Abstract
Iron is an important nutrient for the survival and growth of many organisms. In order to survive, iron uptake from the environment must be strictly regulated and maintained to avoid iron toxicity. The ferric uptake regulator protein (Fur) regulates genes involved in iron homeostasis in many bacteria, including phytopathogens. However, to date, the role played by Fur in the biology of Pectobacterium carotovorum subsp. brasiliense (Pcb1692), an important pathogen of potatoes, has not yet been studied. To this end, we used the lambda recombineering method to generate a fur mutant strain of Pcb1692 and assessed the virulence and fitness of the mutant strain. The results showed that production of siderophores in Pcb1692Δfur increased compared to the Pcb1692 wild-type and the complemented strain Pcb1692Δfur-pfur. However, production of N-acyl homoserine lactone (AHLs), biofilm formation, exopolysaccharide (EPS) production, virulence on potato tubers and swimming motility, were all significantly decreased in Pcb1692Δfur compared to the wild-type and complemented Pcb1692Δfur-pfur strains. The Pcb1692Δfur mutant also demonstrated significant sensitivity to oxidative stress when exposed to H2O2. Consistent with phenotypic results, qRT-PCR results demonstrated that Fur down-regulates genes which encode proteins associated with: iron uptake (HasA-extracellular heme-binding protein and Ferrodoxin-AED-0004132), stress response (SodC-superoxide dismutase), plant cell wall degrading enzymes (PrtA and CelV) and motility (FlhC and MotA). We conclude that the ferric uptake regulator protein (Fur) of Pcb1692 regulates traits that are important to host-pathogens interactions.
Introduction
Pectobacterium carotovorum subsp. brasiliense (Pcb) is a member of the soft rot Enterobacteriaceae (SRE), which consist of members of the Pectobacterium and Dickeya genera [1]. This pathogen is the major causal agent of blackleg and soft rot disease in potato stems and tubers, respectively, in the field and during post-harvest [2]. Pcb strains have been reported in many different countries where they have been shown to cause significant losses to the potato industry and a wide variety of crops in many countries, including Brazil, Kenya, New Zealand and South Africa [2–5]. This pathogen has been identified to be more virulent compared to other Pectobacterium species [2]. Like other SREs, Pectobacterium species use a variety of virulence determinants to adapt, colonize and cause disease in plants. Some of these virulence factors include, amongst other, density-dependent cell-cell communication mediated by acyl homoserine lactones (AHLs), secretion systems, plant cell wall-degrading enzymes (PCWDEs), adhesion, biofilm formation, motility, siderophores and chemotaxis [6–12].
During the host invasion process, pathogenic bacteria encounter different environmental conditions, of which iron limitation and reactive oxygen species produced by the host plant are major factors limiting the ability of the bacteria to spread and colonize the host [13, 14]). Therefore, bacteria must tightly regulate iron uptake and deal with changes occurring during redox conditions [14]. In effect, the host withholds iron, depriving the pathogen of iron and thus limiting its ability to colonize the host plant. However, bacterial pathogens have developed means of acquiring scarce iron through siderophore production [14, 15]. For example, the roles of high-affinity iron siderophores such as chrysobactin and achromobactin in iron uptake during Dickeya dandantii (formerly Erwinia chrysanthemi) colonization of potato tubers, has been demonstrated [16]. In this respect, studies by Franza and colleagues demonstrated that mutagenesis of genes encoding chrysobactin and achromobactin resulted in impaired symptom initiation, suggesting an inability of the pathogen to survive within host intracellular spaces [15, 16].
In addition to the above, SREs can use their PCWDEs to rupture plant cells, thus making nutrients more accessible to the bacteria [17]. In fact, regulation of PCDWEs is often coupled with iron acquisition in Dickeya dandantii [15]. Once nutrients are released, this presents a free-for-all ‘microbes’ situation. Hence, the fittest bacteria will try to assimilate all available iron to themselves. In this respect, Pcb has been shown to use a vast number of ‘antibacterial’ strategies to inhibit growth of some members of the SRE and other bacteria, both in vitro as well as in potato tubers [18]. It can thus be hypothesized that these arsenals of antibacterial factors are necessary for nutrient and/or iron acquisition. In many bacteria, iron homeostasis is regulated by the ferric uptake regulator protein encoded by the fur gene. Notably, mutation of the fur gene in Pseudomonas aeruginosa resulted in defects in iron uptake and reduced virulence [19].
The Fur protein is a transcriptional repressor with the ferrous ion (Fe(II)) as a co-repressor [20, 21]. When levels of iron exceed those required for cellular functions, Fur represses further iron uptake in order to prevent iron overload and toxicity. In many pathogenic bacteria, Fur protein is also implicated in the regulations of virulence determinants unrelated to iron transport such as PCWDEs [16]. In the presence of ferrous ion, Fur binds to the ‘fur box’, a conserved DNA sequence located within the promoter regions of iron-regulated genes [22–25]. The “fur box” in Escherichia coli is predicted to have a consensus sequence of GAT AAT GAT AAT CAT TAT C. However, this exact sequence is rarely found in other prokaryotes. In fact, it has been suggested that the minimum recognition for iron binding is GAT AAT and in prokaryotes the exact mode of recognition is still relatively unknown [26]. Furthermore, the ‘fur box’ can be highly variable even in genes within the fur regulon of a given bacterium. For example, the predicted fur boxes of the pelD and pelE genes in Erwinia chrysanthemi 3937 are GAT AAA ATT AAT CAG CCT C and ATT AAT AAA AAC CAT TGT C, respectively [16, 27].
In this study, the Pectobacterium carotovorum subsp. brasiliense fur gene homolog was identified, a fur mutant strain generated using methods previously described by Datsenko and Wanner [28] and functionally characterized. Its role in virulence and its effect on Pcb1692 virulence factors, including biofilm formation, production of acyl homoserine lactone, swimming motility, extracellular polysaccharide and extracellular enzymes production, was also investigated.
Materials and methods
Strains and growth conditions
All bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown on nutrient agar or in liquid Luria-Bertani (LB) broth and in M9 minimum medium at 37°C [29]. Where necessary, growth media were supplemented with either100 μg/ml Ampicillin (Sigma-Aldrich), 50 μg/ml Kanamycin (Sigma-Aldrich) or 10 mM MnSO4(Sigma-Aldrich).
Table 1. Bacterial strains and plasmids used in this study.
| Bacterial strains | Description | Sources |
|---|---|---|
| Pectobacterium carotovorum subsp. brasiliense 1692 (Pcb1692) | Initially isolated from potato in Brazil, sequenced strain |
[2] |
| Pcb1692Δfur | Pcb1692Δfur, Kanr | This study |
| Pcb1692Δfur-pfur | Pcb1692Δfur expressing the fur gene from the Trc99A plasmid; Kanr, Ampr | This study |
| Pcb1692ΔexpI | Pcb1692ΔexpI, Kanr | [7] |
| Chromobacterium violaceum CV026 | AHL reporter strain | [25] |
| Plasmids | ||
| pKD4 | Plasmid containing a Kanr cassette | [28] |
| pKD46 | Plasmid expressing the lambda red genes | [28] |
| pTrc99A | Bacterial expression vector | [30] |
| pTrc99A-fur | Bacterial expression vector containing the fur gene insert, Ampr | This Study |
Generation of a Pectobacterium carotovorum subsp. brasiliense fur mutant strain
The fur gene of Pectobacterium atrosepticum (ECA1329) was used as a query to identify the fur homolog in Pcb1692. Using the BLASTN alignment tool available on the ASAP database, we identified the fur homolog in Pcb1692 (PcarbP_010200018626) on contig 00060 (coordinates 25375–25824) with 100% identical to ECA1329. The Pcb1692Δfur mutant strain was generated using the strategy described previously by Datsenko and Wanner [28] and is indicated in S1 Fig. In brief, the upstream and downstream regions flanking the Pcb1692 fur gene (approx. 1200bp) were amplified by polymerase chain reaction (PCR). The kanamycin resistance gene was PCR amplified from plasmid pKD4. The three amplicons were fused by overlap extension PCR to produce a gene disruption cassette, as described previously [31]. The fused PCR product was then electroporated into Pcb1692 harboring pKD46 and transformants were selected on nutrient agar supplemented with 50 μg/ml kanamycin. The list of primers used in this study is provided in Table 2.The HiFi HotStart PCR Kit (KAPA Biosystems) was used in all PCR reactions.The PCR thermal cycling conditions were set as follow: initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 98°C for 30 s, annealing at 60–64°C for 15 s (depending on the primer set), extension at 72°C for 3 min and a final extension at 72°C for 5 min. The integrity of the Pcb1692Δfur mutant strain was confirmed by PCR analyses, nucleotide sequencing and Southern blot analysis (results not shown).
