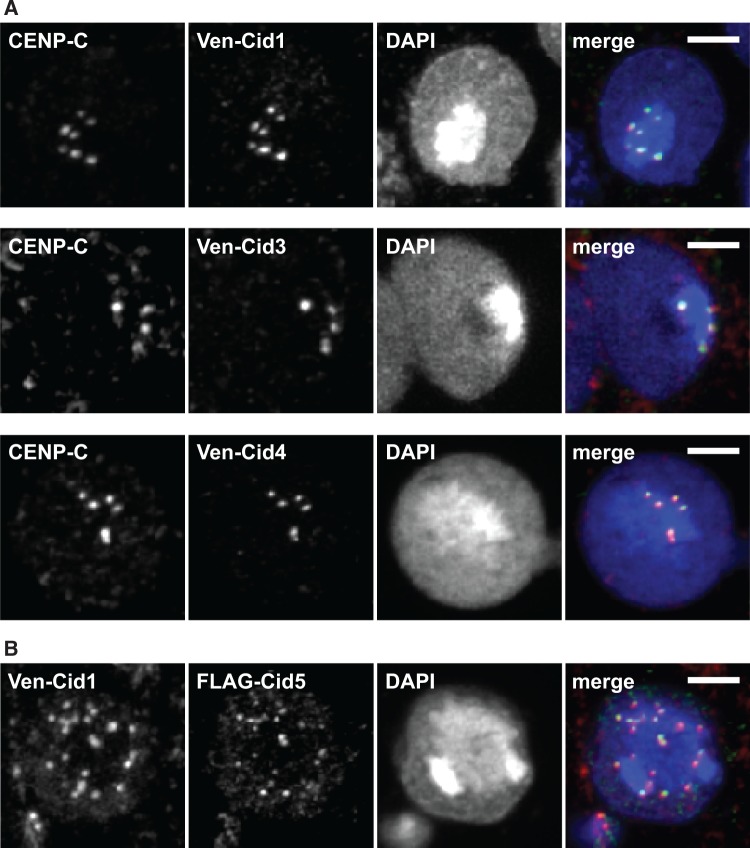
Among all montium subgroup species that contain Cid1, Cid3, and Cid4, cell lines were available only from D. auraria (cell line ML83-68, DGRC). We cloned the Cid1, Cid3, and Cid4 genes from D. auraria and tagged each with an N-terminal Venus tag to aid in visualization. We then transfected these constructs individually into D. auraria cells. We found that each Venus-Cid paralog localized in a similar manner, in punctate foci in a DAPI-intense region of the cells (fig. 4A). This pattern is highly characteristic of centromere localization (van Steensel and Henikoff 2000). To confirm this, we co-stained the cells with an antibody against CENP-C, a constitutively centromeric protein. Since no D. auraria-specific CENP-C antibodies were available, we first confirmed that the D. melanogaster CENP-C antibody appropriately marked centromeres in D. auraria. Indeed, the D. melanogaster CENP-C antibody recognized foci at the primary constriction of D. auraria metaphase chromosomes (supplementary fig. S4, Supplementary Material online). Moreover, we found that Venus-Cid1, Venus-Cid3, and Venus-Cid4 all co-localized with CENP-C in this cell line (fig. 4A). Based on this, we conclude that all the D. auraria Cid paralogs localize to centromeres.
Fig. 4.
Proteins encoded by Cid paralogs localize to centromeres in cell culture. (A) Venus-tagged D. auraria Cid1, Cid3, and Cid4 were transiently transfected in a D. auraria cell line (top, middle, and bottom panels, respectively). Cells were fixed and co-stained with a D. melanogaster CENP-C antibody (red in merged image) and anti-GFP (green in merged image). These data show co-localization of all three montium subgroup Cid proteins with CENP-C. (B) We co-transfected Venus-tagged Cid1 and FLAG-tagged Cid5 from D. virilis into a D. virilis cell line. Venus-Cid1 (red in merged image) and FLAG-Cid5 (green in merged image) both formed co-localized foci in the nucleus. All scale bars indicate a distance of two microns.
We similarly tested the localization of D. virilis Cid1 and Cid5 in a D. virilis cell line (WR Dv-1). Unfortunately, the antibody raised against D. melanogaster CENP-C did not recognize D. virilis centromeres likely due to the high divergence between the CENP-C orthologs from the two species. We therefore co-transfected Venus-Cid1 and FLAG-Cid5. We found that Cid1 and Cid5 co-localize at nuclear foci, in a staining pattern that is typical of centromeric localization (fig. 4B). This suggests that despite their divergence, all Cid duplicates retain the ability to be recognized and deposited at centromeres by the existing machinery including CAL1, the chaperone that deposits Drosophila centromeric histones (Rosin and Mellone 2016). Alternatively, Cid paralog proteins might achieve centromeric co-localization by forming heterodimers with Cid1. Together, these results support the hypothesis that Cid duplicates have been retained to perform a centromeric function. Our cytological findings do not formally rule out the possibility of neofunctionalization; Cid duplicates might have been retained to perform a new centromeric function.
