Abstract
The nocardicins are a family of monocyclic β-lactam antibiotics produced by the actinomycete Nocardia uniformis subsp. tsuyamanensis ATCC 21806. The most potent of this series is nocardicin A, containing a syn-configured oxime moiety, an uncommon feature in natural products. The nocardicin A biosynthetic gene cluster was recently identified and found to encode proteins in keeping with nocardicin A production, including the nocardicin N-oxygenase, NocL, in addition to genes of undetermined function, such as nocK, which bears similarities to a broad family of esterases. The latter was hypothesized to be involved in the formation of the critical β-lactam ring. While previously shown to effect oxidation of the 2′-amine of nocardicin C to provide nocardicin A, it was uncertain whether NocL was the only N-oxidizing enzyme required for nocardicin A biosynthesis. To further detail the role of NocL in nocardicin production in N. uniformis, and to examine the function of nocK, a method for the transformation of N. uniformis protoplasts to inactivate both nocK and nocL was developed and applied. A reliable protocol is reported to achieve both insertional disruption and in trans complementation in this strain. While the nocK mutant still produced nocardicin A at levels near that seen for wild-type N. uniformis, and therefore has no obvious role in nocardicin biosynthesis, the nocL disruptant failed to generate the oxime-containing metabolite. Nocardicin A production was restored in the nocL mutant upon in trans expression of the gene. Furthermore, the nocL mutant accumulated the biosynthetic intermediate nocardicin C, confirming its role as the sole oxime-forming enzyme required for production of nocardicin A.
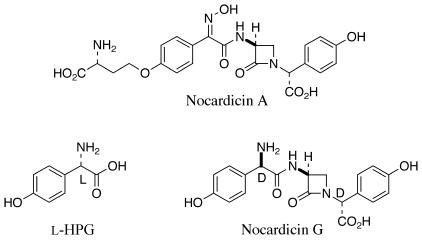
A new family of β-lactam antibiotics was reported in 1976 from the actinomycete Nocardia uniformis subsp. tsuyamanensis ATCC 21806 (2, 12, 13). Nocardicin A (Fig. 1) is the major product, as well as several other structurally related metabolites in smaller amounts, nocardicins B to G (Fig. 1) (2, 12, 16). The nocardicins were the first naturally occurring monocyclic β-lactams that display any significant antibacterial activity to be isolated, and they bear structural and stereochemical similarities to the naturally occurring penicillin N. The notable activity of nocardicin A has been ascribed to the presence of the syn-oxime, an extremely rare functional group in natural products, and to the terminal d configuration of the homoseryl side chain. This unusual feature of nocardicins A to D is known to arise by a 3-amino-3-carboxypropyl transfer from S-adenosylmethionine, a rare metabolic role for this cofactor (33).
FIG. 1.
Structure of the nocardicins.
Nocardicin G (Fig. 1) is the earliest β-lactam-containing intermediate in the pathway (47). The tripeptide core is easily discernible (45), and the presence of the unusual 4-hydroxyphenyglycine (HPG) units, each in the d configuration, suggests that this portion of the molecule arises ultimately through the action of one or more nonribosomal peptide synthetases (NRPSs). The biosynthetic gene cluster thought to be responsible for nocardicin A formation was recently identified (9). This cluster contains two NRPSs, nocardicin 3-amino-3-carboxypropyl transferase (Nat) (33), HPG biosynthetic genes, and the cytochrome P450 NocL (19). There are a number of gene products of the presumed cluster whose functions, however, are not easily predicted on the basis of sequence similarity revealing, proteins either with homology to those of unknown function or with implied chemistry not easily related to nocardicin A biosynthesis. Prominent among these is nocK, whose translated sequence bears similarity to a number of proposed esterases and hydrolases (9). The potential role of this protein in the unknown mechanism of β-lactam formation made the disruption of this gene of central interest.
The cytochrome P450 NocL was recently shown to mediate oxidation of the 2′-amine of nocardicin C to the oxime moiety observed for nocardicin A (19). Due to the inability to detect the oxidation of nocardicin G to form nocardicins E or F by NocL, it was unclear whether this was the only N-oxidizing enzyme required for nocardicin A biosynthesis. The only other previously characterized oxime-forming enzymes are the CYP79 family of cytochromes P450 involved in cyanogenic glucoside and glucosinolate biosynthesis from plants (1, 11, 39, 49). The occurrence of N-oxidized natural products is uncommon, and little is understood about how these oxidized amine species are introduced. The hydroxamate siderophores and azoxy antibiotics have been shown to involve a hydroxylamine intermediate generated by a flavin monooxygenase (5, 30, 42). The nitro group present in the antifungal agent pyrrolnitrin is thought to be the result of a nonheme iron-dependent dioxygenase and a primary amine precursor, whereas the nitro group present in aureothin was recently attributed to an enzyme with no significant similarity to established oxidative systems (15, 22).
With the broader goal in mind to determine the requirement of individual open reading frames for nocardicin A biosynthesis, we report a method for the transformation of N. uniformis. With a reliable protocol in hand, it was then possible to generate insertional disruption mutants of two targeted genes of interest in the nocardicin A biosynthetic cluster, nocK and nocL. In addition to heterologous expression of individual proteins, this approach opens an additional avenue to evaluate the requirement for encoded proteins in the formation of the monocyclic β-lactam metabolite. This avenue becomes particularly valuable for cases where enzyme function cannot be easily rationalized on the basis of sequence similarity. Specifically in the case of NocL, the generation of the disruption mutant enabled further clarification of its critical role in oxime formation and the overall course of nocardicin A biosynthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α (35) and JM110 (51) were used in this work. E. coli SC12155 was obtained from the Squibb Institute for Medical Research (Princeton, N.J.). Vectors pT7Blue-3 (Novagen, Madison, Wis.), pBSIISK(−) (Stratagene, La Jolla, Calif.), and pET24b(+) (New England Biolabs, Beverly, Mass.) were obtained from commercial sources. Vectors pIJ4070 and pULVK2 were generous gifts of Mervyn Bibb (John Innes Center, Norwich, England) and Juan F. Martín (University of León, León, Spain), respectively. Vector pULVK2T was described elsewhere (9). The N. uniformis strains and other plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| N. uniformis strain | ||
| ATCC 21806 | Wild type | 2 |
| nocK::apr | Inactivation of nocK by insertion of apr | This work |
| nocL::apr | Inactivation of nocL by insertion of apr | This work |
| E. coli SC12155 | Supersensitive to β-lactam antibiotics | 2 |
| Plasmid | ||
| pULVK2 | Bifunctional E. coli-Nocardia plasmid, Kmr | 27 |
| pULVK2/KAm | Derivative of pULVK2 containing the nocK fragment with apr insertion | This work |
| pULVK2/LAm | Derivative of pULVK2 containing the nocL fragment with apr insertion | This work |
| pET24b/nocL | Derivative of pET24b(+) containing the native sequence for nocL | This work |
| pIJ4070/nocL | Derivative of pIJ4070 containing nocL behind ermE2 | This work |
| pULVK2T | Derivative of pULVK2, Kmr and Tsr | 11 |
| pULVK2T/nocL | Derivative of pULVK2T containing nocL behind control of the ermE2 promoter | This work |
Biochemicals, chemicals, and media.
