Abstract
The stator of the sodium-driven flagellar motor of Vibrio alginolyticus is a membrane protein complex composed of four PomA and two PomB subunits. PomB has a peptidoglycan-binding motif in the C-terminal region. In this study, four kinds of PomB deletions in the C terminus were constructed. None of the deletion proteins restored motility of the ΔpomB strain. The PomA protein was coisolated with all of the PomB derivatives under detergent-solubilized conditions. Homotypic disulfide cross-linking of all of the deletion derivatives through naturally occurring Cys residues was detected. We conclude that the C-terminal region of PomB is essential for motor function but not for oligomerization of PomB with itself or PomA.
Bacteria can swim by rotating their helical flagellar filaments. The flagellar motor embedded in the cytoplasmic membrane generates torque to turn the flagellum. Energy for rotation is provided by the electrochemical gradient of protons or sodium ions across the cytoplasmic membrane (4, 20, 25, 27, 42). Proton-driven flagellar rotation in Escherichia coli requires the motor proteins MotA and MotB (35). MotA and MotB are cytoplasmic membrane proteins with four and one transmembrane (TM) segments, respectively (12, 13, 36, 45). MotA and MotB form a proton-channel complex and serve as a stator. Successive conformational changes that accompany proton conduction drive the rotation of the MS ring/FliG complex, which comprises the rotor of the flagellar motor (6, 24, 37). A variety of evidence suggests that 8 to 12 torque generators, composed of the MotA/MotB complex, are placed in a circular array around the rotor (5, 8, 22).
Of the sodium-driven motors, the polar flagella of Vibrio species have been the most extensively studied (27, 42). Most of the flagellar genes encode proteins that are similar to those of the proton-driven flagella. Four membrane proteins, PomA, PomB, MotX, and MotY, are essential for torque generation of the sodium-driven motor (for reviews, see references 27 and 42 and the references therein). PomA and PomB are homologs of MotA and MotB, respectively (1), and function as a sodium channel complex (33). PomA and PomB each form homodimers and assemble into a heterohexameric complex containing four PomA and two PomB subunits (34, 43). Thus, PomB associates both with PomA and itself.
MotA and MotB of E. coli support motility of the polar flagellum of Vibrio cholerae and of Vibrio alginolyticus, and certain chimeric motors containing components from both the proton-driven and sodium-driven types function in E. coli (2, 18). These lines of evidence support the hypothesis that the rotation mechanisms of the two types of motors are similar. On the other hand, MotX and MotY, which localize to the outer membrane (30), are unique to the sodium-driven motor. The functions of MotX and MotY are not yet clear.
Gram-negative bacteria, such as E. coli and V. alginolyticus, have a thin but rigid peptidoglycan layer that maintains cell integrity. E. coli MotB and V. alginolyticus PomB and MotY have conserved sequences in their C termini that resemble motifs present in peptidoglycan-associated proteins, such as OmpA and Pal (1, 14, 26, 31) (Fig. 1A). Some paralyzed and slow-swimming mutations in motB alter residues in or near this putative peptidoglycan-binding region (7, 14, 38). Amino acid substitutions and deletions in the putative peptidoglycan-binding region might also influence other functions of PomB (MotB), such as the interactions with PomA and PomB. In the present study, a series of C-terminal deletions in PomB was constructed and characterized.
FIG. 1.
