Abstract
In cervical cancer, the epidermal growth factor receptor (EGFR) is overexpressed in 70–90% of the cases and has been associated with poor prognosis. EGFR-based therapy is currently being explored in cervical cancer. We investigated which EGFR ligand is primarily expressed in cervical cancer and which cell type functions as the major source of this ligand. We hypothesized that macrophages are the main source of EGFR ligands and that a paracrine loop between tumor cells and macrophages is responsible for ligand expression. mRNA expression analysis was performed on 32 cervical cancer cases to determine the expression of the EGFR ligands amphiregulin, β-cellulin, epidermal growth factor (EGF), epiregulin, heparinbinding EGF-like growth factor (HB-EGF) and transforming growth factor α (TGFα). Subsequently, protein expression was determined immunohistochemically on 36 additional cases. To assess whether macrophages are the major source of EGFR ligands, immunohistochemical double staining was performed on four representative tissue slides. Expression of the chemokines granulocyte-macrophage colony-stimulating factor (GM-CSF) and C-C motif ligand 2 (CCL2) was determined by mRNA in situ hybridization. Of the known EGFR ligands, HB-EGF had the highest mRNA expression and HB-EGF and EGFR protein expression were highly correlated. Tumor specimens with high EGFR expression showed higher numbers of macrophages, and higher expression of GM-CSF and CCL2, but only a small subset (9%) of macrophages was found to be HB-EGF-positive. Strikingly, 78% of cervical cancer specimens were found to express HB-EGF. Standardized assessment of staining intensity, using spectral imaging analysis, showed that HB-EGF expression was higher in the tumor compartment than in the stromal compartment. These results suggest that HB-EGF is an important EGFR ligand in cervical cancer and that cervical cancer cells are the predominant source of HB-EGF. Therefore, we propose an autocrine EGFR stimulation model in cervical carcinomas.
Keywords: autocrine expression, cervical cancer, epidermal growth factor receptor, granulocyte-macrophage colony-stimulating factor, heparin-binding EGF-like growth factor, tumor-associated macrophages
Introduction
Cervical cancer is the fourth most common type of cancer among women worldwide, with 527,600 estimated new cases in 2012 (1). Infection with the human papillomavirus (HPV) has been established to be a necessary cause of invasive cervical cancer (2–4). The lifetime risk of acquiring an HPV infection is ~80% among sexually active women, but most women clear the virus uneventfully (5). When infection with HPV persists, this can lead to malignant cellular changes ranging from cervical intraepithelial neoplasia (CIN)1, CIN2 and CIN3 to invasive cervical cancer. Persistent HPV infections have been associated with chronic inflammation, with large numbers of inflammatory cells often surrounding cervical cancer cells (6). Macrophages are a major component of cancer-related inflammation, and they are roughly divided into two main subgroups: the classically activated type 1 macrophages (M1), which are pro-inflammatory (enhanced expression and release of TNF-α, IL-6, IL-12), and the alternative activated type 2 macrophages (M2), which show anti-inflammatory properties (enhanced expression and release of TGF-β1, IL-10, prostaglandin E2) (6–11). Tumors mediate the accumulation and differentiation of macrophages to sustain tumor growth, and increased numbers of tumor-associated macrophages (TAM) are associated with disease progression and poor prognosis in cervical cancer (12–14).
In cervical cancer, high expression of the epidermal growth factor receptor (EGFR) is associated with tumor development, as EGFR expression increases with increasing malignant stage and subsequent EGFR activation leads to cell growth, differentiation, resistance to apoptosis, cell cycle progression and angiogenesis. EGFR-based therapy has established efficacy in selected patients with head and neck squamous cell carcinoma, colorectal carcinoma and non-small cell lung carcinoma (15–17). It is currently being explored as a therapeutic target in cervical cancer, as EGFR expression is overexpressed in 70–90% of the cases and has been associated with poor prognosis (17–22).
Cell line studies and mouse models show that TAM aid tumor progression through a paracrine loop with EGFR. EGFR activation regulates the expression of granulocyte-macrophage colony-stimulating factor (GM-CSF/CFS2) by tumor cells, thus attracting macrophages to EGFR-expressing tumors where these macrophages express and release EGFR ligands, leading to EGFR activation and subsequent tumor cell proliferation (23–29). Although ~70% of cervical cancers show increased EGFR expression, and high EGFR expression is associated with decreased disease-specific survival, this paracrine loop has never been investigated in human cervical cancer (18,30–32).
We hypothesized that macrophages are the main source of EGFR ligands and that a paracrine loop between tumor cells and macrophages is responsible for ligand expression. First, we investigated whether macrophages are the predominant source of EGFR ligands in human cervical cancer. Therefore, we first assessed which of the known EGFR ligands was present in cervical cancer specimens and which ligand was associated with EGFR expression. Subsequently, we tested whether this ligand was predominantly expressed by TAM. Finally we determined whether an association could be found between EGFR expression by cervical cancer cells and the number of cells expressing the EGFR ligand.
Materials and methods
Tissue samples
All tissue samples were collected from the archives of the Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands. Only material from cervical cancer patients who underwent radical hysterectomy with lymphadenectomy between 1985 and 1995, had not received radiotherapy or chemotherapy prior to surgery, had a histopathological diagnosis of squamous cell carcinoma, adenosquamous carcinoma or adenocarcinoma and had enough primary tumor material available was used. For mRNA expression analysis of amphiregulin (AREG), β-cellulin, epidermal growth factor (EGF), epiregulin, heparin-binding EGF-like growth factor (HB-EGF), transforming growth factor a (TGFα) and EGFR, 32 frozen cervical cancer tissue samples were collected. For immunohistochemistry and mRNA in situ hybridization, 36 formalin-fixed, paraffin-embedded cervical cancer tissue samples were collected. The group of 32 frozen cervical cancer specimens did not overlap with the group of 36 formalin-fixed, paraffin-embedded cervical cancer specimens. Tissue samples were used according to the guidelines of the Ethical Committee of the Leiden University Medical Center.
mRNA expression analysis
mRNA extraction and analysis of microarray expression data were performed as previously described (33). In short, mRNA was obtained from 32 frozen cervical squamous cell carcinoma tissue samples, using TRIzol (Life Technologies, Carlsbad, CA, USA). mRNA was subsequently purified, after which cDNA was synthesized and transcribed into cRNA using the Illumina Totalprep RNA amplification kit following the manufacturer's instructions (Ambion, Austin, Tx, USA). Labeled cRNA was hybridized to Illumina Sentrix-human 6 expression bead-chips. Gene expression levels were quantified using beadstudio gene expression module 2.1, and data were subsequently normalized using the VSN method (34).