Testis Restricted Expression of Cid3 and Cid5
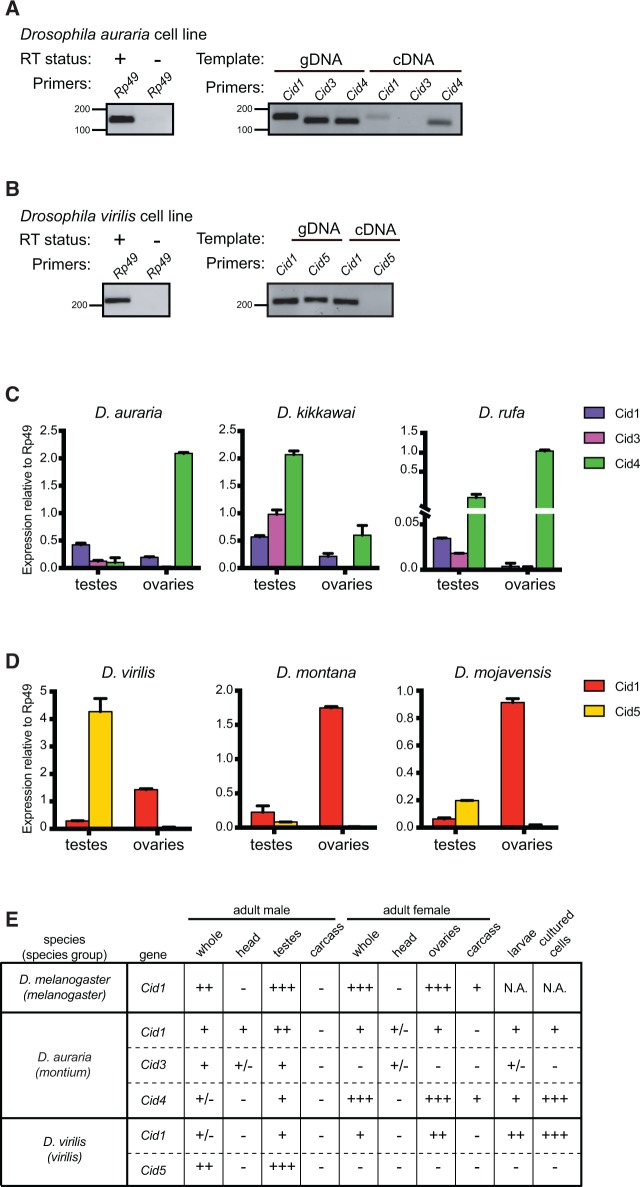
One means by which subfunctionalization can occur is by tissue-specific expression (Force et al. 1999; Lynch and Force 2000). Duplicate genes could retain different subsets of promoter and enhancer elements from their parent gene, requiring both genes’ expression to fully recapitulate parental gene expression (Dorus et al. 2003). We therefore wondered whether any of the Cid duplicates showed tissue-specific expression. We expected that at least one Cid paralog in each species must have maintained mitotic function and would therefore be widely expressed in somatic tissues. To test this, we first looked for expression of Cid paralogs in D. auraria and D. virilis tissue culture cell lines, which are derived from embryonic and larval tissues, respectively. We extracted RNA from both cell lines and performed RT-PCR. After 30 cycles of PCR, we detected a faint Cid1 band in addition to a robust Cid4 band in the D. auraria cell line (fig. 5A). In the D. virilis cell line, we detected expression of Cid1 but not Cid5 after 30 cycles of PCRs (fig. 5B). We did not detect Cid3 (D. auraria) or Cid5 (D. virilis) in this assay, which suggests that both genes are either not expressed or are expressed at low levels in tissue culture cells. From this analysis, we predict that Cid4 (and possibly Cid1) performs somatic Cid function in D. auraria (i.e., mitotic cell divisions for growth) and that Cid1 performs somatic Cid function in D. virilis.
Fig. 5.
Male germline-restricted expression of some Cid paralogs. (A) Left gel: RNA samples used for D. auraria RT-PCR were free of DNA contamination as indicated by performing 35-cycle PCR for Rp49 on cDNA samples generated with (+) and without (−) reverse transcriptase. Right gel: 30-cycle PCR performed with either genomic DNA (gDNA) or cDNA for Cid1, Cid3 and Cid4 from a D. auraria cell line. We detected both Cid1 and Cid4 expression but the Cid4 expression band was more robust than the Cid1 band. We did not detect expression of Cid3 in this cell line. (B) Left gel: as in (A), RNA samples used for D. virilis RT-PCR were free of DNA contamination. Right gel: RT-PCR analyses of Cid1 and Cid5 from a D. virilis cell line at 30 cycles revealed only the expression of Cid1. We did not detect Cid5 by RT-PCR. (C) RT-qPCR for Cid1, Cid3, and Cid4 from dissected tissues from three montium subgroup species revealed that Cid1 and Cid4 are expressed in both the testes and the ovaries whereas Cid3 expression is testis restricted. (D) RT-qPCR from dissected tissues from three species from the Drosophila subgenus revealed that Cid1 is expressed in the testes and ovaries of all three species whereas Cid5 is only expressed in the testes. All RT-qPCR was normalized using Rp49 as a control. Error bars represent standard deviation calculated from three technical replicates. (E) Summary of expression pattern for each Cid paralog in representative species. − = not detected, +/ − = very low expression, + = moderate expression, ++ = high expression, ++ + = very high expression.
To further explore tissue specific expression, we performed RT-qPCR on dissected male and female D. virilis and D. auraria flies (whole fly, head, testes/ovaries, and carcass). We performed the same analysis for D. melanogaster, which only encodes a single Cid1 gene, for comparison. In D. melanogaster, we found that Cid1 expression is highest in testes and ovaries and is relatively low in head and carcass (supplementary fig. S5, Supplementary Material online). This is not unexpected since testes and ovaries contain higher numbers of actively dividing cells than the head and the carcass. Similarly, in D. auraria and D. virilis, we found low expression of Cid paralogs in the head and the carcass of male and female flies (supplementary fig. S5, Supplementary Material online). Interestingly, we found that the expression of Cid3 in D. auraria and Cid5 in D. virilis was primarily restricted to the male germline (fig. 5C and D). We also found that Cid1 and Cid4 in D. auraria as well as Cid1 in D. virilis are expressed in both testes and ovaries.
We wanted to extend our expression analyses of the Cid paralogs to other species containing duplicate Cid genes. We performed RT-qPCR on two additional montium subgroup species (D. kikkawai and D. rufa) and on two additional Drosophila subgenus species (D. montana and D. mojavensis). In all cases, Cid3 or Cid5 expression was detected in testes but not in ovaries. Cid1 and Cid4 expression patterns were similar across species too, with the exception of Cid1 in D. rufa, which expressed at very low levels in ovaries (fig. 5C and D and supplementary fig. S5, Supplementary Material online).
Our findings are consistent with the hypothesis of tissue-specific specialization of the Cid paralogs in both the montium subgroup and the virilis group. These results also suggest that Cid3 and Cid5 were retained to perform a testis-specific function. In contrast, the other Cid paralogs are expressed in both somatic and germline tissues. However, these analyses lack the cellular resolution necessary to conclude whether the expression patterns are mutually exclusive or overlapping in tissues where multiple Cids are expressed. Moreover, in the montium subgroup, Cid4 is expressed broadly in a pattern similar to D. melanogaster Cid1, and it is the primary Cid duplicate expressed in somatic cells. This suggests that Cid4, and not Cid1, performs canonical Cid function in montium subgroup species.
