Abstract
Purpose
Colorectal cancer (CRC) ranks as the third most frequent cancer type, and its incidence continues to rise gradually worldwide, highlighting the need to identify previously unrecognized molecular events that propel development of this malignancy. Recent evidence suggests that dysregulated expression of FOX family of transcription factors may be critical in various genetic disorders as well as cancer; however, the functional and clinical significance of this pathway in CRC remains unclear.
Experimental Design and Results
Herein, we performed a systematic and comprehensive discovery step by evaluating the expression of FOX family members, and identified that FOXM1 and FOXQ1 are frequently overexpressed in CRC. We subsequently confirmed these findings in two large testing cohorts (n=550), and an independent clinical validation cohort (n=134), in which high expression of FOXM1 and FOXQ1 emerged as an independent prognostic factor in CRC patients. We supported these findings by performing functional assays in which knockdown of FOXM1 and FOXQ1 resulted in inhibited cell proliferation, and suppressed migration and invasion in CRC cells. Furthermore, using bioinformatics approaches, we identified miR-342 as a novel regulator of both FOXM1 and FOXQ1. Overexpression or inhibition of miR-342 modulated the expression of both genes and contributed to phenotypic alterations in CRC cells, which was subsequently validated in a xenograft animal model.
Conclusions
Collectively, we have firstly identified FOXM1 and FOXQ1 as promising prognostic biomarkers in CRC patients, and provide novel evidence that therapeutic targeting of these genes or miR-342 may be a potential treatment approach in CRC patients.
Keywords: Colorectal cancer, FOXM1, FOXQ1, microRNA, miR-342-3p, prognosis, biomarker
INTRODUCTION
Colorectal cancer (CRC) is one of leading causes of cancer-related morbidity and mortality worldwide(1). It is now well-established that multiple genetic and epigenetic alterations lead to the result in development of CRC. The classic Vogelgram described the multistep model for CRC pathogenesis outlining few critical events, in which the APC gene mutations permitted adenoma formation, activating mutations in the KRAS oncogene and genomic losses at chromosome 18 loci facilitated adenomatous growth, and inactivation of p53 triggered the final transition of adenoma to carcinoma(2). Mutations in the APC gene occur in about 80–90% of sporadic CRCs, and alterations in the Wnt signaling pathway serve as major molecular drivers of cancer pathogenesis in the colon(3). However, data gathered in recent years have underscored the need for further refinement of these mechanisms since only 5% of adenomatous polyps progress to cancer development; which suggests that additional molecular events must occur for colorectal carcinogenesis to ensue(4, 5). Therefore, identification of previously unrecognized genomic events that propel cancer development would not just be of biological relevance, but may as well be of clinical significance as these will allow development of novel biomarkers and/or drug targets for the management of CRC patients.
Forkhead box (FOX) proteins constitute a superfamily of evolutionarily conserved transcriptional factors, which play an important role in a wide variety of cellular processes. Accumulating evidence indicates that FOX proteins serve as the terminal effectors of multiple signal transduction pathways such as TGF-β cascade, Wnt pathway, Sonic-Hedgehog pathway, and mitogen-activated protein kinase (MAPK) pathway(6). Furthermore, these FOX proteins may function as critical “nodes” in cellular networks, allowing cross-talk between parallel pathways and thus facilitate responses to environmental fluctuations. Considering their key function, it is not surprising that mutations or deregulation of FOX genes has now been associated to the developmental genetic disorders, as well as few human malignancies(7). Till date, mutations in 11 FOX genes have been linked to human hereditary diseases(8). In cancer, several FOX family members have been suggested to act as oncogenes or tumor suppressors, as these proteins have the capability to regulate cell differentiation, proliferation and apoptosis(7). Recently, a few studies have alluded that the expression of Forkhead family members can be regulated by posttranslational modifications such as phosphorylation of serine, threonine or tyrosine residues(9–11). However, the specific mechanism(s) of deregulation of FOX family members seem to context-dependent in cancer progression, and no previous studies have systematically and comprehensively interrogate the functional and clinical relevance of FOX proteins in cancer, particularly CRC.
During the past decade, microRNAs (miRNAs) have emerged as frontiers in gene regulation due to their role in regulation of a broad range of biological processes in various human diseases, particularly cancer(12). Previous work from our group and others have highlighted that specific miRNAs contribute to CRC pathogenesis, and several of these can be used as biomarkers for diagnosis, prognosis and metastasis-prediction in CRC patients(12–17). In fact, a few studies have shown that miRNAs may be able to regulate specific FOX genes in different cancers, including esophageal cancer(18), bladder cancer(19), and hepatocellular carcinoma(20); however, such an interaction between miRNAs and FOX proteins remains largely unclear and not interrogated in CRC.
Based upon this important gap in knowledge in the literature, we envisaged this study to systematically and comprehensively interrogate the molecular contributions of FOX family members in CRC, identify specific miRNAs that modulate their expression, and decipher whether these genes may have translational relevance as clinically-relevant disease biomarkers. Accordingly, we performed a discovery step by querying publically available databases to identify candidate, CRC-specific FOX genes, followed by their validation in multiple cohorts of CRC patients. In addition, we performed functional assays, both in-vitro and animal models, to validate the contribution of specific FOX genes in CRC, and identified a key miRNA that regulated FOX protein expression in this malignancy.
MATERIAL AND METHODS
Patients and Specimen Collection
To identify CRC-specific, differentially expressed FOX genes, we initially queried Oncomine database comprising of 24 independent studies(21). To assess the prognostic impact of FOXM1 and FOXQ1in CRC, we analyzed two publically-available datasets as the testing cohorts, which included a total of 550 colorectal cancer patients; of which 229 patients were from the GSE-17538 dataset (22) and 321 patients from the TCGA dataset(23, 24). In the clinical validation cohort, we analyzed 178 frozen tissues including 134 primary CRC tissues, and 44 matched normal mucosa (NM) tissues, which were collected at the Mie University, Japan. A written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions. Patients with radiotherapy or chemotherapy treatment before surgery were excluded. Survival time was calculated from the date of surgery to the date of death or last follow-up. Clinical information was collected from the medical records of each patient and shown in Supplementary table 1. The Tumor Node Metastasis (TNM) staging was performed according to American Joint Committee on Cancer (AJCC) standards.
Cell Lines
All human CRC cell lines RKO, CaCO2, HCT116, HT29, LoVo, SW480, and SW620 were obtained from the American Type Culture Collection and cultured in Iscove’s modified Dulbecco’s medium (IMDM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 u/ml penicillin and 100 μg/ml streptomycin) at 37°C in 5% humidified CO2 atmosphere. These cell lines were periodically authenticated using a panel of genetic and epigenetic biomarkers.