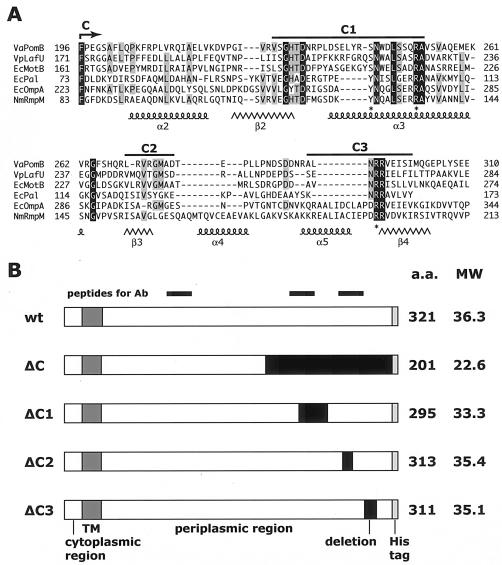
Sequence alignments of the C-terminal regions and the C-terminal deletion derivatives of PomB. (A) Multiple amino acid alignments of V. alginolyticus PomB C-terminal region with PomB homologs and other peptidoglycan-associated proteins. The alignments were constructed by using the Clustal W program (version 1.4). White letters with a black background and black letters with a gray background indicate amino acid residues conserved in all six proteins and in more than four of the six proteins, respectively. The C region (Phe-196 to Gln-315) and the subregions C1 (Val-227 to Ala-252), C2 (Leu-270 to Asp-277), and C3 (Ala-291 to Ile-300) are shown above the sequence. VaPomB, V. alginolyticus PomB; EcMotB, E. coli MotB; EcOmpA, E. coli OmpA; EcPal, E. coli Pal, VpLafU, V. parahaemolyticus LafU; NmRmpM, N. meningitidis RmpM. Amino acid residues with asterisks are proposed to contact peptidoglycan directly (19). Coiled and careted lines indicate α-helices and β-sheets, respectively, as determined for the RmpM protein (19). (B) Schematic diagram of PomB deletion proteins. White areas indicate the N-terminal cytoplasmic region and the C-terminal periplasmic region. Dark gray indicates the TM segment. Light gray indicates the hexahistidine sequence attached to the C terminus. The black boxes indicate deleted residues. The three black bars (top) indicate the regions corresponding to the three peptide sequences used for antibody production. Residue numbering and the predicted molecular weights (MW; in thousands) of the PomB derivatives are shown at the right. wt, wild type; Ab, antibody; a.a., amino acid.
Sequence conservation between PomB and its homologs in the C-terminal region and construction of deletion mutant proteins.
The C-terminal periplasmic region of MotB of E. coli is thought to be involved in interactions with the cell wall (12), and De Mot and Vanderleyden (14) reported conservation between sequences in the C-terminal half of MotB and in outer membrane proteins, such as Pal or OmpA of E. coli, that are known to be associated with peptidoglycan (Fig. 1A). Many missense mutations that inactivate MotB also affect this conserved region. Alignments of sequences at the C termini of V. alginolyticus PomB, E. coli MotB, and Vibrio parahaemolyticus LafU show especially significant conservation. In PomB, which has 315 amino acid residues, these highly conserved sequences include GHTD (from Gly-229 to Asp-232), SNWXLSXXRA (from Ser-243 to Ala-252), VXGM (from Val-272 to Met-275), and NRR (from Asn-293 to Arg-295). Some of these sequences are also conserved in Pal and OmpA (Fig. 1A).
To study the role of the C-terminal conserved regions of PomB (hereafter, the C region), we arbitrarily divided it into subregions C1 (Val-227 to Ala-252), C2 (Leu-270 to Asp-277), and C3 (Ala-291 to Ile-300). We constructed four pomB deletion mutants, ΔC1, ΔC2, ΔC3, and ΔC (ΔPhe-196 to Gln-315), corresponding to the subregions, and the properties of the mutated proteins were assessed (Fig. 1B).
DNA manipulations were carried out according to standard procedures (32). An E. coli strain was used for DNA manipulations (41). The pom genes on the plasmids constructed in this study were expressed from the lac promoter in pSU41 (3). Plasmid pNH21 (pomBΔC) was constructed by PCR with pSK603 (23) as a template. It contains the sequence 5′-(GGT)GCA(CAC)CAT(CAC)CAT(CAC)CAT(TAA)GAGCTC-3′ at the 3′ end of the pomB open reading frame. This sequence encodes Gly194-Ala195-His6 followed by a termination codon (the SacI site is underlined). Plasmid pNH22 (pomBΔC1) was constructed by PCR with pKJ201 (39) as a template and has the sequence 5′-(GTA)CGC(GTA)TCA-3′at the deleted region. This sequence codes for Val225-Arg226-Val253-Ser254 (the MluI site is underlined). Plasmids pNH23 and pNH24 (pomBΔC2 and pomBΔC3) were constructed in the same way as pNH22, with PomBΔC2 [5′-(CAA)CGT(ACG)GAG-3′ (Gln268-Arg269-Thr278-Glu279; the BsiWI site is underlined)] and PomBΔC3 [5′-(AAT)CGC(ATG)CAA-3′ (Asn289-Arg290-Met301-Gln302; the SphI site is underlined)]. The pomB genes containing the various deletions were transferred to pKJ301, which carries pomA and pomB with a His6 tag (39), to make plasmids carrying both pomA and the pomB derivatives: pNH31, pomA-pomBΔC; pNH32, pomA-pomBΔC1; pNH33, pomA-pomBΔC2; and pNH34, pomA-pomBΔC3.