Differential Retention of N-Terminal Tail Motifs and the Evolution of New Motifs following Cid Duplication
Given their sequence divergence and different expression patterns, it seems likely that Cid paralogs may have been retained to perform distinct functions. Unlike the structural constraints that shape the HFD, the N-terminal tail of Cid is highly variable in length and sequence. We speculated that analyses of selective constraint in the N-terminal tail might present an additional opportunity to determine if subfunctionalization had occurred among the Cid paralogs. Although the specific function of the N-terminal tail has yet to be elucidated for Drosophila Cid, studies in humans and fission yeast have shown that the N-terminal tail is important for recruitment and stabilization of inner kinetochore proteins (Fachinetti et al. 2013; Folco et al. 2015; Logsdon et al. 2015). Furthermore, post-translational modifications of the N-terminal tail have been shown to be important for CENP-A mitotic function (Goutte-Gattat et al. 2013) and for facilitating interaction between two CENP-A molecules (Bailey et al. 2013).
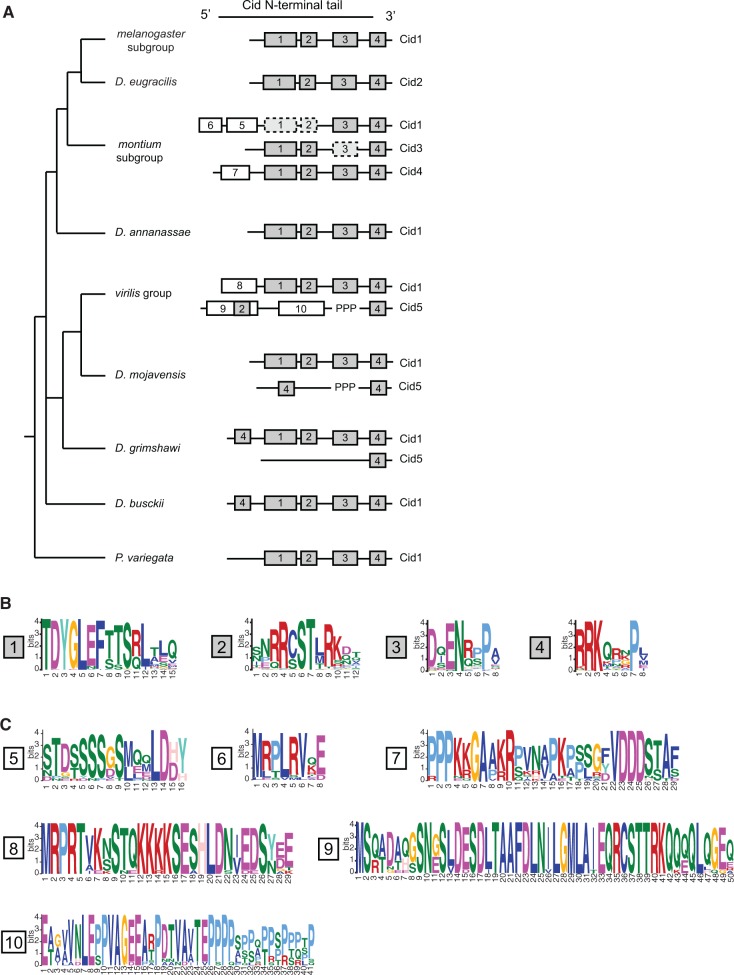
Conserved motifs provide an avenue to evaluate differential selective constraint in the N-terminal tail of different CenH3 paralogs (Maheshwari et al. 2015). Motifs are regions of high similarity among protein sequences. They represent putative sites of protein–protein interaction and post-translational modification. We reasoned that we might be able to use the presence of certain N-terminal tail motifs as a proxy for various functional domains. We therefore used the motif generator algorithm, MEME (Bailey and Elkan 1994), to identify conserved motifs in the N-terminal tail from six different groups of Drosophila Cid proteins: melanogaster group Cid1 (single copy genes only), montium subgroup Cid1, montium subgroup Cid3, montium subgroup Cid4, virilis group Cid1, and virilis group Cid5 (supplementary fig. S6, Supplementary Material online). We then used the motif search algorithm, MAST (Bailey and Gribskov 1998), to search for each motif in all Cid proteins. In total, we found 10 unique motifs (supplementary fig. S6, Supplementary Material online). Finally, we overlaid our motif analysis with the Drosophila species tree to gain insight into the evolution of N-terminal tail motifs (fig. 6A).
Fig. 6.
Evolution of N-terminal motifs among all Cid proteins. (A) A Drosophila species tree with a schematic of N-terminal tail motifs identified by MEME and MAST displayed to right of each species or species group. Each number represents a unique motif that does not statistically match any other motif in the figure with the exception of motif 2 and 9 (see Materials and Methods). Gray boxes indicate “core” motifs 1–4, which are present in all single copy Cid genes. White boxes indicate lineage specific motifs. “PPP” indicates the position of the variable proline-rich region in Cid5. Dashed boxes indicate cases in which a given motif was present in ∼50% of species. (B) Logos generated by MEME for consensus motifs 1–4. (C) Logos generated by MEME for consensus motifs 5–10.
From this analysis, we can make several interesting conclusions. First, motifs 1–4 (fig. 6B) are conserved in every Cid1 protein when it is the only copy encoded in the genome. These motifs correspond nicely to the motifs we previously identified in the melanogaster group using Block Maker (Malik et al. 2002). Although their function remains largely uncharacterized, motif 4 has been shown to be involved in recruitment of mitotic checkpoint protein, BubR1 (Torras-Llort et al. 2010). Motif 4 could also play a role in histone–DNA interaction because it is located in the region where the N-terminal tail exits the nucleosome and passes between the two strands of DNA (Tachiwana et al. 2011). Motif 4 is the only motif present in all Cid paralogs, which suggests that it performs a general function among all Cids. Given their retention in all single copy Cid-containing Drosophila species, we consider motifs 1–4 to be the “core” Cid1 motifs (fig. 6B) and speculate that all are required for Cid1 function when it is the only centromeric histone protein. Indeed, all Drosophila species contain all of these motifs amongst their various Cid paralogs.
