Abstract
Enterobacterial animal pathogens exhibit aggregative multicellular behavior, which is manifested as pellicles on the culture surface and biofilms at the surface-liquid-air interface. Pellicle formation behavior requires production of extracellular polysaccharide, cellulose, and protein filaments, known as curli. Protein filaments analogous to curli are formed by many protein secretion systems, including the type III secretion system (TTSS). Here, we demonstrate that Erwinia chrysanthemi, which does not carry curli genes, requires the TTSS for pellicle formation. These data support a model where cellulose and generic protein filaments, which consist of either curli or TTSS-secreted proteins, are required for enterobacterial aggregative multicellular behavior. Using this assay, we found that hrpY, which encodes a two-component system response regulator homolog, is required for activity of hrpS, which encodes a σ54-dependent enhancer-binding protein homolog. In turn, hrpS is required for activity of the sigma factor homolog hrpL, which activates genes encoding TTSS structural and secreted proteins. Pellicle formation was temperature dependent and pellicles did not form at 36°C, even though TTSS genes were expressed at this temperature. We found that cellulose is a component of the E. chrysanthemi pellicle but that pellicle formation still occurs in a strain with an insertion in a cellulose synthase subunit homolog. Since the TTSS, but not the cellulose synthase subunit, is required for E. chrysanthemi pellicle formation, this inexpensive assay can be used as a high throughput screen for TTSS mutants or inhibitors.
Erwinia chrysanthemi is an economically important enterobacterial plant pathogen that causes soft rot and wilt diseases on numerous species of plants. Aggregative multicellular behavior, which results in the formation of large cell aggregates on the culture surface known as pellicles, was demonstrated in E. chrysanthemi over 40 years ago (32). In related species, pellicle formation requires cellulose and protein filaments, known as aggregative fimbriae or curli (37, 45, 49, 52). The recently sequenced E. chrysanthemi 3937 (Ech 3937) genome revealed homologs of genes required for cellulose production in related enterobacteria, but no homologs of genes required for curli synthesis (N. Perna, personal communication). Ech 3937 also does not encode csgD, a regulatory protein that controls curli and cellulose production in related species (18). Thus, the regulatory proteins controlling pellicle formation and whether or not there are protein filaments that play an analogous role to curli in formation of E. chrysanthemi pellicles were unknown.
In plant-pathogenic bacteria, the type III secretion system (TTSS) is encoded by hrp (hypersensitive response and pathogenicity) and hrc (hypersensitive response conserved) genes. The TTSS functions as a molecular syringe, injecting virulence proteins into host cells; some of these proteins may interfere with host defense machinery (9, 17, 23). The hrc genes, many of which are homologous to flagellar export apparatus, are highly conserved across genera (2). A common feature of TTSSs is the extracellular pilus structure, a 100- to 200-nm conduit through which the secreted proteins travel across the plant cell wall (9). However, whether this filamentous surface appendage is also associated with multicellular processes, reminiscent of the functions of a variety of other surface structures such as flagella, type I pili, type IV pili, curli, and conjugative pili in biofilm formation (14) has not been explored. In this work, we demonstrate that the TTSS, a protein secretion system required for virulence of many gram-negative pathogens, including E. chrysanthemi (51), is required for E. chrysanthemi pellicle formation.
MATERIALS AND METHODS
Bacterial strains and media.
All bacterial strains were routinely grown in Luria-Bertani (LB) medium at 37°C for DNA isolations and mutagenesis. Hrp-inducing minimal medium (48) was used to determine if the mutant strains were able to grow in minimal medium. The growth medium used to induce aggregative multicellular behavior in E. chrysanthemi, SOBG, consisted of SOB plus 2% glycerol (per liter, 20 g of tryptone, 5 g of yeast extract, 0.5 g of NaCl, 2.4 g of MgSO4, 0.186 g of KCl, and 50 ml of 40% glycerol). SOB, which contains 1.5% glycerol, 2% glycerol plus 1% glucose, 2% glycerol plus 1% polygalacturonic acid or 1% glucose, fructose, sucrose, mannitol, or polygalacturonic acid in place of the glycerol, was also tested. For pellicle assays, bacterial cultures were suspended in SOBG to approximately 106 CFU/ml. Cultures were incubated at room temperature (approximately 25°C) unless otherwise noted in stationary 13- and 20-mm glass tubes or 100-ml beakers for at least 3 days. Swimming and swarming assays were performed as previously described by Rashid and Kornberg (38). Chromobacterium violaceum was used as a reporter strain for acyl-homoserine lactone production, essentially as previously described (10).
Mutant and plasmid construction.
Primers were obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa). To construct plasmids for allelic-exchange mutagenesis of the targeted hrp genes, hrp gene regions were PCR amplified with the primer sets listed in Table 1. The products were cloned into pGEMT-Easy (Promega, Madison, Wis.) or pBluescript (Stratagene, La Jolla, Calif.). For some mutants, antibiotic resistance gene cassettes were inserted into the cloned genes; in other cases, the target genes were deleted and replaced by antibiotic resistance gene cassettes (Table 2). These plasmids were electrotransformed into wild-type or mutant Ech 3937 for allelic-exchange mutagenesis following the methods described by Ried and Collmer (39). All mutations were confirmed by PCR and Southern blot analysis. To construct plasmids to complement the mutants, the promoter and coding regions of the hrp genes were PCR amplified and cloned into various expression vectors (Table 2). Some of the vectors used were inducible expression vectors, but no inducers were added, since the hrp gene promoters were included along with the coding regions in these constructs. The plasmids were electrotransformed into mutant strains. When required, antibiotics were used at the following concentrations (in micrograms per milliliter): ampicillin (100), chloramphenicol (50), spectinomycin (50), streptomycin (50), gentamicin (25), and kanamycin (50). Transformation, restriction endonuclease digestion, and other DNA techniques were basically performed as described in Sambrook et al. (41).
TABLE 1.
