Abstract
microRNAs (miRNAs) are a unique class of short endogenous RNAs that became known in the last few decades as major players in gene regulation at the post-transcriptional level. Their regulatory roles make miRNAs crucial for normal development and physiology in several distinct groups of eukaryotes including plants and animals. The common notion in the field is that miRNAs have evolved independently in those distinct lineages, but recent evidence from non-bilaterian metazoans, plants, as well as various algae raise the possibility that already the last common ancestor of these lineages might have employed a miRNA pathway for post-transcriptional regulation. In this review we present the commonalities and differences of the miRNA pathways in various eukaryotes and discuss the contrasting scenarios of their possible evolutionary origin and their proposed link to organismal complexity and multicellularity.
Introduction
MicroRNAs (miRNAs) are short endogenous ~21-24 nucleotides long, single-strand RNAs derived from hairpin transcripts that regulate gene expression in both animals and plants 1–5. miRNAs repress gene expression by binding to a complementary mRNA target thereby mediating translational inhibition, degradation or cleavage 3. They belong to a group of functional small RNAs, which in addition to miRNAs include short interfering RNAs (siRNAs) responsible for RNA interference (RNAi) and Piwi-interacting RNAs (piRNAs) (For review on piRNAs see 6). While siRNAs and miRNAs share important features and are both produced by the ribonuclease Dicer 7, they are responsible for different cellular tasks and can be discerned from one another by unique characteristics (Fig. 1). Since their discovery in Caenorhabditis elegans more than two decades ago 8,9 it has become clear that miRNAs play a role in a broad variety of biological processes and are essential for normal development of animals and plants 2,3,10,11. The apparent rise in the number of miRNAs during the course of animal evolution and their involvement in regulating development raised the possibility that they might have contributed to the evolution of animal complexity 12,13.
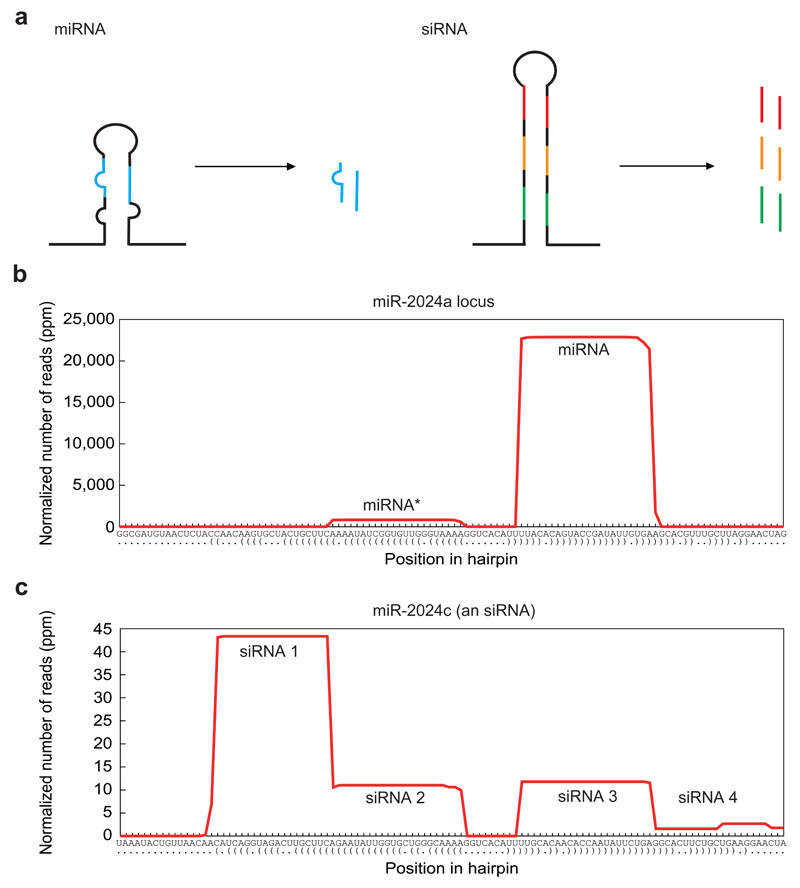
Figure 1.
Differences between miRNAs and siRNAs. a, A scheme of miRNA and siRNA precursors and duplexes. While miRNAs are usually produced from short hairpins carrying mismatches in their stem region, siRNAs are produced from long hairpins with stems of perfect complementarity. miRNA precursors usually give rise to a single duplex whereas siRNA precursors are a source for multiple duplexes. b, Small RNA profiles along a pre-miRNA sequence, here exemplified by miR-2024a of Nematostella vectensis. Note the homogenous product with the dominant guide strand (mature miRNA) and the neglectable passenger strand (miRNA*). c, Small RNA profiles along an siRNA precursor sequence, here exemplified by miR-2024c of N. vectensis. This siRNA locus was originally annotated as miRNA, but later determined to be an siRNA due to the fact it gives rise to multiple small RNAs 45. The x-axis in b) and c) indicates the position along the hairpin sequence with paired (brackets) and unpaired nucleotides (dots). ppm = parts per million. (b and c) modified from 45, with permission.
The lack of sequence homology between miRNA families in plants and animals as well as differences in miRNA biogenesis and mode of action have led to the notion that miRNAs have evolved independently in the two kingdoms from an ancient siRNA mechanism that already existed in the last common ancestor of all eukaryotes. However, recent studies relating plant and animal miRNAs unexpectedly raised again the old questions: Did the common ancestor of all animals have miRNAs? Do miRNAs of plants and animals share a common origin? How many times did miRNAs evolve? This review will attempt to re-address these questions.
Is the lack of miRNA sequence homology between distinct lineages sufficient to determine convergence of the miRNA pathway?
