Abstract
CodY of Lactococcus lactis MG1363 is a transcriptional regulator that represses the expression of several genes encoding proteins of the proteolytic system. These genes include pepN, pepC, opp-pepO1, and probably prtPM, pepX, and pepDA2, since the expression of the latter three genes relative to nitrogen availability is similar to that of the former. By means of in vitro DNA binding assays and DNase I footprinting techniques, we demonstrate that L. lactis CodY interacts directly with a region upstream of the promoter of its major target known so far, the opp system. Our results indicate that multiple molecules of CodY interact with this promoter and that the amount of bound CodY molecules is affected by the presence of branched-chain amino acids and not by GTP. Addition of these amino acids strongly affects the extent of the region protected by CodY in DNase I footprints. Random and site-directed mutagenesis of the upstream region of oppD yielded variants that were derepressed in a medium with an excess of nitrogen sources. Binding studies revealed the importance of specific bases in the promoter region required for recognition by CodY.
Genetic and biochemical research over the past decades has led to a clear picture of the proteolytic system of lactic acid bacteria. To ensure a proper nitrogen balance, several regulators are present that respond to changes in intracellular concentrations of nitrogen-containing compounds (16, 24, 28, 30). The lactic acid bacterium Lactococcus lactis is auxotrophic for several amino acids (6). For optimal growth in milk, it has to degrade milk proteins (e.g., αS1 and β- and κ-casein), because only limited amounts of free amino acids are present in this environment. An elaborate proteolytic system to release amino acids from casein, involving a number of enzymatic activities that are subject to medium-dependent regulation, has evolved in L. lactis. Casein degradation by L. lactis is a process that can be divided into three successive steps (25, 51). First, the extracellular cell wall-bound serine proteinase (PrtP) liberates peptides of various sizes from casein. In the second step, the casein-derived peptides are transported into the cell by the oligopeptide transport system (Opp) or by the di- and tripeptide transport systems (DtpP and DtpT, respectively) (45). In the last step, the internalized peptides are degraded into smaller peptides and free amino acids by a large number of cytoplasmic peptidases. Two major groups of peptidases can be discerned: the endopeptidases (e.g., PepO and PepF), which perform endolytic hydrolysis of their substrates, and the aminopeptidases (e.g., PepN, PepX, and PepC), which cleave off one or two amino acids from the free N termini of their substrates (51).
Transcription of a number of lactococcal genes encoding the proteins that constitute the proteolytic system is regulated similarly in response to peptide availability in the medium (17). Transcriptional luxAB fusions with the promoters of a number of peptidase, protease, and transporter genes were used to show that these genes are down-regulated in peptide-rich medium. More recently, it has been demonstrated that, at least for transcription of the oligopeptide permease genes encoded by opp, a homologue of the nutritional repressor CodY of Bacillus subtilis is responsible for this peptide-dependent regulation (18). Evidence was found that the strength of repression by L. lactis CodY correlated with the intracellular pool of branched-chain amino acids (BCAAs) (18, 39). These findings are supported by observations that the growth in milk of an L. lactis strain lacking the aminotransferases AraT and BcaT, which are involved in the catabolism of BCAAs (42, 55), is severely affected when isoleucine (Ile) or a dipeptide containing this amino acid is added. Since the growth rate of an L. lactis codY mutant was not altered by addition of Ile, inhibition by this amino acid is probably due to CodY-mediated repression of the proteolytic system, which leads to retarded growth (5, 36). In fact, it has been shown recently that in the gram-positive bacterium B. subtilis, BCAAs directly interact with CodY and enhance the affinity for its targets (48). CodY was first identified in this organism, where it serves as a nutritional repressor of the dipeptide permease operon (47, 49) and of genes involved in amino acid metabolism (7, 12, 13, 54), carbon and energy metabolism (23), motility (1), antibiotic production (21), and competence development (38, 46). In addition, the affinity of B. subtilis CodY for its targets is stimulated by its interaction with the cofactor GTP, independently of that with the BCAAs (1, 23, 41, 48). The regulator is thereby thought to sense both the energy state and the nitrogen availability of the cell. In contrast, recent observations imply that lactococcal CodY probably does not respond to GTP, since addition of decoyinine to the medium, which evokes a rapid drop in intracellular GTP levels, did not result in derepression of a CodY target gene (39).