Authentic nocardicin A was a gift of Fujisawa Pharmaceutical Co., Ltd. (Osaka, Japan), and a sample of isonocardicin A was the gift of W. Hofheinz and H.-P. Isenring of Hoffman-La Roche (Basel, Switzerland). Nocardicin C was synthesized as previously described (34). Unless otherwise specified, biochemicals and reagents were from common commercial sources. E. coli strains were grown in Luria-Bertani (LB) medium and selected for their plasmid with the appropriate antibiotic (35). The nocardicin production medium was described previously (16). ISP-2 and Trypticase soy broth (TSB) were purchased from Difco Laboratories (Detroit, Mich.).
DNA isolation, manipulations, and sequencing.
Plasmid DNA isolation from E. coli was carried out using the Qiaprep Spin Miniprep kit (QIAGEN, Valencia, Calif.), and total DNA isolation from N. uniformis strains followed the gram-positive bacterium protocol for the QIAGEN DNeasy Tissue kit. For Southern analysis, 32P-labeled probes were generated by random priming with the RadPrime DNA labeling system (GibcoBRL Life Technologies, Rockville, Md.), and hybridizations were done according to protocol (35). DNA sequencing to confirm constructs was conducted on a Perkin-Elmer ABI 377 DNA sequencer (Foster City, Calif.) at the Biosynthesis and Sequencing Facility at Johns Hopkins Medical School, Baltimore, Md.
Transformation of N. uniformis.
A seed culture of N. uniformis was grown in 10 ml of TSB medium incubating for 24 h at 30°C and 300 rpm. Following this, 2 ml of the seed culture was inoculated into two 500-ml Erlenmeyer flasks with ca. 160 glass beads containing 100 ml of 148G medium (37) (0.85% glycine) supplemented with 1.5 mg of isonicotinic acid hydrazide/liter and 0.1% Tween 80. These cultures were incubated at 30°C and 300 rpm for 13 h. The mycelium was collected at 4,000 × g for 10 min at 4°C, then washed twice with 30 ml of ice-cold 15% sucrose, and finally resuspended in 8 ml of protoplast buffer (20) containing 4 mg of lysozyme/ml. The mixture was incubated at 37°C for 30 min, at which time protoplast formation was found to be 90% complete by microscopic examination, and the digestion continued for an additional 15 min. The suspension was diluted with 10 ml of protoplast buffer, filtered over a sterile cotton plug, and centrifuged at 1,500 × g for 20 min at 4°C. The protoplasts were washed twice with 15 ml of ice-cold protoplast buffer and finally resuspended in 1 ml of ice-cold protoplast buffer. To 100 μl of the protoplast suspension, 10 μl of vector (1 μg) and 300 μl of transformation buffer (20) were added, and the resulting mixture was incubated at room temperature for 1 min. The transformation mixture was diluted into 3 ml of R2L soft agar medium (26) and poured over R2YE (20) solid medium that had been prepared and dried that day for 3 h in a vertical laminar flow hood. After incubation at 30°C for 16 h, an antibiotic solution in 1.2 ml of water was permitted to soak into the solid medium (2.5 mg of apramycin, 5 mg of kanamycin, and 1.25 mg of thiostrepton to result in final concentrations of 100, 200, and 50 μg/ml, respectively). Plates were incubated at 30°C until colonies were well formed, about 10 days.
Construction of nocK::apr N. uniformis.
The nocK gene was amplified by PCR from cosmid DNA (9). The forward primer (5′-AAGGATCCGTGACCATGATCGGAGTCACG-3′) contained a BamHI restriction site (underlined) and the native GTG start codon, while the reverse primer (5′-AAGGATCCTCAGCCGCCGAGCAGGAAGTC-3′) also contained a BamHI restriction site and the native stop codon. The amplified fragment was ligated into the EcoRV site of pT7Blue-3, then excised as the BamHI fragment, and cloned into pBSIISK(−). The resulting vector was linearized with PmlI and treated with shrimp alkaline phosphatase. The EcoRI insert containing apr was treated with Klenow fragment and ligated into the PmlI-digested nocK vector. The resulting nocK::apr fragment was excised with BamHI and cloned into pULVK2 to provide pULVK2/KAm. Following isolation of the vector from E. coli JM110, N. uniformis protoplasts were transformed as described above and selected with 200 μg of kanamycin/ml and 100 μg of apramycin/ml. Eight colonies were picked and streaked onto ISP-2 solid medium containing 100 μg of apramycin/ml. Following growth for several days, spores from a single colony were streaked a second time on ISP-2 solid medium containing apramycin until sporulation occurred. Spores from a single colony from each of the eight plates were used to generate a mycelium stock. Total DNA from wild-type and potential mutant strains was isolated, digested with DraIII, and evaluated by Southern hybridizations to apr and nocK.
Construction of nocL::apr N. uniformis.