An early phylogenetic comparison of miRNAs revealed over 35 conserved families of miRNAs among bilaterian animals, and the pattern of conservation largely appeared to correlate with the accepted phylogeny. This led to the proposal that miRNAs are rarely lost once they have evolved14,15. Consequently, since no sequence similarity was detected between animal and plant miRNAs, they were considered to have evolved independently 16. This line of thought extends also to several other lineages (Fig. 2). For instance, none of the 8 miRNAs of the demosponge Amphimedon queenslandica is shared with other non-poriferan animals. Moreover, no conserved miRNAs could be detected between demosponge, calcareous and homoscleromorph sponges. This led to the extreme possibility that the miRNA pathway evolved convergently multiple times even within sponges 13,17. Further, the traditional view that sponges are the sister group to all other animal lineages is under debate as several studies point to comb jellies (Ctenophora), which lack miRNAs, being the first animal lineage to diverge 18–20. If Ctenophora was indeed the first animal lineage to diverge, an independent origin of animal miRNAs can be supported.
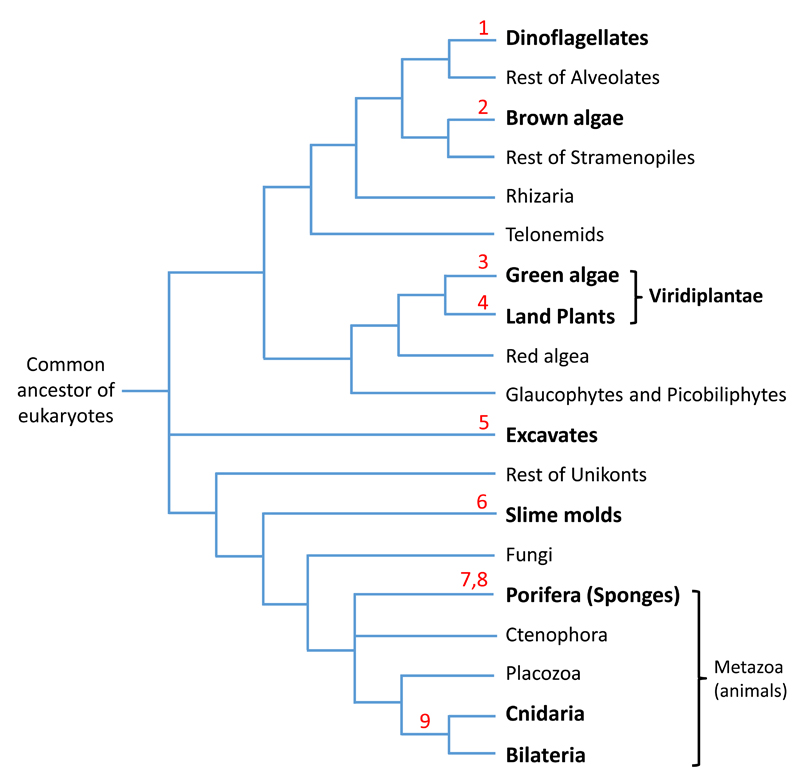
Figure 2.
Phylogenetic tree of the major eukaryotic groups showing presence of miRNA systems. Groups known to possess miRNAs are depicted in bold. Numbers in red count from top to bottom the maximum number of times miRNA systems evolved convergently. The phylogeny is based on 85.
Similarly, until recently, no shared miRNAs were observed between plants and green algae, suggesting convergent origin of the miRNA pathway in these two main groups of Viridiplantae (Fig. 2) 21. However, a new study annotating small RNAs of the liverwort Pellia endiviifolia revealed three miRNAs with high similarity to the green alga Chlamydomonas reinhardtii miRNAs 22,23. This emphasizes the great importance of sampling many species from each phylum in order to fully perceive the extent of miRNA sequence conservation but also highlights the inherent difficulty of annotating miRNAs with high confidence 24. Further, shallow sequencing depth as well as neglecting certain developmental stages and environmental conditions may mask the full miRNA repertoire of an organism and hence possible homology. Regardless of these caveats, the prevailing view is that the miRNA pathways of animals and plants evolved convergently 14,16,21,25–28. In fact, this would mean that the miRNA pathway evolved independently at least 9 times (in bilaterians, in cnidarians; twice in sponges; in land plants; in green algae; in brown algae, in slime molds and in excavates; Fig. 2) 5,8,9,13,17,22,29–32.
However, we suggest an alternative explanation: it might be possible that sequence turnover rates are high in plants and non-bilaterian animals, leaving no trace of shared miRNA sequences between contemporary lineages. In support of this scenario, plant miRNA genes are born and lost at high rates 33,34, hence only a handful of expressed miRNAs are conserved among distant plant lineages 35. Comparison of Arabidopsis thaliana and Arabidopsis lyrata small RNAs has shown that 33% of the miRNA families are not conserved between the two species, hence were gained or lost during the ~10 million years (Myr) since they diverged 33. Analysis based on genome data and small RNA sequencing from Capsella rubella, a very close relative of Arabidopsis, estimated that the net flux rate (birth–death) for miRNA genes in Arabidopsis is 1.2 - 3.3 genes per Myr 34. A high turnover rate of miRNAs is also suggested in green algae as only one miRNA is conserved between the two green algae Chlamydomonas and Volvox that separated about 200 million years ago (MYA) 36,37. Within Bilateria, a recent study demonstrates that despite impressive examples of conservation, miRNA loss is much more common than previously appreciated 38. Further, major losses of miRNAs have been reported within flatworms, a rapidly evolving lineage, perhaps as part of a morphological simplification trend 39. Regardless, while the turnover of most bilaterian miRNAs might be higher than initially estimated, it is still lower than that of plants and estimated at 0.8 to 1.6 genes or possibly even only 0.3 genes per Myr in Drosophila 40,41.
Insights into evolution of miRNAs from understudied eukaryotic groups
As bilaterians have preserved many conserved miRNA families in comparison to plants, it is useful to consider outgroups to bilaterians. Among the non-bilaterians, placozoans and ctenophores do not possess miRNAs 18, while in the sponge Amphimedon only 8 miRNAs have been identified so far 13. The phylum Cnidaria (corals, sea anemones, jellyfish and hydroids) represents the sister group of Bilateria, which has branched off about 600 MYA 19,20. A pioneering study of small RNAs in the sea anemone Nematostella vectensis indicated that this species possesses at least 40 miRNAs, yet only one of them, miR-100, is shared with Bilateria 13. Remarkably, miR-100 is conserved almost over the full length of the miRNA, yet shifted by one nucleotide in the seed region, which was predicted to have significant consequences on the target specificity 13. miR-100 function and its targets are not well conserved in Bilateria 42 making its extreme sequence conservation in the sea anemone puzzling. However, lack of target conservation is true for most of the well-conserved bilaterian miRNAs, as they are frequently rewired into different genetic networks 43. Interestingly, sequencing of small RNAs of the cnidarian Hydra magnipapillata revealed 126 miRNAs with no sequence similarity to those of Bilateria (hence no evidence of miR-100) and only two shared miRNAs with Nematostella 44. These results suggest a rather fast turnover of miRNAs within Cnidaria and highlight the risk of misinterpreting lack of miRNA sequence homology as a lack of homology of the miRNA system.
