Abstract
In Sulfolobus acidocaldarius conjugation assays, recombinant frequency was relatively constant for marker separations from 1,154 bp down to about 50 bp and readily detectable at 10 bp. Three-factor crosses revealed little, if any, genetic linkage over distances of 500 to 600 bp, and large deletion mutants were good donors but poor recipients in matings. The results indicate that most intragenic recombination events occur at one of the mutations, not in the interval between them.
The conjugational mechanism of marker exchange (ME) in Sulfolobus acidocaldarius (8) is so far the only system in which natural exchange and recombination of chromosomal DNA in a hyperthermophilic archaeon can be quantified by genetic selection. This phenomenon occurs between virtually isogenic strains, is not mediated by culture supernatants, is not inhibited by extracellular DNase, and may account for the infectivity of homing introns that have been introduced into S. acidocaldarius cells by electroporation (1, 7, 8). ME appears to differ from bacterial Hfr conjugation with respect to both the requirements of DNA transfer and the manifestation of genetic linkage. Most examples of bacterial conjugation require a self-transmissible plasmid in the donor and no plasmid in the recipient, but in S. acidocaldarius, as in the halophilic archaeon Haloferax volcanii (17), all strains appear equally capable of serving as donor and recipient (8). In addition, ME-promoted recombination yields readily detectable recombination (representing a level of about 5% of the maximal value, for example) for mutations separated by only 28 bp (21), whereas in bacterial Hfr conjugation, recombination at 5% of the maximal frequency usually requires about 25 kbp between markers (16). In the present study, we systematically analyzed the effect of marker separation on the frequency of recombination and related molecular properties of ME to gain insight into its mechanism.
Genetic analyses.
Double mutant strains DG228 and DG229 were spontaneous fluoroorotic acid-resistant derivatives of strain DG64 (9). All other mutants were isolated as spontaneous fluoroorotic acid-resistant derivatives of wild-type strain ATCC 33909. The identity of the pyrE mutation in each strain was determined by PCR amplification and dye terminator sequencing (10, 21). Recombination, reversion, and effects of γ and UV radiation were quantified by determining appropriate plate counts (7, 8, 21, 22).
Recombination frequency as a function of marker separation.
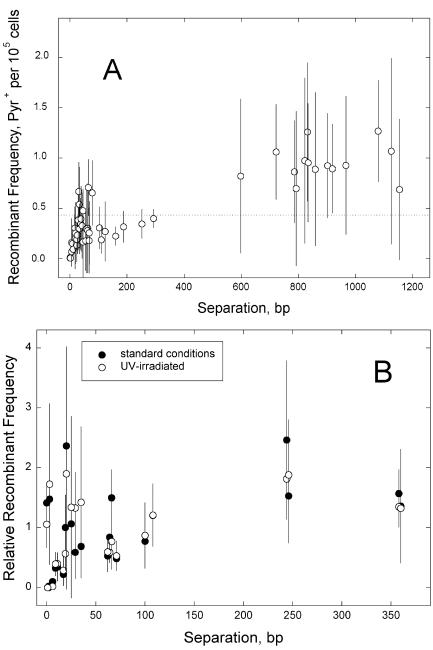
The first set of experiments examined 52 strain pairs defining marker separations of 0 to 1,154 bp. Matings were performed in four to seven independent experiments for each pair, and the median frequencies of recombination were plotted as a function of marker separation (Fig. 1A). Nearly all of the median values fell within a factor of 3 of the average of all medians (Fig. 1A) and were thus more uniform than those for Escherichia coli lacI point mutations (4, 19). Linear regression of the entire set of points yielded a large, negative x-axis intercept, indicating that recombination was not proportional to marker separation over this interval. Indeed, frequencies of recombination fell to consistently low values only for marker separations of less than about 35 bp, which represents less than 3% of the range of marker separations analyzed (Fig. 1A). Similar results were obtained with a slightly different set of mutant pairs defining marker separations of 360 bp or less (Fig. 1B). This second set was further evaluated for the effect of 50 J/m2 UV-C radiation, which was accompanied by an eightfold UV stimulation of ME. As shown in Fig. 1, these experiments yielded similar frequency-versus-distance behaviors, indicating that the characteristics evident in Fig. 1A are not due to the markers chosen or to a fortuitous distribution of initiation sites for normal (i.e., nonstimulated) recombination.
FIG. 1.