The nocL gene was amplified by PCR from cosmid DNA (9). The forward primer (5′-AAGGATCCATGACCAGAACCGACACGCGCT-3′) contained a BamHI restriction site (underlined), and the native GTG start codon was replaced with ATG, while the reverse primer (5′-AAGGATCCTCACCAGGTGACCGCAG-3′) contained a BamHI restriction site and the native stop codon. The fragment was ligated into the EcoRV site of pT7Blue-3, then excised as the BamHI fragment, and cloned into pBSIISK(−). The resulting vector was linearized with Tth111I and then treated with the Klenow fragment and shrimp alkaline phosphatase. The EcoRI insert containing apr was treated with Klenow fragment and ligated into the linearized nocL vector. The resulting nocL::apr fragment was excised with BamHI and cloned into pULVK2 to provide pULVK2/LAm. Following isolation of the vector from E. coli JM110, N. uniformis protoplasts were transformed as described above. Eight colonies were picked and propagated as described above. Total DNA from wild-type and potential mutant strains was isolated, digested with PvuII, and evaluated by Southern hybridizations to apr and nocL.
Evaluation of nocardicin A production in N. uniformis strains.
Mycelium stock of wild-type and mutant strains of N. uniformis was used to inoculate a seed culture of 10 ml of TSB medium (containing 50 μg of apramycin/ml or both apramycin and 25 μg of thiostrepton/ml, if appropriate) and grown at 30°C for 48 h. At this time, a 500-ml Erlenmeyer flask with ca. 160 glass beads containing 100 ml of nocardicin fermentation medium supplemented with 0.5 mM l-methionine and l-HPG was inoculated with 2 ml of the seed culture and grown at 30°C for 120 h. At 72, 96, and 120 h, 1 ml of culture was collected, and mycelia were removed by centrifugation at 4,000 × g for 10 min. Culture supernatant (250 μl) was applied to paper disks placed upon LB solid medium infused with E. coli SC12155. The plates were incubated overnight at 37°C and examined the following morning for the appearance of antibiosis. Additionally, 100 μl of culture supernatant was diluted to 2 ml with 20 mM ammonium acetate (pH to 3.0 with acetic acid) and then applied to approximately 1.5 ml of AG50W-X8 (Bio-Rad, Hercules, Calif.) resin preequilibrated with the loading buffer. The resin was washed, and the bound material was purged from the resin with 3 ml of 1% NH4OH and collected in a tube containing 150 μl of 10% acetic acid to immediately neutralize the basic solution. The solvent was removed by lyophilization, and samples were analyzed by reverse-phase high-pressure liquid chromatography (HPLC) on either a Perkin-Elmer 235C diode array detector and LC410 pump or an Agilent Technologies 1100 diode array detector and quaternary pump by using a Phenomenex (Torrance, Calif.) Luna C18 (2) 250- by 4.6-mm column and monitoring absorbance at 272 nm in 10% acetonitrile-0.1% trifluoroacetic acid (aqueous) at 1 ml/min.
Isolation and characterization of nocardicin A and isonocardicin A from nocK::apr N. uniformis.
A mixture of nocardicin A and isonocardicin A was purified from nocK::apr N. uniformis following previously described protocol (45). A white solid (275 mg) was isolated from 3.2 liters of culture supernatant and found to give a λmax at 272 nm. Electrospray ionization mass spectrometry (ESI-MS) was performed on a Finnigan LCQDECA instrument in positive ion mode of a solution in 60% acetonitrile and 1% acetic acid to provide an [M + H] ion at m/z 501.2, and tandem mass spectrometry (MS/MS) analysis generated fragments at m/z 351.0 and 322.0; mp 210 to 215°C dec (lit [2], 214 to 216°C dec); 400 MHz (Varian UnityPlus 400 Spectrometer) 1H NMR (D2O + K2CO3) δ: 7.48 (2 H, d, J = 8.8 Hz, Ar), 7.20 (2 H, d, J = 8.4 Hz, Ar), 7.01 (2 H, d, J = 8.8 Hz, Ar), 6.84 (2 H, d, J = 8.8 Hz, Ar), 5.30 (1 H, s, H-5), 5.00 (1 H, dd, J = 5.2, 2.4 Hz, H-3), 4.20 (2 H, m, H-7′), 3.84 (1 H, t, J = 5.4 Hz, H-4α), 3.76 (1 H, m, H-9′), 3.23 (1 H, t, J = 2.6 Hz, H-4β), 2.25 (2 H, m, H-8′).
Complementation of nocL::apr N. uniformis.
The nocL gene was PCR amplified from cosmid DNA (9). The forward primer (5′-AACATATGACCAGAACCGACACGCGCT-3′) possessed an NdeI restriction site (underlined), and the native GTG start codon was replaced with ATG. The reverse primer (5′-AAGGATCCTCACCAGGTGACCGGCAG-3′) contained a BamHI restriction site and the native stop codon. The PCR product was ligated into the EcoRV site of pT7Blue-3, then excised as the NdeI-BamHI fragment, and ligated into appropriately digested pET24b(+). The XbaI-HindIII fragment containing nocL behind the ribosomal binding site was excised and cloned into appropriately digested pIJ4070 to provide pIJ4070/nocL. The BglII fragment of this vector containing nocL, the ribosomal binding site, and ermE2 was excised and ligated into BamHI-digested pULVK2T to generate pULVK2T/nocL. The plasmid was isolated from E. coli JM110 and used to transform N. uniformis CAT L4 as described above. Transformants were confirmed by isolation of total DNA from N. uniformis and reisolation of the plasmid from E. coli DH5α.
RESULTS
Protoplast transformation of N. uniformis.
Mycelium was taken from early- to mid-log phase of growth, washed with a sucrose solution, and resuspended in a protoplast generation buffer containing 4 mg of lysozyme/ml. When protoplast formation appeared to be greater than 90% complete by visual inspection, the lysozyme and N. uniformis mixture was filtered over a sterile cotton plug to remove any remaining mycelium and mycelial fragments, and the resulting protoplasts were washed with additional protoplast buffer. Treatment of the protoplasts with a polyethylene glycol (PEG)-containing transformation buffer in the presence of plasmid DNA consistently generated transformants at an efficiency of 102 to 103 transformants per microgram of DNA.