SiRNA, miRNA mimic and inhibitor transfections
Validated silencer select siRNAs for FOXQ1and FOXM1, miR-342 mimic and its inhibitor, negative control for siRNA, mimic and inhibitor were purchased from Ambion (Austin, TX). CRC cells were transfected with siRNA, mimic or inhibitor at a final concentration of 50nmol/L using Lipofectamine RNAi MAX (Invitrogen, Carlsbad, CA) and Opti-MEM (Gibco, Carlsbad, CA) according to the manufacturer’s instructions. The transfection efficiency was evaluated at 48h- and 72h time points for evaluating corresponding changes in the mRNA and protein expression, respectively.
Quantitative RT-PCR (qRT-PCR)
The qRT-PCR assays were performed using QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). For gene-expression analysis, 2μg of total RNA was synthesized to cDNA by using GoScript™ Reverse Transcription System (Promega, Madison, WI) and the Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA). The relative expression of target genes was normalized against β-actin using 2−Δct method as described previously (16, 25, 26). The sequences of all primers used in this study are shown in Supplementary table 2. For miRNA analysis, qRT-PCR was conducted using TaqMan MicroRNA Reverse Transcription Kit and TaqMan® Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Relative expression of miR-342 was determined by 2−Δct method using miR-16 as normalizer. Taqman primers for miR-342 and miR-16 were purchased from Ambion.
Western blot analysis
The total protein extracts from cell lines were collected using RIPA lysis buffer and 30μg protein was loaded per sample on 10% polyacrylamide-SDS gels, which were subsequently transferred onto nitrocellulose membranes. The membranes were then blocked in 5% fat-free milk and incubated with the following primary antibodies overnight at 4°C: rabbit anti-FOXQ1 (1:1000 dilution; Abcam, Cambridge, MA), rabbit-anti-FOXM1 (1:1000 dilution; Abcam, Cambridge, MA), monoclonal mouse anti-β-actin (1:5000 dilution; Sigma-Aldrich, St. Louis, MO). Membranes were thereafter incubated with secondary horseradish peroxidase-conjugated (HRP) conjugated goat anti-rabbit antibody (1:3000 dilution; Santa Cruz, Dallas, TX) and goat anti-mouse antibody (1:3000 dilution; Santa Cruz, Dallas, TX) at room temperature for 1 hour. The proteins were detected using enhanced chemiluminescence system according to the manufacturer’s instructions.
Cell proliferation assays
The SW480 and HT-29 CRC cell lines transfected with siRNA, miRNA mimics, miRNA inhibitors or negative controls were seeded at 1500 cells per well in 96-well plates following which MTT reagent was added at 0h, 24h, 48h and 72h time points. Optical densities were determined using the Infinite 200 Pro multi-readers and i-control 1.10 software (Tecan, Morrisville, NC).
Migration and Invasion assays
Cell migration and invasion assays were performed using Boyden chambers (Corning, Corning, NY) using 8 μm-size pore membrane coated with-matrigel (for invasion assays) or without-matrigel (for migration assays). The transfected cells were seeded onto inserts at 2×105 cells in serum-free medium and transferred to wells with culture medium containing 10% FBS. After 24h incubation, non-invading cells were removed by scraping the top surface of the membrane. Invaded cells on the bottom of the membrane were thereafter fixed and stained by using diff-quick staining kit (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions. The stained cells were counted using a light microscope.
Luciferase Reporter Assays
Oligonucleotides pairs containing miR-342 targeted regions within the 3′-UTR regions of the FOXM1 and FOXQ1 genes were annealed and ligated into the pmirGLO Vector (Promega, Madison, WI) according to the manufacturer’s instructions. The firefly luciferase activity was detected using the dual luciferase assay system (Promega, Madison, WI) according to the manufacturer’s instructions. The relative luciferase activity was quantified using a microplate luminometer (Turner Biosystems, Sunnyvale, CA) and the transfection efficiency was normalized to renilla’s activity.
Xenograft animal studies
Male athymic nude mice were obtained from the Harlan Laboratories (Houston) at 5 weeks of age and kept under controlled conditions (12h light and dark cycles). The animal protocol was approved by the Institutional Animal Care and Use Committee of the Baylor Research Institute. We generated xenograft tumors using SW480 cell line stably over-expressing miR-342, or its corresponding controls. The stable miR-342 overexpressing SW480 cells were established by lentiviral vector infection according to the manufacturer’s instructions (miR-342: pLV-[hsa-mir-342] plasmid; Negative control plasmid: pLV-[mir-control], Biosettia, San Diego, CA). These cancer cells were suspended in PBS and Matrigel at 1:1 ratios (Corning, Corning, NY) and 1×106 cells were subcutaneously injected into the abdominal flanks of each mice. Nine mice were included in each group. The mice were monitored for 20 days following injection, and subcutaneous tumors were measured every two days. Tumor size was measured using calipers and the volume was calculated using the following formula: 1/2LW2, where L represents length, W width, and H height. At 20 days post-injection, all animals were sacrificed. Tumor samples were dissected and stored in RNA-later (Sigma-Aldrich) for subsequent analyses.
Statistical Analysis
All statistical analysis was performed using the GraphPad Prism Ver. 6.0 or Medcalc version 12.3 programs. All data were expressed as mean±SD Statistical differences between groups were determined by Wilcoxon’s signed rank test, the χ2 test or Mann-Whitney U test. Kaplan-Meier analysis and log-rank test were used to estimate and compare survival rates of CRC patients with high and low FOXM1 or FOXQ1 expression. Receiver operating characteristic curves (ROC) were established to determine the cutoff values to discriminate patients with or without death. The Cox’s proportional hazards models were used to estimate hazard ratios (HRs) for death. All P values were 2-sided, and those less than 0.05 were considered statistically significant.