Primers used in this studya
| No. | Primer set | Sequence (5′→3′) (Restriction enzyme sites) | Targeted region |
|---|---|---|---|
| 1a | hrpY-D1 | TGATGGCCAGCATTTCGGC | 2.8-kb hrpY-hrpL |
| 1b | hrpL-f1 | AAGGATCCGATGGAGAGTTAATGAAATG (BamHI) | |
| 2a | hrcJ-D1 | ACCAGGGCTTGCCGAAACGC | 3.4-kb hrcJ-hrpY |
| 2b | hrpY-C1 | GGAAGCTTCGCAGAGTACGCTGATGATCGCTGACGATC (HindIII) | |
| 3a | hrpY-D1 | TGATGGCCAGCATTTCGGC | 3.8-kb hrpY-hrpJ |
| 3b | hrpJ-A1 | TCGTTGCAGCGTACGCAGCC | |
| 4a | hrpL-B2 | GCGTACTCTGCGAAGCTTCCTCCACGGTATTGGCAGGGGT (HindIII) | 2.0-kb hrpL-hrcV |
| 4b | hrcV1 | CCGAACGCCTCCACCACGTG | |
| 5a | hrcJ-D1 | ACCAGGGCTTGCCGAAACGC | 3-kb hrcJ-hrpS |
| 5b | hrpS-f1 | GAGCTCGGATCCAGCGTTTAATCGCCTGA (SacI, BamHI) | |
| 6a | hrpY-C1 | GGAAGCTTCGCAGAGTACGCTGATGATCGCTGACGATC(HindIII) | 3-kb hrpY-hrpA |
| 6b | hrpA-r1 | GTCGACTACCTAGAATTCATCAG (EcoRI) | |
| 7a | hrpXY-f1 | GAGCTCGGATCCGCCAGGCTGTTGATGC (SacI, BamHI) | 2.5-kb hrpX-hrpY |
| 7b | hrpXY-r1 | ACGAAGGGAGGACTCTAGAC (XbaI) | |
| 8a | hrpY-FBamHI | TCTGGAGGAGGATCCGCGAATG (BamHI) | 0.7-kb hrpY |
| 8b | hrpY-RHindIII | TACAAGCTTTCATCAGGCGA (HindIII) | |
| 9a | hrpS-f1 | GAGCTCGGATCCAGCGTTTAATCGCCTGA (SacI, BamHI) | 1.45-kb hrpS |
| 9b | hrpS-r1 | ACAGCCCGATAAATCTAGAGCC (XbaI) | |
| 10a | hrpL-f1 | AAGGATCCGATGGAGAGTTAATGAAATG (BamHI) | 0.55-kb hrpL |
| 10b | hrpL-r1 | TTGAATTCTTATGCATCAACGGCCTGGC (EcoRI) | |
| 11a | hrcJ-f1 | TGCACTAGTCAGGATCAAACCC (SpeI) | 0.97-kb hrcJ |
| 11b | hrcJ-r1 | CATCGGCTACACGTAGAATTCTCA (EcoRI) | |
| 12a | hrpA-f1 | ATCGGGCTGTATACTAGTATCACC (SpeI) | 0.3-kb hrpA |
| 12b | hrpA-r1 | GTCGACTACCTAGAATTCATCAG (EcoRI) | |
| 13a | bcsA-f1 | ACCCGAATTCCTATATTCGCCGTCACC (EcoRI) | 3-kb bcsA |
| 13b | bcsA-r12 | CCCACGCACAGGGACAACA | |
| 14a | RT-hrpLf1 | GGGTATTTGGACTTGCCCTGAATC | 223-bp hrpL |
| 14b | RT-hrpLr2 | GCATCCAGCAGCATCATCAACATC | |
| 15a | hrpN1 | GCGGCATGGCGAAAGAGAT | 226-bp hrpN |
| 15b | hrpN2 | GTGTTGCCGGTATCACCC | |
| 16a | RT-rplUf1 | GTTTGACCAGGTTCTGATGGTTGC | 162-bp rplU |
| 16b | RT-rplUr1 | CCAGCCTGCTTACGGTAGTGTTTA |
Primer sets 1 through 13 were used to amplify hrp and hrc genes for construction of plasmids used for allelic exchange mutagenesis and complementation experiments. Primer sets 14 through 16 were used for RT-PCR. Restriction sites in primers are in boldface type.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (Δ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Clontech |
| E. chrysanthemi | ||
| Ech 3937 | Wild-type Saintpaulia (African violet) isolate | 27 |
| AC4150 | EC16 Nxr derivative, Chrysanthemum morifolium isolate | 12 |
| CUCPB5039 | AC4150 TTSS mutant | 5 |
| WPP42 | outD::kan; Kmr 3937 derivative; type II secretion mutant | This work |
| WPP67 | hrpX::aadA; Spr/Smr 3937 derivative | This work |
| WPP92 | hrpY::kan; Kmr 3937 derivative | This work |
| WPP93 | hrpX::aadA hrpY::kan; Spr/Smr Kmr WPP67 derivative | This work |
| WPP90 | hrpS::cat; Cmr 3937 derivative | This work |
| WPP89 | hrpX::aadA hrpS::cat; Spr Cmr WPP67 derivative | This work |
| WPP96 | ΔhrpL(Δ1-185aa)::aadA; Spr/Smr 3937 derivative | This work |
| WPP98 | hrcJ::kan; Kmr 3937 derivative | This work |
| WPP100 | hrpA::cat; Cmr 3937 derivative | This work |
| WPP101 | bcsA::cat; Cmr 3937 derivative | This work |
| WPP102 | ΔhrpS(Δ46-212aa)::aadA; Spr/Smr 3937 derivative | This work |
| WPP119 | hrpX::aadA bcsA::cat; Spr/Smr Cmr WPP67 derivative | This work |
| Plasmids | ||
| pGEMT-Easy | AprlacZ′ cloning vector | Promega |
| pBluescript SK(+/−) | AprlacZ′ cloning vector | Stratagene |
| pQE31 | Apr His6-tagged expression vector | Qiagen |
| pHP45ΩSp/Sm | Apr Spr/Smr broad-host-range cloning vector | 15 |
| pKD3 | Kmr template plasmid carrying kan cassette | 13 |
| pKD4 | Cmr template plasmid carrying cat cassette | 13 |
| pJN105 | Gmr, arabinose inducible expression vector | 34 |
| pCPP50 | Apr pINIII113-A2-based expression vector | 8 |
| p519ngfp | Kmr broad-host-range vector | 31 |
| pRK415 | Tcr broad-host-range low-copy vector | 24 |
| pThrpYLΩSp | Apr Spr/Smr pGEMT-Easy carrying 4.8-kb hrpY-hrpL region containing hrpX::ΩSp insertion at StuI site | This work |
| pThrpYLKm | Apr Kmr pGEMT-Easy carrying 4.4-kb hrpY-hrpL region containing hrpY::Km insertion at blunt-ended BglII site | This work |
| pThrcJY::cat | Apr Kmr pGEMT-Easy carrying 4.4-kb hrcJ-hrpY region containing hrpS::Cm insertion at PstI site | This work |
| pThrpYJΩSp | Apr Spr 5.2-kb hrpL-deleted hrpY-hrpJ region on pGEMT-Easy; complete hrpL coding region was removed with SmaI and HindIII, blunt ended, and ligated to ΩSp | This work |
| pThrcJYKm | Apr Kmr pGEMT-Easy carrying 5-kb hrcJ-hrpY region containing hrcJ::Km insertion at SmaI site | This work |
| pBShrc/pJSCm | Apr Cmr pBluescript SK(+/−) carrying 3-kb hrcJ-hrpS containing hrpA::Cm insertion at blunt-ended SphI site | This work |
| p519nGFPbcsA | Kmr Cmr p519ngfp with bcsA::cat inserted into EcoRI site; cat is inserted into the BamHI site of bcsA | This work |
| pYAΔSΩSp | Apr Spr 2.0-kb hrpS-deleted hrpY-hrpA region on pGEMT-Easy; hrpS was removed using EcoRV and StuI and replaced with an ΩSp cassette | This work |
| p105XY | Gmr 2.5-kb hrpXY with native promoter cloned into BamHI and XbaI sites of pJN105 | This work |
| pQhrpY | Apr 0.7-kb hrpY in BamHI and HindIII sites of pQE31 | This work |
| p105S | Gmr 1.5-kb hrpS with native promoter cloned into BamHI and XbaI sites of pJN105 | This work |
| p50hrpL | Apr 0.55-kb hrpL in SpeI and EcoRI sites of pCPP50 | This work |
| p50hrcJ | Apr 0.97-kb hrcJ in SpeI and EcoRI sites of pCPP50 | This work |
| p415hrpA | Tcr 0.3-kb hrpA cloned into XbaI and EcoRI sites of pRK415 | This work |
| p50bcsA | Apr 3-kb bcsA with native promoter cloned into pCPP50 | This work |
RT-PCR.
