The E3 ligase PUB22 is phosphorylated by MPK3, thereby inhibiting its autoubiquitination activity, allowing PUB22 to accumulate and dampen the immune response.
Abstract
Crosstalk between posttranslational modifications, such as ubiquitination and phosphorylation, play key roles in controlling the duration and intensity of signaling events to ensure cellular homeostasis. However, the molecular mechanisms underlying the regulation of negative feedback loops remain poorly understood. Here, we uncover a pathway in Arabidopsis thaliana by which a negative feedback loop involving the E3 ubiquitin ligase PUB22 that dampens the immune response is triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3), best known for its function in the activation of signaling. PUB22’s stability is controlled by MPK3-mediated phosphorylation of residues localized in and adjacent to the E2 docking domain. We show that phosphorylation is critical for stabilization by inhibiting PUB22 oligomerization and, thus, autoubiquitination. The activity switch allows PUB22 to dampen the immune response. This regulatory mechanism also suggests that autoubiquitination, which is inherent to most single unit E3s in vitro, can function as a self-regulatory mechanism in vivo.
INTRODUCTION
Signaling networks must be tightly regulated in their timing, amplitude, and duration. This is especially important for immune signaling, as its deregulation can result in the inappropriate activation of downstream processes, resulting in an inefficient defense response against pathogens, deregulated cell death, and increased metabolic penalties. Mechanisms are in place that safeguard an adequate immune response, which is triggered by pathogen-associated molecular patterns (PAMPs) through plasma membrane located receptors. These include negative feedback loops that are inherent to all signaling networks and allow precise gauging of responses, stabilization of signaling circuits, and maintenance of cellular homeostasis (Ferrell, 2013). Ubiquitination and phosphorylation are posttranslational modifications that contribute to this process (Hunter, 2007).
The addition of ubiquitin to target proteins is mediated by an enzymatic cascade composed of the E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and the E3 ubiquitin ligase. The U-box type E3 ubiquitin ligases have been shown to be required for a wide range of responses, including immune reactions (Zeng et al., 2004; González-Lamothe et al., 2006; Yang et al., 2006; Lu et al., 2011), drought (Liu et al., 2011; Seo et al., 2012), reactive oxygen species generation (Woodson et al., 2015), pollen self-incompatibility (Stone et al., 1999), and cell division (Kinoshita et al., 2015).
We previously defined a group of related plant U-box (PUB) type E3 ligases, PUB22, PUB23, and PUB24, as negative regulators of PAMP-triggered immunity in Arabidopsis thaliana (Trujillo et al., 2008). Their function is required for the general dampening of signaling induced by PAMPs (Stegmann et al., 2012). Inactivation of the ligase triplet results in enhanced resistance toward pathogens, such as Pseudomonas syringae pv tomato DC3000 (Pst), Hyaloperonospora arabidopsidis, and the fungus Piriformospora indica (Jacobs et al., 2011; Stegmann et al., 2012). In addition, PUB22 and PUB23 were also reported to negatively regulate responses to drought (Cho et al., 2008). The inactivation of the PUB triplet does not have any apparent consequences for development, indicating that they are mainly dedicated to regulating activated stress signaling.
Downstream signaling includes the activation of canonical mitogen-activated protein kinase (MAPK) cascades. Stimulation of pattern recognition receptors, such as FLS2, results in the activation of MPK3, MPK4, MPK6, and MPK11, which are hallmarks of the immune response. While MPK3 and MPK6 are believed to function as partially redundant positive regulators of immunity, MPK4 has been attributed a negative regulatory role (Rodriguez et al., 2010). However, recent studies have helped to draw a more precise picture, revealing more complex interactions (Popescu et al., 2009; Zhang et al., 2012; Frei dit Frey et al., 2014; Lassowskat et al., 2014).
Ubiquitination can impart regulatory information that can be integrated with phosphorylation to serve as feedback mechanisms. The first indication for the existence of such regulatory circuits in plants was provided by the U-box E3 ligase ARC1, which is phosphorylated by the receptor SRK (Gu et al., 1998). ARC1 is required for the rejection of self-pollen in Brassicaceae, and its phosphorylation was proposed to activate its function during pollen rejection (Stone et al., 1999). Similar interactions were suggested to extend to related E3s and kinases (Samuel et al., 2008). Additional examples include the phosphorylation of PUB1, PUB12, and PUB13 by receptor kinases (Mbengue et al., 2010; Lu et al., 2011). In spite of phosphorylation of PUB E3s being a recurrent phenomenon, it is not known how phosphorylation incorporates mechanistically into the activity of E3 ligases.
E3 ligases carry out dual tasks as both catalysts and in substrate selection, acting as a scaffold bringing together the right E2 with a specific target protein and enhancing the transfer rate of ubiquitin. Even though advances in our understanding of the molecular mechanisms of E3 function and regulation have improved in non-plant systems (Weissman et al., 2011; Berndsen and Wolberger, 2014), little is known about the regulation of single subunit RING/U-box ligases the largest family of E3s in humans (>300), while no information is available for their plant counterparts (Arabidopsis >540).
One ubiquitous observation is the oligomerization of RING/U-box ligases and its proposed involvement in ligase activity (Deshaies and Joazeiro, 2009; Berndsen and Wolberger, 2014). Oligomerization is mediated by the U-box/RING domains through a surface distal to the E2 interacting region, such as in the case of the CHIP U box E3 ubiquitin ligase (Zhang et al., 2005). In several instances, homooligomerization was shown to influence the activity of both self and substrate ubiquitination. Mutant variants that were impaired in their oligomerization displayed reduced activity (Yin et al., 2009; Dueber et al., 2011; Plechanovová et al., 2011; Dou et al., 2012a). Exceptions include the U-box E3s E4B in humans and its ortholog Ufd2 in yeast (Saccharomyces cerevisiae), which function as monomers (Benirschke et al., 2010).
Here, we investigated the regulation of the E3 ligase PUB22 in Arabidopsis and provide evidence for a unique regulatory mechanism for single unit E3 ligases in plants. PUB22 phosphorylation by MPK3 reduces its oligomerization status, which in turn inhibits autoubiquitination, allowing PUB22 to accumulate and subsequently to dampen immune signaling.
RESULTS
PUB22 Degradation Is Regulated by PAMPs
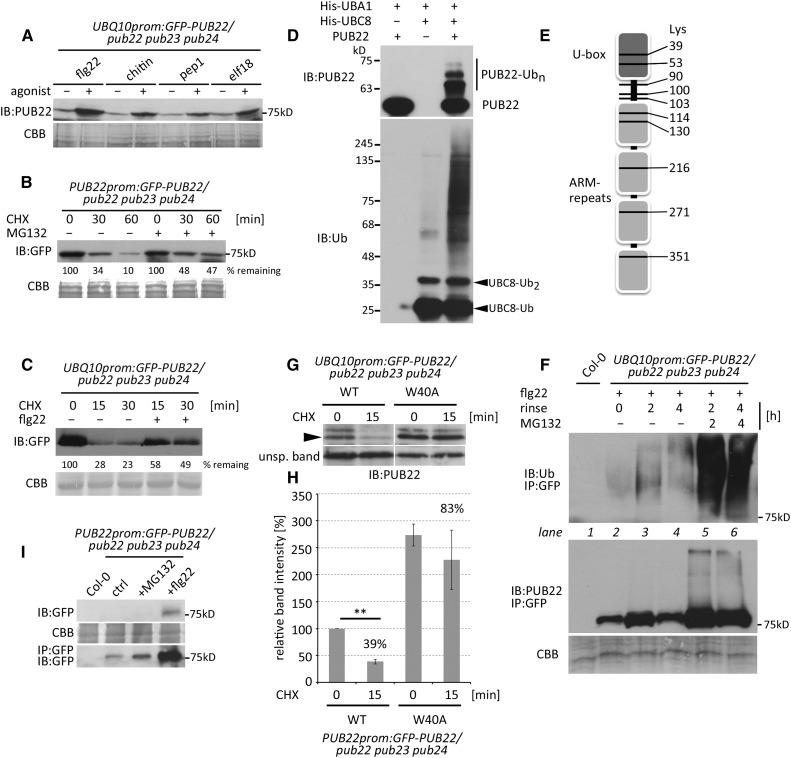
We previously reported that PUB22 is stabilized within minutes after activation of the immune response with the flagellin-derived immunogenic peptide flg22 (Stegmann et al., 2012). This suggested that immune signaling feeds into the activation of PUB22 to activate a negative regulatory loop. We first tested whether stabilization could be triggered by additional agonists including chitin, Pep1, and elf18, which are perceived by different receptors. In all cases, constitutively expressed PUB22, accumulated within 1 h of treatment, as revealed by immunoblot analysis (Figure 1A). Initial stabilization of PUB22 in response to PAMP perception can be detected as early as 5 min after treatment, largely precluding a transcriptional effect (Stegmann et al., 2012). We therefore hypothesized that protein accumulation may result from the inhibition of its degradation. PUB22 protein levels were markedly reduced at 30 min and almost depleted 60 min after cycloheximide (CHX) treatment, indicating active degradation (Figure 1B). Simultaneous treatment with the inhibitor MG132 reduced the degradation rate, confirming degradation by the proteasome (Figure 1B). Moreover, degradation of PUB22 was reduced after flg22 treatment (Figure 1C).
Figure 1.
PUB22 Mediates Its Own Degradation by Autoubiquitination and Is Stabilized upon PAMP Perception.
(A) UBQ10prom:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings were treated with flg22 (1 µM), chitin (200 μg/mL), pep1 (1 µM), and elf18 (1 µM) for 60 min. Total protein extracts were analyzed by immunoblot (IB) using anti-PUB22. Coomassie brilliant blue (CBB) is shown as a loading control.
(B) Transgenic seedlings expressing PUB22prom:GFP-PUB22 in the pub22 pub23 pub24 background were elicited with flg22 (1 µM) to induce protein expression for 2 h, rinsed, and treated with CHX (50 µM) alone or in combination with MG132 (50 µM). The experiment was repeated with similar results.
(C) Transgenic seedlings expressing UBQ10prom:GFP-PUB22 in the pub22 pub23 pub24 background were elicited with flg22 (1 µM) and CHX (20 µM) and samples were taken at the indicated time points. The experiment was repeated three times with similar results.
(D) In vitro autoubiquitination assay using untagged PUB22 (E3), His-UBA1 (E1), and His-UBC8 (E2). Recombinant GST-PUB22 was purified and proteolytically cleaved to obtain tag-free PUB22.
(E) Schematic representation of ubiquitinated residues identified by LC-MS/MS from in vitro autoubiquitinated PUB22. Ten ubiquitination sites were identified with a diGly modification indicative of ubiquitination.
(F) Immunoprecipitation (IP) of GFP-PUB22 from UBQ10prom:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings using anti-GFP beads. Samples were treated with flg22 (1 µM) for 1 h, rinsed with buffer, and harvested after 0, 2, and 4 h. Seedlings were additionally treated with the proteasome inhibitor MG132 (50 µM) for 2 and 4 h, as indicated. The experiment was repeated with similar results.
(G) UBQ10prom:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings expressing PUB22 wild-type or the inactive Trp40Ala variant were treated with CHX (20 µM) and samples were taken at 0 and 15 min. Arrowhead indicates GFP-PUB22. Unspecific band shows equal loading.
(H) Band intensities in (G) were analyzed to determine the relative amounts of PUB22. GFP-PUB22 wild type at 0 min CHX was set to 100%, and relative amounts were calculated using normalized band intensities. Data shown as mean ± sd obtained from three independent experiments. Statistical significance between time points was assessed by Student’s t test, **P < 0.01.
(I) Basal and induced levels of PUB22 protein. IP of GFP-PUB22 was performed using anti-GFP beads from PUB22prom:GFP-PUB22 seedlings treated with DMSO (control), 50 µM MG132, or 1 µM flg22 for 2 h. The experiment was repeated with similar results.
A common trait of most E3 ubiquitin ligases is their ability to autoubiquitinate (Supplemental Figure 1A). In many instances, autoubiquitination results in self-catalyzed destruction (Weissman et al., 2011). To exclude the possibility that the tag deregulated E3 activity or served as an artifactual substrate, we removed GST before the autoubiquitination assay. Untagged PUB22 effectively modified itself, indicating that it possesses true autoubiquitination activity in vitro (Figure 1D). From the in vitro autoubiquitination we identified a diglycine footprint on 10 out of 29 lysines, namely, 39, 53, 90, 100, 103, 114, 130, 216, 271, and 351 (Figure 1E; Supplemental Figure 1B), by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The ubiquitination pattern shows that PUB22 has no specificity toward an individual lysine.