CodY contains a C-terminal helix-turn-helix motif, and the B. subtilus protein has been shown to bind to sequences overlapping the −35 and −10 sequences of its target promoters (13, 47). Although the binding of CodY to several targets has been demonstrated, no consensus recognition sequence for CodY binding has been deduced (12). The present study aims to improve our understanding of L. lactis CodY by studying its binding to one of its DNA targets, the opp region. In order to examine whether repression by CodY occurs by direct protein-DNA interactions, DNA binding and DNase I footprinting studies were performed. By combining a random and a site-directed mutagenesis approach, we show the importance of several nucleotides in the promoter region of opp for recognition by CodY.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in tryptone-yeast extract (TY) medium (Difco Laboratories, West Molesey, United Kingdom) at 37°C with vigorous agitation or on TY medium (2) solidified with 1.5% agar, containing 100 μg of erythromycin per ml where needed. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at a final concentration of 40 μg/ml. L. lactis was grown at 30°C in 0.5× M17 broth (50) or on 0.5× M17 medium solidified with 1.5% agar and supplemented with 0.5% glucose (GM17). When appropriate, erythromycin, chloramphenicol, and X-Gal were added at final concentrations of 5, 5, and 80 μg per ml, respectively. Antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.). A chemically defined medium (CDM) was prepared as described previously (29) and supplemented with Casitone (Difco Laboratories) as a nitrogen source where indicated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. cremoris | ||
| MG1363 | Lac− Prt−; plasmid-free derivative of NCDO712 | 15 |
| LL108 | Cmr; MG1363 derivative containing multiple copies of the pWV01 repA gene in the chromosome | 32 |
| LL302 | RepA+ MG1363, carrying one copy of the pWV01 repA gene in the chromosome | 32 |
| NZ9000 | MG1363 pepN::nisRK | 27 |
| NZ9700 | Nisin-producing transconjugant of MG1363 containing the nisin-sucrose transposon Tn5276 | 26 |
| E. coli EC101 | Kmr; JM101 with repA from pWV01 integrated into the chromosome | 31 |
| Plasmids | ||
| pNZ8048 | Cmr; inducible expression vector carrying PnisA | 27 |
| pNH6CodY | his6-codY of L. lactis MG1363 behind PnisA | This work |
| pORI13 | Emr; integration vector; promoterless lacZ; Ori+ RepA derivative of pWV01 | 44 |
| pORIopp68 | Emr; pORI13 carrying a 160-bp oppD promoter fragment amplified with primers opp1 and opp2 | This work |
| pORIopp111 | Emr; pORI13 carrying a 203-bp oppD promoter fragment amplified with primers opp1 and opp11 | This work |
| pORIopp162 | Emr; pORI13 carrying a 254-bp oppD promoter fragment amplified with primers opp1 and opp3 | This work |
| pORIopp14 | Emr; pORI13 carrying a 170-bp oppD promoter fragment amplified with primers opp1 and opp14 | This work |
| pORIopp15(a) | Emr; pORI13 carrying a 170-bp oppD promoter fragment amplified with primers opp1 and opp15(a) | This work |
| pORIopp15(b) | Emr; pORI13 carrying a 170-bp oppD promoter fragment amplified with primers opp1 and opp15(b) | This work |
DNA preparation, molecular cloning, and transformation.
Routine DNA manipulations were performed as described elsewhere (43). Total chromosomal DNA from L. lactis MG1363 was extracted as described previously (33). Plasmid DNA was isolated by the alkaline lysis procedure as described elsewhere (43). Minipreparations of plasmid DNA from E. coli and L. lactis were essentially made by using the High Pure plasmid isolation kit from Roche Molecular Biochemicals (Mannheim, Germany). Restriction enzymes and T4 DNA ligase were purchased from Roche Molecular Biochemicals. PCR amplifications were carried out using either Pwo DNA polymerase for cloning purposes or Taq DNA polymerase (both from Roche Molecular Biochemicals) for checking plasmid DNA insert sizes from transformants. PCR products were purified with the High Pure PCR product purification kit (Roche Molecular Biochemicals). Electrotransformations of E. coli and L. lactis were performed using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) as described previously (9, 20).
RNA preparation and primer extension.
The opp transcript was subjected to primer extension analysis using oligonucleotide sto14 (see Table 2) essentially as described previously (4). In the reactions, 30 μg of total RNA that was isolated from L. lactis MG1363 cells as described previously (52) was used as a template. A DNA sequence ladder was obtained by using the T7 sequencing kit (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's descriptions.
TABLE 2.
Oligonucleotides used in this study
| Name> | Sequence (5′→3′)a |
|---|---|
| hc-5 | CTAGACCACCATGGGGCATCACCATCACCATCACGTGGCTACATTACTTGAAAAAACACG |
| hc-6 | CTAGTCTAGATTAGAAATTACGTCCAGCAAGTTTATC |
| opp1 | GCTCTAGACACTCACTTGTTTTGCTTCC |
| opp2 | AACTGCAGGAAAATTCATGAACATACC |
| opp3 | AACTGCAGTAAAACAATAATAAAAGCAG |
| opp11 | AACTGCAGCTCCAAAACTTTTGCTTTAC |
| opp14 | AACTGCAGCGTAATGTTCAGAAAATTC |
| opp15(a) | AACTGCAGCGTAATATTTAGAAAATTCATGAACATACC |
| opp15(b) | AACTGCAGCGTACTGTGCCGAAAATTCATGAACATACC |
| sto14 | CTTGCCATGGAATCACCCG |
Restriction enzyme sites are underlined. Italicized sequence in hc-5 encodes the hexahistidine tag. Italicized sequence in hc-6 is the stop codon.
Cloning of oppD promoter fragments.
Oligonucleotides used to amplify oppD promoter fragments are listed in Table 2. Combinations of oligonucleotide opp1 with oligonucleotides opp2, opp3, opp11, opp14, opp15(a), and opp15(b) were used to amplify chromosomal DNA from L. lactis MG1363, in order to obtain various fragments encompassing the oppD promoter. The PCR products were digested with XbaI and PstI and were transcriptionally fused upstream of the promoterless lacZ gene in the pORI13 integration vector (44) digested with the same enzymes. The resulting plasmids, pORIopp68, pORIopp162, pORIopp111, pORIopp14, pORIopp15(a), and pORIopp15(b), respectively, were all isolated by using E. coli EC101, which contains a chromosomal copy of the lactococcal repA gene, needed for pORI13 replication, as the cloning host. After isolation, the plasmids were introduced by electroporation into L. lactis LL302 and/or L. lactis LL108, which contain single and multiple chromosomal copies of repA, respectively, to facilitate replication of the pORI13 derivatives (32).
Random mutagenesis of the oppD promoter region.
PCR fragments encompassing the oppD promoter region containing random base pair substitutions were obtained by using the Diversify PCR random mutagenesis kit (Clontech Laboratories, San Jose, Calif.). L. lactis MG1363 chromosomal DNA was used as a template in the amplification step. Subsequently, the variants obtained were cloned into plasmid pORI13 and introduced into L. lactis LL108 as described above. Mutants showing differential blue coloring (compared to the unmutated oppD region cloned into pORI13) on GM17 plates containing X-Gal were selected and analyzed in more detail as described below.
Determination of β-galactosidase activity in L. lactis.