Next, we observed that some Cid paralogs had evolved and retained “new” N-terminal tail motifs (fig. 6C). We identified three motifs that evolved in Cid paralogs from the montium subgroup; motifs 5 and 6 are found in Cid1 whereas motif 7 is found in Cid4. One might interpret the invention of additional N-terminal tail motifs as evidence of neofunctionalization. Indeed, invention of novel protein–protein interactions to perform new centromeric functions is expected for neofunctionalized paralogs. However, new motifs could also arise in paralogs that have subfunctionalized, to more optimally perform a subset of the pre-existing functions, for example, in the male germline. Thus, formally, even subfunctionalization could lead to the retention of novel motifs, especially if these motifs would be incompatible with all ancestral functions.
More direct evidence of subfunctionalization emerged from our observation of frequent loss of “ancestral” motifs 1–3 from Cid1 and Cid3, despite their completely preservation in Cid4 (fig. 6A, dotted lines indicate motif is absent from ∼50% of queried species). Intriguingly, some Cid1 and Cid3 orthologs in the montium subgroup appear to have differentially retained motifs 1–3; Cid1 has motif 3 and Cid3 has motifs 1 and 2. This differential retention of an ancestrally conserved subset of core motifs is highly suggestive of subfunctionalization (Maheshwari et al. 2015). Furthermore, our findings support the hypothesis that in the montium subgroup, it is the Cid4 paralog rather than the ancestral Cid1, which performs the canonical functions of centromeric histones carried out by Cid1 in other species, because Cid4 contains all core motifs but montium subgroup Cid1 does not. This would also be consistent with our expression analyses, in which Cid4 expresses more robustly than Cid1 in somatic cells (fig. 5A).
This pattern of new motif evolution and ancient motif degeneration is also evident in the Cid paralogs from the virilis group. In this group of species, the Cid1 paralog has retained the core set of motifs 1–4 but added motif 8. In contrast, Cid5 paralogs have added motifs 9 and 10 but lost core motifs 1 and 3. We therefore conclude that the tissue-specific pattern of expression and the differential retention of N-terminal motifs support a general model of subfunctionalization, but that some paralogs may have acquired novel protein-protein interaction motifs perhaps to optimize for new, specialized centromeric functions.
Different Evolutionary Forces Act on Different Cid Duplicates
Tissue specific expression of some Cid paralogs and differential retention of N-terminal tail motifs supports the hypothesis that Cid paralogs may have subfunctionalized. We next considered the possibility that duplicate Cid genes were retained to allow optimization for divergent functions. In the melanogaster group, Cid1 (a single copy Cid gene) has been shown to evolve rapidly (Malik and Henikoff 2001), perhaps due to its interaction with rapidly evolving centromeric DNA and the need for drive suppressors in male meiosis (Henikoff et al. 2001). While this rapid evolution might be required for the “drive suppressor” function, it may be disadvantageous for canonical Cid function (e.g., mitosis). As a result, selection may act differently on Cid in the male germline than on somatic or ovary-expressed Cid. For instance, some Cid paralogs (e.g., those that are expressed primarily in the male germline and may suppress centromere-drive) might evolve under positive selection while others would not.
We used maximum likelihood methods using the PAML suite to test for positive selection on each of the Cid paralogs. For montium subgroup Cid1 and Cid3, we performed each analysis separately on GARD segment 1 and 2 (fig. 3). For all other Cid genes we performed PAML analyses on full-length alignments (supplementary data S4 and S5, Supplementary Material online). Consistent with our prediction, we found that some, but not all, Cid paralogs likely evolve under positive selection (fig. 7A). For example, PAML analyses reveal that Cid3 segment 1 evolved under positive selection (supplementary table S2, M1 vs. M2 P = 0.02 and M8a vs. M8 P = 0.01). However, we did not find evidence that Cid5, another male germline-restricted paralog, evolves under positive selection. We note, however, that we were unable to unambiguously align a highly variable proline-rich segment in Cid5’s N-terminal tail and excluded this segment from our analyses (fig. 7B). If positive selection was occurring in this region, we would be unable to detect it. We also found that Cid4 evolved under positive selection but montium subgroup Cid1 and Cid3 segment 2, and virilis group Cid1, did not (fig. 7A, supplementary table S2, Supplementary Material online). To ensure that recombination in Cid1 and Cid3 segment 2 was not obscuring our ability to detect positive selection in these segments, we re-ran the PAML analyses excluding the species for which we could detect apparent gene conversion events (D. watanabei, D. punjabiensis, D. kanapiae, D. triauraria, D. auraria, and D. rufa). Exclusion of these species did not affect the conclusions from the PAML analyses; we did not detect positive selection in either Cid1 or Cid3 segment 2.
Fig. 7.
Different Cid paralogs evolve under different evolutionary pressures. (A) Summary of tests for positive selection performed on each Cid paralog. Tests that were statistically significant (P < 0.05) are indicated with an asterisk. For the McDonald–Kreitman (MK) test, Neutrality Index (N.I.) is also displayed. N.I. < 1 indicates an excess of nonsynonymous fixed differences between species and suggests positive selection. (B) A protein alignment of Cid5 from virilis group species. The variable, proline-rich region which was excluded from PAML tests for positive selection is highlighted in blue. (C) A schematic of a representative Cid protein, showing sites evolving under positive selection identified by Bayes Empirical Bayes analyses (posterior probability > 0.95).
For those genes that PAML identified as having evolved under positive selection (Cid3 segment 1 and Cid4), Bayes Empirical Bayes analyses identified one amino acid in Cid3 and one amino acid in Cid4 as having evolved under positive selection with a high posterior probability (>0.95). In Cid3, the positively selected site is adjacent to the αN-helix. In Cid4, the positively selected site is in loop 1 of the HFD (fig. 7C, supplementary table S2, Supplementary Material online). Interestingly, these are both places where Cid is predicted to contact centromeric DNA (Tachiwana et al. 2011) although Loop 1 is also the domain that interacts and co-evolves with the centromeric histone chaperone, CAL1 (Rosin and Mellone 2016, 2017). These results are consistent with the hypothesis that both Cid3 and Cid4 are engaged in a genetic conflict involving centromeric DNA.
We next used the McDonald–Kreitman (MK) test to look for positive selection in each of the Cid paralogs. While PAML detects positive selection occurring recurrently at selected amino acid residues across deep evolutionary time, the MK test detects more recent positive selection distributed over entire genes or protein domains. The MK test assumes that if protein constraints have not dramatically altered over evolution, the ratio of nonsynonymous to synonymous fixed differences between species (DN/DS) should approximately equal the ratio of nonsynonymous to synonymous polymorphisms within a species (PN/PS). However, if a higher than expected number of nonsynonymous fixed changes are observed (i.e., DN/DS > PN/PS), this would be indicative of positive selection after the divergence of the species.