Stability and cross-link formation of PomB derivatives.
To test whether the deletion variants were stable, the amount of PomB produced in the presence and absence of coexpressed PomA was compared for each protein. The study used strains NMB192 (Rifr, Laf−, ΔpomB) (40) and NMB191 (Rifr, Laf−, ΔpomAB) (44), which are derivatives of the lateral-flagellar-defective strain of V. alginolyticus, VIO5 (Rifr, Laf−) (31). Plasmids encoding the PomB deletion proteins (pNH21, pNH22, pNH23, and pNH24) (Fig. 1B) were introduced into the ΔpomB strain NMB192 containing the compatible plasmid pMMB206 (28), which carries the lacI gene, by electroporation as described previously (21). Plasmids encoding both wild-type PomA and the PomB derivatives (pNH31, pNH32, pNH33, and pNH34) were introduced into the ΔpomAB strain NMB191 containing pMMB206. The cells were cultured overnight at 30°C on Vibrio complex (VC) medium (0.5% tryptone, 0.5% yeast extract, 0.4% K2HPO4, 3% [513 mM] NaCl, and 0.2% glucose) containing 100 μg of kanamycin/ml and 2.5 μg of chloramphenicol/ml and then diluted in fresh VPG500 medium (1% tryptone, 0.4% K2HPO4, 500 mM NaCl, and 0.5% [wt/vol] glycerol) containing antibiotics.
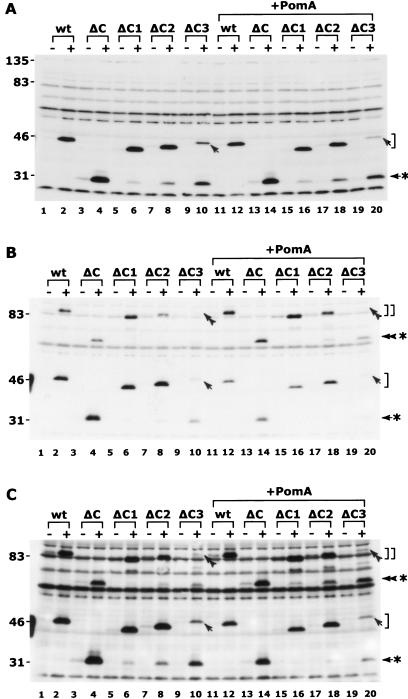
Similar amounts of PomB derivatives were produced in cells with and without coexpression of PomA at levels above those encoded by the chromosomal pomA gene (Fig. 2A). All of the derivatives migrated more slowly than expected from the amino acid sequences during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In the ΔC1, ΔC2, and ΔC3 PomB preparations, a ca. 30-kDa band with mobility similar to that of the ΔC protein was detected. For ΔC3, the 30-kDa band was predominant rather than the full-length ΔC3 protein, which was detected at low levels. These 30-kDa bands (except the ΔC band) were not detected in the sample bound to the affinity resin through the C-terminal His tag (see Fig. 4), suggesting that these bands do not have their normal C termini. All of the mutant PomB proteins were isolated in the membrane fractions (data not shown), and the amounts of the PomA protein found with the different PomB derivatives were almost the same (data not shown).
FIG. 2.