Immunohistochemistry
Four-micrometre tissue sections were deparaffinized, rehydrated, and endogenous peroxidase was blocked with 0.3% hydrogen peroxide (H2O2) for 20 min. For HB-EGF, AREG and TGFα, antigen retrieval was performed in 0.01 M citrate buffer (pH 6.0, 12 min). Subsequently, slides were incubated overnight with polyclonal goat anti-human HB-EGF (1:200 at 4°C, af-259-na, R&D Systems Europe Ltd., Abingdon, UK), polyclonal goat anti-human AREG (1:100, af-262, R&D Systems) and mouse monoclonal anti-human TGFα (1:50, clone P/T1, Abcam, Cambridge, UK) diluted in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). After washing with PBS, tissue sections were incubated with a goat probe and anti-goat horseradish peroxidase (HRP) according to the manufacturer's instructions (Goat HRP-polymer kit, GHP516, Biocare Medical, Concord, CA, USA) for AREG and HB-EGF. Tissue sections were incubated with anti-TGFα with BrightVision-Poly/HRP (Immunologic, Duiven, The Netherlands). Immunoreactions were visualized using a 3.3′-diaminobenzidine-tetrahydrochloryde (DAB)+ chromogen (Dako, Heverlee, Belgium), and counterstained with hematoxylin. The results were scored for tumor compartment staining intensity [negative (0), weak (1), moderate (2) or strong (3)] and for the presence of ligand positive cells in the stroma (positive or negative).
EGFR protein expression was determined as previously described (18). The intensity of EGFR membrane staining was scored as negative (0), weak (1), moderate (2) or strong (3). Strong immunoreactivity was defined as complete membrane staining, producing a thick outline of the cell. Presence of TAM was determined though immunohistochemical staining of CD68 as previously described (25). The total number of TAM in the tumor and stromal compartment was quantified by counting the number of positive cells per six, randomly selected, high-power fields of view.
For immunohistochemical double staining for CD68 and HB-EGF, four representative tissue slides were deparaffinized, rehydrated, and endogenous peroxidase was blocked with 0.3% H2O2 for 20 min. Antigen retrieval was performed with 0.01 M citrate buffer, and slides were incubated overnight with anti-HB-EGF and mouse monoclonal anti-human CD68 (1:50, 514H12, AbD Serotec, Oxford, Uk) diluted in Tris(hydroxymethyl)aminomethane buffered saline (TBS) containing 1% BSA. After washing with TBS, tissue sections were first incubated with a goat probe, subsequent anti-goat HRP and DAB+ to visualize HB-EGF. Then, to visualize CD68, tissue slides were incubated with an alkaline phosphatase (AP)-labeled secondary antibody (1:200, rabbit anti-mouse IgG2a-AP, Southern Biotech, Uithoorn, The Netherlands) and subsequent PermaBlue (Diagnostic Biosystems, Uithoorn, The Netherlands) according to the manufacturer's instructions. No counterstaining was applied. For corresponding single staining of CD68 and HB-EGF the same protocol was used, but only one of the two primary antibodies was applied.
Analysis of immunohistochemically stained slides
The number of CD68 and HB-EGF double positive cells was assessed using spectral imaging (35). In short, five representative spectral images, containing both CD68 and HB-EGF-positive cells, were taken for each tissue specimen with a Leica DM4000 B microscope, equipped with a Nuance FX Multispectral Imaging System. The spectral libraries were created from PermaBlue and DAB single staining, after which spectral images from double stained tissue slides were acquired at the same wavelengths. The obtained spectral images were analyzed using Nuance software version 2.10, allowing unmixing into monochrome PermaBlue and DAB images. Subsequently, a simulated fluorescence composite image was created, after which the appropriate threshold for analysis was determined and the percentage of pixel-based co-localization was calculated for each image.
To measure the HB-EGF expression in the epithelial and stromal compartment, the single stained tissue slides were scanned, using the Pannoramic 250 Flash digital slide scanner (3DHistech), after which five representative images, containing both tumor and HB-EGF-positive stromal cells, were taken from each digitalized slide. Color deconvolution was applied to assess the staining intensity of HB-EGF (DAB) only and not the counterstaining (hematoxylin). The average staining intensity [gray scale range: 0 (black) – 255 (white)] was assessed in a standardized area size (42×42 μm) in three representative parts of the tumor. All measured staining intensities were subsequently inverted as follows for all measurements: 255 - measured staining intensity. Since HB-EGF expression was homogeneous in the tumor compartment (all tumor cells expressed HB-EGF equally), an average of three measured areas was used as a standardized measure to determine the relative HB-EGF amount in the tumor compartment. In the stromal compartment, single cells were positive for HB-EGF. Therefore, first, the staining intensity was determined for three separate DAB-positive cells in the stroma, after which the average of these three measurements was calculated. Then, the percentage of positive cells in the stroma was determined for each image, using the same threshold settings for all analyses. The average signal intensity was multiplied by the percentage of positive cells, to obtain a standardized measure for the relative HB-EGF amount in the stroma. One image was analyzed per slide, as the measurements proved to be consistent. The analyses were performed using ImageJ (version 1.45s, National Institutes of Health, MD, USA, freely available on http://imagej.nih.gov/ij).
mRNA in situ hybridization
Expression of the chemokines GM-CSF and CCL2 was determined by mRNA in situ hybridization as previously described and scored for staining intensity [negative (0), weak (1), moderate (2) and strong (3)] as well as percentage of tumor cells positive for chemokine expression [1–5% (1), 6–25% (2), 26–50% (3), 51–75% (4) and 76–100% (5)]. The sum of the staining intensity score and the percentage of positive cells resulted in an overall score for mRNA expression by tumor cells of either 0 or 2–8, which was then dichotomized into low (0–5) and high expression (6–8) (24,25).
