Abstract
The secreted cysteine proteinase SpeB is an important virulence factor of group A streptococci (GAS), whereby SpeB activity varies widely among strains. To establish the degree to which SpeB activity correlates with disease, GAS organisms were recovered from patients with pharyngitis, impetigo, invasive disease or acute rheumatic fever (ARF), and selected for analysis using rigorous sampling criteria; >300 GAS isolates were tested for SpeB activity by casein digestion assays, and each GAS isolate was scored as a SpeB-producer or non-producer. Highly significant statistical differences (p < 0.01) in SpeB production are observed between GAS recovered from patients with ARF (41.5% SpeB-non-producers) compared to pharyngitis (20.5%), invasive disease (16.7%), and impetigo (5.5%). SpeB activity differences between pharyngitis and impetigo isolates are also significant, whereas pharyngitis versus invasive isolates show no significant difference. The disproportionately greater number of SpeB-non-producers among ARF-associated isolates may indicate an altered transcriptional program for many rheumatogenic strains and/or a protective role for SpeB in GAS-triggered autoimmunity.
Introduction
Group A Streptococcus (GAS) is a human pathogen of global importance that most often causes a superficial self-limiting infection at the throat (pharyngitis) or skin (impetigo), leading to ~750 million infections per year [1]. GAS is also associated with high rates of morbidity and mortality due to invasive and autoimmune disease, although these conditions are far less prevalent. Acute rheumatic fever (ARF) follows an inadequately treated GAS throat infection by a so-called "rheumatogenic" strain, and can often lead to rheumatic heart disease via autoimmune attack of heart valves [2, 3]. The existence of distinct "rheumatogenic" and "non-rheumatogenic" strains of GAS has been long recognized [4–7], but their distinguishing properties are not well-understood. Numerous other GAS strains are often considered to be “disease specialists” [8–12].
The secreted cysteine protease SpeB is a key virulence factor of GAS that acts by degrading human proteins having a protective role in host defense; SpeB also targets many extracellular proteins produced by GAS (reviewed in [13, 14]). Although virtually all GAS isolates harbor a speB gene, SpeB activity can vary widely among strains; speB expression is modulated by several global regulators of transcription and importantly, it is tightly coordinated with expression of a large number of other genes (reviewed in [15]).
Studies by one group of investigators show that SpeB activity correlates with GAS disease, whereby a substantial fraction (~41%) of severe invasive disease isolates harboring emm1 (M-type 1 or M1) fail to produce SpeB in vitro [16]. A direct role in the transition from localized to invasive disease has been elucidated for (a lack of) SpeB and/or co-transcribed genes [13, 17–19]. In a mouse model for invasive disease, mutants with defects in a two-component regulatory system (CovRS) and having decreased SpeB production are more virulent [20]. Possible conflicting findings on the association of SpeB activity with GAS recovered from patients with invasive disease—both the mild and severe forms combined—have also been reported. In this more recent study, the vast majority (>80%) of both invasive and pharyngitis emm1 isolates are SpeB-producers, as are invasive isolates of three other emm types (emm28, emm59, emm89), and there is no significant difference between the invasive versus pharyngitis isolates in terms of SpeB activity [21].
In this report, the relative distribution of a SpeB-producer phenotype is measured for 322 GAS isolates recovered from patients with pharyngitis, impetigo, invasive disease or ARF. Strain sampling follows a strict set of criteria that aims to be representative of the organisms found within patient populations (i.e., population-based). Importantly, the sampling criteria also captures a very wide range of genetic diversity among GAS isolates, as defined by the emm gene marker.
Results
Diversity and characteristics of the GAS strain populations
Study sample sets of GAS isolates associated with four distinct clinical conditions were assembled: ARF, pharyngitis, impetigo and invasive disease. All ARF and pharyngitis isolates were recovered from the upper respiratory tract (URT) of human subjects. Strain sampling followed strict and well-defined criteria, with the goals of assembling a genetically diverse and representative set (Table 1; Supplementary Data S1 Table). Selection of a small number of isolates sharing the same emm type and recovered from the same community reduces the potential skewing effects of highly prevalent clones; this was done for ARF and impetigo isolates, and one pharyngitis collection. All ARF, pharyngitis and invasive disease isolates were recovered from the United States, whereas impetigo isolates had a worldwide distribution.
Table 1. Characteristics and diversity of GAS isolates under analysis.
| Disease group | No. of isolates | Dates | Geographic origin | No. of emm types represented | D, based on emm type a | No. of emm subtypes represented | D, based on emm subtype |
|---|---|---|---|---|---|---|---|
| Pharyngitis | 78 | 2001–2002 | Bristol, CT | 19 | 0.8998 | 27 | 0.9261 |
| Pharyngitis | 68 | 2012 | Chicago, IL | 14 | 0.9166 | 16 | 0.9289 |
| Total pharyngitis | 146 | n/a | n/a | 21 | 0.9125 | 34 | 0.9369 |
| Impetigo | 58 | 1994–1996 | Australia | 28 | 0.9782 | 29 | 0.9794 |
| Impetigo | 16 | 1971–1988 | Worldwide | 16 | 1.0000 | 16 | 1.0000 |
| Total impetigo | 74 | n/a | n/a | 39 | 0.9852 | 41 | 0.9863 |
| Invasive | 60 | 1995 | CT | 20 | 0.8667 | n.d. | n.d. |
| ARF | 42 | 1933–1989 | USA | 20 | 0.9489 | 34 | 0.9907 |
| Total isolates | 322 | n/a | n/a | 69 | 0.9530 | 96 | 0.9811 |
a D, Simpson's diversity index
The genetic markers used for analysis of strain diversity are emm type and emm subtype. Data show that the sampled selections of GAS isolates display very high levels of genetic diversity, with Simpson diversity index (D) values approaching one, signifying that most isolates are distinct (Table 1). The relative diversity D values show impetigo > ARF > pharyngitis > invasive isolates (based on emm type), and ARF > impetigo > pharyngitis isolates (based on emm subtype; emm subtype was not determined for invasive isolates).
For the 322 GAS isolates under study, 69 distinct emm types are represented (Table 1), accounting for ~30% of the known emm types of the Streptococcus pyogenes species [22]. Of the 42 ARF-associated isolates, 57% harbor an emm type that is shared with the pharyngitis isolates (S2 Table), consistent with ARF having extensive overlap with other URT strains. Of the 60 invasive disease isolates (from CT in 1995), 88% have an emm type that is also shared with the pharyngitis isolates, consistent with invasive disease isolates being largely reflective of the prevailing pharyngitis emm types within a region [23, 24]. In sharp contrast, only 19% of the 74 impetigo isolates have an emm type that is shared with the pharyngitis isolates, and even fewer have an emm type present among the ARF-associated (9%) or CT invasive disease (9%) isolates (S2 Table).