Expression of PomB derivatives and their cross-linked dimers. The NMB192(ΔpomB)/pMMB206 strain harboring plasmids carrying the pomB derivatives (lanes 1 to 10), or the NMB191(ΔpomAB)/pMMB206 strains harboring plasmids carrying wild-type (wt) pomA and the pomB derivatives (lanes 11 to 20) were cultivated in liquid VPG500 medium until mid-log phase (optical density at 660 nm of ∼1.0). When necessary, IPTG (1 mM final concentration) was added 2 h prior to harvest. Whole-cell extracts prepared as described previously (40) were dissolved in SDS-PAGE loading buffer containing 5% (vol/vol) 2-mercaptoethanol or 2 mM N-ethylmaleimide and subjected to reducing 12%-acrylamide SDS-PAGE (A) and nonreducing 14%-acrylamide SDS-PAGE (B and C). In the case of the 14% gel, the run time was prolonged until the 31-kDa band of the Kaleidoscope protein standards (Bio-Rad Laboratories) reached the bottom of the slab. Immunoblotting was performed by using anti-PomB serum (PomB93), as described previously (44). Panel C is a prolonged exposure of the gel panel B to enhance the signals. The single square bracket and the double square bracket indicate the positions corresponding to monomer and dimer forms, respectively, of the wild-type, ΔC1, ΔC2, and ΔC3 PomB proteins. Asterisks with an arrow and a double arrowhead indicate the positions of monomer and dimer forms, respectively, of the ΔC protein and a degradation product of PomB. The bands corresponding to the ΔC3 protein and its dimer form are indicated by an arrow and a double arrowhead, respectively. Monomer and dimer forms of degraded ΔC1, ΔC2, and ΔC3 appeared to be similar in size to those of ΔC. The ratio of disulfide-bonded PomB dimer to the monomer increased upon coexpression of PomA (+PomA).
FIG. 4.
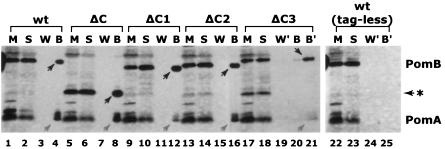
Association of PomB deletion proteins with PomA. The membrane preparation and cosedimentation procedures to assess the association of the PomB derivatives with PomA were performed essentially as described elsewhere (15). Membranes (M fraction) of V. alginolyticus VIO5 (pom+) expressing the PomB derivatives were solubilized with 2.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate; S fraction}. The solubilized materials were incubated with Ni-charged nitrilotriacetic acid-agarose, followed by extensive washing (W fraction). The beads were then treated with SDS-loading buffer (B fraction). All four fractions were subjected to SDS-PAGE, followed by immunoblotting with anti-PomA and anti-PomB. The W′ and B′ fractions were loaded in fivefold-higher amounts than W and B, respectively. The bands derived from PomA and PomB are indicated by gray and black arrows, respectively. The asterisk indicates the stable PomB degradation product (see text). Neither PomB nor PomA was detected in the bound fraction of wild-type (wt) PomB without the His tag (lane 25). All of the PomB derivatives with His tags were detected in the bound fraction.
A study of deletion mutants of Salmonella MotB suggested that some of the residues immediately before position 270, which corresponds to the portion deleted in ΔC3, are involved in maintenance of protein structure (29). The ΔC3 protein was unstable: a 30-kDa band, which corresponds to a C-terminally degraded product, was detected with higher intensity than full-length ΔC3. The corresponding MotB deletion protein, Δ(261-270), is nonfunctional and semistable, and the mutation is recessive. These properties are consistent with those of the ΔC3 protein.
PomB is unstable in the absence of PomA (44). In the present study, however, no apparent differences in the amount of PomB were observed with or without coexpression of PomA. Such an inconsistency is probably due to differences in the expression systems; i.e., proteins were expressed constitutively in the previous study, whereas those in the present study were expressed transiently by the addition of inducer.
Wild-type PomB forms cross-linked dimers via naturally occurring N-terminal cytoplasmic Cys residues, suggesting that the functional unit of PomB is a dimer (9, 40, 43). Cross-linked dimers were detected in all of the PomB proteins subjected to nonreducing SDS-PAGE (Fig. 2B and C). The dimer bands of ΔC2 and ΔC3 were faint, whereas significant proportions of the proteins seemed to be present as monomers (Fig. 2B and C, lanes 8, 10, 18, and 20). The deletions in the C2 and C3 regions may result in some defects in the homotypic PomB-PomB interaction.