RESULTS
FOXM1 and FOXQ1 are frequently overexpressed in CRC
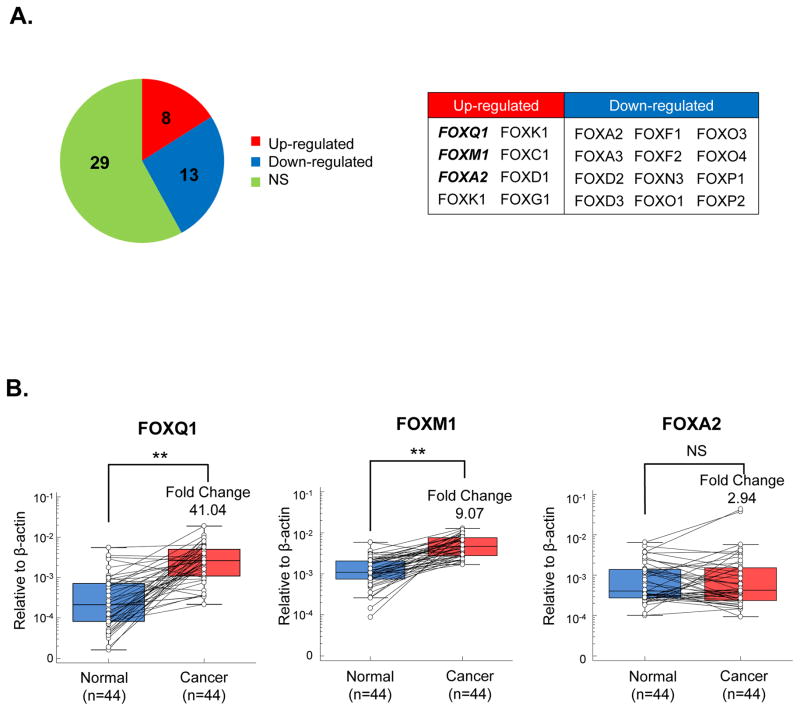
To identify CRC-specific differentially expressed FOX family of genes, we performed a discovery step by comparing the expression levels of various genes in cancer vs. matched normal tissues using the publically-available Oncomine microarray database(21). By systematically analyzing the expression profiles of all available FOX genes across 24 studies, we noted that 8 FOX family members (FOXQ1, FOXM1, FOXA2, FOXK1, FOXK2, FOXC1, FOXD1, FOXG1) were significantly up-regulated (≥2 fold change, and a false discovery rate (FDR) of <1×10−4), 13 were down-regulated, and the remaining were not significantly altered in CRC (Figure 1A, and Supplementary Table 3). Since up-regulated genes are more practical from a diagnostic or therapeutic view point, we thereafter focused our attention on the up-regulated FOX genes. Interestingly, we found three FOX family members (FOXQ1, FOXM1 and FOXA2) were frequently overexpressed in most in silico studies, suggesting that these may play an important role in colorectal carcinogenesis, and also may serve as potential disease biomarkers (Figure 1A and Supplementary Table 3) We next examined the expression levels of these three genes in a subset of 44 CRC and paired normal mucosa specimens by qRT-PCR. Our results revealed that, while we failed to observe any significant differences in the expression of FOXA2, the expression levels of FOXM1 and FOXQ1 were statistically higher in cancer vs. normal tissues, with a 41.04 fold increase (P<0.0001) and 9.07 fold-change (P<0.0001) in each gene respectively (Figure 1B), suggesting these genes to be potential oncogenes in colorectal cancer.
Figure 1.
FOXM1 and FOXQ1 are frequently overexpressed in colorectal cancer. (A) Results analyzed from the Oncomine microarray database revealed that FOX family members are frequently dysregulated in CRC. NS: Non-significant. FOXM1, FOXQ1 and FOXA2 were reported to be up-regulated by most studies. (B) The expression levels of FOXM1, FOXQ1 and FOXA2 were assessed in a subset of 44 matched colorectal cancer and normal tissues. FOXM1 and FOXQ1 were up-regulated in cancer tissues while FOXA2 expression did not demonstrate a significant change between cancer and normal tissues. (*P<0.05, **P<0.01, Wilcoxon paired test)
High FOXM1 and FOXQ1 expression correlated with poor survival in CRC patients
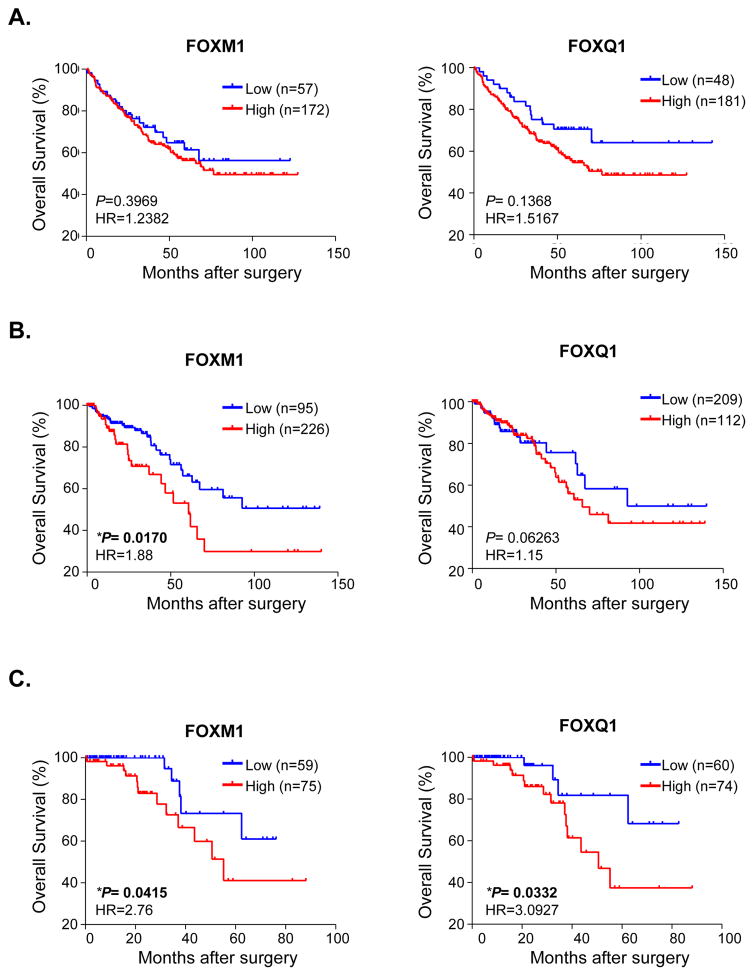
Next, we examined the expression patterns of FOXM1 and FOXQ1 with regards to their prognostic value in two testing cohorts of a combined total of 550 CRC patients from the GSE(22) and TCGA datasets(23, 24). Interestingly, in the GSE dataset, FOXQ1-high expression group demonstrated a marked correlation with poor overall survival (P=0.1368, HR=1.5167), while FOXM1-high expression showed weaker tendency to be associated with poor OS (P=0.3969, HR=1.2382; Figure 2A). In contrast, in the TCGA dataset, FOXM1-high expression group significantly correlated with poor OS (P=0.0170, HR=1.88), while FOXQ1-high expression group demonstrated a strong tendency to correlate with poor prognosis (P=0.06263, HR=1.15; Figure 2B).
Figure 2.