The effect of culture age and growth phase on protoplast transformation and recovery was examined. It was observed that mycelium obtained from the early log phase readily generated protoplasts within 20 to 45 min, whereas mycelium isolated from the late log phase was far more reticent to the lysozyme treatment, requiring nearly 3 h to produce protoplasts. Furthermore, the presence of isoniazide and glycine in the growth medium facilitated protoplast formation (17, 43). Protoplasts obtained from the mixtures exposed to lysozyme for longer than 1 h failed to regenerate to the same extent as those formed in a shorter time period and led to less than 102 transformants per microgram of DNA. In some cases, the addition of dimethyl sulfoxide has been observed to be advantageous for optimal transformation efficiency (50). The inclusion of 10% dimethyl sulfoxide in the transformation reaction with N. uniformis protoplasts, however, did not afford any detectable benefit.
Disruption of nocK and nocL in N. uniformis.
To evaluate the functions of nocK and nocL in nocardicin A biosynthesis, insertion mutants of the two genes were prepared. The apramycin resistance cassette (apr) was inserted into the PmlI restriction site of nocK and the Tth111I restriction site of nocL. The resulting nocK::apr and nocL::apr fragments were each cloned into the E. coli-Nocardia shuttle vector pULVK2 (23) to form pULVK2/KAm and pULVK2/LAm, respectively. The resulting vectors were introduced into N. uniformis by PEG-mediated protoplast transformation. Transformants were replicated in two rounds of sporulation by using only the insertion marker antibiotic apramycin. Potential mutant strains were identified based on resistance to apramycin and acquired sensitivity to kanamycin. Such an antibiotic resistance profile is suggestive of a double crossover event to exchange the wild-type gene with the gene containing the apramycin insertion on the N. uniformis chromosome followed by loss of the residual pULVK2 plasmid.
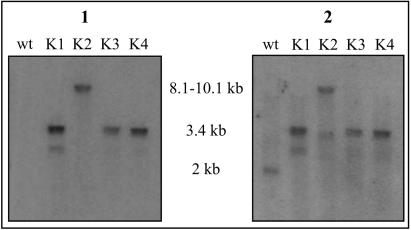
The total DNA from wild-type N. uniformis, three potential nocK insertion mutants (N. uniformis CAT K1, K3, and K4), and a potential mutant resulting from integration of the vector (N. uniformis CAT K2) was isolated and digested with DraIII. This enzyme is predicted to generate a restriction product for the wild-type strain containing nocK on a fragment of about 2 kb and a fragment of approximately 3.4 kb with the apr insertion. A Southern hybridization of the digested total DNA against a 32P-labeled probe of the apramycin resistance gene indicated a single fragment at about 3.4 kb in all three potential mutants, and, as expected, no positive fragments in wild-type DNA (Fig. 2A). The fourth potential mutant displayed a band at 8 to 10 kb, consistent with a single crossover integrating the entire plasmid into the N. uniformis chromosome. A second hybridization against the radiolabeled probe of nocK gave the expected 2-kb fragment for the wild-type strain, while no new bands appeared in any of the potential mutants. The results obtained from the Southern hybridization and the antibiotic resistance profile were indicative of a double crossover event between the vector pULVK2/KAm and the N. uniformis chromosome in three of these strains to produce nocK::apr N. uniformis, and quite likely the fourth strain arose from integration of the vector into the genome.
FIG. 2.
Southern hybridization of DraIII-digested total DNA isolated from wild-type (wt) N. uniformis and potential nocK insertion mutants (K1 to K4) with a 32P probe to the apramycin resistance gene (panel 1) and to nocK (panel 2).
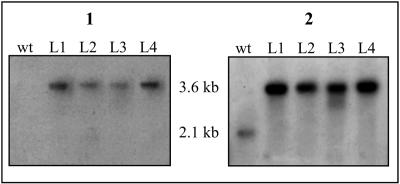
Four potential disruption mutants for nocL (N. uniformis CAT L1 to L4) were also evaluated by Southern hybridization. The isolated total DNA was digested with PvuII to place wild-type nocL on a single fragment of 2.1 kb, while the insertion mutants would provide a single fragment of 3.6 kb. All four potential mutants, when presented with the apr probe, revealed a single band at approximately 3.6 kb (Fig. 3). The second hybridization with nocL gave the expected fragment at 2.1 kb for wild-type N. uniformis and no additional signals in the potential mutants, confirming construction of the nocL insertion mutant.
FIG. 3.
Southern hybridization of PvuII-digested total DNA isolated from wild-type (wt) N. uniformis and potential nocL insertion mutants (L1 to L4) with a 32P probe to the apramycin resistance gene (panel 1) and to nocL (panel 2).
Evaluation of nocardicin A production in nocK::apr N. uniformis.
Following identification of a disruption mutant for nocK, the effect of this mutation on the production of nocardicin A and possibly β-lactam formation by nocK::apr N. uniformis was evaluated. Two separate methods were employed to test for the presence of nocardicin A in N. uniformis culture supernatants: a bioassay and reverse-phase HPLC analysis. The paper disk bioassay utilized a strain of E. coli, SC12155, engineered to be supersensitive to the presence of β-lactam antibiotics (2). Wild-type and nocK::apr N. uniformis were incubated in nocardicin fermentation medium (16) for 120 h, and aliquots of the culture supernatant were obtained at 72, 96, and 120 h of growth and used in the paper disk bioassay. All samples tested from nocK::apr N. uniformis generated a zone of antibiosis surrounding the supernatant-soaked disks with a diameter comparable to that of wild-type N. uniformis (data not shown). The isolated culture supernatants were then partially purified by strong cation exchange chromatography and analyzed by reverse-phase HPLC. Both strains produced a peak in the supernatant, coeluting with an authentic standard of nocardicin A (data not shown). No significant difference was detected between wild-type and nocK::apr N. uniformis nocardicin A production at 72 h, giving 205 mg/liter, and the difference at 96 and 120 h for the mutant strain was within 20% of wild-type levels.