In order to test for positive selection in the montium subgroup using the MK test, we sequenced and compared Cid1, Cid3 and Cid4 paralogs from 26 strains of D. auraria and 10 strains of D. rufa. For virilis group Cids, we sequenced Cid1 and Cid5 paralogs from 10 strains of D. virilis and 21 strains of D. montana (supplementary data S6 and S7, Supplementary Material online). We found an excess of non-synonymous fixed differences between D. auraria and D. rufa Cid1 and Cid3, suggesting that both genes evolve under positive selection (fig. 7A, supplementary table S3, Supplementary Material online). Parsing the signal by performing the MK test on just the N-terminal tail or just the HFD domain revealed that Cid1 and Cid3 HFD domains evolve under positive selection (supplementary table S3, Supplementary Material online). However, we did not find evidence for positive selection in the N-terminal tails. Most of the nonsynonymous fixed differences occur in Loop1, which is predicted to contact centromeric DNA (Tachiwana et al. 2011). Interestingly, even though PAML analyses detected ancient recurrent positive selection in montium group Cid4, we did not find strong evidence for recent positive selection since the D. auraria–D. rufa divergence using the MK test (P = 0.08). We also found no evidence of positive selection having acted on virilis group Cid1 or Cid5 using the MK test (fig. 7A, supplementary table S3, Supplementary Material online).
To summarize our positive selection analyses, we found that Cid3 has experienced both ancient and recent positive selection in protein domains predicted to contact centromeric DNA. Cid4 has also experienced ancient, recurrent positive selection at putative DNA-contacting sites, but we found no evidence of recent positive selection in a MK test comparison. This could suggest that Cid4 was either relieved of its role in such conflict or that the MK test lacks the power to detect selection acting on only a few residues. Similarly, although PAML analyses failed to identify a pattern of ancient, recurrent positive selection, the MK test did reveal positive selection for montium subgroup Cid1 while comparing the entire HFD. In contrast, we did not find evidence for positive selection having acted on Cid1 and Cid5 in the virilis group by either test.
Discussion
The availability of many high-quality sequenced genomes as well as the comprehensive understanding of phylogenetic relatedness between species make Drosophila an ideal system to study gene duplication and evolution. This facilitated our discovery of four ancient Cid duplications in Drosophila. We found that while Cid1 (previously known as just “Cid”) is preserved in its shared syntenic location in all species examined except one, many species encode one or two additional Cid genes. The species of the montium subgroup, including D. kikkawai, have three Cid genes (Cid1, Cid3, and Cid4), which were born from a duplication event ∼15 Ma. The species of the virilis group, as well as D. mojavensis and D. grimshawi (repleta and Hawaiian groups, respectively), have two Cid genes (Cid1 and Cid5), which were born from a duplication event ∼40 Ma. These Cid duplications have been almost completely preserved in extant species. Despite the fact Cid paralogs are divergent from one another at the sequence level, all paralogs have the ability to localize to centromeres when expressed in tissue culture cells. Based on our detailed analysis of two subgenera (Drosophila and Sophophora), we predict that over one thousand Drosophila species encode two or more CenH3 (Cid) genes (Brake and Baechli 2008). We further conclude that D. melanogaster and other Drosophila species that have only one Cid are the minority; most Drosophila species have multiple Cid paralogs.
Our phylogenetic analyses support our synteny-based conclusions, and reveal recurrent recombination between Cid1 and Cid3 in montium subgroup species. This is the first reported case of recombination between CenH3 paralogs. Our results suggest that this recombination results in evolutionary homogenization of the histone fold domain between Cid1 and Cid3, while the N-terminal tails of Cid1 and Cid3 appear to be evolving independently, perhaps maintaining divergent functions. This recombination could be the genetic mechanism by which Cid1 and Cid3 maintain function in the centromeric nucleosome via near-identical HFDs despite having divergent N-terminal tails, which facilitates distinct interactions. This pattern of gene conversion is akin to patterns of recombination seen for paralogous mammalian antiviral proteins, IFIT1 and IFIT1B, in which gene conversion homogenizes the N-terminal oligomerization domain but not the divergent C-terminus, which allows IFIT1 and IFIT1B proteins to have distinct anti-viral specificities (Daugherty et al. 2016).
What is the evidence that Cid paralogs have distinct functions? The strongest evidence is that they have been co-retained in both the montium subgroup and the virilis/repleta/Hawaiian radiation for tens of millions of years. If they performed redundant functions, we predict that one of the paralogs would be lost over this time frame considering the high rate of DNA deletion in Drosophila (Petrov et al. 1996). Indeed, we observed only two instances of Cid duplication followed by pseudogenization (Cid3 pseudogene in D. mayri and Cid1 pseudogene in D. eugracilis) and inferred the possible loss of Cid5 (in D. busckii). Our findings that Cid3 and Cid5 are expressed primarily in the male germline, that N-terminal tail motifs have been differentially retained and that different selective pressures have shaped different Cid paralogs further supports the idea that these Cid paralogs perform nonredundant functions.
Interestingly, our expression and motif analyses strongly suggest that Cid4 has taken over the primary function of somatic centromeric histone function in montium subgroup species. Cid4 is the primary Cid gene expressed in D. auraria tissue culture cells and is the only Cid paralog in this species that contains all four of the “core” N-terminal tail motifs. In contrast, the “ancestral” Cid1 is expressed at lower levels than Cid4, Cid3 is primarily expressed in the male germline, and neither Cid1 nor Cid3 contain all four “core” motifs. This finding has implications for future experiments taking an evolutionary approach to study Cid function. The correct Cid paralog for such studies must be chosen carefully. Further functional experimentation, such as creating genetic knockouts, will be required to determine the specific function of each Cid paralog.