Putative rheumatogenic M protein types in the United States include M-types 1, 3, 5, 6, 14, 18, 19, 24, 27 and 29 [6]. These 10 emm types account for 24 (57%) of the 42 ARF-associated and 59 (40%) of the 146 pharyngitis isolates examined (S1 Table), which is a slight but statistically non-significant enrichment of putative rheumatogenic emm types among the ARF-associated isolates. Collectively, the 10 putative rheumatogenic emm types account for 62% of the invasive disease isolates, similar to the value for ARF-associated isolates but with a strikingly different distribution for emm types 1 and 18, whereby emm1 is highly enriched in the invasive disease set (representing 30% of invasive disease isolates) and emm18 is enriched in the ARF-associated pool (representing 14% ARF isolates). Only 4% of the impetigo isolates had a putative rheumatogenic emm-type as defined by [6].
Clinical correlates of SpeB activity
SpeB activity was measured by digestion of casein following growth of GAS on agar (Table 2, S1 Table). For Columbia agar containing skim milk powder (Columbia-SM), data show highly significant statistical differences (p < 0.01; Fisher's exact test, 2-tailed) for SpeB-non-producer isolates recovered from patients with pharyngitis (20.5% non-producers) versus ARF (41.5%) or impetigo (5.5%). However, the difference in SpeB production for invasive (16.7% non-producers) versus pharyngitis isolates is non-significant. Data also show significant statistical differences for SpeB-non-producer isolates recovered from patients with ARF versus invasive disease (p = 0.011) or impetigo (p < 0.0001).
Table 2. SpeB activity phenotype correlates with GAS disease.
| Columbia agar with skim milk a | C-broth agar with casein (extrapolated values) c | |||||||
|---|---|---|---|---|---|---|---|---|
| Disease group | No. of SpeB- producers | No. of SpeB- non-producers | % SpeB- non-producers | p value, pharyngitis versus b: | No. of SpeB- producers | No. of SpeB-non-producers | % SpeB- non-producers | p value, pharyngitis versus b: |
| Pharyngitis | 116 | 30 | 20.5 | n/a | 106 | 32 | 23.7 | n/a |
| Impetigo | 69 | 4 | 5.5 | 0.0029 | 69 | 4 | 5.5 | 0.0009 |
| Invasive | 50 | 10 | 16.7 | N.S. | 44 | 12 | 21.4 | N.S. |
| ARF | 24 | 17 | 41.5 | 0.0085 | 20 | 20 | 50.0 | 0.0016 |
a One ARF and one impetigo isolate had intermediate ("weak") zones of clearance on Columbia-SM agar (S1 Table), and are excluded from the calculations.
b Fisher's exact test, 2-tailed; N.S., non-significant.
c Data extrapolations for the complete set of GAS organisms are made for impetigo and pharyngitis isolates (italics). Organisms with intermediate ("weak") zones of clearance on CBrothMg-C agar (S1 Table) are excluded from the calculations.
The ARF-associated isolates were collected over a period extending >5 decades. Of the 15 isolates collected before the widespread use of penicillin (pre-1950), 33% (5 of 15) were SpeB-non-producers on Columbia-SM agar, as compared to 46% (12 of 26) of isolates collected between 1950 and 1989; this slight distinction between collection periods is not statistically significant. Similarly, 33% of the ARF-associated isolates collected during the 1980s were SpeB non-producers, as compared to 45% collected prior to 1980 (p = N.S.). Thus, there are no apparent effects of widespread antibiotic usage, long-term laboratory storage and/or shifting epidemiologic patterns on SpeB production by the sample set of ARF-associated organisms.
Of the 74 impetigo isolates, 58 originate from an Aboriginal Australian community located within a larger geographic region that is well-known for its high prevalence of rheumatic heart disease [25, 26]. Of the Australian GAS isolates, 7% were SpeB non-producers on Columbia-SM agar, as compared to none of the 16 impetigo isolates originating from worldwide sources (S1 Table); however, the difference between the two impetigo collections was not statistically significant (p = 0.5699; Fisher’s exact test, 2-tailed). The extent to which an impetigo infection triggers ARF (if at all) is unresolved [3], and the idea remains somewhat speculative. Importantly, the data show that Aboriginal Australian impetigo isolates do not resemble the ARF-associated URT isolates recovered from the United States insofar as SpeB phenotype. It can be difficult to disentangle geography from GAS disease since impetigo is primarily a disease of the tropics, and URT infection is highly prevalent in temperate regions (S2 Table).
Nine of the invasive disease isolates studied were recovered from patients with "severe" disease (i.e., streptococcal toxic shock syndrome, necrotizing fasciitis and/or pyomyositis) [23]. However, only one of these isolates (11%) failed to produce SpeB on Columbia-SM agar (data not shown). Of the invasive disease isolates recovered from the blood, 19% were SpeB-non-producers (data not shown), a value that reflects the larger set of invasive disease strains. Thus, there is no clear evidence for a skewed distribution of SpeB non-producers among clinically-defined subsets of invasive disease isolates, albeit the sample sizes are rather small.
To establish that caseinolytic activity is due to SpeB, and not attributable to other proteases, E64 (a SpeB-specific inhibitor) was added to Columbia-SM agar. All 40 SpeB producer strains tested lack caseinolytic activity in the presence of 10 μM E64 (S1 Table).
C-Broth-based agar containing casein (CBrothMg-C agar) was also used to test for SpeB activity, whereby C-Broth is an optimized formulation for high levels of SpeB production that is protein-rich and carbohydrate-poor [27]. With CBrothMg-C agar, the differences in casein digestion for GAS recovered from pharyngitis versus ARF or impetigo are even more highly significant than that observed for Columbia-SM agar (extrapolated values; Table 2). Data show highly significant statistical differences for SpeB-non-producer isolates recovered from patients with ARF (50%) versus invasive disease (21.4% non-producers; p = 0.0045, Fisher's exact test, 2-tailed) or impetigo (5.5%; p < 0.0001). However, differences between pharyngitis versus invasive disease isolates remain non-significant with CBrothMg-C agar. Although the initial goal in using C-broth-based agar was to further maximize SpeB production, this proved not to be the case: 81% of GAS produced SpeB on Columbia-SM agar, as compared to slightly fewer (78%) isolates on CBrothMg-C agar (Table 2).