High expression of FOXM1 and FOXQ1 correlate with poor prognosis in CRC patients. The prognostic significance of FOXM1 and FOXQ1 was evaluated in CRC patients from 2 testing cohorts (A) testing cohort I: GSE 17538; (B) testing cohort II: TCGA database, and (C) a clinical validation cohort. ROC curve analysis yielded optimal cutoff expression values to discriminate dead or alive patients. CRC patients were thereafter divided into high- and low-expression groups based upon these cutoff values. The overall survival (OS) analysis was performed by Kaplan-Meier test and the log-rank method. (*P<0.05, **P<0.01; HR: Hazard Ratio)
To further validate the prognostic impact of FOXM1 and FOXQ1 in CRC patient survival, we interrogated these associations in an additional cohort of high quality, fresh frozen tissues from 134 CRC patients. The detailed correlations between FOXM1 and FOXQ1 expression and various clinico-pathological variables are shown in Supplementary Table 1, and we were able to successfully validate that both FOXM1 and FOXQ1-high expression groups demonstrated shorter OS (Figure 2C), highlighting their clinical relevance as independent prognostic biomarkers in CRC patients. Furthermore, Cox’s univariate and multivariate analysis also revealed that FOXM1 and FOXQ1 were independent predictors of CRC patient survival (Table 1). Collectively, these findings elucidate that overexpression of FOXM1 and FOXQ1 has clinical significance in terms of serving as potential prognostic biomarkers in CRC patients.
Table 1.
Univariate and multivariate analysis for predictors of overall survival of CRC patients in the clinical validation cohort
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Odds ratio | 95% CI | P value | Odds ratio | 95% CI | P value | |
| Age (≥68) | 1.6413 | 0.6468 – 4.1645 | 0.2994 | |||
| Gender (Female) | 0.597 | 0.2134 – 1.6703 | 0.3282 | |||
| Differentiation (Poor) | 1.8322 | 0.4200 – 7.9924 | 0.4228 | |||
| T stage (pT3/pT4) | 6.4921 | 0.8711 – 48.3824 | 0.0694 | |||
| lymph node (Positive) | 5.0006 | 1.7867 – 13.9954 | 0.0023* | 0.481 | 0.0812 – 2.8492 | 0.4224 |
| Liver Metastasis (Present) | 4.3713 | 1.5551 – 12.2872 | 0.0054* | 3.0106 | 0.8357 – 10.8453 | 0.0936 |
| local invasion (Present) | 4.4019 | 1.5036 – 12.8865 | 0.0071* | 1.3968 | 0.4001 – 4.8766 | 0.6022 |
| Lymphatic invasion (Present) | 2.7398 | 0.6286 – 11.9424 | 0.1819 | |||
| Venous invasion (Present) | 0.955 | 0.3722 – 2.4500 | 0.924 | |||
| Stage (III/V) | 8.4737 | 0.4907 – 3.1461 | 0.0008* | 12.9182 | 1.5694 – 106.3347 | 0.0179* |
| Tumor location (Colon) | 1.2424 | 0.49–3.14 | 0.6487 | |||
| Tumor size (≥42cm) | 0.7191 | 0.2697 – 1.9176 | 0.5121 | |||
| FOXM1-(high) | 2.832 | 1.0028 – 7.9979 | 0.0506 | |||
| FOXQ1 (high) | 3.2067 | 1.0434 – 9.8556 | 0.043* | 4.7409 | 1.2712 – 17.6819 | 0.0211* |
| miR-342-3p(low) | 0.5241 | 0.1194 – 2.2996 | 0.3942 | |||
FOXM1 and FOXQ1 promote migration and invasion in CRC cells
To investigate whether FOXM1 and FOXQ1 expression affects biological characteristics, we performed several functional assays to determine phenotypic alterations following siRNA knockdown of FOXM1 and FOXQ1 in colon cancer cells. In order to select most optimal cell lines for knockdown experiments, we evaluated the expression levels of FOXM1 and FOXQ1 in a panel of CRC cell lines (Supplementary Figure 1A). RKO and HCT-116 displayed low expression, while SW480, SW620 and HT-29 cell lines revealed high expression of FOXQ1. In contrast, the expression levels of FOXM1 were similar across all CRC cell lines. Based upon these findings, we selected SW480 and HT-29 cell lines for siRNA knockdown of both FOXM1 and FOXQ1. We validated this siRNA knockdown efficiency for FOXM1 and FOXQ1 at both transcription and protein levels (Supplementary Figure 1B).
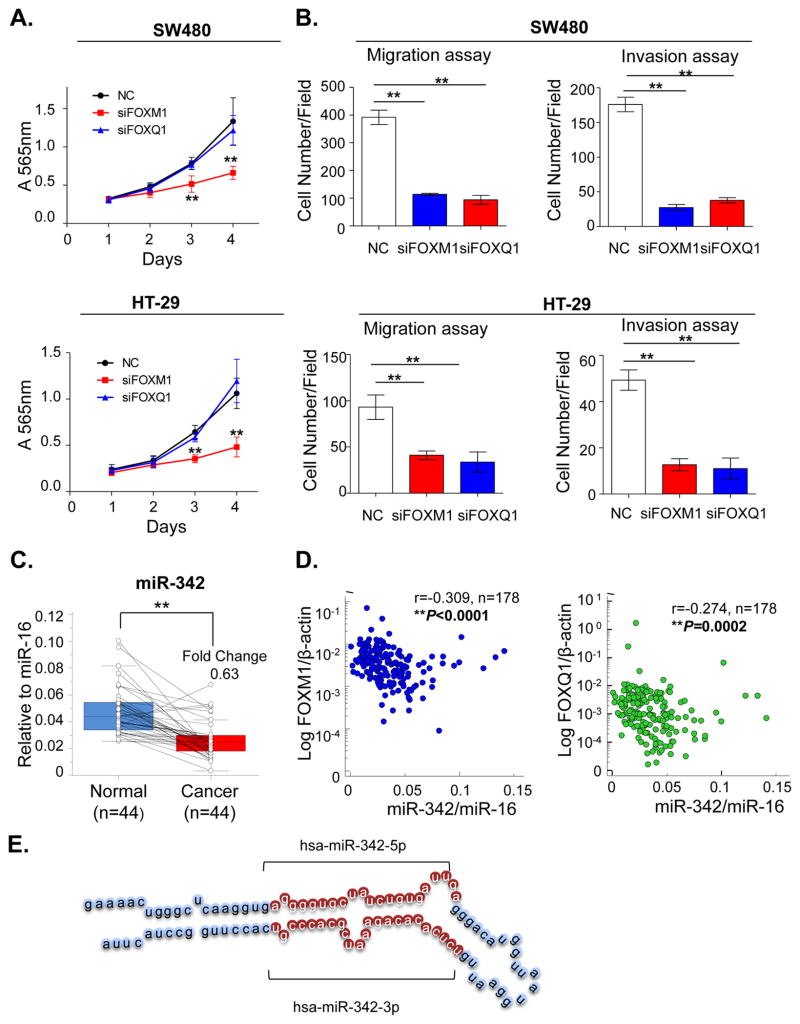
Next we determined whether inhibition of FOXM1 and FOXQ1 results in functional alterations in CRC. The MTT cell proliferation assays revealed that while FOXQ1 inhibition did not have any growth inhibitory effects in either cell line, FOXM1 knockdown significantly suppressed cell growth in both SW480 and HT-29 cell lines (Figure 3A). To determine whether alterations in the expression of FOXM1 and FOXQ1 has any impact on the motility and invasive potential of CRC cells, we performed migration and invasion assays. As shown in Figure 3B and Supplementary Figure 2–3, FOXM1 and FOXQ1 knockdown lead to significantly reduced migration and invasion ability in both cancer cells lines compared to mock transfected control cells.