We propose that in species with a single-copy Cid gene, the same protein must perform multiple functions including mitotic cell division in somatic tissues and drive suppression in the male germline. These functions might require different selective pressures to achieve functional optimality. For example, we have previously proposed that drive suppression results in rapid evolution of Cid to co-evolve with rapidly evolving centromeric DNA (Henikoff et al. 2001) whereas mitotic function might impose purifying selection on Cid, minimizing changes in amino acid sequence. Therefore, it could be advantageous to have two copies of Cid such that each encodes a separate function. Our results suggest that Cid3 and Cid5 are candidate drive suppressors given their male germline-restricted expression. Consistent with this prediction, we detected evidence for positive selection in Cid3. In contrast, we did not find evidence that Cid5 evolves under positive selection. This leaves open the possibility that Cid5 performs an alternative, centromeric, male germline function independent of potential centromere-drive suppression in meiosis.
If it is advantageous to have multiple Cid paralogs, why do not more animal species possess more than one gene encoding centromeric histones? We hypothesize that retention of duplicate Cid genes requires a defined series of evolutionary events and that the cadence of the mutations determines the ultimate fate of the duplicated genes (Ancliff and Park 2014). First, the duplication must not be instantaneously harmful; gene expression must be carefully controlled, as Cid overexpression or expression at the wrong time during the cell cycle can be catastrophic (Heun et al. 2006; Schuh et al. 2007). Even though other kinetochore proteins might limit Cid incorporation into ectopic sites (Schittenhelm et al. 2010), a duplicate Cid gene that acquired a strong or constitutive promoter would almost certainly be detrimental. Furthermore, in order for a duplicate Cid gene to be retained, a series of subfunctionalizing mutations must occur (before pseudogenization of either paralog) such that both paralogs are required for complete Cid function. This model, known as duplication–degeneration–complementation (Force et al. 1999), most often refers to mutations in the promoters of duplicate genes. However, the same principle could be applied to mutations in coding regions. Since it is easier to introduce a mutation that results in a nonfunctional Cid gene than a subfunctionalized Cid, most Cid duplicates probably succumb to pseudogenization early in their evolutionary history and, in Drosophila, are quickly lost from the genome (Petrov et al. 1996).
The existence of Cid duplications in genetically tractable organisms provides an opportunity to study the multiple functions of a gene that is essential when present in a single copy. While we know a lot about the role of Cid in mitosis, its roles in meiosis (Dunleavy et al. 2012) and inheritance of centromere identity through the germline (Raychaudhuri et al. 2012) are less well-characterized. Studying Cid paralogs that may have specialized for different functions (e.g., meiosis) may allow for detailed analysis of these underappreciated Cid functions without the risk of disrupting essential mitotic functions. Future functional studies can now leverage the insight provided by duplicate Cid genes, where evolution and natural selection may have already carried out a “separation of function” experiment.
Materials and Methods
Drosophila Species and Strains
Flies were obtained from the Drosophila Species Stock Center at UC-San Diego (https://stockcenter.ucsd.edu) and from the Drosophila Stocks of Ehime University in Kyoto, Japan (https://kyotofly.kit.jp/cgi-bin/ehime/index.cgi). For a complete list of species and strains used in this study, see supplementary table S4, Supplementary Material online.
Identification of Cid Orthologs and Paralogs in Sequenced Genomes
Drosophila Cid genes were identified in previously sequenced genomes using both D. melanogaster Cid1 and H3 histone fold domain to query the nonredundant database using tBLASTn (Altschul et al. 1997) implemented in Flybase (Attrill et al. 2016) or NCBI genome databases. Since Cid is encoded by a single exon in Drosophila, we took the entire open reading frame for each Cid gene hit. For annotated genomes, we recorded the syntenic locus (3′ and 5′ flanking genes) of each Cid gene hit as indicated by the Flybase genome browser track. For genomes that were sequenced but not annotated (D. eugracilis, D. takahashii, D. ficusphila, D. kikkawai, and P. variegata), we used the 3′ and 5′ nucleotide sequences flanking the putative Cid open reading frame as a query to the D. melanogaster genome using BLASTn. We annotated the syntenic locus according to these D. melanogaster matches. Each Cid gene was named according to its shared syntenic location. It is worth noting that the Flybase gene prediction for D. virilis Cid5 (GJ21033) includes a predicted intron but we found no evidence that Cid5 was spliced in any tissue. The results of all BLAST searches are summarized in supplementary table S1, Supplementary Material online.
Identification of Cid Orthologs and Paralogs in Nonsequenced Genomes
Approximately 10 whole (5 male, 5 female) flies were ground in DNA extraction buffer (10 mM Tris pH 7.5, 10 mM EDTA, 100 mM NaCl, 0.5% SDS) with Proteinase K (New England Biolabs). Ground flies were incubated for 2 h at 55 °C. DNA was extracted using phenol–chloroform (Thermo Fisher Scientific) according to the manufacturer’s instructions. Primers were designed to amplify each Cid paralog based on regions of homology in neighboring genes or intergenic regions. Only Cid paralogs that were predicted to be present in the species based on related species sequenced genomes were amplified. All PCRs were performed using Phusion DNA Polymerase (New England Biolabs). Appropriately sized amplicons were gel isolated and cloned into the cloning/sequencing vector pCR-Blunt (Thermo Fisher Scientific) and Sanger sequenced with M13F and M13R primers plus additional primers as needed to obtain sufficient coverage of the locus. A complete list of primers used in this study can be found in supplementary table S5, Supplementary Material online. A list of primer pairs used to amplify Cid paralogs in nonsequenced genomes can be found in supplementary table S6, Supplementary Material online. Sequences obtained in this study have been deposited in Genbank with the following accession numbers: KY212539-KY212710, KY124384-KY124460. A list of Genbank accession numbers can be found in supplementary table S4, Supplementary Material online.
Phylogenetic Analyses
Cid sequences were aligned using the ClustalW (Larkin et al. 2007) “translation align” function in the Geneious software package (version 6) (Kearse et al. 2012). Alignments were further refined manually, including removal of gaps and poorly aligned regions. Maximum likelihood phylogenetic trees of Cid nucleotide sequences were generated using the HKY85 substitution model in PhyML, implemented in Geneious, using 1000 bootstrap replicates for statistical support. Neighbor-joining trees correcting for multiple substitutions were generated using CLUSTALX (Larkin et al. 2007). We used the GARD algorithm implemented at datamonkey.org to examine alignments for evidence of recombination (Kosakovsky Pond et al. 2006). Pairwise percent identity calculations were made in Geneious. Phylogenies were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) or Dendroscope (Huson et al. 2007)
Cloning Cid Fusion Proteins
Cid genes from D. auraria (Cid1, Cid3, and Cid4) and D. virilis (Cid1 and Cid5) were amplified from genomic DNA and cloned into pENTR/D-TOPO (ThermoFisher). We used LR clonase II (ThermoFisher) to directionally recombine each Cid gene into a destination vector from the Drosophila Gateway Vector Collection, generating either N-terminal Venus (pHVW) or 3XFLAG (pHFW) fusion under the control of the D. melanogaster heat-shock promoter.