Strain diversity within each clinically distinct sub-group of SpeB-producers and non-producers (on Columbia-SM agar) is extensive. The 17 ARF-associated SpeB-non-producers are represented by 11 distinct emm types and 15 emm subtypes (S1 Table); similarly, the 24 ARF-associated SpeB-producers are represented by 13 distinct emm types and 21 emm subtypes. The 10 SpeB-non-producers recovered from cases of invasive disease are also highly diverse, represented by seven distinct emm types. The 30 SpeB-non-producers recovered from patients with pharyngitis are somewhat less diverse than their ARF and invasive counterparts, represented by 10 distinct emm types and 14 emm subtypes. Taken together, a lack of SpeB activity is observed for strains representing a wide range of genotypes for each of the clinical disease groups.
Concordance between different casein digestion assays
The concordance of SpeB phenotypes was examined for GAS strains tested on multiple culture medium types. For 35 GAS isolates that were SpeB-non-producers on Columbia-SM agar, nearly all (33, or 94%) were also SpeB-non-producers when tested on CBrothMg-C agar; one non-producer strain displayed intermediate zones of clearance on CBrothMg-C agar. For 166 isolates that were SpeB-producers on Columbia-SM agar, when tested on CBrothMg-C agar, 91% were also SpeB-producers, 5% had intermediate zones of clearance, and 4% were non-producers. Overall, 201 isolates were tested on both Columbia-SM and CBrothMg-C agar, and 91% were concordant in their SpeB findings. Excluding strains with intermediate zones of clearance, overall concordance between Columbia-SM and CBrothMg-C agar for SpeB production was 98.5%.
SpeB activity was also compared for 152 strains grown both on Columbia-SM agar and in C-Broth liquid medium, via the azocasein assay [27]. Using a cutoff for SpeB-positivity of 3% azocasein digestion activity, relative to a reference strain (Alab49; impetigo isolate) having high levels of SpeB activity, the two assays were concordant for 150 (98.7%) of the isolates tested. The range of % azocasein digestion activity was rather wide among GAS isolates that produced SpeB and shared the same emm type (S3 Table). Taken together, the three assays—Columbia-SM agar, CBrothMg-C agar, C-Broth-azocasein—yield highly concordant findings.
SpeB production among GAS isolates in accordance with emm type
Among the entire study sample of 322 isolates, nine emm types are represented by ≥10 isolates. For these highly prevalent emm types, SpeB-non-producers ranged from 6 to 100% of the isolates (S1 Fig).
Of the ten emm18 URT isolates under study, all are SpeB-non-producers. Yet, these 10 isolates are represented by five emm subtypes and thereby, consist of a mix of distinct genotypes. A critical question is whether the SpeB-non-producer phenotype that is highly prevalent among emm18 strains is a consequence of strong selection. The 150 nt region encoding emm type-specific determinants was analyzed for genetic change among the emm18 isolates. Single nucleotide polymorphisms were identified at 17 of the 150 nt sites, which together yield 12 amino acid substitutions (data not shown). Irrespective of whether the underlying mechanism for genetic change is mutation, recombination or a combination of both, a role for strong (diversifying) selection acting on the emm18 type-specific coding region is evident. Furthermore, the rate of random genetic change observed for emm appears to be sufficiently high to readily allow for reversion to a SpeB-producer phenotype—which could arise via mutation in any one of several genes having many more potential target sites (reviewed in [15])—if indeed such a mutation conferred a strong adaptive advantage to the organism. This evolutionary argument supports the notion that the SpeB-non-producer phenotype is directly or indirectly linked to a strong fitness advantage for emm18 organisms infecting the URT.
Eighteen emm1 invasive disease isolates were examined in this study and of these, 17 were SpeB-producers on Columbia-SM, but fewer (N = 13) were full-fledged SpeB-producers on CBrothMg-C agar (S1 Table). This relatively high discordance between the two casein digestion assays for emm1 invasive isolates—Columbia versus C-broth—may be an indication that SpeB activity in emm1 strains is influenced by unspecified factors in the growth medium. Conceivably, other studies investigating SpeB activity by invasive emm1 isolates, which used different culture medium, are similarly impacted [16, 21, 28].
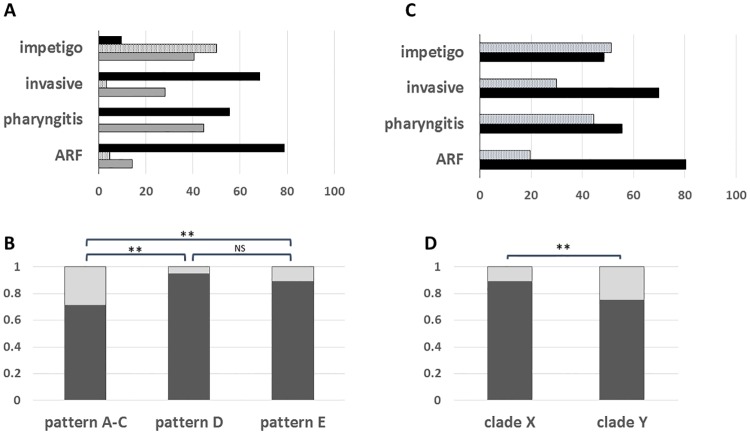
Most emm types can be assigned to an emm pattern group based on the number and arrangement of emm and emm-like genes, as defined by the 3' end region sequence encoding the cell wall-spanning domain of M and M-like proteins [22, 29–31]. Importantly, emm pattern groupings display highly significant correlations with cases of pharyngitis versus impetigo, whereby group emm pattern A-C strains are considered to be "throat specialists", pattern D strains are "skin specialists", and pattern E strains are "generalists" [9, 12]. Based on emm type, an emm pattern group could be predicted for nearly all (99.7%) of the GAS isolates of this study [29] (S1 Table). For this sample set, emm pattern A-C isolates accounted for the majority of pharyngitis isolates (55%), but only a small minority of impetigo isolates (9%; Fig 1A); emm pattern A-C isolates were even more prevalent among ARF and invasive disease isolates (79 and 68%, respectively). Pattern A-C strains had the highest proportion of SpeB-non-producers (29%), as compared to emm pattern D and E strains (5 and 11%, respectively; Fig 1B). The difference in SpeB activity among emm pattern A-C isolates versus pattern D or E isolates is highly significant (p < 0.001; Fisher's exact test, 2-tailed). Previous findings on a smaller sample set of GAS strains (N = 40) showed similar trends, with mean average SpeB activity levels of emm pattern D > pattern E > pattern A-C strains [27].
Fig 1. Study sample composition and SpeB production according to emm pattern group and emm clade.
All GAS isolates under study were assigned to an emm pattern group and emm clade, based on their emm type. Panels A and C: The emm pattern (panel A: pattern A-C, black; pattern D, speckled; pattern E, gray) and clade (panel C: clade X, speckled; clade Y, black) distributions (%) of each disease-defined subset of GAS strains is shown. Panels B and D: The fractional distributions of SpeB producers (dark gray) and SpeB non-producers (light gray) are plotted. SpeB phenotype is based on the Columbia-SM agar assay. The statistical significance of differences was tested using the Fisher exact test (2-tailed; ** for p < 0.01).
