Abstract
Objective
Dysregulated expression of microRNAs (miRNAs) has emerged as a hallmark feature in human cancers. Exportin-5 (XPO5), a karyopherin family member, is a key protein responsible for transporting precursor miRNAs from the nucleus to the cytoplasm. While XPO5 is one of the key regulators of miRNA biogenesis, its functional role and potential clinical significance in colorectal cancer (CRC) remains unclear.
Design
The expression levels of XPO5 were initially assessed in three genomic datasets, followed by determination and validation of the relationship between XPO5 expression and clinicopathological features in two independent CRC patient cohorts. A functional characterization of XPO5 in CRC was examined by targeted gene silencing in colorectal cancer cell lines and a xenograft animal model.
Results
XPO5 is upregulated, both at mRNA and protein levels, in CRCs compared with normal tissues. High-XPO5 expression associated with worse clinicopathological features and poor survival in CRC patient cohorts. The siRNA knockdown of XPO5 resulted in reduced cellular proliferation, attenuated invasion, induction of G1/S cell-cycle arrest, and downregulation of key oncogenic miRNAs in CRC cells. These findings were confirmed in a xenograft animal model wherein silencing of XPO5 resulted in the attenuation of tumor growth.
Conclusion
XPO5 acts like an oncogene in CRC by regulating the expression of miRNAs and may be a potential therapeutic target in CRC.
Keywords: Exportin-5, colorectal cancer, prognostic marker, miRNA, cell cycle
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in the United States (1) and more than 1.2 million patients are diagnosed with CRC annually, with 5-year survival rates of approximately 64% (2, 3). Accumulating evidence indicates that epigenetic modifications play a critical role in progression of CRC. In particular, over the last decade, dysregulated expression of microRNAs (miRNAs) has emerged as a hallmark feature of human cancers, including CRC. MiRNAs are small non-coding RNAs, 19–24 nucleotides in length, which regulate the expression of few to hundreds of downstream target genes post-transcriptionally (4, 5). In addition to their biological role, dysregulated expression of miRNAs is of clinical significance, as it often associates with key cancer-related clinicopathological features (6, 7).
The biogenesis of miRNAs is mediated by a group of proteins including Drosha, Dicer, TRBP, and Exportin-5 (XPO5) (8). Among these, XPO5, a member of the karyopherin family, is a key protein responsible for the transportation of precursor miRNAs (pre-miRNAs) from the nucleus to the cytoplasm (9). While XPO5 is perhaps the most critical regulator of miRNA biogenesis, the clinical significance of altered XPO5 expression in CRC remains unclear. Furthermore, the functional role of altered XPO5 expression, its impact on downstream target miRNAs, and its influence on CRC biology have not been interrogated.
Although no comprehensive studies have been performed in CRC, limited evidence in previous studies has observed overexpression of XPO5 in urothelial (10) and breast cancer (11, 12). Another recent study showed that XPO5 is a critical regulator of cell cycle (13). These studies suggest the oncogenic potential of XPO5, but systematic studies are required to further elucidate its functional and clinical role as a biomarker and as a potential therapeutic target in CRC.
Herein, we systematically and comprehensively characterize the role of XPO5 in CRC by analyzing colorectal cancer cell lines, a xenograft animal model and multiple cohorts of clinical specimens. Our data first show that XPO5 is upregulated in CRCs, and that patients with high XPO5 expression are associated with higher tumor burdens, demonstrate worse clinicopathological features, and poor survival. Furthermore, we studied the functional role of altered XPO5 expression and demonstrated that it functions as an oncogene by up-regulating the expression of important growth-promoting miRNAs in CRC. Taken together, our data highlight that XPO5 is functionally an oncogene, and high expression of XPO5 may serve as a potential therapeutic target in CRC.
MATERIALS AND METHODS
Patient cohorts and specimens
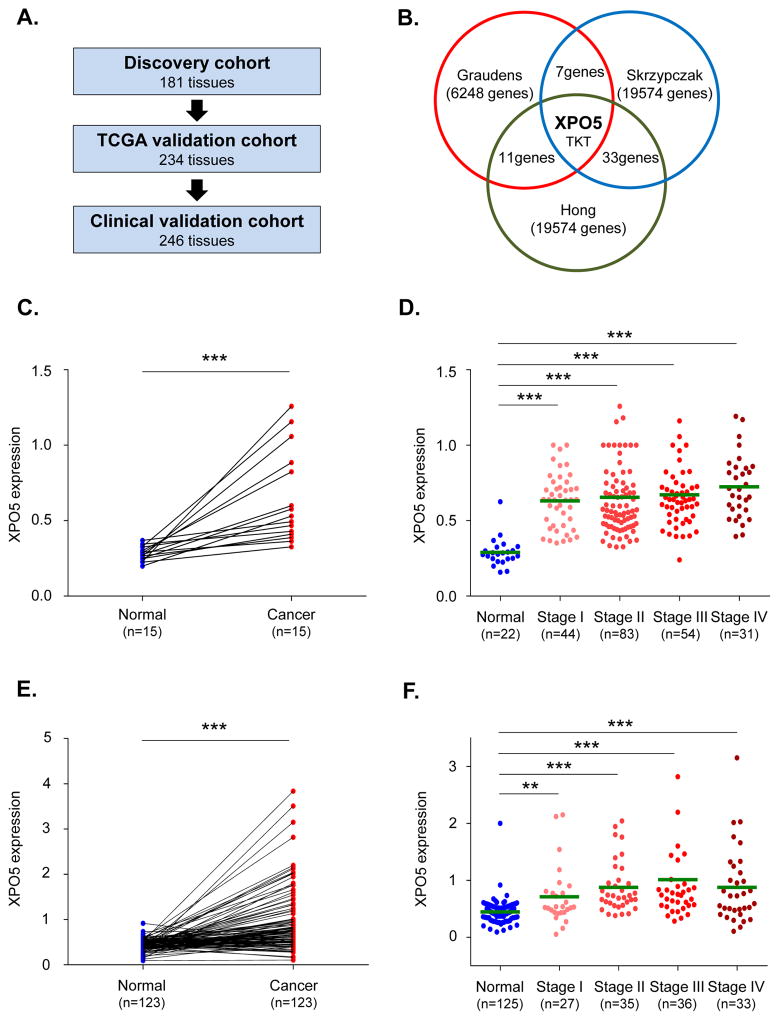
This study included analysis of 661 colorectal tissues, which were analyzed in a three-phase study that included an initial discovery cohort and two subsequent validation cohorts. In the discovery cohort, we analyzed data from the Oncomine microarray database comprised of 181 tissue specimens from normal and CRC tissues (14). This database included results from three independent gene expression profiles obtained from normal and CRC tissues (15–18). The Cancer Genome Atlas (TCGA) validation cohort included data from 234 tissue specimens in the TCGA dataset (19). The information on patient survival was obtained via cBioPortal (20). The Clinical validation cohort included analysis of 246 matched normal and neoplastic tissues from patients enrolled at Mie University, Mie, Japan. Patients with histologically confirmed stage I, II, IIII or IV CRCs who underwent surgical resection of their primary lesions were included in our study. Some stage IV CRC patients that were not candidates for surgical resection were excluded from our study due to unavailability of their clinical specimens. Treatments of patients followed guideline published by NCCN Clinical Practice Guidelines in Oncology. Written informed consent was obtained from each patient, and the study was approved by the institutional review boards of all involved institutions.
