Plants efficiently control the acquisition of cold tolerance using two different signaling pathways in response to gradual and rapid temperature decreases, including seasonal and daily changes.
Abstract
In plants, cold temperatures trigger stress responses and long-term responses that result in cold tolerance. In Arabidopsis thaliana, three dehydration-responsive element (DRE) binding protein 1/C-repeat binding factors (DREB1/CBFs) act as master switches in cold-responsive gene expression. Induction of DREB1 genes triggers the cold stress-inducible transcriptional cascade, followed by the induction of numerous genes that function in the cold stress response and cold tolerance. Many regulatory factors involved in DREB1 induction have been identified, but how these factors orchestrate the cold stress-specific expression of DREB1s has not yet been clarified. Here, we revealed that plants recognize cold stress as two different signals, rapid and gradual temperature decreases, and induce expression of the DREB1 genes. CALMODULIN BINDING TRANSCRIPTION ACTIVATOR3 (CAMTA3) and CAMTA5 respond to a rapid decrease in temperature and induce the expression of DREB1s, but these proteins do not respond to a gradual decrease in temperature. Moreover, they function during the day and night, in contrast to some key circadian components, including CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL, which regulate cold-responsive DREB1 expression as transcriptional activators only during the day. Thus, plants efficiently control the acquisition of freezing tolerance using two different signaling pathways in response to a gradual temperature decrease during seasonal changes and a sudden temperature drop during the night.
INTRODUCTION
Plants have evolved complex systems to adapt to and survive in extreme temperature conditions. Under cold stress conditions, plants induce a large number of genes that function in the stress response and tolerance (Thomashow, 1999; Yamaguchi-Shinozaki and Shinozaki, 2006). The products of these genes function directly to enhance tolerance to freezing stress and to regulate gene expression and signal transduction under cold-stress conditions. The dehydration-responsive element (DRE), which has the same core motif as the C-repeat (A/GCCGAC), has been identified as a cis-acting element that regulates gene expression in response to both cold and dehydration stresses in plants (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994).
DRE binding protein 1A/C-repeat binding factor 3 (DREB1A/CBF3), an APETALA2/ethylene-responsive element binding factor-type transcription factor that binds to the DRE and activates the expression of cold-responsive genes, was isolated in a yeast one-hybrid screen (Stockinger et al., 1997; Liu et al., 1998). Furthermore, two cDNA clones homologous to DREB1A/CBF3 were isolated (DREB1B/CBF1 and DREB1C/CBF2; Liu et al., 1998; Shinwari et al., 1998) and shown to function in cold-responsive gene expression, similar to DREB1A. Overexpression of DREB1/CBFs driven by the Cauliflower mosaic virus (CaMV) 35S promoter improves tolerance to freezing, drought, and high salinity in transgenic Arabidopsis thaliana (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999). Transcriptome analyses have identified more than 100 downstream target genes of DREB1A (Seki et al., 2001; Maruyama et al., 2004, 2009; Park et al., 2015; Zhao et al., 2016). Many of the products of these genes in plants, including late embryogenesis abundant proteins, enzymes for osmoprotectant biosynthesis, transcription factors, and protein kinases, have been shown to function in acquisition of stress tolerance and in further regulation of stress responses. The levels of many metabolites (e.g., carbohydrates, amines, and organic acids) significantly increased in transgenic plants overexpressing DREB1A, similar to cold-treated plants (Kaplan et al., 2007; Maruyama et al., 2009).
Therefore, these three DREB1 transcription factors have been reported to act as master switches in cold-inducible gene expression (Yamaguchi-Shinozaki and Shinozaki, 2006). These genes are tandemly arrayed in the Arabidopsis genome in the order DREB1B, DREB1A, and DREB1C, and they are all rapidly and transiently induced by cold stress (Shinwari et al., 1998). Therefore, induction of DREB1s is thought to be the first step in the cold-responsive transcriptional cascade, followed by the expression of a large number of cold-inducible genes encoding proteins that function in the cold stress response and cold tolerance. Thus, it is important to elucidate the mechanisms of cold-inducible DREB1 expression. The promoter sequences of these three DREB1s, from the transcription initiation sites to −400 bp, contain six highly conserved motifs: boxes I to VI (Shinwari et al., 1998). In addition, two sequences that contribute to cold stress induction, ICEr1 and ICEr2, have been identified in the conserved regions of the CBF/DREB1 promoters (Zarka et al., 2003); these are identical to boxes IV and VI, respectively. Moreover, some transcription factors have been reported to regulate the cold stress induction of DREB1s directly or indirectly. INDUCER OF CBF EXPRESSION1/SCREAM (ICE1/SCRM), ICE2, and MYB15 are basic helix-loop-helix- and Myb-type transcription factors that regulate DREB1A expression under cold stress conditions (Chinnusamy et al., 2003; Agarwal et al., 2006; Kim et al., 2015). CALMODULIN BINDING TRANSCRIPTION ACTIVATOR3/Arabidopsis thaliana SIGNAL-RESPONSIVE GENE1 (CAMTA3/AtSR1), along with CAMTA1 and CAMTA2, have been reported to bind to the CGCG-box in ICEr2 and to activate the expression of DREB1B and DREB1C (Doherty et al., 2009; Kim et al., 2013). SCRM/ICE1 and SCRM2/ICE2 function in stomatal differentiation, and AtSR1/CAMTA3, CAMTA1, and CAMTA2 negatively regulate salicylic acid (SA) biosynthesis at warm temperatures (Kanaoka et al., 2008; Du et al., 2009; Kim et al., 2013).