Figure 3.
MiR-342-3p is a potential regulator of FOXM1 and FOXQ1 expression in CRC (A) MTT assays were performed to evaluate the impact of FOXM1 and FOXQ1 siRNA-mediated knockdown on cell proliferation (n=6, *P<0.05, **P<0.01, independent t-test was used to compare control and treated cells). (B) Migration assay and invasion assays showed FOXM1 and FOXQ knockdown in CRC cell lines impaired their migration and invasive abilities (n=3, *P<0.05, **P<0.01, independent t-test was used to compare control and treated cells).(C) The expression levels of miR-342-3p were evaluated in a subset of 44 matched cancer and normal tissues. MiR-342-3p was down-regulated in cancer vs. normal tissues (*P<0.05, **P<0.01, Wilcoxon paired test). (D) The expression correlation between miR-342 and FOX genes was performed in 178 CRC and normal tissues. Spearman’s correlation was used to calculate the correlation coefficient between expression levels of miR-342-3p and FOXM1/FOXQ1. (r: correlation coefficient; *P<0.05, **P<0.01) (E) The structure of miR-342 precursor. The 99 nucleotide-long stem-loop precursor splits into 2 mature miRNAs: miR-342-3p and miR-342-5p.
MiR-342 targets the 3′-UTR of FOXM1 and FOXQ1, and its expression inversely correlates with both genes in CRC
Considering their important oncogenic role, we were interested in understanding the potential mechanisms for the dysregulated expression of FOXM1 and FOXQ1 in CRC. Considering the central role miRNAs play in carcinogenesis, we hypothesized that overexpression of FOXM1 and FOXQ1 might be caused by aberrant expression of specific miRNAs in CRC. Furthermore, we aimed to identify whether there is a specific miRNA which is capable of regulating both FOX family members in CRC.
Despite complexities underlying miRNA-mRNA interactions, several algorithms are available for predicting genes targeted by specific miRNAs. To identify miRNAs that target both FOXM1 and FOXQ1, we utilized microRNA.org algorithm (www.microRNA.org) and identified 5 miRNAs (miR-342-5p, miR-320a, miR-320b, miR-320c and miR-320d) that can bind to the 3′-UTR region of both genes. However, among these 5 miRNAs, miR-342-3p had the highest mirSVR score for both FOX genes; hence we selected this miRNA as a potential candidate for subsequent interrogation.
Since FOXM1 and FOXQ1 showed oncogenic role in CRC, we first evaluated the expression of miR-342 expression in a subset 44 CRC cancers and matched normal tissues to determine whether miR-342 functions as tumor suppressor in this disease. Consistent with our hypothesis, miR-342 was found to be significantly down-regulated in cancer tissues vis-à-vis adjacent normal tissues (P<0.0001; Figure 3C). Furthermore, we observed a significant inverse correlation between miR-342 expression and both FOXM1 (P<0.0001) and FOXQ1 (P=0.0002) in the primary CRC and normal tissues, suggesting that miR-342 regulates the expression of these genes in CRC (Figure 3D).
MiR-342 is a negative regulator of FOXM1 and FOXQ1 expression in CRC cells
The mature miR-342-3p was generated from 99 nucleotide long pre-miRNAs (structure shown in Figure 3E) and was identified to potentially bind to the 3′-UTR region of FOXM1 and FOXQ1 genes. To determine whether miR-342 actually regulates FOXM1 and FOXQ1 expression, we overexpressed or inhibited miR-342 in CRC cells. To select appropriate cell lines for overexpression or inhibition of miR-342, we examined the expression levels of miR-342 in a panel of CRC cancer cell lines. As shown in Supplementary Figure 1A, miR-342 expression was markedly lower in RKO, SW620, DLD-1 and HT-29 cells, but higher in HCT-116, SW480 and CaCO-2. Based upon these results we selected SW480 for both overexpression and inhibition of miR-342, as well as HT-29 cells for the overexpression of miR-342.
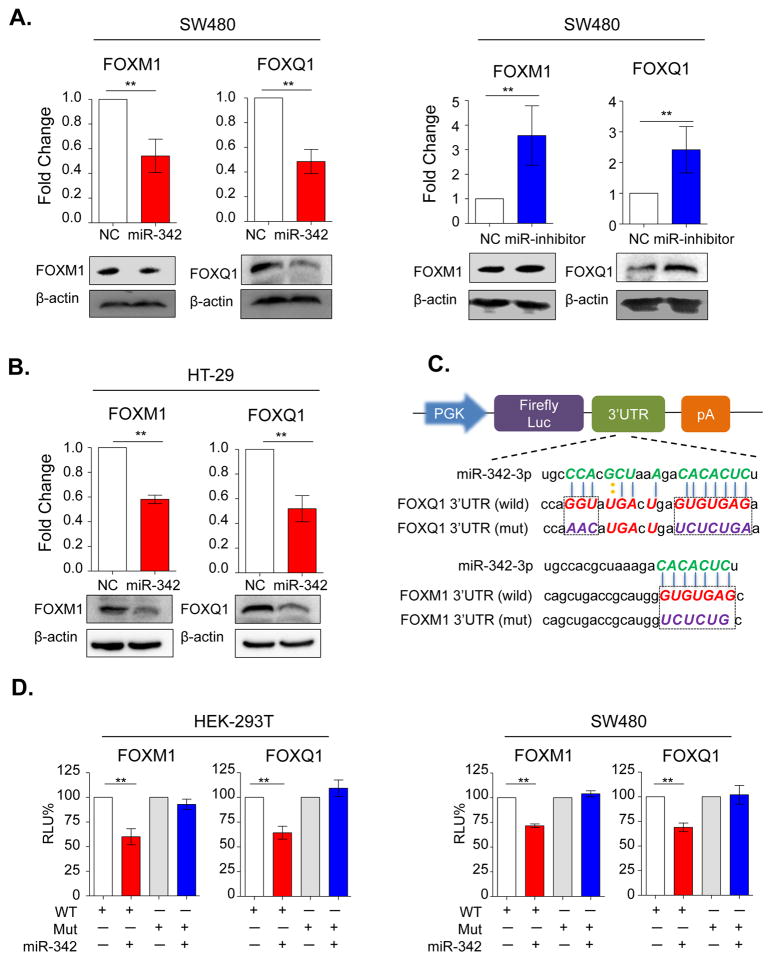
Overexpression of miR-342 by using miRNA mimics in SW480 and HT-29 cells resulted in significant downregulation of FOXM1 and FOXQ1 at both transcriptional and protein levels (Figure 4A, 4B), indicating that miR-342 may regulate both FOXM1 and FOXQ1 expression. Likewise, blocking of the endogenous miR-342 expression through miR-342 inhibitor in SW480 cells resulted in up-regulation of FOXM1 and FOXQ1 (Figure 4A).