Real-time quantitative PCR analysis for XPO5 and miRNAs
Quantitative real-time PCR (qRT-PCR) was performed using the StepOne Real Time PCR System (Applied Biosystems, Foster City, CA, USA). The mRNA expression levels were measured using predesigned XPO5 TaqMan probes (Life Technologies, Carlsbad, CA, USA), and normalized to GAPDH. The relative expression of XPO5 was determined by calculating the differences between cancer and adjacent normal mucosa tissues. Reverse transcription and real-time PCR for miRNAs were performed using TaqMan probes for miR-10b, miR-21, miR-27a, miR-92a, miR-155, miR-182, pri-miR-21, pri-miR-10b, pri-miR-23a~27a~24-2 cluster, and pri-miR-182. A TaqMan primer set for the small nuclear RNA U6 (Life Technologies) was used as a normalization control for miRNAs and GAPDH (Life Technologies) was used for normalization of pri-miRNA expression. The relative expression level of each miRNA was determined using the ΔΔCt method. All assays were performed in duplicate.
Immunohistochemical Analysis
Paraffin-embedded sections derived from ten matched cancer/normal tissues were deparaffinized by xylene and ethanol. Following elimination of endogenous peroxidase activity by H2O2 and antigen retrieval by autoclaving at 121°C for 15 minutes, slides were incubated with anti-XPO5 antibody (EPR8452, Abcam, 1:400 dilution) for 1 hour. The color was developed using EnVision + Dual Link Kit (DAKO, Carpinteria, CA, USA) and hematoxylin. The staining with an isotype anti-body was used as the negative control. The staining levels of XPO5 were analyzed microscopically by two independent experts using staining score (from 1 to 5).
Immunofluorescence analysis
After the fixation of cells by methanol, cultured cells were reacted with anti-XPO5 antibody (EPR8452, Abcam, 1:400 dilution), followed by secondary fluorescence antibody (A-11034, Invitrogen, 10 μg/mL). Fluorescence signal was analyzed using fluorescence microscope.
Cell lines
The microsatellite stable (MSS) CRC cell lines, SW480 and Caco-2 were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). All cell lines were cultured according to the manufacturer’s specifications. Every few months, all cell lines were tested and authenticated using a panel of genetic and epigenetic markers.
Small interfering RNA Knockdown
XPO5-specific validated locked nucleic acid (LNA)-modified Silencer Select siRNA (siXPO5; s33190; Ambion) and control siRNA (siControl; Ambion) were selected for transfection. All experiments were performed by forward transfection according to the manufacturer’s protocol with a mixture of Optimem I (Invitrogen), Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and siRNA oligonucleotides (50 nM for SW480, 100 nM for Caco-2). Cells were incubated in culture media for 48 h after transfection prior to harvesting for analyses. All experiments were conducted in triplicates, and at least three independent experiments were performed.
Western immunoblotting
Western immunoblotting was performed as described previously (21). Anti-XPO5 antibody (EPR8452, Abcam, 1:1000) was used for the XPO5 staining, and anti-β-actin antibody (A5441, Sigma-Aldrich, St. Louis, MO, USA, 1:5000), anti- Vimentin antibody (H-84, Santa Cruz Biotechnology, Dallas, TX, 1:250) was used as the protein loading control.
Cytoplasmic and nuclear protein fraction analysis
NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific) was used for efficient cell lysis and extraction of cytoplasmic and nuclear protein fractions following manufacture’s protocol. The localization of XPO5 was analyzed using Western blotting.
MTT assay
An MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) assay (Sigma-Aldrich) was performed to measure the growth proliferative effects of siXPO5 and siControl, using the methods as described previously (22).
Colony formation assay
A total of 500 cells transfected with siXPO5 or siControl were seeded into 6-well plates and cultured for 10 days in a humidified CO2 incubator at 37°C. The numbers of colonies comprising of >50 cells were counted using GeneTools image analysis software (Syngene, Frederick, MD, USA) after staining of colonies using crystal violet.
Invasion assay
The invasiveness of cancer cells was evaluated using BioCoat Matrigel Invasion Chambers (Corning Life Sciences, Tewksbury, MA, USA) as described previously (22). A total of 5×105 cells transfected with siXPO5 or siControl were seeded into the invasion chambers in serum-free medium. The number of cells that invaded the underside of the membrane was measured.
Cell cycle analysis
The DNA content of siXPO5- and siControl-transfected CRC cells was evaluated using the Muse Cell cycle assay kit and Muse Cell Analyzer (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. The proportions of G0/G1, S and G2/M cells were calculated.
Xenograft studies
Twelve male athymic nude mice were obtained from Harlan Laboratories (Houston, TX USA) at 5 weeks of age and kept under controlled conditions (12 h light and dark cycles). The animal protocol was approved by the Institutional Animal Care and Use Committee of the Baylor Research Institute. Xenograft tumors were generated using SW480 cells with XPO5 siRNA or its controls, in which 3×106 cells were subcutaneously injected in left and right flanks of the mice. The mice were monitored for twelve days following injection, and subcutaneous tumors were measured every two days. At twelve days post-injection, all animals were sacrificed. The expression of XPO5 and target miRNAs in xenograft tissues was confirmed by qRT-PCR.