The circadian clock also regulates the expression of DREB1A (Harmer et al., 2000; Maruyama et al., 2009). Moreover, the expression of many cold-inducible genes, including DREB1s and their downstream genes, is also regulated by the circadian clock at both warm and cold temperatures (Bieniawska et al., 2008; Mikkelsen and Thomashow, 2009). In phytochrome B and phytochrome D mutants, DREB1 expression is upregulated, and one of the phytochrome-interacting factor (PIF) family proteins, PIF7, can repress DREB1B and DREB1C transcription under circadian control (Franklin and Whitelam, 2007; Kidokoro et al., 2009). Moreover, mutations in the genes encoding pseudoresponse regulators (PRR9, PRR7, and PRR5), which are key components of the circadian oscillator, result in the upregulated expression of DREB1s and their downstream genes (Nakamichi et al., 2009, 2012). These three PRRs and a homolog, TIMING OF CAB EXPRESSION1, repress the expression of CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), generating a negative feedback loop for clock function (Nakamichi et al., 2009; Nagel et al., 2015; Kamioka et al., 2016; Liu et al., 2016). CCA1, which encodes a morning-expressed MYB transcription factor, can bind to the promoter regions of DREB1s, and cold-inducible expression of DREB1s has been reported to be significantly reduced in cca1 lhy double mutant plants (Dong et al., 2011), suggesting that CCA1 and its close homolog, LHY, are also important transcriptional activators in the cold-responsive expression of DREB1s.
Thus, many factors have been found to regulate the cold-responsive expression of DREB1 genes. However, most of these factors also function in other key pathways and regulate a variety of genes, and the molecular mechanisms underlying regulation of the cold-specific expression of DREB1s by these factors have not yet been clarified. In this study, we found that plants respond differently to rapid and gradual temperature decreases and induce the expression of DREB1s, which encode main switches of the cold stress response, through two different signaling pathways.
RESULTS
Expression Analyses of Six CAMTA Genes
Our previous study has revealed that the 65-bp fragment (−113 to −47) of the DREB1C promoter, including the conserved regions (boxes V and VI), is sufficient for regulation of its cold-inducible expression in Arabidopsis. Moreover, the G-box in box V functions as a negative regulatory element, and the region around box VI contains positive regulatory elements for the cold-inducible expression of DREB1C (Kidokoro et al., 2009). To screen for positive regulatory factors that interact with this fragment, we performed yeast one-hybrid screens using a cDNA library of Arabidopsis transcription factors (Mitsuda et al., 2010) and isolated a cDNA encoding CAMTA2 as one of the positive clones (Supplemental Table 1). The transcription factors CAMTA1, CAMTA2, and CAMTA3 have been reported to be positive regulators of DREB1 expression at a low temperature (4°C) and negative regulators of SA biosynthesis at a warm temperature (22°C) (Doherty et al., 2009; Du et al., 2009; Kim et al., 2013). The Arabidopsis genome contains six CAMTA family members (Bouché et al., 2002). To understand the roles of all CAMTA family proteins in the cold-responsive expression of DREB1s, we analyzed the phylogenetic relationships among the CAMTA proteins in various plant species. A total of 45 candidate peptide sequences were identified in Phytozome v11.0 genomic database (http://www.phytozome.net/). These proteins were divided into three subtypes, CAMTA1/2/3, CAMTA4, and CAMTA5/6, based on their CG-1 DNA binding domains (Supplemental Figure 1). Physcomitrella patens has only the CAMTA1/2/3 subtype genes, indicating that all six CAMTA genes are likely derived from common ancestors, as recently reported (Rahman et al., 2016).