We confirmed that PUB22 is ubiquitinated in vivo by immunopurification from stable transgenic lines constitutively expressing GFP-PUB22. Due to the inherent low levels of GFP-PUB22 in untreated tissues, which precludes the detection of ubiquitinated forms, we first treated Arabidopsis seedlings with flg22 for 1 h to induce protein accumulation. Subsequently, we analyzed samples at 0, 2, and 4 h after agonist removal. PUB22 accumulated until 2 h, presumably because of remnant flg22 present in the tissues, but dropped at 4 h (Figure 1F, lower panel). Low levels of ubiquitinated PUB22 at 0 h increased 2 h after flg22 removal (upper panel, lanes 2 and 3), proportionally to the unmodified protein (lower panel). However, ubiquitinated species were more abundant after 4 h compared with 0 h (upper panel, lanes 2 and 4), while unmodified amounts were similar (lower panel). Furthermore, overall ubiquitinated levels at 2 and 4 h were comparable (upper panel, lanes 3 and 4), while less unmodified protein was detectable at 4 h (lower panel). This suggests that PUB22 ubiquitination is increased at later time points as a result of flg22 removal. Treatment with MG132 resulted in the strong accumulation of modified PUB22, indicating that the ubiquitinated protein is consequently degraded by the proteasome (Figure 1F, lanes 5 and 6).
To test the function of autoubiquitination in the self-catalyzed degradation of PUB22, we mutated the conserved Trp-40 to alanine, which abrogates most E2 binding but should maintain the overall structure of the U-box (Trujillo et al., 2008). Inhibition of de novo protein synthesis by CHX treatment resulted in rapid degradation of the wild type within 15 min (Figures 1G and 1H). By contrast, the inactive Trp40Ala mutant was more stable (Figure 1H). These results show that autoubiquitination is the main mechanism driving PUB22 degradation.
However, since PUB22 and related genes are induced by infection and PAMP perception (Trujillo et al., 2008; Yee and Goring, 2009), transcriptional regulation may also contribute to elevated protein levels. Using transgenic Arabidopsis lines expressing a functional GFP-PUB22 fusion protein under the control of the native promoter (Supplemental Figure 2A), flg22 elicitation resulted in detectable protein levels only after 60 min, with no apparent basal protein expression (Supplemental Figure 2B). Nevertheless, naïve pub22 pub23 pub24 plants display enhanced early immune signaling (Trujillo et al., 2008), indicating that the PUB22 protein also exerts its function in untreated tissues. Indeed, PUB22 protein was detectable after immuno-enrichment in untreated seedlings (Figure 1I).
Together, these data show that PUB22 mediates its own degradation by autoubiquitination and that agonist-triggered stabilization may be the result of the inhibition of this process, which allows the rapid increase of protein levels. Furthermore, the transcriptional induction may act in concert with protein stabilization to allow PUB22 accumulation.
PUB22 Interacts with and Is Phosphorylated by MPK3
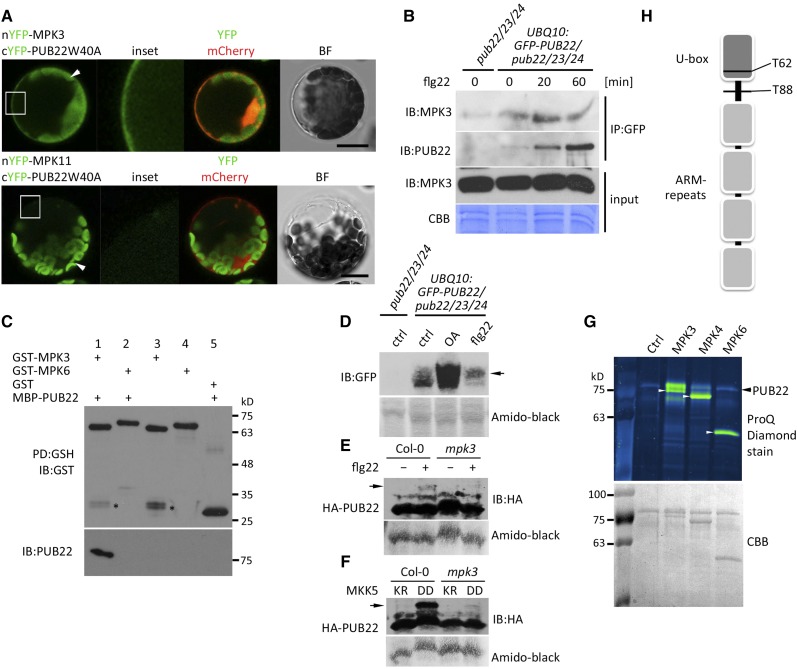
The stabilization of PUB22, which occurs within 5 min after flg22 elicitation and is transient in nature (Stegmann et al., 2012), suggested the involvement of phosphorylation. In addition, the prolonged MPK3 activity in pub22 pub23 pub24 after elicitation implied a PUB22-linked negative feedback loop (Trujillo et al., 2008). Using bimolecular fluorescence complementation, we tested the potential interaction of PUB22 with selected MAPKs. Under our conditions, we only detected fluorescence complementation when PUB22 was coexpressed with MPK3, but not with MPK4, MPK6, MPK8, or MPK11 (Figure 2A; Supplemental Figure 3A). To confirm the interaction between PUB22 and MPK3, we coimmunoprecipitated GFP-PUB22 using the UBQ10prom:GFP-PUB22 line and tested for interaction with the endogenous MPK3. Coimmunoprecipitation of MPK3 with PUB22 confirmed the interaction. The flg22-induced increase of PUB22 did not proportionally correlate with the amounts of interacting MPK3, suggestive of a transient interaction in vivo (Figure 2B). Finally, we showed that recombinant PUB22 physically and specifically interacts with MPK3, but not the closely related MPK6, in an in vitro pull-down assay (Figure 2C).
Figure 2.
PUB22 Interacts with and Is Phosphorylated by MPK3.
(A) Interaction between PUB22 and MPK3 detected by bimolecular fluorescence complementation in Arabidopsis protoplasts. nYFP-MPK3 or nYFP-MPK11 was coexpressed with cYFP-PUB22 W40A as indicated. Free mCherry was coexpressed to label the cytoplasm and nucleus. Shown are representative pictures. Arrows indicate chloroplast autofluorescence. Bar = 50 µm.
(B) UBQ10:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings were treated with flg22 (1 µM) for 20 and 60 min and total protein (input) was subjected to IP with anti-GFP beads. Endogenous coimmunoprecipitated MPK3 was detected with MPK3-antibodies. The experiment was repeated three times with similar results.
(C) MBP-PUB22 pull-down (PD) assay using purified GST-MPK3 and GST-MPK6 on glutathione agarose beads as baits. Asterisk indicates a potential GST cleavage product.
(D) PUB22 mobility shift after flg22 treatment. UBQ10:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings were treated with DMSO (ctrl), flg22 (1 µM), or okadaic acid (OA; 1 µM) for 20 min. Total protein samples were resolved by Phos-tag PAGE. The experiment was repeated with similar results. Mobility shift is indicated by the arrow.
(E) PUB22 mobility shift after flg22 treatment in the wild-type Col-0 and mpk3 backgrounds. HA-PUB22 was transiently expressed and treated with DMSO or flg22 (100 nM) for 20 min. Total protein samples were resolved by Phos-tag PAGE. The experiment was repeated three times with similar results. Mobility shift is indicated by the arrow.
(F) PUB22 mobility shift induction by MKK5 in wild-type Col-0 and mpk3 backgrounds. HA-PUB22 was transiently coexpressed with MKK5 LysArg (inactive) or AspAsp (constitutively active). After overnight incubation, total protein samples were resolved by Phos-tag PAGE. The experiment was repeated with similar results. Mobility shift is indicated by the arrow.
(G) GST-PUB22 was incubated alone or with activated GST-MPK3, GST-MPK4, and untagged MPK6 (white arrowheads). Phosphorylation was visualized with ProQ Diamond stain.
(H) Cartoon depicting the localization of the in vitro phosphorylated sites by MPK3 on PUB22 identified by LC-MS/MS.
The interaction opened the possibility that PUB22 and MPK3 modify each other. However, PUB22 was not able to modify MPK3 in an in vitro ubiquitination assay (Supplemental Figure 3B). Conversely, to demonstrate phosphorylation of PUB22, we treated seedlings expressing the E3 ligase under a constitutive promoter with flg22 and resolved the protein samples using a Phos-tag gel in which phosphorylated proteins specifically migrate more slowly. PUB22 displayed a mobility shift after activation of immune signaling by flg22 treatment (Figure 2D). Inhibition of phosphatase activity with okadaic acid also resulted in the accumulation of PUB22, which included PUB22 species with slower mobility. The slower migrating species after flg22 treatment were absent in the mpk3 background (Figure 2E). In addition, expression of the constitutively active MAPK KINASE5 (MKK5) AspAsp, which activates MPK3, resulted in a slower migrating band, which was absent in the mpk3 background (Figure 2F).
We next tested if MPK3 was able to phosphorylate PUB22 in vitro. Consistent with the direct interaction, PUB22 was readily trans-phosphorylated by MPK3, while phosphorylated levels of PUB22 by MPK4 and MPK6 were comparable to the control (Figure 2G). We confirmed these results using 32P-labeled ATP and included PUB24, which was also phosphorylated by MPK3 in vitro (Supplemental Figure 3C).
Recombinant PUB22 phosphorylated by MPK3 was next analyzed by LC-MS/MS to determine PUB22 phosphorylation sites. We identified two phosphorylation sites: Thr-62 located in the U-box domain and Thr-88 in a predicted disordered region between the U-box and the first ARM domain (Figure 2H; Supplemental Figure 4A). Finally, we were able to confirm both phosphorylation sites in vivo by LC-MS/MS from samples treated 30 min with flg22, which were absent in the controls (Supplemental Figure 4B). Both residues are followed by a proline, thus possessing a typical MAPK phosphorylation motif. No phosphopeptides were identified in controls without kinase. The identified phosphorylation sites are not in the ARM domain that is responsible for substrate recognition, indicating that the modifications are unlikely to directly influence substrate binding.
We analyzed whether the phosphosites were conserved by aligning the predicted amino acid sequences of class II (U-box and ARM repeats) and class III (UND, U-box, and ARM repeats) PUBs and searched for Ser/Thr-Pro motifs. Both residues were conserved in PUB22, PUB23, and PUB24, as well as in PUB6 and PUB7. While Thr-62 was conserved in 40% (17 out of 42), Thr-88 was only present in 21% (9 out of 42) of sequences (Supplemental Figure 5 and Supplemental File 1).
PUB22 Stabilization Is Dependent on MPK3
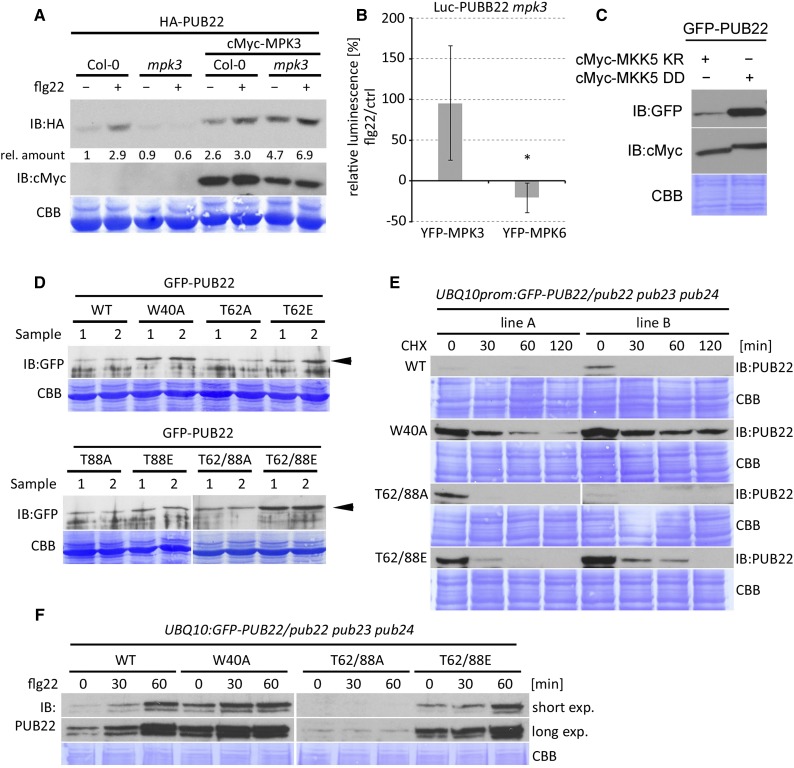
We hypothesized that the stabilization of PUB22 after activation of immune signaling may be dependent on MPK3. To test this, we expressed PUB22 in wild-type and mpk3 genetic backgrounds. As shown before, activation of the immune response resulted in the stabilization of PUB22 in wild-type Columbia-0 (Col-0; Figure 3A). By contrast, in the absence of MPK3, PUB22 levels remained unchanged after flg22 treatment (Figure 3A). However, transient expression of MPK3 in the mpk3 mutant background complemented the stabilization of the PUB22 protein. Furthermore, expression of MPK3 resulted in elevated basal levels of PUB22 (Figure 3A).
