Abstract
This study aimed to identify DNA attributed to Hammondia heydorni oocysts in the feces of wild canids in and around an Ohio wildlife conservation center. Two hundred and eighty-five wild canid fecal samples were analyzed using PCR with melting curve analysis to detect coccidian DNA. Coccidia-positive samples were further subjected to H. heydorni-specific and N. caninum-specific PCR assays. Samples positive by the H. heydorni-specific assay were additionally analyzed with a PCR assay to distinguish H. heydorni from Hammondia triffittae. Coccidian DNA was detected in 51 of the 285 (17.9%) wildlife samples. H. heydorni DNA was detected in three of the coccidia-positive wildlife samples (1.1%) and N. caninum was detected in none. Determining the presence of H. heydorni in wild canids will contribute to a greater understanding of the role these hosts play in the ecology of this parasite.
Keywords: Hammondia, fecal, coccidia, wild canids, PCR
1. Introduction
Hammondia heydorni and Neospora caninum are closely related, tissue-cyst forming members of the Sarcocystidae family. Neospora caninum is an important cause of abortion in cattle worldwide (Dubey and Schares, 2011) and can cause severe neuromuscular disease in young dogs (Reichel et al., 2007). However, H. heydorni generally does not produce clinical signs in either definitive or intermediate hosts (Dubey et al., 2002). Both parasites use domestic and wild canids as definitive hosts and a wide variety of herbivore species as intermediate hosts (Dubey et al., 2002, 2007). Oocysts of these parasites are indistinguishable, with both species producing nearly spherical, thin-walled unsporulated oocysts approximately 11μm in diameter (Lindsay et al., 1999). Therefore, light microscopy cannot be used to distinguish N. caninum and H. heydorni oocysts shed by canids.
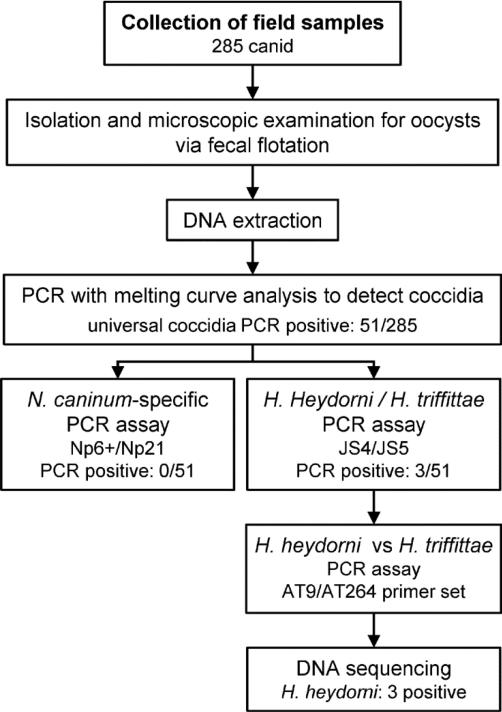
Because of the important economic impact of N. caninum on the cattle industry, much work has been done to understand the transmission dynamics of this disease. Comparatively little is known about the ecology of H. heydorni in wildlife. Surveys of free-ranging wild canids that have detected H. heydorni were unable to genetically distinguish this parasite from N. caninum (Gompper et al., 2003; Elmore et al., 2013). The goal of this study was to use a series of PCR assays to determine the presence of DNA attributed to coccidia, N. caninum or H. heydorni oocysts in wild canid fecal samples (Fig. 1). Assessing the presence of H. heydorni DNA contained in feces from wild canids will provide baseline information for better understanding the ecology of this parasite in free-ranging wildlife.
Fig. 1.
Study design flow chart. Collection of 285 canid fecal samples from the field, followed by fecal flotation for microscopic examination and isolation of parasite ova. DNA isolated from flotation material with subsequent PCR testing, first with a broad universal coccidia PCR assay whereby the 51 positive samples were further analyzed using Neospora caninum and Hammondia PCR assays with DNA sequencing of the three Hammondia heydorni positives.
2. Materials and methods
Two hundred and eighty-five wild canid fecal samples were collected in southeastern Ohio in and around a 10,000-acre private conservation center with a variety of semi-free ranging herbivores from May – August, 2014. Fecal samples were identified as wild canid feces by size, shape, and content (hair, bones, etc.). Samples were kept at 4°C until processing.