Strains were grown at 36 and 25°C in SOBG medium for 2 days until the wild-type pellicle at 25°C had just completely covered the surface of culture. Total RNA was extracted from 1 ml of culture with the RNAqueous kit (Ambion, Inc., Austin, Tex.) and subjected to DNase I treatment with the TURBO DNase kit (Ambion, Inc.) as described in the manufacturer's manual. RNA concentration was measured with the RiboGreen RNA quantitation kit (Molecular Probes, Inc., Eugene, Oreg.). Semiquantitative reverse transcription-PCR (RT-PCR) was performed with One-Step-Ready-to-Go RT-PCR beads (Amersham Biosciences) in a 50-μl reaction mixture containing 50 ng of total RNA, 300 μM primer, 2 U of Taq DNA polymerase, 10 mM Tris-HCl (pH 9.0), 60 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, Moloney murine leukemia virus reverse transcriptase, and RNAguard RNase inhibitor. Primers used are shown in Table 1. rplU was used as an internal control, since it is constitutively expressed under a wide range of conditions (30). RT was performed at 42°C for 30 min, followed by 10 min of reverse transcriptase inactivation at 95°C. The PCR cycling conditions were 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s for a total of 32 cycles and an additional 2-min extension at 72°C. To test for genomic DNA contamination, a negative-control PCR without the RT step was performed by incubation of the rehydrated beads prior to addition of the template and primers at 95°C for 10 min to inactivate the Moloney murine leukemia virus reverse transcriptase prior to the PCR. Equal amounts of RT-PCR products were loaded onto 2% agarose-Tris-borate-EDTA gels and visualized by staining with ethidium bromide. The RT-PCR experiment was repeated three times.
Immunoblot analysis.
Whole-cell lysate and extracellular protein were prepared from the same cultures used to isolate total RNA. Aliquots of the cultures (each, 1.5 ml) were centrifuged to harvest bacterial cells, and the pellets were resuspended in 200 μl of Laemmli buffer (26) and used as the whole-cell lysate. The supernatant was filtered through a 0.2-μm-pore-sized filter to remove the bacterial cells and then concentrated by precipitation in 10% trichloroacetic acid. The proteins were resuspended in 20 μl of Laemmli buffer. Both fractions were subsequently boiled at 100°C for 10 min, separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis, and analyzed by immunoblotting with HrpN-specific polyclonal antiserum (a gift from A. Collmer) with the Immuno-Star AP goat anti-rabbit IgG (immunoglobulin G) detection kit (Bio-Rad Laboratories, Inc., Hercules, Calif.), following the manufacturer's instructions. The immunoblot experiment was repeated twice.
Plant assays.
Plant assays were performed essentially as described by Yang et al. (51). African violet (Saintpaulia ionantha) leaves were infiltrated with a bacterial suspension of 106 CFU/ml. Plants were incubated in a dew chamber at 28°C throughout the course of the assay. Leaf disks were taken from infiltrated leaves and homogenized in sterile water, and the suspension was dilution plated on LB medium plus appropriate antibiotics.
Cellulase assays.
Ech 3937 pellicles were removed from 3-day-old SOBG cultures by pouring the culture into a petri plate. The pellicles were washed twice with 10 ml of sterile water and then placed in 5 ml of 50 mM potassium phosphate (pH 6.0) containing no cellulase, 0.1% cellulase (wt/vol or vol/vol), or 0.1% heat-denatured cellulase. The heat-denatured cellulase was prepared by boiling a 0.1% solution for 1 to 5 min. Several commercial preparations of cellulase were tested, including cellulysin (Calbiochem-Novabiochem Corp., La Jolla, Calif.), cellulase TR (Solvay, Elkhart, Ind.), multifect CS cellulase (Genencor, Rochester, N.Y.), multifect B cellulase (Genencor), multifect cellulase 300 (Bio-cat, Inc., Troy, Va.), and celluclast 1.5L (Novo Nordisk Biochem, Franklinton, N.C.) with Ech 3937 pellicles. Mutifect B cellulase and cellulysin were chosen to examine bacterial aggregates from cultures of the mutant strains. The pellicles were incubated on a rotating shaker (30 rpm) at room temperature for up to 40 min. At least three pellicles were used for each treatment, and the experiments were repeated at least three times.
Microscopy.
Pellicle and biofilm fragments were dried onto glass slides and stained with the DNA stain propidium iodide (Molecular Probes) and the β-glucan stain calcofluor (Bectin, Dickinson and Co., Sparks, Md.) according to the manufacturer's instructions. The stained smears were observed by epifluorescence microscopy with a BX60 epifluorescence microscope (Olympus America, Inc., Melville, N.Y.). Separate filters were used to observed cells stained with propidium iodide (excitation, 556 nm; emission, 592 nm) and β-glucans stained with calcofluor (excitation, 360 nm; emission, 450 nm). Images of the propidium iodide-stained cells and the calcofluor-stained extracellular matrix were captured separately as monochrome images with a Magnafire camera (Optronics, Goleta, Calif.) and Image Pro Plus software (MediaCybernetics, Silver Spring, Md.). The images were merged with Photoshop 5.5 (Adobe Systems, Inc., San Jose, Calif.).
RESULTS
Ech 3937 pellicle formation is temperature and carbon source dependent. We explored the potential for multicellular behavior in E. chrysanthemi and found that some strains, including Ech 3937, could form biofilms and pellicles in a rich medium, SOB plus glycerol (SOBG). For this work, we have used definitions described by other authors (16, 50) where the solid surface-associated biofilms and the pellicles which cover the culture surface are considered distinct. Pellicles also formed in media where the glycerol was replaced by other sugars or sugar alcohols, including glucose, fructose, sucrose, and mannitol (data not shown). Pectate, a major component of plant cell walls and an inducer of E. chrysanthemi virulence genes, did not induce pellicle formation when used in place of glycerol in the culture medium but also did not inhibit pellicle formation when added to SOBG (data not shown). Pellicle, but not biofilm, formation was temperature dependent and occurred at 16 to 32°C but not at 36°C (Fig. 1A). In all cases, pellicle formation was observed visually, and cultures were rated as positive or negative.
FIG. 1.
Pellicle and biofilm formation in SOBG. The pellicles are thick aggregates of cells on the culture surface. The biofilms are aggregates of cells around the edge of the culture tube at the air-liquid-surface interface. (A) Pellicles formed in Ech 3937 SOBG cultures grown at 25°C (tube 1), but only biofilms and not pellicles formed in cultures grown at 36°C (tube 2). (B) All known TTSS regulatory genes were required for wild-type pellicle, but not biofilm, formation. The hrpL mutant WPP96 is shown. Tube 3, Ech 3937; tube 4, WPP96; tube 5, WPP96(p50hrpL). (C) The hrpS and hrpL genes expressed on plasmids restored pellicle formation to the hrpX hrpS mutant WPP89. Tube 6, Ech 3937; tube 7, WPP89; tube 8, WPP89(p105S); tube 9, WPP89(p50hrpL). (D) TTSS structural genes were also required for pellicle, but not biofilm, formation. Tube 10, Ech 3937; tube 11, WPP100; tube 12, WPP100(p415hrpA).