Recombination frequency in ME as a function of the distance between markers. A. Marker separations of 0 to 1,154 bp. Frequencies are numbers of Pyr+ recombinants per cell plated, representing median values from four to seven independent determinations (the horizontal line marks the average of all medians); error bars indicate standard deviations. B. Effect of DNA damage. Washed cell suspensions were divided in half, and one half received 50 J/m2 of UV-C radiation before the mating. Mean numbers of recombinants per CFU and standard deviations (error bars) were normalized to the average of all values for each treatment. Solid symbols, untreated; open symbols, UV irradiated.
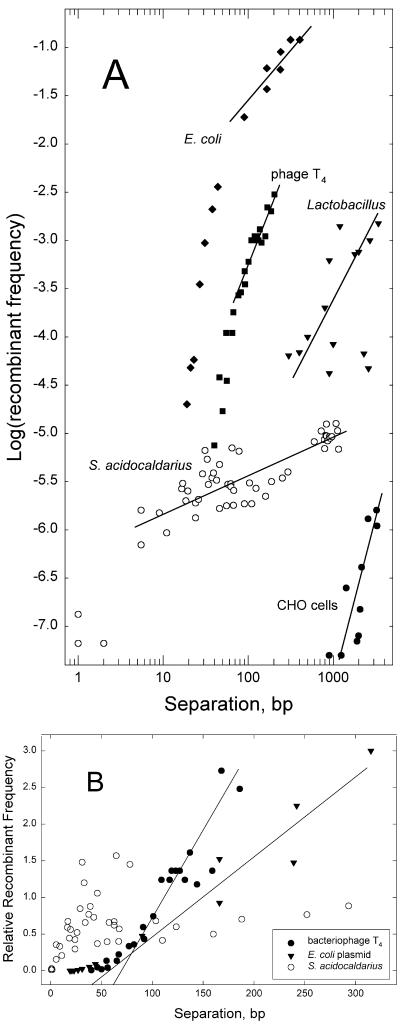
For comparison to other models of homologous recombination, plots of the Sulfolobus data were superimposed on corresponding plots of published data for bacteriophage T4 (26), E. coli (25), Lactobacillus sake (15), and hamster cells (23). Logarithmic plots (Fig. 2A) showed that S. acidocaldarius ME exhibited unusually low dependence on distance compared to that of the other systems. Arithmetic plots (Fig. 2B) confirmed the contrast between S. acidocaldarius ME, which gave no evidence of a threshold or “minimal” efficient processing segment (MEPS) (25), and the other recombination systems. Linear regressions to zero recombination confirm thresholds of 40 to 60 bp for T4 and E. coli, whereas the S. acidocaldarius data indicate no positive x-axis intercept (Fig. 2B). Alternatively, MEPSs can be seen in logarithmic plots as breakpoints between regions of two different slopes (6, 25). In Fig. 2A, these occur at about 50 and 60 bp for T4 and E. coli, respectively, whereas the maximum estimate consistent with the S. acidocaldarius data in Fig. 2B would be about 4 bp (Fig. 2A).
FIG. 2.
Comparison of S. acidocaldarius ME to other systems. A. Logarithmic plot. Straight lines approximate linear regressions of the surrounding points, with the following slopes: E. coli plasmid recombining with bacteriophage λ (25), 1.0; bacteriophage T4 mutants (26), 2.0; plasmid integration into L. sake chromosome (15), 2.2; linearDNA integrating into Chinese hamster ovary cell APRT gene (23), 3.5; S. acidocaldarius conjugational crosses (data of Fig. 1A), 0.5. B. Arithmetic plot. To facilitate comparison, the recombinant frequencies for each system were normalized to the average value over the interval shown. Data are corresponding subsets of those in Fig. 1A. Lines approximate linear extrapolation of higher frequencies to zero recombination.
Tests of genetic linkage.
