Abstract
We aimed to identify novel molecular associations between chronic intermittent hypoxia with re-oxygenation and adverse consequences in obstructive sleep apnea (OSA). We analyzed gene expression profiles of peripheral blood mononuclear cells from 48 patients with sleep-disordered breathing stratified into four groups: primary snoring (PS), moderate to severe OSA (MSO), very severe OSA (VSO), and very severe OSA patients on long-term continuous positive airway pressure treatment (VSOC). Comparisons of the microarray gene expression data identified eight genes up-regulated with OSA and down-regulated with CPAP treatment, and five genes down-regulated with OSA and up-regulated with CPAP treatment. Protein expression levels of two genes related to endothelial tight junction (AMOT P130, and PLEKHH3), and three genes related to anti-or pro-apoptosis (BIRC3, ADAR1 P150, and LGALS3) were all increased in the VSO group, while AMOT P130 was further increased, and PLEKHH3, BIRC3, and ADAR1 P150 were all decreased in the VSOC group. Subgroup analyses revealed that AMOT P130 protein expression was increased in OSA patients with excessive daytime sleepiness, BIRC3 protein expression was decreased in OSA patients with hypertension, and LGALS3 protein expression was increased in OSA patients with chronic kidney disease. In vitro short-term intermittent hypoxia with re-oxygenation experiment showed immediate over-expression of ADAR1 P150. In conclusion, we identified a novel association between AMOT/PLEKHH3/BIRC3/ADAR1/LGALS3 over-expressions and high severity index in OSA patients. AMOT and GALIG may constitute an important determinant for the development of hypersomnia and kidney injury, respectively, while BIRC3 may play a protective role in the development of hypertension.
Background
Obstructive sleep apnea (OSA) has been independently associated with endothelial dysfunction which may explain the increased risk for cardiovascular events, hypertension, and all-cause mortality in this population through chronic intermittent hypoxia with re-oxygenation (IHR) injury[1,2]. A recent meta-analysis has demonstrated that continuous positive airway pressure (CPAP) treatment significantly improved endothelial function as assessed by flow-mediated dilation[3]. It has been observed that circulating leukocytes of OSA patients exhibited markedly enhanced in vitro release of superoxide radical anions and delayed apoptosis, which may in turn lead to endothelial dysfunction[4,5]. However, underlying mechanisms by which IHR leads to endothelial dysfunction and other adverse consequences, such as hypertension, excessive daytime sleepiness (EDS), and chronic kidney disease (CKD), in OSA are largely unknown.
The aim of this study was to explore which expression signatures in peripheral blood could be representative of, or associated with OSA, by investigating the expressions of genes in peripheral blood mononuclear cells (PBMCs) that may be involved in these effects. We hypothesized that the gene expressions of PBMCs involved in the responses to chronic IHR in OSA patients would be markedly different from those in subjects with primary snoring, and that additional differences would be seen between OSA patients with and without hypertension or between OSA patients with and without EDS. Furthermore, we aimed to improve the understanding of the molecular signatures that can correlate endothelial dysfunction with long-term and sufficient CPAP treatment, with the hope that novel genes may be found to be over- or under-expressed after treatment.
A few studies have applied DNA microarray technology to investigate gene expressions in patients with OSA[6–9]. In one of these studies, which focused on gene expressions in the blood leukocytes from adult OSA patients, genes involved in modulation of reactive oxygen species, cell growth, proliferation, and the cell cycle were found to be altered after one night of sleep in four OSA patients free of any co-morbidity[6]. The extent to which the leukocyte genes play a role in OSA patients with adverse consequences, and the effect of nasal CPAP on gene signatures is unclear. Therefore, we extended our investigation into OSA patients with long-term CPAP treatment, hypertension, or EDS by analyzing whole-genome gene expression profiles of PBMC in three comparisons: (1) treatment-naïve moderate to very severe OSA patients versus subjects with primary snoring; (2) moderate to very severe OSA patients with hypertension or EDS versus those without hypertension or EDS, respectively; (3) treatment-naïve very severe OSA patients versus those receiving at least one year of adequate CPAP treatment. We identified a novel association between several genes related to apoptosis or endothelial function and OSA or its clinical phenotypes.
Materials and methods
Subjects
The study was approved by the Institutional Review Board of Chung Gung Memorial Hospital, Taiwan. The study participants were recruited from the pulmonary clinics and health examination center of Kaohsiung Chung Gung Memorial Hospital January 2012 through December 2014. Written informed consent was obtained from each subject participating in the study, who was aged 20 years or older. All study participants underwent full-night in-laboratory polysomnography examinations as described previously, and OSA was diagnosed according to the AASM guideline[10]. The exclusion criteria included ongoing infections, autoimmune disease, use of immunosuppressive agent in the past 6 months, narcolepsy, severe obesity (body mass index, BMI, ≧35 kg/m2), old age (>65 year-old), and those with a BMI < 21 kg/m2. Study cohorts used for the whole-genome microarray gene expression and protein expression experiments included 48 (cohort 1) and 68 subjects (cohort 2: cohort 1 with expanded sample size), respectively. All these participants were classified into the following four groups based on apnea hypopnea index (AHI) and long-term use of CPAP: subjects with primary snoring (PS; AHI<5), treatment-naïve patients with moderate to severe OSA (MSO; 15<AHI≦50), treatment-naïve patients with very severe OSA (VSO; AHI>50), and very severe OSA patients on long-term CPAP treatment (VSOC; AHI>50 and regular use of CPAP: >4 hours/night, > one year). Nocturnal hypoxemia was evaluated in terms of the percentage of total minutes of recording time with oxyhemoglobin saturation <90% (%time <90% SaO2), and the number of dips >4% of basal SaO2%//h (oxygen desaturation index, ODI). The Epworth Sleepiness Scale (ESS) recorded at the PSG exam was used to measure sleep propensity, and EDS was defined as ESS>10. Hypertension was defined as baseline blood pressure>140/90 mmHg. Heart disease included ischemic heart disease, cardiac arrythmia, and congestive heart failure. CKD was defined as estimated glomerular filtration rate <60 mL/min/1.73 m2 for ≥ 3 months.