In addition to emm pattern grouping, emm types can be assigned to phylogenetically-based emm cluster groups, of which there are two major clades [32]. Most pattern A-C emm types (throat specialists) fall into clade Y and most pattern E emm types (generalists) fall into clade X, whereas pattern D isolates (skin specialists) are split between the two clades [9, 32]. Clade Y is the dominant clade for GAS isolates of this study, accounting for 47, 55, 70 and 80% of impetigo, pharyngitis, invasive disease and ARF isolates, respectively (Fig 1C). The proportion of clade Y pharyngitis isolates for the sample set of this study (55%; N = 146) closely parallels the 52% of pharyngitis isolates collected in North America over a recent seven-year period (N = 7,040) that are clade Y [33, 34]. Considering all GAS isolates, the difference between SpeB-producers and non-producers having emm types assigned to clade X versus clade Y is highly significant (p = 0.0022; Fisher's exact test, 2-tailed), whereby 11 and 25% of clade X and Y isolates, respectively, lack SpeB activity (Fig 1D).
Within-patient (lack of) heterogeneity in SpeB phenotype
During experimental infection in mice following large inoculum doses of GAS, SpeB-non-producers can arise from SpeB-producers via mutations in transcriptional regulatory genes, whereby the switch between SpeB-producer and non-producer corresponds to the transition between localized and invasive GAS infection [13, 19, 20, 35]. Combined with the finding of heterogeneity in SpeB phenotype among GAS isolates sharing the same emm type (S1 Fig), it was of interest to determine if a mixture of SpeB phenotypes could be readily detected within individual pharyngitis patients (CMH series of GAS isolates; S1 Table). Data show that all 68 patients having >1 β-hemolytic colony from an oropharyngeal swab yielded homogeneous SpeB phenotypes; 60 patients yielded ≥10 β-hemolytic colonies and 33 patients had ≥25 β-hemolytic colonies (Table 3). In addition, there was no apparent difference between SpeB-producers and non-producers in terms of the number of β-hemolytic colonies recovered from cultured throat swabs (t = 0.68; unpaired Student t-test, 2-tailed). Thus, if SpeB-non-producers arise from infections with SpeB-producing organisms, or vice versa, their numbers are below the limits of detection of this study; those approximate values are calculated as <1 per 1,055 colonies for a SpeB-producer → non-producer transition, and <1 per 341 colonies for a SpeB-non-producer → producer transition (data not shown).
Table 3. Phenotype homogeneity among single colony picks from oropharyngeal swabs taken from pediatric patients with pharyngitis.
| No. of colony picks per patient | No. of patients yielding 100% SpeB-producer colony picks* | No. of patients yielding 100% SpeB-non-producer colony picks* | No. of patients yielding a mixture of SpeB- producer and non-producer colony picks |
|---|---|---|---|
| 2 to 10 | 7 | 1 | 0 |
| 11 to 24 | 21 | 6 | 0 |
| ≥ 25 | 24 | 9 | 0 |
* Columbia-SM agar assay
To address the possibility that bacterial cells within a colony are heterogeneous in their SpeB phenotype, 8 colonies arising from oropharyngeal swabs of 8 different pharyngitis patients were passed through 5 μm filters (which excludes long chains), plated onto blood agar, and colony picks tested for SpeB activity on Columbia-SM agar. All colony picks originating from a single throat swab colony were homogeneous in their SpeB phenotype (Table 4).
Table 4. Phenotypic homogeneity among organisms within single colonies from oropharyngeal swabs taken from pediatric patients with pharyngitis.
| GAS strain | emm subtype | No. of CFUs following colony filtration (x 103) | No. of CFUs screened | Predominant SpeB phenotype | % of CFUs expressing the predominant SpeB phenotype |
|---|---|---|---|---|---|
| CMH100 | 6.4 | 27.4 | 499 | producer | 100 |
| CMH103 | 12.0 | 60.0 | 50 | producer | 100 |
| CMH109 | 87.0 | 6.2 | 940 | producer | 100 |
| CMH113 | 3.91 | 3.9 | 50 | non-producer | 100 |
| CMH119 | 2.0 | 1.2 | 50 | producer | 100 |
| CMH120 | 5.14 | 13.9 | 50 | producer | 100 |
| CMH125 | 1.0 | 13.3 | 49 | producer | 100 |
| CMH135 | 1.0 | 6.8 | 50 | non-producer | 100 |
CFU, colony forming unit
SpeB phenotype differences are due to transcription control
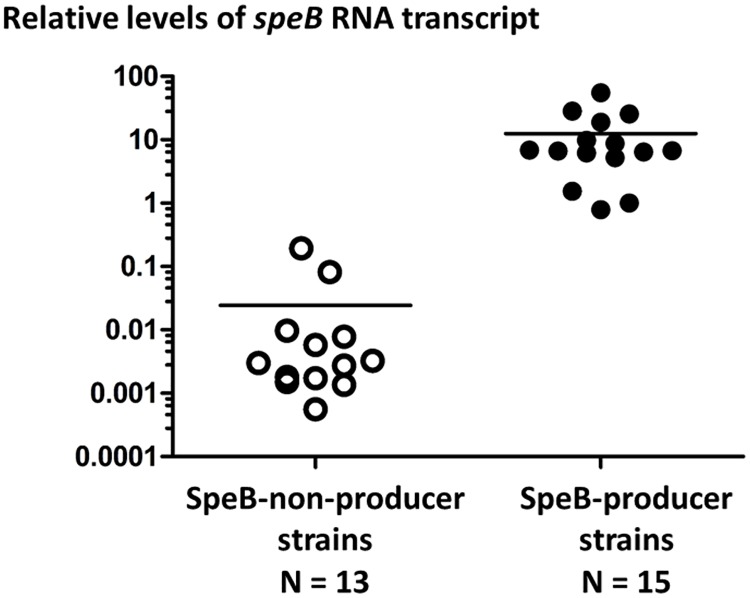
Many studies on naturally-arising mutations affecting SpeB activity (e.g., [13, 36, 37]) in clinical isolates indicate that the molecular basis for differential SpeB casein-digesting activity is typically due to speB transcription; speB is essentially a core gene of GAS. Using quantitative RT-PCR based on RNA recovered from bacterial cultures grown to stationary phase in C-broth culture medium, normalized values for relative RNA levels of speB transcript were compared for 15 strains that displayed caseinolytic activity (on Columbia-SM agar), and 13 strains that were classified as SpeB-non-producers, collectively representing emm3, emm5, emm6 and emm17 isolates of ≥ 17 emm subtypes. The difference in relative speB transcript levels for SpeB-producers versus non-producers is highly significant (Fig 2; t = 0.0047, unpaired Student t-test, 2-tailed, with Welch’s correction; p < 0.0001, Mann-Whitney U-test, 2-tailed). The normalized average mean value for speB RNA transcript levels from SpeB-producer strains exceeds that of non-producer strains by >500-fold. In summary, for the 28 GAS strains examined, SpeB-mediated caseinolytic activity observed after 48 hours of culture on a solid surface (agar) is highly correlated with relative RNA transcript levels for speB following 16 hours of culture in liquid broth medium.