Statistical analyses
Results are expressed as means ± standard errors (SE). JMP software (ver. 10.0, SAS Institute Inc., Cary, NC, USA) was used to perform the statistical analyses. The Wilcoxon’s rank sum test was used to compare continuous variables, and Fisher’s exact test was used to analyze categorical variables. Wilcoxon’s signed rank test was used to compare matched continuous variables. Steel’s test was used for the multiple comparison of XPO5 expression level between normal and each stage. Overall survival (OS) was measured from the date of operation to the date of cancer-related death. Disease-free survival (DFS) was measured from the operation date to the date of recurrence or cancer-related death. The Kaplan–Meier method with Wilcoxon’s test was used to estimate distributions of OS and DFS in each patient group through univariate analyses. In order to determine the cut-off value for XPO5 expression, we first established receiver operating characteristic curves to discriminate patients with or without death. Youden’s index was then used to determine the optimal cutoff threshold for XPO5 expression from each cohort to predict the overall survival or disease free survival (22–25). Cox’s proportional hazard models were used to calculate hazard ratios (HR) with corresponding 95% confidence intervals (CI) for each group in a multivariate analysis. All calculated P values are two-sided, and a P value of <0.05 was considered to indicate statistical significance. All data were represented as mean ± standard error of mean (SEM).
RESULTS
XPO5 is significantly overexpressed in CRC patients
In the discovery cohort, we evaluated the expression level of XPO5 in multiple gene expression databases from CRC patients (Fig. 1A). We observed that among the top 1% overexpressed genes, two genes, XPO5 and TKT, were among the most significantly upregulated (P<0.0001; Fig. 1B, Supplementary Fig. 1A, B, C) in all three databases (15–17). Since a previous study reported TKT as potential oncogene (26, 27), we focused our attention on XPO5 considering its role in miRNA biogenesis and cancer. We discovered that XPO5 was not only overexpressed in CRCs, but was significantly upregulated in colorectal adenomas (P<0.0001; Supplementary Fig. 1D) (18), highlighting its potential oncogenic role during the early steps of the normal-adenoma-carcinoma sequence.
Fig. 1. XPO5 expression in discovery and validation cohorts.
(A) XPO5 expression levels in colorectal cancer (CRC) tissues were analyzed in three microarray databases as the discovery cohort (Oncomine), followed by the TCGA validation cohort and the Clinical validation cohort. (B) Only two genes, XPO5 and TKT, were in the top 1% of overexpressed genes in all three CRC microarray datasets (P<0.0001). (C) XPO5 expression levels were analyzed in 15 matched pairs of cancer and normal samples in TCGA validation cohort. XPO5 levels were significantly higher in CRC tissues than in matched adjacent normal mucosa (P<0.0001). (D) At each tumor stage, XPO5 was more highly upregulated than in normal mucosa (P<0.0001). (E) XPO5 expression levels were significantly higher in cancerous lesions compared with matched normal mucosa (P<0.001) in the Clinical validation cohort. (F) XPO5 expression levels were significantly higher in CRC lesions at all stages relative to normal mucosa (Normal vs. Stage I: P=0.0026, Normal vs. Stage II, III & IV: P<0.0001). *P<0.05, **P<0.01, ***P<0.001
High XPO5 expression is an independent prognostic factor in CRC patients
To confirm the over-expression of XPO5 in CRC tissues, we analyzed a cohort of CRC patients enrolled in TCGA as our first validation cohort and assigned as the “TCGA validation cohort”. As expected, XPO5 expression levels were significantly higher in cancer tissues compared to matched adjacent normal mucosal tissues (P<0.0001; Fig. 1C). Furthermore, XPO5 expression was significantly higher in cancer tissues at all stages (I through IV) relative to normal mucosa (P<0.0001; Fig. 1D). In particular, XPO5 was significantly upregulated in stage III-IV cancers compared to stage I–II tumors (P=0.0395; Supplementary Fig. 2A). In addition, the XPO5 expression levels were higher among patients with distant metastases than in those without (P=0.0388; Supplementary Fig. 2B). These data support the potential involvement of XPO5 during the advanced stages of CRC development. Next, we used an independent cohort, comprised of 246 matched tissue pairs of cancer and adjacent normal mucosa (Clinical validation cohort; Supplementary Table 1) to validate data obtained from the TCGA cohort by RT-qPCR. Consistent with our results from the discovery cohort and the TCGA validation cohort, we observed that XPO5 expression levels were significantly higher in neoplastic lesions compared to matched normal mucosa (P<0.001; Fig. 1E). The expression levels of XPO5 at each cancer stage were higher than the normal mucosa (normal vs. stage I: P=0.0026, normal vs. stage II, III & IV: P<0.0001; Fig. 1F). The XPO5 expression levels were significantly higher in T3 or T4 stages relative to T1 or T2 CRCs (P=0.0483; Supplementary Fig. 2C), and were significantly higher in venous invasion-positive compared to invasion-negative CRCs (P=0.0357; Supplementary Fig. 2D). We evaluated the relationship between XPO5 overexpression and CRC patient survival in the TCGA validation cohort. The high-XPO5 expression group yielded worse DFS (P=0.0774; Fig. 2A) and OS (P=0.0165; Fig. 2B) compared to the low-XPO5 group. Multivariate analysis demonstrated that high-XPO5 expression was an independent prognostic factor in patients with CRC (Table 1). Likewise, the high-XPO5 group had a significantly worse DFS (P=0.0426; Fig. 2C), and OS (P=0.0461; Fig. 2D) compared to the low-XPO5 group in the the Clinical validation cohort. Cox’s proportional hazard analysis revealed that the tumors with high-XPO5 expression was an independent prognostic factor in patients with CRC (P=0.0152; Supplementary Table 2). Taken together, our data consistently demonstrate that the primary tumors with high-XPO5 expression are associated with advanced pathological staging and correlated with poor prognosis, suggesting that XPO5 has a functional role in CRC progression.
Fig. 2. XPO5 as a prognostic marker in CRC patients.