The Ech 3937 TTSS regulatory genes are required for pellicle formation. We decided to test allelic-exchange mutants that had been constructed for other purposes and discovered that the regulatory genes of the TTSS were required for pellicle formation. Additional mutants in other genes were also tested, including (for example) a type II secretion system mutant (3937 outD::kan), which was not impaired in biofilm formation (data not shown). Like many gram-negative pathogens, the Ech 3937 genome encodes a TTSS that plays an important role in its pathogenicity (51). At least four regulatory proteins may control expression of the E. chrysanthemi TTSS structural genes as well as genes encoding proteins secreted through this system, but it is not clear how the regulatory system functions in E. chrysanthemi. Importantly, the E. chrysanthemi TTSS is unlike the tightly regulated TTSS in other plant pathogens in that it is active, at least in some strains, in rich media (22).
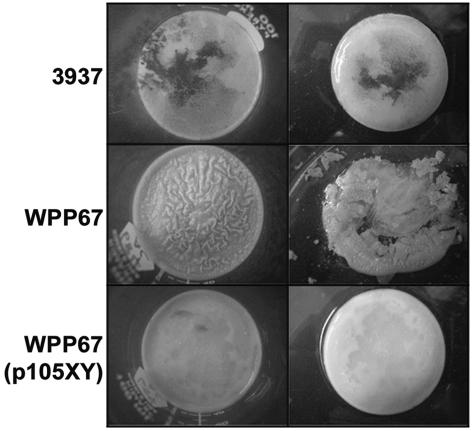
We used allelic exchange to mutate the regulatory genes hrpX, hrpY, hrpS, and hrpL in Ech 3937. The hrpX and hrpY genes are thought to encode a two-component system, with hrpX encoding the sensor kinase and hrpY encoding the response regulator. The hrpS gene encodes a σ54-dependent enhancer-binding protein (EBP) homolog, and hrpL encodes a homolog of a sigma factor. We found that all of the hrp regulatory genes were required for wild-type pellicle formation but that none of them were required for biofilm formation (Fig. 1B and C, 2, and 3). In wild-type Ech 3937, pellicles had a smooth surface and the cells were not dispersed when the aggregates were disturbed. The hrpX mutant strain, WPP67, also formed a biofilm and pellicle, but the pellicle was easily dispersed when disturbed. For additional observations of pellicle formation in the wild type and the hrpX mutant, we grew cultures in larger containers, including 100-ml beakers. The wild-type and WPP67 pellicles in the larger containers were very similar to those that formed in test tubes, except that surface distortions on the WPP67 pellicle surface were more obvious and more easily photographed (Fig. 2).
FIG. 2.
The hrpX mutant WPP67 formed a pellicle that was distorted and fragile compared to the wild-type pellicles (left column). The pigment visible in some of the pellicles is indigoidine, a blue pigment produced by E. chrysanthemi. WPP67 pellicles were much more fragile than those produced in wild-type cultures; when the pellicles were removed from the culture medium by pouring the culture into petri plates, the WPP67 pellicle, but not the wild-type pellicle, fragmented (right). Expression of hrpXY from a plasmid in WPP67 restored the wild-type phenotype.
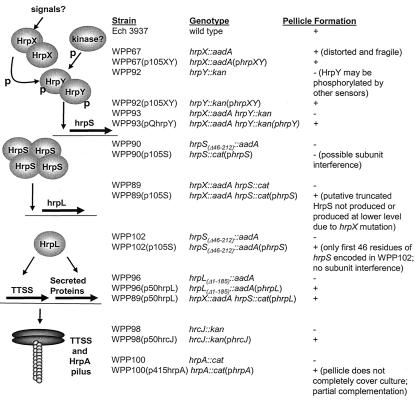
FIG. 3.
Model of TTSS regulatory cascade and table of mutant phenotypes. The TTSS regulatory cascade is shown on the left and the pellicle phenotypes of the mutants are shown on the right. The TTSS regulators HrpX and HrpY (two-component sensor kinase homolog and response regulator homolog, respectively), HrpS (a σ54-EBP homolog), and HrpL (a σ factor homolog) are all required for wild-type pellicle formation. Unlike all other TTSS mutants tested, WPP67 is able to form a pellicle, albeit a weak and distorted one. Although HrpX and HrpY are cognate sensor kinase and response regulator homologs, the phenotypes of WPP67 and WPP92 are not identical. In the diagram, the ovals and horizontal arrows symbolize proteins and genes, respectively. The vertical arrows indicate activation of genes by specific regulatory proteins. p, phosphorylation of HrpY by HrpX and possibly by other kinases. HrpX, HrpY, and HrpS homologs form multimers, which is why these proteins are shown here as multimers, although the multimerization levels for HrpX, HrpY, and HrpS are unknown. Two secretion system structural genes, hrcJ and hrpA, are also required for pellicle formation. Based on homology to other TTSSs, HrcJ is a membrane protein, and HrpA forms the TTSS pilus.
The hrpY, hrpS, and hrpL mutant strains (designated WPP92, WPP90, and WPP96, respectively) formed biofilms but not pellicles (WPP96 is shown in Fig. 1B; the WPP90 and WPP92 phenotypes were identical to that of WPP96). An hrpX hrpY mutant strain, WPP93, which was a derivative of WPP67, also did not form a pellicle (data not shown). With all of our TTSS mutants, the biofilm rings never extended more than approximately 1 mm over the culture surface, even in cultures incubated for up to 1 month, which suggested that biofilms and pellicles were distinct forms of cell aggregation. Additional results described below support this hypothesis. Curiously, the pellicle phenotypes of the hrpX and hrpY mutants were not identical, even though these genes appear to encode a cognate pair of homologs for a two-component system.
Expression of hrpXY from p105XY restored the wild-type pellicle phenotypes to WPP67 (3937 hrpX::aadA) (Fig. 2), but not WPP89 (3937 hrpX::aadA hrpS::cat) (data not shown). Expression of hrpS from p105S restored pellicle formation to WPP89 (Fig. 1C). Expression of hrpL from p50hrpL restored wild-type pellicle formation to all of the regulatory mutants (WPP89 plus p50hrpL is shown in Fig. 1C). We never observed pellicle formation in mutant strains transformed with plasmid vector controls. Thus, the plasmid vectors alone did not promote pellicle formation in Ech 3937.