Overnight cultures of L. lactis grown in GM17 were washed twice in 0.9% NaCl before inoculation to 1% in 50 ml of the appropriate medium containing erythromycin for the maintenance of pORI13 in L. lactis LL108 or LL302. Exponential-phase cells (optical density at 600 nm, ∼1.0) were collected by centrifugation. β-Galactosidase activities were determined in permeabilized cell suspensions as described previously (22). β-Galactosidase enzyme activities, calculated as averages from three independent experiments, were expressed in arbitrary units (37).
Purification of histidine-tagged CodY.
The codY gene of L. lactis MG1363 was amplified from the chromosome by PCR with oligonucleotides hc-5, introducing the NcoI restriction enzyme site upstream of the sequence encoding the hexahistidine tag, and hc-6, introducing the XbaI restriction enzyme site downstream of the stop codon of the resulting His-tagged codY, designated H6-codY. The purified 833-bp PCR product was digested with NcoI and XbaI and ligated into the corresponding sites in pNZ8048, resulting in pNH6CodY. This plasmid was introduced into L. lactis NZ9000 to enable nisin-induced expression from PnisA upstream of H6-codY, as described previously (8, 27). Following induction, H6-CodY was isolated from lysates of induced cells by affinity chromatography in a procedure involving fast-performance protein liquid chromatography (Amersham Pharmacia Biotech) using nickel-nitrilotriacetic acid agarose (QIAGEN GmbH, Hilden, Germany). The concentration of the purified protein was determined by the Bradford procedure (3).
Electrophoretic mobility shift assays (EMSAs).
Gel retardation experiments were carried out essentially as described elsewhere (10). Purified PCR products (2 μg) were end labeled with polynucleotide kinase (Amersham Pharmacia Biotech) for 1 h at 37°C by using 30 μCi of [γ-32P]ATP (Amersham Pharmacia Biotech) in a volume of 20 μl. Reactions were stopped by incubating the mixtures for 10 min at 70°C. Binding studies were carried out in 20-μl reaction volumes containing 20 mM Tris-HCl (pH 8.0), 8.7% (vol/vol) glycerol, 1 mM EDTA (pH 8.0), 5 mM MgCl2, 100 mM KCl, 0.5 mM dithiothreitol, labeled DNA fragment (3,000 cpm), and purified H6-CodY protein (50 to 400 ng). Bovine serum albumin (1 μg) and poly(dI-dC) (Amersham Pharmacia Biotech) were added to the reaction mixtures in order to reduce nonspecific interactions. After incubation for 15 min at 30°C, samples were loaded onto a 4% polyacrylamide gel. Electrophoresis was performed in the Protean II Minigel system (Bio-Rad Laboratories B.V., Veenendaal, The Netherlands) by using a gradient (0.5× to 2×) of Tris-acetate-EDTA buffer (43) at 150 V for 1.5 h. Gels were dried and used for autoradiography at −80°C by using Kodak XAR-5 film and intensifying screens.
DNase I footprinting analysis.
DNase I footprinting was performed essentially according to the description supplied with the Sure Track footprinting kit (Amersham Pharmacia Biotech). The DNA fragments were prepared by PCR using Expand DNA polymerase (Amersham Pharmacia Biotech) and oligonucleotides opp1 and opp3, one of which was first end labeled with T4 polynucleotide kinase (Amersham Pharmacia Biotech) and [γ-32P]ATP as described by the manufacturer. Binding reactions were identical to those used in EMSAs, in a total volume of 40 μl and in the presence of approximately 150,000 cpm of the DNA probe. Subsequently, DNase I footprinting experiments were performed as described previously (19).
RESULTS
Determination of the TSS of oppD.
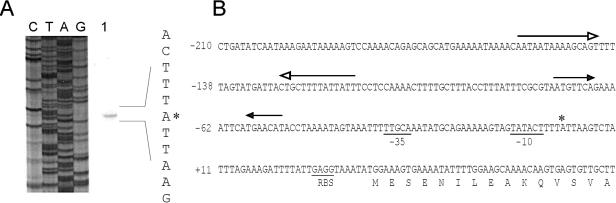
It has been demonstrated that the genes of the oligopeptide permease system, carried on the oppDFBCA-pepO1 locus of L. lactis MG1363, are transcribed polycistronically (17, 51). Upstream of both oppD and oppA are regions that could serve as promoter elements. To precisely determine the location of the promoter upstream of oppD, the transcription start site (TSS) was determined. The opp transcript was analyzed by primer extension using RNA that was isolated from exponentially growing MG1363 cells (Fig. 1A). The mRNA 5′ end corresponds to an adenine residue located 35 bases upstream of the translation start codon (AUG) of oppD. The −35 sequence (TTGCAA) is separated by a consensus 17 bp from the −10 region (TATACT), and a proper lactococcal ribosome binding site (RBS), with the sequence GAGG, is also present (Fig. 1B). Although the distance between the −10 region and the TSS is several base pairs shorter than those for most other lactococcal promoters (53), efficient transcription initiation is allowed, as evidenced by the fact that expression from PoppD was readily detectable (see Fig. 2D). The sequence upstream of oppD contains two regions of dyad symmetry centered on positions −135 (−14.0 kcal/mol) and −62 (−5.6 kcal/mol) relative to the oppD TSS, respectively.
FIG. 1.
Overview of the region upstream of oppD of L. lactis MG1363. (A) Primer extension analysis of PoppD. Primer extension reactions were performed with 30 μg of RNA isolated from exponentially growing L. lactis MG1363 in GM17 and primer sto14. The sequencing ladder (CTAG) and the extension reaction product (lane 1) are shown. The position of the transcription start site, in a part of the sequence given in the right margin, is marked by an asterisk. (B) Detailed view of the region upstream of oppD. The −35, −10, and RBS sequences are underlined. The positions of two regions of dyad symmetry are indicated by arrows with open or closed arrowheads. The position of the transcription start site is marked by an asterisk.