Table 2. Primers used in this study.
| Primer name | Sequence (5’-3’) | Length (bp) |
|---|---|---|
| Mutagenesis primers | ||
| Fur–F | CGATCAACTGCACGCTTATGC | 21 |
| Fur-R | GAATAGTAATGAGCCATTACGC | 22 |
| Test–F | AAGATCTGGCGTCCGGTAAGC | 21 |
| Test -R | TCATTCTGAGACTAAACGCACC | 22 |
| Furkan -R | CGAAGCAGCTCCAGCCTACACATCAACACGATAAATCGACCGC | 43 |
| Furkan -F | CTAAGGAGGATATTCATATGTTAATCCTGTTGCTTACTTATC | 42 |
| Kan–F | GCGGTCGATTTATCGTGTTGATGTGTAGGCTGGAGCTGCTTCG | 43 |
| Kan -R | GATAAGTAAGCAACAGGATTAACATATGAATATCCTCCTTAG | 42 |
| Comp -F | GATTTATCGTGTTGAATCGTC | 21 |
| Comp -R | GCCATTTCGGCTCTGATAATC | 21 |
| Ffh–F | TGGCAAGCCAATTAAATTCC | 20 |
| Ffh–R | TCCAGGAAGTCGGTCAAATC | 20 |
| Fer-F | AGACCCATCATCGGTAGCAC | 20 |
| Fer -R | CTCATCTTCCGCAAAGAAGC | 20 |
| HasA–F | ATTTACGGCCTGATGAGTGG | 20 |
| HasA–R | AACGACGTCAACCACGGTAT | 20 |
| CelV–F | CGTTAAACCGGAACCAACTG | 20 |
| CelV -R | AACCACCGTACTGCCTTTTG | 20 |
| Mot A -F | TTGCCTACGGTTTTGTCTCC | 20 |
| Mot A -R | ACAGCGTTTTACGACCGAAT | 20 |
| SodC- F | TAAATCAGTTCCCGCTCTGG | 20 |
| SodC-R | GCCAGAATTGGGTAGGTTGA | 20 |
| FlhC -F | ATTGCTGCAAAGGGATGTTC | 20 |
| FlhC -R | CCTGTTCATCCAGCAGTTGA | 20 |
| PrtA -F | TGACGCGTTGCATTCATTAT | 20 |
| PrtA–R | TGCCAAATACATTCGAACCA | 20 |
Generation of a complemented Pcb1692Δfur strain
The fur gene and its putative promoter region was PCR amplified from Pcb1692 using primers Comp-F and Comp-R (Table 2). The amplicon was excised from an agarose gel and purified using the Zymo Clean Gel DNA Recovery Kit (Inqaba Biotec, South Africa) according to the manufacturer’s instructions. The amplicon was cloned into pTrc99A to generate pTrc99A-fur (Table 1). The pTrc99A-fur plasmid was electroporated into the Pcb1692Δfur mutant strain and the transformants (Pcb1692Δfur-pfur) were selected on agar plates supplemented with 100 μg/ml Ampicillin.
In vitro growth assays
The in vitro growth properties of the Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur strains were assessed by culturing each bacterial strain in M9 minimal medium and liquid LB broth. The cultures were incubated at 37°C for 16 h with agitation at 370 rpm. The optical density at 600 nm (OD600) of the overnight cultures was adjusted to an OD600 of 0.1, and 1 ml was inoculated into 200 ml of LB broth or M9 minimal medium and grown at 37°C with agitation at 370 rpm. The OD600 was recorded every hour for 16h with a Multiskan GO spectrophotometer (Thermo-Scientific). The experiment was performed in triplicates, three independent times.
Siderophore production
Siderophore production was determined using Chrome Azurol S (CAS) agar plates, as described previously by Louden et al. [32]. Briefly, the Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur strains were grown in LB broth for 16 h with shaking at 370 rpm. The OD600 of the cultures was then adjusted to 1.0,and 50 μl of each strain was spotted onto a CAS agar plate and incubated at 37°C for 48 h. A yellow halo surrounding the inoculation site was taken to indicate production of siderophores. The diameter of each halo was measured in mm. This experiment was performed in triplicates, three independent times.
Resistance against hydrogen peroxide (H2O2)
To determine resistance to H2O2, the Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur strains were grown in LB broth for 16 h in a shaking incubator at 370 rpm. The OD600of each culture was adjusted to 0.4 and 100μl of each bacterial culture was then inoculated into 100 ml of LB broth supplemented with 20 μM H2O2. The cultures were incubated for at 37°Cfor 6h with shaking (370 rpm) and the surviving bacteria were enumerated by serial dilution and plating onto LB agar plates.
Virulence assays
Surface-sterilized potato tubers (cv. Mondial, a susceptible cultivar) were stabbed to a depth of about 1cm with a sterile pipette tip. A 10-μl aliquot of the Pcb1692, Pcb1692Δfur-pfur and Pcb1692Δfur cultures (OD600 = 1) was inoculated into the wounded tubers. For the negative controls, sterile 10 mM MgSO4 was inoculated into the wounded potato tubers. The inoculated potato tubers were placed in moist plastic bags and incubated at 25°C. At 72h post-inoculation, the macerated tissue was scooped and weighed to quantify the extent of tuber maceration by each of the different bacterial strains. This experiment was performed in triplicates, three independent times.
Detection of N-acyl homoserine lactones (AHLs)
The Chromobacterium violaceum (CV026) reporter strain [33] was inoculated into 30 ml of LB broth supplemented with 30 μg/ml kanamycin and grown at 28°C for 16 h with shaking at 270 rpm. A 1-ml aliquot of the overnight CV026 culture was added to 3 ml of filter-sterilized cultures of the Pcb1692, Pcb1692ΔexpI, Pcb1692Δfur and Pcb1692Δfur-pfur strains in different Falcons tubes. The Pcb1692ΔexpI strain was used as negative control. Bacterial cultures were then incubated at 28°C for 48 h. A blue colour indicated the production and presence of AHLs. This experiment was performed in triplicates, three independent times.
Swimming motility assay
Swimming motility assays were performed at 37°C on LB agar plates containing 0.3% (w/v) Bacto agar. The Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur strains were grown overnight in LB broth and the OD600 of each culture was then adjusted to 0.5. A sterile toothpick was dipped into each bacterial culture and then spotted in the middle of the LB agar plates. The agar plates used for inoculation of the Pcb1692Δfur mutant strain were supplemented with 50 μg/ml kanamycin. The agar plates were incubated at 37°C for 24 h. swimming motility was determined by measuring the halos formed by swimming bacteria after 24 h.
Biofilm formation assay
Formation of biofilm was analyzed as described previously by Daniel Perez-Mendoza and colleagues with minimal modifications [34]. In brief, the OD600 of overnight cultures of Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur were adjusted to 0.4. Subsequently, 25 μl of each bacterial culture was inoculated into 40 ml of LB broth and incubated at 37°C for 48h with shaking at 130 rpm. The LB broth was aspirated and biofilm that formed on the walls of the 50-ml Erlenmeyer flasks were visualized by washing three times with double distilled water followed by staining with 0.1% (w/v) crystal violet (Sigma-Aldrich) then incubated for 30 min at room temperature. A violet-coloured ring on the inner wall of the flask indicated biofilm formation. Biofilm formation was also quantitatively assayed by measuring the OD570 of the stained suspensions in 96-well plates with a spectrophotometer.
Extracellular enzyme assays
Semi-quantitative analysis of two plant cell wall-degrading enzymes (PCWDEs), cellulase and proteases, were performed as described by Chatterjee and colleagues [35]. Holes were made on assay plates and the bacterial strains were inoculated into these holes. Cellulase assay plates were stained with 0.1% (w/v) Congo red solution (Sigma-Aldrich), incubated for 30 min and then washed several times with 1M NaCl until a clear zone became visible around the holes. After three days of incubation, Protease plates revealed clear zones around the holes without any further treatment. Enzyme activity was semi-quantified based on the diameter of the haloes around the colonies.