In a recent study of Nematostella small RNAs obtained from several developmental stages the miRNA complement was expanded to 87 miRNAs 45. Interestingly, miR-9422 of Nematostella shares 16 of its positions, including the seed sequence with Arabidopsis miR-156a.. This sequence similarity is unlikely to have occurred randomly as the chance of observing such a sequence identity between Nematostella and shuffled Arabidopsis miRNAs is very small (random sampling P-value < 0.01) 45. As miR-156 is conserved from mosses to higher plants 5,46,47, its similarity to miR-9422 could be an evidence for a miRNA conserved between animals and plants, thus supporting the hypothesis that miRNAs were inherited from the last common ancestor of the two groups. Taken together, in light of the potential high turnover rate, it is questionable whether lack of sequence homology can serve as a proof for multiple cases of convergent evolution of this pathway. Hence, additional features must be taken into account.
Another indication for evolutionary homology of animal and plant miRNA pathways comes from sequencing the genome and the small RNA repertoire of the symbiotic unicellular dinoflagellate Symbiodinium kawagutii, an outgroup to both plants and animals (Fig. 2), which revealed many potential shared miRNAs with plants and animals 48. miRNAs have also been described in excavates, a group of unicellular eukaryotes that includes the parasites Giardia lamblia and Trichomonas vaginalis 32,49. Several putative miRNA families from excavates have sequence homology to conserved animal and plant miRNAs. However, unlike most plant and animal miRNAs those peculiar excavate miRNAs are produced from snoRNAs and tRNAs and exhibit some additional irregular features. Furthermore, the possibility of horizontal gene transfer from a host to its symbiont or parasite could be an alternative explanation for the potential homology in these cases. Thus, further confirmation of these data from dinoflagellate and excavates is required. Yet, if those small RNAs are verified as bona fide miRNAs, this will be of great importance because it will strongly support the placement of the pathway’s origin before the split of plants and animals.
The biogenesis of miRNAs and its potential role in clarifying the pathway’s evolutionary history
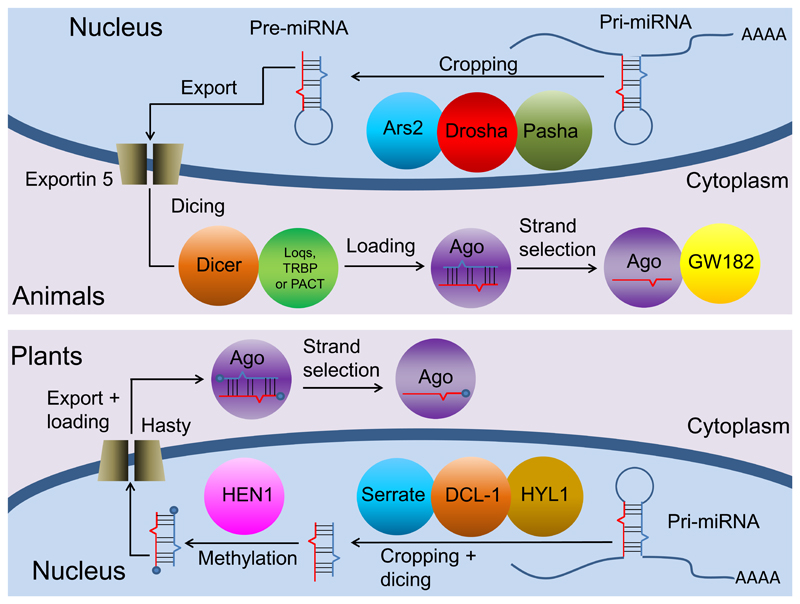
The discussion whether plant and animal miRNA pathways evolved in a common ancestor or independently from an ancient RNAi mechanism must include the differences and similarities in miRNA biogenesis (Fig. 3). miRNAs are synthesized as primary hairpins (pri-miRNAs) and are processed to pre-miRNAs by cutting the hairpin stem, followed by cleaving the hairpin loop 3,7. Next, they are loaded on to an Argonaute (AGO) protein and one RNA strand is selected for complementary target mRNA inhibition or cleavage 3,7. In animals, the first step is conducted by a specialized microprocessor complex comprised of the RNAse III Drosha with the aid of the RNA binding protein Pasha (DGCR8 in vertebrates), while the second step of cleavage is performed by the RNAse III Dicer (Fig. 3) 7. In plants, the Dicer homolog, DICER-LIKE 1 (DCL1), is responsible for both processing events required for miRNA maturation, conducting the two exact same steps in the same order 2,28. In both plants and animals Dicer is crucial for processing the precursor miRNA (pre-miRNA) into mature miR/miR* dsRNA duplexes 2,7,25,50. However, Dicers are common in many eukaryotes and also take part in non-miRNA activities such as RNAi via endogenous siRNA biogenesis and viral defence 25,51 and therefore cannot be used as an argument for the common origin of miRNAs. Phylogenetic and structural analyses indicate that animal Drosha proteins are related to plant and animal Dicers, suggesting that in animals Drosha may have evolved following a duplication of the common ancestor of Dicer and Drosha and further specialised in the first processing step of miRNAs 25,52. A very recent study found that miRNAs in Chlamydomonas are processed by DICER-LIKE3 (DCL3), a Dicer homolog that exhibits a domain organization similar to that of Drosha 53. This finding hints at a possible evolutionary link between miRNA biogenesis in green algae and animals.
Figure 3.
A scheme describing the plant and animal canonical miRNA biogenesis pathways. Homologs carrying similar functions such as Ars2 of animals and Serrate of plants are represented in the same color. This figure is modified after 54 with permission.