Processes of RNA isolation and cRNA synthesis
Peripheral whole blood (20 ml) was collected at AM 6:00 to 8:00 after written informed consent was obtained from all the recruited participants. The PBMCs were isolated by Ficoll-Hypaque gradient centrifugation (HISTOPAQUE®-119, Sigma-Aldrich, Inc., St. Louis, MO USA) within 90 min of drawing blood, washed in PBS, and then stored in RNAlater (Ambion Inc., Austin, TX, USA) at -80°C until RNA isolation. An RNeasy® Plus Mini Kit (Qiagen, Hilden, Germany) was used for isolation of high quality total RNA, and treated with DNase according to the manufacture protocol. RNA samples were run on a RNA 6000 Nano Gel System (Agilent Technologies Inc., Palo Alto, CA, USA) using an Agilent 2100 Bioanalyzer (Agilent) to determine the quality of RNA. Only samples with A260/A280 ratios of 1.9 to 2.1 were used for further analysis. A total of 300 ng RNA was used for synthesis of first strand cDNA and transcription of cRNA using an Illumina Totalprep RNA Amplication kit (Ambion, Inc.).
Gene expression profiling and microarray data analysis in the study cohort 1
Illumina (San Diego, CA) HumanRef-12-version 2 bead microarrays were used with 750 ng labeled cRNA for each sample according to the manufacturer's protocol. Human Ref-12 version 2 arrays consist of 27,455 probes representing 21,910 unique human genes on an eight-strip format array. All expression dataset has been deposited in the NCBI Gene Expression Omnibus with accession number of GSE75097.The preprocessing, quality control, background subtraction, quantile normalization and log2 transformation of array data for cross comparison were performed using BeadStudio software. Statistical analysis of the microarray data was further performed, using GeneSpring software version 11 (Agilent Technologies Inc., Santa Clara, CA, USA) as previously described[11]. Because the sample sizes were relatively small and statistics could be influenced by outliers, a non-parametric U test for unpaired comparisons of the two independent groups was applied. The Benjamini-Hochberg false-discovery rate correction method was used for controlling false positives and a corrected p-value cutoff of 0.05 was used to select the sets of significantly up- and down-regulated genes. The gene sets were then grouped into functional categories according to the Gene Ontology Biological Processes Classification. The pathway enrichment process was used to find direct relationships between genes of interest. This was performed in GeneSpring software with the “simple and direct interaction” algorithm.
Measurement of protein expression levels of five selected genes from PBMC samples in the study cohort 2
PBMCs were lysed in radio-immuno-precipitation assay-buffer containing a protease inhibitor cocktail (Sigma-Aldrich). Protein lysate normalized to 20 ng total protein was used to measure angiomotin (AMOT) P130, pleckstrin homology domain containing, family H member 3 (PLEKHH3), baculoviral IAP repeat-containing 3 (BIRC3; LGALS3), adenosine deaminase RNA-specific (ADAR1) P150, and galectin-3 internal gene (GALIG) levels by a commercial enzyme-linked immunosorbent assay (ELISA) kit, where AMOT kit was from USCN Business Co. (China), PLEKHH3 kit was from Sunlong (China), BIRC3/ADAR kits were from Cusabio (China), and LGALS3 kit was from R&D Systems (Minneapolis, MN). Briefly, 40 μl dye (Bio-Rad Protein Assay Dye Reagent Concentrate #500–0006) was added to 10 μl bovine serum albumin with serial dilution to generate a standard curve for total protein concentration by measuring values at OD 595nm.
In vitro blood cell culture under IHR conditions
PMBC from six healthy subjects (600 μL per well, and adjusted to 1×106 cells per ml) were exposed to normoxia (NOX) or IHR in a custom-designed, incubation chambers which are attached to an external O2-CO2-N2 computer-driven controller, as described previously[12]. Air-phase set point consisted of a 35-min hypoxic period (0% O2 and 5% CO2), followed by 25 min of re-oxygenation (21% O2 and 5% CO2), using the BioSpherix OxyCycler C42 system (BioSpherix, Redfield, NY), 7 hours each day for 4 days. Control cells were maintained in NOX conditions for the same durations. Previous studies have shown that a 30–40% decreases in blood SaO2 could be achieved in the conditioned media by 25 min of continuous exposure of cells to 0%O2 and 5%CO2[13,14]. Protein expression levels of the five selected genes in the PBMC samples were determined using ELISA method, as described above.
Statistical analysis
Continuous values were presented as the mean ± standard deviation (SD). ANOVA test followed by post hoc analysis with Bonferroni test was used for comparing mean values of more than two experimental groups in case of homogeneous data, while Brown-Forsythe test followed by post hoc analysis with Games-Howell test was used in case of non-homogeneous data. Chi-square tests were used to assess the differences of category values between different groups. In subgroup analyses, multivariate linear regression model was used to adjust for confounding factors, including age, gender, BMI, co-morbidities (hypertension, diabetes mellitus, stroke, heart disease, and CKD), smoking and alcoholism history, and to obtain adjusted p values. Pearson correlation test was used to assess the correlation between two continuous variables. All tests were two tailed and the null hypothesis was rejected at p < 0.05. A statistical software package (SPSS, version 15.0, Chicago, IL) was used for all analyses.
Results
Demographic data of the participants
The baseline, sleep, and biochemistry data of the study cohort 1 and 2 are listed in Table 1. The study population was all residents in Taiwan. Age, BMI, male gender ratio, smoking history, alcoholism history, and co-morbidities were all matched among the four subgroups, except that more patients in the VSOC group had diabetes mellitus than those in the other 3 groups in the study cohort 2.
Table 1. Demographic, sleep, and laboratory data of study participants in the microarray gene expression experiment (study cohort 1 and 2).