Manganese resistance assay
Resistance to manganese was determined according to the protocol described previously by Hentke [36]. The Pcb1692, Pcb1692Δfur-pfur and Pcb1692Δfur strains were grown for 16 h in LB broth and the OD600 of each culture was then adjusted to 1.0.The bacterial strains were plated onto LB agar supplemented with 7mM MnSO4.H2O (Sigma-Aldrich), followed by incubation at 37°C for 24 h. Three biological experiments were performed.
Extracellular polysaccharide (EPS) production determination
EPS production was measured as described previously by Tang et al. [37], with minor modifications. Briefly, cultures of Pcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur were grown in 100 ml LB broth at 37°C for 72 h to an OD600 of 2.5. The cultures were then transferred into 50-ml Falcon tubes and centrifuged at 10000 rpm for 10 min. EPS was collected from the supernatant by precipitation with 96% ethanol, dried at 37°C for 3 h and weighed. The experiment was performed in triplicates.
qRT-PCR assays
Bacterial cultures were grown overnight in LB broth and the cells harvested by centrifugation at 14000 rpm for 1 min. The supernatant was carefully discarded and the bacterial cells were suspended in RNA stabilization buffer (Qiagen, Hilden, Germany). Total RNA was extracted with the RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNaseI (Qiagen, Hilden, Germany) was used to remove contaminating genomic DNA from the total RNA samples. The concentration of the total RNA was determined using a NanoDrop1000 spectrophotometer (NanoDrop® Technologies, Wilmington, DE). First-strand cDNA was synthesized from 1 μg of the total RNA samples using the Superscript VI First-Strand Synthesis system kit (Invitrogen).The list of primers used for qRT-PCR is provided in Table 2. The genes targeted for qRT-PCR included: prtA (protease), celV (cellulose),AED-0004132 (ferredoxin) hasA (extracellular heme-binding protein), motA (flagellar motor), flhC (flagellar transcriptional regulator), sodC (copper-zinc superoxide dismutase) and ffh (signal recognition particle subunit). The ffh gene was used as internal normalization gene (Table 2). The cDNA was used in real-time PCR reactions using a Quantstudio 12 flex thermocycler (Applied Biosystems). Experiments were performed in triplicates and three biological replicates were performed for each gene. Comparative 2ΔΔct method was used to analyze the data.
Prediction of putative ‘fur boxes’
Genes previously shown to be under Fur regulation in other organism were selected and homologs in Pcb1692 were identified. Thereafter, using CLC bioinformatics analysis regions upstream of the start codon of each gene sequence were screened for DNA sequences containing the GATAAT signature sequence. These included pelA, expI, flhD, hasA and tonB among others. The “fur box” consensus was manually aligned with selected fur regulated genes to match one of the hexameric repeats. Red boxes were added to indicate all possible alignments within the consensus sequence.
Statistical analysis
In this study, experiments were performed in triplicate and three independent times. Where applicable, a one-way Analysis of Variance (ANOVA) was performed to determine statistical significance and a p-value less than 0.05 (p<0.05) was considered to be a statistically significant difference.
Results
Construction and characterization of Pcb1692Δfur mutant
In order to determine the role of the Fur protein in Pcb1692, we generated a Pcb1692Δfur mutant using the lambda Recombination technique (S1 and S2 Figs), as described in the Material and Methods section. The integrity of Pcb1692Δfur mutant was verified by PCR analyses, nucleotide sequencing and Southern blotting, the results of which confirmed that there was a single insertion of the kanamycin cassette into the genome sequence of Pcb resulting in deletion of the fur gene. Similarly, the integrity of the complemented fur gene (with endogenous promoter) cloned into plasmid Trc99A was also confirmed by nucleotide sequencing and the recombinant plasmid was stably maintained in the Pcb1692Δfur mutant strain (results not shown). In vitro growth assay demonstrated that deletion of the Pcb fur gene does not impair the growth of the mutant strain (results not shown).
The role of Fur in Pcb1692 siderophore production
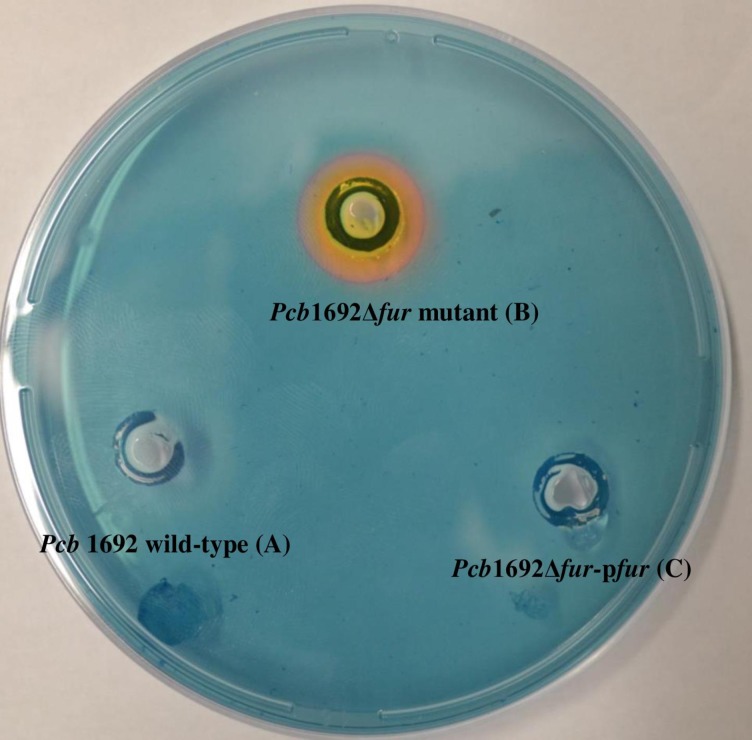
The Pcb1692Δfur mutant strain showed increased siderophore production relative to the Pcb1692 wild-type and Pcb1692Δfur-pfur complemented strain, as indicated by the formation of yellow halos around their colonies (Fig 1). The Pcb1692 wild-type lacked a yellow halo, suggesting that siderophore production was undetectable under the experimental conditions (Fig 1). The results suggest that the increase in siderophore production in the Pcb1692Δfur mutant strain was due to deletion of the fur gene.
Fig 1. Siderophore production in Pcb 1692 wild-type strains compared to Pcb1692Δfur mutant strain using Chrome Azurol S (CAS) plate assay.
A yellow halo indicates siderophore production. Pcb 1692 wild-type (A) and Pcb1692Δfur-pfur strain (C) showed no visible yellow halo while Pcb1692Δfur (B) had a visible yellow halo ring.
Resistance against hydrogen peroxide (H2O2)
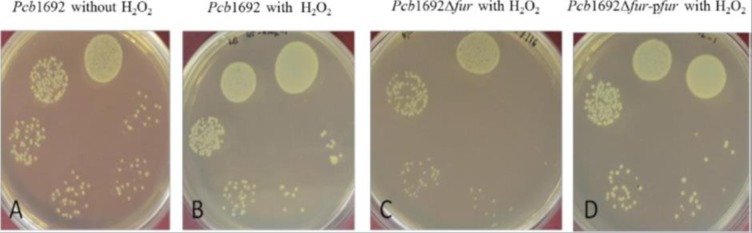
To evaluate the role of Pcb1692 Fur protein in the oxidative stress response, bacterial strains were grown in LB broth supplemented with 20μM H2O2and the bacteria were enumerated (Cfu/ml) after 6 h of incubation. As a control, the Pcb1692 wild-type strain was grown in LB broth lacking H2O2. Addition of H2O2 to the LB broth reduced survival of Pcb1692 by nearly 10%. Interestingly, there was nearly 70% reduction in survival of the Pcb1692Δfur mutant strain when cultured in LB broth supplemented with H2O2 compared to the wild type under similar conditions (Table 3). Trans-complementation of the fur gene in Pcb1692Δfur restored survival of the complemented strain to wild type levels. Our results indicate that the Pcb1692Δfur mutant strain was more sensitive to H2O2 than the wild-type strain (Fig 2), suggesting that the Fur protein of Pcb1692 may play a role in the oxidative stress response in this bacterium.
