Abstract
Much of the research aimed at defining the pathogenesis of Staphylococcus aureus has been done with a limited number of strains, most notably the 8325-4 derivative RN6390. Several lines of evidence indicate that this strain is unique by comparison to clinical isolates of S. aureus. Based on this, we have focused our efforts on two clinical isolates (UAMS-1 and UAMS-601), both of which are hypervirulent in our animal models of musculoskeletal infection. In this study, we used comparative genomic hybridization to assess the genome content of these two isolates relative to RN6390 and each of seven sequenced S. aureus isolates. Our comparisons were done by using an amplicon-based microarray from the Pathogen Functional Genomics Resource Center and an Affymetrix GeneChip that collectively represent the genomes of all seven sequenced strains. Our results confirmed that UAMS-1 and UAMS-601 share specific attributes that distinguish them from RN6390. Potentially important differences included the presence of cna and the absence of isaB, sarT, sarU, and sasG in the UAMS isolates. Among the sequenced strains, the UAMS isolates were most closely related to the dominant European clone EMRSA-16. In contrast, RN6390, NCTC 8325, and COL formed a distinct cluster that, by comparison to the other four sequenced strains (Mu50, N315, MW2, and SANGER-476), was the most distantly related to the UAMS isolates and EMRSA-16.
Staphylococcus aureus is an opportunistic pathogen capable of causing serious human infections. An increasing number of these infections are recalcitrant to antimicrobial therapy. Indeed, the National Nosocomial Infections Surveillance System has seen a rise in oxacillin-resistant S. aureus (ORSA) among hospital isolates from approximately 4% in the 1980s to >50% in the late 1990s (47). Of recent concern is the increasing prevalence of ORSA among patients suffering from community-acquired infections (19). Research comparing these community-acquired ORSA isolates with hospital-acquired ORSA isolates found that the increase in community-acquired ORSA infections can be attributed to the acquisition of a novel SCCmec element (19). Furthermore, both vancomycin-intermediate and vancomycin-resistant S. aureus were recently described (66), confirming that S. aureus has the potential to resist even those antimicrobials often reserved as a “last resort.” This emphasizes the evolutionary adaptability of S. aureus and highlights the importance of finding novel therapeutic approaches in the face of such a readily adaptable pathogen (50).
Much of the research aimed at identifying novel therapeutic targets for the treatment of S. aureus infections has been directed at identification of specific virulence factors and regulatory circuits that are relevant to the disease process. Almost invariably, these studies used the prototypic lab strain NCTC 8325 or its derivatives. The single most widely studied strain is RN6390, which is an 8325-4 strain generated by curing three prophage from NCTC 8325 (1, 12, 45). However, RN6390 has an 11-bp deletion in rsbU, which encodes a positive regulator essential for the activity of the stress response sigma factor SigB (33). This raises the possibility that regulatory networks defined in studies focusing on RN6390 are not representative of the situation observed with clinical isolates, and we recently confirmed that this is, in fact, the case. For instance, we demonstrated that mutation of the staphylococcal accessory regulator (sarA) has a disparate phenotypic effect in RN6390 by comparison to clinical isolates (6). Perhaps more importantly, direct comparisons in our murine septic arthritis model indicate that RN6390 is less virulent than our prototype clinical isolate (7), and preliminary experiments suggest that this is also true with our rabbit model of postsurgical osteomyelitis (data not shown).
Even among clinical isolates, there is both genotypic and phenotypic evidence to suggest that a limited number of clonal variants are of particular significance. For instance, Booth et al. (8) used pulsed-field gel electrophoresis to examine 405 clinical isolates from various types of infection and diverse geographic locations and identified 90 different lineages. Two of these accounted for more than half of all infections. One of the genetic characteristics that best defined the two most prevalent lineages was the presence of cna, which encodes the primary, if not the only, collagen binding adhesin produced by S. aureus (23). Specifically, cna was present in more than 93% of the strains in the two most prevalent lineages. This is significant in that cna is present in less than half of all S. aureus strains (24). In another study, Peacock et al. (49) examined the presence of genes encoding 33 putative virulence factors in 334 S. aureus isolates and found that seven genes (fnbA, ica, sdrE, sej, eta, hlg, and cna) were significantly more common in invasive isolates than in carriage isolates. With respect to phenotypic characteristics, Papakyriacou et al. (48) found that the most prevalent clinical isolates of S. aureus have a phenotype characterized by a high binding capacity for host proteins and limited production of extracellular virulence factors (e.g., toxins and degradative enzymes). It was suggested that such strains may predominate because they have a phenotype that favors the colonization phase of infection (48).
We have focused our efforts on clinical isolates of S. aureus, most notably UAMS-1 and UAMS-601. UAMS-1 is an oxacillin-susceptible strain (OSSA) isolated from the bone of a patient suffering from osteomyelitis (22). UAMS-601 is an ORSA isolate responsible for an outbreak in the neonatal intensive care unit of our hospital (64). Importantly, both of these strains share phenotypic and genotypic properties associated with the predominant clonal lineages discussed above. Specifically, both are cna-positive strains that also contain other genes that Peacock et al. (49) found were associated with invasive isolates, and both have a high binding capacity for host proteins and produce limited amounts of most exoproteins (6, 23). Both of these strains have an intact rsbU locus, and both are highly virulent in our animal models of musculoskeletal infection (6).
The characteristics that define the UAMS isolates and contribute to their virulence in our animal models may be due to unique genetic content and/or differential expression of the regulatory circuits that control the coordinated production of conserved virulence factors. In this study, we address the first of these possibilities. Although survey studies like those reported by Peacock et al. (49) have provided important information toward this goal, such studies are by necessity limited to studying particular loci of interest. In fact, few studies have addressed the overall genomic diversity among clinical isolates of S. aureus. An exception is the work of Fitzgerald et al. (20), who used microarray-based comparative genomic hybridizations (CGH) to define 18 “regions of difference” among several S. aureus strains. The microarray used in these experiments represented >90% of the COL genome. They found that individual strains lacked between 22 and 332 of the COL genes and that 78% of the genome was invariant. However, the converse experiments could not be performed, and the authors noted that “identification of genes present in … test strains but absent in COL is likely to provide new insight into the molecular mechanisms of pathogenic clone adaptation to specialized niches.” To date, little has been done in this regard, but the completed sequencing of additional S. aureus strains, including several clinical isolates, has allowed the development of more-comprehensive microarrays that can potentially be used to address this issue.
To this end, we carried out CGH experiments using two sets of microarrays that collectively represent the genomes of seven sequenced strains of S. aureus (COL, NCTC 8325, Mu50, N315, MW2, EMRSA-16 [SANGER-252], and SANGER-476). Hybridizations were done with genomic DNA from 10 strains, including the two virulent UAMS isolates (UAMS-1 and UAMS-601), the prototypical lab strain RN6390, and all seven sequenced strains. Inclusion of the sequenced strains assisted the development of cutoffs to determine the presence, absence, or divergence of open reading frames (ORFs) in the nonsequenced strains. We also included epidemiological typing methods to place our clinical isolates into well-defined epidemiological paradigms. Collectively, this allowed us to define the unique genetic content among clinically important strains of S. aureus, to cluster strains according to genetic similarity, and to extrapolate our findings to epidemiologically related strains worldwide.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are shown in Table 1. All sequenced strains except COL and NCTC 8325 (8325) were obtained from the culture collection maintained by the Network on Antimicrobial Resistance in Staphylococcus aureus (managed by Focus Technologies, Herndon, Va.). COL and 8325 were obtained from the culture collection of John Iandolo at the University of Oklahoma Health Sciences Center. Cultures were routinely maintained on tryptic soy agar with 10 μg per ml of nafcillin for ORSA isolates. Biofilm formation was assessed using a microtiter plate assay and tryptic soy broth supplemented with glucose and NaCl as previously described (5).
TABLE 1.
Strains
| Strain | Notes | Reference(s) |
|---|---|---|
| UAMS-1 | Osteomyelitis isolate, Little Rock, Ark. | 22 |
| UAMS-601 | Pediatric outbreak strain, Little Rock, Ark. | 64 |
| RN6390 | Laboratory strain | 44 |
| NCTC 8325 | Parent strain of RN6390 | University of Oklahoma Strain Collection, J.J Iandolo |
| SANGER-252 | EMRSA16, dominant ORSA in the UKa | 17, 43 |
| SANGER-476 | Hypervirulent community isolate, UK | 17 |
| MW2 | Community-acquired isolate causing septicemia and septic arthritis, North Dakota | 2 |
| Mu50 | VISA from Japan, surgical wound isolate | 34 |
| N315 | ORSA from Japan, pharyngeal smear isolate | 34 |
| COL | Archaic MRSA strain from UK | University of Oklahoma Strain Collection, J.J Iandolo |
UK, United Kingdom.
Microarray descriptions.
Two genome-scale microarrays that collectively represent all seven sequenced human isolates of S. aureus were used in these experiments. The first was produced by The Institute for Genomic Research (TIGR) as part of the Pathogen Functional Genomics Research Center (PFGRC) initiative. This amplicon-based array, which has been designated version 1.0 by PFGRC, was originally reported to contain PCR products representing 2,576 genes from COL and 117 unique genes from Mu50 (n = 60), MW2 (n = 51), and N315 (n = 6). However, upon examination of our preliminary data and quality control data from TIGR, some of the “unique” genes were found to yield high signals in the COL channel, suggesting that they had high nucleotide homology with genes that are present in the COL genome. This was subsequently confirmed by direct sequence comparisons (Chris Mader and Robin Cline, TIGR, personal communication). Thus, the total number of genes that were present on the array but absent in COL was 111. The total number of ORFs represented among the 2,693 spots on the PFGRC array was therefore 2,687. Each target was printed in triplicate at adjacent sites on the array. Printing pattern and annotation data can be found at http://pfgrc.tigr.org. The second array used in these experiments was a custom-made Affymetrix GeneChip based on the COL, N315, Mu50, EMRSA-16 (SANGER-252), SANGER-476, and 8325 genomes. Also represented were N315 intergenic sequences more than 50 bases in length and unique S. aureus GenBank entries (16). Overall, the GeneChip contained 7,723 S. aureus qualifiers recognizing 4,066 ORFS, 3,343 intergenic regions, and 69 exogenous control probe sets.
DNA isolation, labeling and hybridization.
For the PFGRC arrays, genomic DNA was isolated as described previously (40). The integrity of all DNA preparations was checked by PCR, using strain-specific primers (cna, sarA, or mecA) (Table 2). Genomic DNA was digested to completion with EcoRI (New England Biolabs, Beverly, Mass.) and repurified by using the QIAGEN QIAquick PCR purification kit (QIAGEN, Valencia, Calif.). Digested DNA (1 to 2 μg) was labeled with either Cy3 or Cy5, using the Genisphere Array 350RP kit (Genisphere, Hatfield, Pa.) according to the manufacturer's instructions. Hybridization was carried out for 12 h at 55°C with constant mixing, using the Ventana Discovery System hybridization station (Ventana Medical Systems, Tucson, Ariz.). This was followed by an additional 2-h hybridization at 55°C with Cy3- and Cy5-labeled Genisphere dendrimers as per the manufacturer's protocol and a final wash with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C. Each experiment was run as a competitive hybridization by using Cy3-labeled DNA from one of the 10 strains of interest and Cy-5-labeled DNA from COL. Additionally, experiments focusing on UAMS-1, UAMS-601, RN6390, and N315 were repeated with the opposite fluorophore to rule out any dye-specific differences in hybridization pattern. Finally, we repeated the hybridization of UAMS-1 versus COL a total of four times to assess the reproducibility of our results.