A mixture of nocardicin A and isonocardicin A was isolated from 3 liters of nocK::apr N. uniformis culture and found to coelute with authentic specimens of nocardicin A and isonocardicin A under both chiral HPLC conditions and 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate derivatization (data not shown) (21, 29). ESI-MS of the sample isolated from the disrupted strain was identical to authentic nocardicin A ([M + H] ion peak at m/z 501.2), and MS-MS of this peak produced fragment ions at m/z 351.0 and 322.0. Ideally, a comparison to nocardicin A and the C-5 epimer of nocardicin A would provide a conclusive answer to the stereochemistry at this position. The C-5 epimer of nocardicin A is not readily available; however, the proton signals at C-5 for diastereomeric analogs at this position differ by as much as 0.30 ppm, which would be clearly discernible by 400 MHz 1H nuclear magnetic resonance (4, 14). A mixture of the nocK::apr nocardicin A and isonocardicin A with authentic nocardicin A resulted in a single, well-resolved signal at C-5.
Evaluation of nocardicin A production in nocL::apr N. uniformis.
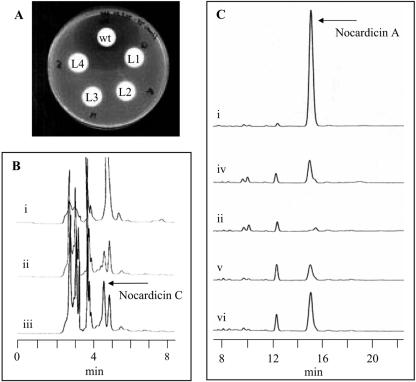
Wild-type and mutant nocL strains of N. uniformis were incubated in nocardicin fermentation medium, and samples of culture supernatant were obtained at 72, 96, and 120 h. Supernatants were soaked into paper disks placed upon LB solid medium infused with E. coli SC12155. Only the supernatant obtained from wild-type N. uniformis was able to exhibit antibiosis (Fig. 4A). All nocL::apr N. uniformis strains failed to inhibit growth of the supersensitive E. coli. This observation is consistent with either a loss of β-lactam production by N. uniformis or a loss of nocardicin A accompanied by accumulation of a less potent β-lactam-containing metabolite.
FIG. 4.
(A) Paper disk bioassay of N. uniformis culture supernatants against E. coli SC12155. (B and C) Reverse-phase HPLC analysis of the N. uniformis supernatants from (i) wild-type N. uniformis, (ii) N. uniformis CAT L4, (iii) L4 with nocardicin C internal standard, (iv) L4 with nocardicin A internal standard, (v) N. uniformis CAT L4 (pULVK2T/nocL), and (vi) N. uniformis CAT L4 (pULVK2T/nocL) with nocardicin A internal standard.
The isolated culture supernatants were partially purified by strong cation exchange chromatography and analyzed by reverse-phase HPLC (Fig. 4B and C). In the nocL insertion mutants, there was a complete absence of nocardicin A. Further examination of these chromatograms did reveal a peak present in the nocL::apr strains that coeluted with a synthetic standard of nocardicin C and possessed an identical UV-visible spectrum. Analysis of this peak by ESI-MS produced an [M + H] ion at m/z 487.3, the expected [M + H] peak for nocardicin C. An MS-MS analysis of this peak produced fragment ions at m/z 291.1, 292.1, 319.9, and 337.0, in a pattern identical to that of synthetic nocardicin C.
Complementation of nocL::apr N. uniformis.
To verify that the loss of nocardicin A production in nocL::apr N. uniformis was due entirely to the absence of nocL and was not the result of a polar mutation introduced by the insertion, in trans expression of nocL in the N. uniformis mutant strain was performed. Expression of nocL was placed under control of the ermE2 promoter in the E. coli-Nocardia shuttle vector pULVK2T to provide pULVK2T/nocL. The complementation vector was introduced into N. uniformis CAT L4 by PEG-mediated protoplast transformation. The total DNA from the potential transformants was prepared and used to transform E. coli DH5α. The plasmid was then isolated and analyzed by restriction digestion to confirm nocL::apr N. uniformis (pULVK2T/nocL).
Culture supernatants of nocL::apr N. uniformis (pULVK2T/nocL) grown in nocardicin fermentation medium were analyzed for restoration of nocardicin A production by both bioassay against E. coli SC12155 and reverse-phase HPLC. None of the potential complemented N. uniformis strains produced a discernible antibiosis zone (data not shown). However, examination of the supernatants by reverse-phase HPLC did reveal the presence of nocardicin A, which was absent in the nocL disruption mutant (Fig. 4C). Isolation of this peak and analysis by ESI-MS demonstrated a mass and fragmentation pattern identical to that of authentic nocardicin A, providing an [M + H] ion at m/z 501.3 and MS-MS fragment ions of this peak at m/z 351.1 and 322.0.
DISCUSSION
While much attention has centered on transformation protocols for Streptomyces, relatively little has been devoted to other actinomycete species, despite the ability of many of these organisms to biosynthesize clinically relevant antibiotics and other natural products of interest. For example, Amycolatopsis mediterranei LBG A3136 produces the antitubercular agent rifamycin (38), Amycolatopsis orientalis ATCC 19795 generates vancomycin (28), which is used in the treatment of methicillin-resistant Staphylococcus aureus, and Nocardia lactamdurans yields the β-lactam antibiotic cephamycin C (40). The approach taken here is based upon the methods described for A. mediterranei and A. orientalis (27, 37). The PEG-mediated transformation of N. uniformis protoplasts then facilitated the evaluation of the requirement for two different genes, nocK and nocL, in nocardicin A production. Furthermore, development of a robust protocol will enable further studies of the genes of undetermined function in the nocardicin A biosynthetic pathway through the generation of insertional disruption mutants and complementation experiments.