Table 3. Percentage survival of Pcb1692 wild-type, Pcb1692Δfur mutant and Pcb1692Δfur-pfur complemented strains inoculated into LB broth supplemented with H2O2 compared to the Pcb1692 wild-type strain in LB without H2O2.
| Strain | Averaged CFU/ml (6hpi) | Survival (%) |
|---|---|---|
| Pcb1692 wild-type (no H2O2) | 34 | 100% |
| Pcb1692wild-type (+H2O2) | 31 | 91.17±2.82% |
| Pcb1692Δfur mutant (+H2O2) | 8 | 23.53±1.41% |
| Pcb1692Δfur-pfur (+H2O2) | 29 | 85.29±2.82% |
Fig 2. The effect of H2O2 on Pcb1692 wild-type and mutant strain survival.
(A) Pcb 1692 wild-type cultured in LB medium (B) Pcb1692 wild-type in LB medium supplemented with H2O2 (C) Pcb1692Δfur mutant strain cultured in LB supplemented with H2O2 (D) Pcb1692Δfur-pfur cultured in LB supplemented with H2O2.
Swimming motility was impaired in the Pcb1692Δfur mutant strain
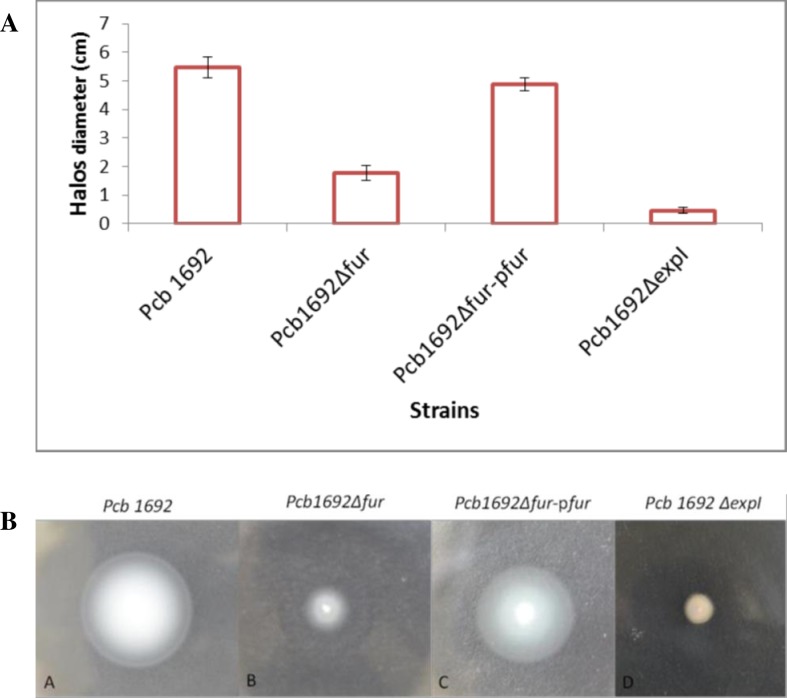
Swimming motility assays indicated that halos formed by the Pcb1692 wild-type and the complimented Pcb1692Δfur-pfur strains had diameters of 5.467±0.36cm and 4.867±0.229cm, respectively, at 48 h post-inoculation. On the contrary, the halo diameter of the Pcb1692Δfur mutant strain (1.767±0.269cm) was significantly (p< 0.05) reduced compared to that of Pcb1692 wild-type strain (Fig 3A). The Pcb1692ΔexpI mutant strain was used as a negative control since this strain is completely impaired in swimming motility [7]. As expected, the Pcb1692ΔexpI mutant strain showed a complete loss of swimming motility (0.456±0.101 cm) (Fig 3B). Together, our results implicate the Pcb1692 Fur protein in swimming motility.
Fig 3. Analysis of swimming motility of the Pcb1692 wild-type and mutant strain.
In the figure, A represents quantitative while B represents qualitative comparison of swimming motility between the wild type and mutant strain. The diameters of halos formed by each strain on LB agar for each strain was measured in triplicates (three biological replicates) and mean values plotted. Error bars represent the standard deviation of the mean. Statistically significant difference were determined by the one way ANOVA and p-values less than 0.05 (p<0.05) were considered to be statistically significant.
The Pcb1692Δfur mutant strain is attenuated in AHL production
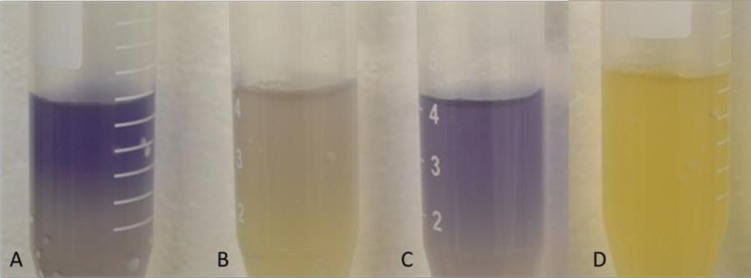
To investigate a role for the Pcb1692 Fur protein in N-acyl homoserine lactones (AHLs) synthesis, we compared the Pcb1692, Pcb1692Δfur-pfur, Pcb1692Δfur and Pcb16920ΔexpI strains with respect to their ability to synthesize and produce AHLs. To this end, the C. violaceum CV026 reporter strain was used. As expected, Pcb1692 (Fig 4A) produced a strong blue colour, indicative of AHL production. On the contrary, the Pcb1692Δfur mutant strain produced a faint blue colour, indicating minimal AHL production by this strain. Trans-complementation of the mutant strain with the wild-type fur gene in the mutant strain (Pcb1692Δfur-pfur) restored AHL production (Fig 4C). A Pcb1692ΔexpI mutant strain that lacks the ability to produce AHLs was used as a negative control [7] and accordingly showed no colour change (Fig 4D). These results implicate the Pcb1692 Fur protein, either directly or indirectly, in AHL production.
Fig 4. Effect of Fur on synthesis of AHLs in Pcb1692Δfur mutant relative to the Pcb1692 wild type strain.
Production of AHLs was indicated by formation of blue color ring 48 hours after each strain was co-inoculated with CV026 reporter strain at 28°C. (A) Pcb1692 wild-type strain (B) Pcb1692Δfur mutant (C) Pcb1692Δfur-pfur complemented mutant strain and (D) Pcb1692ΔexpI mutant strain.
Contribution of Fur in the virulence of Pcb1692
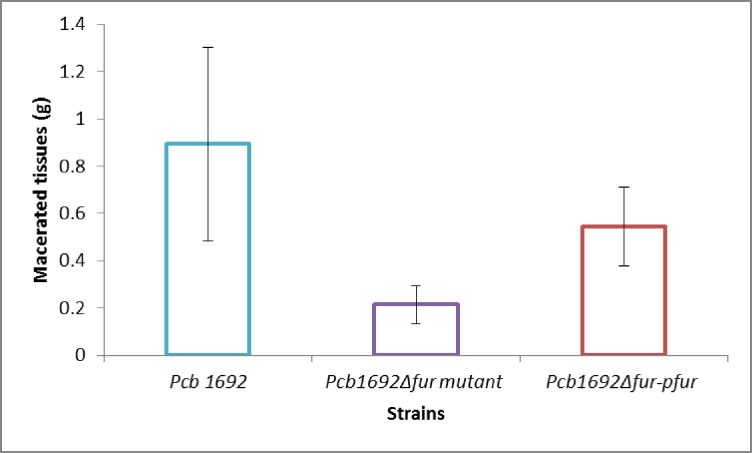
To determine whether the Fur protein plays a role in the ability of Pcb1692 to macerate potato tubers, surface-sterilized potato tubers were inoculated with standardized cultures of thePcb1692, Pcb1692Δfur and Pcb1692Δfur-pfur strains. As a control, potato tubers were inoculated with MgSO4 buffer. At 72 h post-inoculation, macerated tissue was scooped and weighed. The results showed that the average weight of the macerated tissue due to thePcb1692 wild-type strain was significantly (p< 0.05) higher compared to that of the Pcb1692Δfur mutant (Fig 5). In addition, trans-complementation of the fur gene in the Pcb1692Δfur mutant strain (Pcb1692Δfur-pfur strain) restored the virulence similar to the wild-type strain (Fig 5). Potato tubers mock-inoculated with 10 mM MgSO4 showed no tissue maceration. The findings suggest that the Fur regulon of Pcb1692 may include several virulence factors involved in maceration of potato tubers.