Figure 3.
PUB22 Stabilization Is Dependent on MPK3.
(A) HA-PUB22 was expressed alone or with cMyc-MPK3 in Col-0 and mpk3 background and treated with DMSO or flg22 (100 nM) for 45 min in Arabidopsis protoplasts. Band intensities were determined and amounts relative to HA-PUB22 in Col-0 calculated. The experiment was repeated with similar results.
(B) Luciferase (Luc) PUB22 fusion was coexpressed with MPK3 or MPK6 in mpk3 protoplasts. Luminescence was measured in control (water) and flg22 (100 nM) treated samples after 30 min. Relative luminescence to control samples after flg22 is shown as average ± sd obtained from three independent experiments. Statistical significance was assessed by Student’s t test, *P < 0.05.
(C) GFP-PUB22 was coexpressed with cMyc-MKK5 AspAsp (active) or cMyc-MKK5 LysArg (inactive) versions in Arabidopsis protoplasts. Samples were taken after 8 h incubation. The experiment was repeated three times with similar results.
(D) GFP-PUB22 wild-type and mutant variants were constitutively expressed in Arabidopsis protoplasts to test protein accumulation. Shown are samples from two independent transformations. Arrowhead highlights PUB22 and mutant variants.
(E) UBQ10:GFP-PUB22/pub22 pub23 pub24 independent transgenic lines (A and B) constitutively expressing GFP-PUB22 wild-type and mutant variants were treated with CHX (50 µM), and samples were taken at the indicated times. The experiment was repeated three times with similar results.
(F) UBQ10:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings constitutively expressing GFP-PUB22 wild-type and variants were treated with flg22 (1 µM) for the indicated times to determine protein stabilization. The experiment was repeated three times with similar results.
To validate MPK3’s role in PUB22 accumulation, we coexpressed a Luciferase (Luc) PUB22 fusion with either MPK3 or MPK6 in the mpk3 background. Treatment with flg22 resulted in an increase of the luminescence compared with the control only in the presence of MPK3 (Figure 3B; Supplemental Figure 6). Furthermore, we tested whether MKK5, which activates MPK3, was also able to mediate the stabilization of PUB22. Coexpression of the constitutively active MKK5 AspAsp resulted in an increase in PUB22 protein levels, in comparison to samples expressing the inactive MKK5 LysArg version (Figure 3C) (Lee et al., 2004). Together, these results indicate that MPK3 can induce PUB22 stabilization.
Given that PUB22 is phosphorylated by MPK3 and that its stabilization is MPK3 dependent, we tested whether the identified phosphosites are responsible for this process. We generated constructs in which we replaced Thr-62 and/or Thr-88 with an alanine, which cannot be phosphorylated (phosphonull), or into glutamic acid, a negatively charged amino acid, to mimic phosphorylation (phosphomimetic). To assay the effect of each residue, we transiently expressed all PUB22 variants under the control of a constitutive promoter. Expression of wild-type PUB22 resulted in low levels of protein accumulation. By comparison, the Trp40Ala protein, which is unable to autoubiquitinate, displayed distinctly higher amounts of protein (Figure 3D; samples from two independent experiments). Protein accumulation of the Thr62Ala phosphonull mutant was comparable to the wild type. By contrast, expression of the phosphomimetic Thr62Glu resulted in higher protein amounts. A similar trend was observed for the Thr-88 variants. Nevertheless, the impact on the protein stability of each phosphomimetic was modest. The Thr62/88Glu double phosphomimetic resulted in a marked increase in protein stability in comparison to the double phosphonull, which still behaved like the wild type (Figure 3D). These results indicate that the phosphorylation of both Thr-62 and Thr-88 contributes additively to the stabilization of PUB22.
To further confirm these results, we generated transgenic Arabidopsis lines carrying wild-type, inactive Trp40Ala, double phosphonull Thr62/88Ala, and phosphomimetic Thr62/88Glu mutants. Two independent lines were selected for each construct expressing similar transcript levels (Supplemental Figure 7A). In agreement with the results obtained from the transient transformation, protein amounts of Trp40AlaPUB22 and the phosphomimetic Thr62/88Glu were higher than in lines expressing the wild type or the phosphonull Thr62/88Ala (Figure 3E; Supplemental Figures 7B and 7C). Notably, mutant variants were still degraded, including the inactive Trp40Ala mutant, suggesting that E3 degradation is mediated by additional factors.
To investigate the function of these residues in the stabilization of PUB22 during the immune response, we treated transgenic seedlings with flg22. The wild-type protein gradually accumulated within an hour after treatment and was less abundant than the inactive Trp40Ala (Figure 3F). Importantly, protein levels of the Thr62/88Ala phosphonull variant were unaffected and remained low, showing that both residues are essential to initiate stabilization. Protein levels of PUB22 Trp40Ala and Thr62/88Glu still increased after flg22 treatment, indicating that additional factors contribute to protein accumulation.
These results support a role for MPK3 in the phosphorylation of PUB22’s Thr-62 and Thr-88, which results in the stabilization of the protein.
PUB22 Autoubiquitination Activity Is Controlled by Thr-62
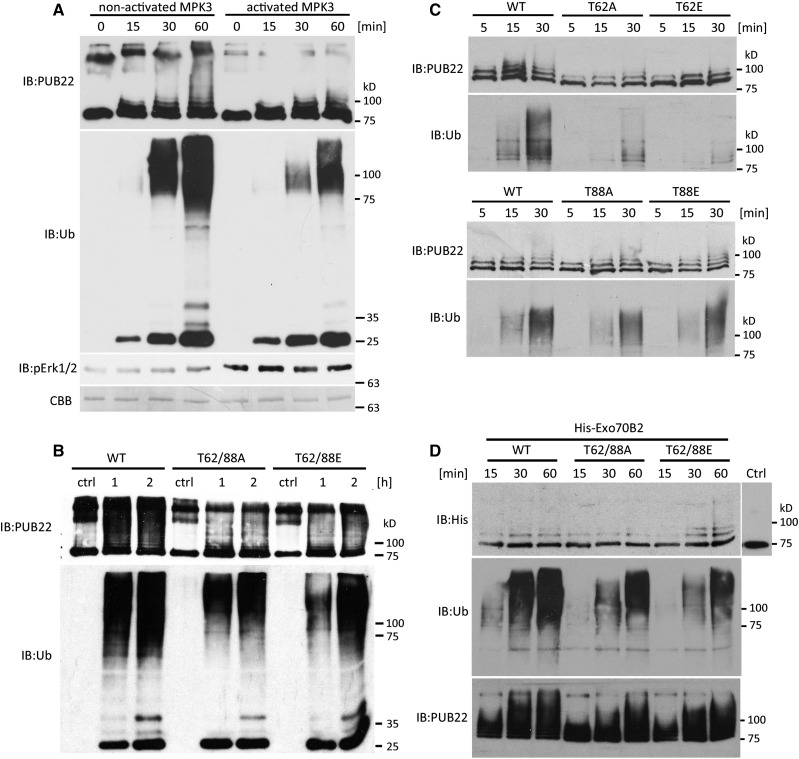
Given that the identified phosphorylated residues of PUB22 modulate its stability, it is possible that the modification impinges on the autoubiquitination activity of the ligase. To test this, we performed in vitro autoubiquitination assays. As shown before, PUB22 wild type displays a robust autoubiquitination activity (Figure 1D; Supplemental Figure 1A). We first assayed the impact of MPK3 activity on PUB22’s autoubiquitination by incubating it with either activated or nonactivated MPK3. Autoubiquitination activity was reduced in the presence of activated MPK3 (Figure 4A), showing that MPK3 activity inhibits PUB22 autoubiquitination in vitro. Consistent with the increased stability in vivo (Figures 3D to 3F), the Thr62/88Glu phosphomimetic variant displayed reduced autoubiquitination (Figure 4B). However, the Thr62/88Ala phosphonull also showed less activity than the wild-type E3 ligase. Nevertheless, general ubiquitination was indeed lower for Thr62/88Glu when compared with the phosphonull variant, as was anticipated (Figure 4B, IB:Ub).
Figure 4.
PUB22 Thr-62 Mediates a Reduction in Autoubiquitination Activity.
(A) In vitro autoubiquitination of MBP-PUB22 in the presence of Arabidopsis His-UBA1 and His-UBC8 and incubated with activated or nonactivated MPK3. Activation of MPK3 was detected using anti-pERK1/2.
(B) In vitro autoubiquitination using MBP-PUB22 and mutant variants in the presence of Arabidopsis His-UBA1 and His-UBC8.
(C) In vitro autoubiquitination using MBP-PUB22 and single mutant variants in the presence of Arabidopsis His-UBA1 and His-UBC8.
(D) In vitro ubiquitination of the substrate His-Exo70B2 by MBP-PUB22 and mutant variants in the presence of Arabidopsis His-UBA1 and His-UBC8. Control (ctrl) reaction as above but in the absence of ATP.
Thr-62 is located in the U-box, the domain mediating the interaction with the E2, while Thr-88 is in a predicted disordered region between the U-box and the first ARM repeat, with modifications at each phosphorylation site potentially affecting different aspects of ligase activity. To test this, we performed autoubiquitination assays with the single mutants. As predicted, only mutation Thr-62 inhibited the autoubiquitination activity, while mutation of Thr-88 had no effect (Figure 4C). Also, the Thr62Ala single mutant showed unexpectedly reduced autoubiquitination. However, the ubiquitination activity detected in the anti-Ub immunoblot, consistently showed a stronger inhibition of Thr62Glu activity compared with Thr62Ala (Figure 4C, IB:Ub).
We next tested whether mimicking phosphorylation of PUB22 also resulted in changes in substrate ubiquitination. For this purpose, we used Exo70B2, a subunit of the exocyst complex, which is ubiquitinated by PUB22 (Stegmann et al., 2012). As shown previously, PUB22 was able to ubiquitinate Exo70B2 in vitro (Figure 4D). The ubiquitination of Exo70B2 by Thr62/88Ala was slightly reduced in comparison to the wild type (Figure 4D). By contrast, the ubiquitination activity of the phosphomimetic Thr62/88Glu toward the substrate was slightly increased.
Together, our results are consistent with the notion that Thr-62 phosphorylation inhibits autoubiquitination, while not having a major effect on substrate ubiquitination activity and thus, that mutations do not result in a general impairment of PUB22 activity. Even though Thr62Ala mutation affected autoubiquitination (detected by anti-PUB22), inhibition of PUB22 ubiquitination activity was highest in the Thr62Glu variant (detected by anti-Ub; Figures 4B and 4C), in line with the effects observed for PUB22 stability in vivo (Figures 3D and 3E; Supplemental Figures 7B and 7C).
Phosphorylation of Thr-62 Regulates Dimerization
To dissect the potential function of Thr-62 phosphorylation, we generated a structural model based on the zebra fish (Danio rerio) CHIP (PDB ID code 2F24) (Xu et al., 2006). Thr-62 is predicted to be located on the opposite side of the E2 interaction surface and is therefore unlikely to affect E2 binding. However, dimerization of E3 ligases via the backside of the E2 interacting surface has been shown in many instances, including for the U-box ligases CHIP (Zhang et al., 2005) and Prp19 (Vander Kooi et al., 2006).