Cell Culture
Cell lines (D. auraria cell line ML83-68 and D. virilis cell line WR DV-1) were obtained from the Drosophila Genomics Resource Center in Bloomington, Indiana (https://dgrc.bio.indiana.edu). D. auraria cells were grown at room temperature in M3 + BPYE + 12.5%FCS and D. virilis cells were grown in M3 + BPYE + 10%FCS.
Transfection Experiments
Two micrograms plasmid DNA was transfected using Xtremegene HP transfection reagent (Roche) according to the manufacturer’s instructions. 24 hrs after transfection, cells were heat shocked for 1 hr to induce expression of the Cid fusion protein.
Imaging
Cells were transferred to a glass coverslip 48 h after heatshock. Cells were treated with 0.5% sodium citrate for 10 min and then centrifuged on a Cytospin III (Shandon) at 1900 rpm for 1 min to remove cytoplasm. Cells were fixed in 4% PFA for 5 min and blocked with PBSTx (0.3% Triton) plus 3% BSA for 30 min at room temperature. Coverslips with cells were incubated with primary antibodies at 4 °C overnight at the following concentrations: mouse anti-FLAG (Sigma F3165) 1:1000, chicken anti-GFP (Abcam AB13970) 1:1000, rabbit anti-CENP-C (gift from Aaron Straight) 1:1000. Coverslips with cells were incubated with secondary antibodies for 1 h at room temperature at the following concentrations: goat anti-rabbit (Invitrogen Alexa Fluor 568, A-11011) 1:2000, goat anti-chicken (Invitrogen Alexa Fluor 488, A-11039) 1:5000, goat anti-mouse (Invitrogen Alexa Fluor 568, A-11031) 1:2000. Images were acquired from the Leica TCS SP5 II confocal microscope with LASAF software.
Expression Analyses
RNA was extracted from D. auraria cell line ML83-68 and D. virilis cell line WR DV-1 using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To investigate expression profiles in adult tissues, RNA was extracted from whole bodies, and dissected tissues (heads, germline, and the remaining carcasses) from D. auraria, D. rufa, D. kikkawai, D. virilis,D.montana, and D. mojavensis flies. All samples were DNase treated (Ambion) and then used for cDNA synthesis (SuperScript III, Invitrogen). During cDNA synthesis, a “No RT” control was generated for each RNA extraction in which the reverse transcriptase was excluded from the reaction. For RT-PCR experiments, the presence of genomic DNA contamination was ruled out by performing PCR that amplified the housekeeping gene, Rp49, on each cDNA sample as well as each “No RT” control. 25- (data not shown) and 30-cycle PCRs were performed with primers specific to each Cid paralog and samples were run on an agarose gel for visualization. RT-qPCR was performed according to the standard curve method using the Platinum SYBR Green reagent (Invitrogen) and primers designed to each Cid paralog and to Rp49. Reactions were run on an ABI QuantStudio 5 qPCR machine using the following conditions: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of (95 °C for 15 s, 60 °C for 30 s). We ensured that all primer pairs had similar amplification efficiencies using a dilution series of genomic DNA. Three technical replicates were performed for each cDNA sample. Transcript levels of each gene were normalized to Rp49. For all primers used in RT-PCR and RT-qPCR experiments, see supplementary tables S5 and S6, Supplementary Material online.
Motif Analyses
Motifs were identified in six different groups of Cid proteins (supplementary fig. S6, Supplementary Material online) using the motif generator algorithm MEME (Bailey and Elkan 1994) implemented on http://meme-suite.org/(Bailey et al. 2009). Several motifs identified in different groups were similar to one another. For example, the motif “TDYLEFTTS” appeared in melanogaster group Cid1s, montium subgroup Cid3s and Cid4s and virilis group Cid1s (supplementary fig. S6, underlined residues, Supplementary Material online). To determine which motifs were the same, we used the motif search algorithm MAST (Bailey and Gribskov 1998) to search for the top four motifs from each group against all 86 sequences used for motif generation. In total, we found 10 unique motifs (fig. 6B and 6C). The only instance in which the motifs were not totally independent was for motif 2 and motif 9. Motif 2 was contained within motif 9, but motif 9 was significantly longer than motif 2 so we considered it to be an independent motif. We mapped all 10 motifs to the Cid genes in the six groups plus D. eugracilis Cid2, D. mojavensis and D. grimshawi Cid1 and Cid5, D. busckii, and the outgroup species P. variegata Cid1. We considered a motif to be present in a given protein if the MAST P-value was <10−5.