Fig 5. Effect of Fur in Pcb1692 virulence on potato tubers.
Susceptible potato tuber (cv Mondial) were inoculated with Pcb1692 wild type strain, Pcb1692Δfur mutant and Pcb1692Δfur-pfur complemented strain (OD600 equivalent to 1). MgSO4 was used as negative control. Macerated tissue was weighed at 72 hpi.
Biofilm formation
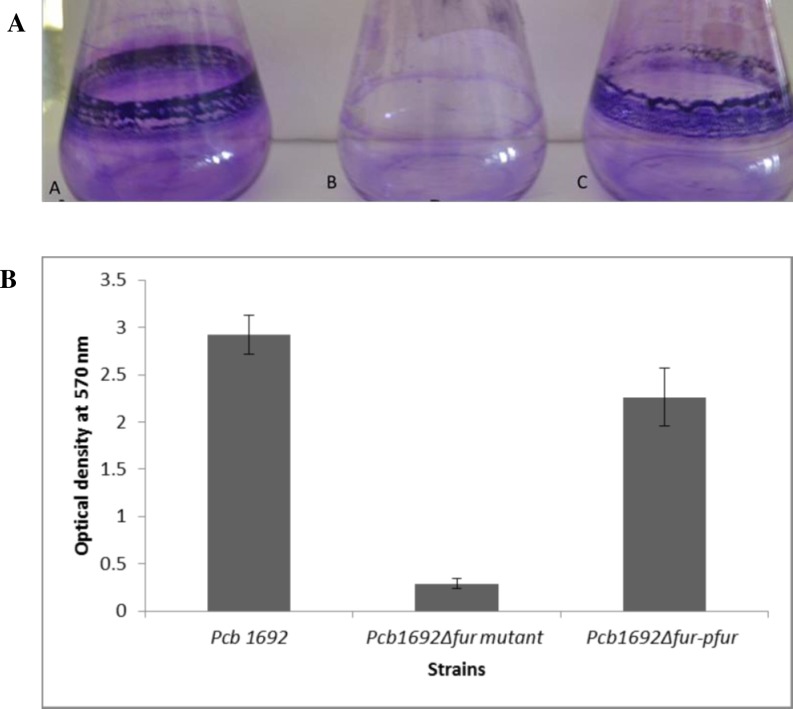
Biofilm formation is an important virulence determinant of many phytopathogens and animal bacterial pathogens. In Pcb, biofilm formation may contribute to the colonization of potato tubers and eventually disease development. We thus investigated whether the Fur protein affects biofilm formation by Pcb1692 using crystal violets (CV) staining assay. The results indicated that thePcb1692 wild-type and Pcb1692Δfur-pfur complemented strains both formed a biofilm on the inner surface of a conical flask, whereas the Pcb1692Δfur mutant did not form a biofilm under the same assay conditions (Fig 6A). These results were further confirmed with a quantitative biofilm assay, the results of which demonstrated that the absorption mean value of the biofilm by the Pcb1692Δfur mutant was significantly reduced (p<0.05) with 2 fold reduction compared to those of the Pcb1692 wild-type and Pcb1692Δfur-pfur strains (Fig 6B, Table 4).
Fig 6. Effect of fur mutation on biofilm formation in Pcb1692.
A. Qualitative biofilm formation showing biofilms in conical flasks after crystal violet staining. Flask A, B, and C represents Pcb1692 wild-type, Pcb1692Δfur mutant strain, and Pcb1692Δfur-pfur strain respectively. B. Quantitative biofilm formation by Pcb1692 wild-type, Pcb1692Δfur mutant and Pcb1692Δfur-pfur complement strain. Standard deviation and averaged mean values represent optical densities (OD570) from three independent experiments. Statistically significant difference were determined by the one way ANOVA and p-values less than 0.05 (p<0.05) were considered to be statistically significant.
Table 4. Quantitative biofilm formation between Pcb1692 wild-type, Pcb1692Δfur mutant and Pcb1692Δfur-pfur complement strain.
Standard deviation and averaged mean values represent optical densities (OD570) from three independent experiments. Statically significant differences were determined by the one way ANOVA and p-values less that 0.05 (p<0.05) were considered to be statistically significant (p<0.05).
| Strain | Optical Density (OD570) |
|---|---|
| Pcb1692 wild-type | 2.92± 0.204 |
| Pcb1692Δfur mutant | 0.289± 0.052 |
| Pcb1692Δfur-pfur | 2.26± 0.307 |
The Pcb1692Δfur mutant strain is attenuated in extracellular enzyme production
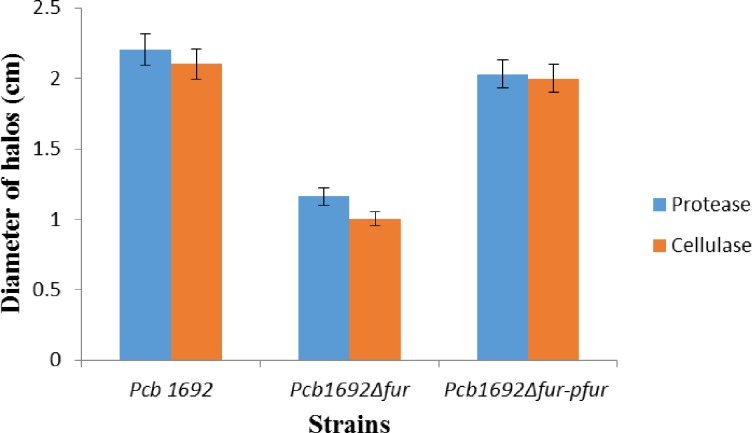
Pcb1692, like other brute-force pathogens, secretes several plant cell wall-degrading enzymes (PCWDEs) that macerate plant tissues, thereby releasing nutrients and iron. Given that Fur regulates iron homeostasis and secretion of some PCWDEs in soft rot Enterobacteriaceae [16], we reasoned that the reduced virulence associated with the Pcb1692Δfur mutant strain in potato tubers may be associated with reduced synthesis and secretion of PCWDEs. To test this hypothesis, we assayed the production of two different PCWDEs, namely cellulase and protease, as described in the Materials and Methods. Consistent with our hypothesis, the results showed that both the cellulase and protease activities were significantly reduced in the Pcb1692Δfur mutant strain compared to the Pcb1692 wild-type and Pcb1692Δfur-pfur complemented strains, thus implicating Fur, directly or indirectly, in the synthesis and production of some PCWDEs in Pcb1692 (Fig 7).
Fig 7. Quantitative cellulase and protease activity of Pcb1692, Pcb1692Δfur mutant and Pcb1692Δfur-pfur complement strain.
Halo diameter in Pcb1692Δfur was significantly reduced compared to Pcb1692 wild-type and Pcb1692Δfur-pfur (p<0.05).
Involvement of Fur in Pcb1692 EPS production
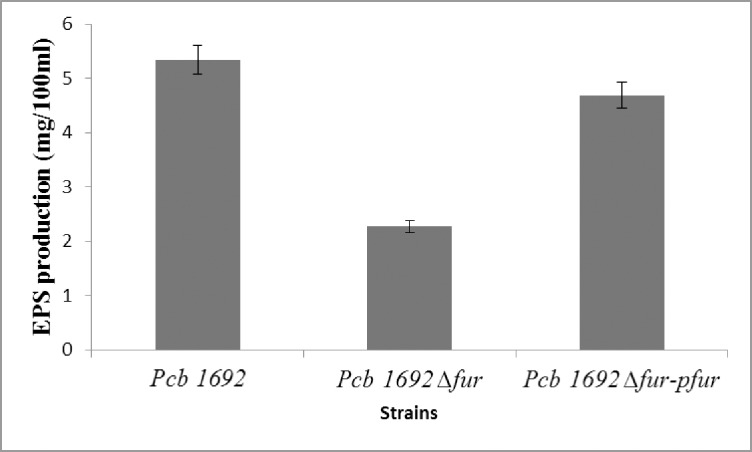
Pcb1692 causes stem rot and eventual wilting of potato plants [2]. This phenomenom has been associated with the production of copious amounts of EPS which occludes xylem vessels, resulting in wilting and die-back of plants [38]. In addition, EPS is a major component of biofilms that aids in protecting bacteria from UV light, plant antimicrobial agents, and serves as a source of nutrients to the bacteria, thus making EPS production an important virulence determinant in several bacteria, including phytopathogens [38]. EPS production was determined by weighing the amount of EPS produced by the wild-type, Pcb1692Δfur and Pcb1692Δfur-pfur strains grown in LB broth for 72 h. Our results showed that wild-type Pcb1692 produced 5.34 mg/100ml of EPS compared to 2.27 mg/100ml for Pcb1692Δfur mutant and 4.69 mg/100ml for the complemented Pcb1692Δfur-pfur strain (Fig 8). These findings implicate the Pcb1692 Fur protein in the regulation of EPS production.