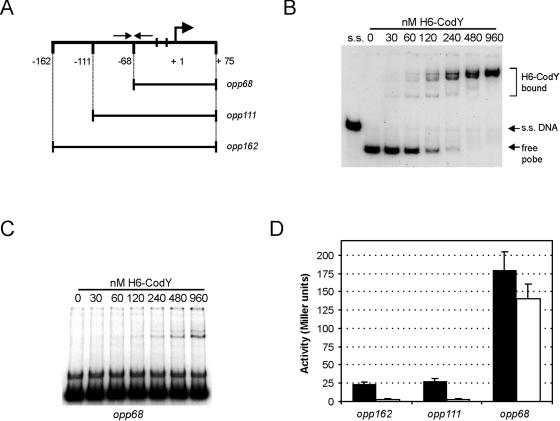
FIG. 2.
Interaction of H6-CodY with fragments of the oppD promoter region. (A) Schematic view of the probes used in panels B and C. Probes opp162, opp111, and opp68 were obtained by PCR on chromosomal DNA of L. lactis MG1363, using a combination of oligonucleotide opp1 with oligonucleotide opp3, opp11, or opp2, respectively. Arrows indicate the position of the region of dyad symmetry closest to the −35 sequence (see Fig. 1B). Vertical bars indicate the positions of the −35 and −10 sequences. Nucleotide positions are relative to the transcriptional start site (right-turn arrow at +1). (B) Interaction of H6-CodY with the upstream region of oppD. The radioactively labeled probe opp162 was incubated alone or with increasing amounts of purified H6-CodY. The first lane contains the same probe, which was boiled in a 95% formamide solution in order to obtain single-stranded (s.s.) DNA fragments. (C) H6-CodY binding to probe opp68. The same conditions were used as for probe opp162 (B). (D) In vivo activities of PoppD variants. L. lactis LL108 strains carrying lacZ reporter plasmids fused to the opp fragments depicted schematically in panel A were grown in CDM containing either 0.2 or 2% Casitone (solid and open bars, respectively). Cells were harvested in the exponential phase of growth, and β-galactosidase activity was measured. Assays were carried out twice, in triplicate each time. Error bars, standard deviations.
H6-CodY interacts with the region upstream of oppD.
In order to examine whether L. lactis CodY directly interacts with its main target known so far, the opp-pepO1 region, in vitro DNA binding studies were performed using probes spanning PoppD. For this purpose, CodY carrying a histidine tag at its N terminus (H6-CodY) was overexpressed in L. lactis by using the nisin-inducible gene expression system (8) and was subsequently purified to apparent homogeneity (data not shown). A 254-bp radioactively labeled DNA probe (opp162), spanning the oppD promoter from position −162 to +75 relative to the TSS, was prepared by PCR as described in Materials and Methods (Fig. 2A). EMSAs clearly showed that purified H6-CodY was capable of binding directly to this fragment (Fig. 2B). Multiple retarded fragments were present, indicating that several molecules of H6-CodY can bind to the oppD fragment. When the amount of H6-CodY was gradually increased from 0 to almost 1,000 nM, at least three distinct bands could be discerned, possibly reflecting a multimeric state of the protein or binding of CodY to multiple independent binding sites. In most of our DNA binding experiments, a band corresponding to the single-stranded probe (Fig. 2B) was observed irrespective of the presence of H6-CodY. The occurrence of this denatured DNA probably resulted from the high AT content of the opp promoter region. No H6-CodY binding occurred when DNA fragments with similar AT contents that were obtained from internal gene segments (e.g., from comG of B. subtilis) were used, indicating that H6-CodY binding to PoppD is specific (data not shown).
To get a better indication of which part of the region upstream of oppD is important for interaction with CodY, deletion analysis was performed. Two truncated fragments, shortened from the 5′ end of the oppD upstream region contained in opp162, were obtained by PCR and examined for H6-CodY binding (Fig. 2A and C). Probe opp111, spanning the region from −111 to + 75, was bound by H6-CodY similarly to opp162 (data not shown), suggesting that nucleotides that are critical for CodY binding must reside downstream of the 5′ end of this probe. When a 160-bp DNA fragment (probe opp68), the 5′ end of which coincides with the center of the inverted repeat (IR) closest to the RBS of oppD (Fig. 1A), was used, binding of H6-CodY was almost completely abolished, since the probe would shift only at high concentrations of protein and labeled DNA, and fewer molecules of H6-CodY appeared to bind (Fig. 2A and C).
To verify that the in vitro binding of the three probes by CodY reflected the in vivo situation, the fragments were fused upstream of the promoterless lacZ gene in plasmid pORI13 (44) and introduced into L. lactis LL108. Subsequently, β-galactosidase activities in cells growing exponentially in media differing in their peptide contents were determined. In CDM supplemented with 2% Casitone, where strong CodY-mediated repression is ensured (18), expression driven from promoter constructs derived from the two longer probes (opp162 and opp111) seemed to be strongly repressed, in contrast to the behavior of the strain carrying the opp68-derived reporter fusion, which displayed approximately 30- to 40-fold-higher expression. In CDM containing 0.2% Casitone, where CodY-mediated repression is strongly relieved, expression from the opp162 and opp111 fragments was derepressed about ninefold whereas regulation of the opp68 fragment was much lower. Thus, the strength of regulation of PoppD-driven expression seemed to correlate with the observed binding pattern (Fig. 2D), suggesting that at least part of an operator site for CodY must be located in a region between positions −111 and −68 relative to the oppD TSS.