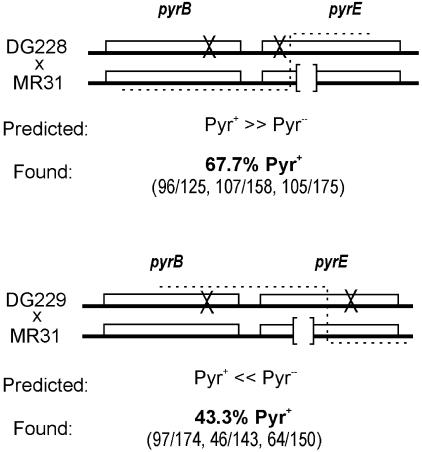
In addition to demonstrating no measurable MEPS, the S. acidocaldarius results further suggested that the rate of recombination was not limited by marker separation beyond about 50 bp, which would predict a concomitant lack of genetic linkage over this and longer distances. To test this prediction, we used the method of 3-factor crosses to evaluate recombination between pyrE and the neighboring pyrB gene. The rationale of the experiment, outlined in Fig. 3, is that reciprocal crossovers between a pyrB pyrE chromosome and a pyrE chromosome yielding PyrE+ recombinants should preferentially incorporate either the wild-type or the mutant pyrB allele, depending on the relative orientation of the pyrE mutations. We generated pyrB pyrE double mutants DG228 and DG229 and identified the sites of pyrE mutation by sequencing. When mated to a distinct pyrE strain (e.g., MR31), these strains allowed PyrE+ recombinants to be selected without constraint on the pyrB allele by plating cells on medium containing orotic acid as the sole pyrimidine source. To control for the possibility of a weak selection biasing recovery of the pyrB4 mutation among recombinants, this analysis was performed for two double mutants in which the pyrE mutations lie on opposite sides of the pyrE mutation in the reference strain. In both cases, the Oro+ recombinants were then scored for the pyrB allele by a form of replica plating.
FIG. 3.
Tests for genetic linkage. Three-factor crosses used to evaluate genetic linkage by ME. The ×'s symbolize point mutations, whereas the open brackets represent the 18-bp deletion in strain MR31. Predicted frequencies of Pyr+ recombinants are based on corresponding distances in E. coli Hfr crosses (16).
Instead of the strong bias for the unselected (pyrB) marker predicted by rare reciprocal events (i.e., crossing over), both strain pairs yielded similar proportions of Pyr+ and Pyr− recombinants (Fig. 3). For comparison, analogous Hfr matings with this arrangement of markers are expected to yield >99% Pyr+ mutants in the first cross and <1% Pyr+ mutants in the second one (16).
Reciprocal versus nonreciprocal modes of homologous recombination.
A negligible MEPS, an extremely short region of proportionality to marker separation, and a lack of genetic linkage over short distances are not characteristics of reciprocal homologous recombination (25, 28). These features may, however, be accommodated by a nonreciprocal mode of recombination, reflecting the fact that such events occur at one of the mutations, not in the interval between them (2, 4, 19, 20). This observation prompted us to test S. acidocaldarius ME for other special characteristics of nonreciprocal homologous recombination with a small patch size. For example, a large deletion in a parental strain should impose an asymmetry on the crosses, due to the fact that deletions larger than the average patch size would lack the flanking complementarity needed to incorporate donor markers by a nonreciprocal mechanism, whereas this would not pose a problem for reciprocal recombination.
A technical obstacle to testing this property is that S. acidocaldarius ME lacks the natural donor-recipient directionality imposed by most bacterial conjugation systems (8). However, the organism is fully sensitive to γ radiation (26). We therefore used γ irradiation of one of the two parental strains to prevent it from serving as the recipient in matings but allow it to serve as donor, at least for a brief period of time. Reversion controls confirmed that spontaneous mutation was undetected and that any mutagenic effect of the γ radiation (26) was masked by the extremely low survival (Table 1). However, an irradiated deletion mutant mated to an untreated point mutant generated recombinants at high frequencies, confirming that an irradiated, and thus moribund, cell could successfully transfer DNA to a recipient cell under these conditions. In contrast, no mating in which both parents had been irradiated yielded recombinants, confirming the inability of irradiated cells to serve as ME recipients under these conditions. In the asymmetric crosses between point mutant donors and deletion mutant recipients, the yields of recombinants were consistently low (Table 1). This effect could not be attributed simply to the failure of γ radiation to stimulate recombination in these particular crosses, as the recombinant frequencies were also lower than those of the corresponding untreated strain pairs (Table 1).
TABLE 1.