(A) The high-XPO5 expression group had slightly worse disease-free survival than the low-XPO5 group (P=0.0774) in the TCGA validation cohort. (B) The high-XPO5 expression group exhibited worse overall survival than the low-XPO5 group (P=0.0165) in the TCGA validation cohort. (C) The high-XPO5 expression group had a significantly worse disease-free survival (DFS) than the low-XPO5 expression group (P=0.0426) in the Clinical validation cohort. (D) The high-XPO5 expression group had significantly worse overall survival (OS) than the low-XPO5 expression group (P=0.0461) in the Clinical validation cohort. (E) XPO5 protein expression levels were analyzed using immunohistochemical analyses in the Clinical validation cohort. XPO5 protein was mainly detected in the nucleus (brown: XPO5, blue: hematoxylin, ×200). Staining intensity was significantly higher compared to adjacent normal mucosa (P= 0.0267). *P<0.05, **P<0.01, ***P<0.001
Table 1.
Multivariate analysis for the predictors of overall survival in patients with colorectal cancer (TCGA validation cohort)
| Characteristic | HR | 95% CI | P value | |
|---|---|---|---|---|
| Gender | Female/Male | 0.92 | 0.47 – 1.81 | 0.8134 |
| Age (years) | ≥70/<70 | 1.83 | 0.91 – 3.73 | 0.0917 |
| Location | Rectum/Colon | 0.77 | 0.33 – 1.64 | 0.5009 |
| Stage | III or IV/I or II | 4.00 | 2.02 – 8.39 | < 0.0001 *** |
| MSI status | MSI/MSS | 1.22 | 0.42 – 3.16 | 0.6932 |
| XPO5 expression | High/Low | 2.19 | 1.06 – 4.38 | 0.0342 * |
P<0.05,
P<0.01,
P< 0.001 HR, hazard ratio; CI, confidence interval
High XPO5 protein expression is primarily confined to the nuclear compartment in CRC
In order to further confirm the significance of our gene expression data, XPO5 expression was examined using immunohistochemical staining. We observed that XPO5 protein expression was primarily confined within the nuclear compartment, and its staining intensity was significantly higher compared to adjacent normal mucosa (staining score by experts; P=0.038; Fig. 2E). This result confirms that alterations in XPO5 protein are manifested at the protein level, consistent with a putative oncogenic role in CRC.
High XPO5 expression correlated with worse clinicopathological features in microsatellite stable CRCs
Next, we analyzed the relationship between microsatellite instability status and XPO5 expression in the TCGA validation cohort. Only 13.9% of all CRCs were MSI and the remaining 86.1% were MSS (Supplementary Fig. 3A). XPO5 expression was significantly lower in MSI vs. MSS CRCs (P=0.0020; Supplementary Fig. 3B). In MSS CRCs, XPO5 was upregulated in a stage-dependent manner (stage I/II vs. III/IV; P=0.0118, Supplementary Fig. 3C), while no significant differences were observed in MSI-positive patients (Supplementary Fig. 3D). Patients with high-XPO5 showed worse overall survival only in the MSS group (P=0.0219; Supplementary Fig. 3E), highlighting the important role of XPO5 in microsatellite stable CRCs.
Inhibition of XPO5 results in decreased cell proliferation, attenuated tumorigenicity, lower invasive potential and cell cycle arrest in CRC cells
We first determined the expression of XPO5 in several CRC cell lines including Caco-2, LOVO, HCT116, SW480, SW620, and HT29 to clarify the role of XPO5 in cancer cell lines. Since the relative expression levels of XPO5 were relatively low and did not differ significantly between cell lines (data not shown), we decided to use additional criteria for selecting cell lines. Because one of aims of our study was also to clarify the significance of XPO5 expression in MSS CRC, we selected SW480 (MSS, KRAS G12V, BRAF wild) and Caco2 (MSS, KRAS wild, BRAF wild) cells for further experiments.
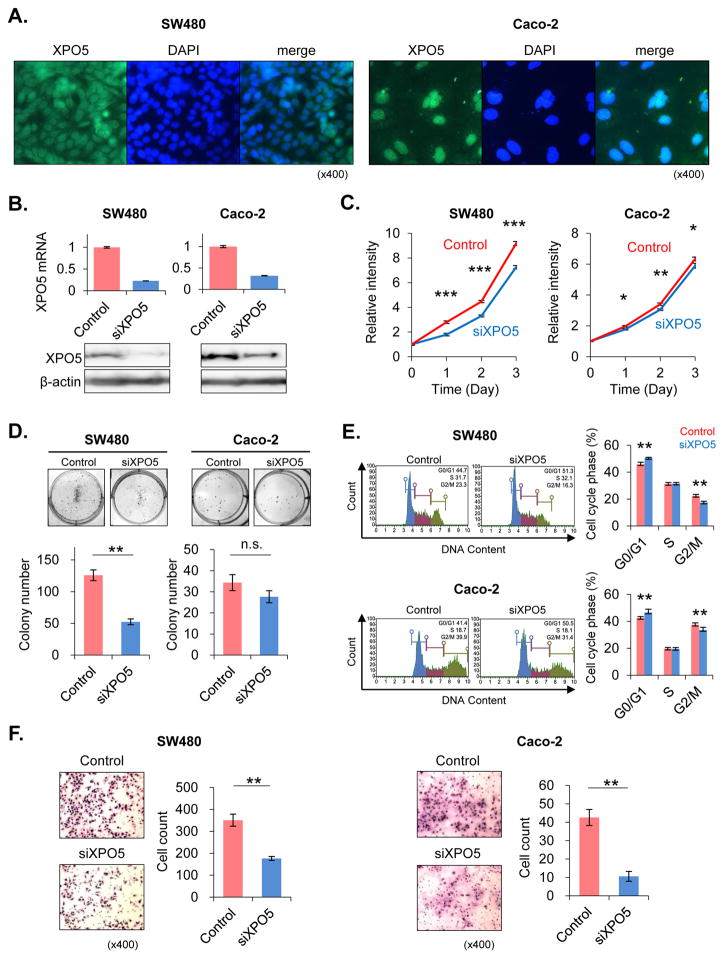
To evaluate the functional consequences of altered XPO5 expression and determine whether XPO5 modulates the biological characteristics of CRC cells, we performed siRNA transfection to knockdown XPO5 expression in SW480 and Caco-2 cell lines. XPO5 was primarily detected in nuclear compartment in SW480 and Caco-2 cells by immunofluorescence staining analysis (Fig. 3A). These results were further confirmed by performing western immnoblotting using nuclear and cytoplasmic fractions (Supplementary Fig. 4). Both cell lines revealed effective inhibition in XPO5 mRNA expression post-transfection, with siXPO5-transfected cells displaying <30% expression vs. siControl-transfected cells. These results were further validated at the protein level using western blotting, where both CRC cell lines showed significant suppression of XPO5 protein expression following XPO5 knockdown (Fig. 3B).