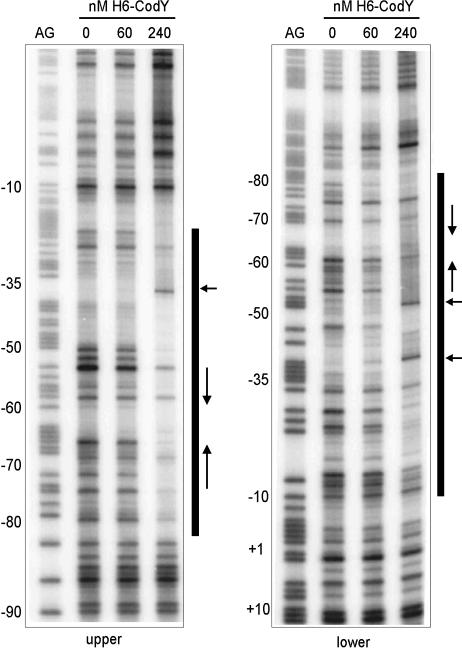
H6-CodY protects an extended region of the oppD promoter.
Although the EMSA experiments using truncated PoppD DNA fragments pointed to a specific region that is important for CodY binding, the actual area facilitating binding was rather large (positions −68 to −111). Moreover, the assays did not exclude the possibility that more downstream sequences could contribute to CodY binding. Therefore, formation of a complex by H6-CodY and the oppD promoter region was investigated by DNase I footprinting experiments using the labeled promoter fragment opp162 and the binding conditions used for the EMSAs (Fig. 3). H6-CodY binding resulted in the protection of bases extending from position −80 to −20 and position −80 to −10 relative to the TSS of the upper and lower strands, respectively. In addition to these protected regions, both DNA strands contained hypersensitive sites when they were incubated with DNase I in the presence of H6-CodY. These results show that H6-CodY binds to a region encompassing the −35 to −10 sequences of the promoter of oppD.
FIG. 3.
DNase I footprinting analysis of H6-CodY binding to the oppD promoter region. The left and right panels show the footprints of the upper and lower strands, respectively. Footprints, obtained in the absence or presence of 60 or 240 nM H6-CodY by using radioactively labeled probe opp162, are flanked by Maxam and Gilbert A+G sequence ladders (AG) on the left. Numbers on the left indicate base pair positions relative to the TSS. Protected regions are marked with bars, and horizontal arrows indicate the positions of hypersensitive bonds. Vertical arrows indicate the region of dyad symmetry closest to the −35 sequence (see Fig. 1B).
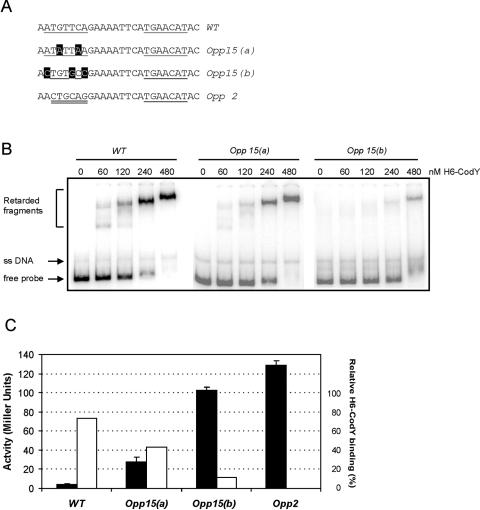
A region in the oppD promoter containing an IR is important for CodY-mediated regulation.
A closer inspection of the area in the oppD promoter to which CodY binds revealed the presence of a short sequence (ATGTTCA) that is inversely repeated, with a spacing of 9 bp between the partners (Fig. 1). This region of dyad symmetry is located 18 bp upstream of the −35 sequence and is entirely present in probes opp162 and opp111, to which H6-CodY binds (Fig. 2A and B). The fact that H6-CodY was hardly able to form a protein-DNA complex when a probe lacking the upstream arm of this IR (probe opp68 [Fig. 2A and C]) was used implies that this area might serve as an operator site for CodY on PoppD. To study this region in more detail, site-directed mutations were introduced by PCR using combinations of oligonucleotide opp1 with oligonucleotide opp14 (wild type), opp15(a), opp(15b), or opp2 (Fig. 4A). The PCR products were inserted upstream of the promoterless lacZ reporter gene in pORI13 and introduced into L. lactis LL108. Introducing base substitutions into the upstream arm of the repeat [opp15(a) and opp15(b)] resulted in both weaker binding of H6-CodY (Fig. 4B) and derepression of PoppD-driven lacZ expression (Fig. 4C). When the unchanged half-site was replaced by an unrelated sequence (an XbaI endonuclease site, present in opp2), repression was reduced approximately 30-fold (in cells growing exponentially in a rich medium) and H6-CodY binding was almost completely abolished (as shown in Fig. 2C). Converting both the C and G residues in this region to adenines [opp15(a)] resulted in a ∼7-fold reduction of repression, whereas changing 3 out of 6 bases [opp15(b)] led to a >20-fold derepression of expression. These results were in accordance with those obtained from gel retardation analyses (Fig. 4B), where the affinity of H6-CodY was highest for the wild-type probe, intermediate for the opp15(a) probe, and lowest for the opp15(b) probe. In the case of the opp15(b) probe, hardly any protein-DNA complexes were present and all intermediate complexes that were observed with the wild-type and opp15(a) fragments were absent.
FIG. 4.
Site-directed mutagenesis of the region of dyad symmetry closest to the RBS in the oppD promoter region. (A) Three variants of PoppD that contain mutations in the left arm of the IR (underlined) were obtained by PCR and cloned upstream of lacZ in pORI13 as described in Materials and Methods. The WT fragment contains no substitutions; variants opp15(a) and opp15(b) contain 2- and 3-bp substitutions, respectively (highlighted). In opp2 an XbaI endonuclease site (double underlined) preceded by two adenine residues replaced the left arm. (B) EMSA using H6-CodY and the indicated radioactively labeled PoppD variants. Binding reactions were performed as described in the legend to Fig. 2. (C) Promoter activities of PoppD variants. L. lactis LL108 strains carrying the lacZ reporter plasmids were grown in GM17. Cells were harvested in the exponential phase of growth, and β-galactosidase activity was measured (solid bars). The experiments were carried out in triplicate. Error bars, standard deviations. Open bars, quantitative representation of the DNA binding assay for which results are shown in panel B; the relative binding affinities of H6-CodY for the PoppD variants were calculated by comparing the intensity of the shifted, H6-CodY bound complexes with the total radioactive signal in each lane in the presence of 240 nM protein.