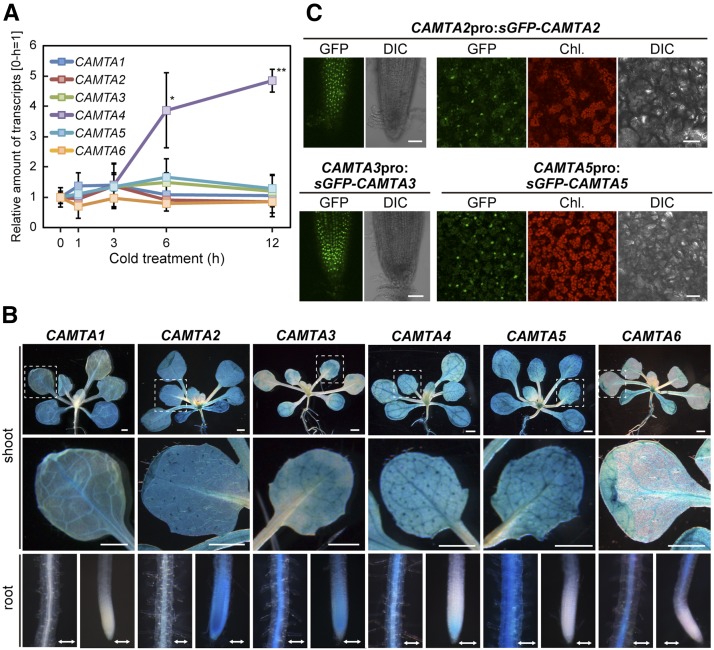
We next measured expression of the CAMTA genes under cold conditions in the plants by RT-qPCR. CAMTA4 expression was induced by the cold treatment after 6 h, and that of the other genes was not induced (Figure 1A). To determine the tissue-specific expression profiles of CAMTAs, we generated transgenic Arabidopsis expressing the GUS reporter gene under the control of each CAMTA promoter. GUS staining was detected in the cotyledons and rosette leaves of all transgenic Arabidopsis plants (Figure 1B). The staining in the roots differed depending on the gene examined. The nuclear localization of GFP-fused CAMTA3 and CAMTA5 driven by the CaMV 35S promoter has been previously demonstrated using transient transformation (Yang and Poovaiah, 2002; Mitsuda et al., 2003). We analyzed the subcellular localization of the six CAMTA proteins fused to synthetic GFP (sGFP) (Chiu et al., 1996) and driven by their own promoters and the CaMV 35S promoter in transgenic plants. The sGFP fusion proteins of CAMTA2, CAMTA3, and CAMTA5 localized to the nuclei of the Arabidopsis plants at room temperature (Figure 1C; Supplemental Figure 2). However, no GFP fluorescence of the fusion proteins of CAMTA1, CAMTA4, and CAMTA6 was observed at room temperature or at 4°C in any part of the cell. These CAMTA proteins might be unstable under these conditions.
Figure 1.
Expression of the Six CAMTA Family Genes.
(A) Expression of the CAMTA family genes under cold conditions in Arabidopsis grown in soil pots. Three-week-old plants grown in soil pots under a 12-h-light/12-h-dark cycle at 22°C were immediately cooled to 4°C beginning at ZT2. The transcript level of each gene in the plants treated for the indicated duration (0, 1, 3, 6, or 12 h) was measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript level of each gene before the cold treatment (0 h) was set to 1. The asterisks indicate significant differences (*P < 0.05 and **P < 0.01 according to Student’s t test) compared with the expression in plants before the cold treatment in three biological replicates using the plants sampled at different times.
(B) Patterns of GUS expression driven by the CAMTA promoter in 3-week-old seedlings. The aerial part (shoot), root mature zone (root, left), and root tip (root, right) are shown. Bars: line = 1 mm, arrow = 0.2 mm.
(C) Expression of the CAMTA-GFP fusion gene driven by its own promoter in 2-week-old seedlings. Root tips (CAMTA2 and CAMTA3) and rosette leaves (CAMTA2 and CAMTA5) are shown. GFP fluorescence, chlorophyll (Chl.) autofluorescence, and differential interference contrast (DIC) images are presented. Bars = 20 μm.