Fig. 3. Knockdown of XPO5 in colorectal cancer cells.
(A) XPO5 was detected in nuclear compartment in SW480 and Caco-2 cells in immunofluorescence staining. (B) XPO5 was knocked down in SW480 and Caco-2 cell lines using siRNA. The XPO5 knockdown level was analyzed using real-time PCR. XPO5 expression levels in XPO5 siRNA (siXPO5)-transfected cells were <30% of the levels measured in control siRNA (siControl)-transfected cells. The knockdown effect was also analyzed using western blotting. In each cell line, XPO5 protein expression was significantly suppressed by siXPO5 knockdown. (C) In the SW480 cell line, siXPO5-transfected cells exhibited significantly suppressed proliferation compared to siControl-transfected cells (P<0.0001). Similarly, XPO5 suppression resulted in reduced proliferation in Caco-2 cells (P=0.0489). (D) Colony-formation assays were performed to examine the effect of siXPO5 knockdown on the colony-forming abilities of single cells plated in vitro. siXPO5-transfected SW480 cells formed a significantly fewer colonies than siControl-transfected cells (P=0.0039). The same tendency occurred in Caco-2 cells, although this result was not significant. (E) Cell cycle analyses revealed a significant increase in the G0/G1 phase fraction after siXPO5 knockdown in both SW480 and Caco-2 cells (P=0.0090 SW480; P=0.0090 Caco-2). (F) To determine whether XPO5 knockdown inhibited cell invasion, in vitro invasion chamber assays were performed. In both SW480 and Caco-2 CRC cells, siXPO5 transfection significantly reduced invasiveness compared to siControl-transfected cells (P=0.0088 SW480; P=0.0088 Caco-2). *P<0.05, **P<0.01, ***P<0.001
To determine whether downregulation of XPO5 results in suppression of cell proliferation in human cancer cell lines, we analyzed cell proliferation rates by the MTT assay. Although the degree of proliferation inhibition varied between both cell lines, siXPO5-transfected cells showed a significant inhibition of cellular proliferation compared with siControl-transfected cells (P<0.0001 SW480; P=0.0489 Caco2; Fig. 3C). We performed colony formation assays to evaluate the effectiveness of siXPO5, and found that both SW480 and Caco-2 cells transfected with siXPO5 produced fewer colonies compared with siControl-transfected cells, and this effect was more pronounced in SW480 cells (P=0.0039; Fig. 3D).
Since a previous report suggested an important function of XPO5 in cell cycle regulation (13), we investigated whether inhibition of XPO5 alters cell cycle dynamics in CRC cell lines. As expected, flow cytometry-based cell cycle analyses revealed significant increases in the G0/G1 fraction after siXPO5 knockdown in both SW480 (P=0.0090) and Caco-2 (P=0.0090) cells (Fig. 3E). These data indicate that inhibition of XPO5 induces G1/S cell cycle arrest, which further supports our observations of reduced proliferation rates in CRC cells.
Our clinical data indicated that XPO5 overexpression significantly correlated with invasive and metastatic potential. Therefore, we tested the hypothesis that XPO5 knockdown would inhibit the invasive capacity of CRC cells using in-vitro transwell invasion assays. The siXPO5 transfection resulted in significantly decreased invasiveness relative to siControl-transfected cells in both SW480 (P=0.0088) and Caco-2 (P=0.0088) CRC cells (Fig. 3F), supporting our observations in clinical specimens from CRC patients.