| Study Cohort 1 | Study Cohort 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subjects with primary snoring (PS) N = 6 | Moderate-severe OSA patients (MSO) N = 15 |
Very severe OSA patients(VSO) N = 12 | Very severe OSA patients on CPAP (VSOC) N = 15 |
P value | Subjects with primary snoring(PS) N = 16 |
Moderate-severe OSA patients (MSO) N = 18 |
Very severe OSA patients (VSO) N = 18 |
Very severe OSA patients on CPAP (VSOC) N = 16 |
P value | |
| Age, years | 51.0±11.3 | 46.9±11.9 | 47±11.4 | 50.4±8 | 0.592 | 49.7±9.3 | 47.6±11.5 | 47.5±10.2 | 50.4±8.0 | 0.766 |
| Male Sex, n(%) | 4 (66.7) | 12 (80) | 9 (81.8) | 12 (75) | 0.871 | 8 (50) | 14 (77.8) | 16 (88.9) | 12 (75) | 0.075 |
| BMI, kg/m2 | 25.1±2 | 27.2±3.8 | 28±3.6 | 28.4±4.5 | 0.231 | 26.6±3.4 | 27.2±3.8 | 26.9±4.5 | 28.3±4.5 | 0.647 |
| AHI,events/hour | 4.2±2.1 | 34.1±8.8 | 67.4±12.3 | 66.2±24.3 | <0.001 | 3.1±2.5 | 34.8±9.5 | 70.3±12.1 | 64.5±24.5 | <0.001 |
| ODI,events/hour | 2.4±1.4 | 24.9±13.8 | 62.1±11.9 | 58.1±26.4 | <0.001 | 2.1±2.0 | 23.9±13.2 | 59.1±22.1 | 58.1±26.4 | <0.001 |
| Mean SaO2, % | 96.8±0.4 | 95.2±1.9 | 93.3±2.2 | 93.2±3.6 | <0.001 | 97.2±0.7 | 95.3±1.8 | 93.6±2.3 | 93.2±3.6 | <0.001 |
| Minimum SaO2, % | 88.5±1.8 | 78.3±8.5 | 65.5±12.7 | 72±11.3 | <0.001 | 89.3±3.4 | 78.9±8.2 | 67.6±12.9 | 72.0±11.3 | <0.001 |
| Arousal index | 10.9±9.6 | 30.5±24.4 | 46.6±29.5 | 63.6±41.9 | 0.002 | 13.9±10.5 | 32.9±25.1 | 46.9±27.3 | 63.6±41.9 | <0.001 |
| ESS | 12.3±5.9 | 8.7±5.4 | 10.5±4.7 | 14.1±4.7 | 0.026 | 6.8±6.2 | 8.4±5.4 | 11.1±5.1 | 14.1±5.7 | 0.003 |
| Smoking, n(%) | 5 (83.3) | 9 (60) | 6 (54.5) | 11 (68.8) | 0.612 | 2 (14.3) | 7 (38.9) | 7 (38.9) | 5 (31.3) | 0.421 |
| Alcoholism, n(%) | 5 (83.3) | 7 (46.7) | 8 (72.7) | 11 (68.8) | 0.322 | 0 (0) | 2 (11.1) | 2 (11.1) | 0 (0) | 0.315 |
| Cholesterol, mg/dl | 190.2±36.2 | 203.1±47.3 | 174.6±59.3 | 164.4±71.5 | 0.356 | 190.2±39.6 | 187.6±66.4 | 184.6±53.8 | 164.5±71.5 | 0.621 |
| Triglycerides, mg/dl | 130.8±67 | 199±126.4 | 155.5±59.9 | 194.6±151.9 | 0.499 | 120.6±56.8 | 194.8±121.1 | 155.4±60.1 | 194.6±151.9 | 0.153 |
| HDL, mg/dl | 59.8±14.5 | 52.7±10.9 | 49.9±4.7 | 48.8±18.4 | 0.193 | 57.5±11.2 | 49.1±16.1 | 52.7±9.5 | 48.7±13.4 | 0.321 |
| LDL, mg/dl | 106.7±33.6 | 110.1±31.6 | 102.5±31.7 | 100.4±40.1 | 0.956 | 102.2±31.4 | 110.7±33.1 | 107.2±38.1 | 100.4±40.1 | 0.866 |
| Glucose, mg/dl | 96.3±5.8 | 101.3±15.7 | 101.6±12.3 | 107.9±25.5 | 0.355 | 94.8±5.7 | 99.7±15.7 | 97.2±13.7 | 107.9±25.4 | 0.194 |
| Co-morbidity,n(%) | ||||||||||
| Hypertension | 2 (33.3) | 6 (40) | 2 (18.2) | 8 (50) | 0.271 | 4 (25) | 7 (38.9) | 6 (33.3) | 8 (50) | 0.516 |
| Diabetes mellitus | 0 (0) | 0 (0) | 0 (0) | 3 (18.8) | 0.071 | 0 (0) | 0 (0) | 0 (0) | 3 (18.8) | 0.017 |
| Cardiac disease | 1 (16.7) | 1 (6.7) | 1 (9.1) | 2 (12.5) | 0.752 | 2 (12.5) | 2 (11.1) | 2 (11.1) | 2 (12.5) | 0.999 |
| Stroke | 1 (16.7) | 0 (0) | 0 (0) | 0 (0) | 0.067 | 1 (6.3) | 0 (0) | 0 (0) | 0 (0) | 0.348 |
| Chronic kidney disease | 1 (16.7) | 1 (6.7) | 1 (8.3) | 2 (13.3) | 0.384 | 1 (6.3) | 1 (5.6) | 2 (11.1) | 2 (12.5) | 0.86 |
BMI = body mass index; AHI = apnea hypopnea index; ODI = oxygen desaturation index; HDL = high density lipoprotein; LDL = low density lipoprotein
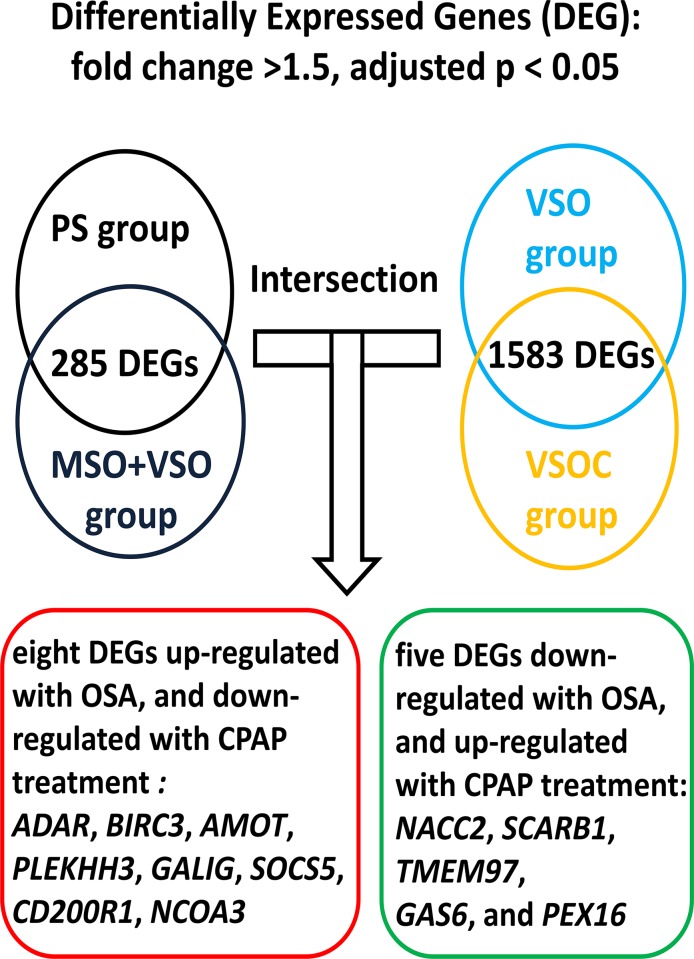
Differentially expressed genes (DEG) identified in the whole-genome gene expression experiment in the validation cohort 1
Microarrays from the 48 subjects that passed the quality-control filters were included in this study. From the 22,150 probes, 285 transcripts were differentially expressed between the treatment-naïve OSA patients (MSO and VSO groups) and PS group using a Mann-Whitney unpaired test (comparison I). To assess the effect of long-term CPAP treatment on PBMC, gene expression profiles in the VSOC group was compared with that in the VSO group, and differential expression of 1583transcripts was identified (comparison II). The intersection of comparison I and II result in eight DEG up-regulated with OSA and down-regulated with CPAP treatment, and five DEG down-regulated with OSA and up-regulated with CPAP treatment (Fig 1 and Table 2). Gene Ontology and gene interaction analyses revealed that AMOT, BIRC3, and ADAR are all involved in anti-apoptosis signaling, and LGALS3 serves as pro-apoptosis molecule, while AMOT, BIRC3, and PLEKHH3 are all involved in angiogenesis or tight junction (Fig 2A and 2B). To further clarify the effects of hypertension or EDS on the gene signature of PBMCs, gene expression profiles in the treatment-naïve OSA patients with hypertension or EDS was compared with those without hypertension or EDS, respectively. Six genes, including AMOT and PLEKHH3, were up-regulated (Table 3) in the treatment-naïve OSA patients versus PS subjects and further up-regulated in OSA patients with EDS as compared to that in those without EDS.LGALS3(fold change 2.23, p = 0.003) and metallopeptidase inhibitor 2 (TIMP2; fold change 4.44, p = 0.043) were up-regulated in the treatment-naïve OSA patients versus PS subjects, and further up-regulated in OSA patients with hypertension(LGALS3: fold change 4.37, p = 0.044; TIMP2; fold change 5.42, p = 0.018) as compared to that in those without hypertension (Table 3).