The dimer forms of all of the PomB derivatives, including the wild type, increased with respect to the monomer upon coexpression of PomA. According to one recent model for the arrangement of transmembrane segments in the (MotA2/MotB)2 or (PomA2/PomB)2 complex, eight transmembrane segments of MotA or PomA surround the two transmembrane segments of the MotB or PomB dimer (4, 10). It is thus reasonable that coexpression of PomA might affect the geometry and/or stability of the PomB dimer.
Motility of cells with PomB derivatives.
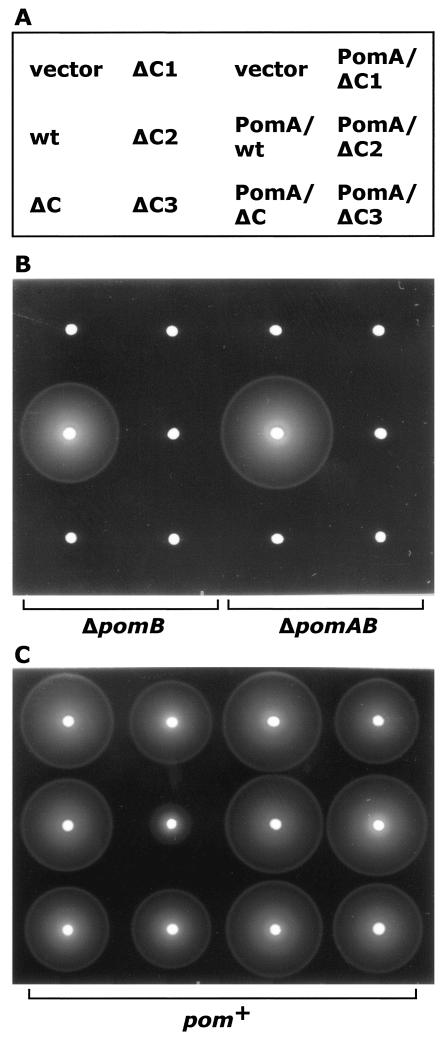
We investigated the swarming abilities of strains expressing the PomB derivatives with or without coexpression of wild-type PomA from the plasmid (Fig. 3). None of the PomB derivatives supported swarming in either case (Fig. 3B), nor did the strains swim in liquid medium (data not shown).
FIG. 3.
Swarms of the strains expressing the PomB deletion proteins. (A) Pattern of strains spotted. wt, wild type. (B) Aliquots (1 μl) of overnight cultures of V. alginolyticus NMB192 (ΔpomB; left) harboring plasmids carrying the pomB derivatives in VC medium were spotted on soft agar VPG500 plates containing 1 mM IPTG and 0.26% agar, 100 μg of kanamycin/ml, and 2.5 μg of chloramphenicol/ml and incubated at 30°C for 8 h. Similarly, V. alginolyticus NMB191 (ΔpomAB; right) harboring plasmids carrying wild-type pomA and the pomB derivatives were spotted. (C) V. alginolyticus VIO5 (pom+) harboring plasmids carrying the pomB derivatives (left) or wild-type pomA and the pomB derivatives (right).
None of the PomB-deletion proteins generated in this study complemented the motility defect of the ΔpomB strain, although all were detected and localized to the membrane. The deleted regions of PomB have high similarity to peptidoglycan-associated proteins and PomB homologs (1, 29) (Fig. 1). Presumably, deletions in the C terminus of PomB impair association with peptidoglycan and prevent proper anchoring and/or localization of the stator elements around the rotor.
Based on in silico docking of the crystal structure of the OmpA-like domain of the putative peptidoglycan binding protein RmpM from Neisseria meningitidis, Tyr-127, Arg-135, and Arg-197 may be involved in peptidoglycan binding (19) (Fig. 1A). According to a multiple sequence alignment (Fig. 1), Tyr-127 is not conserved in MotB homologs, and there are gaps in the sequence in its vicinity. On the other hand, Arg-135 and Arg-197 are conserved and correspond to Arg-251 (C1 region) and Arg-294 (C3 region) of PomB, respectively. In MotB of E. coli, amino acid replacements R217W, R258C, and R258H at the corresponding residues result in loss of function (7). Therefore, the C1 and C3 regions may directly participate in binding to peptidoglycan.