TABLE 2.
Primers
| ORF name or description | PFGRC locus no.a | Template strain | Primer 1 | Primer 2 | Product length (bp) | Reference |
|---|---|---|---|---|---|---|
| seh | NA | MW2 | ATTACTTTCATTCACATCATATGC | GTCACATTTTTCTTATTTGTTCG | 431 | This study |
| sej | NA | U1 | TTGCAATATGTAGTGTAAACAATGC | CAGCTACTCCTATATTTTTAGGCTCG | 421 | This study |
| sarT | SA2506 | RN6390 | GTAAGGGATGAACTCGAGATGAATGATT | ACGGGGATCCAAAAATACATTTAACTGC | 420 | 61 |
| sarU | SA2507 | RN6390 | TGACGATTTCGGCTGAACTTC | TGGAACACGAAATGGTGAAC | 2,300 | 39 |
| trap | SA1891 | RN6390 | CGCGCGGATCCCAACTATTCCAATTTTCAG | CGCGAAGCTTCTTAAAGTCTTCGTATG | 317 | 3 |
| cna | NA | U1 | CAAGCAGTTATTACACCAGACGG | CACCTTTTACAGTACCTTCAATACC | 688 | This study |
| sdrE | SA0610 | SANGER476 | CAGTAAATGTGTCAAAAGA | TTGACTACCAGCTATATC | 767 | 49 |
| bbp | NA | SANGER252 | CAGTAAATGTGTCAAAAGA | TACACCCTGTTGAACTG | 1,055 | 49 |
| clfB | SA2652 | U1 | TGCAAGATCAAACTGTTCCT | TCGGTCTGTAAATAAAGGTA | 596 | 49 |
| epiC | SA1876 | COL | CAAAAGCATCTACTGAAACC | CAACGTCGTCATTACACC | 779 | This study |
| isaA | SA2660 | COL | TTGACTTAGCGCATAATCACC | TTAGAATCCCCAAGCACC | 584 | This study |
| isaB | SA2584 | COL | GTGTAGCAGCAACATTAGC | TATTATGATCAACGACAAACC | 456 | This study |
| LPXTG motif protein | SA2676 | COL | CCTACAAATAACACGTTCC | GCACTATCGTAACTTAATCC | 602 | This study |
| lukD | SA1880 | COL | CAATTGCACTGCTTTTGC | ATTTTGATCCATTTAATCCACC | 447 | This study |
| Transcriptional regulator, GntR family | SA0091 | COL | ACTTATAGCGAAAGACATAATATTCC | AGACTAACCATACAAAATAACCTCC | 415 | This study |
| DNA-binding response regulator, putative | SA2646 | COL | TGATGATATACTGTCTATCTTTAACC | CATCTATGAAGTTTTCTGATTCC | 464 | This study |
| aur | SA2659 | COL | TTACGCATTATACATTGCAACC | CCAAGTACACCTTTTCCTTTACC | 413 | This study |
| lukM | SA2006 | COL | AGATAAAAATTCAACAGCACC | ACCATACTTCAAGTCATTCG | 514 | This study |
| sdrD | SA0609 | U1 | GGAAATAAAGTTGAAGTTTC | ACTTTGTCATCAACTGTAAT | 500 | 49 |
| ABC transporter, ATP-binding protein | SA1893 | COL | AAGTAGAACAGCTTACAGGTGG | CGGGATTAACGATAAACG | 447 | This study |
| LPXTG-motif protein | SA0119 | COL | CAAAATGTACAAAATCATCGTCC | CGTCGTCTAAATGCTAAGAATAACC | 590 | This study |
| fnbA | SA2511 | U1 | ATCTTAACTTTTCATTAACTCGCT | AATTTCCTCGACTGGTCCTTGTGC | 2,200 | 64 |
| fnbB | SA2509 | COL | ATGTTGAAACTCATGGTATCTCAAC | GAACGCCTTCATAGTGTCATTGAG | 2,277 | 64 |
NA, none available.
For hybridization to the Affymetrix GeneChips, S. aureus strains were grown overnight in brain heart infusion broth at 37°C with vigorous aeration. Cells were harvested by centrifugation and resuspended in 0.5 ml of Tris-EDTA buffer (pH. 8.0). Cell suspensions were lysed, using the FastPrep DNA procedure, in a FP120 reciprocating shaker (Q-BIOgene, Carlsbad, Calif.). Genomic DNA was then isolated by using a DNeasy tissue kit, following the manufacturer's instructions for bacterial DNA (QIAGEN, Valencia, Calif.). For DNA labeling, 5 μg of purified DNA was incubated at 90°C for 3 min and then plunged into an ice bath followed by standard DNA fragmentation and labeling procedures according to the manufacturer's instructions (Affymetrix Inc., Santa Clara, Calif.) for labeling mRNA for antisense prokaryotic arrays.
Array scanning and analysis.
Hybridized PFGRC arrays were scanned using the Perkin-Elmer ScanArray 5000 and read using ScanArray Express software (Perkin-Elmer, Boston, Mass.). Raw fluorescence intensity values were corrected for background by using ScanArray Express software and subsequently normalized by using the Lowess algorithm. Normalized values were expressed as a ratio of test strain intensity to COL strain intensity. These ratios were imported into Spotfire DecisionSite software (Spotfire, Somerville, Mass.) for graphical visualization and further analysis. The data were filtered for low spot signal intensity by using a threshold of the mean plus one standard deviation of the intensity observed from all empty and control spots on the array. Spots having COL channel normalized signal intensity less than this value were excluded from further analysis. However, the 111 unique spots from strains MW2, N315, and Mu50 were not subjected to this threshold, since these would be expected to have low signal intensity in the COL channel.
To develop cutoffs to determine “present” versus “absent” or “divergent” ORFs in our experiments, we included comparisons of each of the sequenced S. aureus strains versus COL. This allowed us to compare our microarray data with the results of BLAST comparisons. Specifically, BLASTN was used to compare the sequence of each ORF on the PFGRC array to the genomes of Mu50, N315, and MW2. This allowed for categorization of each ORF according to percent identity to COL, Mu50, N315, and MW2 sequences. Each ORF on the array was then categorized as present (≥90% nucleotide sequence identity), divergent (70 to 90% nucleotide sequence identity), highly divergent (≤70% nucleotide sequence identity), or absent (no hit based on a minimum alignment of 40 bp). For our analysis, the “absent” and “highly divergent” categories were subsequently combined under the heading of “absent.” Using this information, the following cutoffs were determined based on the ratio of normalized test strain intensity to normalized COL strain intensity: “absent,” ratio of <0.3; “divergent,” ratio of ≥0.3 but <0.5; “present,” ratio of ≥0.5. However, because the signal in the COL channel was low for those ORFs from strains other than COL, the criteria above were modified for these ORFs as follows: “present,” ratio of ≥2.0; “absent,” ratio of <2.0. Error rates were calculated based on the agreement of results from the BLAST-based versus the array-based categorization (see results).
Hybridized Affymetrix GeneChips were scanned as previously described (15). Signal intensities for elements tiled onto each chip were normalized to account for loading errors and differences in labeling efficiencies by dividing the log-transformed signal intensity of each qualifier (putative ORF or intergenic region) by the mean signal intensity for each individual chip. Results were analyzed with GeneSpring version 6.0 (Silicon Genetics, Redwood, Calif.) and Spotfire Decision Site. Based on the normalized signal intensities of each element, adjusted-call predictions were made to determine if an element was present, absent, or indeterminate (marginal) for UAMS-1, UAMS-601, and each of the sequenced S. aureus strains as previously described (16). Using this methodology, ∼97% of ORFs were correctly determined to be absent or present for each of the sequenced strains.
Hierarchical clustering.
Clustering was performed with PFGRC microarray data, using Spotfire Decision Site software. Consensus “present,” “absent,” and “divergent” calls for all CGH experiments were converted to integers and subjected to the complete linkage clustering method with the correlation similarity measure and average value ordering function. Calls for COL were based on the updated annotation of the microarrays as discussed above. Hierarchical clustering with the Affymetrix GeneChip data was performed by using GeneSpring version 6.0's Pearson correlation for all samples to develop a dendrogram that simultaneously compared the normalized signal intensity of each qualifier for a given strain to the signal intensity of the same qualifier across all strains analyzed.
Southern blotting for verification of microarray data.
DNA probes for Southern blotting were generated by PCR, using the primers shown in Table 2. Amplified fragments were labeled by using the Roche DIG-High Prime kit (Roche, Basel, Switzerland). Genomic DNA preparations were digested to completion with either EcoRI or EcoRI and AluI (sarT and sarU blots). Blots were then transferred, hybridized, and developed as previously described (64).
Epidemiological typing.
Multilocus sequence typing (MLST) was performed for strains UAMS-1, UAMS-601, and RN6390 according to the method of Maiden et al. (38). Primer sequences (Table 2) and specific protocols can be viewed at www.mlst.net. Amplification products were sequenced in triplicate to ensure their identity. The MLST sequence types for all of the sequenced strains were kindly provided by B.N. Kreiswirth (Public Health Research Institute, Newark, N.J.). SCCmec typing (46) was performed via multiplex PCR to identify polymorphisms in the staphylococcal chromosomal cassette region containing the mec element. Typing based on polymorphisms within the staphylococcal protein A gene (spa) was performed as previously described (62).
RESULTS
Genomic analysis of UAMS-1.
Based on previous work in our laboratory demonstrating that UAMS-1 is highly virulent in our animal models of musculoskeletal infection (7), we analyzed the results of our CGH experiments in the context of this strain. Of the 2,693 spots on the PFGRC array, 2,224 (82.6%) were present in UAMS-1, and 256 (9.7%) were absent. It was not possible to make a definitive present/absent call on the remaining 213 spots. Analysis of the distribution of these genes among the other strains included in our experiments demonstrated that UAMS-1 is most similar to UAMS-601 and EMRSA-16. Specifically, of 2,224 spots on the PFGRC arrays that were present in UAMS-1, 2,184 (98.2%) and 2,192 (98.6%) were present in UAMS-601 and EMRSA-16, respectively. This was the highest percentage observed among the nine strains examined with the exception of COL, which is to be expected since COL is the reference strain for the PFGRC array. Similarly, of 256 genes on the PFGRC array that were absent in UAMS-1, 203 (79.3%) and 202 (78.9%) were also absent in UAMS-601 and EMRSA-16, respectively. To further evaluate the relationships among all 10 strains, we performed hierarchical clustering, using microarray results for all spots on the PFGRC array (Fig. 1). Clustering not only confirmed that UAMS-1, UAMS-601, and EMRSA-16 are most closely related to each other but also revealed that RN6390, 8325, and COL represent a distinct cluster and that this cluster is the most distantly related with respect to the UAMS strains and EMRSA-16. Furthermore, our clustering data also linked Mu50 and N315, which is consistent with the work of Baba et al. (2), who noted 99.7% identity between these two strains. Our clustering also linked MW2 and SANGER-476, which is consistent with the observation that both of these strains are community-acquired S. aureus isolates. Importantly, all of the clustering relationships defined by PFGRC data were also confirmed by hierarchical clustering of Affymetrix data (data not shown).