Fig 1. Flow chart for identifying candidate differentially expressed genes through the intersection of comparison I and II.
Differentially expressed genes were identified from the intersection of the comparisons between treatment-naïve OSA versus PS (comparison I) and treatment-naïve OSA versus OSA patients with long-term CPAP treatment (comparison II).Microarray identified 13 differentially expressed genes (DEG) regulated in the opposite direction for the two comparisons.
Table 2. Differentially expressed genes associated with OSA and reversed with CPAP treatment in the microarray gene expression experiment.
| Gene Name | Probe ID | Treatment-naïve OSAversus PS subjects | VSOC versus VSO patients | Description | ||
|---|---|---|---|---|---|---|
| Fold Change | P value | Fold change | P value | |||
| ADAR | 5130612 | 6.8036 | 0.00561 | 0.29877 | 0.04467 | Adenosine deaminase, RNA-specific variant 4. |
| BIRC3 | 3060255 | 1.5465 | 0.02985 | 0.71495 | 0.04965 | Baculoviral IAP repeat-containing 3, variant 2. |
| AMOT | 3290646 | 2.00353 | 0.01754 | 0.77545 | 0.03214 | Angiomotin, variant 2. |
| PLEKHH3 | 1070709 | 4.23447 | 0.03055 | 0.31054 | 0.04834 | Pleckstrin homology domain containing, family H member 3. |
| LGALS3 | 1660647 | 3.87876 | 0.02832 | 0.29394 | 0.0462 | Galectin-3 internal gene. |
| SOCS5 | 4880750 | 1.51511 | 0.03722 | 0.15028 | 0.00193 | Suppressor of cytokine signaling 5, variant 2. |
| CD200R1 | 4280523 | 2.10105 | 0.02045 | 0.35276 | 0.00151 | CD200 receptor 1, variant 3. |
| NCOA3 | 2760390 | 1.31953 | 0.01321 | 0.73542 | 0.0086 | Nuclear receptor coactivator 3, variant 1. |
| NACC2 | 4220739 | 0.36967 | 0.0464 | 2.5467 | 0.00732 | NACC family member 2, BEN and BTB (POZ) domain containing. |
| SCARB1 | 1170338 | 0.59278 | 0.01744 | 1.6449 | 0.02541 | Scavenger receptor class B, member 1. |
| TMEM97 | 3890561 | 0.70155 | 0.02856 | 1.31361 | 0.01644 | Transmembrane protein 97. |
| GAS6 | 70730 | 0.53071 | 0.0155 | 1.63026 | 0.0131 | Growth arrest-specific 6. |
| PEX16 | 2750358 | 0.57187 | 0.01781 | 1.38059 | 0.01495 | Peroxisomal biogenesis factor 16, variant 2. |
OSA = obstructive sleep apnea; CPAP = continuous positive airway pressure; VSO = very severe OSA patients; VSOC = very severe OSA patients on long-term CPAP treatment; PS = primary snoring subjects
Fig 2. OSA-regulated signaling pathways and gene interactions based on the analyses of the microarray gene expression data.
(A) OSA-regulated signaling pathways were involved in apoptosis, angiogenesis, and tight junctions. Differentially expressed genes up-regulated with OSA and down-regulated with CPAP treatment are shown in green colored circles. (B) Gene interactions among the candidate differentially expressed genes.
Table 3. Differentially expressed genes up-regulated with obstructive sleep apnea (OSA) and further up-regulated with excessive daytime sleepiness (EDS) or hypertension (HT).