To test whether the deletion proteins inhibit motility in the wild-type strain, the swarming and swimming of VIO5 (pom+) strains expressing the PomB derivatives with (pNH31, pNH32, pNH33 and pNH34) or without (pNH21, pNH22, pNH23 and pNH24) plasmid-encoded PomA were assessed in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Fig. 3C). The ΔC1, ΔC3, and ΔC proteins had almost no effect on the motility of VIO5. When the ΔC2 protein was overproduced in VIO5, both the swarming ability (Fig. 3C) and the swimming (data not shown) of VIO5 were significantly inhibited, and the inhibition was correlated with the concentration of IPTG (data not shown). However, upon coexpression of plasmid-encoded PomA and the ΔC2 protein (pNH33), the swarming and swimming were only slightly inhibited, indicating that the inhibitory effect of the ΔC2 protein is compensated by overexpression of PomA.
Association of PomB derivatives with PomA.
We examined whether the PomB deletion proteins associate with PomA in wild-type cells. We detected the chromosomally encoded PomA bound to all of the histidine-tagged PomB derivatives after detergent solubilization (Fig. 4, lanes 4, 8, 12, 16, and 21). The amount of ΔC3 protein was significantly lower, presumably because of degradation of the C-terminal region to which the His tag was attached (Fig. 4, asterisk). PomA was detected in the bound fraction of wild-type PomB and all of the PomB derivatives, indicating that PomA associates with the PomB derivatives. Similar levels of PomA were detected with ΔC, ΔC2, and wild-type PomB, suggesting that chromosomally encoded PomA associates with ΔC and ΔC2 as well as with wild-type PomB. The amounts of PomA bound to ΔC1 and ΔC3 were significantly lower than those bound to the wild type, ΔC, and ΔC2. For ΔC3 in particular, a faint band of PomA was detected only when a fivefold amount of bound material was loaded onto the gel (lane 21). Essentially the same results were obtained by using the ΔpomAB strains expressing plasmid-encoded PomA and PomB (data not shown).
The ΔC protein formed a cross-linked homodimer, dimer formation increased upon coexpression with PomA, and PomA associated with ΔC and wild-type PomB equally well. These results suggest that the putative peptidoglycan-binding region is not necessary for oligomerization of PomA and PomB. This conclusion is reasonable because the TM segments of PomA and PomB are close to each other (40), and a similar association was proposed for the TM segments of MotA and MotB in the proton-driven motor (10). Some proteins associated with peptidoglycan have been described as dimers (11, 19). Although the peptidoglycan-binding region of N. meningitidis RmpM is proposed to be responsible for dimerization (19), it does not appear to be solely responsible for dimerization of PomB.
While complete deletion of the C-terminal region preserves the association with PomA or with itself, certain smaller deletions weaken it, indicating a possible linkage between the conformation of the C-terminal region and the TM region. The amount of PomA associated with the ΔC1 and ΔC3 proteins seems low compared to the wild type, ΔC, and ΔC2, suggesting that the ΔC1 and ΔC3 proteins have some defects in the association with PomA (Fig. 4). On the other hand, the amounts of PomB homodimer were specifically reduced for ΔC2 and ΔC3, although similar levels of homodimer were detected in the other PomB derivatives (Fig. 2). It is noteworthy that the amount of PomA that coprecipitated with ΔC and disulfide bond formation of ΔC were comparable to those of the wild-type PomB. Garza et al. (16, 17) reported that motB missense mutations P159I and G164D in or immediately preceding the C region can be suppressed by mutations in motA or in fliG, which suggests that mutations in the C region can influence the arrangements of the TM region where MotA is presumably mounted. The slow-swimming phenotype of G240D substitution (C2 region) in MotB, which was suppressed by mutations in motA but not in fliG (16, 17), may have resulted in misassociation with MotA. Based on various mutations that affect the C region (7, 38), the structure of this region may in part determine the conformation around the TM region of PomB. The linker region between the C and TM regions (29) may have a rigid rather than a flexible structure to transmit the structural change. This assumption is consistent with results showing that the degradation products of ΔC1, ΔC2, and ΔC3 are similar in size to the intact ΔC protein, suggesting that the linker region assumes a conformation that is resistant to proteolytic degradation (Fig. 2 and 4).