Both Drosha and Pasha homologs were found in Cnidaria 13,54 and in two sponge lineages 13,17, for which independent evolution of the miRNA pathway has been proposed . Thus, Drosha and Pasha were already present in the common ancestor of all sponges, Cnidaria and Bilateria and likely served miRNA-related functions, although their involvement in the miRNA pathway in the first two lineages remains to be shown. If Drosha and Pasha have an ancestral function in miRNA processing in these lineages, the only explanation for the lack of homology of miRNA sequences in distinct sponge lineages would be high turnover rates of miRNA genes (see above). Nevertheless, it should be noted that Drosha and Pasha are known to be involved also in non-miRNA related functions in mammalian cells that range from ribosomal RNA maturation to cleavage of specific mRNAs (reviewed in 55). The tendency to associate the presence of the microprocessor components with the presence of miRNAs in basally-branching lineages may thus be misleading.
The site of miRNA biogenesis seems to differ between plants and animals. In plants, both processing steps by DCL1 take place in the nucleus, while in animals the first step performed by Drosha occurs in the nucleus and the second cleavage by Dicer occurs in the cytoplasm 3,16. However, several studies in animals reported the presence of Dicer in the nucleus (reviewed in 55). Whether the nuclear localization of Dicer in animals is a relic of an ancient miRNA-processing pathway or a secondary adaptation is an open question.
In plants and animals Dicer requires protein partners in order to accurately cleave pre-miRNAs (Fig. 3) 2,7. In plants, DCL1 is assisted by the RNA binding proteins SERRATE (SE) and HYPONASTIC LEAVES1 (HYL1), both crucial for miRNA biogenesis and development 2,56,57. While a SE homolog, Ars2, is known in animals as a partner of the microprocessor and Dicer 58, no HYL1 homologs were found in bilaterian animals. These differences in the biogenesis were taken as additional evidence for an independent evolution of plant and animal miRNAs. However, homologs for both SE and HYL1 were recently reported in the sponge Amphimedon and in several cnidarians including Nematostella, all possessing dsRNA binding motifs 54. This suggests that a HYL1-like protein was present in the last common ancestor of plants and animals and was lost in multiple lineages, including Bilateria 54. The lack of any known animal Dicer partners such as Loquacious (Loqs), TRBP or PACT 7,59–61 in Nematostella 54 suggests that HYL1 may constitute a Dicer partner in Cnidaria.
The miR/miR* duplexes in both plants and animals are similar: they are ~22nt-long with imperfect complementarity between the two strands and a 2nt 3'-overhang2,3,7. However, unlike in bilaterian animals, the stem-loop precursor in plants is long and variable 16. Interestingly, sponges and slime molds present longer pre-miRNAs than their bilaterian counterparts 13,30. Those longer and more variable miRNA precursors are reminiscent of siRNA precursors. Interestingly, the miR-2024 family in Nematostella has members that are bona fide miRNAs whereas other members of the family show processing variability that is typical for siRNAs (Fig. 1b and c) 45. This suggests that transformation of a miRNA into a siRNA or vice versa can happen in animals like in plants 62 and that the siRNA-like features of some miRNAs are not sufficient in order to rule out a common origin for plant and animal miRNAs.
Differences can also be found in the genetic structure of miRNA genes in plants and animals. In animals, roughly 50% of miRNA genes are located in clusters, often comprised of different mature miRNAs 16,63. In plants, fewer cases of miRNA clusters are found, mostly encoding miRNAs of the same family with noticeable homology 16,64. Nevertheless, a few clusters of non-homologous miRNAs were discovered in plants, predicted to target related proteins 64. Interestingly, the sea anemone Nematostella has only two repetitive clusters, both of miR-2024, similarly to plants 45. Hence, it seems that while having different frequencies, the same genomic topologies can be found in both kingdoms.
Another genomic pattern separating animal miRNAs from those of plants is the location of miRNA genes. Approximately 30% of animal miRNA genes are located in introns of pre-mRNAs 16,65. In contrast, only three tested examples of intronic miRNAs are known so far in plants 66–68. The hypothesis that this distinction supports an independent origin of miRNA is undermined by a recent study on the green alga Chlamydomonas. This study found that unlike in plants, 50% of the miRNAs in Chlamydomonas are embedded within introns of protein-coding genes, similar to animals 53. These results can support a common origin of the animal and plant miRNA pathways, assuming higher plants lost this genomic feature.
In both plants and animals, binding of small RNAs to extensively complementary target RNAs induces degradation of the small RNA by adenosine or uracil addition (‘tailing’) and 3ʹ-to-5ʹ exonucleolytic decay (‘trimming’) 1,69. siRNAs in flies and PIWI interacting RNAs (piRNAs) in germ cells of all animals are protected from these processes by 2ʹ-O-methylation of their 3ʹ ends performed by the methyltransferase Hua enhancer 1 (HEN1) 1,27. In plants, HEN1 is responsible for the methylation of both siRNAs and miRNAs 1,2,70, while bilaterian miRNAs do not undergo this modification 27. Interestingly, HEN1 was found to be expressed throughout the body in Nematostella 54 and not only in germ cells, suggesting a non-restricted function. Moreover, periodate treatment proves that a major fraction of the Nematostella miRNAs is methylated, similar to plants 13,45. One can speculate that the common ancestor of animals and plants possessed methylated miRNAs, a feature later lost in bilaterians possibly due to the loss of high complementarity between miRNAs and their targets (see next section).