| Gene Name | Probe ID | Treatment-naïve OSA versus primary snoring subjects | OSA patients with EDS or HT versus those without EDS or HT | Description | ||
|---|---|---|---|---|---|---|
| Fold Change | P value | Fold change | P value | |||
| Genes up-regulated with both OSA and EDS | ||||||
| AMOT | 3290646 | 2.00353 | 0.01754 | 1.66391 | 0.03214 | Angiomotin, transcript variant 2. |
| LAMB3 | 730040 | 4.29633 | 0.02958 | 3.40345 | 0.0451 | Laminin, beta 3, transcript variant 1. |
| SEC14L2 | 4890341 | 3.76125 | 0.02942 | 3.73724 | 0.04485 | SEC14-like 2 (S. cerevisiae). |
| ITFG3 | 6250553 | 1.34628 | 0.03237 | 1.29703 | 0.04873 | Integrin alpha FG-GAP repeat containing 3. |
| HIF1A | 2680722 | 1.31563 | 0.04622 | 2.62267 | 0.04917 | Hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor), transcript variant 2. |
| PLEKHH3 | 107079 | 4.23447 | 0.03055 | 2.75882 | 0.02976 | Pleckstrin homology domain containing, family H (with MyTH4 domain) member 3. |
| Genes up-regulated with both OSA and HT | ||||||
| LGALS3 | 1660647 | 2.23 | 0.03 | 4.37 | 0.044 | Galectin-3 internal gene. |
| TIMP2 | 5420743 | 4.44 | 0.043 | 5.42 | 0.018 | metallopeptidase inhibitor 2 |
Differential protein expression levels of the five selected genes in the study cohort 2
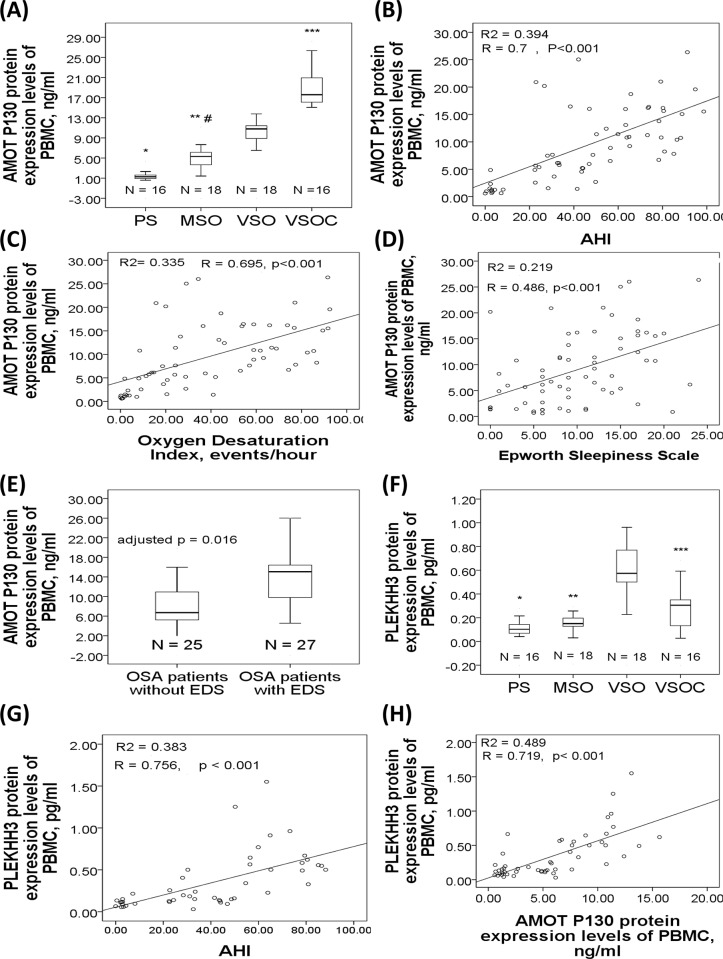
Analysis of the variance revealed significant between-group differences in AMOT (p<0.001), PLEKHH3 (p<0.001), BIRC3 (p = 0.001), ADAR1 (p<0.001), and LGALS3 (p = 0.017) protein expression levels. Post hoc analyses with corrections for multiple comparisons revealed that patients in both the MSO (4.97±1.92 ng/ml, p<0.001) and VSO (10.55±29.72.38 ng/ml, p<0.001) groups had significantly increased AMOT P130 protein expression than the PS group (1.46±1.01 ng/ml), and that patients in the VSO group had higher AMOT P130 protein expression than those in the MSO group (p<0.001), while patients in the VSOC group (19.1±3.86 ng/ml, p<0.001) had even higher AMOT P130 protein expression than those in the VSO group (Fig 3A).AMOT P130 protein expression was positively correlated with AHI (r = 0.7, p<0.001, Fig 3B), ODI (r = 0.695, p<0.001, Fig 3C), percentage time with SaO2<90% (r = 0.585, p<0.001), and ESS (r = 0.476, p<0.001, Fig 3D). Subgroup analysis showed that OSA patients with EDS had significantly increased AMOT P130 protein expression than those without EDS (14.05±6.19 versus 8.23±5.22 ng/ml, adjusted p = 0.016, Fig 3E). PLEKHH3 protein expression in the VSO group (0.67±0.33 pg/ml) was significantly increased as compared to that either in the PS (0.15±0.16 pg/ml, p<0.001), MSO (0.18±0.11 pg/ml, p<0.001), or VSOC (0.29±0.17 pg/ml, p<0.001) group (Fig 3F).PLEKHH3 protein expression level was positively correlated with AHI (r = 0.737, p<0.001, Fig 3G), ODI (r = 0.681, p<0.001), percentage time with SaO2<90% (r = 0.631, p<0.001), and AMOT P130 protein expression (r = 0.719, p<0.001, Fig 3H) in all the treatment-naïve subjects (PS, MSO, and VSO groups).
Fig 3. AMOT P130 and PLKHH3 protein expression levels in the study cohort 2.
(A) AMOT P130 protein expression was increased in treatment-naïve moderate to severe (MSO) and very severe OSA (VSO) patients as compared to that in primary snoring (PS) subjects, and further increased in very severe OSA patients with long-term CPAP treatment (VSOC). AMOT P130 protein expression was positive correlated with (B) apnea hypopnea index, (C) oxygen desaturation index, and (D) Epworth Sleepiness Scale. (E) AMOT P130 protein expression was increased in OSA patients with excessive daytime sleepiness. (F) PLEKHH3 protein expression was increased in the VSO group and decreased in the VSOC group. PLEKHH3 protein expression was positively correlated with (G) AHI and (H) AMOT P130 protein expression. *p<0.001 compared between the VSO and PS groups. **P<0.001, compared between the VSO and MSO groups. ***P<0.001, compared between the VSO and VSOC groups. #p<0.001, compared between the MSO and PS groups.