Figure 4.
MiR-342 post-transcriptionally regulates FOXM1 and FOXQ1 expression in CRC cell lines. (A) The mRNA and protein expression levels of FOXM1 and FOXQ1 were evaluated in SW480 cells following miR-342 mimics (Left figure) and inhibitors (Right figure). (B) The expression levels of FOXM1 and FOXQ1 were measured in HT-29 cells using miR-342 mimics and inhibitors. (C) Schematic representation of luciferase reporter constructs. The PGK promoter drives constitutive transcription of a chimeric mRNA containing the Firefly luciferase coding sequence fused to the wide-type or mutated FOXM1 and FOXQ1 3′-UTRs. (D) Relative activity of the luciferase gene fused with the wild-type or mutant FOXM1 and FOXQ1 3′-UTR in HEK-293T and SW480 cells. The data was normalized to renilla luciferase activity. The data are represented as means±SD from separate transfections (n=3). Statistical analysis was performed using independent t-test to compare fold changes between transfection and control groups. RLU%: percentage of relative luminescence; WT: wild type; Mut: mutant type; All statistical tests were two-sided. *P< 0.05, **P<0.01.
To further determine whether miR-342 regulates FOXM1 and FOXQ1 expression directly or indirectly, we conducted luciferase reporter assays. As shown in Figure 4C, we cloned miR-342 binding region of FOXM1 and FOXQ1 into luciferase report plasmid to establish wild type of 3′-UTR report plasmids. In addition, we also mutated miR-342 binding sites and established mutant 3′-UTR regions of FOXM1 and FOXQ1. HEK-293T, SW480 and HT-29 were transiently transfected with these constructs along with miR-342 mimics or negative controls (NC). In line with our previous results, miR-342 overexpression significantly suppressed luciferase activity of the reporter genes containing wild type 3′-UTR regions of FOXM1 and FOXQ1, but no inhibitory effects were observed in mutated cell lines (Figure 4D, Supplementary Figure 4). Collectively, these results indicate that miR-342 suppressed expression of FOXM1 and FOXQ1 through direct binding within the putative 3′-UTR region binding sites of these genes.
MiR-342 inhibits proliferation, migration and invasion CRC cells and xenograft animal models
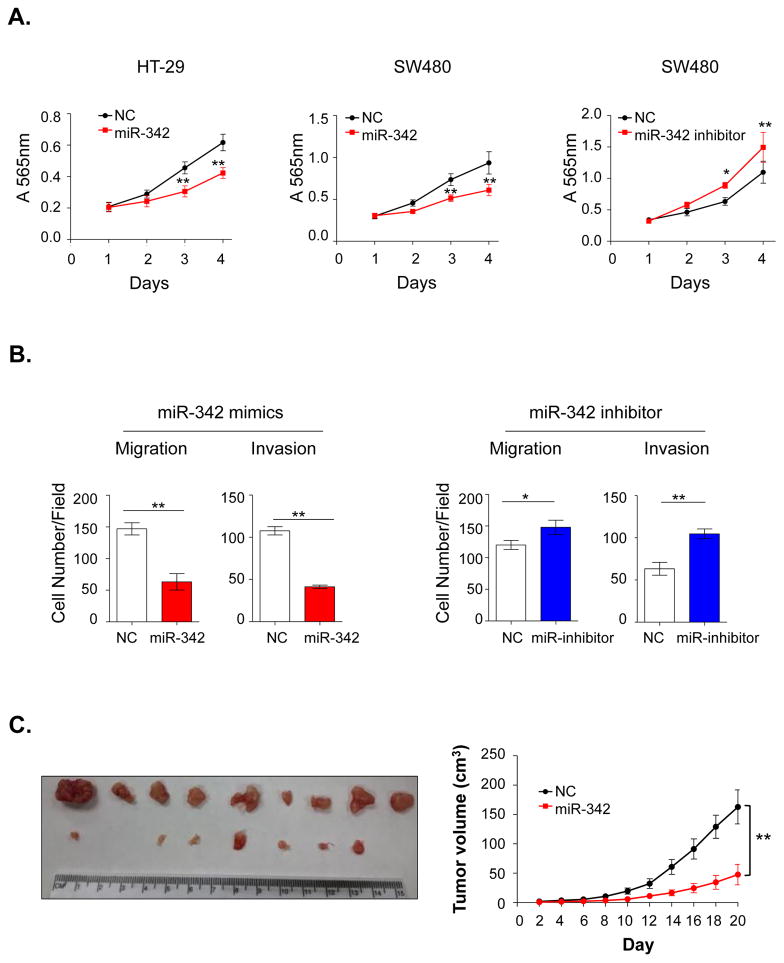
Subsequent to our observation that miR-342 is a direct regulator of FOXM1 and FOXQ1 expression, we next examined whether modulation of miR-342 expression contributes to phenotypic alterations in CRC cells. To test this hypothesis, we first overexpressed miR-342 in SW480 and HT-29 cells using miRNA mimics and conducted MTT assays. As expected, miR-342 overexpression inhibited cellular proliferation in both CRC cell lines. Likewise, inhibition of miR-342 expression in SW480 by using inhibitors resulted in increased cellular proliferation (Figure 5A). To determine whether miR-342 also functions as a metastasis suppressor, we overexpressed miR-342 in SW480 and HT-29 cells and observed dramatic suppression of cell motility and invasiveness in both cell lines. In line with these findings, inhibition of miR-342 in SW480 cells resulted in enhanced migration and invasion capacity (Figure 5B and Supplementary Figure 5–7).