The open reading frame nocK codes for a protein with similarity to ferulic acid esterases and polyhydroxyalkanoate (PHA) depolymerases of the α/β hydrolase structural family, suggesting a possible hydrolytic function in nocardicin biosynthesis (6, 18). These classes of esterases are modular in nature, each harboring a distinct domain responsible for catalysis (7, 8). At a predicted 346 amino acids, NocK is considerably shorter than the typical PHA depolymerase or ferulic acid esterase, which are generally over 500 amino acids in length. The similarity of NocK to these proteins is localized to the catalytic domain, notable in regions thought to comprise the catalytic triad and oxyanion hole (18). NocK appears to possess all of these required components: the oxyanion hole, the nucleophilic serine, the second histidine residue, and the first of the conserved aspartic acid residues. The second conserved aspartate of the PHA depolymerases has been replaced by a glutamate in NocK. It is conceivable that glutamate could form the third component of a catalytic triad for NocK: serine-histidine-glutamate triads have been observed in certain lipases, esterases, and proteases (10, 36, 41).
The role such an enzyme would play in nocardicin biosynthesis, however, was not obvious despite its prominent location in the cluster. In consideration of the potential function of NocK, no candidate protein is evident to account for either β-lactam formation or epimerization of the C-5 stereocenter (9). Earlier biosynthetic experiments suggested that the serine hydroxyl must be activated to enable β-lactam ring formation to occur (44, 46). An esterase can be visualized to take part in such a process. In the event, the function of NocK was examined by insertional mutagenesis. It was determined that its presence is not required for nocardicin A production, as the antibiotic is still produced by the disrupted strain at a level comparable to that of the wild type. Therefore, if NocK does have a role in nocardicin formation, it is not an essential one. Bioinformatic analysis of the NRPSs of the nocardicin A biosynthetic cluster hints that a pentapeptide could be produced as an intermediate (9). It was also thought possible that NocK could serve as a protease to process this putative pentapeptide intermediate to a tripeptide, and another cellular protease or a self-catalyzed process could replace this function in its absence. NocK might also participate in an as yet undetermined regulatory or self-resistance mechanism or have no function at all. Further assignment of NocK function awaits future studies.
It was demonstrated that NocL is capable of oxidizing the C-2′ primary amine of nocardicin C to the oxime present in nocardicin A in vitro, but no activity was observed when NocL was presented with nocardicin G to form nocardicin E and/or its oxime configurational isomer, nocardicin F (19). It is certainly possible that nocardicin G is a substrate for NocL but that it escaped the detection limit in the assays employed for NocL activity. An alternative explanation for the lack of observable activity is that a second oxidative enzyme located outside of the putative biosynthetic cluster is responsible for the synthesis of nocardicins E and F. The lack of nocardicin A produced by the nocL disruption mutant argues against this possibility, however. If a second enzyme were necessary for nocardicin E formation, the subsequent action of Nat, which attaches the homoseryl side chain to both nocardicin G and nocardicin E to produce isonocardicin C and isonocardicin A, respectively, would have then facilitated the synthesis of nocardicin A (33). Notwithstanding, nocardicin C, not nocardicin A, accumulated in the fermentation broth of the nocL disruptant, supporting the contention that NocL is the only N-oxygenase required for nocardicin A biosynthesis.
At least two factors could contribute to the low level of nocardicin A production in the nocL::apr N. uniformis complementation strain relative to the wild type. It was observed that the transformants were unable to achieve the same cell density as either the insertionally inactivated strain or the wild type, which could conceivably affect secondary metabolism in the bacterium. Additionally, thiostrepton has been demonstrated to alter expression levels of cellular proteins in various species of Streptomyces, Nocardia, and Amycolatopsis (23, 24), and deviation from a normal protein expression pattern could also affect production levels of nocardicin A. Although thiostrepton was omitted from the production medium, it was present in the seed culture. Finally, while the ermE2 promoter has been successfully employed in complementation experiments with A. mediterranei and in protein expression of various Streptomyces species, its efficiency in N. uniformis is unknown (3, 31, 32, 48). Ideally, a promoter from the N. uniformis biosynthetic pathway should be employed for this purpose, but none has been characterized at this time. It is quite possible that ermE2 is poorly recognized in N. uniformis or that expression is not optimally coordinated with the remaining nocardicin A biosynthetic machinery.
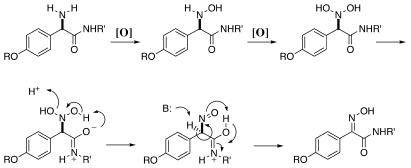
NocL is the first example of a prokaryotic cytochrome P450 catalyzing N oxygenation, specifically oxime formation. It is tempting to speculate that the greater abundance of the syn-oxime (nocardicin A) relative to the anti-oxime (nocardicin B) isolated from N. uniformis fermentation broth arises from intramolecular hydrogen bonding during the course of oxidation by NocL (Fig. 5) (2, 25). We propose successive hydroxylation of nocardicin C to 2′-N,N-dihydroxynocardicin C, in which the neighboring amide bond could facilitate elimination of water to form a nitroso species and then, owing to the principle of least motion, tautomerization to the syn-oxime. The oxyanion amide resonance form could be visualized to remove the N-hydroxyl proton and initiate elimination of the second N-hydroxyl as water. Additionally, the same intramolecular interaction could facilitate preferential formation of the syn-oxime by stabilizing this conformation of the nitroso group, locking its position with a hydrogen bond. Furthermore, the resulting protonated amide tautomer could donate a proton as the syn-oxime is generated. Similar intramolecular hydrogen-bonding interactions have been invoked to account for syn or anti stereochemistry in synthetic oxime-forming reactions (34).
FIG. 5.
Proposed mechanism of NocL-catalyzed oxime formation.