Duplicate tubes of five grams of each sample were processed by standard double centrifugation fecal flotation using Sheather's sugar solution. One tube was used microscopically examination for the presence of oocysts and the other tube had the top 2 mL of the flotation solution removed into 6 mL of 2.5% potassium dichromate to inhibit fungal and bacterial growth for storage at 4°C (7-13 months) until processed for DNA extraction (Fig. 1). To extract DNA, the flotation material was washed with phosphate buffered saline and centrifuged to remove the potassium dichromate. The resulting pellet was homogenized in 1.5 mL tubes containing ceramic beads using a Precellys® 24 tissue lyser/homogenizer (Bertin Technologies, USA). DNA was extracted using a Qiagen DNeasy® Blood & Tissue kit (Qiagen, Germany). Control oocysts of Hammondia heydorni (UCD 2011 isolate provided by P. Conrad) underwent similar processing. The Qiagen DNeasy® Blood & Tissue kit was also used to extract DNA from cell cultures of Neospora caninum (VMDL isolate, Hyun et al. 2003). Other controls for the PCR assays included DNA from the following parasites: Neospora hughesi, Besnoitia, Caryospora, Frenkelia, or S. neurona
As an initial screening tool for the presence of small coccidian oocysts, PCR with melting curve analysis was performed using the universal coccidian primer cocktail developed by Lalonde and Gajadhar (2011) to amplify a ~315 bp region of 18S rDNA. Originally, this assay was published and applied to detect Cryptosporidium parvum, Toxoplasma gondii, Cyclospora cayetanensis, and several species of Eimeria, Isospora, and Sarcocystis in food sources and clinical samples. Based on the target area and DNA sequence alignment analysis, this assay was hypothesized to be applicable to a wide range of coccidia that might be found in wild canid fecal samples. The PCR mixture contained 2X Power SYBR® green PCR Master Mix (Applied Biosystems, USA), the universal coccidian primer cocktail (500 nM Crypto-F, Crypto-R, Cyclo-F, Cyclo-R, Sarco-R, Toxo-F, and Toxo-R), 1μl template DNA, and dH2O. Assays were performed using a StepOne™ Real-Time PCR system (Applied Biosystems). The cycling conditions consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 sec and annealing and elongation at 60°C for 1 min. Melting curve analysis began immediately after the final cycle and consisted of a 15 sec hold at 95°C followed by a gradual increase from 55°C to 95°C in 0.1°C increments. DNA extracted from control H. heydorni oocysts and culture-derived control N. caninum tachyzoites were used as positive controls; distilled water served as a negative control. To evaluate the application and analytic sensitivity of the universal coccidian PCR and melting curve analysis to Hammondia and Neospora, ten-fold dilutions of 2 - 2 × 104 H. heydorni control oocysts and 2 - 2 × 105 cultured-derived N. caninum control tachyzoites underwent DNA extraction and universal coccidian amplification as described above.
The H. heydorni-specific and N. caninum-specific PCR assays were applied to the samples considered positive by the universal coccidian PCR assay, showing amplicons with a distinct melting curve and threshold peak values (melting temperature of 76°-86°C with -Rn’ values >0.1). The JS4/JS5 primers target the ITS-1 region of H. heydorni (Slapeta et al., 2002) whereas the Np6+/Np21+ primers target the Nc5 gene of N. caninum (Müller et al., 1996; Spencer et al., 2000). Both assays were performed concurrently using a StepOne™ Real-Time PCR system (Applied Biosystems). The final reaction mixtures contained 2X iQ™ SYBR® green Supermix (Bio-Rad, USA), 1μl template DNA, dH2O, and 500nM JS4 and JS5 for the H. heydorni-specific assay and 500nM Np6+ and Np21+ for the N. caninum-specific assay. The cycling conditions consisted of initial denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 30 sec and annealing and elongation at 60°C for 1 min. All assays included positive controls (H. heydorni and N. caninum DNA for the JS4/JS5 and Np6+/Np21+ respectively), dH2O and other parasite DNA as used in the universal coccidia PCR assay as a negative controls. Amplification products (7 μl) for both assays were visualized in 4% agarose gels stained with SYBR® Safe DNA gel stain (Thermo Fisher Scientific) using a Typhoon™ 9410 imaging system (Amersham Biosciences, USA) and compared to the positive controls for proper size amplification asscociated with either Neospora caninum or Hammondia identification.