FIG. 1.
Dendrogram illustrating genetic relationships between strains. The dendrogram was derived from PFGRC array data based on Spotfire hierarchical clustering. MLST for UAMS-1, UAMS-601, and RN6390 was performed according to the method of Maiden et al. (38). The spa genes for UAMS-1, UAMS-601, and RN6390 were amplified, sequenced, and typed according to the method of Shopsin et al. (62). Agr subgroups were determined by direct sequence analysis of the agr locus. The percentage of genes present or absent in each strain by comparison to UAMS-1 was determined from final PFGRC microarray results.
Comparative genomics of UAMS-1 and RN6390.
RN6390 is the prototype laboratory strain, and previous experiments from our laboratory have confirmed the existence of important differences between RN6390 and the UAMS isolates (6, 7). To investigate the genetic basis for these differences, we analyzed the genomic differences between UAMS-1 and RN6390, using data from the Affymetrix GeneChips. We chose to focus on the Affymetrix GeneChip for this comparison because these arrays include ORFs from EMRSA-16, which, as noted above, is the most closely related of the sequenced strains to the UAMS isolates. GeneChip qualifiers for intergenic regions were excluded, as were those qualifiers that gave ambiguous results for both strains or for which annotation information was not available. Of the 3,543 qualifiers remaining, 2,343 (65.7%) were present in both UAMS-1 and RN6390, whereas 677 (19%) were absent in both strains. The remaining 523 (14.8%) were present in one strain but not the other. These 523 ORFs are detailed in supplemental Tables S1 and S2. Included among these genes were ORFs predicted to encode proteins that fall into every functional category defined by the COGs (cluster of orthologous groups) database. However, 296 of the 523 ORFs are annotated only as encoding hypothetical proteins, and it is not possible to make functional predictions in these cases. Of the remaining 227 ORFs, 54 were predicted to have metabolic or transport functions. An additional 38 were associated with mobile elements and pathogenicity islands, and 11 were associated with drug or heavy metal resistance. The remaining 124 ORFs were predicted to encode surface or secreted proteins, including toxins and exoenyzmes (n = 102) or regulatory elements (n = 22). Because many of these are virulence factors, and based on our interest in the pathogenesis of musculoskeletal infection, we chose to focus on differences involving these genes as described below.
Adhesins.
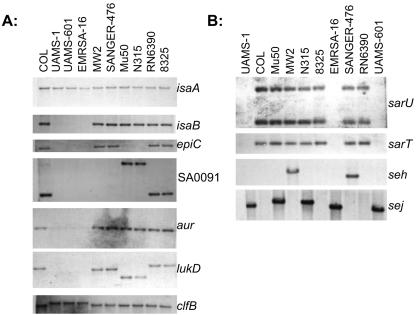
The PFGRC array used in these experiments included spots corresponding to 15 recognized adhesin genes. Unfortunately, the collagen adhesin gene cna was not included. Since the presence of cna correlates well with virulence (8, 49), we assessed its presence in our strains by Southern blotting. The cna gene was present only in UAMS-1, UAMS-601, EMRSA-16, MW2, and SANGER-476 (data not shown). These results were confirmed by Affymetrix data and correspond to our hierarchical clustering (Fig. 1), suggesting that cna may, in fact, represent a marker of overall genetic relatedness. Other adhesin genes that were not represented on the PFGRC array included those encoding the plasminogen, vitronectin, and thrombospondin binding proteins. Of the adhesin genes that were included on the PFGRC array, the gene encoding clumping factor B (clfB) was unambiguously present in 8 of 10 strains by microarray analysis. However, Southern blotting confirmed the presence of clfB in all 10 strains (Fig. 2). Similarly, the gene encoding clumping factor A (clfA) was classified by microarray as absent in N315 and Mu50, but direct sequence analysis confirmed that clfA is present in both of these strains. These results are consistent with experiments done with the Affymetrix GeneChips, which contains three clfA alleles and four clfB alleles. More directly, hybridizations done with the Affymetrix arrays confirmed that clfA and clfB were present in all 10 strains. Furthermore, RN6390, 8325, COL, MW2, and SANGER-476 were found to contain the same clfA allele (BAB94629), as did the Mu50/N315 cluster (BAB56973.1) and the UAMS-1/UAMS-601/EMRSA-16 cluster (86.7% amino acid identity to BAB41975). All 10 strains also encoded the fibrinogen binding protein COL-SA1168; however, the fibrinogen-binding precursor-related protein (COL-SA1169) was absent or divergent in UAMS-1, UAMS-601, and EMRSA-16.
FIG. 2.
Southern hybridization for validation of array data. Genomic DNA isolated from the strain indicated above each lane was hybridized with probes corresponding to the genes shown to the right of each panel. The sarU probe used in these experiments also included a region corresponding to sarT, while the sarT probe was limited to sarT itself.
The sdr loci encode proteins with similarity to clfA and clfB. Specifically, SdrC, SdrD, and SdrE contain variable numbers of serine-aspartate repeats, which are also present in both ClfA and ClfB (31). Based on results with the PFGRC arrays, sdrC was present in all strains, while sdrD was present in all strains except UAMS-1 and EMRSA-16. However, Affymetrix data indicated that both sdrC and sdrD were present in all 10 strains. This suggests that UAMS-1 and EMRSA-16 encode a divergent form of sdrD by comparison to the other strains. In contrast, sdrE was absent from RN6390, 8325, and UAMS-1 based on results from both the PFGRC and Affymetrix microarrays. The Affymetrix array also included bbp, and our hybridization data indicate that this variant was present in all strains including UAMS-1. This suggests that both sdrE and bbp are present in some if not most strains, and this is not consistent with the results of Peacock et al. (49), who concluded that sdrE and bbp are different alleles of the same locus. To address this issue, we performed BLAST searches of the sequenced S. aureus strains. This confirmed that the ORFs surrounding sdrE were conserved in all seven strains. Five of these (COL, MW2, SANGER-476, Mu50, and N315) contained ORFs with a high percent identity (95 to 99%) to sdrE. In contrast, the corresponding ORF in EMRSA-16 was identical to bbp and had only 87% identity to sdrE. PCR analysis using the bbp-specific primers described by Peacock et al. (49) suggested that UAMS-601 also encodes the bbp allele, and this was subsequently confirmed by DNA sequencing (data not shown). This is interesting in light of reports indicating that the presence of bbp and the consequent ability to bind bone sialoprotein contributes to the pathogenesis of musculoskeletal infection (55). However, PCR analysis also indicated that bbp is either absent in UAMS-1 or is highly divergent by comparison to EMRSA-16 and UAMS-601. BLAST analysis also indicated that while 8325 encodes sdrC and sdrD, it does not encode either of the sdrE or bbp alleles. PCR analysis suggests that this is also true of RN6390, which would be expected since RN6390 is an 8325-4 strain derived from 8235.
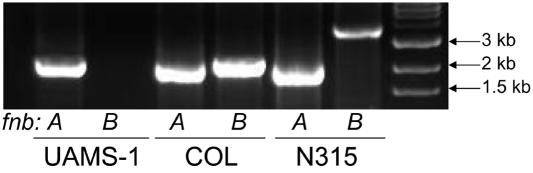
Peacock et al. (49) also found that fnbA, which encodes a fibronectin-binding protein, was significantly more common in invasive isolates. This is one of two very similar genes (the other is designated fnbB) that, in those strains in which both are present, are contiguous within the S. aureus chromosome. The fact that fnbA, but not fnbB, was more common among invasive isolates therefore suggests that such strains do not encode fnbB. Although two strains (UAMS-601 and SANGER-476) initially gave a “divergent” result for fnbA, it was subsequently confirmed by PCR that fnbA was present in all 10 strains (Fig. 3). In contrast, fnbB was classified as absent by microarray in UAMS-1, UAMS-601, EMRSA-16, SANGER-476, and Mu50. Analysis of the TIGR Comprehensive Microbial Resource indicated that Mu50 does indeed contain fnbB (NT02SA2496); however, the specific allele present in Mu50 was not represented on the array. Direct sequence analysis also indicated that MW2 and N315 have genes that share 80 and 84% nucleic acid identity with fnbB, respectively. We therefore undertook a PCR-based approach to confirm the presence of fnbB in each strain, using primers designed to amplify regions specific to the fnbB allele (Table 2). By PCR, fnbB was present in 8325, RN6390, COL, MW2, and SANGER-476 (Fig. 3). Mu50 and N315 also had a positive PCR, although the region amplified was larger. However, fnbB was absent in UAMS-1, UAMS-601, and EMRSA-16 based on PCR results (Fig. 3), as well as sequence analysis in the case of EMRSA-16. Affymetrix data confirmed these results, since all strains except UAMS-1, UAMS-601, and EMRSA-16 contained one of the three fnbB alleles present on the Affymetrix GeneChip.
FIG. 3.
PCR verification of fnbA and fnbB. Genomic DNA isolated from the indicated strains was used as a template in PCRs with primers that amplify unique regions of fnbA or fnbB. The results observed with UAMS-1, COL, and N315 are representative of the results observed with the other seven strains as described in the text.
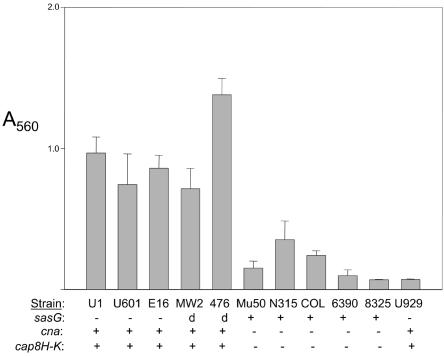
Other adhesin genes were conserved among all strains, including those encoding protein A (spa), the elastin binding protein (ebpS), a secretory extracellular matrix and plasma binding protein (SA0858), and the intracellular adhesion (ica) locus. Several additional LPXTG motif cell wall anchor domain proteins were also included on the array, and most of these were conserved among all strains (Table 3). However, one of the LPXTG motif proteins, SA2505, was absent in UAMS-1, UAMS-601, EMRSA-16, MW2, and SANGER-476 based on our array data. Interestingly, these same strains formed a more robust biofilm in our microtiter plate assay (Fig. 4). SA2505 encodes a pls homolog, and Savolainen et al. (60) reported that expression of pls is associated with reduced bacterial adhesion in vitro. Roche et al. (54) also described an LPXTG-containing gene (sasG) that also encodes a pls homolog, and a BLAST search confirmed that SA2505 and sasG are identical (data not shown). Roche et al. (53) subsequently reported that MW2 and SANGER-476 do encode sasG but that it is a highly divergent allele by comparison to other strains. These two strains formed a robust biofilm comparable to that of strains that lack sasG, which suggests either that the presence or absence of sasG is not responsible for the observed biofilm phenotype or that the divergent form of sasG is functionally distinct from the sasG gene encoded by other strains. We also found that those strains that formed a robust biofilm were all cna positive and capsular serotype 8 (Fig. 4).
TABLE 3.