Deletion mutants as recipients in ME
| γ-Ray dose (kilorads) | Avg survival | Parental strain irradiateda
|
Relative efficiency of deletion mutant | Parental strain irradiateda
|
Relative efficiency of deletion mutant | ||||
|---|---|---|---|---|---|---|---|---|---|
| JDS10 | MR311 | Neither | JDS21 | MR103 | Neither | ||||
| 100 | 6.8 × 10−4 | 0.77 ± 0.25b (8) | 4.33 ± 2.46 (10) | 3.94 ± 0.66 (2) | 0.18 | <0.007 (8) | 9.80 ± 4.22 (6) | 2.96 ± 0.67 (4) | <0.001 |
| 150 | 5.6 × 10−5 | 1.34 ± 0.97 (8) | 4.51 ± 1.31 (8) | 2.99 ± 1.05 (8) | 0.30 | 0.007 (8) | 12.2 ± 2.6 (8) | 0.71 ± 0.45 (4) | 0.001 |
| 200 | 1.6 × 10−4 | 0.81 ± 0.41 (8) | 2.58 ± 2.64 (8) | 10.1 ± 3.2 (4) | 0.31 | 0.22 ± 0.22 (8) | 70.1 ± 64.0 (8) | 4.64 ± 1.94 (4) | 0.003 |
The decreased yields of recombinants indicate that the deletion mutants could not serve as recipients in a significant fraction of the intragenic recombination events normally observed between two viable parental cells. This result could not be attributed to an intrinsic inability of deletion mutants to serve as recipients in marker exchange, as crosses among unirradiated suspensions of deletion mutants JDS22, MR31, MR123, and MR311 under similar conditions yielded frequencies of 2 × 10−6 to 3 × 10−6 Pyr+ recombinants per CFU. The differential effect of irradiating the point mutant versus the deletion mutant therefore provides evidence that much of the intragenic recombination in these crosses involved donor DNA fragments shorter than about 300 bp. The dominant role of these short donor fragments could not be attributed to chromosome fragmentation by γ irradiation, since (i) these γ-ray doses introduce fewer than about 10 double-strand breaks per S. acidocaldarius genome (22), (ii) the differential effect of the choice of irradiated parent did not increase with radiation dose (Table 2), and (iii) the magnitude of the effect was reproducibly greater for one strain pair than for the other.
Conclusions.
Multiple, independent assays of 63 S. acidocaldarius mutant pairs, defining a total of 56 intervals in the pyrEF region ranging from 0 to 1,154 bp in length, revealed a reproducible pattern of recombination versus distance that contrasts dramatically with the systems of reciprocal recombination that have been analyzed at this level of resolution. Whereas bacteriophage T4, E. coli plasmid, Bacillus subtilis, Saccharomyces cerevisiae, and murine hybridoma cells exhibit MEPSs of about 40, 70, 70, 250, and 2,000 bp, respectively (3, 12, 14, 25, 26), no statistical support was seen for an MEPS in S. acidocaldarius ME. Furthermore, recombinant frequency remains approximately proportional to marker separation up to at least 2 kbp in bacteriophage T4, 20 kbp in E. coli, and 30 kbp in S. cerevisiae (16, 18, 26) but only to about 0.05 kbp for ME in S. acidocaldarius. Thus, the distance-frequency parameters of recombinant formation by S. acidocaldarius ME fall well outside the range established by assays of reciprocal recombination in bacteriophages, bacterial plasmids, bacterial chromosomes, and eukaryotic chromosomes.
Our current hypothesis, that intragenic recombination by ME is dominated by nonreciprocal events involving a small average patch size, remains consistent with (i) the results shown in Fig. 3, in which there is little evidence of genetic linkage in 3-factor crosses over distances of 500 to 600 bp, and (ii) the results shown in Table 1, which show that ME does not incorporate donor DNA efficiently at deletions larger than about 300 bp, despite no such impairment for deletions of 18 to 27 bp. It should be noted, however, that the average patch size suggested by Fig. 1 is well below that observed for other models of nonreciprocal recombination (5, 13, 20), making it difficult to identify extensive quantitative similarities with these systems. We also noted that the single-strand annealing mode of double-strand break repair in S. cerevisiae increases sharply in frequency with repeat length up to about 400 bp and remains constant thereafter (27), similar to the behavior we observed for S. acidocaldarius.
It must be emphasized that our results relate to the most numerous events under these conditions and do not rule out all reciprocal recombination in S. acidocaldarius. Figure 2A, for example, suggests a slight dependence on marker separation which may, in principle, reflect an increasing contribution from reciprocal events over larger distances. It must also be emphasized that S. acidocaldarius ME measures the combined effects of DNA transfer followed by homologous recombination, and it has not been feasible to resolve these two processes experimentally (24). However, both the high efficiency and small patch size of intragenic recombination via ME indicated by our analyses have important implications for the population genetics of S. acidocaldarius and for the use of homologous recombination to alter Sulfolobus genomes (29).
Acknowledgments
This work was supported by grant MCB 9733303 from the National Science Foundation.
We thank H. Boeing, Department of Nuclear Engineering, for providing the γ irradiation and J. Drake, NIEHS, for communicating unpublished results.