There is evidence that the Hammondia organisms shed by foxes are genetically distinct from H. heydorni and constitute a separate species: H. triffittae. Gjerde and Dahlgren (2011) found the ITS-1 sequence of H. triffittae to be identical to that of H. heydorni. Therefore, the JS4/JS5 primers used in this study to detect H. heydorni are inadequate to differentiate these species. To distinguish them, the AT9/AT264 primers developed by Abel et al. (2006) were applied to the samples that tested positive with the JS4/JS5 primer set. These primers target the intron 1 region of the alpha tubulin gene which distinguishes these species on the basis of a 9 bp repetitive insertion present in H. triffittae but lacking in H. heydorni (Abel et al. 2006). The PCR mixture contained 2X Power SYBR® green PCR Master Mix (Applied Biosystems), 500 nM AT9 and AT264 primers, 1μl template DNA, and dH2O. DNA from H. heydorni control oocysts served as a positive control and dH2O served as a negative control in each assay. Assays were performed using a StepOne™ Real-Time PCR system (Applied Biosystems). The cycling conditions consisted of initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 15 sec and annealing and elongation at 56°C for 30 sec.
To confirm the identity of the amplicons, DNA sequencing was performed on the following amplified products: three JS4/JS5-positive amplicons, three AT9/AT264-positive amplicons, and three universal coccidia-positive but Neospora- and Hammondia-negative amplicons. The templates and applicable primers were submitted to The Ohio State University Plant-Microbe Genomics Facility for BigDye Terminator Cycle Sequencing on a 3730 DNA Analyzer (Applied Biosystems). The resulting sequences were aligned using Vector NTI® software (Thermo Fisher Scientific). The DNA sequences were compared to published sequences using BLAST analysis available through the NCBI database.
3. Results
Of the 285 samples examined, 59 (20.7%) showed the presence of small oocysts-like structures (10-15 um) by fecal flotation. Other parasite ova seen included Eimeria, Sarcocystis, Cystoisospora, Capillaria, Stongyle-type ova, Trichuris, and Giardia. To better identify the small oocyst-like structures, initial screening with the universal coccidia PCR assay yielded 51 coccidia-positive samples (Fig. 1). This assay was sensitive enough to detect DNA from 20 N. caninum tachyzoites and two H. heydorni oocysts, and consistently detected both H. heydorni and N. caninum controls. Hammondia heydorni and N. caninum both produced melting curve peaks at approximately 82.9°C (82.91°-82.96°C).
The H. heydorni-specific assay was performed on the 51 coccidia-positive samples. This assay was able to detect DNA from two H. heydorni oocysts and did not amplify the N. caninum control. Three of the samples (1.1%) had positive amplification above threshold (Fig. 1). Sequencing results showed 99-100% identity to the ITS-1 region of H. heydorni (GenBank GQ984218) for these three samples. These three samples also showed above threshold amplification using the AT9/AT264 primers. Sequences derived from the AT9/AT264 amplicons confirmed the presence of H. heydorni rather than H. triffittae. Comparing the sequences of the samples to H. heydorni and H. triffittae illustrates the lack of the H. triffittae-specific 9 bp repetitive insertion in these samples (Table 1).
Table 1.
Comparison of the intron 1 region of the alpha tubulin published sequences of H. triffittae (GenBank GQ984228) and H. heydorni (GenBank GQ984230) to the sequenced samples, tentatively identified as Hammondia based on JS4/JS5 PCR results. The sample sequences lack the 9 bp repetitive insertion thereby confirming their identity as H. heydorni and not H. triffittae.
| Sample | Nucleotides sequenced | Identity | Sequence (3’-5’) |
|---|---|---|---|
| HtRtVulpes-1981-N (H. triffittae) | GenBank GQ984228 | -- | ATGTGTGCATGGAGTGCATGGTAAA |
| HhCanis-2000-1-N (H. heydorni) | GenBank GQ984230 | -- | ATGTGTGCATGG– – – – – – – – –TAAA |
| Wild canid 1 | 162 | 97% H. heydorni | ATGTGTGCATGG– – – – – – – – –TAAC |
| Wild canid 2 | 136 | 98% H. heydorni | ATGTGTGCATGG– – – – – – – – –TAAG |
| Wild canid 3 | 146 | 99% H. heydorni | ATGTGTGCATGG– – – – – – – – –TAAG |
The N. caninum-specific assay was also performed on the 51 coccidia-positive samples. All samples were negative for N. caninum (Fig. 1). This assay was able to detect DNA from two N. caninum tachyzoites and did not amplify the H. heydorni control.