Positive Selection Analyses
We used the PAML suite of programs (Yang 1997) to test for positive selection on each Cid paralog across deep evolutionary time. Alignments for each Cid paralog were generated and manually refined as described above. Alignments (supplementary data S5, Supplementary Material online) and Cid gene trees were used as input into the CODEML NSsites model of PAML. To determine whether each Cid paralog evolves under positive selection, we compared two models that do not allow dN/dS to exceed 1 (M7 and M8a) to a model that allows dN/dS > 1 (M8). Positively selected sites were classified as those sites with a M8 Bayes Empirical Bayes posterior probability > 95%. We used the MK test (McDonald and Kreitman 1991) implemented in the DnaSP program v5.10.1 (Librado and Rozas 2009) to look for more recent positive selection at the population level. To implement the MK test for montium subgroup Cid paralogs we compared Cid sequences in 26 strains of D. auraria to 10 strains of D. rufa. In the virilis group, we compared Cid sequences in 10 strains of D. virilis to 20 strains of D. montana.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Rick McLaughlin, Antoine Molaro, Courtney Schroeder, Janet Young, Tera Levin and Rini Kasinathan for their comments on the manuscript and past and present members of the Malik lab for valuable discussions. We thank Frances Welsh and Tobey Casey for help with the PCR analyses to confirm the presence or absence of potential Cid paralogs. We thank the San Diego and Ehime Species stock centers for the use of Drosophila strains, and Aaron Straight for sharing the Drosophila CENP-C antibody. This work was supported by funding from the National Institutes of Health training grants T32 HG000035 and T32 GM007270 (to L.E.K.) and R01 GM074108 (to H.S.M.). The funders played no role in study design, data collection and interpretation, or the decision to publish this study. H.S.M. is an Investigator of the Howard Hughes Medical Institute.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancliff M, Park JM.. 2014. Evolution dynamics of a model for gene duplication under adaptive conflict. Phys Rev E Stat Nonlin Soft Matter Phys. 89:062702.. [DOI] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ, FlyBase C.. 2016. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44:D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aul RB, Oko RJ.. 2001. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis. Dev Biol. 239:376–387. [DOI] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. 2013. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci U S A. 110:11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Thomas GE, Rusch DW, Merkel AW, Jeppesen CD, Carstens JN, Randall CE, McClintock WE, Russell JM.. 2009. Phase functions of polar mesospheric cloud ice as observed by the CIPS instrument on the AIM satellite. J Atmos Solar-Terrest Phys. 71:373–380. [Google Scholar]
- Bailey TL, Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M.. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW.. 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell 25:309–322. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH.. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 3:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake L, Baechli G.. 2008. Drosophilidae (Diptera) In: World catalogue of insects, Vol. 9 Stenstrup: Apollo Books, p. 412. [Google Scholar]
- Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA.. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 24:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel A. 2002. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A). Am J Med Genet. 111:450–452. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Schaller AM, Geballe AP, Malik HS.. 2016. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife 5:14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Gilbert SL, Forster ML, Barndt RJ, Lahn BT.. 2003. The CDY-related gene family: coordinated evolution in copy number, expression profile and protein sequence. Hum Mol Genet. 12:1643–1650. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, deYoung D, Henikoff S, Malik HS.. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife 3:03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH.. 2012. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 10:e1001460.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N.. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, et al. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 15:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finseth FR, Dong Y, Saunders A, Fishman L.. 2015. Duplication and adaptive evolution of a key centromeric protein in Mimulus, a genus with female meiotic drive. Mol Biol Evol. 32:2694–2706. [DOI] [PubMed] [Google Scholar]
- Fishman L, Saunders A.. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322:1559–1562. [DOI] [PubMed] [Google Scholar]
- Folco HD, Campbell CS, May KM, Espinoza CA, Oegema K, Hardwick KG, Grewal SI, Desai A.. 2015. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr Biol. 25:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J.. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Betran E.. 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 26:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte-Gattat D, Shuaib M, Ouararhni K, Gautier T, Skoufias DA, Hamiche A, Dimitrov S.. 2013. Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function. Proc Natl Acad Sci U S A. 110:8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P.. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2:280–291. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik H.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science (New York, N.Y.) 293:1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B.. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A. 97:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH.. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA.. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A. 97:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R.. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Karimi-Ashtiyani R, Banaei-Moghaddam AM, Schubert V, Fuchs J, Houben A.. 2015. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23:277–284. [DOI] [PubMed] [Google Scholar]
- Kawabe A, Nasuda S, Charlesworth D.. 2006. Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences. Genetics 174:2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD.. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 23:1891–1901. [DOI] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2016. Centromeres. Curr Biol. 26:R487–R490. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J.. 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A. 102:11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang JF.. 2008. Identification and molecular evolution of cow CENP-A gene family. Mammal Genome 19:139–143. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE.. 2015. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol. 208:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Brutlag DL.. 1987. Identical satellite DNA sequences in sibling species of Drosophila. J Mol Biol. 194:161–170. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A.. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154:459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Tan EH, West A, Franklin FC, Comai L, Chan SW.. 2015. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 11:e1004970.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2003. Phylogenomics of the nucleosome. Nat Struct Biol. 10:882–891. [DOI] [PubMed] [Google Scholar]
- Malik HS, Vermaak D, Henikoff S.. 2002. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A. 99:1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci U S A. 25:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- Monen J, Hattersley N, Muroyama A, Stevens D, Oegema K, Desai A.. 2015. Separase cleaves the N-tail of the CENP-A related protein CPAR-1 at the Meiosis I metaphase-anaphase transition in C. elegans. PLoS One 10:e125382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C-elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 7:1248–1255. [DOI] [PubMed] [Google Scholar]
- Moraes IC, Lermontova I, Schubert I.. 2011. Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol. 75:253–261. [DOI] [PubMed] [Google Scholar]
- Neumann P, Navratilova A, Schroeder-Reiter E, Koblizkova A, Steinbauerova V, Chocholova E, Novak P, Wanner G, Macas J.. 2012. Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet. 8:e1002777.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Pavlikova Z, Koblizkova A, Fukova I, Jedlickova V, Novak P, Macas J.. 2015. Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 Gene in Fabeae species. Mol Biol Evol. 32:1862–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL.. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 88:3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL.. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384:346–349. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF.. 2012. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 10:e1001434.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin L, Mellone BG.. 2017. Centromeres drive a hard bargain. Trends Genet. doi:10.1016/j.tig.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin L, Mellone BG.. 2016. Co-evolving CENP-A and CAL1 domains mediate centromeric CENP-A deposition across Drosophila species. Dev Cell 37:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CAM, Mello B, Frazao A, Voloch CM.. 2013. Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool J Linn Soc Lond. 169:765–775. [Google Scholar]
- Sanei M, Pickering R, Kumke K, Nasuda S, Houben A.. 2011. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A. 108:E498–E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut E, Miller CA, Nickoloff JA.. 2005. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 33:1574–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Althoff F, Heidmann S, Lehner CF.. 2010. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci. 123:3768–3779. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF.. 2001. Genomic and genetic definition of a functional human centromere. Science 294:109–115. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Program NCS, Green ED.. 2010. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 27:1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S.. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 17:237–243. [DOI] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgeraldhayes M.. 1995. A mutation in Cse4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell-cycle arrest at mitosis. Genes Dev. 9:573–586. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476:232–235. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S.. 2004. Adaptive evolution of centromere proteins in plants and animals. J Biol. 3:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S.. 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras-Llort M, Medina-Giro S, Moreno-Moreno O, Azorin F.. 2010. A conserved arginine-rich motif within the hypervariable N-domain of Drosophila centromeric histone H3 (CenH3) mediates BubR1 recruitment. PLoS One 5:e13747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S.. 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 18:424–428. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S.. 2002. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 22:7553–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.