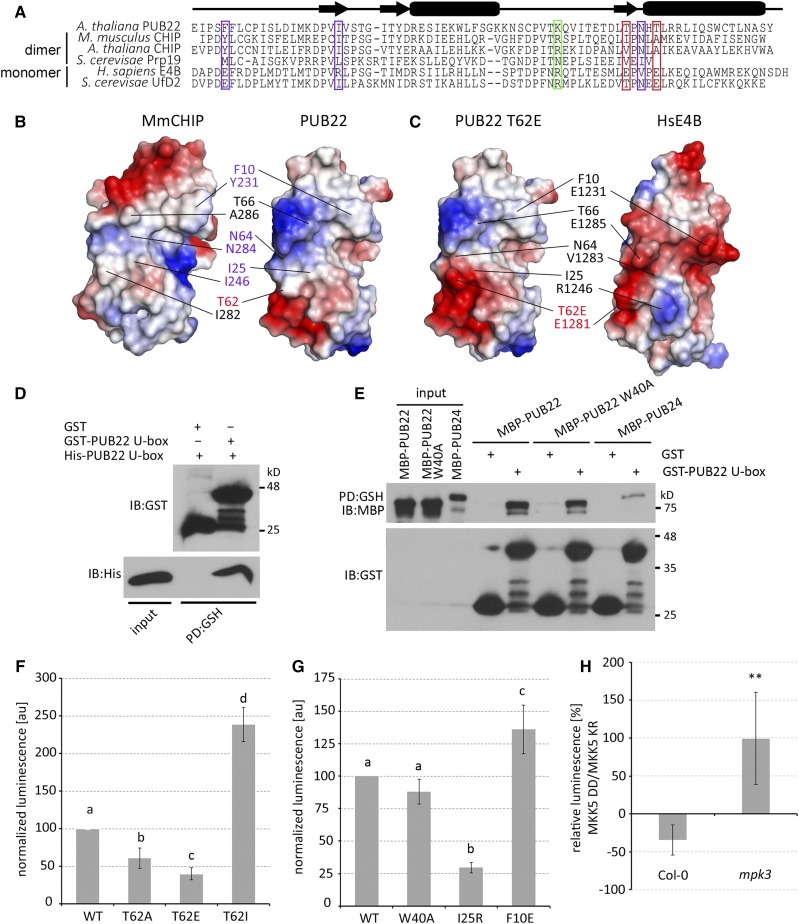
Dimer formation in the mouse CHIP is mediated by the hydrophobic residues Tyr-231, Ile-246, Ile-282, and Ala-286, in addition to Asn-284, which forms H-bonds with the same residue on the complementary protomer (Zhang et al., 2005). To examine the conservation of these residues in PUB22, we performed a sequence alignment with E3s shown to dimerize, including CHIP and Prp19 (Zhang et al., 2005; Vander Kooi et al., 2006). The first two corresponding hydrophobic residues Phe-10 and Ile-25, as well as Asp-64, are conserved in PUB22 (Figure 5A). However, residues Ile-282 and Ala-286 in CHIP are replaced by threonines in PUB22, and include Thr-62. All residues are conserved in the Arabidopsis ortholog of the mouse (Mus musculus) CHIP. By contrast, E3s such as the human E4B and its yeast ortholog Ufd2, which function as monomers, possess charged residues at most of these positions (Figure 5A).
Figure 5.
PUB22 Forms a Dimer/Oligomer through Its U-Box Domain.
(A) Sequence alignment of U-box domains from different E3 ubiquitin ligases highlighting key conserved residues in PUB22 that are required for CHIP dimerization (violet boxes) and variants including Thr-62 (red box left) or that may play a role in the priming of the E2-ubiquitin conjugate (green). Locations of secondary-structure elements are shown by the diagram above the alignment; arrows indicate α-helices and boxes β-strands.
(B) Electrostatic surface potentials of mouse CHIP U-box monomer (PDB ID code 2C2V) and the structural model of PUB22 highlighting the residues important for dimerization described in (A).
(C) Electrostatic surface potentials of human E4B (PDB ID code 3L1X) and the structural model of PUB22 Thr62Glu.
(D) In vitro pull-down assay to test U-box oligomerization using purified GST-PUB22 U-box as bait on glutathione agarose beads and His-PUB22 U-box as prey.
(E) In vitro pull-down assay to test PUB22 oligomerization using purified GST-PUB22 U-box as baits on glutathione agarose beads and MBP-PUB22, MBP-PUB22 Trp40Ala, or MBP-PUB24 as preys. Protein samples were analyzed by IB using anti-GST and anti-MBP.
(F) Split luciferase assay to test oligomerization of PUB22 U-box wild-type and mutant variants Thr62Glu, Thr62Ile, and Thr62Ala fused to N and C termini of luciferase and coexpressed in Arabidopsis protoplasts.
(G) Split luciferase assay to test interaction between PUB22 U-box wild type and Trp40Ala, Ile25Arg, and Phe10Glu U-box variants as described in (F).
(F) and (G) Signal intensity was normalized to PUB22 U-box wild type. Shown are the mean signal intensities ± sd 10 min after adding substrate. Different letters indicate significantly different values between wild-type and mutant variants obtained from at least three independent experiments at P < 0.05 (one-way ANOVA, Tukey post hoc test).
(H) Split luciferase assay to test MPK3-dependent regulation of oligomerization. Wild-type PUB22 U-box fused to N and C termini of luciferase was coexpressed with either MKK5 LysArg (inactive) or MKK5 AspAsp (constitutive active) to activate immune signaling in the Col-0 or mpk3 backgrounds. Shown is the relative luminescence of MKK5 AspAsp to MKK5 LysArg expressing samples ± sd obtained from three independent experiments. Statistical significance was assessed by Student’s t test, **P < 0.01.
The hydrophobic nature of the interaction surface in CHIP is in stark contrast to the charged surface in E4B, likely precluding the formation of a dimer (Benirschke et al., 2010) (Figures 5B and 5C). The electrostatic charge of the corresponding surface in PUB22 resembles that of CHIP (Figure 5B), while mimicking phosphorylation results in a negative patch, also found in E4B (Figure 5C).
We first tested whether PUB22 was able to oligomerize. As predicted, GST-PUB22 U-box was able to interact with His-PUB22 U-box (Figure 5D). We also performed a pull-down assay using full-length MBP-PUB22, which was precipitated by GST-PUB22 U-box (Figure 5E). The Trp40Ala mutation did not affect oligomerization (Figure 5E). Furthermore, we show that PUB22 U-box can interact with PUB24 in vitro, indicating that it may form heterooligomers (Figure 5E).
To substantiate our results, we performed in vivo split-luciferase complementation assays, which allow the analysis of dynamic protein-protein interactions (Stefan et al., 2007). As expected, the wild-type U-box domains were able to interact, as evidenced by the luminescence due to luciferase reconstitution (Figure 5F; Supplemental Figure 8A). Both Thr62Glu and Thr62Ala versions displayed a reduced oligomerization. However, the Thr62Glu phosphomimetic was considerably more impaired than Thr62Ala, which is in accordance with the reduced autoubiquitination activity (Figures 4B and 4C). Thr-62 is predicted to be located next to a negatively charged patch (Figure 5B). It is therefore possible that the exchange by a smaller amino acid such as alanine allows an expanded influence of this charged surface, inhibiting dimerization. Replacing Thr-62 with an isoleucine, a larger hydrophobic residue, resulted in enhanced interaction, potentially by limiting the influence of the negative patch and increasing hydrophobicity (Figure 5F). Confirming the importance of hydrophobic residues for oligomerization, replacement of the conserved Ile-25 by a positively charged arginine reduced oligomerization (Figure 5G; Supplemental Figure 8B). These results are also in agreement with a modeled PUB22 dimer (Supplemental Figure 8C). We challenged the structural model by replacing Phe-10, which faces a positively charged patch on the complementary monomer, by a glutamic acid that should create an electrostatic interaction (Supplemental Figure 8C). The Phe10Glu variant displayed increased interaction, supporting the validity of our model (Figure 5G). Finally, we also confirmed that the mutation Trp40Ala, which inhibits the interaction with the E2, has no effect on dimer/oligomer formation (Figure 5G).
To confirm that MPK3 influences PUB22 oligomerization, we expressed the U-box domains in Col-0 and mpk3 backgrounds. We used the MKK5 AspAsp to activate the immune response and the inactive LysArg as a control. While oligomerization was inhibited in Col-0, strong interaction was detected in the mpk3 background (Figure 5H; Supplemental Figure 8D).
Together, we show that PUB22 forms a dimer/oligomer via its U-box domain through hydrophobic interactions and that Thr-62 phosphorylation by MPK3 may inhibit the oligomeric status of the protein.
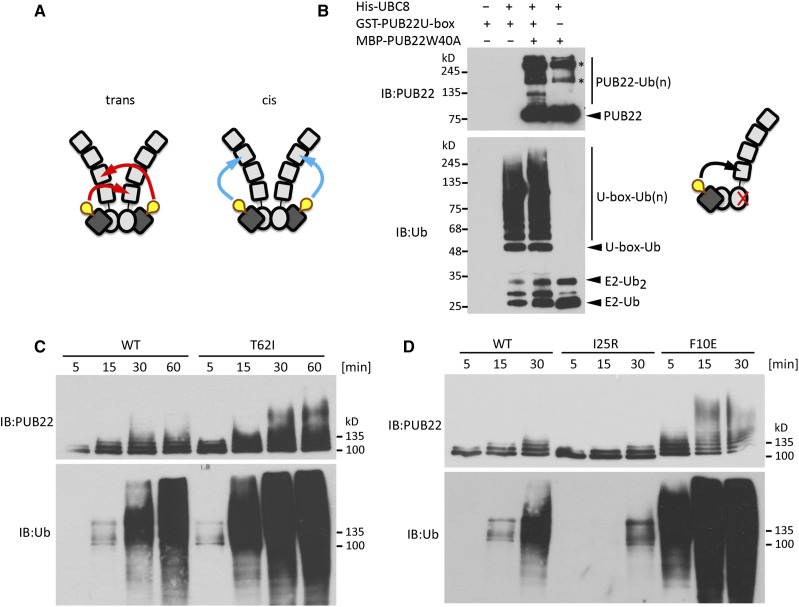
PUB22 Autoubiquitination Occurs in trans and Is Influenced by Oligomerization
A prerequisite for the oligomerization state of PUB22 to impinge on the autoubiquitination activity is that each PUB22 protomer ubiquitinates the other in trans; autoubiquitination in cis is less likely to be affected (Figure 6A). In order to test autoubiquitination in trans, we performed an in vitro ubiquitination assay using an active U-box, alone or in combination with an inactive full-length Trp40Ala PUB22. Full-length Trp40Ala and U-box proteins are able to form heterooligomers (Figure 5E), and if the inactive Trp40Ala is modified, it can only be the result of autoubiquitination in trans (Figure 6B). When both were mixed, the U-box efficiently ubiquitinated the Trp40Ala mutant protein, indicating that ubiquitination occurs in trans (Figure 6B, upper panel). The PUB22 U-box, which forms homooligomers, also displayed autoubiquitination both alone and in the presence of the Trp40Ala mutant (Figure 6B, lower panel).
Figure 6.
Autoubiquitination Activity of PUB22 Is Proportional to the Interaction Strength.
(A) Schematic illustrations of potential mechanisms of self-catalyzed ubiquitination by PUB22 dimer occurring in trans or cis.
(B) Trans-autoubiquitination assay employing GST-PUB22 U-box and MBP-PUB22 Trp40Ala as indicated. Samples were analyzed by IB using anti-PUB22 (recognizing the C terminus) and anti-ubiquitin. Schematic illustration of trans-autoubiquitination. Asterisks indicate potentially oligomeric forms of PUB22.
(C) Autoubiquitination assays with wild-type MBP-PUB22 and Thr62Ile variant in the presence of Arabidopsis His-UBA1 and His-UBC8.
(D) Autoubiquitination assays with wild-type MBP-PUB22, and Ile25Arg and Phe10Glu variants in the presence of Arabidopsis His-UBA1 and His-UBC8.
To gain further insight into the mechanism regulating PUB22 stability, we investigated the connection between oligomerization and autoubiquitination. The replacement of Thr-62 by an isoleucine increased oligomerization (Figure 5F). We tested whether the increased interaction resulted in increased self-catalyzed ubiquitination. In comparison to the wild-type PUB22, the Thr62Ile variant displayed increased activity (Figure 6C). By contrast, reduced interaction led to a reduced autoubiquitination when the conserved Ile-25 residue was replaced by an arginine (Figure 6D). Accordingly, replacing Phe-10 by a glutamic acid, which creates an electrostatic interaction, increased autoubiquitination (Figure 6D).