The mode of action of miRNAs varies between plants and animals
In both plants and animals siRNAs and miRNAs require a class of AGO proteins to execute gene regulatory functions 71–73. The mature miRNA duplex is loaded onto AGO, a core component of the RNA induced silencing complex (RISC) (Fig. 3) 3,74–76. The guide RNA strand is selected by AGO and directs RISC to the complementary RNA transcript 3. AGO proteins are conserved throughout all classes of life, from archaea and bacteria to eukaryotes, where they participate mostly in small non-coding RNA related mechanisms 25,71,77–80. Furthermore, the four structural domains of AGO proteins and their endonuclease and RNA-binding activity are highly conserved 71,74. Target slicing by siRNA-guided AGO is used in a wide variety of organisms to protect cells from viruses and transposons 81. In plants, in addition to those activities AGO proteins can cleave mRNA targets, thus controlling their expression. However, this gene regulation mechanism requires a miRNA guide loaded into the AGO protein 3. Similar to siRNAs, plant miRNA target binding requires a nearly-full complementarity between the miRNA and its target, leading to the endonucleolytic cleavage of the target by AGO between position 10 and 11 of the miRNA, often resulting in a strong effect on a small number of targets 1,82–84. The AGO proteins of prokaryotes that usually target foreign DNA also cleave their targets at the same position as plant AGOs, suggesting this is an ancient mode of action 77,85,86. In contrast, in bilaterians, every miRNA potentially regulates a large number of targets because miRNA-mRNA recognition requires only a 7 nucleotides "seed" sequence, residing in positions 2-8 of the miRNA (Fig. 4) 4. Unlike in plants, the vast majority of animal AGOs do not induce miRNA target cleavage. Instead, animal RISC induces translational repression of targets by blocking translation initiation r elongation or by deadenylation 1,74. This mode of action results in relatively weak modulation of less than 2-fold both at the RNA and protein levels 87,88. An advantage of translational repression is its reversibility that allows rapid expression of existing blocked mRNAs in a given condition, time or location 89,90. The destabilization and translational inhibition of targets is not performed directly by AGO, but by a partner protein known as GW182 in Drosophila or TNRC6 in vertebrates 91–94. Interestingly, GW182 proteins evolve very rapidly, making detection of homology even within bilaterians quite challenging 95.
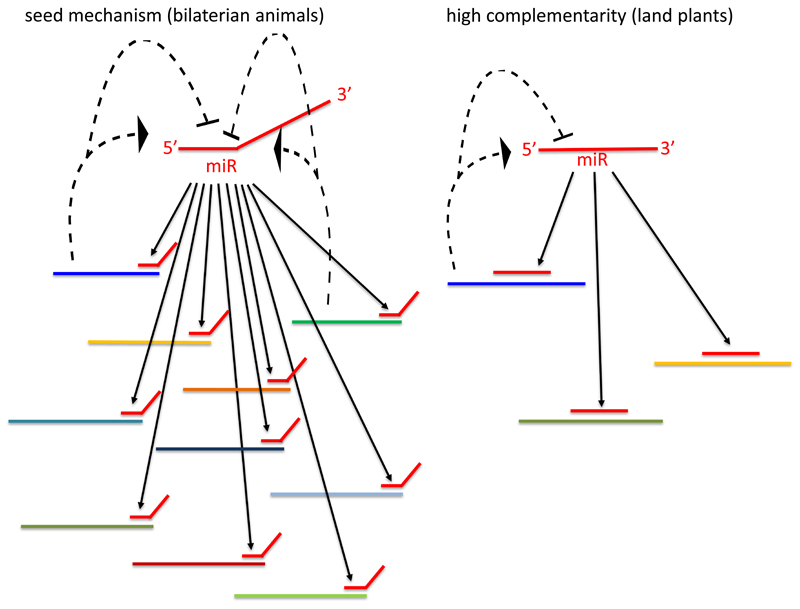
Figure 4.
Schematic comparison of miRNA network topology in land plants and bilaterian animals. Solid lines represent inhibition of targets by miRNA either by seed match (bilaterian animals) or by nearly-full complementarity (land plants). Dotted lines represent reciprocal effect of targets on miRNAs.
The degree of complementarity between the miRNA and its mRNA target has been considered as a major factor in determining the mode of target repression. High complementarity, as seen in plants, promotes target cleavage by AGO, while seed-matching leads to translational inhibition and is the common mode of action of bilaterian miRNAs 74,92,96. The boundaries between plant and animal miRNA mode of action are becoming somewhat more blurry as there is more translation inhibition in plants than previously appreciated 83,84,97,98. However, even this type of inhibition requires nearly full complementarity as short matches limited to the seed region were shown to confer no target inhibition in plants 83,84. Surprisingly, miRNA-mediated translational inhibition in plants also depends on a GW-repeat protein called SUO 99. GW182 of animals and SUO of plants do not share detectable homology. However, it is tempting to consider the possibility of a common origin of those two proteins that cannot be detected anymore due to the extremely fast evolution of the GW182 family.
In Bilateria there are only a handful of examples for miRNA mediated target cleavage due to full complementarity of the miRNA to the target 100,101. For instance, in a genome-wide screen, only a single transcript was shown to be cleaved by a perfectly-matching miRNA in C. elegans 6. Further, the majority of AGO proteins in Bilateria have lost their endonucleolytic activity 74,102,103. However, endonucleolytic activity of the vertebrate AGO2 protein was shown to be evolutionary conserved due to its role in processing a single miRNA, miR-451, which cannot be detected by Dicer 43,104,105. Thus, it is plausible that the conservation of this AGO activity in vertebrates is unrelated to target regulation. These are strong indications that target cleavage is far from being a major mechanism of action of bilaterian miRNAs. Recently, Nematostella miRNAs were reported to induce target cleavage by nearly perfect complementarity, more resembling siRNAs and plant miRNAs than bilaterian miRNAs 45. Unlike in Bilateria, the target cleavage mechanism seems to be common in Cnidaria as it is also present in Hydra, a cnidarian that diverged from Nematostella at least 550 MYA 45. The fact that both Cnidaria and plants use target cleavage mechanism combined with the existence of siRNAs in fungi, plants and animals suggest that cleavage was the ancestral form of small RNA action. The presence of a cleavage mechanism in sponges, the only other non-bilaterian lineage with miRNAs, would strongly support this view. While experimental evidence is still missing, at least several of the eight miRNAs identified in Amphimedon queenslandica show high complementarity to putative target mRNAs. Additionally, the origin of the miRNA systems in the siRNA mechanism, which is based on slicing, by itself suggests that high complementarity is the ancestral state of the miRNA mode of action. This is also supported by the fact that seed matching does not promote target inhibition in plants 83,84. Those mechanistic differences indicate that AGO holds the guide strand very differently in plants and bilaterian animals leading to different binding characteristics 106. It seems that seed matching is a derived state that appeared in the last common ancestor of all bilaterians. It is likely that this mode of action allowed animals to expand their regulatory networks as one miRNA can potentially inhibit hundreds of targets (Fig. 4) 4,32. Those complex networks involving miRNAs might be a reason for the lower turnover rates of miRNAs in bilaterians since the loss of a single miRNA might affect the expression level of numerous targets.