Adhesins
| ORF name or description | PFGRC/Affymetrix locus no. | Presence of ORF in strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UAMS-1 | UAMS-601 | EMRSA-16 | SANGER-476 | MW2 | Mu50 | N315 | RN6390 | NCTC 8325 | COL | ||
| Clumping factor A (clfA) | COL-SA0856 | + | + | + | + | + | − | − | + | + | + |
| clfA allele | WAN014GQG | − | − | − | + | + | − | − | + | + | + |
| clfA allele | WAN014GQH_s | − | − | − | − | − | + | + | − | − | − |
| clfA allele | WAN014GQJ | + | + | + | − | − | − | − | − | − | − |
| Clumping factor B (clfB) | COL-SA2652 | + | + | + | + | + | + | + | + | + | + |
| sdrC | COL-SA0608 | + | + | + | + | + | + | + | + | + | + |
| sdrD | COL-SA0609 | − | + | − | + | + | + | + | + | + | + |
| sdrD allele | WAN014IPR | − | − | − | − | − | + | + | − | − | − |
| sdrD allele | WAN01BPA1_x | + | + | + | + | + | + | + | + | + | + |
| sdrE | COL-SA0610 | − | + | + | + | + | + | + | − | − | + |
| sdrE allele | WAN014IPS_x | − | + | + | + | + | + | + | − | − | + |
| sdrE allele | WAN014IPT | − | − | − | + | + | − | − | − | − | + |
| Fibronectin Binding protein (fnbA) | COL-SA2511 | + | + | + | + | + | + | + | + | + | + |
| Fibronectin Protein A related | COL-SA1220 | + | + | + | + | + | + | + | + | + | + |
| fnbA allele | WAN014IPC | − | − | − | − | − | − | − | + | + | + |
| fnbA allele | WAN014IPD | − | − | − | − | − | + | + | − | − | − |
| Fibronectin binding protein (fnbB) | COL-SA2509 | − | − | − | − | − | − | − | + | + | + |
| fnbB allele | WAN014IPE | − | − | − | − | − | − | − | + | + | + |
| fnbB allele | WAN014IPF | − | − | − | − | − | + | + | − | − | − |
| fnbB allele | WAN01BYXJ | − | − | − | + | + | − | − | − | − | − |
| Fibrinogen-binding protein | COL-SA1168 | + | + | + | + | + | + | + | + | + | + |
| Fibrinogen-binding protein precursor-related protein | COL-SA1169 | − | − | − | + | + | + | + | + | + | + |
| Protein A precursor | COL-SA0095 | + | + | + | + | + | + | + | + | + | + |
| Elastin binding protein | COL-SA1522 | + | + | + | + | + | + | + | + | + | + |
| Secretory extracellular matrix and plasma binding protein | COL-SA0858 | + | + | + | + | + | + | + | + | + | + |
| icaA to -D | COL-SA2689-SA2692 | + | + | + | + | + | + | + | + | + | + |
| icaR | COL-SA2688 | + | + | + | + | + | + | + | + | + | + |
| cna | WAN014HM2 | + | + | + | + | + | − | − | − | − | − |
FIG. 4.
Microtiter plate assay of biofilm formation. Biofilm formation was assessed as described by Beenken et al. (5). Results are shown as absorbance at 560 nm (A560) and represent the average and standard errors of the means from nine independent experiments. Strain U929 (UAMS-1 sarA mutant) was included as a negative control. Distribution of cna and sasG among each of the 10 strains was confirmed by microarray results, Southern hybridization, and BLAST searches. Capsular serotype was determined from microarray results for all strains, and confirmed in UAMS-1 by serotyping analysis (Chia Y. Lee, personal communication). The designation “d” indicates that MW2 and SANGER-476 contain a divergent form of sasG (53).
Exoenzymes.
A large number of the genes encoding exoenzymes were conserved among all strains examined. However, we did observe some differences that were closely correlated with the hierarchical clusters defined by our microarray experiments. Characterization of individual genes in this group was highly dependent on the specific array used. For example, based on the results obtained with the PFGRC arrays, the serine protease genes splA and splB were absent in the virulent UAMS isolates and EMRSA-16 but present in all other strains (Table 4). These genes are part of a previously characterized region of difference (RD1), which also encodes six other serine protease genes, two leukocidin genes, and the genes for bacteriocin synthesis (see below). While the Affymetrix data confirmed the absence of splA, analysis of Affymetrix array data indicated the presence of a gene with 97.7% predicted amino acid sequence identity to the splB gene of MW2 (GenBank accession no. BAB95619) in UAMS-1, UAMS-601, and EMRSA-16, suggesting that these strains contain an allelic variant of the splB gene. The serine protease gene splE was absent in SANGER-476, Mu50, MW2, and N315 yet present in all other strains. Interestingly, the splE gene was also part of RD1, emphasizing that strain-dependent differences occur within these large regions of difference and even within a cluster of genes.
TABLE 4.
Exoenzymesa
| ORF description | PFGRC locus no. | Presence of ORF in strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JAMS-1 | UAMS-601 | EMRSA-16 | SANGER-476 | MW2 | Mu50 | N315 | RN6390 | NCTC 8325 | COL | ||
| Probable 5′ nucleotidase | SA0024 | + | + | + | + | + | + | + | + | + | + |
| 1-Phosphatidyl phosphodiesterase (plc) | SA0078 | + | + | + | + | + | + | + | + | + | + |
| Staphylocoagulase (coa) | SA0209 | Ab | A | A | A | + | + | + | + | + | + |
| Lipase (geh) | SA0317 | + | + | + | + | + | + | + | + | + | + |
| Probable lipase | SA0712 | + | + | + | + | + | + | + | + | + | + |
| Possible staphylocoagulase | SA0857 | + | + | + | + | + | + | + | + | + | + |
| Staphylococcal nuclease | SA0860 | + | + | + | + | + | + | + | + | + | + |
| Serine protease | SA1028 | + | + | + | + | + | + | + | + | + | + |
| Cysteine protease (sspB) | SA1056 | + | + | + | + | + | + | + | + | + | + |
| V8 protease (sspA) | SA1057 | + | + | + | + | + | + | + | + | + | + |
| Thermonuclease (nuc) | SA1357 | + | + | + | + | + | + | + | + | + | + |
| Serine protease (splF) | SA1864 | + | + | + | + | + | + | + | + | + | + |
| Serine protease (splE) | SA1865 | + | + | + | − | − | − | − | + | + | + |
| Serine protease (splD) | SA1866 | + | + | + | + | + | + | + | + | + | + |
| Serine protease (splC) | SA1867 | + | + | + | + | + | + | + | + | + | + |
| Serine protease (splB) | SA1868 | − | − | − | + | + | + | + | + | + | + |
| Serine protease (splA) | SA1869 | − | − | − | + | + | + | + | + | + | + |
| Staphopain, cysteine protease | SA1970 | + | + | + | + | + | + | + | + | + | + |
| Hyaluronidase (hysA) | SA2194 | + | + | + | + | + | + | + | + | + | + |
| Probable lipase | SA2549 | + | + | + | + | + | + | + | + | + | + |
| Zinc metalloproteinase aureolysin (aur) | SA2659 | A | A | A | + | + | + | + | + | + | + |
| Triacylglycerol lipase (lip) | SA2694 | + | + | + | + | + | + | + | + | + | + |
| Phospholipase C (plc) precursor | SA2003 | + | + | + | + | + | + | + | + | + | + |
Only microarray results are given, not confirmatory Southern hybridization results.
A, allelic variant.
The zinc metalloproteinase aureolysin gene (aur) was classified as divergent in UAMS-1 and EMRSA-16 by PFGRC microarrays. This was somewhat surprising, since Karlsson et al. (32) analyzed 92 strains and found that four major S. aureus protease genes, including aur, were present in all strains, including those with a protease-negative phenotype. Based on this, we amplified an internal fragment of the aur gene (Table 2) and used this as a probe in a Southern blot of genomic DNA from all 10 strains. The results indicated that both UAMS-1 and EMRSA-16 encode the aur gene (Fig. 2). This suggests that these strains encode an allelic variant by comparison to the strains used to generate the PFGRC array. Indeed, Sabat et al. (57) previously reported the existence of aur allelic variants, and the Affymetrix GeneChip includes a second aur allele that was indeed present in UAMS-1, UAMS-601, and EMRSA-16. Similarly, the coagulase gene (COL-SA0209) was classified as absent in UAMS-1, UAMS-601, EMRSA-16, and SANGER-476 based on the PFGRC arrays, which included only one coa allele. In contrast, the Affymetrix GeneChips included 17 different coa alleles, four of which collectively accounted for the coa alleles for all 10 strains we examined. The distribution of coa alleles was correlated with our epidemiological clustering data with each of our four primary clusters (RN6390/8325/COL, N315/Mu50, MW2/SANGER-476, and UAMS-1/UAMS-601/EMRSA-16) having a distinct coa allele (data not shown). Other exoenzymes that were represented on the PFGRC and/or Affymetrix arrays were conserved, including the cysteine proteases sspA to -C, phospholipase C (plc), lipase (lip and geh), hyaluronidase (hysA), staphylococcal secretory antigen (ssaA), elastase (sepA), and thermonuclease (nuc).
Toxins.
The genetic analysis of toxin genes is complicated by the occurrence of a large number of variants that share considerable homology with each other (42). Four enterotoxin ORFs (seb, sei, ent, and an ORF for an enterotoxin family protein) were represented on the version 1 PFGRC arrays used in these experiments. The Affymetrix GeneChip included probe sets representing a total of 19 enterotoxin genes, many of which were represented by multiple alleles. We found that only COL encoded seb, as represented on both the PFGRC array and Affymetrix GeneChip (Table 5). Both sei (COL-SA0887) and ent (COL-SA0886) were present only in MW2, SANGER-476, and COL, according to the PFGRC arrays. However, the Affymetrix arrays also included a second allele for sei (N315-SA1646) that was present in UAMS-1, UAMS-601, EMRSA-16, Mu50, and N315. A search of the TIGR Comprehensive Microbial Resource database shows that the protein with highest homology to SEI in MW2 is designated SEG2. Similarly, the protein with highest homology to ENT in MW2 is designated SEK. It is therefore unclear whether our microarray analysis has failed to discern the genes encoding two antigenic types of enterotoxin or if the annotation is simply inconsistent.
TABLE 5.