Transactivation Activity and DNA Binding Specificity of CAMTA Proteins
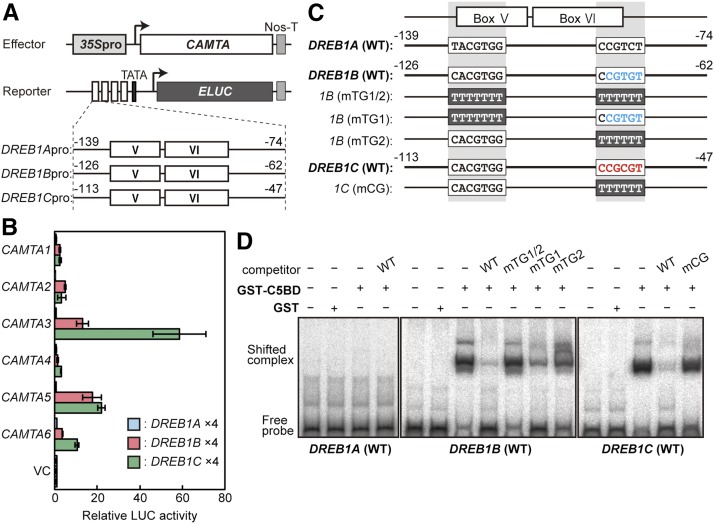
To select candidate CAMTA proteins that function in the regulation of DREB1 expression, we performed transactivation assays using Arabidopsis protoplasts. Quadruple repeats of each promoter fragment, including boxes V and VI of DREB1A (−139 to −74), DREB1B (−126 to −62), and DREB1C (−113 to −47), were fused to the ELUC reporter gene (Figure 2A). The six CAMTA genes driven by the CaMV 35S promoter were used as effectors. Among them, CAMTA3 and CAMTA5 clearly enhanced the activities of the reporter genes driven by the DREB1B and DREB1C promoters, but none of the CAMTA proteins activated expression of the reporter gene driven by the DREB1A promoter (Figure 2B). Similar results were obtained using the 1-kb promoters of DREBs to drive reporter gene expression (Supplemental Figure 3).
Figure 2.
Transactivation and DNA Binding Activities of the CAMTA Proteins to the DREB1 Promoters.
(A) Schematic diagram of the effector and reporter constructs used in transactivation analyses. The effector constructs contained the CaMV 35S promoter fused to the CAMTA1-6 coding sequences. Nos-T indicates the polyadenylation signal of the nopaline synthase gene. The reporter construct contained the emerald LUC gene fused to four tandem repeats of fragments, including the conserved regions (boxes V and VI) of the promoters of DREB1A (−139 to −74), DREB1B (−126 to −62), or DREB1C (−113 to −47) and the minimal promoter of DREB1C.
(B) Transactivation activities of the CAMTA family proteins in Arabidopsis mesophyll protoplasts. The relative activity indicates the multiples of expression compared with the value obtained with the vector control. The error bars indicate the standard deviations of three biological replicates.
(C) Comparison of the sequences of the fragments containing boxes V and VI among the DREB1 promoters.
(D) Electrophoresis mobility shift assay of binding of the recombinant CAMTA5BD protein to the DREB1 promoters. Radioactive probes of DREB1A, DREB1B, and DREB1C were incubated with or without a 1000-fold excess of competitors in the presence of the recombinant CAMTA5BD protein (GST-C5BD).
In the DREB1C promoter, the CGCG-box sequence (CCGCGT), which is a binding site of CAMTA proteins [(A/C/G)CGCG(G/T/C)] (Yang and Poovaiah, 2002), was identified, but this sequence was not found in the DREB1A or DREB1B promoter. A rice (Oryza sativa) CAMTA homolog, OsCBT, has been shown to preferentially bind to the CG(C/T)GT sequence, similar to the CGCG-box (Choi et al., 2005). We found this CGTGT sequence in the 3′ region of box VI of the DREB1B promoter, as well as a similar sequence, CGTGG, in the G-box in box V of all three DREB1 promoters (Figure 2C). To determine whether CAMTA directly binds to the CGCG-box and similar sequences in the DREB1 promoters, we performed electrophoresis mobility shift assays (Figure 2D). A fragment (amino acids 1–160) of CAMTA5, including the CG-1 DNA binding domain (CAMTA5BD), was expressed as a GST fusion protein in Escherichia coli. Following incubation of CAMTA5BD with the promoter fragments of DREB1B and DREB1C, shifted bands were detected, and the addition of each promoter fragment as a competitor (wild type) led to a decrease in the intensity of the shifted bands. However, no shifted bands were observed following incubation with the promoter fragment of DREB1A. Substitutions in the CGCG-box of the DREB1C promoter fragment abolished the competition activity. In the DREB1B promoter, substitutions in both the CGTGT and CGTGG sequences (mTG1/2) completely abolished the competition activity. Only slight decreases in the competition activity were observed with substitutions in CGTGG (mTG1), but moderate decreases were detected with substitutions in CGTGT (mTG2). These results indicate that CAMTA5 binds to the CGCG-box in the DREB1C promoter and the CGTGT sequence in the DREB1B promoter. In addition, it might weakly bind to the CGTGG sequence in the DREB1B promoter. We obtained similar results with a fragment (amino acids 1–150) of CAMTA3 (CAMTA3BD; Supplemental Figure 4).