miRNAs and the evolution of complex organisms
In both animals and plants gene regulation by miRNAs allowed organisms to develop more complex gene regulatory networks 4,107. Further, miRNAs were theorized by various authors to drive evolution to a multicellular state, making it possible for complex organisms to evolve 3,12–15. To assess this theory, one needs to look into the evolution of multicellularity and the correlation between the miRNA repertoire and the complexity of an organism. Multicellular life forms had evolved from a unicellular ancestor independently in multiple lineages leading to fungi, animals, plants and various algae 108,109. In all cases, it was speculated that this transition required the evolution of cell adhesion molecules, cell communication, co-operation between cells and cellular differentiation (division of labour) 108,110,111. miRNAs in both plants and animals are known to be involved in pathways related to multicellularity such as development timing 112–114, differentiation 36,115–118 and morphogenesis 10. Additionally, in multicellular organisms "fine tuning" of gene expression by miRNAs can lower phenotypic variation (“canalization”) among individuals in a population and even among cells, thus reducing conflicts between cells of different genetic background 12,110,119,120. Nevertheless, their connection to the evolution of multicellularity is at best a mere correlation. Indeed, many multicellular organisms including plants and animals possess miRNA regulatory mechanisms. However, miRNAs were found in unicellular organisms such as the green alga Chlamydomonas reinhardtii 22,29 and the reef-building coral dinoflagellate endosymbionts Symbiodinium microadriaticum 121 and Symbiodinium kawagutii 48. This proves that the presence of miRNAs does not necessarily predict multicellularity. It can be hypothesized that miRNAs may be part of the genetic toolkit allowing the transition to multicellularity under the right environmental conditions. For example, it was demonstrated that under strong artificial settlement selection Chlamydomonas can develop a multicellular life stage 122, suggesting that ecological conditions are as important as genetic potential on the way to multicellularity.
miRNAs are not mandatory for multicellularity as there are multicellular organisms that do not possess miRNAs such as fungi 123,placozoans 13 and ctenophores 18. In the case of ctenophores, it is possible that their extraordinary large collection of RNA-binding proteins substitutes the functions that miRNAs cover in other animals, allowing this phylum to produce complex cell types such as muscles and neurons 20. Is the loss of complex, allegedly integral features feasible? For example, it was suggested that sponges and Placozoa might have lost their nervous system for an adaptive advantage 72. A similar logic can explain the possible loss of miRNAs in organisms such as fungi, Placozoa or choanoflagellates, An example for advantageous loss of the RNAi components in fungi can be found in a study on the yeast Saccharomyces cerevisiae. The study demonstrated that several strains that had lost RNAi became more susceptible to killer virus infection when RNAi components were restored 73. If losing the entire RNAi machinery is possible, we propose that losing only miRNAs must be a possibility as well. Despite of all this, the alternative scenario arguing that miRNAs evolved independently in plants, animals and other clades cannot be ruled out.
It has been suggested that the relative number of miRNAs is in correlation with the relative level of organism morphological complexity. While it is generally difficult to define complexity, miRNA complements might have contributed to the complexity of gene regulation 13–15,124–126. For example, sea anemones and humans possess ~80 and ~500-1000 miRNAs, respectively (but see also a recent higher count for humans in 127) 13,24,45. The abundance of miRNAs in human in relation to the sea anemone may help explain how despite high similarity in their genome and protein-coding genes humans have evolved a much more complex body form and cell type diversity than the relatively simple sea anemone 13,128. However, on closer inspection it seems that no strong correlation actually exists between the number of miRNAs and organismal complexity. For example, sea anemones, annelids and fruit flies have a similar number of miRNA families (~80, ~105 and ~110, respectively) despite differences in morphological complexity 14,45,126,129. Moreover, the thalliform multicellular brown alga Ectocarpus possesses over 60 miRNA families despite its low complexity 14. Yet, instead of the number of miRNAs, one should consider the number of targets per miRNA. We hypothesize that the "seed" target recognition approach in bilaterian animals, where each miRNA recognizes and regulates many targets, allowed the evolution of highly complex and interwoven regulatory networks (Fig. 4), which may have contributed to cell type diversification during evolution.
Conclusion and future prospects
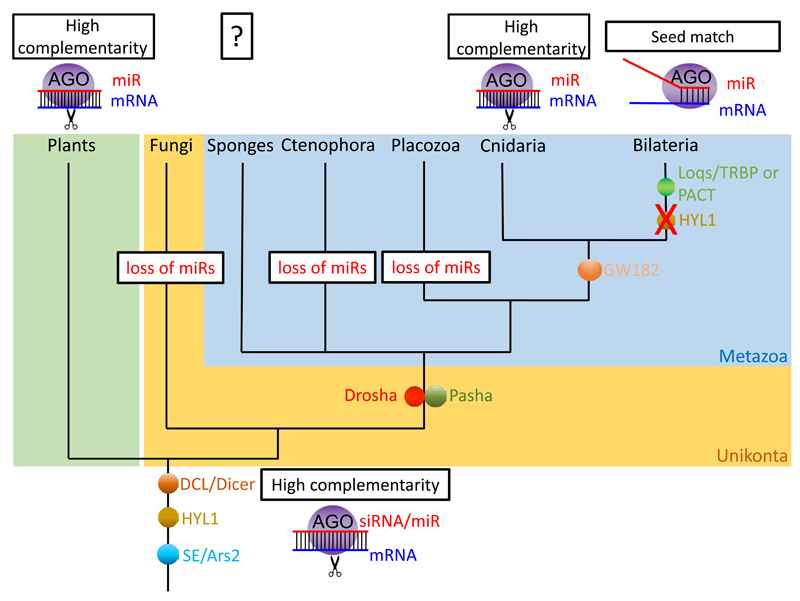
The rapidly expanding field of non-coding RNA research allows us to constantly re-evaluate the evolutionary origin of miRNAs. This review presents claims for and against a common origin of miRNA in plants and animals. Given the fast turnover in miRNA sequences and significant loss of miRNAs in both plants and animals 33,34,38, we consider the lack of miRNA sequence homology known today as not sufficient to conclude on convergent evolution. Instead, the available information provides support for the possibility that the last common ancestor of plants and animals did possess a miRNA system (Fig. 5).