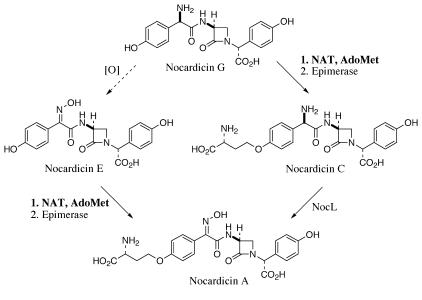
Application of the N. uniformis transformation procedure described herein enabled a functional analysis of two genes in the nocardicin A biosynthetic cluster. While NocK was found to be dispensable, NocL was established as the only N-oxygenase required for nocardicin A biosynthesis. Additionally, the bias of recombinant NocL to effect oxime formation from nocardicin C over nocardicin G complements the in vitro function of Nat, for which nocardicin G is the preferred substrate for 3-amino-3-carboxypropyl group transfer (33). It now appears the preferred biosynthetic route taken for the late stages of nocardicin A biosynthesis from nocardicin G first requires the attachment of the homoseryl side chain and is followed by, in an as yet undetermined order, inversion of stereochemistry at the C-9′ position and oxime formation (Fig. 6). Further studies of NocL will reveal whether oxidation preferentially occurs on nocardicin C or isonocardicin C to delineate the exact timing of the final steps.
FIG. 6.
The late stages of nocardicin A biosynthesis. AdoMet, S-adenosylmethionine.
Acknowledgments
We are pleased to acknowledge the financial support of the NIH (AI14937) and the Henry Sonneborn III Fellowship through the auspices of the Department of Chemistry of the Johns Hopkins University (W.L.K.).
We thank Rongfeng Li for his invaluable advice and discussions concerning actinomycete microbiology and transformation techniques, as well as Jacques Ravel.
REFERENCES
- 1.Andersen, M. D., P. K. Busk, I. Svendsen, and B. L. Moller. 2000. Cytochromes P-450 from cassava (Manihot esculenta crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. Cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J. Biol. Chem. 275:1966-1975. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., H. Sakai, M. Kohsaka, T. Konomi, and J. Hosoda. 1976. Nocardicin A, a new monocyclic beta-lactam antibiotic. I. Discovery, isolation and characterization. J. Antibiot. 29:492-500. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, Y. Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiba, K., M. Mori, and Y. Ban. 1985. A novel synthesis of (+/-)-3-aminonocardicinic acid. Tetrahedron 41:387-392. [Google Scholar]
- 5.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fillingham, I. J., P. A. Kroon, G. Williamson, H. J. Gilbert, and G. P. Hazlewood. 1999. A modular cinnamoyl ester hydrolase from the anaerobic fungus Piromyces equi acts synergistically with xylanase and is part of a multiprotein cellulose-binding cellulase-hemicellulase complex. Biochem. J. 343:215-224. [PMC free article] [PubMed] [Google Scholar]
- 7.Fukui, T., T. Narikawa, K. Miwa, Y. Shirakura, T. Saito, and K. Tomita. 1988. Effect of limited tryptic modification of a bacterial poly(3-hydroxybutyrate) depolymerase on its catalytic activity. Biochim. Biophys. Acta 952:164-171. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert, H. J., G. P. Hazlewood, J. I. Laurie, C. G. Orpin, and G. P. Xue. 1992. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol. Microbiol. 6:2065-2072. [DOI] [PubMed] [Google Scholar]
- 9.Gunsior, M., S. D. Breazeale, A. J. Lind, J. Ravel, J. W. Janc, and C. A. Townsend. 2004. The biosynthetic gene cluster for a monocyclic β-lactam antibiotic, nocardicin A. Chem. Biol. 11:927-938. [DOI] [PubMed] [Google Scholar]
- 10.Hakansson, K., A. H. Wang, and C. G. Miller. 2000. The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad. Proc. Natl. Acad. Sci. USA 97:14097-14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, C. H., U. Wittstock, C. E. Olsen, A. J. Hick, J. A. Pickett, and B. A. Halkier. 2001. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276:11078-11085. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, M., T. Komori, and T. Kamiya. 1976. Nocardicin A, a new monocyclic beta-lactam antibiotic. II. Structure determination of nocardicins A and B. J. Antibiot. 29:890-901. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto, M., T.-A. Komori, and T. Kamiya. 1976. Nocardicin A and B, novel monocyclic β-lactam antibiotics from a Nocardia species. J. Am. Chem. Soc. 98:3023-3025. [DOI] [PubMed] [Google Scholar]
- 14.Hatanaka, M., N. Noguchi, and T. Ishimaru. 1982. Synthesis of 3-aminonocardicinic acid, the basic nucleus of nocardicins. Bull. Chem. Soc. Jpn. 55:1234-1237. [Google Scholar]
- 15.He, J., and C. Hertweck. 2004. Biosynthetic origin of the rare nitroaryl moiety of the polyketide antibiotic aureothin: involvement of an unprecedented N-oxygenase. J. Am. Chem. Soc. 126:3694-3695. [DOI] [PubMed] [Google Scholar]
- 16.Hosoda, J., T. Konomi, N. Tani, and H. Aoki. 1977. Isolation of new nocardicins from Nocardia uniformis subs. tsuyamanensis.. Agric. Biol. Chem. 41:2013-2020. [Google Scholar]
- 17.Jang, K.-H., D. Pierotti, G. W. Kemp, G. R. Best, and M. L. Britz. 1997. Mycolic acid composition of Corynebacterium glutamicum and its cell surface mutants: effects of growth with glycine and isonicotinic acid. Microbiology 143:3209-3221. [DOI] [PubMed] [Google Scholar]
- 18.Jendrossek, D., A. Frisse, A. Behrends, M. Adermann, H. D. Kratzin, T. Stanislawski, and H. G. Schlegel. 1995. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J. Bacteriol. 177:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, W. L., and C. A. Townsend. 2002. Role of the cytochrome P450 NocL in nocardicin A biosynthesis. J. Am. Chem. Soc. 124:8186-8187. [DOI] [PubMed] [Google Scholar]
- 20.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 21.Kinoshita, T., Y. Kasahara, and N. Nimura. 1981. Reversed-phase high-performance liquid chromatographic resolution of non-esterified enantiomeric amino acids by derivatization with 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate and 2,3,4-tri-O-acetyl-α-d-arabinopyranosyl isothiocyanate. J. Chromatogr. 210:77-81. [Google Scholar]