These results indicate that the oligomerization level regulates the self-catalyzed trans-ubiquitination activity of a PUB22 dimer/oligomer in a directly proportional manner.
Phosphorylation of PUB22 Contributes to the Dampening of MAPK Signaling and the Immune Response
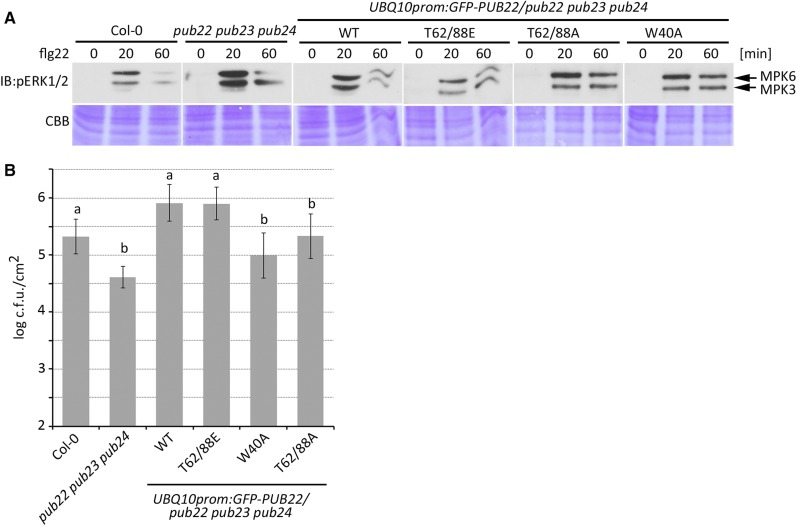
Finally, we investigated the biological significance of PUB22 stabilization by MPK3. The activation of the MAPK cascade is a hallmark of the immune responses triggered by pattern recognition receptors such as FLS2. This response is increased and prolonged in the pub22 pub23 pub24 triple mutant (Figure 7A). To test the in vivo effect of PUB22 phosphorylation, we analyzed MAPK phosphorylation in the triple mutant complemented with the wild-type PUB22, the phosphomimetic Thr62/88Glu, phosphonull Thr62/88Ala, and the inactive Trp40Ala variants. Introduction of PUB22 into the pub22 pub23 pub24 background resulted in partial complementation, as reflected by the reduced MAPK phosphorylation (Figure 7A). However, the phosphomimetic Thr62/88Glu dampened MAPK activation more efficiently than the wild-type E3.
Figure 7.
PUB22 Phosphorylation Is Required to Dampen Immune Responses.
(A) Phosphorylation of MPK3 and MPK6 in different genetic backgrounds. Two-week-old seedlings from Col-0, pub22 pub23 pub24, and transgenic lines constitutively expressing GFP-PUB22 wild-type and mutant variants in the pub22 pub23 pub24 background were treated with flg22 (100 nM) and harvested at the indicated time points. Blots were edited for presentation. The experiment was repeated with similar results.
(B) Infection assays with the bacterial pathogen Pst ΔAvrPto/AvrPtoB. Six-week-old plants were syringe inoculated with a bacterial suspension of 1 × 105 colony-forming units (c.f.u.)/mL and analyzed 2 d after inoculation. Data shown as mean ± sd (n = 5). Similar results were obtained in three independent experiments. Different letters indicate significantly different values P < 0.05 compared with pub22 pub23 pub24 (one-way ANOVA and Tukey post hoc test).
The inactive Trp40Ala and the Thr62/88Ala mutants were also able to partially complement the triple mutant phenotype, although less effectively than the Thr62/88Glu phosphomimetic (Figure 7A). This is not surprising, as PUB22 mutant variants are impaired in specific functions, while maintaining others. For example, the ligase inactive Trp40Ala is fully capable of forming heterooligomers and may stabilize related PUBs. Thr62/88Ala, on the other hand, even though present in low levels, is an active E3 that can mediate the ubiquitination of substrates. Indeed, PUB22 is present at low concentrations in naïve plants (Figure 1I). Nevertheless, as exemplified by the phenotypes of the PUB22 mutant lines, the absence of these low concentrations influences early responses (Trujillo et al., 2008).
To corroborate the relevance for plant immunity, we performed pathogen infection assays using Pst lacking the two effector proteins AvrPto and AvrPtoB. Reduced virulence of this strain improves the detection of PAMP-related phenotypes. The introduction of the wild-type PUB22 resulted in complementation of the enhanced resistance phenotype in the pub triple mutant (Figure 7B), i.e., resulting in plants being again more susceptible than the triple mutant toward PstΔAvrPto/AvrPtoB infection. The suppression of the enhanced resistance phenotype in plants expressing the Thr62/88Glu phosphomimetic was as high as that of wild-type protein (Figure 7B). By contrast, Thr62/88Ala resulted in no significant change in resistance. As a control, we used the inactive Trp40Ala mutant, which similarly to Thr62/88Ala, did not result in an enhanced susceptibility compared with Thr62/88Glu or the wild type.
Thus, we showed that the phosphomimetic PUB22 actively dampens the immune response, which as a consequence renders plants more susceptible to infection when introduced into the pub22 pub23 pub24 background.
DISCUSSION
Negative Regulatory Loops in the Immune Response
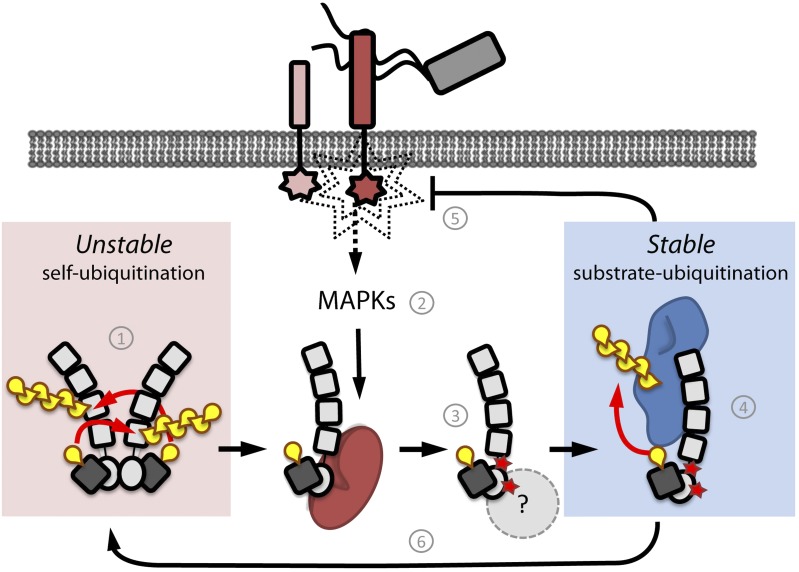
Negative feedback loops are inherent to all signaling networks and play several functions that include buffering perturbations and determining the kinetics of a signaling event (Kiel and Serrano, 2012; Ferrell, 2013). The PUB22 ubiquitin ligase belongs to a group of proteins shown to collectively dampen immune signaling. Here, we identified MPK3 as an upstream signaling component controlling PUB22’s activity to trigger a negative feedback loop (Figure 8).
Figure 8.
Model of PUB22 Regulation and Dampening of the Immune Response.
(1) PUB22 exists as a dimer that mediates constant autoubiquitination in trans, mediating its degradation and keeping levels low. (2) Upon infection, immune signaling is triggered and MAPK cascades are activated. (3) Activated MPK3 (red) phosphorylates PUB22 at residues Thr-62 and Thr-88, inhibiting dimerization. This results in a nonautoubiquitinating and stable monomeric PUB22. Additional factors may contribute to PUB22 stabilization. (4) Increased stability of the monomeric PUB22 allows it to engage substrates such as Exo70B2 (blue), which are likely to control signaling upstream of MPK3. (5) Ubiquitination of PUB22 targets leads to dampening of the immune response. (6) Finally, depletion of substrates and/or dephosphorylation may allow dimerization and, thus, autoubiquitination and finally degradation.
MAPKs are highly conserved signaling modules that regulate fundamental cellular processes, including immune signaling. MAPK activity has a major impact on cellular reprogramming by targeting transcription factors and other substrates (Rodriguez et al., 2010). Because MPK3 activation is prolonged in pub22 pub23 pub24, our initial hypothesis was that PUB22 negatively regulated MPK3 by ubiquitination. In line with this notion, the cIAP1 and XIAP E3s conjugate K63-linked ubiquitin chains to MEKK2 and MEKK3, directly impeding the MEK5-ERK5 interaction and downregulating the activity of the MAPK ERK5 (Takeda et al., 2014). However, we did not detect any ubiquitination of MPK3 by PUB22 (Supplemental Figure 3B) under our conditions, suggesting that the prolonged activation of MPK3 originates from a different mechanism. Enhanced MAPK activity in the pub22 pub23 pub24 background may instead be the result of the accumulation of substrates such as Exo70B2 (Stegmann et al., 2012), which potentially control the levels of signaling components upstream of MAPK activation.
Instead, we found that MPK3 phosphorylates PUB22. Both MPK3 and MPK6 are commonly described as partially redundant positive regulators, while MPK4 is appreciated as a negative regulator. However, recent studies have helped to draw a more precise picture, indicating that MPK3 and MPK6 are far more functionally distinct than initially assumed (Frei dit Frey et al., 2014; Lassowskat et al., 2014). At the same time, MPK3 displays an overlap in transcriptome, substrates, and functions with MPK4 (Popescu et al., 2009; Frei dit Frey et al., 2014). Unlike mpk4 mutants, which display a constitutively active immunity phenotype, the negative regulatory function of mpk3 is restricted to pathways activated during the immune response, in analogy to the function of PUB22 and its homologs (Trujillo et al., 2008; Frei dit Frey et al., 2014). In line with this, mpk3 phenocopies the pub22 pub23 pub24 triple mutant, both of which display increased responses (Trujillo et al., 2008; Frei dit Frey et al., 2014). However, MPK4 function during immunity needs to be reassessed, as phenotypes in mpk4 may originate from the activation of the NOD-like receptor SUMM2 (Zhang et al., 2012).
The function of MPK3 as a convergence point of signaling mediated by different receptors explains the stabilization of PUB22 by treatment with various agonists. Indeed, the interaction between U-box type E3 ligases and kinases, especially receptor kinases with functions in diverse processes such as symbioses, self-incompatibility, and immunity, is a common theme (Gu et al., 1998; Samuel et al., 2008; Mbengue et al., 2010; Lu et al., 2011). Our study doesn’t exclude the possibility that PUB22 is also controlled by additional kinases nor that MPK3 may regulate the activity of additional PUB E3s. Efforts to generate a multiple mutant have been unsuccessful because MPK3 (At3g45640) is genetically linked to PUB24 (At3g11840) and PUB22 (At3g52450).
It is conceivable that autoubiquitination, a trait common to all PUB E3s investigated to date, is regulated by phosphorylation, which in consequence, impinges on their dimerization status and, thus, stability.
Dimerization and E3 Ligase Activity
Our results indicate that PUB22 displays ubiquitination activity (activity being defined as auto- or substrate ubiquitination), both as a dimer/oligomer and a monomer. This raises a fundamental question: How is the conformational restriction of the E2-ubiquitin conjugate achieved? This process requires the E3 and primes ubiquitin for catalysis (Berndsen and Wolberger, 2014).
In several instances, it has been shown that dimeric E3s require dimerization for ubiquitination activity (Dueber et al., 2011; Dou et al., 2012a; Plechanovová et al., 2012). Elegant experiments by Plechanovová et al. (2012) and Dou et al. (2012b) showed that the E2 contacts a single protomer of the dimeric E3 (RNF4 and Birc7, respectively), while ubiquitin is folded back onto the E2 via contacts from both RING molecules. Mutational analysis of the involved residues supported a mechanism by which both protomers cooperate for ubiquitin priming (Plechanovová et al., 2011). However, impairing dimerization of PUB22 by mutating Thr-62 did not reduce Exo70B2 ubiquitination or its activity in dampening the immune response.