To further confirm the lack of Neospora and Hammondia in the universal PCR positive sample group, three samples were selected for sequencing with the DNA sequencing results indicating the presence of a Sarcocystis sp., an Eimeria sp., and a Cystoisospora sp. (Table 2), thereby confirming the broad application of the universal primers to the coccidia group of parasites.
Table 2.
Sequencing results of the coccidia-positive, H. heydorni- and N. caninum-negative wildlife samples (coccidia + samples 1-3) indicate the presence of a variety of other coccidia species which seen by microscopy and amplified using the coccidia universal primers yet no Hammondia spp or Neospora caninum DNA detected.
4. Discussion
The low detection of H. heydorni DNA in wild canid feces in this study is similar to previous surveys. Gompper et al. (2003) reported a prevalence of 2% in coyote feces in New York, and Elmore et al. (2013) found a single Neospora/Hammondia-like positive sample out of 95 arctic fox feces in Canada (1.1%). However, neither of these studies confirmed the identity of these oocysts using molecular methods. Evaluation of dogs also show very low prevalence: 0.8% in dogs in China (Jie et al., 2013), 0.05% in dogs in Germany (Schares et al., 2005), and 0.2% in dogs in the Czech Republic (Slapeta et al., 2002). Li et al. (2014) reported a seemingly unusual high prevalence in dogs in rural China (17.5%).
Since each fecal sample in this study was collected from the environment and not from individual animals, the proportion of H. heydorni-positive samples reported here does not represent a true prevalence but rather estimates environmental contamination of this parasite within the study area. All three H. heydorni-positive samples had Neospora/Hammondia-like oocysts visualized microscopically, so the positive DNA amplification seen likely derives from oocysts rather than tissue cyst stages present in intermediate host prey passing through the intestines of the wild canids.
The limited sequencing results from the three coccidia-positive but H. heydorni- and N. caninum-negative samples suggest that the 48 fecal samples that were coccidia-positive but negative for H. heydorni and N. caninum could harbor a number of other coccidia species that use canids as definitive hosts such as Sarcocystis or Cystoisospora. It is also possible that these coccidia-positive samples represent felid-specific coccidian oocysts such as T. gondii and Hammondia hammondi which contaminated the samples through consumption of felid feces or were domestic cat feces that were visually similar to canid feces and mis-identified. Other coccidian parasites not known to infect canids likely were detected as a result of tissue cyst stages or gastrointestinal coccidia present in the prey animals, passing through the gut of the predator and into the collected samples as several of the microscopy results found sporulated Eimeria. These results highlight that universal primers although good for amplifying a number of coccidia for screening purposes, this assay should be followed up with specific DNA detection methods if species identification is needed. This study also highlights the limitations of visual microscopy for field collected samples and the need for confirmatory testing by another testing modality such as PCR followed by DNA sequencing of the amplicon.
Although it is to-date considered non-pathogenic and low in prevalence, more fully understanding the ecology of H. heydorni may provide useful information in the future. For example, its interactions with other gastrointestinal parasites could potentially impact the transmission of these pathogens. Rynkiewicz et al. (2015) suggest parasites that share space and resources and alter the immune environment that other parasites experience. Rynkiewicz et al. (2015) also suggest that these co-habiting parasites will likely face a high level of competition, citing the relationship between the nematode Heligmosomoides polygyrus and the coccidian Eimeria hungaryensis in the intestines of wild wood mice as an example. Because of their similarities in life cycles and biological niches, it is possible that H. heydorni could be acting as a competitor with other more pathogenic endoparasites, thereby negatively affecting their transmission. However, Moreno et al. (2013) found a synergistic effect on oocyst shedding in wild capybaras and guanacos co-infected with multiple Eimeria species. Further studies are needed to better understand the potential effects of multi-parasite infections and competition on transmission.