XPO5 inhibition alters the expression of key oncogenic miRNAs in CRC cells
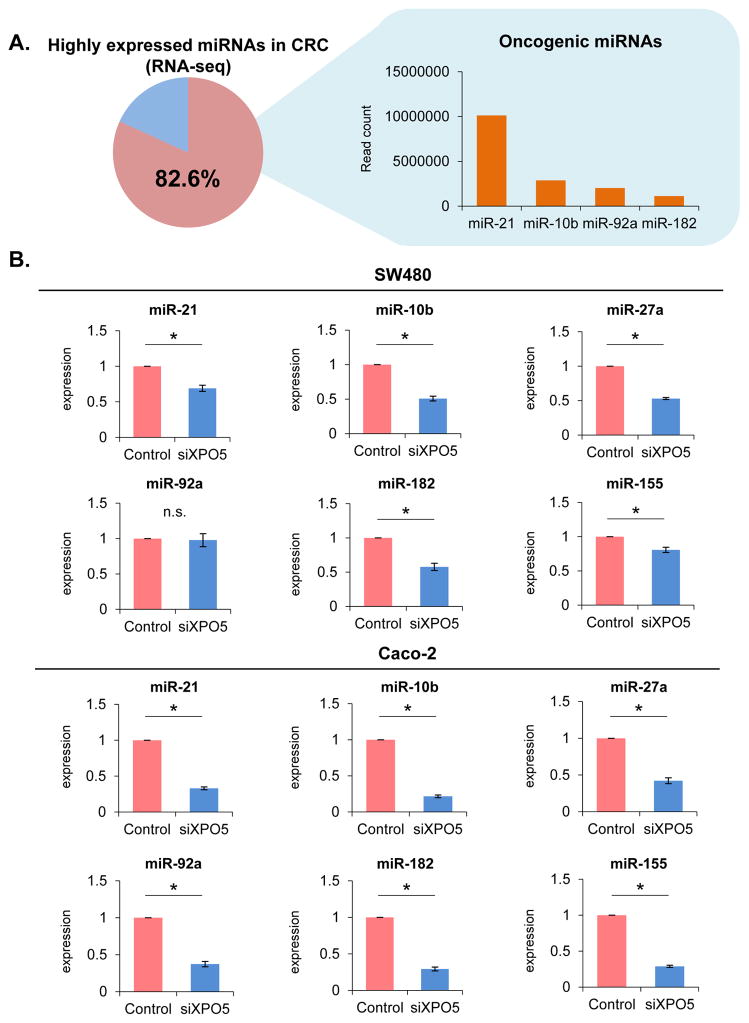
Many oncogenic miRNAs (onco-miRs) are overexpressed in several human cancers, including CRC. One of the critical functional roles of XPO5 is regulation of miRNA biogenesis, and we hypothesized that XPO5 inhibition might result in suppressing the expression of critical onco-miRs in CRC. At first we selected 4 overexpressed onco-miRs (miR-21, miR-10b, miR-92a and miR-182) belonging to a panel of the top 20 most overexpressed miRNAs based upon more than 80% of read counts as reported in a next-generation sequencing database of CRCs (28). In addition, two more oncogenic miRNAs, miR-27a and miR-155, which have been shown to play a key role in CRC, were also selected as the candidates for analysis (Fig. 4A). In support of our hypothesis, we observed that expression of the most oncogenic miRNAs was significantly downregulated in siXPO5 vs. siControl transfected SW480 and Caco-2 CRC cells (Fig. 4B; P<0.05). On the other hand, pri-miR-21 was significantly overexpressed in siXPO5 SW480 cells (P<0.05) compared with that of siControls. There were no significant differences in the expression levels of pri-miR-10b, pri-miR-23a~27a~24-2 cluster, and pri-miR-182 between siXPO5 cells and siControls. In Caco-2 cells, pri-miR-21 and pri-miR-182 were significantly overexpressed in siXPO5 cells (P<0.05) compared with that of siControls. However, there were no difference in the expression levels of pri-miR-10b and pri-miR-23a~27a~24-2 cluster between Caco-2 siXPO5 cells and siControl cells (Supplementary Fig. 5). These results clearly highlight that the downregulation of miRNA by siXPO5 is not due to the transcriptional suppression, but most likely because of the post-transcriptional interference of pre-miRNA transportation. Hence, oncogenic potential of XPO5 may be at least in part mediated through its ability to regulate the expression of important onco-miRs in CRC.
Fig. 4. Down-regulation of key oncogenic miRNAs by knockdown of XPO5.
(A) Four overexpressed oncogenic miRNAs (miR-21, miR-10b, miR-92a and miR-182) were selected from the top 20 overexpressed miRNAs occupying 80% of all read counts in the next-generation sequencing database of CRCs. (B) The expression of most oncogenic miRNAs were significantly downregulated in siXPO5-transfected vs. siControl-transfected CRC cells (P<0.05).
Reduced XPO5 expression inhibited tumor growth in a xenograft animal model
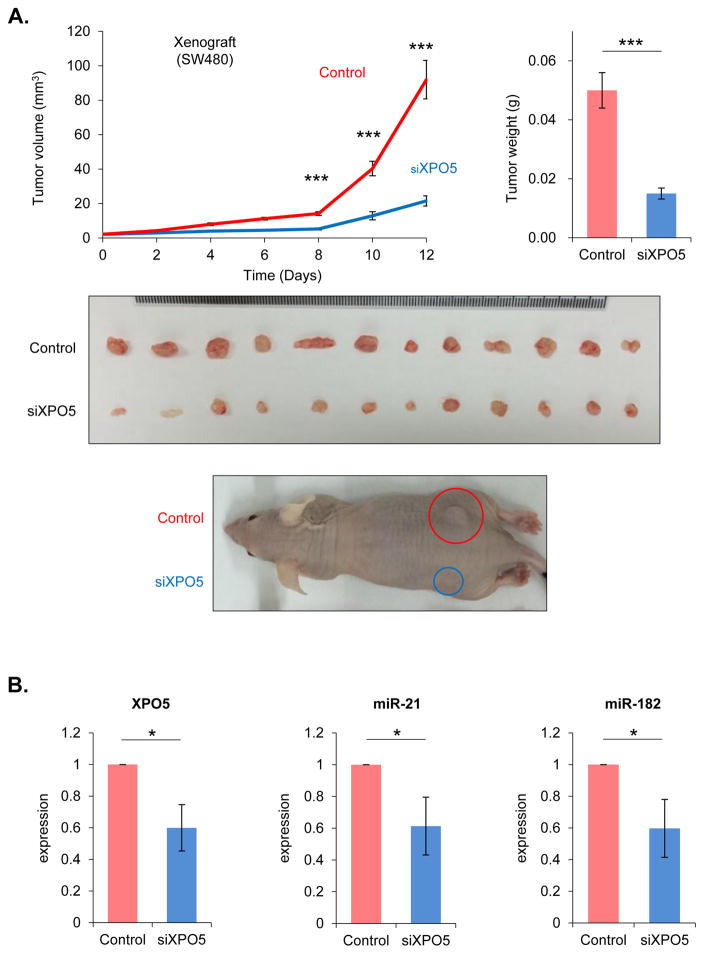
To confirm our in vitro findings, we generated xenograft tumors using SW480 cells transfected with either siXPO5 or siControl. Twelve days following the initial injection, the tumor volume and weight were significantly lower in animals that were implanted with siXPO5-transfected cells compared with siControl-transfected cells (P=0.0002 volume; P=0.0004 weight; Fig. 5A). To further confirm that the reduced tumor burden was as a result of lower XPO5 levels, we determined expression levels and found they were significantly lower in siXPO5 tumors at the end of the experimental duration. The oncogenic miRNAs including miR-21 and miR-182 were also downregulated in tumors (P<0.05; Fig. 5B). Taken together, these results highlight the oncogenic role of XPO5 in colorectal cancer.
Fig. 5. XPO5 expression in xenograft studies.