Toxins and superantigensa
| ORF name or description | Array | PFGRC/ Affymetrix locus | Presence of ORF in strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UAMS-1 | UAMS-601 | EMRSA-16 | SANGER-476 | MW2 | Mu50 | N315 | RN6390 | NCTC 8325 | COL | |||
| Exotoxin, putative | PFGRC | COL-SA0470 | − | − | − | + | + | + | + | + | + | + |
| Exotoxin 1 | PFGRC | COL-SA0469 | + | + | − | + | + | + | + | + | + | + |
| Exotoxin, putative exotoxin 2 | PFGRC/Affymetrix | COL-SA0472/WAN014GWB | − | − | − | + | + | − | + | + | + | + |
| Exotoxin 3 | PFGRC | COL-SA0468 | − | − | − | − | − | + | + | + | + | + |
| set1 | PFGRC | COL-SA1178 | − | − | − | + | + | + | + | + | + | + |
| set1 | Affymetrix | WAN014IVU | + | + | + | − | − | − | − | − | − | − |
| set2 | Affymetrix | WAN014HIS | + | + | + | − | − | − | − | − | − | − |
| set3 | PFGRC | COL-SA1180 | + | + | + | + | + | + | + | + | + | + |
| set3 | Affymetrix | WAN014HIT | + | + | + | − | − | − | − | − | − | − |
| set4 | PFGRC | COL-SA1179 | − | − | − | + | + | + | + | + | + | + |
| set4 | Affymetrix | WAN014INI | + | + | + | − | − | − | − | − | − | − |
| set5 | Affymetrix | WAN014INH | + | + | + | − | − | − | − | − | − | − |
| set6 | Affymetrix | WAN014G2B | − | − | − | − | − | + | + | − | − | − |
| set7 | Affymetrix | WAN014IV4 | + | + | + | + | + | + | + | + | + | + |
| set8 | Affymetrix | WAN014GW6 | − | − | − | + | + | + | + | + | + | + |
| set9 | Affymetrix | WAN014GWD | − | − | − | + | + | − | + | + | + | + |
| set10 | Affymetrix | WAN014GKN | − | − | − | + | + | + | + | + | + | − |
| set11 | Affymetrix | WAN014GKP | − | − | − | + | − | + | + | + | + | − |
| set12 | Affymetrix | WAN014GKQ | − | − | − | + | + | + | + | + | + | − |
| set13 | Affymetrix | WAN014IMJ | − | − | − | + | + | + | + | + | + | + |
| set13 (N315), set5 (COL) | PFGRC | COL-SA0473 | − | + | − | + | + | + | + | + | + | + |
| set14 | Affymetrix | WAN014IMK | − | − | − | + | + | + | + | + | + | + |
| set14 (N315), set4 (COL) | PFGRC | COL-0474 | − | − | − | + | + | + | + | + | + | + |
| set15 | Affymetrix | WAN014G2E | − | − | − | − | − | + | + | − | − | − |
| set15 (N315), exotoxin 3 (COL) | PFGRC | COL-SA0478 | − | − | − | − | − | + | + | + | + | + |
| set16 | Affymetrix | WAN014HQV | + | + | + | + | + | − | − | − | − | − |
| set19 | Affymetrix | WAN014GWE | − | − | − | + | + | − | − | − | − | − |
| set21 | Affymetrix | WAN014FU6 | − | − | − | + | + | − | − | − | − | − |
| set26 | Affymetrix | WAN014FUA | − | − | − | + | + | − | − | − | − | − |
| Similar to set19 | Affymetrix | WAN014HQY | + | + | + | − | − | − | − | − | − | − |
| Similar to set21 | Affymetrix | WAN014GKO | − | − | − | − | − | − | − | + | + | − |
| Similar to set26 | Affymetrix | WAN014HQZ | + | + | + | − | − | − | − | − | − | − |
| seb | PFGRC/Affymetrix | COL-SA0907/WAN014HGZ | − | − | − | − | − | − | − | − | − | + |
| sec3 | Affymetrix | WAN014IW4 | − | − | − | − | + | + | + | − | − | − |
| sei | Affymetrix | WAN014HJJ | + | + | + | − | − | + | + | − | − | − |
| seg | Affymetrix | WAN014HHY | + | + | + | − | − | + | + | − | − | − |
| seh | Affymetrix | WAN014HMA | − | − | − | + | + | − | − | − | − | − |
| sei (COL), seg2 (MW2) | PFGRC/Affymetrix | COL-SA0887/WAN014ITN | − | − | − | + | + | − | − | − | − | + |
| sek, ent (COL) | PFGRC/Affymetrix | COL-SA0886/WAN014ITM | − | − | − | + | + | − | − | − | − | + |
| sel | Affymetrix | WAN014IRW | − | − | − | − | + | + | + | − | − | − |
| sem | Affymetrix | WAN01BUE4 | + | + | + | − | − | − | − | − | − | − |
| sem | Affymetrix | WAN01CAT8 | − | − | − | − | − | + | + | − | − | − |
| sen | Affymetrix | WAN014HK5 | + | + | + | − | − | + | + | − | − | − |
| seo | Affymetrix | WAN014HK2 | + | + | + | − | − | + | + | − | − | − |
| sep | Affymetrix | WAN014GK5 | − | − | − | − | − | − | + | − | − | − |
| entP | Affymetrix | WAN014HLS | + | + | + | + | + | + | − | − | − | − |
| yent1 | Affymetrix | WAN014HK3 | + | + | − | − | − | + | + | − | − | − |
| yent2 | Affymetrix | WAN014HK4 | + | + | + | − | − | + | + | − | − | − |
| Staphylococcal enterotoxin, putative | PFGRC | COL-SA0442 | − | − | − | + | + | + | + | + | + | + |
| Enterotoxin family protein | PFGRC | COL-SA1657 | + | + | + | + | + | + | + | + | + | + |
| Diarrheal toxin-like protein | PFGRC | COL-SA0265 | + | + | + | + | + | + | + | + | + | + |
| Probable hemolysin | PFGRC | COL-SA0762 | + | + | + | + | + | + | + | + | + | + |
| Probable hemolysin | PFGRC | COL-SA2160 | + | + | + | + | + | + | + | + | + | + |
| Gamma hemolysin A (hlgA) | PFGRC | COL-SA2419 | + | + | + | + | + | + | + | + | + | + |
| Gamma hemolysin B (hlgB) | PFGRC | COL-SA2422 | + | + | + | + | + | + | + | + | + | + |
| Gamma hemolysin C (hlgC) | PFGRC | COL-SA2421 | + | + | + | + | + | + | + | + | + | + |
| Leukotoxin lukD | PFGRC | COL-SA1880 | − | − | − | + | + | + | + | + | + | + |
| Leukotoxin E (lukS) | PFGRC | COL-SA1881 | − | − | − | + | + | + | + | + | + | + |
| Possible leukocidin lukM | PFGRC | COL-SA2006 | − | − | − | + | + | + | + | + | + | + |
| Leukocidin E subunit precursor | PFGRC | COL-SA2004 | − | − | − | + | + | + | + | + | + | + |
Only microarray results are given, not Southern hybridization results.
The Affymetrix arrays included the enterotoxin H gene (seh), which was previously reported to be present only in MW2 based on comparison to the genomes of COL, N315, Mu50, 8325, and EMRSA-16 (2). Both Southern blotting and Affymetrix data demonstrated that seh was also present in SANGER-476 (Fig. 2). This is consistent with our hierarchical clustering linking MW2 and SANGER-476 (Fig. 1) and suggests that seh may be specifically associated with community-acquired isolates. The sej gene was also included on the Affymetrix arrays, but surprisingly no strains were predicted to contain this gene based on Affymetrix algorithms. Importantly, sej is one of the seven genes that Peacock et al. (49) found were more commonly associated with invasive isolates. We therefore analyzed the sej gene by Southern hybridization and found that it was present in UAMS-1, UAMS-601, EMRSA-16, Mu50, and N315 (Fig. 2). In contrast to seh, sej may therefore be more specific to hospital-acquired isolates. The distribution of other enterotoxin genes included on the Affymetrix arrays is shown in Table 5; however, due to considerable homology among these genes, it is unlikely that microarray-based methods can account for the small sequence differences that define antigenic specificity. A multiplex PCR approach may be more suitable in this regard (41, 42).
Eleven exotoxin-like ORFs were represented on the PFGRC arrays. Similar to the enterotoxin genes, the exotoxin-like (set) genes of S. aureus also share considerable homology and a confusing nomenclature. The annotation for these genes on the PFGRC microarray is confusing as well. For example, three ORFs on the version 1.0 PFGRC arrays were described as encoding exotoxin 3 (COL-SA0468, COL-SA0478, and COL-SA1180). However, products of SA0468 and SA0478 were most closely related to exotoxin 6 and exotoxin 15 of N315, respectively. Similarly, two ORFs (COL-SA0469 and COL-SA1178) were described as encoding exotoxin 2, and the product of one of these (SA0469) was homologous to exotoxin 7 of N315. Exotoxin-like protein genes set13 and set14 were homologous to set5 and set4 of COL, respectively. Microarray data from such homologous genes can complicate interpretation; nevertheless, the exotoxin-like genes 1, 2, 4, 6, 8, 9, 14, and 15 were all classified by the PFGRC microarray as absent or divergent in UAMS-1, UAMS-601, and EMRSA-16. However, the Affymetrix GeneChips were more comprehensive than the PFGRC arrays and included a total of 25 exotoxin-like genes. Multiple alleles were included for some of these, and alleles of exotoxin-like genes 1, 2, and 4 were present in UAMS-1, UAMS-601, and EMRSA-16. Given the functional similarity among the products of these genes and the presence of multiple exotoxin-like genes in every strain, it is certainly possible that these genotypic differences are phenotypically transparent and merely represent allelic variations. More directly, it seems likely based on our data for both the enterotoxin and exotoxin-like genes that all of the strains examined in these experiments have the capacity to produce at least one superantigen. As with the enterotoxin genes, a PCR-based approach may be more efficient for accurately determining the presence or absence of specific exotoxin-like alleles.
The genes encoding exfoliative toxins A (eta) and B (etb) are not present in COL and were not present on the version 1.0 PFGRC array. Both were present on the Affymetrix GeneChip, and our hybridization experiments indicated that neither eta or etb was present in any of the 10 strains studied. However, a gene (COL-SA1184) that encodes a protein with 80% amino acid identity to the product of an S. hyicus gene that is described as an exfoliative toxin was present in all strains. This gene was previously incorrectly designated an exfoliative toxin A gene in the COL genome (and PFGRC arrays), but this designation has since been corrected.
S. aureus may also encode the genes for several bicomponent leukotoxins, including the genes encoding Panton-Valentine leukocidin (PVL), gamma hemolysin, and lukD/E. This is important because leukotoxin production has been associated with some specific disease manifestations including life-threatening pneumonia (25, 36). The PFGRC arrays contained ORFs corresponding to lukDE and gamma hemolysin but not the PVL genes. The genes encoding PVL (lukF-PV and lukS-PV) were included on the Affymetrix arrays and were present only in MW2. This is consistent with previous reports linking the PVL genes with community-acquired isolates (14, 25). The genes encoding gamma hemolysin, component A, and several other putative hemolysins contained on the array were present in all 10 strains. However, lukDE was classified as absent in UAMS-1, UAMS-601, and EMRSA-16, and this was subsequently confirmed by Southern blotting (Fig. 2). This is not surprising considering that lukD and lukE are contained on RD1 (20), which contains other genes that are also absent in the UAMS strains and EMRSA-16 (see above). However, as noted above, the redundant nature of S. aureus virulence factors makes it difficult to predict whether the absence of either or both of these genes would have a significant impact on the phenotype of these strains.
Notably, the gene encoding toxic shock syndrome toxin 1 (tst) was not included on the PFGRC microarray. However, Affymetrix data reveal that this gene is present in UAMS-1, UAMS-601, N315, and Mu50 but not EMRSA-16. This is one of the few differences observed between the UAMS isolates and EMRSA-16.
Regulatory elements.