Additionally, H. heydorni may serve as a sentinel of climate change and habitat loss, with alterations in precipitation and water runoff impacting the spread of oocysts and thereby the transmission and prevalence of this parasite. Transmission via surface water has been previously well documented for other gastrointestinal protozoan parasites. Miller et al. (2002) identified freshwater runoff as a likely cause of movement of T. gondii oocysts from inland felid feces to coastal waters in California. Surface water runoff from melting snow has also been implicated in the spread of T. gondii oocysts from land to sea in arctic habitats (Simon et al. 2013). The salinity and vegetation of wetlands has been shown to significantly remove fecal pathogens such as T. gondii, C. parvum, and Giardia duodenalis and prevent their spread via surface water (Hogan et al., 2013), but loss of these habitats removes this barrier to transmission. Hammondia heydorni likely behaves in a manner similar to these other protozoal parasites and too may serve as an indicator of environmental changes, especially in areas with large populations of dogs or wild canids and susceptible prey animals.
Acknowledgments
We would like to thank Emily Morehouse, Kat Rossos, Pallavi Oruganti, and Cathy Bremer for assisting with collection and processing of the fecal samples; the staff at The Wilds and the residents of our study area for allowing us to collect samples on their properties; Dr. Josh Daniels at OSU-VMC for his guidance with the Applied Biosystems light cycler; and Dr. Patricia Conrad at the University of California-Davis School of Veterinary Medicine for providing canine Hammondia (UCD 2011) control oocysts. Funding for this project was provided by The Ohio State University Department of Veterinary Preventive Medicine, the Columbus Zoo and Aquarium Cooperative Grant Program, Duncan Alexander (Geneva, IL) and NIH T35 Training Grant OD010977.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Abel J, Schares G, Orzeszko K, Gasser RB, Ellis JT. Hammondia isolated from dogs and foxes are genetically distinct. Parasitology. 2006;132:187–19-2. doi: 10.1017/S0031182005008814. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Barr BC, Barta JR, Bjerkas I, Bjorkman C, Blagburn BL, Bowman DD, Buxton D, Ellis JT, Gottstein B, Hemphill A, Hill DE, Howe DK, Jenkins MC, Kobayashi Y, Koudela B, Marsh AE, Mattsson JG, McAllister MM, Modry D, Omata Y, Sibley LD, Speer CA, Trees AJ, Uggla A, Upton SJ, Williams DJL, Lindsay DS. Redescription of Neospora caninum and its differentiation from related coccidia. Int. J. Parasitol. 2002;32:929–94-6. doi: 10.1016/s0020-7519(02)00094-2. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Schares G, Ortega-Mora LM. Epidemiology and control of neosporosis and Neospora caninum. Clin. Microbiol. Rev. 2007;20:323–36-7. doi: 10.1128/CMR.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Schares G. Neosporosis in animals—The last five years. Vet Parasitol. 2011;180:90–10-8. doi: 10.1016/j.vetpar.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Elmore SA, Lalonde LF, Samelius G, Alisauskas RT, Gajadhar AA, Jenkins EJ. Endoparasites in the feces of arctic foxes in a terrestrial ecosystem in Canada. Int. J. Parasitol. Parasites Wildl. 2013;2:90–9-6. doi: 10.1016/j.ijppaw.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerde B, Dahlgren SS. Hammondia triffittae n. comb. of foxes (Vulpes spp.): biological and molecular characteristics and differentiation from Hammondia heydorni of dogs. Parasitology. 2011;138:303–32-1. doi: 10.1017/S0031182010001265. [DOI] [PubMed] [Google Scholar]
- Gompper ME, Goodman RM, Kays RW, Ray JC, Fiorello CV, Wade SE. A survey of the parasites of coyotes (Canis latrans) in New York based on fecal analysis. J. Wildl. Dis. 2003;39:712–71-7. doi: 10.7589/0090-3558-39.3.712. [DOI] [PubMed] [Google Scholar]