Acknowledgments
We thank Ikuro Kawagishi and Masaru Kojima for stimulating discussions and encouragement. We are grateful to Terry A. Krulwich and David F. Blair for carefully reading the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan; the Japan Science and Technology Corporation (to M.H. and T.Y.); and from the Soft Nano-Machine Project of the Japan Science and Technology Agency (to T.Y. and M.H.).
REFERENCES
- 1.Asai, Y., S. Kojima, H. Kato, N. Nishioka, I. Kawagishi, and M. Homma. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327:453-463. [DOI] [PubMed] [Google Scholar]
- 3.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACY184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 4.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 6.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 7.Blair, D. F., D. Y. Kim, and H. C. Berg. 1991. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 173:4049-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block, S. M., and H. C. Berg. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470-472. [DOI] [PubMed] [Google Scholar]
- 9.Braun, T., and D. Blair. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 40:13051-13059. [DOI] [PubMed] [Google Scholar]
- 10.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 13.Dean, G. D., R. M. Macnab, J. Stader, P. Matsumura, and C. Burks. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 15.Fukuoka, H., T. Yakushi, and M. Homma. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J. Bacteriol. 186:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garza, A., L. Harris-Haller, R. Stoebner, and M. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garza, A. G., R. Biran, J. A. Wohlschlegel, and M. D. Manson. 1996. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 258:270-285. [DOI] [PubMed] [Google Scholar]
- 18.Gosink, K. K., and C. C. Häse. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grizot, S., and S. K. Buchanan. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51:1027-1037. [DOI] [PubMed] [Google Scholar]
- 20.Hirota, N., M. Kitada, and Y. Imae. 1981. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+. FEBS Lett. 132:278-280. [Google Scholar]
- 21.Kawagishi, I., I. Okunishi, M. Homma, and Y. Imae. 1994. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology 140:2355-2361. [Google Scholar]
- 22.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 23.Kojima, S., Y. Asai, T. Atsumi, I. Kawagishi, and M. Homma. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J. Mol. Biol. 285:1537-1547. [DOI] [PubMed] [Google Scholar]
- 24.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 25.Manson, M., P. Tedesco, H. Berg, F. Harold, and C. Van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarter, L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 29.Muramoto, K., and R. M. Macnab. 1998. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 29:1191-1202. [DOI] [PubMed] [Google Scholar]
- 30.Okabe, M., T. Yakushi, M. Kojima, and M. Homma. 2002. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46:125-134. [DOI] [PubMed] [Google Scholar]
- 31.Okunishi, I., I. Kawagishi, and M. Homma. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 34.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:20223-20228. [DOI] [PubMed] [Google Scholar]
- 35.Silverman, M., P. Matsumura, and M. Simon. 1976. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc. Natl. Acad. Sci. USA 73:3126-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stader, J., P. Matsumura, D. Vacante, G. E. Dean, and R. M. Macnab. 1986. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Togashi, F., S. Yamaguchi, M. Kihara, S. I. Aizawa, and R. M. Macnab. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakushi, T., M. Kojima, and M. Homma. 2004. Isolation of Vibrio alginolyticus sodium-driven flagellar motor complex composed of PomA and PomB solubilized by sucrose monocaprate. Microbiology 150:911-920. [DOI] [PubMed] [Google Scholar]
- 40.Yakushi, T., S. Maki, and M. Homma. 2004. Interaction of PomB with the third transmembrane segment of PomA in the Na+-driven polar flagellum of Vibrio alginolyticus. J. Bacteriol. 186:5281-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Yorimitsu, T., and M. Homma. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82-93. [DOI] [PubMed] [Google Scholar]
- 43.Yorimitsu, T., M. Kojima, T. Yakushi, and M. Homma. 2004. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. 135:43-51. [DOI] [PubMed] [Google Scholar]
- 44.Yorimitsu, T., K. Sato, Y. Asai, I. Kawagishi, and M. Homma. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, J. D., R. T. Fazzio, and D. F. Blair. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237-242. [DOI] [PubMed] [Google Scholar]