Figure 5.
A possible scenario of miRNA evolution in plants and animals where their last common ancestor possessed a miRNA system. Appearances and losses of proteins and traits are depicted on the relevant branches.
Of course, we cannot rule out convergent evolution. If indeed miRNAs have evolved independently at least 9 times among eukaryotes (Fig. 2), it is intriguing to consider what is so unique about them that will support the evolutionary pressures required to allow convergence of such a magnitude. Ctenophores certainly demonstrate that an animal with a wide variety of cell types, including neurons and muscles can exist in the absence of this class of small RNAs 18 and single-celled green algae demonstrate that miRNAs are not exclusive for “complex” multicellular organisms 29. Further, the lack of classic RNAi components in some fungi and the loss of DGCR8/Pasha in Placozoa 13,105 suggests that losing the protein machinery of the miRNA system is possible (Fig. 5) 13.
Taken together, we can suggest two alternative hypotheses regarding the evolution of miRNAs: 1) The miRNA pathways and their mode of action via slicing originated convergently from siRNA mechanisms in the plant and several animal lineages and a system based on seed-matching of miRNA to its targets evolved later, after bilaterians diverged from the rest of animals. 2) The common ancestor of plants and animals already had a miRNA system acting via slicing that was recruited from the siRNA system. Still, also in this scenario bilaterian animals have evolved a derived system based on seed-matching. In both scenarios the siRNA system evolved to protect cells from viruses and transposons and was adapted to post-transcriptional regulation of gene expression by targeting mRNAs.
Given the recent new findings, we favor the hypothesis of a common origin of miRNAs in plants and animals (Fig. 5). To further test this idea, biochemical and genetic methods are required to map the mode of action and the biogenesis pathway of miRNAs in groups beyond bilaterian animals and higher plants. We believe that some of the key points to study in the near future should be:
Test whether miRNAs in basally-branching lineages such as sponges or algae cleave their targets.
Test whether the cnidarian HYL1 proteins, known to be involved in miRNA biogenesis in plants, are also involved in biogenesis of miRNAs in Cnidaria.
Assess the impact of the slicing versus the seed-matching mechanism on the gene regulatory networks in animals and plants.
Test whether homologous miRNA-related proteins in algae are indeed functional in the miRNA pathway.
Sequence small RNAs at an adequate depth from more eukaryotic lineages and annotate them as miRNAs according to widely accepted guidelines to reveal pattern of evolutionary turnover.
When those five points are met, we might get a better understanding of the evolution of the miRNA pathway in eukaryotes and answer the question of how ancient is this system.
Acknowledgements
Small RNA research in the Moran lab is supported by a European Research Council Starting Grant (CNIDARIAMICRORNA, 637456) and a Young Investigator Grant by the German-Israeli Foundation for Scientific Research and Development (I-1058-203.7-2013). Research in the Technau group is supported by grants of the Austrian Research Fund FWF (P24858 and P22618).
Footnotes
Author's contributions:
Y.M. and U.T. conceived the manuscript, Y.M., M.A., D.P. and U.T. wrote the paper.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nature reviews Molecular cell biology. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & development. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annual review of biochemistry. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 9.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 10.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 11.Chen X. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 13.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarver JE, et al. microRNAs and the evolution of complex multicellularity: identification of a large, diverse complement of microRNAs in the brown alga Ectocarpus. Nucleic acids research. 2015;43:6384–98. doi: 10.1093/nar/gkv578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erwin DH, et al. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 16.Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson JM, et al. The identification of microRNAs in calcisponges: independent evolution of microRNAs in basal metazoans. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2013;320:84–93. doi: 10.1002/jez.b.22485. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell EK, Ryan JF, Schnitzler CE, Browne WE, Baxevanis AD. MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi. BMC genomics. 2012;13:714. doi: 10.1186/1471-2164-13-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan JF, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarver JE, Donoghue PC, Peterson KJ. Do miRNAs have a deep evolutionary history? Bioessays. 2012;34:857–866. doi: 10.1002/bies.201200055. [DOI] [PubMed] [Google Scholar]
- 22.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 23.Alaba S, et al. The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytologist. 2015;206:352–367. doi: 10.1111/nph.13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromm B, et al. A Uniform System for the Annotation of Human microRNA Genes and the Evolution of the Human microRNAome. Annual Review of Genetics. 2015;49 doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Current genetics. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Functional & integrative genomics. 2005;5:129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 27.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Reviews Genetics. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 29.Zhao T, et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes & development. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avesson L, Reimegård J, Wagner EGH, Söderbom F. MicroRNAs in Amoebozoa: Deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012;18:1771–1782. doi: 10.1261/rna.033175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinas A, et al. The small RNA repertoire of Dictyostelium discoideum and its regulation by components of the RNAi pathway. Nucleic acids research. 2007;35:6714–6726. doi: 10.1093/nar/gkm707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang P-J, et al. Identification of putative miRNAs from the deep-branching unicellular flagellates. Genomics. 2012;99:101–107. doi: 10.1016/j.ygeno.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. The Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fahlgren N, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. The Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. The Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Wu Y, Qi Y. MicroRNAs in a multicellular green alga Volvox carteri. Science China Life Sciences. 2014;57:36–45. doi: 10.1007/s11427-013-4580-3. [DOI] [PubMed] [Google Scholar]
- 37.Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proceedings of the National Academy of Sciences. 2009;106:3254–3258. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson RC, Plachetzki DC, Mahler DL, Moore BR. A critical appraisal of the use of microRNA data in phylogenetics. Proceedings of the national academy of sciences. 2014;111:E3659–E3668. doi: 10.1073/pnas.1407207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fromm B, Worren MM, Hahn C, Hovig E, Bachmann L. Substantial loss of conserved and gain of novel microRNA families in flatworms. Molecular Biology and Evolution. 2013;30:2619–2628. doi: 10.1093/molbev/mst155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, et al. The birth and death of microRNA genes in Drosophila. Nature genetics. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 41.Berezikov E, et al. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nature genetics. 2010;42:6–9. doi: 10.1038/ng0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw WR, Armisen J, Lehrbach NJ, Miska EA. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics. 2010;185:897–905. doi: 10.1534/genetics.110.117515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 44.Krishna S, et al. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic acids research. 2013;41:599–616. doi: 10.1093/nar/gks1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran Y, et al. Cnidarian microRNAs frequently regulate targets by cleavage. Genome research. 2014;24:651–663. doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arazi T, et al. Cloning and characterization of micro-RNAs from moss. The Plant Journal. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 47.Schwab R, et al. Specific effects of microRNAs on the plant transcriptome. Developmental cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 48.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 49.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Jong D, et al. Multiple dicer genes in the early-diverging metazoa. Molecular Biology and Evolution. 2009;26:1333–1340. doi: 10.1093/molbev/msp042. [DOI] [PubMed] [Google Scholar]