Fig 8. Comparison of extracellular EPS production by Pcb1692Δfur and Pcb1692 wild type strains.
EPS production in Pcb1692Δfur was significantly reduced compared to Pcb1692 wild-type and Pcb1692Δfur-pfur (p<0.05). The error bars represent EPS mean values from the three experiments and three biological experiments.
Gene expression
Given that Fur has been implicated in the regulation of bacterial genes associated with virulence, and fitness [36], we next investigated a role for the Pcb1692 Fur protein in regulating the expression of randomly selected virulence and fitness genes. The genes selected in this study have been shown previously to be under regulation the Fur protein. The genes selected for qRT-PCR included: AED-0004132 (encoding ferredoxin), hasA (encoding an extracellular heme-binding protein) and sodC (encoding a copper-zinc superoxide dismutase). We also included genes encoding PCWDEs such as prtA (encoding a protease) and celV (encoding cellulase), as well as motA (encoding the flagellar motor protein) and fliC (encoding a flagellar transcriptional activator).
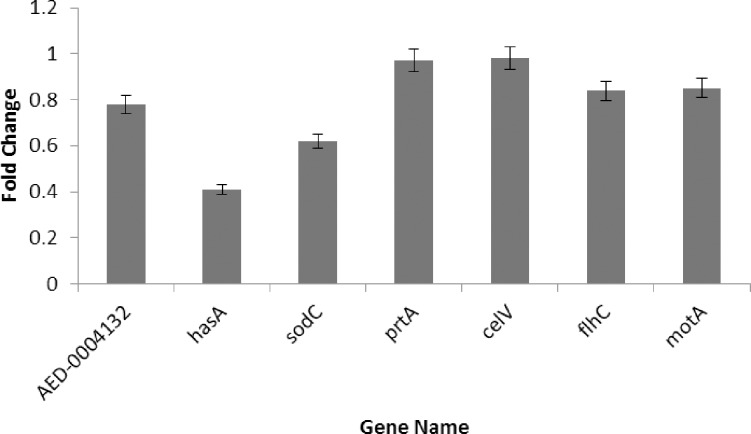
The qPCR results indicated that the relative expression of AED-0004132, hasA and sodC was significantly reduced in the Pcb1692Δfur mutant strain (0.78-, 0.41- and 0.62-fold, respectively) compared to the Pcb1692 wild-type strain. These findings confirm that the Fur protein of Pcb1692 is involved in iron homeostasis and the stress response. In addition, motA (0.84-fold) and flhC (0.85-fold) were significantly down-regulated in the Pcb1692Δfur mutant strain compared to Pcb1692 wild-type, indicating that expressions of some flagella-related genes are also regulated by Fur (Fig 9). Conversely, both the celV and prtA gene expression levels were significantly reduced in the Pcb1692Δfur mutant strain (0.97-and 0.98-fold, respectively) compared to Pcb1692 wild-type confirming that Fur regulates production of some PCWDEs based on our results.
Fig 9. Differentially expressed candidate pathogenicity genes involved in iron uptake.
(AED-004132, hasA), oxidative stress tolerance (sodC), plant cell wall degrading enzymes (prtA, celV) and motility (flhC, motA) in Pcb1692Δfur mutant strain relative to Pcb1692 wild-type. The ffh gene encoding signal recognition particle subunit was used as housekeeping gene to normalize gene expression.
Putative ‘fur boxes’ identified upstream of a selected number of genes in Pcb1692
In an attempt to establish whether the observed phenotypes of Fur mutant relative to the wild type strain are indeed a consequence of Fur regulation, we searched for presence of fur boxes upstream of a selected number of genes responsible for some of the observed phenotypes. Our bioinformatics analysis indicated that genes regulated by Fur contained a GATAAT iron binding sequence indicated by red boxes (S3 Fig). These sequences were all found upstream of the selected genes within or near the promoter regions. The consensus sequence shows that at least one repeat of conserved GATAAT sequence is present within the selected genes with AT rich regions prior to/or following the repeat. The exact GATAAT hexameric repeat and corresponding palindrome sequences were not identified here.
Discussion
The function of the ferric uptake regulator (Fur) protein has been studied in a number of bacteria, including plant pathogens such as Erwinia chrysanthemi, Pantoea sterwartii, Xanthomonas vesicatoria and Xanthomonas campestris pv. campestris [15, 16, 39, 40]. However, the role of fur in Pcb1692, an important emerging pathogen of potatoes and other crops, has not been investigated before. In addition, the mode of action of the Fur protein has been extensively studied in E. coli, where it has been shown that in the presence of ferric ion (Fe2+), the Fur protein binds and forms a complex with Fe2+ [25]. The resulting Fur-Fe2+ complex binds to a conserved consensus sequence called the ‘fur box’, which is located within the promoter region of many genes. This binding results in transcriptional repression of genes involved in iron acquisition and storage [25]. Given that Fur cannot bind to the ‘fur box’ sequence in the absence of iron, transcriptional repression of target genes is relieved [41].
In the current study, we deleted the fur gene from the genome of Pcb1692 and functionally characterized its role in virulence, fitness and host-pathogen interactions. Our data demonstrated that siderophore production was undetectable in Pcb1692 wild-type, while the Pcb1692Δfur mutant strain produced statistically significant higher levels of siderophore. These findings demonstrate that Fur of Pcb1692 negatively regulates the synthesis and production of siderophores. Siderophores are iron scavengers that are produced by many Gram-negative bacteria as a crucial means of acquiring iron in iron-limiting environments [42]. Consistent with our findings, deletion of the fur gene in many bacteria, including X. vesicatoria, resulted in an increased production of siderophores [39]. In addition, the Pcb1692Δfur mutant was more sensitive to oxidative stress and was attenuated in EPS production, biofilm formation, swimming motility and virulence in potato tubers. Together, these findings indicate that the Fur protein of Pcb1692 plays a vital role in regulating many virulence factors, directly or indirectly.
Fur plays a pivotal role in preventing damage caused by the production of reactive oxygen species (ROS) within the host plants by regulating genes involved in reactive oxygen detoxification [14]. In many Gram-negative bacteria, deletion of the fur gene results in cell death due to oxidative stress and damage [19]. Consistent with these findings, our results indicated that survival of the Pcb1692Δfur mutant strain was severely reduced compared to the Pcb1692 wild-type strain and the complemented strain Pcb1692Δfur-pfur when cultured in the presence of 20μM of H2O2. These findings demonstrate that the Pcb1692Δfur mutant strain was impaired in its ability to resist oxidative stress and damage. It is possible that the Pcb1692Δfur mutant strain may have lost the ability to regulate genes involved in degradation of ROS. This hypothesis is supported by gene expression analysis, which showed that the sodC gene (encoding for superoxide dismutase) was significantly down-regulated in the Pcb1692Δfur mutant strain compared to the wild-type (Fig 9).
Motility is an important virulence determinant during the epiphytic and invasion stages in many pathogenic bacteria, including Pectobacterium carotovorum subsp. carotovorum, and enables bacterial cells to swim to nutrient-rich niches or avoid environmental stresses [43]. Moleleki and colleagues observed that quorum sensing-defective Pcb1692 was not motile and suggested that quorum sensing regulates genes involved in flagella synthesis [7]. Our results demonstrated that motility of the Pcb1692Δfur mutant strain was significantly reduced compared to the wild-type strain, suggesting that the Pcb1692 Fur protein may be involved in regulating motility. Consistent with these findings, gene expression levels of flhC and motA were significantly reduced in the Pcb1692Δfur mutant strain compared to the wild-type strain. It is currently not clear how Fur regulates flagella and quorum sensing-related genes in Pcb1692 and therefore, needs to be investigated in the future studies.