Statistical analysis
Statistical analyses were performed using the SPSS program (Version 17.0 for Windows; SPSS Inc. Chicago, Ill, USA). Significance tests were two-sided and statistical significance was assumed when P<0.05, corresponding to 95% confidence intervals (CI). For correlation analysis, the Spearman's correlation coefficient (ϱ, rho) was determined using ungrouped mRNA or protein expression scores. For crosstab analyses, odds ratios (OR) were calculated and the Fisher's exact test was used for calculation of P-values. In case of empty cells, 0.5 was added to each cell for OR calculation.
Results
Expression of HB-EGF in cervical cancer
To assess which EGFR ligands are expressed in cervical cancer specimens, a genome-wide mRNA expression array of 32 cervical cancer specimens was analyzed for mRNA expression of EGFR and EGFR ligands. Median normalized mRNA expression was found to be substantial for AREG, HB-EGF, TGFα and EGFR, while expression of EGF, β-cellulin and epiregulin was low or absent (Table I). To assess protein expression of HB-EGF, AREG and TGFα in cervical cancer and to determine the primary site of ligand expression, immunohistochemistry was performed on 36 specimens. Expression of all three ligands was observed in both the tumor and stromal compartment (Table II). Representative examples of positive tumor compartment staining are shown in Fig. 1. The correlation between EGFR ligand expression and EGFR expression was determined, to assess which ligand is most likely the primary EGFR ligand in cervical cancer and whether this ligand was primarily expressed in the epithelial or stromal compartment (Table III). Both stromal compartment (ϱ=0.58, P<0.001) and tumor compartment HB-EGF expression (ϱ=0.33, P=0.049) were correlated to EGFR expression. No correlations were observed between EGFR and AREG or TGFα.
Table I.
Epidermal growth factor receptor (EGFR) ligand and EGFR mRNA expression in cervical cancer.
| mRNA expressiona
|
||
|---|---|---|
| Median | (25th–75th percentile) | |
| AREG | 355 | 149–870 |
| BTC | 15 | 0–43 |
| EGF | −6 | −14–3 |
| EREG | 15 | 7–27 |
| HB-EGF | 233 | 145–461 |
| TGFα | 145 | 99–196 |
| EGFR | 63 | 37–94 |
Normalized mRNA expression was previously determined using expression array analysis of 32 frozen cervical cancer specimens, as described in Materials and methods. AREG, amphiregulin; BTC, β-cellulin; EGF, epidermal growth factor; EREG, epiregulin; HB-EGF, heparin-binding EGF-like growth factor; TGFα, transforming growth factor a; EGFR, epidermal growth factor receptor.
Table II.
Tumor and stromal protein expression of HB-EGF, AREG and TGFα in cervical cancer patients.
| Na (%) | |
|---|---|
| AREG | |
| Tumor | |
| Negative | – |
| Weak | 13 (36) |
| Moderate | 11 (31) |
| Strong | 12 (33) |
| Stroma | |
| Negative | 9 (25) |
| Positive | 27 (75) |
| HB-EGF | |
| Tumor | |
| Negative | – |
| Weak | 8 (22) |
| Moderate | 18 (50) |
| Strong | 10 (28) |
| Stroma | |
| Negative | 12 (33) |
| Positive | 24 (67) |
| TGFα | |
| Tumor | |
| Negative | – |
| Weak | 19 (53) |
| Moderate | 10 (28) |
| Strong | 7 (19) |
| Stroma | |
| Negative | 10 (28) |
| Positive | 26 (72)\ |
Total number of 36 formalin-fixed, paraffin-embedded cervical cancer tissue samples. AREG, amphiregulin; HB-EGF, heparinbinding EGF-like growth factor; TGFα, transforming growth factor-α.
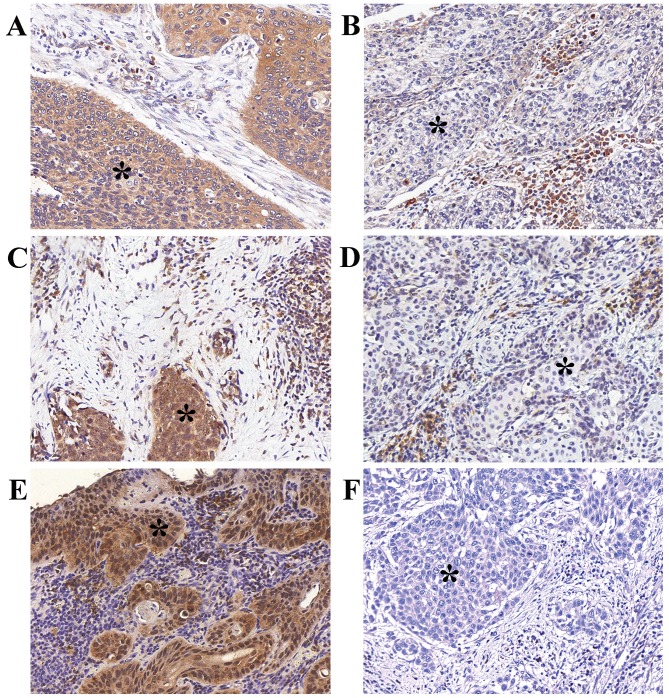
Figure 1.