- 51.Ding S-W. RNA-based antiviral immunity. Nature Reviews Immunology. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 52.Kwon SC, et al. Structure of Human DROSHA. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 53.Valli AA, et al. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Research. 2016;26:519–529. doi: 10.1101/gr.199703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moran Y, Praher D, Fredman D, Technau U. The evolution of microRNA pathway protein components in Cnidaria. Molecular Biology and Evolution. 2013;30:2541–2552. doi: 10.1093/molbev/mst159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burger K, Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nature reviews Molecular cell biology. 2015 doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- 56.Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Current Biology. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 57.Han M-H, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabin LR, et al. Ars2 regulates both miRNA-and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y, et al. The role of PACT in the RNA silencing pathway. The EMBO journal. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS biology. 2005;3:1187. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature genetics. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 63.Kim VN, Nam J-W. Genomics of microRNA. TRENDS in Genetics. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Merchan F, Boualem A, Crespi M, Frugier F. Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol. 2009;10:R136. doi: 10.1186/gb-2009-10-12-r136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome research. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & development. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Q-H, et al. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome research. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joshi PK, et al. Identification of mirtrons in rice using MirtronPred: a tool for predicting plant mirtrons. Genomics. 2012;99:370–375. doi: 10.1016/j.ygeno.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Ameres SL, et al. Target RNA–directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swarts DC, et al. The evolutionary journey of Argonaute proteins. Nature structural & molecular biology. 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryan JF, Chiodin M. Where is my mind? How sponges and placozoans may have lost neural cell types. Phil Trans R Soc B. 2015;370:20150059. doi: 10.1098/rstb.2015.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592–1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature reviews Molecular cell biology. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 75.Meister G. Argonaute proteins: functional insights and emerging roles. Nature Reviews Genetics. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 76.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & development. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swarts DC, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Molecular cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 83.Iwakawa H-o, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Molecular cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 84.Liu Q, Wang F, Axtell MJ. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. The Plant Cell. 2014;26:741–753. doi: 10.1105/tpc.113.120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burki F, Okamoto N, Pombert J-F, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proceedings of the Royal Society of London B: Biological Sciences. 2012 doi: 10.1098/rspb.2011.2301. rspb20112301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 88.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Molecular cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 91.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 92.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 93.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nature structural & molecular biology. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meister G, et al. Identification of novel argonaute-associated proteins. Current biology. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 95.Kuzuoğlu-Öztürk D, Huntzinger E, Schmidt S, Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic acids research. 2012;40:5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 97.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 98.Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nature Plants. 2015;1 doi: 10.1038/nplants.2014.27. [DOI] [PubMed] [Google Scholar]
- 99.Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proceedings of the national academy of sciences. 2012;109:315–320. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karginov FV, et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Molecular cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shin C, et al. Expanding the microRNA targeting code: functional sites with centered pairing. Molecular cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell reports. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hauptmann J, et al. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nature structural & molecular biology. 2013;20:814–817. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 104.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakayashiki H, Kadotani N, Mayama S. Evolution and diversification of RNA silencing proteins in fungi. Journal of molecular evolution. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- 106.Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 108.Niklas KJ, Newman SA. The origins of multicellular organisms. Evolution & development. 2013;15:41–52. doi: 10.1111/ede.12013. [DOI] [PubMed] [Google Scholar]
- 109.Trillo IR, Nedelcu AM. Evolutionary Transitions to Multicellular Life: Principles and mechanisms. Vol. 2 Springer; 2015. [Google Scholar]
- 110.Michod RE, Roze D. Cooperation and conflict in the evolution of multicellularity. Heredity. 2001;86:1–7. doi: 10.1046/j.1365-2540.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 111.Michod RE, Viossat Y, Solari CA, Hurand M, Nedelcu AM. Life-history evolution and the origin of multicellularity. Journal of theoretical Biology. 2006;239:257–272. doi: 10.1016/j.jtbi.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 112.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Developmental biology. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 113.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 114.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer research. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 116.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosa A, et al. The interplay between the master transcription factor PU. 1 and miR-424 regulates human monocyte/macrophage differentiation. Proceedings of the national academy of sciences. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schoolmeesters A, et al. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nature Reviews Genetics. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hornstein E, Shomron N. Canalization of development by microRNAs. Nature genetics. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 121.Baumgarten S, et al. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC genomics. 2013;14:1. doi: 10.1186/1471-2164-14-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ratcliff WC, et al. Experimental evolution of an alternating uni-and multicellular life cycle in Chlamydomonas reinhardtii. Nature communications. 2013;4 doi: 10.1038/ncomms3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee H-C, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Molecular cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proceedings of the National Academy of Sciences. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sempere LF, Cole CN, Mcpeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 126.Wheeler BM, et al. The deep evolution of metazoan microRNAs. Evolution & development. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 127.Londin E, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate-and tissue-specific microRNAs. Proceedings of the National Academy of Sciences. 2015;112:E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 129.Sperling EA, et al. MicroRNAs resolve an apparent conflict between annelid systematics and their fossil record. Proceedings of the Royal Society of London B: Biological Sciences. 2009;276:4315–22. doi: 10.1098/rspb.2009.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]