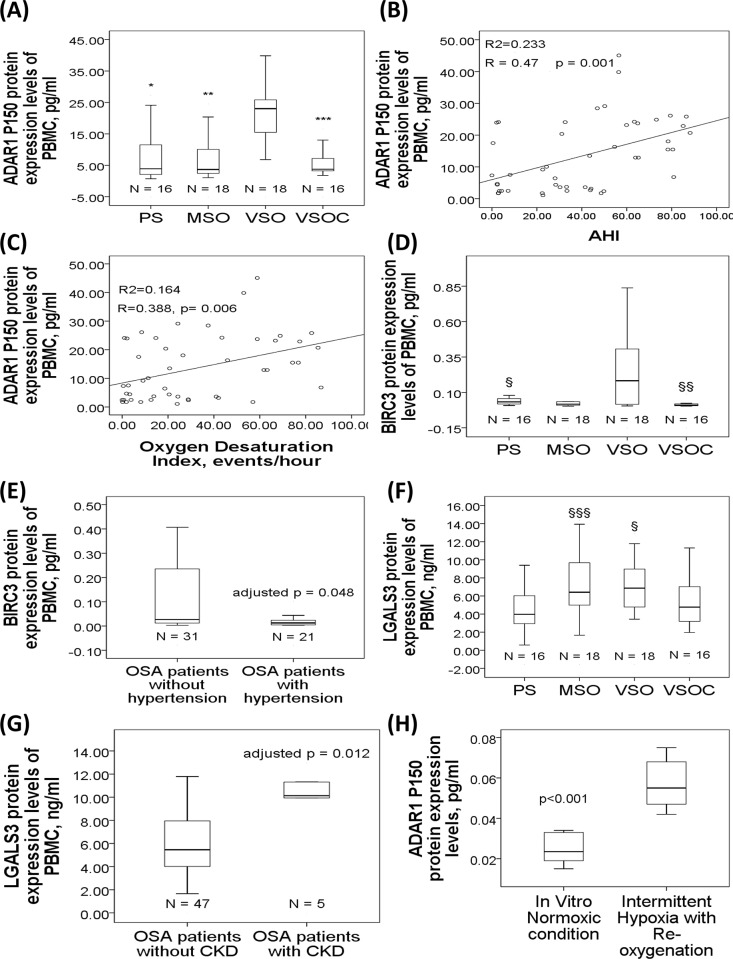
ADAR1 P150 protein expression level was significantly increased in the VSO group (22.42±9.33 pg/ml) as compared to that either in the PS (7.6±8.03 pg/ml, p<0.001), MSO (7.92±8.33 pg/ml, p<0.001), or VSOC (6.29±6.83 pg/ml, p<0.001) group (Fig 4A). ADAR1 P150 protein expression level was positively correlated with AHI (r = 0.47, p = 0.001, Fig 4B) and ODI(r = 0.388, p = 0.006, Fig 4C). BIRC3 protein expression was significantly increased in the VSO group (0.31±0.39 pg/ml) as compared to that in the PS (0.05±0.05 pg/ml, p = 0.031) or VSOC (0.03±0.05 pg/ml, p = 0.033) group (Fig 4D). Subgroup analysis showed that OSA patients with hypertension had significantly deceased BIRC3 protein expression levels as compared to those without hypertension (0.04±0.06 versus 0.21±0.33 pg/ml, adjusted p = 0.048, Fig 4E). LGALS3 protein expression level was significantly increased in both the MSO (7.04±3.18 ng/ml, p = 0.036) and VSO groups (4.52±1.07 ng/ml, p = 0.035) as compared to that in the PS group (4.24±2.35 ng/ml) (Fig 4F). Subgroup analysis showed that OSA patients with CKD had significantly increased LGALS3 protein expression levels than those without CKD (10.67±3.51 versus 6.4±3.44 ng/ml, adjusted p = 0.012, Fig 4G).
Fig 4. ADAR1 P150, BIRC3, and LGALS3 protein expression levels in the study cohort 2.
(A) ADAR1 P150 protein expression was increased in the VSO group, and decreased in the VSCO group. ADAR1 P150 protein expression was positively correlated with (B) AHI and (C) ODI. (D) BIRC3 protein expression was increased in the VSO group and decreased in the VSOC group. (E) BIRC3 protein expression was decreased in OSA patients with hypertension as compared to that in those without hypertension. (F) LGALS3 protein expression was increased in the MSO and VSO groups as compared to that in the PS group. (G) LGALS3 protein expression was increased in OSA patients with chronic kidney disease. (H) ADAR1 P150 protein expression was increased under short-term in vitro intermittent hypoxia with re-oxygenation condition. §p<0.05, compared between the VSO and PS groups. §§p<0.05, compared between the VSO and VSOC groups. §§§p<0.05, compared between the MSO and PS groups.
Effects of in vitro IHR on protein expressions of the five selected genes
To determine whether IHR per se can affect expressions of the five selected genes, PBMC from six healthy subjects were exposed in vitro to either 7 cycles of IHR per day for 4 days or 4 days of continuous normoxic condition. Short-term IHR treatment in vitro resulted in a significant increase of ADAR1 protein expression (0.057±0.013 versus 0.025±0.008, p value<0.001, Fig 4H) as compared with normoxic condition, whereas no significant change was found for the other 4 protein expressions.
Discussion
In this study, we identified a novel association between several genes related to apoptosis or endothelial function and OSA or its clinical phenotypes through whole-genome gene expression microarray analyses. Moreover, we verified AMOT, PLEKHH3, ADAR, BIRC3, and LGALS3 protein over-expressions in the treatment-naïve OSA patients, and discovered a correlation between AMOT/BIRC3/LGALS3 protein expression and the presence of EDS/hypertension/CKD, respectively. Furthermore, we found an immediate effect of in vitro IHR on ADAR1 P150 protein over-expression in a cell culture model.
Endothelial tight junctions form a seal between polar cells, isolating the lumen of the blood vessel from the surrounding tissue and restricting the diffusion of solutes from the blood to the surrounding cells. AMOT is a trans-membrane receptor for the angiostatic factor angiostatin. Alternative splicing of AMOT mRNA results in two protein isoforms (p80 and p130), which exert very distinct roles during angiogenesis[15]. AMOT p80 stimulates endothelial cell migration and angiogenesis in response to angiostatin, whereas AMOT p130 localizes to actin and tight junctions[16,17]. AMOT is also involved in the maturation of epithelial tight junctions as part of the zonular signalosome formation[17,18]. In this study, AMOT gene and P130 protein expressions were both up-regulated in the treatment-naïve OSA patients, whereas AMOT gene expression was down-regulated and P130 protein expression was further up-regulated in the OSA patients with long-term CPAP treatment. We speculate that post-transcriptional modifications, maturation, alternative splicing, and degradation processes, may contribute to this discrepancy. The further increase of AMOT P130 protein expression in the VSOC group may be attributed to long-term air flow stimulation of upper airway epithelial cells with CPAP use. In line with our findings, AMOT expression has been shown to be a good indicator of plasticity of the vascular network in skeletal muscle, and P130 expression can be reduced with exercise training in an obese rat model[19].
Pleckstrin homology (PH) domains family function as a versatile protein–protein interaction platform and are integrated in an increasing number of available multidomain structures[20]. For example, PLEKHA7 is a recently identified protein of the epithelial zonula adhaerens, and stabilizes it by modulating the dynamics of assembly and disassembly of the tight junction barrier, through E-cadherin protein complex- and microtubule-dependent mechanisms[21]. Zonula adhaerens are topologically associated with tight junctions in the apical junctional complex at the apicolateral border of both epithelial and endothelial polar cells. In this study, we found a positive correlation between AHI and PLEKHH3 expression level, which was reversed with CPAP treatment. We think that PLEKHH3, another PH family member, may play a pivotal role in chronic IHR-related endothelial dysfunction in OSA patients through regulating tight junction formation of endothelial cells specifically.