Analyses of Plants with Quintuple and Sextuple Mutants of CAMTA Family Genes
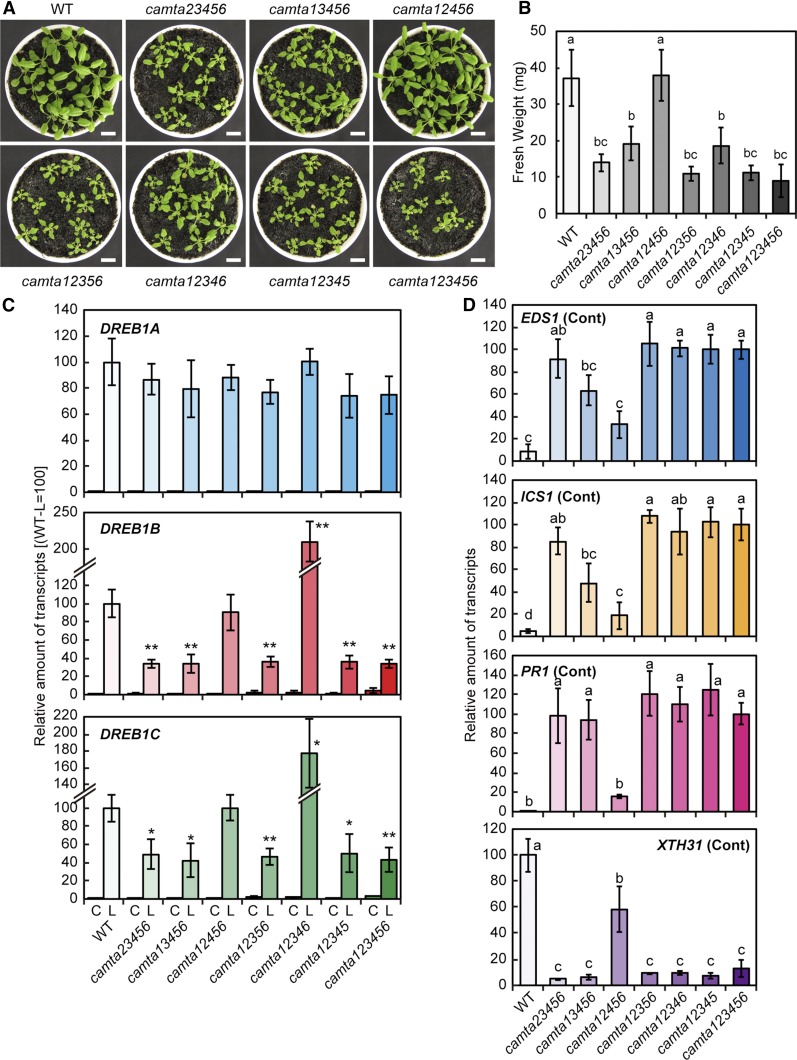
To elucidate the functions of the CAMTA family proteins in DREB1 expression, we generated quintuple and sextuple mutants of the six CAMTA genes, camta23456, camta13456, camta12456, camta12356, camta12346, camta12345, and camta123456, by crossing six T-DNA insertion mutants of each gene to one another. RT-PCR analysis confirmed the decreased expression of CAMTA2 and lack of expression of the other CAMTAs in these mutants (Supplemental Figure 5). The sextuple and quintuple mutant plants grown in soil pots, except for camta12456, exhibited growth retardation and early leaf senescence (Figures 3A and 3B). Expression of three DREB1 genes was measured under control (22°C) and cold stress conditions (4°C) for 3 h in these camta mutants. In the camta123456 plants, the cold induction of DREB1B and DREB1C was clearly decreased compared with that in the wild-type plants, but no significant changes in DREB1A expression were observed in the mutant plants (Figure 3C). Among the quintuple mutants, the cold-inducible expression of DREB1B and DREB1C in camta12456 was similar to that in the wild-type plants, and that in camta12346 was increased. Among the other quintuple mutants, the cold-inducible expression of DREB1B and DREB1C was lower than that in the wild type and similar to that in the sextuple mutants. No camta quintuple mutants showed significantly altered DREB1A expression. These results clearly indicate that CAMTA3 and CAMTA5 are active transcription factors that regulate the cold-inducible expression of DREB1B and DREB1C. However, low levels of the cold-inducible expression of both genes were still detected, even in the sextuple mutant, suggesting that transcription factors other than the CAMTA proteins are involved in the cold-responsive expression of DREB1B and DREB1C.
Figure 3.
Analyses of Quintuple and Sextuple Mutants of the CAMTA Family Genes.
(A) Growth of quintuple and sextuple mutants of the CAMTA family genes. The seedlings were grown in soil pots at 22°C for 3 weeks. Bars = 1 cm.
(B) Fresh weights of quintuple and sextuple mutants of the CAMTA family genes grown under the conditions in (A), n = 10. The letters above the bars indicate significant differences among the seedlings (P < 0.01 according to Games-Howell’s multiple range test).