Expression of hrpS from p105S did not restore the wild-type phenotype to the initial hrpS mutant strain WPP90, which encodes the N-terminal half of HrpS, but did restore the wild-type phenotype to both WPP89 (3937 hrpX::aadA hrpS::cat) (Fig. 1C), which was constructed using the WPP90 mutant background, and WPP102 (3937 hrpSΔ46-212::aadA), which has nearly the entire hrpS coding region deleted (data not shown). Prior to the construction of WPP102, an additional completely independent mutant strain was constructed by the same strategy used for WPP90. As with WPP90, this mutant strain was not complemented by p105S, demonstrating that the lack of complementation was probably not due to a second, unintended spontaneous mutation. These data suggest that the N-terminal half of hrpS carried on the chromosome of WPP90 was interfering with the function of the full-length hrpS carried on p105S. Since HrpX is required for the expression of hrpS, the partial HrpS fragment would not be expressed in WPP89, allowing this mutant to be complemented by p105S. The Ech 3937 TTSS regulatory cascade deduced from the pellicle formation assay is shown in Fig. 3.
TTSS structural genes are required for Ech 3937 pellicle formation. To determine if the TTSS was required for pellicle formation or if the TTSS regulators were activating other genes required for pellicle formation, two TTSS structural genes, hrcJ and hrpA, were mutated by allelic-exchange to construct WPP98 and WPP100, respectively. HrcJ is a membrane protein required for a functional TTSS (2). HrpA multimers form the TTSS pilus, which delivers virulence proteins to host cells (23). Neither WPP98 (data not shown) nor WPP100 (Fig. 1D) formed pellicles, demonstrating that a functional TTSS is required for pellicle formation. The plasmid p50hrcJ restored pellicle formation to WPP98 (data not shown). WPP100 was partially complemented by p415hrpA; the pellicles in these cultures never completely covered the culture surface (Fig. 1D). WPP100(p415hrpA) was the only complemented mutant strain in our study with this phenotype. Other investigators have also had difficulty fully complementing hrpA mutants in other species (43). The hrcJ and hrpA mutants demonstrate that a functional TTSS is required for aggregative multicellular behavior and suggest that in Ech 3937, the HrpA pilus may play an analogous role to curli in other enterobacteria, since both types of protein filament contribute to aggregative multicellular behavior in culture (45).
Importantly, none of the TTSS mutants was impaired in growth in LB, Hrp-inducing minimal medium, or SOBG when grown in a shaking incubator at 37 or 30°C, demonstrating that the inability to form a pellicle in culture is not due to a reduced ability to grow in liquid culture. All of the mutants were also tested for swimming and swarming motility (38) and ability to produce acyl-homoserine lactones (10), all functions that could affect bacterial aggregation. None of the mutants were impaired in these assays (not shown).
Not all E. chrysanthemi strains tested formed pellicles in SOBG medium, although all formed biofilms. Notably, E. chrysanthemi AC4150, another model E. chrysanthemi strain that encodes a functional TTSS system, only formed biofilms. As with the Ech 3937 TTSS mutants, an AC4150 TTSS mutant, CUCPB5039 (5), retained the ability to form biofilms on culture tubes. Similar variation in pellicle formation among strains is seen in other enterobacterial species (52) and has been tied in some cases to point mutations in a regulatory gene (44).
We did not explore bacterial growth in plants for the majority of our mutants because others have already conclusively demonstrated that the TTSS contributes to growth of E. chrysanthemi in plants (29, 51). Because unlike all of the other TTSS mutants tested, the hrpX mutant strain formed a pellicle, we examined bacterial growth of WPP67 in African violets. The wild type and the hrpS mutant strain lacking a pellicle, WPP90, were used as controls. WPP90 and WPP67 were equally reduced in growth in African violet leaves in comparison to the wild type (data not shown). Thus, although WPP67 is still able to form a pellicle, albeit a fragile one, suggesting that HrpY is still active in the absence of HrpX, this activity is not sufficient to promote wild-type levels of bacterial growth in plants.
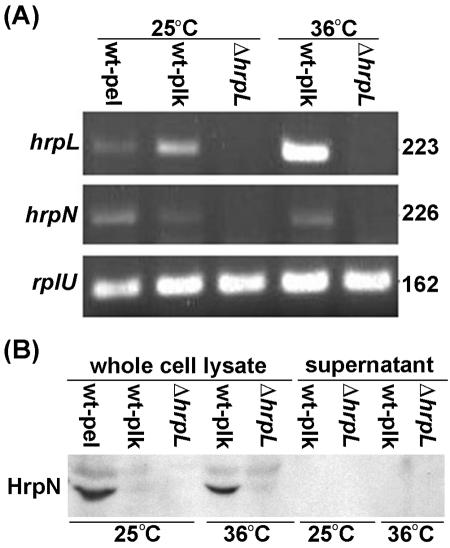
TTSS genes are expressed and HrpN is produced at 36°C, even though pellicle formation does not occur. Since wild-type cultures grown at 36°C formed only biofilms and not pellicles, similarly to the TTSS mutants grown at lower temperatures (Fig. 1A and B), we suspected that the TTSS genes were not expressed at 36°C. The gene expression of both hrpL and hrpN were examined. The hrpN gene encodes a harpin and is predicted to be regulated by HrpL. Harpins, which are glycine-rich, cysteine-lacking, heat-stable proteins capable of eliciting a plant hypersensitive response, are encoded by most gram-negative plant pathogens and are secreted via the TTSS. We found that both hrpL and hrpN were expressed in planktonic and pellicle cells at 25°C and in planktonic cells at 36°C (Fig. 4A). As expected, hrpN was not expressed in the hrpL mutant WPP96.
FIG. 4.
RT-PCR and immunoblot analysis of Ech 3937 and WPP96 under pellicle-inducing and noninducing conditions. (A) RT-PCR performed with total RNA isolated from strains grown in SOBG at 25 and 36°C for 2 days. The pellicle only formed in the wild-type culture at 25°C. Both hrpL and hrpN were expressed in all samples from wild-type cultures grown at 25 and at 36°C; neither was detected in WPP96 cultures. The amount of RNA template was monitored by using constitutively expressed rplU gene as the internal control. In all cases, no product was detected in the control samples amplified without the RT step (data not shown). The amplified mRNA is indicated on the left, and the amplicon sizes (in base pairs) are indicated on the right. (B) Immunoblot analysis performed with the same cultures as in panel A and with an anti-HrpN antibody. Twenty microliters of protein sample was loaded in each lane. HrpN was only detected in pellicle cells grown at 25°C, although hrpN mRNA was detected in both planktonic and pellicle cells. HrpN was detected in planktonic cells grown at 36°C. The RT-PCR experiment was repeated three times, and the immunoblot experiment was repeated twice. Similar results were obtained each time. Abbreviations: wt, wild-type Ech 3937; pel, pellicle; plk, planktonic cells.
Although both hrpL and hrpN are expressed at 36°C, it is possible that TTSS-secreted proteins are not produced at 36°C, due to posttranscriptional regulation. To determine if any TTSS-associated proteins are produced at 36°C, immunoblots were used to examine HrpN accumulation. Although hrpL and hrpN were both expressed in planktonic and pellicle cells at 28°C, we were only able to detect HrpN in the pellicle (Fig. 4B). Conversely, with cultures grown at 36°C, we were able to detect HrpN in the planktonic cells (Fig. 4B). We were unable to detect HrpN in the supernatant at either temperature. These results clearly show that the TTSS genes hrpL and hrpN are expressed and that HrpN protein accumulates at 36°C, even though a pellicle does not form. These results also show that HrpN protein only accumulates in the pellicle fraction of cultures grown at 25°C, even though the gene is also expressed in the planktonic cells.