Figure 5.
MiR-342 inhibits proliferation, migration and invasion in both cancer cell lines and xenograft animal models. (A) MTT assays were performed to evaluate the proliferation alterations in SW480 and HT-29 cells after treatment with miR-342 mimics or inhibitors (n=6). (B) In SW480 cells, overexpression or inhibition of miR-342 expression dramatically suppressed or enhanced cell motility and invasion respectively. (C) Stable overexpression of miR-342 inhibited CRC cell growth in nude mice. The left figure depicts images of miR-342 overexpressing tumors and controls. The right figure illustrates the tumor growth curves of the negative and untreated control groups. For the MTT, migration and invasion assays, independent t-test was used to compare fold changes between treatment and control groups. For the tumor growth analysis, paired t-test was used. All statistical tests were two-sided. *P<0.05, **P<0.01.
Finally, to determine whether ectopic expression of miR-342 inhibits tumorigenicity in an animal model, we established SW480 cells with stable overexpression of miR-342, and subcutaneously injected test and control cells into nude mice. As shown in Figure 5C, during the initial 10 days after injection, there were no significant differences in tumor size. However, 12 days after injection, controls cells reported an accelerated tumor growth while miR-342 overexpressing cells grew much slowly, highlighting the tumor-suppressive role of miR-342 in CRC. Furthermore, the decreased level of FOXM1 and FOXQ1 in miR-342 overexpressing xenograft tissues supported the hypothesis that miR-342 exerts functions in CRC, at least in part, through inhibition of FOXM1 and FOXQ1 expression (Supplementary Figure 8).
DISCUSSION
CRC is one of the most common cancers worldwide. Although patients with early stage CRC have relatively better prognosis, most patients at first presentation of disease are diagnosed late, and the outcome of such patients is usually poor(1). Therefore, elucidating the molecular mechanisms underlying CRC progression is critical for the development of new therapeutic strategies for the improvement of prognosis of patients with advanced CRC. Herein, in this study, we for the first time demonstrate that FOXM1 and FOXQ1 are involved in the colorectal carcinogenesis, their expression is regulated by a miR-342 tumor suppressive miRNA, and the high-expression of these genes serves as an important prognostic biomarker in CRC.
In view of recent limited evidence that FOX transcription factors play an important role in gene regulation and carcinogenesis, in the current study, we first identified key differentially expressed FOX genes in CRC. Following a systematic analysis of CRC microarray data, we identified FOXM1 and FOXQ1 as the most up-regulated genes in CRC tissues. Furthermore, by measuring the expression levels of these genes in two different testing cohorts, and an independent validation cohort, we demonstrated that high expression of FOXM1 and FOXQ1 correlated with poor survival in CRC patients. Our data, which are first of its kind in CRC, are consistent with some of the other reports which showed that FOXM1 is overexpressed and associates with worse prognosis in other human cancers(27–31).
Similarly, FOXQ1 has also been reported to be up-regulated in several cancer types (32–35). However, none of the previous studies have systematically explored the prognostic significance of these genes in CRC. We addressed this important gap in knowledge by evaluating the expression of these genes in two large testing cohorts (n=553), and another independent clinical validation cohort (n=134) to firstly identify the prognostic value of FOXM1 and FOXQ1 in CRC. However, in order to fully validate the prognostic potential of FOXM1 and FOXQ1 in CRC, additional prospective studies are required.
Recently, a few studies have suggested that FOXM1 and FOXQ1 play a key role in regulation of epithelial-to-mesenchymal transition (EMT) in various types of cancers (34, 36–39). In this regard, our data are consistent with this hypothesis, since we observed that inhibition of FOXM1 and FOXQ1 caused reduced mobility and invasive capability in CRC cells. It is well-established that invasion-metastasis cascade drives CRC progression, which eventually leads to significantly worse patient survival. Several studies have shown that liver metastasis is a manifestation in nearly 20% patients with stage II CRC, and this rate increases to 50% in patients with stage III disease(40). Moreover, metastasis is responsible for more than 90% of CRC-associated deaths(41). Based upon our findings that these two FOX family members are often over-expressed in CRC, and associate with poor patient survival, therapeutic targeting of FOXM1 and FOXQ1 may be a potential strategy in attenuating metastasis and improving survival of CRC patients in future.
MiRNAs have emerged as major players in the complex network of gene regulation, and dysregulation of miRNA expression has been implicated in carcinogenesis. In this study not only we have uncovered the oncogenic role of FOXM1 and FOXQ1, we have also firstly identified that miR-342 tumor-suppressor directly targets the 3-UTR of both genes, and regulates their expression in CRC. MiR-342 is encoded within an intron of the EVL gene, and is frequently silenced due to aberrant hyper-methylation in CRC(42). Consistent with this finding, our study revealed that miR-342 is down-regulated in colorectal cancer tissues relative to adjacent normal mucosa, indicating that DNA methylation may be the potential mechanism for its downregulation in CRC. In the current study, we also noted that the expression levels of miR-342 negatively correlated with those of FOXM1 and FOXQ1 in CRC tissues, and overexpression or inhibition of miR-342 modulated the expression of both FOX genes in CRC cells. In further support of these findings, our luciferase reporter assays confirmed that miR-342 over-expression led to inhibition of FOXM1 and FOXQ1 through direct binding of the 3′-UTR region of these genes. Ours is the first study to demonstrate the underlying mechanisms by which miR-342 regulates the oncogenic function of both FOXM1 and FOXQ1 in CRC.
Taken together, ours is the first study that has systematically and comprehensively dissected the role of FOX family of transcription factors in CRC, both from a functional and clinical standpoint. Herein, we provide novel evidence that FOXM1 and FOXQ1 are frequently overexpressed in CRCs, and contribute to cancer pathogenesis. Furthermore, high expression of these genes significantly correlated with tumor growth, metastasis and poor prognosis in CRC. From a functional perspective, we identified that miR-342, a tumor suppressive miRNA, which directly binds and regulates the expression of both FOXM1 and FOXQ1 expression in CRC cells. Down-regulation of miR-342, resulted in upregulation of both genes, and these findings were subsequently validated in a xenograft animal model. We conclude that FOXM1 and FOXQ1 are promising prognostic biomarkers for CRC patients and modulation of miR-342 expression or therapeutic targeting of FOXM1 and FOXQ1 may be a potential treatment option for patients with colorectal cancer.
Supplementary Material
Translational Relevance.