Fig 2. Relative speB RNA transcript levels for SpeB-producer and non-producer GAS strains.
Normalized (log10) relative levels of speB transcript for SpeB-producer and non-producer GAS strains are plotted. A single RNA/cDNA sample of the SpeB-producer strain CT02-99 (emm6) was chosen as the reference for normalization; its relative speB transcript level was adjusted to 1. Mean average values for each group are indicated (bars).
Discussion
As a species, GAS are extraordinary in the complexity and diversity of their genotypes, phenotypic expression, and the wide range of human diseases they can cause. Untangling these relationships, via identifying epidemiologic correlates between microbial genotypes, phenotypes and clinical disease, can often lead to a greater understanding of the pivotal molecular interactions between agent and host. The findings of this report on the strong correlations between the SpeB activity phenotype and different human diseases caused by GAS, although imperfect, may provide such a foundation. For ARF, the defining attributes of a “rheumatogenic” strain have remained elusive, and the finding for a relative lack of SpeB activity among ARF-associated isolates may provide a molecular handle.
The highly significant association between SpeB-non-producers and ARF provides support for a novel hypothesis: That the ability of GAS to trigger ARF is due, in part, to a phenotype that is related to depressed SpeB activity. However, many ARF-associated isolates produce SpeB and therefore, a lack of SpeB can, at best, provide only a partial explanation for rheumatogenic potential. It is noteworthy that the emm18 (100% SpeB-non-producers) and emm3 (50% SpeB-non-producers) organisms recovered from the URT in this study (i.e., pharyngitis and ARF-associated combined; S1 Table) represent emm types that were highly prevalent in ARF outbreaks within the United States during the 1980s [38, 39].
A relative lack of SpeB activity may have direct and indirect effects on the pathogenesis of ARF. An indirect effect might arise from the orchestrated expression of other genes having a role in ARF (e.g., CovRS, RopB) (reviewed in [13, 15]). A direct effect for a lack of SpeB might entail the sparing of extracellular GAS proteolytic targets (reviewed in [13, 14]). M protein elicits an autoimmune response to cardiac myosin through antigenic mimicry [40], but it is also cleaved by SpeB; conceivably, unmodified M protein may yield a more potent cross-reactive immune response. SpeB degrades superantigens produced by GAS [17], and it has been postulated that superantigens play a role in autoimmune processes.
SpeB also cleaves streptolysin O (SLO) and deoxyribonucleases (DNases) produced by GAS. The revised Jones criteria [3] used for establishing a diagnosis of ARF requires evidence for a recent GAS infection, which is usually satisfied by elevated anti-SLO (ASO) or anti-DNaseB (ADB) titers when GAS is not recovered from oropharyngeal swabs, as is very often the case. A plausible consequence of infections with SpeB-non-producers is that ASO and ADB titers are elevated because SLO and DNaseB remain fully intact. This argument raises the possibility that cases of ARF triggered by SpeB-producers are under-diagnosed. Since the URT infection preceding ARF is often clinically inapparent, the possibility should also be considered that a lack of SpeB production results in fewer overt symptoms of pharyngitis. If correct then this, in turn, would provide an adaptive advantage to SpeB-non-producers because they would be more likely to escape diagnosis and therefore antibiotic treatment.
Well-documented ARF-associated GAS isolates are not readily available and difficult to obtain, largely because the inciting GAS infection is usually resolved by a host immune response well before the first acute clinical signs of autoimmune disease appear. The ARF-associated organisms selected for this analysis (S1 Table) were derived from institutional or community outbreaks that were well-studied, or individual patients that were carefully tracked; like the pharyngitis and invasive disease isolates, all were recovered from the United States. However, ARF is highly prevalent in many resource-poor regions of the world [1], where the circulating GAS strains differ markedly in emm type from GAS of resource-rich regions, and impetigo is often endemic [41, 42]. Whether ARF-associated organisms from resource-poor regions exhibit significantly lower levels of SpeB activity, when compared to the larger pool of circulating strains, remains to be established.
The findings of this report on SpeB activity and disease association confirm some previous studies. That impetigo isolates have significantly higher levels of SpeB activity than pharyngitis isolates had been noted in a small-scale study, wherein the role of SpeB in impetigo was experimentally validated as well [43]. In the present study, ~20% of both pharyngitis and invasive disease isolates, each represented by ≥ 20 emm types, lacked SpeB activity, a finding that closely parallels the data on several thousand pharyngitis and/or invasive disease GAS isolates represented by four emm types [21]. In a prior study, emm1 isolates that were stratified according to severe versus non-severe invasive disease showed statistically significant differences in SpeB production, whereby ~40% of severe invasive disease isolates were SpeB-deficient [16]. The data of this study were unable to replicate those latter findings, however, the sample size for emm1 invasive disease isolates may have been too small to distinguish among clinical subgroups. A role for a lack of SpeB in GAS dissemination to systemic tissue sites has also been experimentally validated [13, 18–20].
Population studies by other investigators studying SpeB activity focused on invasive and/or URT isolates sharing the same emm type [16, 21]. A distinct advantage of the genetically diverse collection of isolates used in this study is that biological tendencies which are broadly associated with a disease group may be more readily captured. For many emm types, organisms tend to cause only a subset of the numerous GAS disease types; for e.g., the highly rheumatogenic emm18 organisms are (relatively) rarely recovered from cases of pharyngitis or invasive disease [34, 44–47].
Among the SpeB-producer strains, the biological implications of “lower” versus “higher” levels of SpeB activity, if any, are not known. It is plausible that relatively low levels of SpeB activity leads to degradation of GAS extracellular proteins while leaving more distal host tissue proteins largely intact. Although the subset of 152 strains tested for azocasein digestion do not fully reflect the larger set of 322 isolates in terms of genetic diversity (Table 1), some insights may be gleaned from the data. Comparisons of % azocasein digestion activity for SpeB-producer strains reveal no significant difference for ARF versus pharyngitis isolates, but do show statistically significant differences for impetigo versus either ARF or pharyngitis isolates (t < 0.05; unpaired student t-test with Welch’s correction, 2-tailed). Thus, the absolute level of SpeB activity may further distinguish URT from superficial skin infections, consistent with previous findings [27]. Future studies can more carefully explore biologically significant thresholds for SpeB activity and/or speB transcript levels.