Representative examples of heparin-binding EGF-like growth factor [(HB-EGF); (A) strong, (B) weak], amphiregulin [(AREG)/0; (C) strong, (D) weak] and transforming growth factor-α staining [(TGFα); (E) strong, (F) weak)] immunohistochemical staining in the epithelial compartment (asterisk) of squamous cell carcinoma of the cervix.
Table III.
Correlations between protein expression of epidermal growth factor receptor (EGFR), EGFR ligands and tumor associated macrophages.
| EGFR expression
|
||
|---|---|---|
| ϱ | P-value | |
| AREG | ||
| Tumor | 0.11 | 0.521 |
| Stromal | 0.15 | 0.392 |
| HB-EGF | ||
| Tumor | 0.33 | 0.049 |
| Stromal | 0.58 | <0.001 |
| TGFα | ||
| Tumor | −0.06 | 0.751 |
| Stromal | −0.02 | 0.927 |
| TAM | ||
| Tumor | 0.43 | 0.009 |
| Stromal | 0.56 | <0.001 |
ϱ, Spearman's correlation coefficient. TAM, tumor associated macrophages; EGFR, epidermal growth factor receptor; AREG, amphiregulin; HB-EGF, heparin-binding EGF-like growth factor; TGFα, transforming growth factor-α; TAM, tumor associated macrophages (CD68-positive cells).
Association between intensity of EGFR expression, number of TAM and expression of chemotactic factors by tumor cells
Next, we assessed the correlation between EGFR expression and the number of CD68-positive TAM (cluster of differentiation 68, membrane glycoprotein that binds low density lipoprotein, a marker predominantly expressed by monocytes/macrophages). High EGFR expression was correlated to a high number of stromal TAM (ϱ=0.56, P<0.001) and a high number of TAM in the tumor compartment (ϱ=0.43, P=0.009, Table III). To assess whether EGFR expression was associated with macrophage recruitment, the association between CD68+ TAM, EGFR, M-CSF, GM-CSF and CCL2 expression by cervical cancer cells was determined. First, the Spearman's correlation between mRNA expression of CD68 and M-CSF, GM-CSF, CCL2 and their receptors M-CSFR, GM-CFSR and CCR was determined. CD68 was correlated with M-CSFR (P=0.003) and CCR2 (P=0.018). However, EGFR was not associated with M-CSF, GM-CSF, CCL2, M-CSFR, GM-CFSR or CCR. Next, GM-CSF and CCL2 mRNA expression, assessed through mRNA in situ hybridization was divided into low expression (combined intensity and percentage scores 0–5) and high expression (combined intensity and percentage scores 6–8). EGFR expression was divided into low (0–1) and high (2–3) intensity scores. High EGFR expression was associated with high GM-CSF and CCL2 expression (OR, 11; P=0.039 and OR, 16; P=0.006, respectively).
Expression of HB-EGF in tumor-associated macrophages
We further investigated whether TAM could be an important source of HB-EGF in cervical cancer. Therefore, the correlation between HB-EGF expression and the number of TAM was determined. Stromal HB-EGF expression was positively correlated to the number of stromal TAM (ϱ=0.34, P=0.044), but no correlation was observed between tumor HB-EGF expression and the number of TAM in the tumor compartment (ϱ=0.24, P=0.163). To investigate whether the observed HB-EGF-positive cells in the stroma were indeed macrophages, immunohistochemical double staining was performed for CD68 and HB-EGF. CD68 and HB-EGF-double-positive cells were observed, i.e., the cervical cancer specimens showed macrophages expressing HB-EGF (Fig. 2), but analysis using spectral imaging showed that a subset of 4–14% of all observed macrophages expressed HB-EGF, with a mean of 9%. Furthermore, the majority of HB-EGF-positive cells in the stroma were CD68-negative, most likely fibroblasts or B-cells, suggesting that macrophages are not the predominant stromal source of HB-EGF in cervical cancer.
Figure 2.
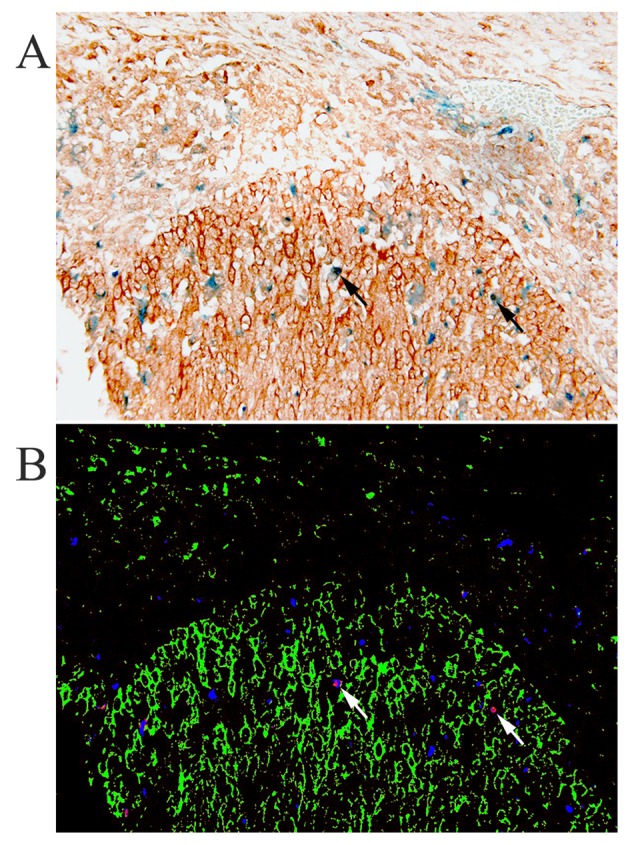
Immunohistochemical double staining of heparin-binding EGF-like growth factor (HB-EGF, DAB) and macrophages (CD68, PermaBlue), without nuclear counterstaining (A), and the simulated fluorescence composite image obtained through spectral imaging (B), showing not only macrophages (arrow), but also other cells in the stroma and tumor cells strongly expressing HB-EGF in cervical cancer.