Regulatory circuits in S. aureus are complicated and involve a number of interactive elements. Most notable among these is the staphylococcal accessory regulator (sarA) and the accessory gene regulator (agr), both of which have been correlated with virulence (7, 10). However, recent reports have described a number of other loci (Table 6), most of which appear to function, at least in part, by modulating the activity of sarA and/or agr (9, 45). The agr locus is known to have strain-dependent differences in nucleotide sequence, particularly in agrD and specific regions of agrB and agrC, and these differences define agr subtypes (29). Importantly, these subtypes have functional significance as well, since components of the agr locus from one group may inhibit the agr response in another group (29). Consistent with these observations, we found that the agrB and agrC genes were classified as divergent or absent in several strains by PFGRC microarray analysis. All of the agr genes were classified as present in UAMS-1, RN6390, and 8325, which could indicate that these strains share the same agr subtype as COL whereas the other strains represent different groups. However, for UAMS-1 two of four replicate experiments gave ambiguous results for the agrC gene (SA2025), and results from the Affymetrix GeneChip experiments indicated that SA2025 was not present in UAMS-1 (data not shown). Since COL is agrC subtype I, this is consistent with our previous agr subgroup typing data indicating that both UAMS-1 and UAMS-601 are agrC subtype III (6). The Affymetrix GeneChips were more comprehensive than the PFGRC arrays with respect to the agr locus, since they included representative alleles from each of the four major agr subgroups. We found the Affymetrix data to be correct in predicting agr subtype, since it was identical to subgroup delineation by direct sequence comparison (Fig. 1).
TABLE 6.
| ORF name or description | PFGRC/Affymetrix locus no. | Presence of ORF in strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UAMS-1 | UAMS-601 | EMRSA-16 | SANGER-476 | MW2 | Mu50 | N315 | RN6390 | NCTC 8325 | COL | ||
| agrA | Affymetrix | + | + | + | + | + | + | + | + | + | + |
| agrD-1 | Affymetrix | − | − | − | − | − | − | − | + | + | + |
| agrD-2 | Affymetrix | − | − | − | − | − | + | + | − | − | − |
| agrD-3 | Affymetrix | + | + | + | + | + | − | − | − | − | − |
| agrD-4 | Affymetrix | − | − | − | − | − | − | − | − | − | − |
| saeR | SA0766 | + | + | + | − | + | + | + | + | + | + |
| saeS | SA0765 | + | + | + | + | + | + | + | + | + | + |
| arlS | SA1450 | + | + | + | + | + | + | + | + | + | + |
| Rot | SA1812 | + | + | + | + | + | + | + | + | + | + |
| sarA | SA0672 | + | + | + | + | + | + | + | + | + | + |
| sarS (sarH1) | SA0096 | + | + | + | + | + | + | + | + | + | + |
| sarR | SA2287 | + | + | + | + | + | + | + | + | + | + |
| sarT | SA2506 | − | − | − | + | + | + | + | + | + | + |
| sarU | SA2507 | − | − | − | + | + | + | + | + | + | + |
| sarV | SA2258 | + | + | + | + | + | + | + | + | + | + |
| svrA | SA0416 | + | + | + | + | + | + | + | + | + | + |
| srrAB | SA1534 | + | + | + | + | + | + | + | + | + | + |
| Trap | SA1891 | − | − | − | + | + | + | + | + | + | + |
| yycG | SA0020 | + | + | + | + | + | + | + | + | + | + |
| lrgB | SA0248 | + | + | + | + | + | + | + | + | + | + |
| Lytic regulatory protein | SA2140 | + | + | + | + | + | + | + | + | + | + |
| vraR | SA1905/1942 | + | + | + | + | + | + | + | + | + | + |
| deoR family transcriptional regulator | SA0757/1964 | + | + | + | + | + | + | + | + | − | + |
| lysR family transcriptional regulators | SA0072/0074 | − | − | − | + | + | − | − | + | + | + |
| marR family transcriptional regulator | SA2524 | − | − | − | − | − | − | − | + | + | + |
| gntR family transcriptional regulator | SA0091 | − | − | − | − | − | + | + | + | + | + |
Several transcriptional regulator protein families were conserved among all study strains, according to PFGRC microarray data: Cro/Cl family (SA0420); rpiR family (SA0179); luxR family (SA2389); lacl family (SA1552); argR family (SA1565); sir2 family (SA2189); merR family (SA2193/SA2388/SA2517); deg family (SA2086); fur family (SA1541/SA1611/SA1919); and tetR family (SA2349/SA2374/SA2587/SA2593/SA2610).
Only microarray results are given, not confirmatory Southern hybridization results.
The global regulator SarA and several of its homologues (SarH1 [SarS], SarR, and SarV) were conserved among all strains (Table 6). However, UAMS-1, UAMS-601, and EMRSA-16 all lacked the sarT and sarU genes based on both microarray analysis and Southern hybridization (Fig. 2). UAMS-1, UAMS-601, and EMRSA-16 were also negative for the gene encoding TRAP by microarray analysis (4). Southern hybridization confirmed these results, but upon overexposure of the membrane, a faint hybridization signal was observed for TRAP in all three strains (data not shown). This suggests that UAMS-1, UAMS-601, and EMRSA-16 either do not encode trap or encode a divergent form. Gilot et al. (26) has described trap subtypes similar to those described for agr, and Gov et al. (27) recently reported that EMRSA-16 (SANGER-252) does in fact encode a highly divergent form of trap by comparison to RN6390. Based on this, we designed primers corresponding to the trap variant present in EMRSA-16 and confirmed by PCR and DNA sequencing that both UAMS-1 and UAMS-601 encode a trap variant identical to EMRSA-16 (data not shown).
Most of the other recognized regulatory components were conserved among all strains. This includes rot, yyc, arl, sae, lrg, vra, svr, and srr. Several families of putative transcriptional regulators were also represented on the array, and the majority of these were also conserved among all strains (Table 6). However, some strain-specific differences were seen. For example, COL-SA0072 and COL-SA0074 are members of the LysR family of transcriptional regulators, and both of these ORFs were absent in UAMS-1, UAMS-601, EMRSA-16, Mu50, and N315. A similar pattern was seen with the MarR and GntR families of transcriptional regulators. For example, COL-SA2524 encodes a transcriptional regulator of the MarR family, and was present only in COL, 8325, and RN6390. Similarly, SA0091 encodes a transcriptional regulator of the GntR family and was present only in COL, 8325, RN6390, Mu50, and N315 (Fig. 2). Other members of the GntR and MarR families of transcriptional regulators were represented on the array but were conserved among all strains.
Other virulence factors.
Fitzgerald et al. (20) found eight genes that are required for bacteriocin (epidermin) synthesis in the same region of difference (RD1) as the spl genes and lukD/E. Consistent with this observation, the array data from both PFGRC microarrays and Affymetrix GeneChips confirms that UAMS-1, UAMS-601, and EMRSA-16 lack the epidermin biosynthesis genes (Table 7). Epidermin is a type A lantibiotic (58), which is an antimicrobial peptide produced by gram-positive organisms and active against other gram-positive organisms. The epidermin ORFs were also absent in Mu50 and N315, indicating a possible trend among hospital-acquired isolates. The major histocompatibility complex class II-like molecules represented by SA0089 (antigen, 67 kDa), SA0985, and SA2197 (putative map protein) were conserved among all strains. Another map protein ORF, SA2002, was classified as absent among SANGER-476, Mu50, MW2, and N315. Strain-specific differences were also seen with the gene encoding immunodominant antigen B (isaB), which was absent in UAMS-1, UAMS-601, and EMRSA-16 (FIG. 2). In contrast, the gene encoding immunodominant antigen A (isaA) was conserved among all strains. These proteins were identified as immunodominant based on Western blots done with sera from humans with methicillin-resistant S. aureus sepsis (37). The absence of isaB in the UAMS strains and EMRSA-16 is therefore intriguing in that it could be correlated with immune evasion.
TABLE 7.
Other virulence factorsa
| ORF name or description | PFGRC locus no. | Presence of ORF in strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UAMS 1 | UAMS 601 | EMRSA 16 | SANGER 476 | MW2 | Mu50 | N315 | RN6390 | NCTC 8325 | COL | ||
| Iantibiotic gallidermin precursor (epiA) | SA1878 | − | − | − | + | + | − | − | + | + | + |
| Iantibiotic epidermin biosynthesis protein (epiB) | SA1877 | − | − | − | + | + | − | − | + | + | + |
| Epidermin biosynthesis protein (epiC) | SA1876 | − | − | − | + | + | − | − | + | + | + |
| Epidermin biosynthesis protein (epiD) | SA1875 | − | − | − | + | + | − | − | + | + | + |
| Epidermin immunity protein F | SA1873 | − | − | − | + | + | − | − | + | + | + |
| Epidermin leader peptide processing serine protease (epiP) | SA1874 | − | − | − | + | + | − | − | + | + | + |
| Antigen, 67 kDa (myosin cross-reactive homologue) | SA0089 | + | + | + | + | + | + | + | + | + | + |
| Map protein, putative | SA0985 | + | + | + | + | + | + | + | + | + | + |
| Map protein, putative | SA2197 | + | + | + | + | + | + | + | + | + | + |
| Map protein | SA2002 | + | + | + | − | − | − | − | + | + | + |
| Capsular polysaccharide synthesis enzyme (cap8H) | NTL03SA0131 (MW2) | + | + | + | + | + | − | − | − | − | − |
| Capsular polysaccharide synthesis enzyme (cap8J) | NTL03SA0133 (MW2) | + | + | + | + | + | − | − | − | − | − |
| cap5A to -F | SA0136-SA0141 | + | + | + | + | + | + | + | + | + | + |
| UDP-N-acetylglucosamine 2-epimerase (cap5G) | SA0142 | + | + | + | + | + | + | + | + | + | + |
| cap5H to -K protein | SA0143-SA0146 | − | − | − | − | − | + | + | + | + | + |
| cap5L | SA0147 | + | + | + | + | + | + | + | + | + | + |
| Protein/glycosyltransferase, putative capsular polysaccharide biosynthesis galactosyltransferase Cap5M | SA0148 | + | + | + | + | + | + | + | + | + | + |
| cap5N protein/UDP-glucose 4-epimerase, putative | SA0149 | + | + | + | + | + | + | + | + | + | + |
| cap5O protein/UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase | SA0150 | + | + | + | + | + | + | + | + | + | + |
| IgG-binding protein SBI | SA2418 | + | + | + | + | + | + | + | + | + | + |
| Siderophore biosynthesis protein, lucA family | SA0104 | + | + | + | + | + | + | + | + | + | + |
| Siderophore biosynthesis protein, lucC family | SA0102, SA0105 | + | + | + | + | + | + | + | + | + | + |
| Iron compound ABC transporter, ATP-binding protein | SA0704 | + | + | + | + | + | + | + | + | + | + |
| Iron compound ABC transporter, ATP-binding protein, putative | SA0798 | − | − | − | + | + | + | + | + | + | + |
| Iron compound ABC transporter, permease protein (sirB) | SA0098 | + | + | + | + | + | + | + | + | + | + |
| Iron compound ABC transporter, permease protein (sirC) | SA0097 | + | + | + | + | + | + | + | + | + | + |
| Iron compound ABC transporter, substrate-binding protein (sirA) | SA0099 | + | + | + | + | + | + | + | + | + | + |
| Iron compound ABC transporter, substrate-binding protein | SA2010 | − | − | − | + | + | + | − | + | + | + |
| Immunodominant antigen A (isaA) | SA2584 | + | + | + | + | + | + | + | + | + | + |
| Immunodominant antigen B (isaB) | SA2660 | − | − | − | + | + | + | + | + | + | + |
Only microarray results are given, not confirmatory Southern hybridization results.