Monomeric E3s, such as the U-box type E4B, are also able to prime ubiquitin through a mechanism that has not yet been fully elucidated (Benirschke et al., 2010; Pruneda et al., 2012; Soss et al., 2013). The residue Arg-1143 in E4B plays a key role in bringing ubiquitin into a closed state required for hydrolysis (Pruneda et al., 2012). Notably, the equivalent residue is conserved and critical for catalysis in the dimeric RNF4 (Arg-181) and is also present in PUB22 and dimerizing U-box E3s (Figure 5A, green box). Arg-181 in RNF4 and Arg-1143 in E4B have contacts with both the E2 and Ub at the junction of the E3-E2-Ub heterotrimer, emphasizing their importance (Plechanovová et al., 2012; Pruneda et al., 2012). This indicates that monomeric as well as dimeric E3s share elements required to achieve the priming of Ub, while dimeric ligases may require additional interactions. Therefore, our results suggest that in contrast to dimeric E3s, PUB22’s ubiquitination activity and oligomerization status are uncoupled. Hence, even if PUB22 can exist in a dimeric state, it is likely to rely on the monomeric E3 priming mechanism.
Autoubiquitination, a Mechanism of Self-Regulation?
Autoubiquitination is a general feature of the majority of E3 ligases in vitro and it is commonly used to assess their activity. Nevertheless, the functional consequences of self-catalyzed ubiquitination in vivo are not entirely understood.
Although in vivo autoubiquitination has not been previously shown for single subunit E3 ligases, some examples are available from other systems (Weissman et al., 2011). These include the human E3 cIAP1, which is thought to occur as a monomer in which the RING domain is sequestered (Dueber et al., 2011). Binding of cIAP1 antagonists induces a conformational rearrangement that enables dimerization through the RING domain. Subsequently, cIAP1 autoubiquitination leads to its proteasomal degradation and releases inhibition of apoptosis (Varfolomeev et al., 2007). In addition, cIAP1 and related E3s play central roles in the activation of the innate immune response (Vandenabeele and Bertrand, 2012). However, independently of the signaling pathway (e.g., apoptosis or innate immunity), an intact RING mediating dimerization, and thus also autoubiquitination, is required. Similarly, activation of cCBL triggers its ubiquitinating activity toward its substrates and itself, leading to a rapid decrease of both its levels and the substrates (Ryan et al., 2006; Dou et al., 2012b).
In contrast to cCBL and cIAP1, which need to be relieved from an autoinhibited state, our results indicate that PUB22 constitutively autoubiquitinates to maintain low basal levels and that a shift to the monomeric state enables it to carry out its function by allowing it to accumulate. Similarly, inhibition of autoubiquitination may allow the E3 ligase Mdm2 to target p53. Mdm2 associates with the related MdmX, which lacks appreciable E3 activity. The RING-dependent heterodimerization enhances p53 ubiquitination and at the same time stabilizes Mdm2 by reducing autoubiquitination (Uldrijan et al., 2007; Okamoto et al., 2009). F-boxes, which are substrate adaptors for Cullin-RING-Ligases (CRLs), have also been shown to be unstable and may underlie a similar regulatory principle (Galan and Peter, 1999). Ubiquitination requires all core components of the CRL, suggesting that it occurs within the complex by an autocatalytic mechanism.
Genes encoding PUB22 and related ligases are transcriptionally induced during the immune response. However, even with expression driven by a strong constitutive promoter, PUB22 protein levels were barely detectable (Figure 3E). Together with the absence of stabilization after flg22 stimulation in the phosphonull variant (Figure 3F), this indicates that inhibition of the autoubiquitination activity through Thr-62 phosphorylation is a prerequisite for PUB22 accumulation. The same holds true for Thr-88, which additively contributes to stabilization, likely through a different mechanism. Thr-88 is in a predicted disordered region (amino acids 81–92), and phosphorylation may trigger a transition to a structured form, the consequences of which could be twofold. First, it may alter the geometry of the molecule so that it is less prone to self-modification. Indeed, three of the identified autoubiquitination sites (Lys-90, -100, and -103) are located in or next to the predicted disordered region. Second, proteasomal degradation is enhanced by the presence of an unstructured site (Prakash et al., 2004). Thus, phosphorylation may result in PUB22 being less suitable as a substrate for the proteasome.
It can be argued that in most cases, the outcome of autoubiquitination is a self-regulatory feedback through which activity is tightly regulated. Phosphorylation of PUB22 most likely reduces autoubiquitination by shifting a dynamic balance from oligomeric to a monomeric state. However, the inability of the nonphosphorylatable variant Thr62Ala to stabilize after the activation of immune signaling in vivo, although it displays reduced autoubiquitination in vitro, may suggest an additional layer of regulation. We showed that PUB22 can heterooligomerize with PUB24, opening the possibility that heterooligomerization with related PUBs is not significantly affected in the Thr62/88Ala variant in vivo; thus, its instability is maintained. Heterooligomerization may also account for the remnant degradation levels of the inactive Trp40Ala variant. A second possibility is that the phosphorylated variant is recognized by an additional stabilizing factor such as a ubiquitin-specific protease that contributes to its stabilization, as shown for TRIM25 and TRAF6 (Pauli et al., 2014; Lin et al., 2015a).
Once stabilized, PUB22 may engage substrates such as Exo70B2 and contribute to the downregulation of the immune response upstream of MPK3 (Figure 8). The interaction with substrates may also contribute to stabilization, and their degradation will ultimately result in the E3s following their fate, terminating a signaling cycle. Together, our observations uncover the multilayered regulation of ubiquitination by a single unit E3 that allows fine-tuning of its activity. The inherent tendency of E3 ligases to autoubiquitinate may thus act as a built-in fail-safe system to ensure their self-catalyzed neutralization in the absence of a stimulus or substrates.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the control in the complementation assays. The pub22 pub23 pub24 triple mutant plants (SALK_07261, SALK_133841, and SALK_041046, respectively) and mpk3 single mutant plants (SALK_151594) were described previously (Wang et al., 2007; Trujillo et al., 2008). Primers used for genotyping are listed in Supplemental Table 1. Transgenic lines were generated using Agrobacterium tumefaciens GV3101 for transformation. Plants for protoplast isolation and disease resistance assay were grown for 6 weeks in phytochambers at 21°C in 8 h light (combination of cool white fluorescent lamps and incandescent lamps, 250 μmoles/m2/s irradiance) and 16 h dark at 60% humidity. Seedlings for biochemical analysis were grown for 2 weeks in 0.5× Murashige and Skoog (MS) medium with vitamins supplemented with 0.25% sucrose and 500 mg/L MES, pH 5.6, in sterile 6-well plates, 12-well plates, or 50-mL flasks. All seeds were first stratified for 2 d at 4°C in the dark. Two independent lines were chosen for further characterization, and B lines were used for the assays. The following concentrations were used for treatments as indicated: 100 nM or 1 µM flg22, 20 or 50 µM CHX, 50 µM MG132, and 1 µM okadaic acid. After the specified treatments, seedlings were dried on paper, flash-frozen in liquid nitrogen, and ground in a mortar. The frozen powder was resuspended in protein extraction buffer (50 mM Tris-HCl, pH 6.8, 4% SDS, 8 M urea, 30% glycerol, 0.1 M DTT, and 0.005% bromophenol blue), incubated for 10 min at 68°C, and centrifuged for 5 min at 6000g before protein gel blot analysis.
Generation of Transgenic Lines
The cloning of PUB22 coding sequence into the pENTR/D-TOPO entry vector (Invitrogen) was described by Stegmann et al. (2012). Site-directed mutagenesis was performed on the entry vector, adapting the Type II’s restriction digest-based mutagenesis method described in Palm-Forster (Palm-Forster et al., 2012) using the oligonucleotides listed in Supplemental Table 2. The mutation event was confirmed by sequencing. The PUB22prom:GFP-PUB22 construct was generated using the pGWB406 (Nakagawa et al., 2007) vector backbone. The 35S promoter was replaced using HindIII-XbaI restriction digest by a USER cloning cassette and a 2-kb promoter fragment of PUB22 was inserted employing USER cloning. PUB22 was cloned into the modified vector via LR reaction (Invitrogen). For constitutive expression, PUB22 and PUB22 mutants were cloned into the plant expression vector pUBN-GFP-Dest (Grefen et al., 2010) under the control of the UBIQUITIN10 (UBQ10) promoter via LR reaction (Invitrogen). The transgenic lines were generated via Agrobacterium-mediated transformation in pub22 pub23 pub24 plants. Positive transformants were screened with 200 mg/L BASTA and confirmed via immunoblot using anti-GFP antibodies. The homozygous lines were selected based on the survival rate on full-strength MS plates supplemented with 1% sucrose and 8 g/L agar containing 50 mg/L kanamycin or 10 mg/L glufosinate. Stability analyses were performed on T3 homozygous seedlings or on T2 seedlings segregating 1:2:1 using n ≥ 100.
Protoplast Transformation for Bimolecular Fluorescence Complementation and PUB22 Stabilization Assay
PUB22 and W40A variant were cloned into pE-SPYCE (HA-cYFP) (Ehlert et al., 2006) via LR reaction (Invitrogen). Constructs of MPKs in pE-SPYNE (myc-nYFP) were previously published (Pecher et al., 2014). Each plasmid was used for cotransformation with an mCherry-expressing vector at a final concentration of 50 and 30 µg/mL, respectively. For microscopy analysis, an LSM710 (Zeiss) was employed with the following settings: YFP excitation, 488 nm; emission, 510 to 560 nm; mCherry excitation, 594 nm; emission, 600 to 640 nm.
The HA-tagged PUB22 construct and the cMyc-tagged MKK5 AspAsp and MKK5 LysArg constructs from Petroselinum crispum were previously described (Stegmann et al., 2012; Lassowskat et al., 2014).
For the luciferase-based assays, PUB22 was cloned into a modified pUGW15 vector. After the insertion of an additional KpnI site following the HA tag, the luciferase gene was amplified and ligated using KpnI-KpnI restriction ends. PUB22 was cloned into pUGW15-HA-LUC via LR reaction to generate an N-terminal HA-Luciferase fusion protein (HA-Luc-PUB22).
For protein stabilization analysis, the following DNA concentrations were used: 50 µg/mL GFP-PUB22, 50 µg/mL Luc-PUB22, 30 µg/mL HA-PUB22, 30 µg/mL cMyc-nYFP-MPK3, 40 µg/mL YFP-MPK3, 40 µg/mL YFP-MPK6, and 10 µg/mL cMyc-MKK5. Specified treatments were performed 1 d after transformation. For immunoblot analysis, protoplast cells were harvested by spinning down for 1 min at 200g, flash frozen in liquid nitrogen, resuspended in protein extraction buffer, and incubated for 10 min at 68°C. Mesophyll protoplasts were isolated as previously described (Wu et al., 2009).
Split-Luciferase Assay
The PUB22 U-box coding sequence for C-terminal fusion was amplified using the primers in Supplemental Table 3 and cloned by USER-cloning (NEB) into a modified pENTR3C-U. LR reactions were performed with the destination vectors pCAMBIA/des/cLuc and pCAMBIA/des/nLuc (Chen et al., 2008; Lin et al., 2015b). Protoplasts were generated as previously described (Wu et al., 2009) from pub22 pub23 pub24 or mkp3 mutant plants and transformed with 25 µg/mL of each construct. Split luciferase assays were performed 1 d after transformation. Luciferase activity was assessed by adding 1 mM d-Luciferin substrate to the protoplast solution and measuring the luminescence on the Tecan Infinite F200 Pro plate reader using 20,000-ms integration time.
In Vivo Coimmunoprecipitation
For immunoprecipitation, 500 mg of seedling material was ground in liquid nitrogen using a mortar. The ground material was resuspended in lysis buffer (dilution buffer supplemented with 1% Nonidet P-40) and cleared by centrifugation for 10 min at 14,000g. The supernatant was diluted 1:1 with dilution buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM EDTA, 5 mM DTT, 50 µM MG132, 1 mM 4-benzenesulfonyl fluoride hydrochloride, 1 mM NaF, 0.5 mM Na3VO4, 15 mM β-glycerophosphate, and 1% Protease Inhibitor Cocktail) and incubated for 3 h at 4°C with GFP-Trap-A beads (Chromotek). Beads were washed four times with dilution buffer, and the bound fraction was eluted by adding SDS-PAGE sample buffer (240 mM Tris-HCl, pH 6.8, 0.5 M DTT, 10% SDS, and 50% glycerol) and incubating beads for 10 min at 95°C. Samples were analyzed by immunoblot with the specified antibodies.