Anti-apoptotic marker, BIRC3, has been shown to mediate the pro-survival and inflammatory responses induced by the docosahexaenoic acid / neuroprotectin D1 pathway under oxidative stress in an ischemia-reperfusion stroke model[22].BIRC3 has been demonstrated to be a downstream effector of HIF-1 signaling involved in the survival response of endothelial cells to hypoxia[23]. BIRC3 is also a negative regulator of the non-canonical NF-κB signaling pathway and mutated primarily in patients with aggressive chronic lymphocytic leukemia[24]. In this study, BIRC3 was up-regulated in the treatment-naive OSA patients, and down-regulated with CPAP treatment. BIRC3 under-expression was associated with the occurrence of hypertension in OSA patients. We speculate that BIRC3 may play a role in protecting from hypertension in OSA patients through inhibiting endothelial cell apoptosis and NF-κB signaling.
Adenosine deaminase acting on RNA1 (ADAR1) catalyzes the C6 deamination of adenosine (A) to produce inosine (I) in regions of double-stranded RNA, known as A-to-I RNA editing. Alternative splicing gives rise to transcripts that encode twoADAR1 protein size isoforms. ADAR1 p150 is an interferon-inducible dsRNA adenosine deaminase found in the cytoplasm and nucleus, mounting pro-viral and anti-apoptotic responses, whereas ADAR1 p110 is constitutively expressed in the nucleus[25]. Among the biologically relevant substrates of ADAR1 that result in amino acid coding changes following editing are transcripts for the 5-HT2c-R neurotransmitter receptor for serotonin[26]. In recent years, this modification has been discovered to occur not only in coding RNAs but also in non-coding RNAs, such as microRNAs, small interfering RNAs, transfer RNAs, and long non-coding RNAs[27].The malfunction of this editing machinery is associated with various human diseases, such as neurodegenerative, cardiovascular, and carcinogenic diseases. For the first time, we found an association between ADAR1 up-regulation and AHI, and demonstrated a direct link between IHR and ADAR1 P150 protein over-expression in vitro.
LGALS3 (GALIG) is a novel cell death gene encoding mitogaligin, a protein promoting cytochrome c release upon direct interaction with the mitochondria or nucleus[28,29].LGALS3 pro-apoptotic gene is up-regulated during neutrophils apoptosis and under-expressed in acute myeloid leukemia cells[30]. In this study, we found that LGALS3 was up regulated in moderate to very severe OSA patients, especially in those with CKD. Impaired renal function has been observed in OSA patients with metabolic syndrome, hypertension or heart failure, possibly through reduced endothelial nitricoxide synthase expression[31,32]. Our findings suggest that LGALS3 may play a crucial role in mediating renal dysfunction in OSA patients.
There are several limitations to the present study. First, the cause and effect relationship could not be determined in this cross-sectional clinical study design, and further studies are required to elucidate underlying mechanisms for these five novel biomarkers. However, the in vitro experiment demonstrated immediate over-expression of ADAR1 P150 under short-term IHR stimuli. Second, gene expression levels of the five selected genes were not examined in the PBMC samples because of inadequate RNA samples. However, their protein expressions showed corresponding changes, indicating a functional role of these novel genes in mediating pathogenesis of OSA and its adverse consequences. Third, the sample size of each subgroup is relatively small, and many confounding factors may affect the expression levels. Further studies with sufficiently large sample sizes are required for the internal and external validity and the reliability of the results.
In summary, we reported a novel association of increased AHI in OSA patients of Asian origin with over-expressions of the AMOT, PLEKHH3, ADAR1, BIRC3, and LGALS3 genes in blood immune cells. The findings extend reports linking AMOT with EDS in OSA patients, and provide direct evidence that perturbation of BIRC3 and LGALS3 signaling may play an important role in the mediation of hypertension and CKD in OSA patients, respectively.
Acknowledgments
The authors acknowledge the technical support provided by the Genomic and Proteomic Core Laboratory, and the Internal Medicine Core Facility of the Kaohsiung Chang Gung Memorial Hospital. We also acknowledge the support of bioinformatics analysis from Professor Petrus Tang, PhD (Molecular Medicine Research Center, and Bioinformatics Center of Chang Gung University, Taiwan).
Data Availability
All expression dataset has been deposited in the NCBI Gene Expression Omnibus with accession number of GSE75097.
Funding Statement
This work was supported by grants from the Ministry of Science and Technology, Taiwan (NMRPG8D6131/103-2314-B-182A-094-MY2 & NMRPG8B6192/101-2314-B-182A-130-MY2 to M.C. Lin) and grants from Chang Gung Memorial Hospital (CMRPG8C1181-1182 to M.C. Lin), Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marin JM, Agusti A, Villar I, Forner M, Nieto D, et al. (2012) Association between treated and untreated obstructive sleep apnea and risk of hypertension. JAMA 307: 2169–2176. 10.1001/jama.2012.3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jennum P, Tonnesen P, Ibsen R, Kjellberg J (2015) All-cause mortality from obstructive sleep apnea in male and female patients with and without continuous positive airway pressure treatment: a registry study with 10 years of follow-up. Nat Sci Sleep 7: 43–50. 10.2147/NSS.S75166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Wang Y, Guan J, Yi H, Yin S (2015) Effect of CPAP on Endothelial Function in Subjects With Obstructive Sleep Apnea: A Meta-Analysis. Respir Care 60: 749–755. 10.4187/respcare.03739 [DOI] [PubMed] [Google Scholar]
- 4.Dyugovskaya L, Lavie P, Lavie L (2002) Increased adhesion molecules expression and production of reactive oxygen species in leukocytes of sleep apnea patients. Am J Respir Crit Care Med 165: 934–939. 10.1164/ajrccm.165.7.2104126 [DOI] [PubMed] [Google Scholar]
- 5.Dyugovskaya L, Polyakov A, Cohen-Kaplan V, Lavie P, Lavie L (2012) Bax/Mcl-1 balance affects neutrophil survival in intermittent hypoxia and obstructive sleep apnea: effects of p38MAPK and ERK1/2 signaling. J Transl Med 10: 211 10.1186/1479-5876-10-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann MS, Singh P, Wolk R, Romero-Corral A, Raghavakaimal S, et al. (2007) Microarray studies of genomic oxidative stress and cell cycle responses in obstructive sleep apnea. Antioxid Redox Signal 9: 661–669. 10.1089/ars.2007.1589 [DOI] [PubMed] [Google Scholar]