The cap8 genes H to K are specific to capsule type 8, while cap5 genes H to K are likewise specific to capsule type 5 (59). In contrast, cap genes A to D and L to O are conserved among both capsule types. Our array data confirm that strains COL, RN6390, 8325, Mu50, and N315 are type 5, since they contain the cap5H-K genes but lack the cap8H and cap8J genes. In contrast, UAMS-1, UAMS-601, EMRSA-16, SANGER-476, and MW2 contain the cap8H and cap8J genes but lack the cap5H-K genes, indicating that they are capsule type 8. These results are consistent with serotyping data (Chia Y. Lee, personal communication) and provide further support for the accuracy of our microarray data.
DISCUSSION
UAMS-1 is a clinical isolate obtained directly by biopsy from the bone of a patient suffering from osteomyelitis. We have demonstrated in previous experiments that UAMS-1 is highly virulent in our animal models of musculoskeletal infection (7, 63). For this reason and because it has become clear that RN6390 has specific attributes (e.g., the rsbU mutation) that distinguish it from clinical isolates, we have focused our efforts on UAMS-1. However, in the interest of putting our results in the context of previous studies, we have carried out a limited set of experiments comparing UAMS-1 and RN6390 in our animal models, and these experiments clearly indicate that UAMS-1 is more virulent than RN6390. For example, in our murine model of septic arthritis, intravenous injection of equivalent numbers of the respective strains resulted in clear evidence of arthritis in more than 96% of mice infected with UAMS-1 but only 75% of those infected with RN6390 (7). Overall inflammation and arthritic index scores were also higher in mice infected with UAMS-1. Similarly, in preliminary experiments using our rabbit model of postsurgical osteomyelitis, we observed very little evidence of disease in rabbits infected with 105 CFU of RN6390 even after introducing cna into the chromosome and conferring on RN6390 the ability to bind collagen (unpublished observations). In contrast, UAMS-1 caused infection in >75% of all rabbits and significant pathological changes in the bone even when introduced at a dose as low as 103 CFU (64).
Both to investigate the genetic basis for these differences and to facilitate our ability to genetically manipulate UAMS-1, we used CGH with a genome-scale, amplicon-based microarray to assess the genetic relationships between two clinical isolates (UAMS-1 and UAMS-601), the laboratory strain RN6390, and seven sequenced strains of S. aureus. A central concern in studies of this nature is the assignment of cutoffs to define “present” versus “absent” ORFs based on the hybridization ratio of a test strain to the reference strain used to generate the array. An emerging theme in CGH experiments that include sequenced strains is the use of BLAST comparisons to define empirical cutoffs for microarray data (51, 67). We utilized this approach to develop empirical cutoffs and found a high level of agreement (93.6 to 97.6% depending on the strain under study) between our empirical cutoffs and the BLAST-based comparison data.
It is not possible to eliminate all ambiguity in microarray-based CGH experiments. Some of this ambiguity arises from sequence divergence between the strains used to generate the array and the strains under study. Other limitations include errors generated from poor microarray printing, inefficient labeling of genomic DNA, improper data normalization, or poor hybridization of labeled probes. Indeed, we found 104 spots (4.04%) in which the majority of experiments failed the low signal intensity threshold in the COL channel; that is, the presence or absence of the ORF could not be unambiguously determined in multiple strains because the COL channel intensity failed to meet the mean-plus-one-standard-deviation requirement. An additional 125 (4.64%) spots on the array failed the low signal threshold in at least one experiment. For those strains with available genome sequence, these spots could be examined by direct sequence comparison. However, for strains RN6390, UAMS-1, and UAMS-601, it was necessary to use alternative means to address these discrepancies. We first attempted to account for microarray failures through the analysis of comparative data from Affymetrix GeneChip experiments. Although the Affymetrix GeneChip generally included a greater number of allelic variants, allowing for enhanced specificity relative to the PFGRC arrays, the results from corresponding ORFs on each set of arrays compared very favorably (93.3 to 98.5%). For strain UAMS-1, 2,444 spots from the PFGRC arrays were also represented on the Affymetrix GeneChip, and 2,281 (93.3%) of these had identical calls between PFGRC and Affymetrix. For strain UAMS-601, 2,473 PFGRC spots were represented on the Affymetrix arrays, and of these, 2,321 (93.85%) had identical calls. For strain RN6390, 2,422 of 2,460 detection calls (98.5%) were in agreement between the PFGRC and Affymetrix arrays. The increased accuracy observed with RN6390 by comparison to the UAMS strains may reflect the fact that both the PFGRC and Affymetrix arrays are based primarily on sequence data from the COL genome and the close relationship between COL and RN6390 (52). Indeed, COL and other isolates with the MLST sequence type ST250 are thought to be descendant from isolates with an MLST sequence type of ST8, which includes RN6390 and NCTC8325 (18).
Importantly, the experiments utilizing the PFGRC microarrays were performed independently of those utilizing the Affymetrix GeneChips, with unique methods for data analysis and cutoff determination. This approach allowed for not only a more comprehensive analysis but also a greater level of confidence in the appropriateness of our empirical cutoffs. We also tried to resolve any remaining ambiguities by using Southern hybridization and/or PCR. Indeed, we believe the results of our experiments confirm the need to combine the high-throughput advantage of CGH with confirmation using more gene-specific approaches, particularly in those cases involving divergent genes thought to be of particular clinical significance.
We found that 2,029 (78.4%) of the 2,589 spots on the PFGRC array that yielded results above the low signal threshold were present in all strains and thus represent the core genetic content of S. aureus. This number is essentially identical to the results from the work of Fitzgerald et al. (20), who found that 78% of the S. aureus genome was invariant. Given our focus on clinical isolates in general and musculoskeletal isolates in particular, we chose to analyze our results in the specific context of the genomic content of UAMS-1. Our data clearly demonstrate that UAMS-1 is most similar to UAMS-601 and EMRSA-16. Importantly, preliminary testing has confirmed that both UAMS-601 and EMRSA-16 are also hypervirulent in our models (data not shown), and the enhanced virulence of UAMS-601 has also been independently confirmed with a rabbit endocarditis model (Arnold Bayer, personal communication). It is therefore noteworthy that clustering based solely on genotypic information could be correlated to relative virulence, at least with respect to the two clusters that defined the extremes in our analysis (e.g., those that included UAMS-1 and RN6390, respectively). Indeed, hierarchical clustering based on microarray data fit very well with the clusters defined by other epidemiological typing methods and with studies suggesting the existence of specific clonal variants of particular clinical significance. For example, EMRSA-16 and the closely related strain EMRSA-15 are the dominant ORSA clones in the United Kingdom (43). These clones account for more than 95% of all ORSA bacteremias in the United Kingdom (30). EMRSA-16 has a core gene profile virtually identical to previously characterized OSSA clones, leading to the hypothesis that EMRSA-16 emerged from an OSSA strain after acquiring an SCCmecA element (43). Interestingly, UAMS-1 and UAMS-601 appear to share a similar relationship. Specifically, UAMS-601 is an ORSA isolate with the same MLST sequence type (ST36) as EMRSA-16, while UAMS-1 is an oxacillin-susceptible strain with an MLST sequence type of 30 (ST30). It has been postulated that ST30 is the ancestral sequence type of ST36 (17). Enright et al. also analyzed 155 isolates in the United Kingdom and found that most ORSA isolates were ST36 and that 87% of the OSSA isolates were closely related to EMRSA-16 and had an MLST genotype of ST30 (17). It is therefore important to focus not only on the dominant ORSA clones but also on the most prevalent OSSA isolates that give rise to these ORSA strains. We believe the data we report clearly indicate that UAMS-1 is a prototype example of such an OSSA isolate. It is also of interest to note that RN6390, COL, and 8325 all clustered closely together and were the most distantly related stains with respect to the UAMS strains and EMRSA-16. This is an important consideration given the experimental emphasis placed on RN6390. Collectively, we believe the results reported here clearly demonstrate that this emphasis is misplaced and that it should be refocused on more relevant strains like the UAMS isolates and EMRSA-16.
Our experiments were strictly genotypic and do not imply phenotypic differences, and they cannot account for point mutations or other small changes that may produce large differences in the expression level and/or function of individual loci. Nevertheless, we believe that our data have important implications with respect to the phenotype of the predominant clinical isolates of S. aureus. We base this belief on the presumed impact such genotypic differences would have on phenotypes previously reported from studies done with other strains of S. aureus. For instance, both sarT and sarU are absent in UAMS-1, UAMS-601, and EMRSA-16. SarT and SarU are reportedly members of a secondary amplification loop that modulates agr expression. Specifically, Manna and Cheung (39) reported that production of RNAIII is elevated in an RN6390 sarT mutant. This observation alone would suggest that the UAMS strains would be expected to produce elevated levels of RNAIII based on the absence of sarT. However, previous results from our laboratory have confirmed that UAMS-1 produces reduced amounts of RNAIII by comparison to RN6390 production (6). At the same time, Schmidt et al. (61) proposed a model in which sarT represses expression of sarU. Since sarU induces production of RNAIII, the absence of sarU in the UAMS strains would be consistent with their reduced expression of agr. Moreover, in this model, mutation of sarT would result in increased expression of agr, but only in the presence of sarU (i.e., in the absence of SarT-mediated repression, SarU would be free to up-regulate expression of RNAIII). In the absence of both sarT and sarU, the result would presumably be defined by the absence of sarU and the inability to induce RNAIII production, at least through this alternative pathway.
SarT is also reported to repress production of alpha-toxin and to induce sarS transcription (9, 45, 61). SarS, in turn, is an activator of protein A production (12, 58). This is not consistent with the observation that the UAMS isolates lack sarT but transcribe sarS at high levels (data not shown) and make an abundance of protein A (6). However, to the extent that the UAMS isolates produce reduced amounts of RNAIII, the increased transcription of sarS in these strains is consistent with the fact that sarS transcription is repressed by agr (11). It is also well established that induction of agr expression results in reduced expression of spa (1, 9, 45). Once again, this would suggest that the phenotype of UAMS-1 and other clinical isolates is defined by the absence of sarT and sarU rather than the impact of sarT alone. At the same time, the absence of sarT alone could explain why mutation of sarA results in decreased hemolytic activity in RN6390 and increased hemolytic activity in the UAMS isolates. Specifically, in the model of Schmidt et al. (61), mutation of sarA represses hla transcription indirectly by virtue of increased expression of sarT. Perhaps in the absence of SarT, SarA is able to bind the hla promoter and repress transcription. This is consistent with reports indicating that purified SarA binds the hla promoter at least under in vitro conditions (13, 65). Assuming that the binding of SarA would result in repression of transcription as suggested by Arvidson and Tegmark (1), mutation of sarA in strains that lack sarT would result in increased transcription of hla and increased hemolytic activity as was observed in both UAMS-1 and UAMS-601.