Cellulose is part of the Ech 3937 pellicle, but cellulose production is not required for pellicle formation. In Escherichia coli and Salmonella enterica, cellulose also contributes to pellicle formation. Four operons, bcsABZC, bcsEFG, adrA, and csgD, are required for cellulose production in these species (40, 42, 44, 53). CsgD activates both the expression of curli and the production of cellulose. AdrA activates cellulose production on the posttranscriptional level. The functions of most of the Bcs proteins are unknown; BcsA is a cellulose synthase and BcsZ is a cellulase. The Ech 3937 genome carries bcsABZC and adrA genes, but the arrangement of the bcs operon differs in Ech 3937 compared to other enterobacteria. Ech 3937 does not carry bcsEFG or csgD.
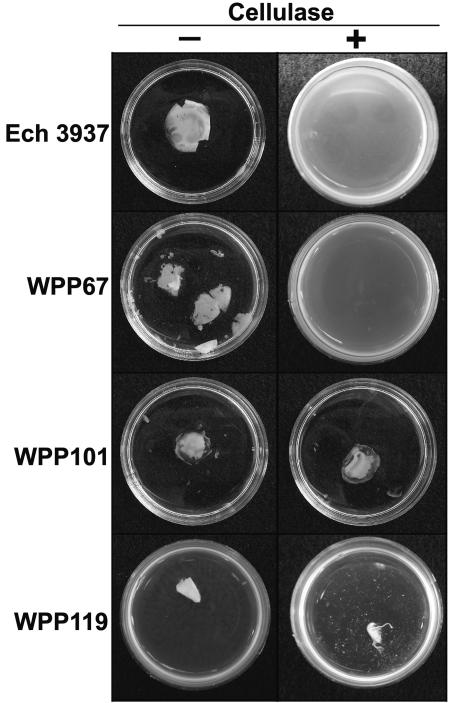
To determine if cellulose was also a component of the Ech 3937 pellicle, pellicles were removed from culture tubes and treated with six different commercial cellulase preparations. In all cases, the cellulase-treated pellicles disintegrated, while the untreated pellicles and pellicles treated with heat-denatured cellulase remained intact (Fig. 5). Two commercial cellulases, multifect B and cellulysin, were chosen for further experiments. A cellulose synthesis mutant, WPP101 (3937 bcsA::cat), was constructed to determine if the Ech 3937 bcsA gene contributed to pellicle formation. WPP101 formed biofilms similar to those of wild-type cells, but the pellicles were irregular and more fragile and translucent than those formed by the wild type (Fig. 5). The pellicles formed by this mutant were not degraded by cellulase, suggesting that the matrix holding the WPP101 (3937 bcsA::cat) pellicles together was not cellulose (Fig. 5).
FIG. 5.
Cellulase digestion of Ech 3937 pellicles. Cellulase digests Ech 3937 and WPP67 (3937 hrpX::aadA) pellicles but has no effect on WPP101 (3937 bcsA::cat) pellicles or WPP119 (3937 hrpX::aadA bcsA::cat) aggregates.
In contrast, WPP67 (3937 hrpX::aadA) pellicles treated with cellulase disintegrated (Fig. 5). These results suggest that the WPP67 pellicle consisted mainly of cellulose, while the WPP101 pellicle consisted mainly of other matrix components, including possibly HrpA protein filaments or other proteins secreted via the TTSS. Since the WPP101 pellicles were more resistant to cellulase than wild-type pellicles, these results also suggest that the remaining matrix components interact differently with each other in the absence of cellulose. WPP119 (3937 hrpX::aadA bcsA::cat) was constructed from WPP67 to determine if pellicle formation would be completely eliminated. This mutant formed biofilms that sometimes had small, viscous, cellulase-resistant aggregates protruding from them (Fig. 5). Although the wild-type and WPP67 pellicles were degraded by cellulase, the solid surface-associated biofilm ring remained intact in the cellulase-treated pellicles, suggesting that this biofilm was not composed of cellulose (data not shown). Biofilms from mutants (such as WPP96) and culture conditions (such as Ech 3937 grown in SOBG at 36°C) (data not shown) where pellicles do not form were also resistant to cellulase.
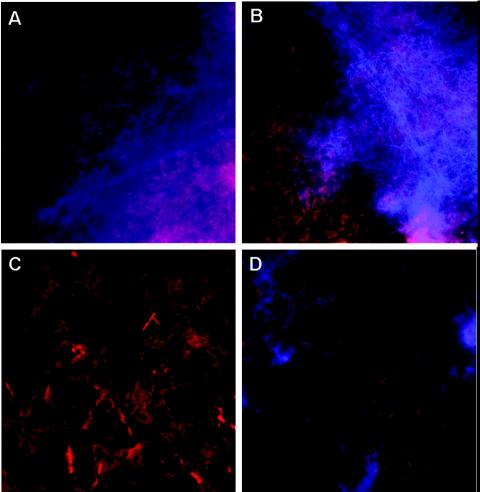
Pellicles and/or biofilms from Ech 3937, WPP67, WPP96, WPP101, and WPP119 cultures grown in SOBG were smeared onto slides and stained with the DNA stain propidium iodide and the β-glucan stain calcofluor. Calcofluor-stained fibers were clearly visible in pellicle fragments from Ech 3937 (Fig. 6A) and WPP67 (Fig. 6B), but not in biofilms from WPP96 (Fig. 6C). Similarly to WPP96, calcofluor-stained fibers were not visible in biofilms from Ech 3937 cultures grown at 36°C (not shown). Surprisingly, calcofluor-stained fibers were present in some but not all smears from WPP101 (Fig. 6D) and WPP119 (data not shown) cultures, which suggested that the bcsA mutant still produces a small amount of a β-glucan. These calcofluor-stained fibers in WPP101 were also present in smears from cellulase-treated WPP101 pellicles (data not shown).
FIG. 6.
Micrographs of pellicle and biofilm aggregates stained with the red fluorescent DNA stain propidium iodide and the blue β-glucan stain calcofluor. (A and B) Ech 3937 and WPP67 pellicles, respectively. In both cases, fibrous β-glucan-stained fibers are visible surrounding the cell aggregates. (C) Solid surface-associated biofilms from WPP96 did not stain with calcofluor. (D) β-Glucan-stained fibers were visible surrounding cell aggregates in some smears from WPP101 cultures.
DISCUSSION
In total, these results demonstrate that the TTSS contributes to multicellular behavior, which manifests as pellicle formation, in Ech 3937. Neither the TTSS pilus nor the TTSS regulatory proteins were required for bacterial cells to form a biofilm ring around the culture tube at the air-liquid interface, demonstrating that there are two genetically distinguishable forms of E. chrysanthemi aggregative multicellular behavior. Unlike some of the pellicles, the solid surface-associated biofilms were resistant to cellulase. Biofilms but not pellicles also formed at 36°C, demonstrating that biofilms and pellicles are also induced under different conditions. Neither biofilms nor pellicles formed in SOB cultures. Biofilm formation always preceded pellicle formation, and a solid surface-associated biofilm is probably required for pellicle formation, since the pellicles are not buoyant and require attachment to the test tube surface to remain on top of the culture. Because the biofilms and pellicles can be separated genetically, enzymatically, and by culture conditions, we like others (16, 50) define them as separate phenomena. The genes required for Ech 3937 solid surface-associated biofilm formation remain to be discovered.