- 22.Kirner, S., P. E. Hammer, D. S. Hill, A. Altmann, I. Fischer, L. J. Weislo, M. Lanahan, K.-H. van Pée, and J. M. Ligon. 1998. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, C. V., J.-J. R. Coque, and J. F. Martin. 1994. Efficient transformation of the cephamycin C producer Nocardia lactamdurans and development of shuttle and promoter-probe cloning vectors. Appl. Environ. Microbiol. 60:4086-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, C. V., and J. F. Martin. 1994. Thiostrepton induced proteins in Streptomyces, Amycolatopsis and Nocardia species. FEMS Microbiol. Lett. 118:107-111. [Google Scholar]
- 25.Kurita, M., K. Jomon, T. Komori, N. Miyairi, and H. Aoki. 1976. Isolation and characterization of nocardicin B. J. Antibiot. 29:1243-1245. [DOI] [PubMed] [Google Scholar]
- 26.Madon, J., and R. Hutter. 1991. Transformation system for Amycolatopsis (Nocardia) mediterranei: direct transformation of mycelium with plasmid DNA. J. Bacteriol. 173:6325-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushima, P., M. A. McHenney, and R. H. Baltz. 1987. Efficient transformation of Amycolatopsis orientalis (Nocardia orientalis) protoplasts by Streptomyces plasmids. J. Bacteriol. 169:2298-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick, M. H., W. M. Stark, G. E. Pittenger, R. C. Pittenger, and J. M. McGuire. 1956. Vancomycin, a new antibiotic. I. chemical and biologic properties. Antibiot. Annu. 1955-1956:606-611. [PubMed] [Google Scholar]
- 29.Nimura, N., H. Ogura, and T. Kinoshita. 1980. Reversed-phase liquid chromatographic resolution of amino acid enantiomers by derivatization with 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate. J. Chromatogr. 202:375-379. [Google Scholar]
- 30.Parry, R. J., W. Li, and H. N. Cooper. 1997. Cloning, analysis and overexpression of the gene encoding isobutylamine N-hydroxylase from the valanimycin producer Stretptomyces viridifaciens. J. Bacteriol. 179:409-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quiros, L. M., I. Aguirrezabalaga, C. Olano, C. Mendez, and J. A. Salas. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1185. [DOI] [PubMed] [Google Scholar]
- 32.Recktenwald, J., R. Shawky, O. Puk, F. Pfennig, U. Keller, W. Wohlleben, and S. Pelzer. 2002. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiology 148:1105-1118. [DOI] [PubMed] [Google Scholar]
- 33.Reeve, A. M., S. D. Breazeale, and C. A. Townsend. 1998. Purification, characterization, and cloning of an S-adenosylmethionine-dependent 3-amino-3-carboxypropyltransferase in nocardicin biosynthesis. J. Biol. Chem. 273:30695-30703. [DOI] [PubMed] [Google Scholar]
- 34.Salituro, G. M., and C. A. Townsend. 1990. Total syntheses of (-)-nocardicins A-G: a biogenetic approach. J. Am. Chem. Soc. 112:760-770. [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schrag, J. D., Y. Li, S. Wu, and M. Cygler. 1991. Ser-His-Glu traid forms the catalytic site of the lipase from Geotrichum candidum. Nature 351:761-764. [DOI] [PubMed] [Google Scholar]
- 37.Schupp, T., and M. Divers. 1986. Protoplast preparation and regeneration in Nocardia mediterranei. FEMS Microbiol. Lett. 36:159-162. [Google Scholar]
- 38.Sensi, P., and J. E. Thiemann. 1967. Production of rifamycins. Prog. Ind. Microbiol. 6:21-60. [Google Scholar]
- 39.Sibbesen, O., B. Koch, B. A. Halkier, and B. L. Moller. 1995. Cytochrome P-450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of l-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J. Biol. Chem. 270:3506-3511. [DOI] [PubMed] [Google Scholar]
- 40.Stapley, E. O., M. Jackson, S. Hernandez, S. B. Zimmerman, S. A. Currie, S. Mochales, J. M. Mata, H. B. Woodruff, and D. Hendlin. 1972. Cephamycins, a new family of β-lactam antibiotics. I. Productions by actinomycetes, including Streptomyces lactamdurans sp. n. Antimicrob. Agents Chemother. 2:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sussman, J. L., M. Harel, F. Frolow, C. Oefner, A. Goldman, L. Toker, and I. Silman. 1991. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253:872-879. [DOI] [PubMed] [Google Scholar]
- 42.Thariath, A., D. Socha, M. A. Valvano, and T. Viswanatha. 1993. Construction and biochemical characterization of recombinant cytoplasmic forms of the IucD protein (lysine:N6-hydroxylase) encoded by the pColV-K30 aerobactin gene cluster. J. Bacteriol. 175:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomiyasu, I., and I. Yano. 1984. Isonicotinic acid hydrazide induced changes and inhibition in mycolic acid synthesis in Nocardia and related taxa. Arch. Microbiol. 137:316-323. [DOI] [PubMed] [Google Scholar]
- 44.Townsend, C. A., and A. M. Brown. 1982. Nocardicin A biosynthesis: stereochemical course of monocyclic beta-lactam formation. J. Am. Chem. Soc. 104:1748-1750. [Google Scholar]
- 45.Townsend, C. A., and A. M. Brown. 1983. Nocardicin A: biosynthetic experiments with amino acid precursors. J. Am. Chem. Soc. 105:913-918. [Google Scholar]
- 46.Townsend, C. A., A. M. Brown, and L. T. Nguyen. 1983. Nocardicin A: stereochemical and biomimetic studies of monocyclic beta-lactam formation. J. Am. Chem. Soc. 105:919-927. [Google Scholar]
- 47.Townsend, C. A., and B. A. Wilson. 1988. The role of nocardicin G in nocardicin A biosynthesis. J. Am. Chem. Soc. 110:3320-3321. [Google Scholar]
- 48.Vara, J., M. Lewandowska-Skarbek, Y. G. Wang, S. Donadio, and C. R. Hutchinson. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wittstock, U., and B. A. Halkier. 2000. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 275:14659-14666. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, H., K. H. Maurer, and C. R. Hutchinson. 1986. Transformation of Streptomyces erythraeus. J. Antibiot. 39:1304-1313. [DOI] [PubMed] [Google Scholar]
- 51.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]