HB-EGF is expressed on tumor cells
Immunohistochemical staining showed high HB-EGF expression in 78% of the cervical tumor specimens, suggesting that HB-EGF is utilized in an autocrine manner. Fig. 3 shows a representative example of HB-EGF staining and EGFR staining within the same tumor specimen. To estimate the amount of HB-EGF in the tumor and stromal compartment, we measured the HB-EGF staining intensity in the cervical cancer samples, and adjusted the HB-EGF staining intensity for the number of HB-EGF-positive cells in the stroma and the epithelial compartment (relative HB-EGF amount). The average staining intensities as determined using ImageJ corresponded to the previously appointed immunohistochemical (weak, moderate or strong) intensity scores (data not shown). Subsequently, the relative HB-EGF expression in the tumor and stromal compartment was determined in each tissue slide. All individual relative HB-EGF amount ratios (epithelial cancer cells/stromal cells) were >1, except for 2 cases (N=36), suggesting that even in cases with weak HB-EGF expression in the tumor compartment, the tumor compartment showed higher expression of HB-EGF than the stromal compartment (Fig. 4).
Figure 3.
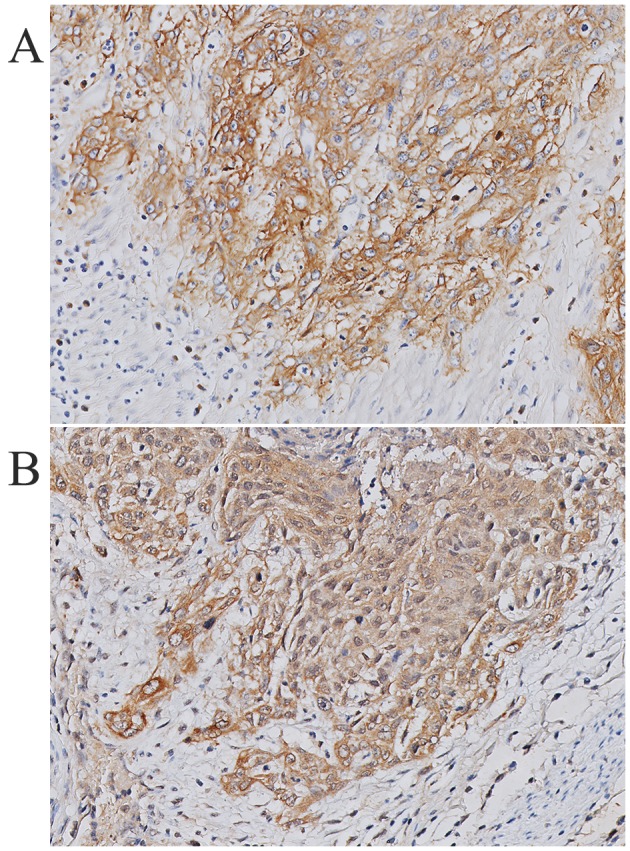
Representative example of epidermal growth factor receptor (EGFR) (A) and heparin-binding EGF-like growth factor (HB-EGF) (B) immunohistochemical staining within the same squamous cell carcinoma tumor specimen, showing membranous staining of EGFR and cytoplasmic staining of HB-EGF.
Figure 4.
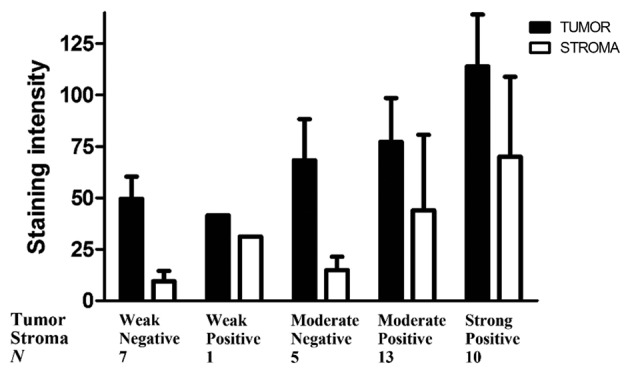
Average staining intensities for heparin-binding EGF-like growth factor (HB-EGF) for each subgroup in the tumor and stromal compartment.
Discussion
In this study, we investigated which EGFR ligands are expressed in cervical cancer and whether macrophages are the predominant source of these EGFR ligands. The results obtained suggest HB-EGF as the predominant EGFR ligand. Although EGFR expression is associated with macrophage counts and the expression of CCL2 and GM-CSF, and macrophage numbers are associated with the expression of M-CSFR and CCR2, macrophages do not appear to be the predominant source of HB-EGF since the relative amount of HB-EGF measured in the epithelial compartment was on average four times higher than the relative amount of HB-EGF in the stromal compartment. These results suggest that, in cervical cancer, the tumor cells are the major source of HB-EGF.
mRNA expression analysis showed that three EGFR ligands were expressed in cervical cancer tissue, namely HB-EGF, AREG and TGFα. In previous studies in pancreatic cancer, where mRNA transcription of several EGFR ligands was observed, only HB-EGF was shown to be of significance at protein level (36). To substantiate the findings on mRNA level in our study, we performed immunohistochemical staining on an independent group of cervical cancer patients, to determine protein expression of HB-EGF, AREG and TGFα. All three ligands were strongly expressed both in tumor stroma and in the epithelial compartment. However, only HB-EGF expression was associated with EGFR expression on the tumor cell membrane, indicating HB-EGF as the primary ligand for EGFR in cervical cancer.