Recombinant Protein Purification and in Vitro Ubiquitination Assay
PUB22 and PUB24 were cloned using SalI-PstI and EcoRI-PstI restriction digestion, respectively, into pMal-c2X (New England Biolabs) (Supplemental Table 3). MBP-PUB22 and MBP-PUB24 were purified by affinity chromatography using amylose resin (New England Biolabs). The cloning of PUB22 U-box in pENTR/D-TOPO (Invitrogen) entry vector was previously described (Stegmann et al., 2012). Entry clone for PUB22 U-box was used for cloning into pDest15 Gateway vector via LR reaction (Invitrogen) to generate recombinant GST-PUB22 U-box, which was purified using Protino Glutathione Agarose 4B (Macherey Nagel). Exo70B2 was amplified from the entry clone (Stegmann et al., 2012) using primers in Supplemental Table 3 and cloned into pET28b (Novagen) using EcoRI and XhoI restriction enzymes. Recombinant His-Exo70B2 was purified using Protino Ni-Ted resin (Macherey Nagel). Recombinant His-UBA1 and His-UBC8 were purified using Protino Ni-Ted resin (Macherey Nagel) and stored at −80°C. All recombinant proteins were expressed in Escherichia coli Rosetta 2(DE3) pLysS.
The E3 ligase enzyme was used for ubiquitination assays on the same day it was purified. In a total of 30 µL, 0.2 µg of His-UBA1 and 1.2 µg of His-UBC8 were combined with 2 µg of E3 ligase in ubiquitination buffer (40 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 mM KCl, 1 mM DTT, 2 mM ATP, and 2 µg ubiquitin from bovine erythrocytes) and incubated at 30°C for the indicated time. For substrate ubiquitination, all of the components, including 0.3 µg of His-Exo70B2, were mixed and preincubated at 30°C for 1 h in the absence of ubiquitin. The reaction was stopped by the addition of 10 μL of SDS-PAGE sample buffer and incubated at 68°C for 10 min. The samples were analyzed by protein gel blot analysis.
For the untagged PUB22 ubiquitination assay, GST-PUB22 fusion protein expressed from pGEX-4T-1 (Trujillo et al. 2008) was purified using Protino Glutathione Agarose 4B (Macherey Nagel). After washing, the loaded agarose beads were incubated in PBS buffer in the presence of 1 unit of thrombin for 4 h at 22°C. The supernatant containing the untagged PUB22 was then transferred into a new tube and incubated in ubiquitination buffer with His-UBA1 and His-UBC8 overnight at 22°C.
To assay MPK3-dependent inhibition of PUB22 autoubiquitination, GST-MPK3 bound to glutathione resin was first used to pull down MBP-PUB22 as described below. The constitutively active PcMKK5-AspAsp or inactive PcMKK5-LysArg (Lee et al., 2004) was added to the MPK3-PUB22 complex and incubated for 1 h at 30°C in ubiquitination buffer lacking ubiquitin. The samples were harvested at the indicated time points after the addition of ubiquitin.
In Vitro Pull-Down
Recombinant GST-MPK3 and GST-MPK6 were expressed from pGEX-4T-1 (Feilner et al., 2005) in E. coli BL21(DE3). Free GST was expressed from the empty pGEX-4T-1. PUB22 U-box was cloned in pDest17 Gateway vector (Thermo Fisher Scientific) for His-tagged fusion protein expression via LR reaction (Invitrogen).
Lysates from E. coli containing GST-tagged recombinant proteins were immobilized for 1 h at room temperature on Protino Glutathione Agarose 4B (Macherey Nagel) in PBS buffer. The loaded agarose beads were then incubated with lysate containing the interacting partner for 1 h at room temperature in PBS buffer. The bound protein was washed four times and eluted with SDS-PAGE sample buffer and analyzed by immunoblotting.
In Vitro Phosphorylation Assay
In vitro phosphorylation assays were performed in kinase buffer (20 mM HEPES, pH 7.5, 15 mM MgCl2, 5 mM EGTA, 1 mM DTT, 0.1 mM ATP, and 2 µCi [γ-32P]ATP) using recombinant full-length GST-PUB22, MBP-PUB22, and MBP-PUB24 and active GST-MPK3 and GST-MPK4 or nontagged MPK6 (preactivation was performed using constitutively active PcMKK5-DD) (Lee et al., 2004). Samples were incubated for 30 min at 37°C; reactions were stopped by the addition of SDS-PAGE sample buffer and separated by 10% SDS-PAGE. Gels were stained with Coomassie Brilliant Blue and analyzed by autoradiography. The Phos-tag gels were made as described (Kinoshita et al., 2009)
Structure Modeling
Model building was performed using the web-based SWISS-MODEL for protein structure homology modeling (Arnold et al., 2006) using the zebra fish (Danio rerio) CHIP (PDB ID code 2F42). Model quality was assessed by determining the QMEAN score: monomer Q-mean 0.647, z-score −0.55; dimer Q-mean 0.692, z-score −0.8. Images were generated using PyMol (Schrodinger, 2010).
Bacterial Infection Assay
For bacterial infection assays, 6-week-old plants were used. Pseudomonas syringae pv tomato ΔavrPto/ΔavrPtoB were streaked out on Kings B media, grown overnight at 28°C, and subsequently resuspended in 10 mM MgCl2 to a concentration of 1 × 105 colony-forming units/mL. The bacterial solution was used for leaf infiltration using a needle-less syringe. Leaf discs were taken 2 d postinoculation using a cork borer (0.25 cm2) from three leaves per plant and six plants per genotype. Leaf discs were ground in water, diluted, and plated on Luria-Bertani agar with appropriate selection. Plates were incubated at 28°C and colonies were counted 2 d later.
Root Growth Inhibition Assay
T3 generation homozygous seedlings were used. Seedlings were placed onto solid 0.5× MS medium (Duchefa) and stratified for 3 d at 4°C in a vertical position and subsequently grown for 5 d under long-day conditions. The seedlings were transferred onto solid 0.5× MS medium square plates supplemented with 1 µM flg22 or the vehicle DMSO and grown vertically for another 7 d under long-day conditions. Root length was measured using ImageJ software.
Immunoblot Analysis and Antibodies Used in This Study
Protein samples were analyzed on 10% bis-acrylamide Tris-glycine SDS-PAGE. Following the separation on SDS-PAGE and blotting on PVDF membranes (GE Healthcare), immunoblot analyses were performed with the following antibodies: anti-GFP 1:3000 (Santa Cruz Biotech; cat. no. Sc-8334), anti-AtMPK3 1:5000 (Sigma-Aldrich; cat. no. M8318, lot 104M4841V), anti-phospho-p44/42 MAPK 1:1000 (Cell Signaling Technology; cat. no. 9101, lot 28), anti-cMyc 1:5000 (Sigma Aldrich; cat. no. C3956), anti-HA 1:1000 (Eurogentec; cat. no. MMS-101P, lot HA16031), anti-ubiquitin P4D1 1:5000 (Santa Cruz Biotechnology; cat. no. Sc-8017), anti-His 1:10000 (MACS; cat. no. 120-003-811), anti-MBP 1:1000 (Sigma-Aldrich; cat. no. M1321, lot 103M4825V), anti-GST 1:5000 (GE Healthcare; cat. no. 27-4577, lot 9523210), and anti-luciferase 1:5000 (Sigma-Aldrich; cat. no. L0159). The anti-PUB22 antibody was developed against the C-terminal peptide RVWRESPCVPRNLYDSYPA and diluted 1:1000 for use (Thermo Fisher Scientific). For comparison of proteins from different transgenic lines, PAGE was performed in parallel, and gel sections containing the protein of interest were blotted onto a single membrane and developed by exposing films to chemiluminescence. Quantification of the bands’ intensities was performed using ImageJ by integrating area of the peak corresponding to the band of PUB22 and normalizing the value to an unspecific band (anti-PUB22) or Coomassie (anti-GFP).
cDNA Synthesis and Quantitative RT-PCR
Total RNA was prepared from adult Arabidopsis leaves using a Plant RNA mini kit (E.Z.N.A. Omega bio-tek), followed by a DNaseI digestion (Thermo Fisher Scientific). For first-strand synthesis, 1 μg of total RNA was converted into cDNA with the Maxima first-strand cDNA synthesis kit for RT-qPCR (Thermo Fisher Scientific) according to the manufacturer’s protocol. The cDNA was diluted to fixed quantities (9.5 ng per reaction of reverse transcribed total RNA).
Quantitative PCR was performed in 20 μL reaction volume, including 9.5 ng of reverse transcribed total RNA, 0.3 µM of each gene-specific primer (Supplemental Table 4), and Maxima SYBR Green qPCR Master Mix 2X (Thermo Fisher Scientific). Corresponding minus reverse transcriptase and no template controls were performed with each primer pair. The RT-qPCR was performed using a Bio-Rad CFX device with the following protocol: 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and a subsequent standard dissociation protocol to validate the presence of a unique PCR product.
In order to calculate relative transcription levels, the delta of threshold cycle (ΔCt) values were calculated by subtracting the arithmetic mean Ct values of the target PUB22 from the arithmetic mean Ct value of the normalizing PP2A, which was obtained from the three technical replicates. The relative transcription level (2−ΔCt) was calculated from samples obtained from three independent experiments.
Proteomics
Identification of Phosphopeptides by LC-MS/MS
Site-specific phosphorylation of PUB22 by MPK3 and MPK4 was studied in vitro by kinase assay followed by liquid chromatography online with high-resolution accurate mass spectrometry. Proteins were separated with SDS-PAGE. Following in-solution protein digestion with trypsin, phosphorylated peptides were enriched using TiO2 affinity chromatography. Peptides were separated using C18 reverse phase chemistry employing a precolumn (EASY column SC001, length 2 cm, i.d. 100 µm, particle size 5 µm) in line with an EASY column SC200 with a length of 10 cm, an i.d. of 75 µm, and a particle size of 3 µm (both from Thermo Fisher Scientific). Peptides were eluted into a Nanospray Flex ion source (Thermo Fisher Scientific) with a 30-min gradient increasing from 5 to 40% acetonitrile in double distilled water and electrosprayed into an Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific). The source voltage was set to 1.9 kV and the S-Lens RF level to 50%. The delta multipole offset was −7.00. The phospho-peptide fraction was measured with a data-dependent acquisition scan strategy with inclusion list to specifically target PUB22 peptides bearing an MAPK phosphorylation site motif potentially phosphorylated by MPK3, MPK4, and MPK6 for MS/MS peptide sequencing. The AGC target value was set to 1e06 and the maximum injection time (max IT) to 500 ms in the Orbitrap. The parameters were set to 1e04 and 100 ms in the LTQ with an isolation width of 2 D for precursor isolation and MS/MS scanning. Multistage activation was applied to further fragment ion peaks resulting from neutral loss of the phosphate moiety by dissociation of the high energy phosphate bond to generate b- and y- fragment ion series rich in peptide sequence information. MS/MS spectra were used to search the TAIR10 database with the Mascot software v.2.5 integrated in Proteome Discoverer v.1.4. The enzyme specificity was set to trypsin, and two missed cleavages were tolerated. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine and phosphorylation of serine and threonine as variable modifications. The precursor tolerance was set to 7 ppm, and the product ion mass tolerance was set to 0.8 D. A decoy database search was performed to determine the peptide false discovery rate. The phosphoRS module was used to localize the phosphorylation site in the peptide’s primary structure.
To identify the phosphorylation sites in vivo, UBQ10:GFP-PUB22/pub22 pub23 pub24 seedlings were grown 12 d in liquid medium and treated +/− flg22 (1 µM) for 30 min. GFP-PUB22 was immunoprecipitated using GFP-trap beads and separated with SDS-PAGE and in-gel digested using a combination of Glu-C and trypsin. Phosphorylated peptides were enriched as above and separated as above on an EASY-nLC 1000 LC system with a column length of 50 cm, i.d. of 75 µm, and a particle size of 2 µm using a 90-min gradient and a flow rate of 250 nL/min. Peptides were electrosprayed online into a QExactive Plus mass spectrometer from Thermo Fisher Scientific. The spray voltage was 1.9 kV, the capillary temperature 275°C, and the Z-Lens voltage 240 V. A full MS survey scan was performed with chromatographic peak width set to 15 s, resolution 70,000, automatic gain control 3E+06, and a max injection time of 200 ms. MS/MS peptide sequencing was performed using a Top10 data-dependent acquisition inclusion list scan strategy as above with high-energy collisional dissociation fragmentation. MS/MS scans were acquired with resolution 17,500, automatic gain control 5E+04, injection time of 150 ms, isolation width 1.6 m/z, normalized collision energy 28, under fill ratio 3%, and an intensity threshold of 1E+04. MS/MS spectra were searched as above with enzyme specificity set to trypsin + Glu-C tolerating three missed cleavages, a precursor tolerance of 5 ppm, and a product ion mass tolerance of 0.02 D.