Our results show that the TTSS regulatory and structural genes play a critical role in cell aggregation in culture in the absence of host cells. These results expand the list of secretion systems that can contribute to enterobacterial cell aggregation and pellicle formation and support a model where cellulose and generic proteins, which consist of either curli or TTSS-secreted proteins, are required for pellicle formation in enterobacteria. The TTSS pilus itself may contribute to pellicle formation, since in other systems, curli, type I pili, or conjugative pili are required for pellicle and/or biofilm formation (19, 36, 37, 45, 49, 52). However, other TTSS-secreted proteins may also be required; this possibility remains to be explored.
Notably, both curli and the TTSS contribute to bacterial virulence and interact intimately with host cells (6, 17, 20, 23, 35). However, none of the genes required for formation of the TTSS or curli are homologous. Currently, it is not clear if Ech 3937 bacterial aggregation in hosts is mediated by the TTSS or if this sort of aggregation plays an important role in E. chrysanthemi virulence.
Since the pellicle phenotype is easily and inexpensively reproduced, this phenotypic assay is useful as a preliminary high throughput screen for mutations or compounds affecting the expression and function of the E. chrysanthemi TTSS. Inhibitors found through this assay may be broadly useful antimicrobial compounds, since many of the TTSS structural and regulatory proteins are conserved across many gram-negative plant and animal pathogens. We used the pellicle assays to explore the regulatory cascade controlling the TTSS in E. chrysanthemi. At least four regulatory proteins (HrpX, HrpY, HrpS, and HrpL) may control expression of the E. chrysanthemi TTSS structural genes as well as genes encoding proteins secreted through this system. The regulatory cascades deduced in two species related to E. chrysanthemi are not identical even though the regulatory proteins are homologous. In Erwinia amylovora, HrpX and HrpY, two-component system homologs, and HrpS, a σ54 EBP homolog, both activate the expression of hrpL. HrpL, a σ factor homolog, activates the expression of genes encoding the TTSS and proteins secreted through the TTSS (46). In Pantoea stewartii, HrpX and HrpY activate hrpS, and HrpS activates hrpL (33). Our results support an E. chrysanthemi TTSS regulatory model similar to that in P. stewartii.
WPP67 (3937 hrpX::aadA) and WPP92 (3937 hrpY::kan) did not have identical phenotypes; WPP67 produced a pellicle and WPP92 did not. A double mutant strain, WPP93 (3937 hrpX::aadA hrpY::kan), did not produced a pellicle and expression of hrpY from a plasmid restored pellicle production to this mutant, demonstrating that HrpY retains some activity in the absence of HrpX. Like other response regulators, HrpY may be phosphorylated by noncognate sensor proteins (7). If so, HrpY would retain the ability to activate downstream genes in the absence of HrpX, although the level of activation is likely to be lower than in wild-type cells. Notably, WPP67 was still impaired in growth on African violets. Thus, the apparent residual HrpY activity in WPP67 is not sufficient for promoting virulence, and pellicle formation is not completely correlated with virulence.
WPP90 (3937 hrpS::cat) was the first TTSS mutant tested for the pellicle phenotype. Because this mutant could not be complemented with plasmid-expressed hrpS, we initially suspected that a secondary, unintended mutation was responsible for the loss of pellicle formation. However, since mutations in hrpX, hrpY, hrpL, hrcJ, and hrpA all affected pellicle formation, we decided to further explore why the hrpS mutant could not be complemented. Based on protein homology, HrpS belongs to the NtrC family and is likely to encode a multidomain protein, with the N-terminal half expressing oligomerization and σ54 interaction domains and the C-terminal half expressing a DNA-binding motif. To construct WPP90, an antibiotic resistance gene was inserted in the middle of hrpS; thus, WPP90 retains the ability to produce the N-terminal half of HrpS. This HrpS fragment may interfere with the oligomerization or σ54 interactions of the full-length protein expressed from the plasmid. Similar subunit interference occurs in homologous proteins (28).
The hrpX hrpS double mutant WPP89 was derived from WPP90, and this mutant can be complemented with hrpS expressed from a plasmid. If HrpX and HrpY are required to activate hrpS in Ech 3937, then in WPP89 the partial hrpS gene would not be expressed at high levels due to a lack of HrpX. This could explain how WPP89, but not WPP90, was complemented by plasmid-carried hrpS. A second hrpS mutant, WPP102, which only encodes the first 46 amino acids of HrpS, was constructed. This mutant was unable to form pellicles in SOBG, and expression of plasmid-carried hrpS did restore pellicle formation to WPP102. This supports the hypothesis that the N-terminal half of HrpS causes subunit interference. These data also provide additional support for a linear regulatory cascade model in E. chrysanthemi similar to that in P. stewartii where HrpX and HrpY activate hrpS.
Because a functional TTSS system was required for pellicle formation at 25°C and because pellicles did not form in wild-type cultures at 36°C, we suspected that TTSS genes were not expressed at 36°C. Surprisingly, our RT-PCR and immunoblot data demonstrated that at least two TTSS genes, hrpL and hrpN, are active at both 25 and 36°C. We found that HrpN protein was only detectable in the pellicles and not planktonic cells of cultures grown at 25°C, even though hrpL and hrpN mRNA were detected in planktonic cells. In contrast, HrpN was detectable in planktonic cells of cultures grown at 36°C. These results suggest that HrpN production and/or accumulation is affected by a posttranscriptional temperature dependent phenomenon.
Harpins, such as HrpN, HrpW, and PopA, produced by plant pathogenic Pseudomonas and Ralstonia species are easily detected in culture supernatants (for example, see references 1, 4, and 11). In contrast, the initial report of a harpin, the E. amylovora HrpN, described the protein as cell associated (47). Similarly to the initial E. amylovora findings, we were unable to detect HrpN in culture supernatants. We do not know why pellicle formation does not occur at 36°C, but it is not simply because the TTSS genes are not expressed. It is possible that a second set of genes required for pellicle formation is not expressed at 36°C or that a protein or carbohydrate required for pellicle formation is not produced, is not stable, or is not folded correctly at 36°C.
Since Ech 3937 pellicles could be degraded by cellulase, cellulose appears to be an important component of the pellicle matrix, which is intriguing for a plant pathogen better known for producing cellulase (3, 21, 25) than cellulose. The bcsA gene has been implicated in cellulose production in other enterobacterial pathogens, such as S. enterica (42, 53). Therefore, we mutated this gene to determine if it was required for Ech 3937 pellicle formation. Although the cellulase experiments suggested that cellulose was a critical component of the Ech 3937 pellicle and bcsA is thought to encode a cellulose synthase subunit, the bcsA mutant still formed pellicles. Cell aggregates from these pellicles fluoresce after being stained with calcofluor, which binds to β-glucans such as cellulose and chitin. However, the Ech 3937 bcsA mutant pellicles are resistant to cellulose, and cellulase-treated pellicles still bind calcofluor, suggesting that the β-glucans in these pellicles are not cellulose. These observations may lead to new avenues of inquiry into the physiology of this model plant pathogen, such as determining the identity and function of the β-glucan produced by the Ech 3937 bcsA mutant, discovering if TTSS-secreted proteins interact with β-glucans synthesized by bacteria, and revealing if bacterial cellulase and cellulose production are coregulated in this soft rot pathogen.