Pollard (32) and Vlaicu et al (37), have shown through cell line research and mouse models, that in breast cancer, macrophage production of EGF induces EGFR activation on tumor cells, which in turn leads to M-CSF (CSF-1) production, consequently attracting more macrophages. M-CSF and its receptor have been shown to be upregulated in cervical cancer both on the mRNA and protein level, when compared to normal cervical tissue (38). In turn, the inhibition of M-CSF prevents tumor progression in a cervical cancer mouse model (39). In cervical cancer HeLa cells and macrophages, an analogous paracrine loop has been described with GM-CSF and HB-EGF expression (26,27). In addition to cervical cancer cell lines, HB-EGF, AREG and TGFα expression and EGFR activation have previously been shown to induce GM-CSF release in several human cell lines, such as airway epithelial cells, keratinocytes and colon adenocarcinoma cells (DLD-1) (27,28,40). The present study analyzed whether this interdependent paracrine signaling between tumor cells and macrophages through HB-EGF and GM-CSF was present in human cervical cancer. We observed that EGFR expression was associated with high numbers of TAM in the tumor and stromal compartment, and with high GM-CSF and CCL2 expression. Previously, increased EGFR expression has been shown to downregulate CCL2, while EGFR inhibition has been shown to upregulate CCL2 expression in vitro in head and neck squamous cell carcinoma and primary cultures of normal human keratinocytes (41,42). Only in osteoblasts and vascular smooth muscle cells has EGFR activation been shown to increase CCL2 expression (43,44). Our results are concordant with the findings that EGFR expression is associated with GM-CSF and CCL2 expression by tumor cells, thus attracting macrophages to the tumor site. However, although EGFR expression was associated with macrophage recruitment, double staining for CD68 and HB-EGF showed that only a limited number (9%) of the macrophages were HB-EGF-positive. Furthermore, only a small proportion of HB-EGF-positive cells in the stroma was CD68-positive, as fibroblasts and B-cells also appeared to be HB-EGF-positive, suggesting that macrophages are not the predominant source of HB-EGF in cervical cancer stroma.
It has been suggested that HB-EGF is expressed by cervical cancer-associated stromal fibroblasts, thus promoting cancer cell proliferation in a paracrine manner (45). However, although our study showed that tumor stroma was indeed positive for HB-EGF, the largest relative amount of HB-EGF was detected in the cytoplasm of cervical cancer cells, as the relative amount of HB-EGF in the epithelial compartment exceeded the relative amount of HB-EGF in the stromal compartment by a factor of four. Thus, we conclude that HB-EGF is expressed in an autocrine manner in cervical cancer. In vitro experiments with head and neck squamous cell carcinoma cells show that this HB-EGF autocrine loop is associated with invasive processes through the EGFR-Src-cortactin cascade (46).
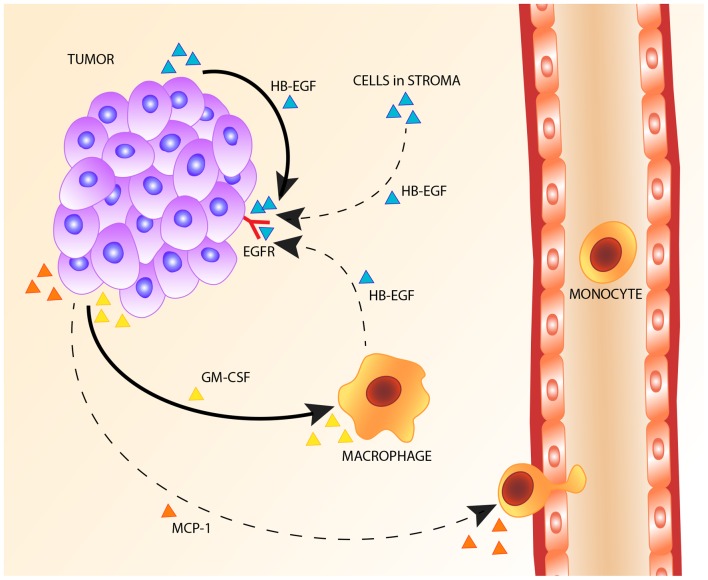
Based on our results, we propose an autocrine EGFR stimulation model in cervical carcinoma, with cervical cancer cells being the predominant source of HB-EGF. Cervical cancer cells are shown to express GM-CSF to attract macrophages to the tumor environment. Although macrophages and other stromal cells express the primary EGFR ligand HB-EGF, they do not appear to be the major source of HB-EGF, as the relative amount of HB-EGF measured in the tumor cell compartment was on average four times higher than the relative amount of HB-EGF in the stromal compartment. The proposed autocrine mechanism of HB-EGF expression is shown in Fig. 5.
Figure 5.
Proposed mechanism of heparin-binding EGF-like growth factor (HB-EGF) expression and macrophage recruitment in cervical cancer.
Acknowledgments
We gratefully acknowledge Natalja T. ter Haar for her technical support and Patty M. Jansen for her helpful suggestions concerning the HB-EGF-positive cells in the stroma.