Each GAS strain appears to have a strong tendency towards causing only a limited subset of the human diseases that have been associated with this species. The genotypic diversity among GAS isolates is partly manifest by a very large number of accessory genes, several of which also display extensive nt sequence divergence [12, 48, 49]. Equally complex is the vast array of intersecting transcriptional networks that coordinate the expression of virulence factor genes. Because speB expression is regulated via numerous pathways, SpeB activity provides a simple output for measuring the transcriptional state(s) of the cell. Strong tendencies for SpeB activity versus SpeB deficits among collections of clinically-defined isolates, as revealed in this report, may in turn reflect distinguishing molecular pathways that contribute to disease phenotypes and pathogenesis.
Materials and methods
Strain sampling
GAS isolates were chosen using a strategy that aimed to include strains representative of the human host clinical sub-populations, coupled with maximizing genetic diversity. For pharyngitis isolates from a population-based collection (N = 78; Bristol, CT, 2001–2002) [44], one isolate of each emm subtype [49] was selected; if there were multiple isolates of a given emm subtype, ~33% of isolates (range, 25–40%) of that emm subtype were chosen; overall ≥30% of the isolates of a given emm type were sampled. For pharyngitis isolates whose emm types were unknown at the onset of the study (N = 68 patients; Chicago, 2012), between 2 and 53 colony picks of β-hemolytic colonies derived from pediatric throat swabs plated on blood agar were collected for analysis; group A carbohydrate was confirmed by latex bead agglutination, and the emm type and subtype was ascertained by sequence-based typing of representative colony picks; one representative isolate from each patient was used for further study, unless otherwise noted; the phenotypic homogeneity of multiple colony picks from the same throat swab is demonstrated in Table 3. All ARF-associated isolates (N = 42; USA, 1933–1989) were recovered from the upper respiratory tract (URT; most were previously characterized [50–52]); sources for ARF-associated isolates are The Rockefeller University Hospital (RS or RP, rheumatic patient series; N = 15, NY), the Great Lakes Naval Training Station (N = 8; IL), others from the Lancefield collection (N = 4; NY), WHO-Minneapolis (N = 12; USA) and the C.D.C. (N = 3); ≤ 2 ARF-associated isolates from the same collection (i.e., strain source and sample type) and sharing an emm type were chosen. For impetigo isolates of a population-based collection (N = 58; tropical Australia, 1994–1996) [26, 53], ≥ 1 isolate of each emm subtype was sampled; if there were 2 to 6 isolates of a given emm subtype, 2 isolates were sampled, and if there were 7 to 10 isolates of a given emm subtype, 3 isolates were sampled. An additional 16 impetigo isolates, each having a distinct emm type, are also included (from USA, Trinidad and Czech Republic; 1971–1988) [50]. Invasive disease isolates were collected from normally sterile tissue sites of patients in CT hospitals over a 6-month period in 1995 as previously described [23]; included in this study are 60 of the 64 original GAS isolates that were reported (4 cultures had been lost); all available isolates were included in the analysis.
emm type determination
emm subtype (which is roughly equivalent to partial allele) was assigned according to [49].
Agar-based casein digestion assays
Modified Columbia agar with skim milk (Columbia-SM agar) was prepared as follows: 0.5X Columbia agar base (Difco-BBL), 3% w/v skim milk powder (Difco-BBL) and additional Bacto-agar for 1.5% w/v final concentration. Following GAS inoculation of agar (by short stab of a colony pick derived from a blood agar plate, or surface plating of 2 μl of 10-fold concentrated C-broth culture that had been grown overnight) and 48 h incubation at 37°C, SpeB-producers had average zones of clearance ≥6.0 mm, whereas SpeB-non-producers had average zones of clearance of 0 to 1.0 mm. All GAS isolates were tested in multiple replicates.
For CBrothMg-C agar, C-broth was prepared by the standard method [27], except with 7.5 mM MgSO4, 3% w/v casein (Acros Organics) and 1.5% w/v Bacto-agar added. Following GAS inoculation of agar plates (done by plating 2 μl of a 10-fold concentrated C-broth culture grown overnight) and 48 h incubation at 37°C, SpeB-producers had average zones of clearance ≥4.0 mm, whereas SpeB-non-producers had average zones of clearance of 0 to 1.0 mm. Only a subset of pharyngitis and impetigo isolates were tested using CBrothMg-C agar (S1 Table), and the % of SpeB-non-producers was extrapolated to the whole sample set. Intermediate zones of clearance were scored as "weak.”
Azocasein digestion assay
The azocasein digestion assay was performed using culture supernatants, as previously described [27], following growth of GAS in C-broth for 16 h at 37°C; data is expressed as % SpeB activity relative to the control strain Alab49. Organisms were tested in multiple replicates, and % SpeB activity values averaged.
Quantitative PCR (qPCR)
RNA was purified from cells grown 16 h at 37°C in C-broth, and the cDNA generated was used as a template for quantitative PCR, according to [54]. Both the recA and gyr housekeeping genes were used to calculate relative RNA transcript levels for the speB target gene, and values were averaged. One SpeB-producer strain was chosen as the reference for normalization of relative speB RNA transcript levels for all other strains of the dataset. Each gene was tested in triplicate; independent RNA preparations (up to four) were generated for many strains, and relative speB transcript levels (normalized values) were reported as the mean average. Oligonucleotide primers are as follows: recA-forward, 5’-ATTGATTGATTCTGGTGCGG; recA-reverse, 5’-ATTTACGCATGGCCTGACTC; gyr-forward, 5’-CGATGCCAGTCAAATTCAGG; gyr-reverse, 5’-CCCAGACTAAATGATGCAAACCC; speB-forward, 5’-TGTCGGTAAAGTAGGCGGAC; speB-reverse, 5’-GAGCTGAAGGGTTTAGTGCG.
Supporting information
(XLSX)
(PDF)
(PDF)
The fractional distribution of SpeB-producers (dark gray) and SpeB-non-producers (light gray) is plotted in accordance with emm type, for all isolates of the most common emm types: emm1 (N = 46 isolates); emm2 (N = 11); emm3 (N = 29); emm4 (N = 11); emm6 (N = 20); emm12 (N = 27); emm18 (N = 11); emm28 (N = 17); emm89 (N = 10). SpeB phenotype is based on the Columbia-SM agar assay.