Random mutation analysis of the oppD promoter area.
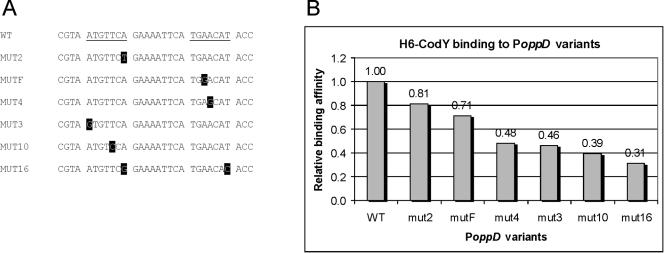
As shown above, a region close to the −35 sequence seems to be important for the binding of CodY to the oppD regulatory region. Since we could not find any obviously similar sequences in the upstream regions of any of the other CodY-regulated genes identified so far, it is possible that structural determinants are required for CodY recognition of a promoter region. Therefore, PCR-based random mutagenesis was carried out on the smallest oppD promoter fragment still showing strong repression of transcription by CodY (probe opp111 [Fig. 2]). PCR fragments containing random base pair substitutions or small deletions were restricted by using the appropriate restriction enzymes and ligated upstream of the promoterless lacZ in pORI13. The ligation mixture was used to transform L. lactis LL108. Cells carrying a pORI13 construct with a PoppD fragment containing a derepressing mutation formed light-blue or blue colonies on CDM plates containing X-Gal and excess nitrogen sources (in the form of Casitone), as opposed to the whitish color of colonies formed by cells harboring pORI13 with wild-type PoppD. The oppD fragments in cells with a derepressed phenotype were amplified by PCR, radioactively labeled, and tested by an EMSA for their abilities to form complexes with H6-CodY. The relative strength of H6-CodY binding was examined by comparing the amount of the retarded mutated DNA fragment resulting from H6-CodY binding to that of the unchanged PoppD fragment (Fig. 5). Weaker binding to all of the mutated promoter fragments was observed. Sequence analyses of the PoppD variants obtained in this study revealed that all of them carried one or more base pair substitutions, at least one of which was located in the region from −82 to −56 relative to the TSS of oppD. This finding, again, is an indication of the importance of this region for CodY binding.
FIG. 5.
H6-CodY binding to derepressed variants of PoppD. (A) Positions in PoppD of the base pair substitutions (highlighted) that led to distorted repression by CodY relative to the wild-type (WT) fragment. Underlining marks the inverted repeat closest to the −35 sequence of PoppD. (B) Binding of H6-CodY to labeled PCR products encompassing the mutated promoter regions presented in panel A in an in vitro binding assay, relative to its binding to the wild-type PoppD fragment. The relative affinity was calculated by comparing the amount of H6-CodY required to shift 50% of the labeled DNA in the binding assay.
BCAAs stimulate stimulate H6-CodY binding.
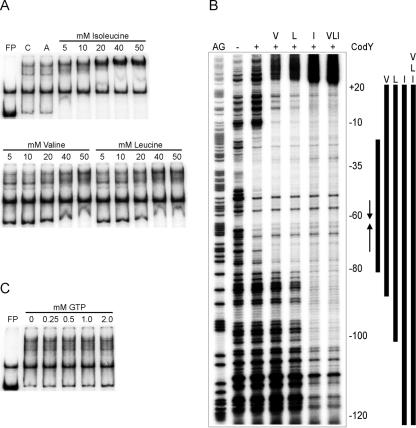
Evidence has been presented that CodY senses the nitrogen supply of the cell as a function of the BCAA pool (18, 39). Although BCAAs act as direct effectors of CodY activity in B. subtilis (48), the exact nature of this signal in L. lactis remains to be established. Therefore, in vitro DNA binding of H6-CodY was examined in the presence or absence of the three BCAAs Val, Leu, and Ile (Fig. 6A). Addition of any of the three BCAAs resulted in increased binding of H6-CodY to the PoppD probe opp162 compared to a situation in which no amino acid or another, aliphatic amino acid (i.e., alanine) was present. Stimulation of H6-CodY binding by Val and Leu was observed at concentrations of these amino acids above 10 mM. In a titration of Ile and a constant amount of H6-CodY, most of the probe DNA was already retarded in the presence of 5 mM Ile, showing that this BCAA stimulates the binding of H6-CodY to the highest extent.
FIG. 6.
EMSA and DNase I footprinting analysis of the effects of BCAAs and GTP on H6-CodY binding to PoppD. EMSA reactions were carried out as described in the legend to Fig. 2B in the presence of probe opp162 and 120 nM H6-CodY and with varying concentrations of (A) BCAAs or (C) GTP. All reactions contain H6-CodY except lanes FP, which contain free probe. Lane A contains 50 mM Ala. (B) Footprinting reactions were carried out as described in the legend to Fig. 3 in the absence (−) or presence (+) of 120 nM H6-CodY and 20 mM Val, Leu, Ile, or a combination of these three (lanes V, L, I, and VLI, respectively). A Maxam and Gilbert A+G sequence ladder is present on the left (AG). Numbers on the right indicate base pair positions relative to the TSS. Protected regions are indicated by bars. The vertical arrows indicate the region of dyad symmetry closest to the −35 sequence (see Fig. 1B).