Acknowledgments
Support was provided by Hatch Project 4605, the Department of Plant Pathology, the College of Agricultural and Life Sciences, the University of Wisconsin—Madison Graduate School; and the U.S. Department of Agriculture-Agricultural Reseach Service (ARS CRIS 5325-42000-040-00D).
We thank Alan Collmer for providing the HrpN polyclonal antibody, Thomas Jeffries for providing the cellulase enzymes, Russell Spear for aid with the microscopy experiments, Holly Simon for aid in data interpretation and manuscript preparation, and Robert Goodman and Andrew Bent for reviewing the manuscript.
REFERENCES
- 1.Alfano, J. R., D. W. Bauer, T. M. Milos, and A. Collmer. 1996. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19:715-728. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andro, T., J.-P. Chambost, A. Kotoujansky, J. Catteneo, Y. Bertheau, F. Barrase, F. Van Gejsegem, and A. Colenon. 1984. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gisegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer, D. W., A. J. Bogdanove, S. V. Beer, and A. Collmer. 1994. Erwinia chrysanthemi hrp genes, and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant Microbe Interact. 7:573-581. [DOI] [PubMed] [Google Scholar]
- 6.Bian, Z., Z. Q. Yan, G. K. Hansson, P. Thoren, and S. Normark. 2001. Activation of inducible nitric oxide synthase/nitric oxide by curli fibers leads to a fall in blood pressure during systemic Escherichia coli infection in mice. J. Infect. Dis. 183:612-619. [DOI] [PubMed] [Google Scholar]
- 7.Bijisma, J. J. E., and E. A. Groisman. 2003. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 11:359-366. [DOI] [PubMed] [Google Scholar]
- 8.Bogdanove, A. J., D. W. Bauer, and S. V. Beer. 1998. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol. 180:2244-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttner, D., and U. Bonas. 2003. Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Plant Biol. 6:312-319. [DOI] [PubMed] [Google Scholar]
- 10.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 11.Charkowski, A. O., J. R. Alfano, G. Preston, J. Yuan, S. Y. He, and A. Collmer. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee, A. K., K. K. Thurn, and D. A. Feese. 1983. Tn5-induced mutations in the enterobacterial phytopathogen Erwinia chrysanthemi. Appl. Environ. Microbiol. 45:644-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 17.Gauthier, A., N. A. Thomas, and B. B. Finlay. 2003. Bacterial injection machines. J. Biol. Chem. 278:25273-25276. [DOI] [PubMed] [Google Scholar]
- 18.Gerstel, U., and U. Römling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154:659-667. [DOI] [PubMed] [Google Scholar]
- 19.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 20.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiseppi, A., J. L. Aymeric, B. Cami, F. Barras, and N. Creuzet. 1991. Sequence analysis of the cellulase encoding celY gene of Erwinia chrysanthemi: a possible case of interspecies gene transfer. Gene 106:109-114. [DOI] [PubMed] [Google Scholar]
- 22.Ham, J.-H., C. Yaya, J. R. Alfano, P. Rodríguez-Palenzuela, C. M. Rojas, A. K. Chatterjee, and A. Collmer. 2004. Analysis of Erwinia chrysanthemi EC16 pelE::uidA, pelL::uidA, and hrpN::uidA mutants reveals strain-specific atypical regulation of the Hrp type III secretion system. Mol. Plant Microbe Interact. 17:184-194. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Q. L., and S. Y. He. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294:2556-2558. [DOI] [PubMed] [Google Scholar]
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Kotoujansky, A., A. Diolez, M. Boccara, Y. Bertheau, T. Andro, and A. Coleno. 1985. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 4:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lemattre, M., and J. P. Narcy. 1972. Une infection bacterienne nouvelle du Saintpaulia due a Erwinia chrysanthemi. C.R. Acad. Sci. 58:227-231. [Google Scholar]
- 28.Leonhartsberger, S., A. Ehrenreich, and A. Böck. 2000. Analysis of the domain structure and the DNA binding sites of the transcriptional activator FhlA. Eur. J. Biochem. 267:3672-3684. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Solanilla, E., A. Llama-Palacios, A. Collmer, F. Garcia-Olmedo, and P. Rodriguez-Palenzuela. 2001. Relative effects on virulence of mutations in the sap, pel, and hrp loci of Erwinia chrysanthemi. Mol. Plant Microbe Interact. 14:386-393. [DOI] [PubMed] [Google Scholar]
- 30.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 31.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 32.McFadden, L. A. 1961. Bacterial stem and leaf rot of Dieffenbachia in Florida. Phytopathology 51:663-667. [Google Scholar]
- 33.Merighi, M., D. R. Majerczak, E. H. Stover, and D. L. Coplin. 2003. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the hrp regulon in Pantoea stewartii subsp. stewartii. Mol. Plant Microbe Interact. 16:238-248. [DOI] [PubMed] [Google Scholar]
- 34.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 35.Olsen, A., M. J. Wick, M. Morgelin, and L. Bjorck. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 37.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 38.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ried, J. L., and A. Collmer. 1987. An nptI-SacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker-exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 40.Römling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 43.Taira, S., J. Tuimala, E. Roine, E.-L. Nurmiaho-Lassila, H. Savilahi, and M. Romantschuk. 1999. Mutational analysis of the Pseudomonas syringae pv. tomato hrpA gene encoding Hrp pilus subunit. Mol. Microbiol. 34:736-744. [DOI] [PubMed] [Google Scholar]
- 44.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejuene. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, Z. M., J. F. Kim, and S. V. Beer. 2000. Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two-component system, and HrpS. Mol. Plant Microbe Interact. 13:1251-1262. [DOI] [PubMed] [Google Scholar]
- 47.Wei, Z.-M., R. J. Laby, C. H. Zumoff, D. W. Bauer, S.-Y. He, A. Collmer, and S. V. Beer. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257:85-88. [DOI] [PubMed] [Google Scholar]
- 48.Wei, Z.-M., B. Sneath, and S. V. Beer. 1992. Expression of Erwinia amylovora hrp genes in response to enviromental stimuli. J. Bacteriol. 174:1875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White, A. P., D. L. Gibson, S. K. Collinson, P. A. Banser, and W. W. Kay. 2003. Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar Enteritidis. J. Bacteriol. 185:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 51.Yang, C.-H., M. Gavilanes-Ruiz, Y. Okinaka, R. Vedel, I. Berthuy, M. Boccara, J. W. T. Chen, N. T. Perna, and N. T. Keen. 2002. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant-Microbe Interact. 15:472-480. [DOI] [PubMed] [Google Scholar]
- 52.Zogaj, X., W. Bokranz, M. Nimtz, and U. Römling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]