(C) Expression levels of the DREB1 genes in response to cold stress in plants grown in soil pots. Three-week-old plants grown in soil pots under a 12-h-light/12-h-dark cycle at 22°C were immediately cooled to 4°C for 3 h beginning at ZT2. The transcript levels of each gene in the plants before (C) and after (L) the cold treatment were measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript level in the wild-type (WT) plants after the cold treatment was set to 100. The asterisks indicate significant differences (*P < 0.05 and **P < 0.01 according to Student’s t test) in the expression of each gene compared with that in the wild-type plants after the cold treatment in three biological replicates using the plants sampled at different times.
(D) Expression levels of the EDS1, ICS1, PR1, and XTH31 genes in plants grown in soil pots at 22°C. The transcript level of each gene under unstressed conditions was measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript levels in the wild type (XTH31) or camta sextuple mutant plants (others) were set to 100. The letters above the bars indicate significant differences (P < 0.01 according to the Tukey-Kramer method) in the expression of each gene in three biological replicates using the plants sampled at different times.
We also analyzed the expression of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and ISOCHORISMATE SYNTHASE1 (ICS1), which encode important positive regulators of SA biosynthesis because the camta3 mutant has been reported to increase EDS1 expression, leading to SA accumulation and growth retardation (Du et al., 2009). In the sextuple mutant, the expression of both genes was strongly elevated compared with that in the wild type (Figure 3D). Furthermore, EDS1 and ICS1 expression in camta12456 was clearly suppressed compared with that in the sextuple mutant. The expression of these genes was also slightly decreased in camta13456. We further examined the expression of PATHOGENESIS-RELATED GENE1 (PR1), an SA-induced gene, and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE31 (XTH31), an SA-repressed gene that encodes xyloglucan endotransglycosylase/hydrolase (Miura et al., 2010). PR1 expression was strongly suppressed in camta12456 compared with that in the sextuple mutant, but its level was higher than that in the wild type. In contrast, XTH31 expression was increased in camta12456 compared with that in the sextuple mutant, but its level was lower than that in the wild type. All of these findings indicate that CAMTA3 plays a major role and that CAMTA2 has a minor role in the suppression of SA biosynthesis.
Comprehensive Analysis of Up- and Downregulated Genes in camta Sextuple Mutant Plants under Cold Stress Conditions
To identify the roles of CAMTA proteins in cold-responsive gene expression, we assessed the gene expression profiles in camta sextuple mutant plants subjected to cold stress (4°C) for 3 h by RNA-seq analysis. A total of 213 and 971 genes were upregulated and downregulated, respectively, in the mutant compared with the wild type (fold change > 2 or fold change < 0.5, P < 0.01). Next, the downregulated genes in the mutant plants were compared with the upregulated genes in Arabidopsis plants overexpressing DREB1B (fold change > 2, P < 0.05; Park et al., 2015). Thirty-five genes were found to overlap (Supplemental Figure 6A and Supplemental Data Sets 1A and B), including direct target genes of DREB1s, such as COLD-REGULATED 15A (COR15A) and GALACTINOL SYNTHASE3 (GolS3) (Liu et al., 1998; Maruyama et al., 2004). On the other hand, the expression of many cold-inducible DREB1 target genes, such as COR15B and KIN2, was unchanged in the mutant. Expression of these genes was confirmed by RT-qPCR (Supplemental Figure 6B). Then, the downregulated genes in the camta123456 mutant were compared with the upregulated genes in the plants subjected to cold stress for 1 d identified by microarray analyses (Maruyama et al., 2009). A total of 94 genes were found to overlap that were not downstream of DREB1B (Supplemental Figure 6A and Supplemental Data Set 1C).
Furthermore, we examined the expression patterns of the top 100 upregulated and downregulated genes in the sextuple mutant using Genevestigator microarray database (https://www.genevestigator.com/gv/) (Zimmermann et al., 2004) and found that most of the upregulated genes in the mutant were highly responsive to SA treatment and cold treatment for more than 3 d. However, these genes were not responsive to cold treatment for less than 3 d (Supplemental Figure 6C). Because SA accumulation significantly increases in response to long-term cold treatment of more than 1 week (Kim et al., 2013; Scott et al., 2004), long-term cold treatment might have the same effect on Arabidopsis plants as SA. Indeed, most of the genes responsive to cold treatment for more than 3 d were also SA-inducible genes. Next, we analyzed the enrichment of hexamer motifs in the promoters of the top 100 of up- and downregulated genes in the sextuple mutant (Maruyama et al., 2012). Among the top 10 overrepresented hexamer sequences in the promoters of the upregulated genes, two sequences contained a TGAC core sequence in the W-box involved in SA-responsive gene expression. In contrast, among the top 10 overrepresented hexamer sequences in the downregulated genes, two contained the CGCG core sequence of the CGCG-box (Supplemental Table 2).