The role of BCAAs in CodY binding was also investigated by means of DNase I footprinting experiments, in which the opp162 probe was incubated with H6-CodY either alone; in the presence of 20 mM Val, Leu, or Ile; or with a combination of these three BCAAs (Fig. 6B). Addition of any of these BCAAs resulted in extended protection of PoppD. As in the EMSAs, the effect of Ile addition was most severe. These results show that the binding of lactococcal CodY to the target region of oppD is directly stimulated by BCAAs and that, as in B. subtilis, Ile is most effective.
Similarly, we also tested whether GTP could stimulate the binding of H6-CodY to lactococcal PoppD, since GTP serves as an effector molecule that enhances the binding of B. subtilis CodY to a number of its targets. As can be seen from Fig. 6C, the binding of L. lactis H6-CodY was not affected by the presence of GTP at a range of concentrations between 0.25 and 2.0 mM. These results are in good agreement with recent evidence showing that lactococcal CodY activity is independent of GTP at physiological concentrations, which do not exceed 0.55 mM in a medium containing Casitone (39).
DISCUSSION
L. lactis possesses a number of genes whose products are involved in the utilization of proteins present in milk, such as an extracellular protease, peptide transport systems, and intracellular peptidases (24). Most of these genes have been cloned and sequenced, and their enzymes have been biochemically characterized (24). Recently, it has been established that CodY plays a pleiotropic role in the regulation of a number of these genes in response to nitrogen availability (17, 18).
The present study was conducted to gain insight into the role of CodY in the regulation of the opp system and, more specifically, into the molecular interactions between the cis site and CodY. By combining data from in vivo and in vitro experiments, we have clearly demonstrated that repression by CodY is mediated by direct binding of this protein to the oppD promoter region. For this to occur most efficiently, binding of several molecules of CodY is probably required, since several protein-DNA complexes were discerned in all EMSAs in which variants (in size or base composition) of this promoter fragment were retarded by H6-CodY. Formation of multiple retarded DNA fragments was also observed in EMSAs using B. subtilis CodY with promoter DNA fragments of the B. subtilis dipeptide permease operon, dpp (47). Similar results were obtained in EMSAs that we performed using lactococcal H6-CodY with probes encompassing upstream regions of the peptidase genes pepN and pepC (data not shown) and an intergenic region containing the divergently transcribed promoters of prtP and prtM (14), which encode the proteinase and proteinase maturase of L. lactis, respectively (34, 35).
The observations that several molecules of CodY are able to interact with target DNA (Fig. 2), together with the DNase I footprinting data (Fig. 3) showing that a region encompassing the −35 area of the promoter is protected, lead to a tentative model in which CodY binding is thought to prevent access of RNA polymerase to the promoter, thereby hampering transcription initiation. DNA binding studies using reconstituted lactococcal RNA polymerase would help to elucidate such a mechanism.
As mentioned, the intracellular pool of BCAAs exerts an influence on the activity of CodY in L. lactis, but the exact nature of this signal remains to be elucidated (18). In addition, as was also demonstrated for B. subtilis CodY, lactococcal CodY appears to bind quite well to its targets in the absence of any cellular components in in vitro DNA binding studies, although we cannot fully exclude the possibility that cofactors were copurified with H6-CodY. Here we show that BCAAs alone could function as cofactors for CodY activity, since the binding properties of H6-CodY are greatly altered by the addition of any of these amino acids (Fig. 6). Ile in particular strongly enhances the binding of H6-CodY to PoppD, which suggests that the intracellular pool of this amino acid is important for the modulation of CodY activity. Recently, intracellular concentrations of BCAAs have been determined in exponentially growing cells (40). It was shown that when Casitone was added to the growth medium, the BCAA concentration increased to almost 10 mM and CodY-mediated repression occurred. These data are consistent with our DNA binding assays, which show that CodY binding is stimulated by BCAAs at this concentration (Fig. 6) and support the view that these amino acids might directly activate lactococcal CodY (39). Thus, B. subtilis and L. lactis respond similarly to intracellular BCAA levels.
Whereas GTP, which is a marker of the energy state of the cell, has a modulating effect in B. subtilis on the activity of CodY by enhancing its affinity for its targets (1, 23, 38, 41), such a stimulatory effect on the L. lactis repressor seems to be absent. In our in vitro DNA binding experiments, using probes encompassing PoppD, addition of GTP did not enhance binding of CodY (Fig. 6), a result in agreement with the observation that a drop in the intracellular GTP level does not result in derepression of a CodY target gene (39). These results support the possibility that L. lactis CodY does not sense the energy state of the cell, unlike its B. subtilis counterpart. It is tempting to speculate that this would explain why, thus far, the L. lactis CodY regulon seems to comprise only genes involved in nitrogen metabolism, while B. subtilis CodY appears to serve as a factor that couples nitrogen to carbon metabolism. However, DNA binding experiments using promoter DNA fragments of other members of the lactococcal CodY regulon will have to be performed, since it is possible that, as in B. subtilis, GTP does stimulate L. lactis CodY binding to some of its other targets (38).
Although several CodY-regulated genes of B. subtilis and L. lactis have been described, a consensus binding site, if any, remains to be elucidated for both regulators (11, 47, 49). It is likely that the two proteins, which share 67% similarity on the amino acid level (18), recognize similar binding sites, since we observed using EMSAs (data not shown) that purified lactococcal H6-CodY was capable of binding to the upstream region of B. subtilis comK, which is a known, direct member of the B. subtilis CodY regulon (46). Part of the present study was aimed at gaining insight into the sequence requirements for L. lactis CodY recognition of its target promoters. Random and site-directed mutagenesis revealed that a region required for recognition of a promoter area by the CodY repressor, at least in the case of PoppD, contains an inversely repeated nucleotide sequence. Therefore, it would be tempting to speculate that this 7-bp IR of the nucleotides ATGTTCA is needed for CodY binding, since regions of dyad symmetry often serve as operator sites for transcriptional regulators.