(PDF)
Acknowledgments
The authors thank the many investigators who have provided bacterial strains over past years. This work was supported by The National Institutes of Health (AI065572 and AI117088, to D.E.B.).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by NIH AI065572 and AI117088 (www.nih.gov) to D.E.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. The Lancet infectious diseases. 2005;5(11):685–94. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- 2.Cunningham MW. Streptococcus and rheumatic fever. Current opinion in rheumatology. 2012;24(4):408–16. Epub 2012/05/24. 10.1097/BOR.0b013e32835461d3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gewitz MH, Baltimore RS, Tani LY, Sable CA, Shulman ST, Carapetis J, et al. Revision of the jones criteria for the diagnosis of acute rheumatic Fever in the era of Doppler echocardiography: a scientific statement from the american heart association. Circulation. 2015;131(20):1806–18. Epub 2015/04/25. 10.1161/CIR.0000000000000205 [DOI] [PubMed] [Google Scholar]
- 4.Stollerman GH. Rheumatogenic streptococci and autoimmunity. Clinical immunology and immunopathology. 1991;61:131–42. [DOI] [PubMed] [Google Scholar]
- 5.Bisno AL, Pearce IA, Stollerman GH. Streptococcal infections that fail to cause recurrences of rheumatic fever. JInfectDis. 1977;136:278–85. [DOI] [PubMed] [Google Scholar]
- 6.Shulman ST, Stollerman G, Beall B, Dale JB, Tanz RR. Temporal changes in streptococcal M protein types and the near-disappearance of acute rheumatic fever in the United States. Clin Infect Dis. 2006;42(4):441–7. 10.1086/499812 [DOI] [PubMed] [Google Scholar]
- 7.Kuttner AG, Krumwiede E. Observations on the effect of streptococcal upper respiratory infections on rheumatic children: A three year study. JClinInvest. 1941;20:273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, et al. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. The Journal of infectious diseases. 2005;192(5):760–70. Epub 2005/08/10. 10.1086/430618 [DOI] [PubMed] [Google Scholar]
- 9.Bessen DE. Tissue tropisms in group A Streptococcus: what virulence factors distinguish pharyngitis from impetigo strains? Current opinion in infectious diseases. 2016;29(3):295–303. 10.1097/QCO.0000000000000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter EV, Svartman M, Mohammed I, Cox R, Poon-King T, Earle DP. Tropical acute rheumatic fever and associated streptococcal infections compared with concurrent acute glomerulonephritis. JPediatr. 1978;92:325–33. [DOI] [PubMed] [Google Scholar]
- 11.Wannamaker LW. Differences between streptococcal infections of the throat and of the skin. N Engl J Med. 1970;282:23–31. 10.1056/NEJM197001012820106 [DOI] [PubMed] [Google Scholar]
- 12.Bessen DE, McShan WM, Nguyen SV, Shetty A, Agrawal S, Tettelin H. Molecular epidemiology and genomics of group A Streptococcus. Infect Genet Evol. 2015;33:393–418. 10.1016/j.meegid.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nature reviews Microbiology. 2011;9(10):724–36. 10.1038/nrmicro2648 [DOI] [PubMed] [Google Scholar]
- 14.Nelson DC, Garbe J, Collin M. Cysteine proteinase SpeB from Streptococcus pyogenes—a potent modifier of immunologically important host and bacterial proteins. Biological chemistry. 2011;392(12):1077–88. 10.1515/BC.2011.208 [DOI] [PubMed] [Google Scholar]
- 15.Carroll RK, Musser JM. From transcription to activation: how group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol. 2011;81(3):588–601. 10.1111/j.1365-2958.2011.07709.x [DOI] [PubMed] [Google Scholar]
- 16.Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, et al. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Molecular microbiology. 2004;51(1):123–34. [DOI] [PubMed] [Google Scholar]
- 18.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, et al. Trigger for group A streptococcal M1T1 invasive disease. Faseb J. 2006;20(10):1745–7. 10.1096/fj.06-5804fje [DOI] [PubMed] [Google Scholar]
- 19.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nature medicine. 2007;13(8):981–5. 10.1038/nm1612 [DOI] [PubMed] [Google Scholar]
- 20.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS pathogens. 2006;2(1):e5 10.1371/journal.ppat.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen RJ, Raghuram A, Cantu C, Hartman MH, Jimenez FE, Lee S, et al. The Majority of 9,729 Group A Streptococcus Strains Causing Disease Secrete SpeB Cysteine Protease: Pathogenesis Implications. Infection and immunity. 2015;83(12):4750–8. Epub 2015/09/30. 10.1128/IAI.00989-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bessen DE. Molecular Basis of Serotyping and the Underlying Genetic Organization of Streptococcus pyogenes In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK)2016. [PubMed] [Google Scholar]
- 23.Fiorentino TR, Beall B, Mshar P, Bessen DE. A genetic-based evaluation of the principal tissue reservoir for group A streptococci isolated from normally sterile sites. The Journal of infectious diseases. 1997;176(1):177–82. [DOI] [PubMed] [Google Scholar]
- 24.Haukness HA, Tanz RR, Thomson RB Jr., Pierry DK, Kaplan EL, Beall B, et al. The heterogeneity of endemic community pediatric group a streptococcal pharyngeal isolates and their relationship to invasive isolates. The Journal of infectious diseases. 2002;185(7):915–20. 10.1086/339407 [DOI] [PubMed] [Google Scholar]
- 25.Carapetis JR, Wolff DR, Currie BJ. Acute rheumatic fever and rheumatic heart disease in the Top End of Australia's Northern Territory. MedJAust. 1996;164:146–9. [DOI] [PubMed] [Google Scholar]
- 26.Bessen DE, Carapetis JR, Beall B, Katz R, Hibble M, Currie BJ, et al. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. The Journal of infectious diseases. 2000;182(4):1109–16. Epub 2000/09/09. 10.1086/315842 [DOI] [PubMed] [Google Scholar]
- 27.Svensson MD, Scaramuzzino DA, Sjobring U, Olsen A, Frank C, Bessen DE. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Molecular microbiology. 2000;38(2):242–53. [DOI] [PubMed] [Google Scholar]