camta Mutant Plants Grown on Agar Plates and in Soil Pots Showed Different Characteristics
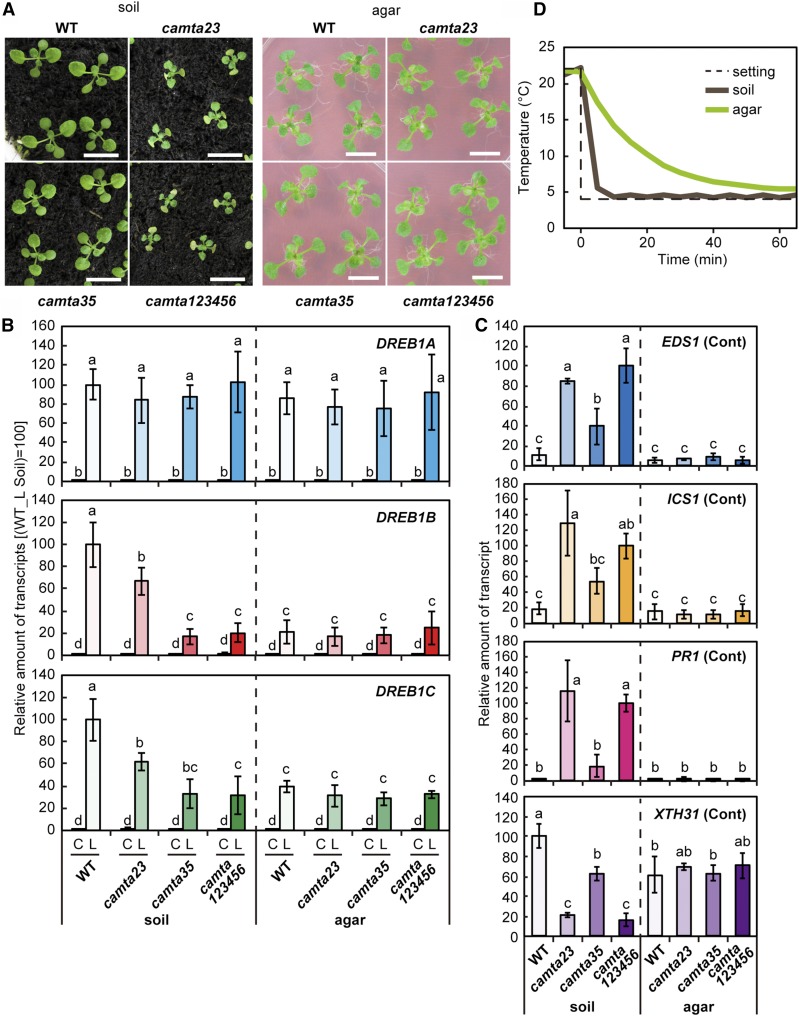
The camta123456 and camta23 mutants grown in soil pots exhibited severe growth retardation and early leaf senescence, but camta35 did not. In contrast, none of the tested mutants grown on agar plates had any visible phenotypic alterations (Figure 4A). We examined the expression of the 6 CAMTA genes in wild-type plants and found that these genes were expressed in both plants grown in soil pots and plants grown on agar plates (Supplemental Figure 7). Then, the expression of DREB1s was assessed under cold conditions in wild-type and mutant plants grown in soil pots and on agar plates (Figure 4B). In camta123456 and camta35 mutant plants grown in soil pots, the cold-inducible expression of DREB1B was markedly decreased compared with that in the wild type, but in those grown on agar plates, expression of DREB1B under cold conditions remained at a low level in all wild-type and mutant plants. Similar results were obtained with the cold induction of DREB1C, but its relative expression levels in the camta mutants were higher than those of DREB1B. By contrast, DREB1A was strongly induced by cold stress in both the wild-type and mutant plants, regardless of the growth conditions. These results indicate that the CAMTA3 and CAMTA5 proteins function as positive regulators of DREB1B and DREB1C expression under cold conditions in plants grown in soil pots but not in those grown on agar plates. We also analyzed EDS1, ICS1, PR1, and XTH31 expression in wild-type and camta mutant plants grown on agar plates. Expression of EDS1, ICS1, and PR1 in all plants grown on agar plates remained low and expression of XTH31 in those plants was high, in contrast with their expression in the mutant plants grown in soil pots (Figure 4C). Thus, EDS1, ICS1, and PR1 were repressed and XTH31 was expressed in the plants grown on agar plates, regardless of the presence of the CAMTA genes. Considering that the CAMTA proteins did not function in the plants grown on agar plates, it is possible that the closed small space affected their activity. Therefore, we next measured the time courses of the temperature decreases around the plants grown in soil pots and on agar plates after transferring them from 22°C to 4°C. The temperature around the plants grown in soil pots rapidly decreased, and that around the plants grown on agar plates slowly decreased (Figure 4D).