- Hogan JN, Daniels ME, Watson FG, Oates SC, Miller MA, Conrad PA, Shapiro K, Hardin D, Dominik C, Melli A, Jessup DA, Miller WA. Hydrologic and vegetative removal of Cryptosporidium parvum, Giardia lamblia, and Toxoplasma gondii surrogate microspheres in coastal wetlands. Appl. Environ. Microbiol. 2013;79:1859–186-5. doi: 10.1128/AEM.03251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun C, Gupta GD, Marsh AE. Sequence comparison of Sarcocystis neurona surface antigen from multiple isolates. Vet. Parasitol. 2003;112:11–2-0. doi: 10.1016/s0304-4017(02)00392-8. [DOI] [PubMed] [Google Scholar]
- Jie HJ, Yu M, Fen YY, Mei GY, Yan Y, Esch GW, QingTuan F. First isolation of Hammondia heydorni from dogs in China. Vet. Parasitol. 2013;197:43–4-9. doi: 10.1016/j.vetpar.2013.04.035. [DOI] [PubMed] [Google Scholar]
- Lalonde LF, Gajadhar AA. Detection and differentiation of coccidian oocysts by real-time PCR and melting curve analysis. J. Parasitol. 2011;97:725–73-0. doi: 10.1645/GE-2706.1. [DOI] [PubMed] [Google Scholar]
- Li J, He P, Yu Y, Du L, Gong P, Zhang G, Zhang X. Detection of Neospora caninum-DNA in feces collected from dogs in Shenyang (China) and ITS1 phylogenetic analysis. Vet. Parasitol. 2014;205:361–36-4. doi: 10.1016/j.vetpar.2014.06.036. [DOI] [PubMed] [Google Scholar]
- Lindsay DS, Upton SJ, Dubey JP. A structural study of the Neospora caninum oocyst. Int. J. Parasitol. 1999;29:1521–152-3. doi: 10.1016/s0020-7519(99)00121-6. [DOI] [PubMed] [Google Scholar]
- Miller MA, Gerdner IA, Kreuder C, Paradies DM, Worcester KR, Jessup DA, Dodd E, Harris MD, Ames JA, Packham AE, Conrad PA. Coastal freshwater runoff is a risk factor for Toxoplasma gondii infection of southern sea otters (Enhydra lutris nereis). Int. J. Parasitol. 2002;32:997–1006. doi: 10.1016/s0020-7519(02)00069-3. [DOI] [PubMed] [Google Scholar]
- Müller N, Zimmermann V, Hentrich B, Gottstein B. Diagnosis of Neospora caninum and Toxoplasma gondii infection by PCR and DNA hybridization immunoassay. J. Clin. Microbiol. 1996;34:2850–285-2. doi: 10.1128/jcm.34.11.2850-2852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno PG, Eberhardt MAT, Lamattina D, Previtali MA, Beldomenico PM. Intra-phylum and inter-phyla associations among gastrointestinal parasites in two wild mammal species. Parasitol. Res. 2013;112:3295–330-4. doi: 10.1007/s00436-013-3509-x. [DOI] [PubMed] [Google Scholar]
- Spencer JA, Witherow AK, Blagburn BL. A random amplified polymorphic DNA polymerase chain reaction technique that differentiates between Neospora species. J. Parasitol. 2000;86:1366–136-8. doi: 10.1645/0022-3395(2000)086[1366:ARAPDP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Reichel MP, Ellis JT, Dubey JP. Neosporosis and hammondiosis in dogs. J. Small Anim. Pract. 2007;48:308–31-2. doi: 10.1111/j.1748-5827.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- Rynkiewicz EC, Pedersen AB, Fenton A. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends Parasitol. 2015;31:212–22-1. doi: 10.1016/j.pt.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Schares G, Pantchev N, Barutzki D, Heydorn AO, Bauer C, Conraths FJ. Oocysts of Neospora caninum, Hammondia heydorni, Toxoplasma gondii and Hammondia hammondi in faeces collected from dogs in Germany. Int. J. Parasitol. 2005;35:1525–153-7. doi: 10.1016/j.ijpara.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Simon A, Rousseau AN, Savary S, Bigras-Poulin M, Ogden NH. Hydrological modelling of Toxoplasma gondii oocysts transport to investigate contaminated snowmelt runoff as a potential source of infection for marine mammals in the Canadian Arctic. J. Environ. Manage. 2013;127:150–16-1. doi: 10.1016/j.jenvman.2013.04.031. [DOI] [PubMed] [Google Scholar]
- Slapeta JR, Koudela B, Votypka J, Modry D, Horejs R, Lukes J. Coprodiagnosis of Hammondia heydorni in dogs by PCR based amplification of ITS-1 rRNA: Differentiation from morphologically indistinguishable oocysts of Neospora caninum. Vet. J. 2002;163:147–154. doi: 10.1053/tvjl.2001.0599. [DOI] [PubMed] [Google Scholar]