Abbreviations
- AREG
amphiregulin
- CCL2
C-C motif ligand 2
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- HB-EGF
heparin-binding EGF-like growth factor
- HPV
human papillomavirus
- TAM
tumor-associated macrophages
- TGFα
transforming growth factor α
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 4.Joura EA, Ault KA, Bosch FX, Brown D, Cuzick J, Ferris D, Garland SM, Giuliano AR, Hernandez-Avila M, Huh W, et al. Attribution of 12 high-risk human papillomavirus genotypes to infection and cervical disease. Cancer Epidemiol Biomarkers Prev. 2014;23:1997–2008. doi: 10.1158/1055-9965.EPI-14-0410. [DOI] [PubMed] [Google Scholar]
- 5.Koutsky L. Epidemiology of genital human papillomavirus infection. Am J Med. 1997;102A:3–8. doi: 10.1016/S0002-9343(97)00177-0. [DOI] [PubMed] [Google Scholar]
- 6.Utrera-Barillas D, Castro-Manrreza M, Castellanos E, Gutiérrez-Rodríguez M, Arciniega-Ruíz de Esparza O, García-Cebada J, Velazquez JR, Flores-Reséndiz D, Hernández-Hernández D, Benítez-Bribiesca L. The role of macrophages and mast cells in lymphangiogenesis and angiogenesis in cervical carcinogenesis. Exp Mol Pathol. 2010;89:190–196. doi: 10.1016/j.yexmp.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Boccardo E, Lepique AP, Villa LL. The role of inflammation in HPV carcinogenesis. Carcinogenesis. 2010;31:1905–1912. doi: 10.1093/carcin/bgq176. [DOI] [PubMed] [Google Scholar]
- 8.Lepique AP, Daghastanli KR, Cuccovia IM, Villa LL. HPV 16 tumor associated macrophages suppress antitumor T cell responses. Clin Cancer Res. 2009;15:4391–4400. doi: 10.1158/1078-0432.CCR-09-0489. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani A, Germano G, Marchesi F, Locatelli M, Biswas SK. Cancer-promoting tumor-associated macrophages: New vistas and open questions. Eur J Immunol. 2011;41:2522–2525. doi: 10.1002/eji.201141894. [DOI] [PubMed] [Google Scholar]
- 10.Mantovani A, Sica A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 11.de Vos van Steenwijk PJ, Ramwadhdoebe TH, Goedemans R, Doorduijn EM, van Ham JJ, Gorter A, van Hall T, Kuijjer ML, van Poelgeest MI, van der Burg SH, et al. Tumor-infiltrating CD14-positive myeloid cells and CD8-positive T-cells prolong survival in patients with cervical carcinoma. Int J Cancer. 2013;133:2884–2894. doi: 10.1002/ijc.28309. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto J, Sakaguchi H, Aoki I, Tamaya T. Clinical implications of expression of interleukin 8 related to angiogenesis in uterine cervical cancers. Cancer Res. 2000;60:2632–2635. [PubMed] [Google Scholar]
- 13.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 14.Petrillo M, Zannoni GF, Martinelli E, Pedone Anchora L, Ferrandina G, Tropeano G, Fagotti A, Scambia G. Polarisation of tumor-associated macrophages toward M2 phenotype correlates with poor response to chemoradiation and reduced Survival in patients with locally advanced cervical cancer. PLoS One. 2015;10:e0136654. doi: 10.1371/journal.pone.0136654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong W, Xiao Y, Wei Z, Yuan Y, Qiu M, Sun C, Zeng X, Liang X, Feng M, Chen Q. Toward the use of precision medicine for the treatment of head and neck squamous cell carcinoma. Oncotarget. 2017;8:2141–2152. doi: 10.18632/oncotarget.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 17.Greenhalgh J, Dwan K, Boland A, Bates V, Vecchio F, Dundar Y, Jain P, Green JA. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Database Syst Rev. 2016;5:CD010383. doi: 10.1002/14651858.CD010383.pub2. [DOI] [PubMed] [Google Scholar]
- 18.Schrevel M, Gorter A, Kolkman-Uljee SM, Trimbos JB, Fleuren GJ, Jordanova ES. Molecular mechanisms of epidermal growth factor receptor overexpression in patients with cervical cancer. Mod Pathol. 2011;24:720–728. doi: 10.1038/modpathol.2010.239. [DOI] [PubMed] [Google Scholar]
- 19.Narayanan R, Kim HN, Narayanan NK, Nargi D, Narayanan B. Epidermal growth factor-stimulated human cervical cancer cell growth is associated with EGFR and cyclin D1 activation, independent of COX-2 expression levels. Int J Oncol. 2012;40:13–20. doi: 10.3892/ijo.2011.1211. [DOI] [PubMed] [Google Scholar]
- 20.Reyes HD, Thiel KW, Carlson MJ, Meng X, Yang S, Stephan JM, Leslie KK. Comprehensive profiling of EGFR/HER receptors for personalized treatment of gynecologic cancers. Mol Diagn Ther. 2014;18:137–151. doi: 10.1007/s40291-013-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassouf E, Tabchi S, Tehfe M. Anti-EGFR therapy for metastatic colorectal cancer in the era of extended RAS gene mutational analysis. BioDrugs. 2016;30:95–104. doi: 10.1007/s40259-016-0166-5. [DOI] [PubMed] [Google Scholar]
- 22.Sacco AG, Worden FP. Molecularly targeted therapy for the treatment of head and neck cancer: A review of the ErbB family inhibitors. Onco Targets Ther. 2016;9:1927–1943. doi: 10.2147/OTT.S93720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leek RD, Hunt NC, Landers RJ, Lewis CE, Royds JA, Harris AL. Macrophage infiltration is associated with VEGF and EGFR expression in breast cancer. J Pathol. 2000;190:430–436. doi: 10.1002/(SICI)1096-9896(200003)190:4<430::AID-PATH538>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Zijlmans HJ, Fleuren GJ, Baelde HJ, Eilers PH, Kenter GG, Gorter A. The absence of CCL2 expression in cervical carcinoma is associated with increased survival and loss of heterozygosity at 17q11.2. J Pathol. 2006;208:507–517. doi: 10.1002/path.1918. [DOI] [PubMed] [Google Scholar]
- 25.Zijlmans HJ, Fleuren GJ, Baelde HJ, Eilers PH, Kenter GG, Gorter A. Role of tumor-derived proinflammatory cytokines GM-CSF, TNF-alpha, and IL-12 in the migration and differentiation of antigen-presenting cells in cervical carcinoma. Cancer. 2007;109:556–565. doi: 10.1002/cncr.22428. [DOI] [PubMed] [Google Scholar]