To analyze whether XPO5 knockdown suppresses colorectal cancer (CRC) tumorigenicity, SW480 cells transfected with either XPO5 siRNA (siXPO5) or control siRNA (siControl) were injected subcutaneously in nude mice (106 cells per mouse) to generate a xenograft model. (A) At 12 days post-injection, tumor volume and weight in recipients of siXPO5-transfected cells were significantly decreased relative to those in recipients of siControl-transfected cells (P=0.0002 volume; P=0.0004 weight). (B) XPO5, miR-21, and miR-182 expression levels were significantly lower in siXPO5 tumors than in scramble-control transfected tumors at the end of the experiment (P<0.05). *P<0.05, **P<0.01, ***P<0.001
DISCUSSION
MiRNAs are a class of small non-coding RNAs (18–25 nucleotides) that post-transcriptionally regulate gene expression through RNA interference (RNAi) (29). Accumulating evidence indicates that dysregulation of miRNAs is associated with metastatic processes in CRC (25, 30). XPO5 mediates the transport of pre-miRNAs from the nucleus to the cytoplasm and the final processing into mature miRNA by Dicer (29, 31). Accordingly, XPO5 functions as a key regulator of miRNA biogenesis, and facilitates the export of miRNAs from the nucleus to cytoplasm (29, 32–36). Despite its critical and well-recognized function, not much is known about its biological role or clinical significance in CRC. To fill this important gap in knowledge, the present study for the first time systematically interrogated the role of XPO5 in CRC by evaluating its expression in multiple independent cohorts of patients with CRC, followed by extensive analysis for its clinical significance in this cancer. Thereafter, we performed a series of functional validation studies in CRC cell lines and xenograft animal model, and identified and validated that XPO5 acts like an oncogene in CRC.
In this report, we have made several important and previously unrecognized discoveries regarding the role of XPO5 in CRC. First, we observed that expression of XPO5 is frequently upregulated in CRC compared to normal mucosa by analyzing multiple independent clinical specimen cohorts of patients with CRC. Second, we identified that high expression of XPO5 correlated with advanced disease, increased invasion and metastasis, associated with poor OS and DFS in CRC patients, and is an independent prognostic factor. Third, we found that XPO5 protein expression is primarily confined within the nuclear compartment in the cells. Fourth, we demonstrated that dysregulated expression of XPO5 leads to altered expression of various oncogenic miRNAs in CRC. Fifth, we performed a series of functional studies to demonstrate that downregulation of XPO5 expression in CRC cells inhibited not only its invasive capacities, but also suppressed cell proliferation and induced cell cycle arrest. Finally, we were able to validate our in-vitro findings in a xenograft animal model, wherein we demonstrated that siXPO5 reduced the growth of tumors, further highlighting its oncogenic role in CRC.
In our original cohort of CRC patients, XPO5 expression was frequently upregulated, both at mRNA and protein levels, in tumors compared to adjacent normal mucosae. Furthermore, high-XPO5 expression correlated with greater depth of tumor invasion and venous invasion, as well as worse DFS and OS. In a multivariate analysis, XPO5 expression remained an independent prognostic factor for survival, highlighting an oncogenic role of XPO5 in CRC. In a recent study, mutated XPO5 was shown to have an oncogenic role in MSI-positive cell lines (37), however, since majority of CRCs are MSS, a systematic and comprehensive analysis for the clinical significance of XPO5 in CRC is warranted. To the best of our knowledge, this is the first study to systematically explore the role of XPO5 in CRC patients. Our results are consistent with previous data reported in breast (11) and urothelial cancers (10). From a functional standpoint, a prior study suggested that XPO5 plays a role during cell cycle progression (13). Collectively, these data are consistent with our findings in CRC and support the oncogenic role of XPO5 in cancer.
We evaluated the relationship between XPO5 expression and CRC cells through in vitro and in vivo experimental models. The knockdown of XPO5 led to reduced invasiveness and suppression of proliferation in CRC cell lines, as well as suppressed tumor growth in the xenograft animal model. In line with a previous report (13), we observed G1/S cell cycle arrest following XPO5 inhibition in cancer cells, suggesting that XPO5 is a regulator of cell cycle in normal as well as cancer cells. Interestingly, many key oncogenic miRNAs were significantly suppressed by siXPO5, suggesting the importance of XPO5 in the maintenance of over-expressed oncogenic miRNAs in CRC. Even though knockdown of XPO5 induces inhibition of all miRNAs, most of the key tumor suppressor-miRs such as miR-34a (38) and miR-143/145 cluster (39) are known to be downregulated in cancers, indicating that further inhibition of these miRNAs are less relevant and aberrantly overexpressed oncogenic miRNAs appear to be more important.
Based on these results, we conclude that XPO5 acts as oncogene, and may serve as a promising prognostic biomarker in CRC. Furthermore, since XPO5 expression is much higher in MSS compared to MSI CRCs, overexpression of this oncogene is likely more critical in the background of microsatellite stable phenotype. Considering that XPO5 inhibition leads to reduced growth proliferation in cultured cancer cells and the xenograft model, this miRNA transporter may also be a potential therapeutic target in cancer. However, a previous study suggested that the underlying mechanisms for XPO5 in cancer may be more complex (37). There appears to be different affinity between XPO5 and each pre-miRNA and further studies using immunoprecipitation and next-generation sequencing are needed to clearly establish the role of XPO5 in cancer.
One of the potential limitations with the invasion assays that we conducted was that we observed reduced proliferation in siXPO5 cells. Therefore, it is possible that the observed differences in proliferation may have affected the invasive potential in these cells. Nevertheless, even with the small differences in proliferation, siXPO5 cells had significantly reduced cellular invasiveness compared to the control cells.
In summary, this study provides a novel evidence for the oncogenic role of XPO5 in CRC. Our study highlighted the clinical and biological significance of XPO5 in CRC through a series of in vitro and in vivo experiments. In addition, analysis of multiple cohorts of clinical specimens demonstrated that high XPO5 expression correlated with worse clinicopathological features and poor patient survival. Collectively, this is the first systematic and comprehensive demonstration for the biological and clinical significance of XPO5 and our data indicates that XPO5 may serve as a possible therapeutic target in CRC.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Exportin-5 (XPO5) plays a key role in microRNA (miRNA) biogenesis, and is known to transport precursor miRNAs from nucleus to cytoplasm. However, the clinical significance of XPO5 expression in colorectal cancer (CRC) remains unclear. Herein, we systematically and comprehensively characterize the role of XPO5 in CRC by analyzing colon cancer cell lines, a xenograft animal model and multiple cohorts of clinical specimens. We demonstrate that high XPO5 expression resulted in worse clinicopathological features in two independent CRC clinical cohorts. XPO5 showed oncogenic potential in a series of in vitro and in vivo experimental models by control of miRNA expression. Knockdown of XPO5 showed significant anti-tumor effect. The present study identified previously unrecognized oncogenic role and clinical significance of XPO5 in CRC. Our data suggest that XPO5 may be a potential therapeutic target in CRC.