- 7.Khalyfa A, Capdevila OS, Buazza MO, Serpero LD, Kheirandish-Gozal L, et al. (2009) Genome-wide gene expression profiling in children with non-obese obstructive sleep apnea. Sleep Med 10: 75–86. 10.1016/j.sleep.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 8.Khalyfa A, Gharib SA, Kim J, Dayyat E, Snow AB, et al. (2010) Transcriptomic analysis identifies phosphatases as novel targets for adenotonsillar hypertrophy of pediatric obstructive sleep apnea. Am J Respir Crit Care Med 181: 1114–1120. 10.1164/rccm.200909-1398OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song CM, Lee CH, Rhee CS, Min YG, Kim JW (2012) Analysis of genetic expression in the soft palate of patients with obstructive sleep apnea. Acta Otolaryngol 132 Suppl 1: S63–68. [DOI] [PubMed] [Google Scholar]
- 10.Berry RB, Budhiraja R, Gottlieb DJ, Gozal D, Iber C, et al. (2012) Rules for scoring respiratory events in sleep: update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. Deliberations of the Sleep Apnea Definitions Task Force of the American Academy of Sleep Medicine. J Clin Sleep Med 8: 597–619. 10.5664/jcsm.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YC, Hsiao CC, Chen KD, Hung YC, Wu CY, et al. (2013) Peripheral immune cell gene expression changes in advanced non-small cell lung cancer patients treated with first line combination chemotherapy. PLoS One 8: e57053 10.1371/journal.pone.0057053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YC, Su MC, Liou CW, Liu SF, Chen CJ, et al. (2015) Co-upregulation of Toll-like receptors 2 and 6 on peripheral blood cells in patients with obstructive sleep apnea. Sleep Breath 19: 873–882. 10.1007/s11325-014-1116-4 [DOI] [PubMed] [Google Scholar]
- 13.Dyugovskaya L, Polyakov A, Lavie P, Lavie L (2008) Delayed neutrophil apoptosis in patients with sleep apnea. Am J Respir Crit Care Med 177: 544–554. 10.1164/rccm.200705-675OC [DOI] [PubMed] [Google Scholar]
- 14.Dyugovskaya L, Polyakov A, Ginsberg D, Lavie P, Lavie L (2011) Molecular pathways of spontaneous and TNF-{alpha}-mediated neutrophil apoptosis under intermittent hypoxia. Am J Respir Cell Mol Biol 45: 154–162. 10.1165/rcmb.2010-0025OC [DOI] [PubMed] [Google Scholar]
- 15.Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, et al. (2005) Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem 280: 34859–34869. 10.1074/jbc.M503915200 [DOI] [PubMed] [Google Scholar]
- 16.Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, et al. (2006) p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J 273: 2000–2011. 10.1111/j.1742-4658.2006.05216.x [DOI] [PubMed] [Google Scholar]
- 17.Citi S, Guerrera D, Spadaro D, Shah J (2014) Epithelial junctions and Rho family GTPases: the zonular signalosome. Small GTPases 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, et al. (2011) A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19: 527–540. 10.1016/j.ccr.2011.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roudier E, Chapados N, Decary S, Gineste C, Le Bel C, et al. (2009) Angiomotin p80/p130 ratio: a new indicator of exercise-induced angiogenic activity in skeletal muscles from obese and non-obese rats? J Physiol 587: 4105–4119. 10.1113/jphysiol.2009.175554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheffzek K, Welti S (2012) Pleckstrin homology (PH) like domains—versatile modules in protein-protein interaction platforms. FEBS Lett 586: 2662–2673. 10.1016/j.febslet.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 21.Paschoud S, Jond L, Guerrera D, Citi S (2014) PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers 2: e28755 10.4161/tisb.28755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calandria JM, Asatryan A, Balaszczuk V, Knott EJ, Jun BK, et al. (2015) NPD1-mediated stereoselective regulation of BIRC3 expression through cREL is decisive for neural cell survival. Cell Death Differ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martorell L, Gentile M, Rius J, Rodriguez C, Crespo J, et al. (2009) The hypoxia-inducible factor 1/NOR-1 axis regulates the survival response of endothelial cells to hypoxia. Mol Cell Biol 29: 5828–5842. 10.1128/MCB.00945-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Y, Fan L, Wang L, Gale RP, Wang M, et al. (2015) Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget 6: 5426–5434. 10.18632/oncotarget.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George CX, John L, Samuel CE (2014) An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1). J Interferon Cytokine Res 34: 437–446. 10.1089/jir.2014.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaniewska M, Alenina N, Wydra K, Frohler S, Kusmider M, et al. (2015) Discovering the mechanisms underlying serotonin (5-HT) and 5-HT receptor regulation following nicotine withdrawal in rats. J Neurochem. [DOI] [PubMed] [Google Scholar]
- 27.Nigita G, Veneziano D, Ferro A (2015) A -to-I RNA Editing: Current Knowledge Sources and Computational Approaches with Special Emphasis on Non-Coding RNA Molecules. Front Bioeng Biotechnol 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duneau M, Boyer-Guittaut M, Gonzalez P, Charpentier S, Normand T, et al. (2005) Galig, a novel cell death gene that encodes a mitochondrial protein promoting cytochrome c release. Exp Cell Res 302: 194–205. 10.1016/j.yexcr.2004.08.041 [DOI] [PubMed] [Google Scholar]
- 29.Robinet P, Mollet L, Gonzalez P, Normand T, Charpentier S, et al. (2010) The mitogaligin protein is addressed to the nucleus via a non-classical localization signal. Biochem Biophys Res Commun 392: 53–57. 10.1016/j.bbrc.2009.12.162 [DOI] [PubMed] [Google Scholar]
- 30.Mollet L, Robinet P, Dubois M, Aurouet A, Normand T, et al. (2013) Opposing Mcl-1, the GALIG proapoptotic gene is upregulated as neutrophils die and underexpressed in Acute Myeloid Leukemia cells. Mol Immunol 56: 123–128. 10.1016/j.molimm.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 31.Uyar M, Davutoglu V, Gundogdu N, Kosovali D, Sari I (2015) Renal functions in obstructive sleep apnea patients. Sleep Breath. [DOI] [PubMed] [Google Scholar]
- 32.Bruno RM, Rossi L, Fabbrini M, Duranti E, Di Coscio E, et al. (2013) Renal vasodilating capacity and endothelial function are impaired in patients with obstructive sleep apnea syndrome and no traditional cardiovascular risk factors. J Hypertens 31: 1456–1464; discussion 1464. 10.1097/HJH.0b013e328360f773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All expression dataset has been deposited in the NCBI Gene Expression Omnibus with accession number of GSE75097.