REFERENCES
- 1.Aagaard, C., J. Z. Dalgaard, and R. A. Garrett. 1995. Intercellular mobility and homing of an archaeal rDNA intron confers a selective advantage over intron-minus cells of Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 92:12285-12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati, P., and M. Meselson. 1964. Localized negative interference in bacteriophage λ. Genetics 51:369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, M. D., L. H. Read, B. G. Beatty, and P. Ng. 1996. Requirements for ectopic homologous recombination in mammalian somatic cells. Mol. Cell. Biol. 16:7122-7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulondre, C., and J. H. Miller. 1977. Genetic studies of the lac repressor III: additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 117:525-575. [DOI] [PubMed] [Google Scholar]
- 5.Fishel, R. A., E. C. Siegel, and R. Kolodner. 1986. Gene conversion in Escherichia coli: resolution of heteroallelic mismatched nucleotides by co-repair. J. Mol. Biol. 188:147-157. [DOI] [PubMed] [Google Scholar]
- 6.Fujitani, Y., K. Yamamoto, and I. Kobayashi. 1995. Dependence of frequency of homologous recombination on the homology length. Genetics 140:797-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghané, F., and D. W. Grogan. 1998. Chromosomal marker exchange in the archaeon Sulfolobus acidocaldarius: physiological and cellular aspects. Microbiology (Reading) 144:1649-1657. [DOI] [PubMed] [Google Scholar]
- 8.Grogan, D. W. 1996. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grogan, D. W., and R. P. Gunsalus. 1993. Sulfolobus acidocaldarius synthesizes UMP via a standard de novo pathway: results of a biochemical-genetic study. J. Bacteriol. 175:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grogan, D. W., G. T. Carver, and J. W. Drake. 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 98:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grogan, D. W., and J. E. Hansen. 2003. Molecular characteristics of spontaneous deletions in the hyperthermophilic archaeon Sulfolobus acidocaldarius. J. Bacteriol. 185:1266-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayden, M. S., and B. Byers. 1993. Minimal extent of homology required for completion of meiotic recombination in Saccharomyces cerevisiae. Dev. Genet. 13:498-512. [DOI] [PubMed] [Google Scholar]
- 13.Jinks-Robertson, S., M. Michelitch, and S. Ramcharan. 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3937-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khasanov, F. K., D. J. Zvingila, A.A Zainullin, A. A. Prozorov, and V. I. Bashirov. 1992. Homologous recombination between plasmid and chromosomal DNA in Bacillus subtilis requires approximately 70 bp of homology. Mol. Gen. Genet. 234:494-497. [DOI] [PubMed] [Google Scholar]
- 15.Leloupe, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Low, K. B. 1991. Conjugational methods for mapping with Hfr and F-prime strains. Methods Enzymol. 204:43-62. [DOI] [PubMed] [Google Scholar]
- 17.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol 162:461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortimer, R. K., D. Schild, C. R. Contopoulou, and J. A. Kans. 1989. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast 5:321-403. [DOI] [PubMed] [Google Scholar]
- 19.Norkin, L. C. 1970. Marker-specific effects in genetic recombination. J. Mol. Biol. 51:633-655. [DOI] [PubMed] [Google Scholar]
- 20.Orr-Weaver, T., and J. Szostak. 1985. Fungal recombination. Microbiol. Rev. 49:33-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reilly, M. S., and D. W. Grogan. 2001. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reilly, M. S., and D. W. Grogan. 2002. Biological effects of DNA damage in the hyperthermophilic archaeon Sulfolobus acidocaldarius. FEMS Microbiol. Lett. 208:29-34. [DOI] [PubMed] [Google Scholar]
- 23.Scheerer, J. B., and G. M. Adair. 1994. Homology dependence of targeted recombination at the Chinese hamster APRT locus. Mol. Cell. Biol. 14:6663-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt, K. J., K. Beck, and D. W. Grogan. 1999. UV-stimulation of chromosomal marker exchange in Sulfolobus acidocaldarius: implications for DNA repair, conjugation, and homologous recombination at extremely high temperatures. Genetics 152:1407-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, P., and H. V. Huang. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer, B. W., L. Gold, P. Gauss, and D. H. Doherty. 1982. Determination of the amount of homology required for recombination in bacteriophage T4. Cell 31:25-33. [DOI] [PubMed] [Google Scholar]
- 27.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watt, V. M., C. J. Ingles, M. S. Urdea, and W. J. Rutter. 1985. Homology requirements for recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:4768-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worthington, P., V. Hoang, F. Perez-Pomares, and P. Blum. 2003. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]