- 26.Edwards JP, Zhang X, Mosser DM. The expression of heparin-binding epidermal growth factor-like growth factor by regulatory macrophages. J Immunol. 2009;182:1929–1939. doi: 10.4049/jimmunol.0802703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigo A, Gottardi M, Zamo A, Mauri P, Bonifacio M, Krampera M, Damiani E, Pizzolo G, Vinante F. Macrophages may promote cancer growth via a GM-CSF/HB-EGF paracrine loop that is enhanced by CXCL12. Mol Cancer. 2010;9:273. doi: 10.1186/1476-4598-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascia F, Cataisson C, Lee TC, Threadgill D, Mariani V, Amerio P, Chandrasekhara C, Souto Adeva G, Girolomoni G, Yuspa SH, et al. EGFR regulates the expression of keratinocyte-derived granulocyte/macrophage colony-stimulating factor in vitro and in vivo. J Invest Dermatol. 2010;130:682–693. doi: 10.1038/jid.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu YY, Nergard JC, Barnette NK, Wang YL, Chai KX, Chen LM. Proteasome inhibition augments cigarette smoke-induced GM-CSF expression in trophoblast cells via the epidermal growth factor receptor. PLoS One. 2012;7:e43042. doi: 10.1371/journal.pone.0043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 31.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 32.Pollard JW. Macrophages define the invasive microenvironment in breast cancer. J Leukoc Biol. 2008;84:623–630. doi: 10.1189/jlb.1107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloth JN, Gorter A, Fleuren GJ, Oosting J, Uljee S, ter Haar N, Dreef EJ, Kenter GG, Jordanova ES. Elevated expression of SerpinA1 and SerpinA3 in HLA-positive cervical carcinoma. J Pathol. 2008;215:222–230. doi: 10.1002/path.2347. [DOI] [PubMed] [Google Scholar]
- 34.Huber W, Gentleman R. matchprobes: A Bioconductor package for the sequence-matching of microarray probe elements. Bioinformatics. 2004;20:1651–1652. doi: 10.1093/bioinformatics/bth133. [DOI] [PubMed] [Google Scholar]
- 35.Lehr HA, van der Loos CM, Teeling P, Gown AM. Complete chromogen separation and analysis in double immunohistochemical stains using Photoshop-based image analysis. J Histochem Cytochem. 1999;47:119–126. doi: 10.1177/002215549904700113. [DOI] [PubMed] [Google Scholar]
- 36.Sloss CM, Wang F, Palladino MA, Cusack JC., Jr Activation of EGFR by proteasome inhibition requires HB-EGF in pancreatic cancer cells. Oncogene. 2010;29:3146–3152. doi: 10.1038/onc.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlaicu P, Mertins P, Mayr T, Widschwendter P, Ataseven B, Hogel B, Eiermann W, Knyazev P, Ullrich A. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer. 2013;13:197. doi: 10.1186/1471-2407-13-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirma N, Hammes LS, Liu YG, Nair HB, Valente PT, Kumar S, Flowers LC, Tekmal RR. Elevated expression of the oncogene c-fms and its ligand, the macrophage colony-stimulating factor-1, in cervical cancer and the role of transforming growth factor-beta1 in inducing c-fms expression. Cancer Res. 2007;67:1918–1926. doi: 10.1158/0008-5472.CAN-06-1991. [DOI] [PubMed] [Google Scholar]
- 39.Strachan DC, Ruffell B, Oei Y, Bissell MJ, Coussens LM, Pryer N, Daniel D. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8(+) T cells. OncoImmunology. 2013;2:e26968. doi: 10.4161/onci.26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rumelhard M, Ramgolam K, Hamel R, Marano F, Baeza-Squiban A. Expression and role of EGFR ligands induced in airway cells by M2.5 and its components. Eur Respir J. 2007;30:1064–1073. doi: 10.1183/09031936.00085907. [DOI] [PubMed] [Google Scholar]
- 41.Pastore S, Mascia F, Mariotti F, Dattilo C, Mariani V, Girolomoni G. ERK1/2 regulates epidermal chemokine expression and skin inflammation. J Immunol. 2005;174:5047–5056. doi: 10.4049/jimmunol.174.8.5047. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann TK, Schirlau K, Sonkoly E, Brandau S, Lang S, Pivarcsi A, Balz V, Müller A, Homey B, Boelke E, et al. A novel mechanism for anti-EGFR antibody action involves chemokine-mediated leukocyte infiltration. Int J Cancer. 2009;124:2589–2596. doi: 10.1002/ijc.24269. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Jia X, Xiao G, Kang Y, Partridge NC, Qin L. EGF-like ligands stimulate osteoclastogenesis by regulating expression of osteoclast regulatory factors by osteoblasts: Implications for osteolytic bone metastases. J Biol Chem. 2007;282:26656–26664. doi: 10.1074/jbc.M705064200. [DOI] [PubMed] [Google Scholar]
- 44.Singh NK, Wang D, Kundumani-Sridharan V, Van Quyen D, Niu J, Rao GN. 15-Lipoxygenase-1-enhanced Src-Janus kinase 2-signal transducer and activator of transcription 3 stimulation and monocyte chemoattractant protein-1 expression require redox-sensitive activation of epidermal growth factor receptor in vascular wall remodeling. J Biol Chem. 2011;286:22478–22488. doi: 10.1074/jbc.M111.225060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murata T, Mizushima H, Chinen I, Moribe H, Yagi S, Hoffman RM, Kimura T, Yoshino K, Ueda Y, Enomoto T, et al. HB-EGF and PDGF mediate reciprocal interactions of carcinoma cells with cancer-associated fibroblasts to support progression of uterine cervical cancers. Cancer Res. 2011;71:6633–6642. doi: 10.1158/0008-5472.CAN-11-0034. [DOI] [PubMed] [Google Scholar]
- 46.Hayes KE, Walk EL, Ammer AG, Kelley LC, Martin KH, Weed SA. Ableson kinases negatively regulate invadopodia function and invasion in head and neck squamous cell carcinoma by inhibiting an HB-EGF autocrine loop. Oncogene. 2013;32:4766–4777. doi: 10.1038/onc.2012.513. [DOI] [PMC free article] [PubMed] [Google Scholar]