A sequence comparison of the upstream regions of all known CodY-regulated promoters in L. lactis revealed that a subset of these (e.g., pepC, pepN, araT, and prtPM) contain an area of dyad symmetry close to their (predicted) promoters. However, these repeats do not seem to have any obvious mutual sequence similarity, and therefore it is possible that they do not serve a role in CodY-mediated regulation. As postulated (47), it could also be that CodY does not recognize a specific nucleotide stretch but that, rather, a topological structure (e.g., bent DNA) in its targets is required for binding.
Further mutational analysis would be of great importance in gaining a better understanding of the sequence requirements for CodY binding. In addition, DNA microarray experiments are currently being performed in order to identify possible new members of the lactococcal CodY regulon, which would provide more cis sequence information as well.
Acknowledgments
We are grateful to Anne de Jong for skillful technical assistance in determining the transcription start site of oppD.
This work was funded by a BTS grant (SENTER) to Friesland Coberco Dairy Foods.
REFERENCES
- 1.Bergara, F., C. Ibarra, J. Iwamasa, J. C. Patarroyo, R. Aguilera, and L. M. Marquez-Magana. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J. Bacteriol. 185:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswal, N., A. K. Kleinschmidt, H. C. Spatz, and T. A. Trautner. 1967. Physical properties of the DNA of bacteriophage SP50. Mol. Gen. Genet. 100:39-55. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Buist, G., H. Karsens, A. Nauta, D. van Sinderen, G. Venema, and J. Kok. 1997. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl. Environ. Microbiol. 63:2722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambellon, E., and M. Yvon. 2003. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids. Appl. Environ. Microbiol. 69:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-37. [DOI] [PubMed] [Google Scholar]
- 7.Débarbouillé, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebbole, D. J., and H. Zalkin. 1989. Interaction of a putative repressor protein with an extended control region of the Bacillus subtilis pur operon. J. Biol. Chem. 264:3553-3561. [PubMed] [Google Scholar]
- 11.Eda, S., T. Hoshino, and M. Oda. 2000. Role of the DNA sequence downstream of the Bacillus subtilis hut promoter in regulation of the hut operon. Biosci. Biotechnol. Biochem. 64:484-491. [DOI] [PubMed] [Google Scholar]
- 12.Fisher, S. H. 1999. Regulation of nitrogen metabolism in. Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, S. H., K. Rohrer, and A. E. Ferson. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajic, O. 2003. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 15.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guedon, E., E. Jamet, and P. Renault. 2002. Gene regulation in Lactococcus lactis: the gap between predicted and characterized regulators. Antonie Leeuwenhoek 82:93-112. [PubMed] [Google Scholar]
- 17.Guédon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guédon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 19.Hamoen, L. W., A. F. Van Werkhoven, J. J. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 21.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 22.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kok, J., and G. Buist. 2003. Genetics of proteolysis in Lactococcus lactis, p. 189-224. In B. J. B. Wood and W. M. de Vos (ed.), Genetics of lactic acid bacteria. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 25.Kok, J., and W. M. de Vos. 1993. The proteolytic system of lactic acid bacteria, p. 169-210. In M. J. Gasson and W. M. de Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Blackie Academic and Professional, London, United Kingdom.
- 26.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 27.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 28.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 29.Kunji, E. R., I. Mierau, B. Poolman, W. N. Konings, G. Venema, and J. Kok. 1996. Fate of peptides in peptidase mutants of Lactococcus lactis. Mol. Microbiol. 21:123-131. [DOI] [PubMed] [Google Scholar]
- 30.Larsen, R., G. Buist, O. P. Kuipers, and J. Kok. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 33.Leenhouts, K. J., J. Kok, and G. Venema. 1991. Replacement recombination in Lactococcus lactis. J. Bacteriol. 173:4794-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marugg, J. D., W. Meijer, R. van Kranenburg, P. Laverman, P. G. Bruinenberg, and W. M. de Vos. 1995. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J. Bacteriol. 177:2982-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marugg, J. D., R. van Kranenburg, P. Laverman, G. A. Rutten, and W. M. de Vos. 1996. Identical transcriptional control of the divergently transcribed prtP and prtM genes that are required for proteinase production in Lactococcus lactis SK11. J. Bacteriol. 178:1525-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mierau, I., E. R. Kunji, K. J. Leenhouts, M. A. Hellendoorn, A. J. Haandrikman, B. Poolman, W. N. Konings, G. Venema, and J. Kok. 1996. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J. Bacteriol. 178:2794-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613-621. [DOI] [PubMed] [Google Scholar]
- 40.Petranovic, D., and I. Mijakovic. 2004. Photometric assay for measuring the intracellular concentration of branched-chain amino acids in bacteria. J. Microbiol. Methods 56:133-136. [DOI] [PubMed] [Google Scholar]
- 41.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rijnen, L., S. Bonneau, and M. Yvon. 1999. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 65:4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Sanders, J. W., G. Venema, J. Kok, and K. Leenhouts. 1998. Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 257:681-685. [DOI] [PubMed] [Google Scholar]
- 45.Sanz, Y., F. C. Lanfermeijer, P. Renault, A. Bolotin, W. N. Konings, and B. Poolman. 2001. Genetic and functional characterization of dpp genes encoding a dipeptide transport system in Lactococcus lactis. Arch. Microbiol. 175:334-343. [DOI] [PubMed] [Google Scholar]
- 46.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843-852. [DOI] [PubMed] [Google Scholar]
- 48.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 49.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 50.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tynkkynen, S., G. Buist, E. Kunji, J. Kok, B. Poolman, G. Venema, and A. Haandrikman. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175:7523-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Asseldonk, M., A. Simons, H. Visser, W. M. de Vos, and G. Simons. 1993. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J. Bacteriol. 175:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Guchte, M., J. Kok, and G. Venema. 1992. Gene expression in Lactococcus lactis. FEMS Microbiol. Rev. 8:73-92. [DOI] [PubMed] [Google Scholar]
- 54.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]