Identification of Ubiquitinated Proteins by LC-MS/MS
In-gel digestions were performed as described previously (Shevchenko et al., 2006). Digested peptides in gel pieces were recovered by adding 5% formic acid/acetonitrile, desalted using StageTips (Rappsilber et al., 2003), dried in a vacuum evaporator, and dissolved in 5% acetonitrile containing 0.1% trifluoroacetic acid. An LTQ-OrbitrapXL coupled with an EASY-nLC1000 (Thermo Fisher Scientific) was used for LC-MS/MS analyses. Self-pulled needle packed with C18 resin was used as an analytical column (Ishihama et al., 2002). Spray voltage of 2400 V was applied. Mobile phase consisted of 0.5% acetic acid (A) and 0.5% acetic acid and 80% acetonitrile (B). Two-step linear gradient of 0 to 40% B in 30 min, 40 to 100% B in 5 min, and 100% B for 10 min was employed at a flow rate of 500 nL/min. MS scan range was m/z 300 to 1400. Top 10 precursor ions were selected in MS scan by Orbitrap with 100,000 resolution and for subsequent MS/MS scans by ion trap in automated gain control mode, where automated gain control values of 5.00e+05 and 1.00e+04 were set for full MS and MS/MS, respectively. Normalized collision-induced dissociation was set to 35.0. Lock mass function was used to obtain constant mass accuracy during gradient analysis (Olsen et al., 2005). Database searching was performed as described previously (Nakagami et al., 2010). Peptides were identified by means of automated database searching using Mascot version 2.5 (Matrix Science) in The Arabidopsis Information Resource database (TAIR10_pep_20101214, ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/) containing protein sequence information for GST-fused PUB22 with a precursor mass tolerance of 3 ppm, a fragment ion mass tolerance of 0.8 D, and strict trypsin specificity (Olsen et al., 2004), allowing for up to two missed cleavages. Carbamidomethylation of Cys was set as a fixed modification, and oxidation of Met and diGly modification of Lys were allowed as variable modifications.
Phylogenetic Analysis
Forty-two PUB and one AtCHIP amino acid sequence from Arabidopsis were aligned using the L-INS-i option in MAFFT (Katoh et al., 2005). For visualization and editing of the alignment, Jalview was used (Waterhouse et al., 2009). LG+G was selected as the best fitting amino acid substitution model according to the Bayesian Information Criterion in MEGA-CC Model Selection analysis (Kumar et al., 2012). To reconstruct the phylogeny, the maximum likelihood (ML) algorithm with a bootstrap test (1000 replications) implemented in MEGA-CC was used (additional settings: no. of discrete gamma categories = 5, site coverage cutoff (%) = 95, ML heuristic method = nearest-neighbor-interchange, initial tree for ML = make initial tree automatically, branch swap filter = none, gaps/missing data treatment = partial deletion). To root the tree, AtCHIP was used as an outgroup because it possesses a conserved U-box, but ARM repeats are replaced by tetratricopeptide repeat. For visualization of the tree, FigTree (version 1.4.3) was used.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: PUB22 (At3G52450), PUB23 (At2G35930), PUB24 (At3G11840), UBC8 (At5G41700), UBA1 (AT2G30110), MPK3 (At3G45640), MPK4 (At4G01370), MPK6 (At2G43790), MPK11 (At1G01560), MKK5 (AY533302), Exo70B2 (At1G07000), PP2A (At1G13320), HsE4B (NM_001105562), MmCHIP (NM_019719), and DrCHIP (BC051775).
Supplemental Data
Supplemental Figure 1. PUB22 displays autoubiquitination activity in vitro.
Supplemental Figure 2. PUB22prom:GFP-PUB22 complements the pub22 pub23 pub24 phenotype.
Supplemental Figure 3. Posttranslational modification of PUB22 and PUB24.
Supplemental Figure 4. In vitro and in vivo phosphorylation sites of PUB22.
Supplemental Figure 5. Phylogenetic relationships between Arabidopsis PUB proteins class II and III.
Supplemental Figure 6. PUB22 stabilization is MPK3 dependent.
Supplemental Figure 7. Transgenic lines express comparable levels of transcripts of PUB22 wild-type and mutant variants.
Supplemental Figure 8. PUB22 homo- and heterooligomerizes.
Supplemental Table 1. Primers used for genotyping.
Supplemental Table 2. Primers used for SDM on PUB22 and PUB22 U-box in entry vectors.
Supplemental Table 3. Primers used for cloning.
Supplemental Table 4. Primers used for qRT-PCR.
Supplemental File 1. Alignment used to produce the phylogenetic tree shown in Supplemental Figure 5.
Supplementary Material
Acknowledgments
We thank Vardis Ntoukakis, Patrick Schäfer, and Maria Klecker for critical reading of the manuscript and all the members of the Trujillo Lab and Frederik Faden for fruitful discussions and valuable comments. We thank Marcel Quint and Phillipp Janitza for help with the phylogenetic analysis and Jörn Klinkenberg for the generation of the PUB22prom:GFP-PUB22/pub22 pub23 pub24 lines. This research was funded by the Leibniz association and the state of Saxony-Anhalt (G.F., K.K., P.M., W.H., and M.T.), the BMBF project ProNET-T3 03ISO2211B (J.L. and L.E.-L.), JSPS KAKENHI Grants 24688007 and 15H01247 (H.N.), and German-Israeli Foundation Grant I-1192-149.12/2012 (X.J.).
AUTHOR CONTRIBUTIONS
G.F., J.L., W.H., and M.T. designed experiments. G.F. generated lines, performed genetic analyses, ubiquitination assays, and interaction assays. L.E.-L. performed phosphorylation assays. K.K. performed RT-qPCR. X.J. performed the Phos-tag gels. W.H., H.N., and P.M. performed MS analyses. M.T. wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- MAPK
mitogen-activated protein kinase
- CHX
cycloheximide
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MS
Murashige and Skoog
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Benirschke R.C., Thompson J.R., Nominé Y., Wasielewski E., Juranić N., Macura S., Hatakeyama S., Nakayama K.I., Botuyan M.V., Mer G. (2010). Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure 18: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen C.E., Wolberger C. (2014). New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21: 301–307. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Song C., Kwak J.M., Kim W.T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Dou H., Buetow L., Sibbet G.J., Cameron K., Huang D.T. (2012a). BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19: 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H., Buetow L., Hock A., Sibbet G.J., Vousden K.H., Huang D.T. (2012b). Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19: 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber E.C., et al. (2011). Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 334: 376–380. [DOI] [PubMed] [Google Scholar]
- Ehlert A., Weltmeier F., Wang X., Mayer C.S., Smeekens S., Vicente-Carbajosa J., Droge-Laser W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46: 890–900. [DOI] [PubMed] [Google Scholar]
- Feilner T., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4: 1558–1568. [DOI] [PubMed] [Google Scholar]
- Ferrell J.E., Jr (2013). Feedback loops and reciprocal regulation: recurring motifs in the systems biology of the cell cycle. Curr. Opin. Cell Biol. 25: 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey N., et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.M., Peter M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96: 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R., Tsitsigiannis D.I., Ludwig A.A., Panicot M., Shirasu K., Jones J.D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007). The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28: 730–738. [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Rappsilber J., Andersen J.S., Mann M. (2002). Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979: 233–239. [DOI] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., Kogel K.H., Schäfer P. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156: 726–740. Erratum. Plant Physiol. 157: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C., Serrano L. (2012). Challenges ahead in signal transduction: MAPK as an example. Curr. Opin. Biotechnol. 23: 305–314. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., et al. (2015). A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 142: 444–453. [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Koike T. (2009). Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4: 1513–1521. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Peterson D., Tamura K. (2012). MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28: 2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I., Böttcher C., Eschen-Lippold L., Scheel D., Lee J. (2014). Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front. Plant Sci. 5: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Rudd J.J., Macioszek V.K., Scheel D. (2004). Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J. Biol. Chem. 279: 22440–22448. [DOI] [PubMed] [Google Scholar]
- Lin D., Zhang M., Zhang M.X., Ren Y., Jin J., Zhao Q., Pan Z., Wu M., Shu H.B., Dong C., Zhong B. (2015a). Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl. Acad. Sci. USA 112: 11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z.J., Liebrand T.W., Yadeta K.A., Coaker G. (2015b). PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 169: 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Wu Y.R., Huang X.H., Sun J., Xie Q. (2011). AtPUB19, a U-Box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana Mol. Plant. 4: 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153: 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Taya Y., Nakagama H. (2009). Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 583: 2710–2714. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Ong S.E., Mann M. (2004). Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3: 608–614. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., de Godoy L.M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005). Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4: 2010–2021. [DOI] [PubMed] [Google Scholar]
- Palm-Forster M.A., Eschen-Lippold L., Lee J. (2012). A mutagenesis-based screen to rapidly identify phosphorylation sites in mitogen-activated protein kinase substrates. Anal. Biochem. 427: 127–129. [DOI] [PubMed] [Google Scholar]
- Pauli E.K., Chan Y.K., Davis M.E., Gableske S., Wang M.K., Feister K.F., Gack M.U. (2014). The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 7: ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecher P., Eschen-Lippold L., Herklotz S., Kuhle K., Naumann K., Bethke G., Uhrig J., Weyhe M., Scheel D., Lee J. (2014). The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol. 203: 592–606. [DOI] [PubMed] [Google Scholar]
- Plechanovová A., Jaffray E.G., Tatham M.H., Naismith J.H., Hay R.T. (2012). Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489: 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A., Jaffray E.G., McMahon S.A., Johnson K.A., Navrátilová I., Naismith J.H., Hay R.T. (2011). Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18: 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Tian L., Ratliff K.S., Lehotzky R.E., Matouschek A. (2004). An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11: 830–837. [DOI] [PubMed] [Google Scholar]
- Pruneda J.N., Littlefield P.J., Soss S.E., Nordquist K.A., Chazin W.J., Brzovic P.S., Klevit R.E. (2012). Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47: 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75: 663–670. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Ryan P.E., Davies G.C., Nau M.M., Lipkowitz S. (2006). Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 31: 79–88. [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L.L.C. (2010). The PyMOL Molecular Graphics System, Version 1.3r1. (New York: Schrödinger). [Google Scholar]
- Seo D.H., Ryu M.Y., Jammes F., Hwang J.H., Turek M., Kang B.G., Kwak J.M., Kim W.T. (2012). Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160: 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Soss S.E., Klevit R.E., Chazin W.J. (2013). Activation of UbcH5c∼Ub is the result of a shift in interdomain motions of the conjugate bound to U-box E3 ligase E4B. Biochemistry 52: 2991–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan E., Aquin S., Berger N., Landry C.R., Nyfeler B., Bouvier M., Michnick S.W. (2007). Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. USA 104: 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Ichimura K., Pecenkova T., Reuter P., Žársky V., McDowell J.M., Shirasu K., Trujillo M. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731. [DOI] [PubMed] [Google Scholar]
- Takeda A.N., Oberoi-Khanuja T.K., Glatz G., Schulenburg K., Scholz R.P., Carpy A., Macek B., Remenyi A., Rajalingam K. (2014). Ubiquitin-dependent regulation of MEKK2/3-MEK5-ERK5 signaling module by XIAP and cIAP1. EMBO J. 33: 1784–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Uldrijan S., Pannekoek W.J., Vousden K.H. (2007). An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 26: 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P., Bertrand M.J. (2012). The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling. Nat. Rev. Immunol. 12: 833–844. [DOI] [PubMed] [Google Scholar]
- Vander Kooi C.W., Ohi M.D., Rosenberg J.A., Oldham M.L., Newcomer M.E., Gould K.L., Chazin W.J. (2006). The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry 45: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E., et al. (2007). IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131: 669–681. [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A.M., Shabek N., Ciechanover A. (2011). The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12: 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Joens M.S., Sinson A.B., Gilkerson J., Salomé P.A., Weigel D., Fitzpatrick J.A., Chory J. (2015). Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Devlin K.I., Ford M.G., Nix J.C., Qin J., Misra S. (2006). Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry 45: 4749–4759. [DOI] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D., Sadanandom A. (2006). The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D., Goring D.R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Yin Q., et al. (2009). E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S.M., Peggie M., Cohen P., Prodromou C., Pearl L.H. (2005). Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20: 525–538. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.