Iron acquisition has been shown to be an important virulence factor for many pathogenic bacteria; iron uptake is tightly regulated by the ferric uptake regulator (fur) [14]. In this study we found that the Pcb1692Δfur mutant strain was significantly reduced in virulence compared to the wild-type strain, suggesting that Fur contributes to Pcb1692 virulence on potato tubers. We hypothesized that disrupting the fur gene, which is a metal-sensing system in Pcb, resulted in high uptake of iron thereby causing toxicity and hence, cell death. High intracellular concentrations of iron results in the generation of reactive oxygen species, causing oxidative stress and eventually damage cells [14]. Previous studies have shown that mutation of the fur gene in X. vesicatoria and X. campestris pv. campestris resulted in reduced virulence, biofilm formation, EPS and AHL production; similar to the results observed for the Pcb1692Δfur mutant in this study [16, 39]. Expression of the AED-0004132 (ferredoxin) and hasA (extracellular heme-binding protein) genes, which are involved in iron uptake, were significantly down-regulated in the Pcb1692Δfur mutant strain. It is tempting to speculate that the Fur protein of Pcb1692 is may therefore be an important regulator of iron uptake during in planta infection and may be involved directly or indirectly in regulating other virulence factors such as motility, which is important for successful pathogenesis. These findings strongly suggest that Fur is more likely to be a regulator of the expression of virulence determinants in Pcb1692.
Production of PCWDEs such as proteases, cellulases and pectinases by many phytopathogens has been shown to be important for plant disease symptom development [10]. In Pectobacterium atrosepticum (Pba), production and secretion of PCWDEs and other virulence factors is tightly regulated by a quorum sensing mechanism [39]. Quorum sensing is only one of several complex regulatory networks that modulate the expression of virulence factors in bacteria. Lamont and colleagues showed that iron availability not only controls the production and secretion of siderophores, but also regulates production of PCWDEs (protease) and antibiotics in P. aeruginosa [44]. It is interesting to note that production of cellulase and proteases were significantly lower in Pcb1692Δfur compared to the wild-type strain and the complemented Pcb1692Δfur-pfur strain. These phenotypes were confirmed using gene expression analysis, which demonstrated that the celV and prtA expression levels were significantly reduced in the Pcb1692Δfur mutant compared to the Pcb1692 wild-type strain. A link between quorum sensing and the Fur protein was investigated by evaluating the production of N-acyl homoserine lactones (AHLs) in a quorum sensing-deficient mutant (Pcb1692ΔexpI) and the Pcb1692Δfur mutant strain. It was observed that while Pcb1692ΔexpI completely lacked the ability to produce AHLs, the Pcb1692Δfur mutant displayed reduced production of AHLs compared to the Pcb1692 and Pcb1692Δfur-pfur strains. This, it appears that Fur may be directly or indirectly involved in the regulation of virulence factors under quorum sensing control.
The Fur protein in Escherichia coli has been well characterized and shown to act as a transcriptional repressor to iron-related genes by binding to ‘fur boxes’ found in the promoter region [22]. The complex prevents the entry of RNA polymerase therefore inhibiting initiation of transcription [22]. Our bioinformatics analysis revealed predicted ‘fur boxes’ in a number of genes involved in iron uptake, oxidative stress, motility and plant cell wall degrading enzymes. These results suggested that Fur protein in Pcb1692 represses not only iron-regulated genes but many genes coding for other virulence factors. Indicating fur regulon in Pcb1692 plays a more important role in virulence than previously thought. This argument is supported by results obtained by Franza and colleagues where they found that some genes coding for PCWDEs harbour ‘fur boxes’ [16].
In conclusion, Pcb1692 fur gene was characterized and shown to regulate the expression of genes involved in iron acquisition and iron storage systems. Regulation of iron uptake could be pivotal for survival and virulence of Pcb1692 in potato tubers, given that in the early stages of infection iron is limiting in this environment and Pcb1692 must therefore compete with other bacteria for the limited iron. At the later stage of infection when Pcb1692 reaches a high cell density (quorum) the pathogen synthesizes and secretes PCWDEs which macerate potato tubers thereby releasing nutrients and iron. It may therefore be pivotal for Pcb1692 to coordinate production of PCWDE with availability of iron. Conversely, the presence of excess iron leads to oxidative burst generated from the Fenton reaction. To survive this oxidative stress Pcb1692 produces proteins such as superoxide dismutase (SodC) which neutralize the deleterious effects of the reactive oxygen species. Furthermore, when nutrients and iron are depleted from macerated tissue the Pcb1692 Fur protein then, directly, or indirectly up-regulates flagella genes allowing the pathogen to move to a nutrient rich environment. Together, the data presented here shows that the Pcb1692 Fur regulon is not limited to iron metabolism but also regulates genes which encode protein associated with virulence factors, motility, oxidative stress and quorum sensing.
Supporting information
A) Using specific set of primers, PCR amplifications of the fur upstream and downstream regions were generated as indicated in S1A Fig. Kanamycin cassette was amplified from pKD4 plasmid with primers Kan F and Kan R. B) Primers Fur F and R, were used in a PCR reaction consisting of, the fur upstream kanamycin and downstream PCR fragment to generate a PCR fusion product. C) The fusion product was electroporated into electrocompetent Pcb1692 to generate the Pcb1692Δfur mutant strain (S1D Fig). Both electrocompetent Pcb1692 and Pcb1692Δfur mutant strain were electroporated with empty pTrc99A.
(TIF)
Lane 1. DNA ladder, 2. fur downstream PCR fragment, 3. kanamycin cassette PCR product, 4. fur upstream PCR fragment, 5. Fusion product consisting of the downstream, kanamycin and upstream fragment. 6. The fragment used for complementation which contains the fur gene and its promoter region. 7. Control.
(TIF)
Based on our qRT-PCR results, some of the genes under the Pcb1692 Fur regulon were aligned to the consensus fur box and the putative fur boxes for each gene is indicated by red boxes.