Acknowledgments
Funding: The present work was supported by grants R01 CA72851, CA18172, CA184792 and U01 CA187956 from the National Cancer Institute, National Institutes of Health, funds from the Baylor Research Institute and a pilot grant from Charles A Sammons Cancer Center. This work was also supported by grant from Uehara Memorial Foundation.
Footnotes
Competing Interests: The authors have no competing interests to disclose.
Author Contributions: Conceived and designed experiments: KS, YO, ST, CRB, AG; performed experiments: KS, ST; Analyzed data: KS, AG; Contributed reagents, materials and other analytical tools: KS, YO; wrote the manuscript: KS, ST, CRB and AG.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA: a cancer journal for clinicians. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochemical and biophysical research communications. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. The Journal of pathology. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph A, Shi H, Forsti A, Hoffmeister M, Sainz J, Jansen L, et al. Repeat polymorphisms in ESR2 and AR and colorectal cancer risk and prognosis: results from a German population-based case-control study. BMC cancer. 2014;14:817. doi: 10.1186/1471-2407-14-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammerman A, Greenberg-Dotan S, Battat E, Feldhamer I, Bitterman H, Brenner B. The ‘real-life’ impact of adding bevacizumab to first-line therapy in metastatic colorectal cancer patients: a large Israeli retrospective cohort study. Acta Oncol. 2015;54:164–70. doi: 10.3109/0284186X.2014.958532. [DOI] [PubMed] [Google Scholar]
- 8.Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Science signaling. 2015;8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- 9.Ristau J, Staffa J, Schrotz-King P, Gigic B, Makar KW, Hoffmeister M, et al. Suitability of circulating miRNAs as potential prognostic markers in colorectal cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:2632–7. doi: 10.1158/1055-9965.EPI-14-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Y, Liu Y, Gui Y, Cai Z. Inducing cell proliferation inhibition and apoptosis via silencing Dicer, Drosha, and Exportin 5 in urothelial carcinoma of the bladder. Journal of surgical oncology. 2013;107:201–5. doi: 10.1002/jso.23214. [DOI] [PubMed] [Google Scholar]
- 11.Leaderer D, Hoffman AE, Zheng T, Fu A, Weidhaas J, Paranjape T, et al. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. International journal of molecular epidemiology and genetics. 2011;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- 12.Vaidyanathan S, Thangavelu PU, Duijf PH. Overexpression of Ran GTPase Components Regulating Nuclear Export, but not Mitotic Spindle Assembly, Marks Chromosome Instability and Poor Prognosis in Breast Cancer. Targeted oncology. 2016 doi: 10.1007/s11523-016-0432-y. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki YW, Kiga K, Kayo H, Fukuda-Yuzawa Y, Weise J, Inada T, et al. Global microRNA elevation by inducible Exportin 5 regulates cell cycle entry. RNA. 2013;19:490–7. doi: 10.1261/rna.036608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [Accessed in August 2014];Oncomine_database. https://www.oncomine.org/
- 15.Graudens E, Boulanger V, Mollard C, Mariage-Samson R, Barlet X, Gremy G, et al. Deciphering cellular states of innate tumor drug responses. Genome biology. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PloS one. 2010;5 doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clinical & experimental metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 18.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, et al. Transcriptome profile of human colorectal adenomas. Molecular cancer research : MCR. 2007;5:1263–75. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 19.TCGA_Research_Network. [Accessed in May 2014]; http://cancergenome.nih.gov/
- 20.cBioPortal. [Accessed in August 2014]; http://www.cbioportal.org/index.do.
- 21.Toden S, Okugawa Y, Buhrmann C, Nattamai D, Anguiano E, Baldwin N, et al. Novel Evidence for Curcumin and Boswellic Acid-Induced Chemoprevention through Regulation of miR-34a and miR-27a in Colorectal Cancer. Cancer Prev Res (Phila) 2015;8:431–43. doi: 10.1158/1940-6207.CAPR-14-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka S, Hattori N, Ishikawa N, Shoda H, Takano A, Nishino R, et al. Krebs von den Lungen-6 (KL-6) is a prognostic biomarker in patients with surgically resected nonsmall cell lung cancer. International journal of cancer Journal international du cancer. 2012;130:377–87. doi: 10.1002/ijc.26007. [DOI] [PubMed] [Google Scholar]
- 24.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute. 2013;105:849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. Journal of the National Cancer Institute. 2015:107. doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du MX, Sim J, Fang L, Yin Z, Koh S, Stratton J, et al. Identification of novel small-molecule inhibitors for human transketolase by high-throughput screening with fluorescent intensity (FLINT) assay. Journal of biomolecular screening. 2004;9:427–33. doi: 10.1177/1087057104263913. [DOI] [PubMed] [Google Scholar]
- 27.Langbein S, Zerilli M, Zur Hausen A, Staiger W, Rensch-Boschert K, Lukan N, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. British journal of cancer. 2006;94:578–85. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schee K, Lorenz S, Worren MM, Gunther CC, Holden M, Hovig E, et al. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PloS one. 2013;8:e66165. doi: 10.1371/journal.pone.0066165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muqbil I, Bao B, Abou-Samra AB, Mohammad RM, Azmi AS. Nuclear export mediated regulation of microRNAs: potential target for drug intervention. Current drug targets. 2013;14:1094–100. doi: 10.2174/1389450111314100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neerincx M, Sie DL, van de Wiel MA, van Grieken NC, Burggraaf JD, Dekker H, et al. MiR expression profiles of paired primary colorectal cancer and metastases by next-generation sequencing. Oncogenesis. 2015;4:e170. doi: 10.1038/oncsis.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 32.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends in cell biology. 2004;14:156–9. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 34.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Current opinion in structural biology. 2010;20:782–90. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–7. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 37.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. Journal of hematology & oncology. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamatani A, Nakagawa Y, Akao Y, Maruyama N, Nagasaka M, Shibata T, et al. Downregulation of anti-oncomirs miR-143/145 cluster occurs before APC gene aberration in the development of colorectal tumors. Medical molecular morphology. 2013;46:166–71. doi: 10.1007/s00795-013-0020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.