- 28.Olsen RJ, Sitkiewicz I, Ayeras AA, Gonulal VE, Cantu C, Beres SB, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):888–93. Epub 2010/01/19. 10.1073/pnas.0911811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, Beall B, et al. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. Journal of bacteriology. 2004;186(13):4285–94. Epub 2004/06/19. 10.1128/JB.186.13.4285-4294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMillan DJ, Dreze PA, Vu T, Bessen DE, Guglielmini J, Steer AC, et al. Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin Microbiol Infect. 2013;19(5):E222–9. Epub 2013/03/08. 10.1111/1469-0691.12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingshead SK, Readdy TL, Yung DL, Bessen DE. Structural heterogeneity of the emm gene cluster in group A streptococci. Molecular microbiology. 1993;8(4):707–17. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. The Journal of infectious diseases. 2014;210(8):1325–38. Epub 2014/05/07. 10.1093/infdis/jiu260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shulman ST, Tanz RR, Dale JB, Steer AC, Smeesters PR. Added value of the emm-cluster typing system to analyze group A Streptococcus epidemiology in high-income settings. Clin Infect Dis. 2014;59(11):1651–2. Epub 2014/08/15. 10.1093/cid/ciu649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulman ST, Tanz RR, Dale JB, Beall B, Kabat W, Kabat K, et al. Seven-year surveillance of north american pediatric group a streptococcal pharyngitis isolates. Clin Infect Dis. 2009;49(1):78–84. 10.1086/599344 [DOI] [PubMed] [Google Scholar]
- 35.Kansal RG, Datta V, Aziz RK, Abdeltawab NF, Rowe SL, Kotb M. Dissection of the Molecular Basis for Hypervirulence of An In Vivo Selected Phenotype of the Widely Disseminated M1T1 Strain of Group A Streptococcus Bacteria. J Infect Dis. 2010;201:855–65. 10.1086/651019 [DOI] [PubMed] [Google Scholar]
- 36.Carroll RK, Beres SB, Sitkiewicz I, Peterson L, Matsunami RK, Engler DA, et al. Evolution of diversity in epidemics revealed by analysis of the human bacterial pathogen group A Streptococcus. Epidemics. 3(3–4):159–70. 10.1016/j.epidem.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 37.Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, et al. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS pathogens. 2010;6(4):e1000832 10.1371/journal.ppat.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veasy LG, Wiedmeier SE, Orsmond GS, Ruttenberg HD, Boucek MM, Roth SJ, et al. Resurgence of acute rheumatic fever in the intermountain area of the United States. The New England journal of medicine. 1987;316(8):421–7. Epub 1987/02/19. 10.1056/NEJM198702193160801 [DOI] [PubMed] [Google Scholar]
- 39.Veasy LG, Tani LY, Daly JA, Korgenski K, Miner L, Bale J, et al. Temporal association of the appearance of mucoid strains of Streptococcus pyogenes with a continuing high incidence of rheumatic fever in Utah. Pediatrics. 2004;113(3 Pt 1):e168–72. Epub 2004/03/03. [DOI] [PubMed] [Google Scholar]
- 40.Quinn A, Kosanke S, Fischetti VA, Factor SM, Cunningham MW. Induction of autoimmune valvular heart disease by recombinant streptococcal M protein. Infection and immunity. 2001;69:4072–8. 10.1128/IAI.69.6.4072-4078.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. The Lancet infectious diseases. 2009;9(10):611–6. 10.1016/S1473-3099(09)70178-1 [DOI] [PubMed] [Google Scholar]
- 42.Williamson DA, Smeesters PR, Steer AC, Steemson JD, Ng AC, Proft T, et al. M-Protein Analysis of Streptococcus pyogenes Isolates Associated with Acute Rheumatic Fever in New Zealand. Journal of clinical microbiology. 2015;53(11):3618–20. Epub 2015/08/21. 10.1128/JCM.02129-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ringdahl U, Svensson HG, Kotarsky H, Gustafsson M, Weineisen M, Sjobring U. A role for the fibrinogen-binding regions of streptococcal M proteins in phagocytosis resistance. Molecular microbiology. 2000;37(6):1318–26. [DOI] [PubMed] [Google Scholar]
- 44.Shulman ST, Tanz RR, Kabat W, Kabat K, Cederlund E, Patel D, et al. Group A streptococcal pharyngitis serotype surveillance in North America, 2000–2002. Clin Infect Dis. 2004;39(3):325–32. Epub 2004/08/13. 10.1086/421949 [DOI] [PubMed] [Google Scholar]
- 45.O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, et al. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis. 2007;45(7):853–62. 10.1086/521264 [DOI] [PubMed] [Google Scholar]
- 46.O'Brien KL, Beall B, Barrett NL, Cieslak PR, Reingold A, Farley MM, et al. Epidemiology of invasive group a streptococcus disease in the United States, 1995–1999. Clin Infect Dis. 2002;35(3):268–76. 10.1086/341409 [DOI] [PubMed] [Google Scholar]
- 47.Lynskey NN, Banerji S, Johnson LA, Holder KA, Reglinski M, Wing PA, et al. Rapid Lymphatic Dissemination of Encapsulated Group A Streptococci via Lymphatic Vessel Endothelial Receptor-1 Interaction. PLoS pathogens. 2015;11(9):e1005137 Epub 2015/09/10. 10.1371/journal.ppat.1005137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bessen DE, Kumar N, Hall GS, Riley DR, Luo F, Lizano S, et al. Whole-genome association study on tissue tropism phenotypes in group A Streptococcus. Journal of bacteriology. 2011;193(23):6651–63. Epub 2011/09/29. 10.1128/JB.05263-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beall B. http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm Atlanta2016.
- 50.Bessen DE, McGregor KF, Whatmore AM. Relationships between emm and multilocus sequence types within a global collection of Streptococcus pyogenes. BMC Microbiol. 2008;8:59 10.1186/1471-2180-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bessen D, Jones KF, Fischetti VA. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. The Journal of experimental medicine. 1989;169(1):269–83. Epub 1989/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bessen DE, Sotir CM, Readdy TL, Hollingshead SK. Genetic correlates of throat and skin isolates of group A streptococci. The Journal of infectious diseases. 1996;173(4):896–900. [DOI] [PubMed] [Google Scholar]
- 53.McGregor KF, Bilek N, Bennett A, Kalia A, Beall B, Carapetis JR, et al. Group A streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources. The Journal of infectious diseases. 2004;189(4):717–23. Epub 2004/02/10. 10.1086/381452 [DOI] [PubMed] [Google Scholar]
- 54.Lizano S, Luo F, Tengra FK, Bessen DE. Impact of orthologous gene replacement on the circuitry governing pilus gene transcription in streptococci. PLoS ONE. 2008;3(10):e3450 10.1371/journal.pone.0003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(PDF)
(PDF)
The fractional distribution of SpeB-producers (dark gray) and SpeB-non-producers (light gray) is plotted in accordance with emm type, for all isolates of the most common emm types: emm1 (N = 46 isolates); emm2 (N = 11); emm3 (N = 29); emm4 (N = 11); emm6 (N = 20); emm12 (N = 27); emm18 (N = 11); emm28 (N = 17); emm89 (N = 10). SpeB phenotype is based on the Columbia-SM agar assay.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.