(TIF)
Acknowledgments
This research study was funded by the National Research Foundation (NRF), South Africa through Competitive Funding for Rated Researchers (CFRR) 98993; NRF Bioinformatics and Functional Genomics (BFG 93685) and NRF Research Technology and Transfer Fund (RTF) 98654. CKT was funded by University of Pretoria Bursary and SLP funded by the NRF Grant Holder Linked Bursary. SLP was also funded by Potatoes South Africa Transformation Bursary.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research study was funded by the National Research Foundation (NRF), South Africa through Competitive Funding for Rated Researchers (CFRR) 98993; NRF Bioinformatics and Functional Genomics (BFG 93685) and NRF Research Technology and Transfer Fund (RTF) 98654. CKT was funded by University of Pretoria Bursary and SLP funded by the NRF Grant Holder Linked Bursary. SLP was also funded by Potatoes South Africa Transformation Bursary. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Charkowski AO. The soft rot Erwinia. Plant-associated bacteria: Springer; 2007. p. 423–505. [Google Scholar]
- 2.Duarte V, De Boer S, Ward L, Oliveira A. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. Journal of Applied Microbiology. 2004;96(3):535–45. [DOI] [PubMed] [Google Scholar]
- 3.Onkendi E, Maluleke L, Moleleki L. First Report of Pectobacterium carotovorum subsp. brasiliense Causing Soft Rot and Blackleg of Potatoes in Kenya. Plant Disease. 2017;101(2):279–287. [DOI] [PubMed] [Google Scholar]
- 4.Panda P, Fiers M, Armstrong K, Pitman A. First report of blackleg and soft rot of potato caused by Pectobacterium carotovorum subsp. brasiliensis in New Zealand. New Dis Rep. 2012;26:15. [Google Scholar]
- 5.van der Merwe JJ, Coutinho TA, Korsten L, van der Waals JE. Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. European Journal of Plant Pathology. 2010;126(2):175–85. [Google Scholar]
- 6.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS pathogens. 2008;4(6):e1000093 doi: 10.1371/journal.ppat.1000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moleleki L, Pretorius R, Tanui C, Mosina G, Theron J. A quorum sensing‐defective mutant of Pectobacterium carotovorum subsp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Molecular Plant Pathology. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubheka GC, Coutinho T, Moleleki N, Moleleki LN. Colonisation patterns of a mCherry-tagged Pectobacterium carotovorum subsp. brasiliense strain in potato plants. Phytopathology. 2013;(ja). [DOI] [PubMed] [Google Scholar]
- 9.Alfano JR, Collmer A. Type III secretion system effector proteins: Double Agents in Bacterial Disease and Plant Defense. Annual Review of Phytopathology.2004;42(1):385–414. [DOI] [PubMed] [Google Scholar]
- 10.Barras F, van Gijsegem F, Chatterjee AK. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annual Review of Phytopathology. 1994;32(1):201–34. [Google Scholar]
- 11.Collmer A, Schneider DJ, Lindeberg M. Lifestyles of the effector rich: genome-enabled characterization of bacterial plant pathogens. Plant Physiology. 2009;150(4):1623–30. doi: 10.1104/pp.109.140327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Toole R, Milton DL, Wolf‐Watz H. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Molecular Microbiology. 1996;19(3):625–37. [DOI] [PubMed] [Google Scholar]
- 13.Fones H, Preston GM. Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS Microbiology letters. 2012;327(1):1–8. doi: 10.1111/j.1574-6968.2011.02449.x [DOI] [PubMed] [Google Scholar]
- 14.Fones H, Preston GM. The impact of transition metals on bacterial plant disease. FEMS Microbiology Reviews. 2013;37(4):495–519. doi: 10.1111/1574-6976.12004 [DOI] [PubMed] [Google Scholar]
- 15.Franza T, Expert D. Iron uptake in soft rot Erwinia. Iron Uptake and Homeostasis in Microorganisms. 2010:101–15. [Google Scholar]
- 16.Franza T, Sauvage C, Expert D. Iron regulation and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Molecular Plant-microbe Interactions. 1999;12(2):119–28. doi: 10.1094/MPMI.1999.12.2.119 [DOI] [PubMed] [Google Scholar]
- 17.Charkowski A, Blanco C, Condemine G, Expert D, Franza T, Hayes C, et al. The role of secretion systems and small molecules in soft-rot enterobacteriaceae pathogenicity. Annual Review of Phytopathology. 2012;50:425–49. doi: 10.1146/annurev-phyto-081211-173013 [DOI] [PubMed] [Google Scholar]
- 18.Marquez-Villavicencio MdP Groves RL, Charkowski AO. Soft rot disease severity is affected by potato physiology and Pectobacterium taxa. Plant Disease. 2011;95(3):232–41. [DOI] [PubMed] [Google Scholar]
- 19.Hassett DJ, Sokol PA, Howell ML, Ma J- F, Schweizer HT, Ochsner U, et al. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. Journal of Bacteriology. 1996;178(14):3996–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagg A, Neilands J. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26(17):5471–7. [DOI] [PubMed] [Google Scholar]
- 21.Escolar L, Lorenzo Vd, Pérez‐Martíín J. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Molecular Microbiology. 1997;26(4):799–808. [DOI] [PubMed] [Google Scholar]
- 22.La Escolar, Pérez-Martín J, de Lorenzo Vc. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. Journal of Molecular Biology. 1998;283(3):537–47. doi: 10.1006/jmbi.1998.2119 [DOI] [PubMed] [Google Scholar]
- 23.Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. Journal of Bacteriology. 1993;175(17):5428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stojiljkovic I, Bäumler AJ, Hantke K. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. Journal of Molecular Biology. 1994;236(2):531–45. doi: 10.1006/jmbi.1994.1163 [DOI] [PubMed] [Google Scholar]
- 25.Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiology and Molecular Biology Reviews. 1997;61(3):319–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baichoo N, Helmann JD. Recognition of DNA by Fur. Journal of Bacteriology. 2002; 184(21):5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauvage C, Franza T, Expert D. Analysis of the Erwinia chrysanthemi ferrichrysobactin receptor gene: resemblance to the Escherichia coli fepA-fes bidirectional promoter region and homology with hydroxamate receptors. Journal of Bacteriology. 1996;178(4):1227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 2000;97(12):6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: Cold spring harbor laboratory press; New York; 1989. [Google Scholar]
- 30.Velkov T, Chuang S, Prankerd R, Sakellaris H, Porter CJ, Scanlon MJ. An improved method for the purification of rat liver-type fatty acid binding protein from Escherichia coli. Protein expression and purification. 2005;44(1):23–31. doi: 10.1016/j.pep.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 31.Shyntum DY, Theron J, Venter SN, Moleleki LN, Toth IK, Coutinho TA. Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Molecular Plant-Microbe Interactions. 2015;28(4):420–31. doi: 10.1094/MPMI-07-14-0219-R [DOI] [PubMed] [Google Scholar]
- 32.Louden BC, Haarmann D, Lynne AM. Use of blue agar CAS assay for siderophore detection. Journal of Microbiology &Biology Education: JMBE. 2011;12(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(12):3703–11. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Mendoza D, Coulthurst SJ, Sanjuán J, Salmond GP. N-Acetylglucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiology. 2011;157(12):3340–8. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee A, Cui Y, Chatterjee AK. RsmC of Erwinia carotovora subsp. carotovora negatively controls motility, extracellular protein production, and virulence by binding FlhD and modulating transcriptional activity of the master regulator, FlhDC. Journal of Bacteriology. 2009;191(14):4582–93. doi: 10.1128/JB.00154-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K12: fur not only affects iron metabolism. Molecular and General Genetics MGG. 1987;210(1):135–9. [DOI] [PubMed] [Google Scholar]
- 37.Tang J- L, Liu Y- N, Barber C, Dow J, Wootton J, Daniels M. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Molecular and General Genetics MGG. 1991;226(3):409–17. [DOI] [PubMed] [Google Scholar]
- 38.Marcotte L, Kegelaer G, Sandt C, Barbeau J, Lafleur M. An alternative infrared spectroscopy assay for the quantification of polysaccharides in bacterial samples. Analytical Biochemistry. 2007;361(1):7–14. doi: 10.1016/j.ab.2006.11.009 [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Dong C, Zhao T, Han J, Wang T, Wen X, et al. Functional Analysis of the Ferric Uptake Regulator Gene fur in Xanthomonas vesicatoria. PloS One. 2016;11(2):e0149280 doi: 10.1371/journal.pone.0149280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burbank L, Mohammadi M, Roper MC. Siderophore-mediated iron acquisition influences motility and is required for full virulence of the xylem-dwelling bacterial phytopathogen Pantoea stewartii subsp. stewartii. Applied and Environmental Microbiology. 2015;81(1):139–48. doi: 10.1128/AEM.02503-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagg A, Neilands J. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiological Reviews. 1987;51(4):509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touati D. Iron and oxidative stress in bacteria. Archives of Biochemistry and Biophysics. 2000;373(1):1–6. doi: 10.1006/abbi.1999.1518 [DOI] [PubMed] [Google Scholar]
- 43.Hossain MM, Shibata S, Aizawa S-I, Tsuyumu S. Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiological and Molecular Plant Pathology. 2005;66(4):134–43. [Google Scholar]
- 44.Lamont IL, Konings AF, Reid DW. Iron acquisition by Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Biometals. 2009;22(1):53–60. doi: 10.1007/s10534-008-9197-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Using specific set of primers, PCR amplifications of the fur upstream and downstream regions were generated as indicated in S1A Fig. Kanamycin cassette was amplified from pKD4 plasmid with primers Kan F and Kan R. B) Primers Fur F and R, were used in a PCR reaction consisting of, the fur upstream kanamycin and downstream PCR fragment to generate a PCR fusion product. C) The fusion product was electroporated into electrocompetent Pcb1692 to generate the Pcb1692Δfur mutant strain (S1D Fig). Both electrocompetent Pcb1692 and Pcb1692Δfur mutant strain were electroporated with empty pTrc99A.
(TIF)
Lane 1. DNA ladder, 2. fur downstream PCR fragment, 3. kanamycin cassette PCR product, 4. fur upstream PCR fragment, 5. Fusion product consisting of the downstream, kanamycin and upstream fragment. 6. The fragment used for complementation which contains the fur gene and its promoter region. 7. Control.
(TIF)
Based on our qRT-PCR results, some of the genes under the Pcb1692 Fur regulon were aligned to the consensus fur box and the putative fur boxes for each gene is indicated by red boxes.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.