While our studies were limited to 10 strains, other investigators have found that sarT is absent in many if not most clinical isolates (P. M. Dunman, personal communication). Indeed, Fitzgerald et al. (20) found that a significant number of clinical isolates of S. aureus lack a 5-kb chromosomal region that they referred to as RD5. This region was originally reported to encode an FnbA homologue, two SarA homologues, and a protein with homology to a multidrug resistance protein made by Pyrococcus abyssi. Based on BLAST analysis, the two sarA homologs correspond to sarT and sarU. This emphasizes not only that a significant number of clinical isolates lack sarT but also that these same isolates almost certainly lack the sarU gene as well. This would suggest that such isolates would exhibit phenotypic traits like those observed with the UAMS isolates. This would include both quantitative (e.g., high expression of sarS and spa and low expression of agr and hla) and qualitative (e.g., the impact of mutating sarA on transcription of hla) differences as defined by comparison to RN6390. This is consistent with the results reported by Papakyriacou et al. (48), who found that the most prevalent clinical isolates of S. aureus have a phenotype characterized by a high binding capacity for host proteins and limited production of extracellular virulence factors (e.g., toxins and degradative enzymes). While this could be interpreted to suggest that the role of agr in S. aureus pathogenesis has been overinterpreted based on studies done in RN6390, it must be emphasized that mutation of agr in UAMS-1 has been shown to result in a reduced capacity to cause both osteomyelitis and septic arthritis (6, 22). Nevertheless, it is possible that RN6390 expresses agr at levels that have a negative impact on certain attributes that ultimately contribute to the ability of S. aureus to cause at least some forms of infection. Indeed, Vuong et al. (68) found that strains that produce high levels of RNAIII have a reduced capacity to form a biofilm, and recent studies from our laboratory confirmed that mutation of agr in RN6390 results in an enhanced capacity to form a biofilm (5). Importantly, this was not true for UAMS-1 or UAMS-601. To the extent that biofilm formation is an important attribute in the pathogenesis of S. aureus infection, this is consistent with the hypothesis that agr is functionally overexpressed in RN6390.
Our results suggest that the absence of RD5 may also have a more direct impact on biofilm formation. Specifically, the 10 strains we examined could be divided into two distinct groups, one of which (UAMS-1, UAMS-601, EMRSA-16, MW2, and SANGER-476) formed a more robust biofilm than the other (RN6390, COL, 8325, Mu50, and N315). All of the biofilm-forming strains were serotype 8 strains that encode cna, and three of five (UAMS-1, UAMS-601, and EMRSA-16) also lacked sasG (SA2505) while the other two (MW2 and SANGER-476) encode a divergent form of sasG. The fact that the three strains that lacked any form of sasG also lacked sarT and sarU is consistent with the results reported by Roche et al. (53), who demonstrated that sasG is also included within RD5. With respect to the association between cna and serotype, our results are consistent with the results of Ryding et al. (56), who found that cna was present almost exclusively in serotype 8 strains. However, it is unlikely that either of these is directly associated with biofilm formation based on previous studies from our laboratory demonstrating that at least some cna-negative, serotype 5 strains form a biofilm comparable to that of the UAMS isolates (5). In contrast, sasG encodes an LPXTG-containing protein that is a homolog of Pls, and Savolainen et al. (60) demonstrated that expression of pls is associated with a reduced capacity to bind fibrinogen and fibronectin. Because biofilm formation by UAMS-1 and UAMS-601 in vitro is dependent on coating the substrate with plasma proteins (5), this suggests that the absence of sasG may be directly responsible for the increased capacity of these strains to form a biofilm. If so, then the divergent form of sasG present in MW2 and SANGER-476 would presumably have reduced function by comparison to strains that encode other forms of sasG. SasG has also been associated with an enhanced capacity to colonize nasal epithelial cells (54). This suggests that strains that either lack sasG or encode a functionally divergent form may have a reduced capacity to colonize nasal mucosal surfaces. Alternatively, these strains may utilize an alternative means of colonizing the nasal mucosa. For instance, Weidenmaier et al. (69) has suggested that cell wall teichoic acids are both necessary and sufficient for nasal colonization.
Overall, our results support the hypothesis that UAMS-1, UAMS-601, and EMRSA-16 are closely related strains that are genetically distinct from the more commonly studied laboratory strains, including RN6390. Indeed, Holden et al. (28) noted that EMRSA-16 is the most genetically diverse sequenced strain of S. aureus to date, since approximately 6% of the genome is novel compared with other published genomes. These results, together with the observation that RN6390 carries at least one relevant mutation (e.g., rsbU), clearly emphasize the need to reassess the relevance of studies focusing on 8325-4 strains like RN6390 with use of clinical isolates. Moreover, mounting evidence from other laboratories indicates that certain of the genotypic properties shared by EMRSA-16 and the UAMS strains are characteristic of the most predominant clinical isolates worldwide. These results strongly suggest that these strains are appropriate choices for these experiments. At present, it is difficult to determine which of the specific characteristics shared by these strains make a direct contribution to virulence. This will require mutagenesis and gain-of-function studies, both of which are somewhat complicated by the recalcitrance of clinical isolates to genetic manipulation. Nevertheless, the work presented here establishes a background for future experiments aimed at identifying the relevant virulence factors and the regulatory networks that control them, and having access to a complete genome sequence for a representative strain from this group will greatly facilitate these experiments.
Supplementary Material
Acknowledgments
This work was supported by R01-AI043356 from the National Institute of Allergy and Infectious Diseases (NIAID) to M.S.S. and by the NIAID-sponsored Pathogen Functional Genomics Resource Center at The Institute for Genomic Research.
We thank Robin Cline, Chris Mader, Joseph Gilbert, and Scott Peterson for technical support regarding the PFGRC arrays. The availability of strains from the NIAID-sponsored Network on Antimicrobial Resistance in Staphylococcus aureus is also gratefully acknowledged. We also thank the UAMS Microarray Core Facility for technical support, Alan Gies of the UAMS Core Sequencing Facility for DNA sequencing, Mark Enright for providing MLST profiles of all sequenced strains, and Barry Kreiswirth for providing the spa types of all sequenced strains and for generating the spa profile from sequence data for UAMS-1, UAMS-601, and RN6390.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N., T. Goldkorn, Y. Gov, M. Hirshberg, N. Koyfman, H. R. Matthews, R. T. Nhan, B. Singh, and O. Uziel. 2001. Regulation of Staphylococcus aureus pathogenesis via target of RNAIII-activating protein (TRAP). J. Biol. Chem. 276:2658-2667. [DOI] [PubMed] [Google Scholar]
- 4.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. Von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins, J. S., M. O. Elasri, S. D. Allmendinger, K. E. Beenken, R. A. Skinner, J. R. Thomas, and M. S. Smeltzer. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:d1825-d1842. [DOI] [PubMed] [Google Scholar]
- 13.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 14.Diep, B. A., G. F. Sensabaugh, N. S. Somboona, H. A. Carleton, and F. Perdreau-Remington. 2004. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J. Clin. Microbiol. 42:2080-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, and S. Wu. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunman, P. M., W. Mounts, F. McAleese, F. Immermann, D. Macapagal, E. Marsilio, L. McDougal, F. Tenover, P. A. Bradford, P. J. Petersen, S. J. Projan, and E. Murphy. 2004. Uses of Staphylococcus aureus GeneChips in genotyping and genetic composition analysis. J. Clin. Microbiol. 42:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert, P. M. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacometti, A., O. Cirioni, Y. Gov, R. Ghiselli, M. S. Del Prete, F. Mocchegiani, V. Saba, F. Orlando, G. Scalise, N. Balaban, and G. Dell'Acqua. 2003. RNA III inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillaspy, A. F., C. Y. Lee, S. Sau, A. L. Cheung, and M. S. Smeltzer. 1998. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect. Immun. 66:3170-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillaspy, A. F., J. M. Patti, F. L. Pratt, J. J. Iandolo, and M. S. Smeltzer. 1997. The Staphylococcus aureus collagen adhesin-encoding gene (cna) is within a discrete genetic element. Gene 196:239-248. [DOI] [PubMed] [Google Scholar]
- 25.Gillet, Y,. B. Issartel, P. Vanhems, J. C. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, N., D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 26.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gov, Y., I. Borovok, M. Korem, V. K. Singh, R. K. Jayaswal, B. J. Wilkinson, S. M. Rich, and N. Balaban. 2004. Quorum sensing in staphylococci is regulated via phosphorylation of three conserved histidine residues. J. Biol. Chem. 279:14665-14672. [DOI] [PubMed] [Google Scholar]
- 28.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 31.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu, K. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 35.Ladhani, S., C. L. Joannou, D. P. Lochrie, R. W. Evans, and S. M. Poston. 1999. Clinical, microbial, and biochemical aspects of the exfoliative toxins causing staphylococcal scalded-skin syndrome. Clin. Microbiol. Rev. 12:224-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 37.Lorenz, U., K. Ohlsen, H. Karch, A. Thiede, and J. Hacker. 2000. Immunodominant proteins in human sepsis caused by methicillin resistant Staphylococcus aureus. Adv. Exp. Med. Biol. 485:273-278. [DOI] [PubMed] [Google Scholar]
- 38.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt, B. G. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manna, A. C., and A. L. Cheung. 2003. SarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason, W. J., J. S. Blevins, K. Beenken, N. Wibowo, N. Ojha, and M. S. Smeltzer. 2001. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 39:3332-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterization of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51:516-521. [DOI] [PubMed] [Google Scholar]
- 44.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 48.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 49.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Projan, S. J. 2002. New (and not so new) antibacterial targets—from where and when will the novel drugs come? Curr. Opin. Pharmacol. 2:513-522. [DOI] [PubMed] [Google Scholar]
- 51.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, and K. E. Nelson. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 52.Robinson, D. A., and M. C. Enright. 2004. Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 10:92-97. [DOI] [PubMed] [Google Scholar]
- 53.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 54.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759-2767. [DOI] [PubMed] [Google Scholar]
- 55.Ryden, C., A. I. Yacoub, I. Maxe, D. Heinegard, A. Oldberg, A. Franzen, A. Ljungh, and K. Rubin. 1989. Specific binding of bone sialoprotein to Staphylococcus aureus isolated from patients with osteomyelitis. Eur. J. Biochem. 184:331-336. [DOI] [PubMed] [Google Scholar]
- 56.Ryding, U., J. I. Flock, N. Flock, B. Soderquist, and B. Christensson. 1997. Expression of collagen-binding protein and types 5 and 8 capsular polysaccharide in clinical isolates of Staphylococcus aureus. J. Infect. Dis. 176:1096-1099. [DOI] [PubMed] [Google Scholar]
- 57.Sabat, A., K. Kosowska, K. Poulsen, A. Kasprowicz, A. Sekowska, and B. van Den Burg. 2000. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect. Immun. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saris, P. E., T. Immonen, M. Reis, and H. G. Sahl. 1996. Immunity to lantibiotics. Antonie Leeuwenhoek 69:151-159. [DOI] [PubMed] [Google Scholar]
- 59.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395-2405. [DOI] [PubMed] [Google Scholar]
- 60.Savolainen, K., L. Paulin, B. Westerlund-Wikstrom, T. J. Foster, T. K. Korhonen, and P. Kuusela. 2001. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 69:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smeltzer, M. S., J. R. Thomas, S. G. Hickmon, R. A. Skinner, C. L. Nelson, D. Griffith, T. R. Parr, Jr., and R. P. Evans. 1997. Characterization of a rabbit model of staphylococcal osteomyelitis. J. Orthop. Res. 15:414-421. [DOI] [PubMed] [Google Scholar]
- 64.Smeltzer, M. S., F. L. Pratt, A. F. Gillaspy, and L. A. Young. 1996. Genomic fingerprinting for epidemiological differentiation of Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 34:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sterba, K. M., S. G. Mackintosh, J. S. Blevins, B. K. Hurlburt, and M. S. Smeltzer. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 69.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.