Dysregulated expression of FOX family of transcription factors is being recognized as a critical event in several genetic disorders and cancer, but the functional and clinical significance of this pathway in colorectal cancer (CRC) remains unexplored. Herein, we performed a systematic and comprehensive discovery phase to identify FOXM1 and FOXQ1 as novel prognostic biomarkers in CRC, which was subsequently validated in two large testing cohorts and another clinical validation cohort. Functional assays identified miR-342 as a novel regulator of both FOXM1 and FOXQ1, and overexpression or inhibition of miR-342 modulated the expression of both genes and contributed to phenotypic alterations in CRC cells, which was subsequently validated in a xenograft animal model. Collectively, we have firstly identified FOXM1 and FOXQ1 as promising prognostic biomarkers in CRC patients, and provide novel evidence that therapeutic targeting of these genes or miR-342 may be a potential treatment approach in CRC patients.
Acknowledgments
Funding: The present work was supported by the grants R01 CA72851, CA 181572, CA184792 and U01 CA187956 from the National Cancer Institute, National Institute of Health, pilot grants from the Baylor Sammons Cancer Center and Foundation, as well as funds from the Baylor Research Institute.
Footnotes
Conflict of Interest: None of the authors have any potential conflicts to disclose
Author contributions: Study concept and design, analysis and interpretation of data and statistical analysis: WWH and AG; Specimen provider: YO, YT and MK; Acquisition of clinical data: YO; Animal experiments: ST and WWH; Drafting of the manuscript: WWH and AG.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Weng W, Feng J, Qin H, Ma Y. Molecular therapy of colorectal cancer: progress and future directions. Int J Cancer. 2015;136:493–502. doi: 10.1002/ijc.28722. [DOI] [PubMed] [Google Scholar]
- 5.Sillars-Hardebol AH, Carvalho B, de Wit M, Postma C, Delis-van Diemen PM, Mongera S, et al. Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol. 2010;31:89–96. doi: 10.1007/s13277-009-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (Review) Oncol Rep. 2014;32:1327–34. doi: 10.3892/or.2014.3357. [DOI] [PubMed] [Google Scholar]
- 7.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–32. doi: 10.1016/j.tig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–88. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 10.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 11.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–16. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng W, Feng J, Qin H, Ma Y, Goel A. An update on miRNAs as biological and clinical determinants in colorectal cancer: a bench-to-bedside approach. Future Oncol. 2015;11:1791–808. doi: 10.2217/fon.15.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308737. Epub Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res. 2015;21:4234–42. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308455. Epub Ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/dju492. Epub Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64:203–14. doi: 10.1136/gutjnl-2013-306640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Yu X, Bai Q. miR-204 inhibits invasion and epithelial-mesenchymal transition by targeting FOXM1 in esophageal cancer. Int J Clin Exp Pathol. 2015;8:12775–83. [PMC free article] [PubMed] [Google Scholar]
- 19.Song GQ, Zhao Y. MicroRNA-211, a direct negative regulator of CDC25B expression, inhibits triple-negative breast cancer cells’ growth and migration. Tumour Biol. 2015;36:5001–9. doi: 10.1007/s13277-015-3151-6. [DOI] [PubMed] [Google Scholar]
- 20.Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng HP, Shen G, et al. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer. 2014;14:909. doi: 10.1186/1471-2407-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2015-309359. Epub Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735–43. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao DC, Lu ZD, Qiao JH, Yan M, Cui SD, Liu ZZ. Expression of CDCA8 correlates closely with FOXM1 in breast cancer: public microarray data analysis and immunohistochemical study. Neoplasma. 2015;62:464–9. doi: 10.4149/neo_2015_055. [DOI] [PubMed] [Google Scholar]
- 28.Kong FF, Qu ZQ, Yuan HH, Wang JY, Zhao M, Guo YH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31:2660–8. doi: 10.3892/or.2014.3129. [DOI] [PubMed] [Google Scholar]
- 29.Kocarslan S, Guldur ME, Ekinci T, Ciftci H, Ozardali HI. Comparison of clinicopathological parameters with FoxM1 expression in renal cell carcinoma. J Cancer Res Ther. 2014;10:1076–81. doi: 10.4103/0973-1482.137988. [DOI] [PubMed] [Google Scholar]
- 30.Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY, Le XF, et al. Overexpression of forkhead box protein M1 (FOXM1) in ovarian cancer correlates with poor patient survival and contributes to paclitaxel resistance. PLoS One. 2014;9:e113478. doi: 10.1371/journal.pone.0113478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Tang D, Yao Y, Qi W, Liang J. Clinical significance and positive correlation of FoxM1 and Her-2 expression in gastric cancer. Clin Exp Med. 2014;14:447–55. doi: 10.1007/s10238-013-0261-6. [DOI] [PubMed] [Google Scholar]
- 32.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–33. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 33.Meng F, Speyer CL, Zhang B, Zhao Y, Chen W, Gorski DH, et al. PDGFRalpha and beta play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res. 2015;75:584–93. doi: 10.1158/0008-5472.CAN-13-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J, Xu L, Ni S, Gu J, Zhu H, Wang H, et al. Involvement of FoxQ1 in NSCLC through regulating EMT and increasing chemosensitivity. Oncotarget. 2014;5:9689–702. doi: 10.18632/oncotarget.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng XH, Huang HR, Lu J, Liu X, Zhao FP, Zhang B, et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer. 2014;13:186. doi: 10.1186/1476-4598-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C, Chen L, Yie L, Wei L, Wen T, Liu Y, et al. Targeting FoxM1 inhibits proliferation, invasion and migration of nasopharyngeal carcinoma through the epithelialto-mesenchymal transition pathway. Oncol Rep. 2015;33:2402–10. doi: 10.3892/or.2015.3834. [DOI] [PubMed] [Google Scholar]
- 37.Chiu WT, Huang YF, Tsai HY, Chen CC, Chang CH, Huang SC, et al. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2015;6:2349–65. doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao L, Xiong X, Lin Y, Cheng Y, Lu J, Zhang J, et al. Down-regulation of FoxM1 leads to the inhibition of the epithelial-mesenchymal transition in gastric cancer cells. Cancer Genet. 2014;207:75–82. doi: 10.1016/j.cancergen.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Fan DM, Feng XS, Qi PW, Chen YW. Forkhead factor FOXQ1 promotes TGF-beta1 expression and induces epithelial-mesenchymal transition. Mol Cell Biochem. 2014;397:179–86. doi: 10.1007/s11010-014-2185-1. [DOI] [PubMed] [Google Scholar]
- 40.Penna C, Nordlinger B. Surgery of liver metastases from colorectal cancer: new promises. Br Med Bull. 2002;64:127–40. doi: 10.1093/bmb/64.1.127. [DOI] [PubMed] [Google Scholar]
- 41.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–8. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.