Figure 4.
Analyses of Sextuple Mutants of the CAMTA Family Genes Grown on Agar Plates or in Soil Pots.
(A) Growth of sextuple mutants of the CAMTA family genes grown on agar plates or in soil pots. The seedlings were grown for 2 weeks in soil pots or on agar plates. Bars = 1 cm.
(B) Expression levels of the DREB1 genes under unstressed or cold stress conditions. Three-week-old plants grown in soil pots (soil) or on agar plats (agar) under a 12-h-light/12-h-dark cycle at 22°C were immediately cooled to 4°C for 3 h beginning at ZT2. The transcript levels of each gene in the plants before (C) and after (L) the cold treatment were measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript levels in the wild-type plants after the cold treatment were set to 100. The letters above the bars indicate significant differences (P < 0.01 according to the Tukey-Kramer method) in the expression of each gene in three biological replicates using the plants sampled at different times.
(C) Expression levels of the EDS1, ICS1, PR1, and XTH31 genes under unstressed conditions. The transcript level of each gene was measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript levels in the wild type (XTH31) or camta sextuple mutant plants (others) grown in soil pots were set to 100. The letters above the bars indicate significant differences (P < 0.01 according to the Tukey-Kramer method) in the expression of each gene in three biological replicates using the plants sampled at different times.
(D) Temperatures around the plants grown in soil pots and on agar plates.
CAMTA Proteins Function in Cold-Inducible Expression of DREB1B in Response to Rapid Temperature Decrease
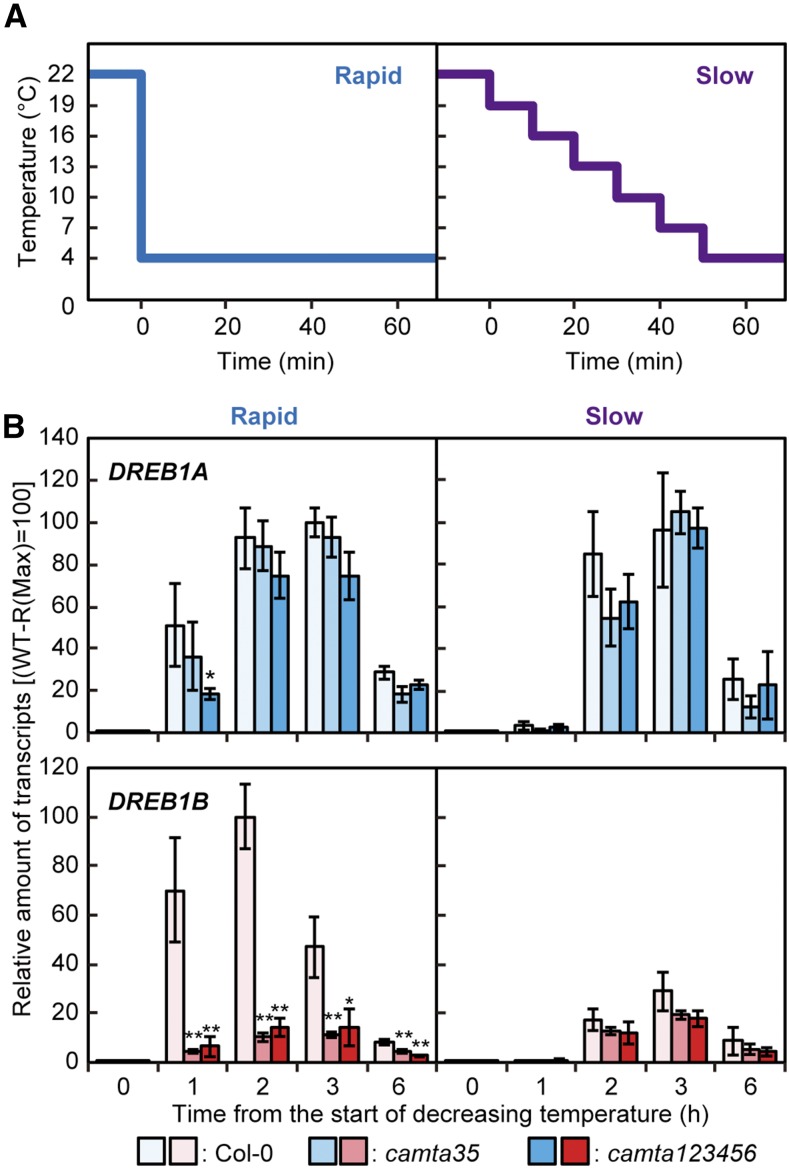
To confirm the effect of the rate of temperature decrease on CAMTA activity, wild-type and mutant plants were subjected to two different cold stress treatments (Figure 5A). For one, plants grown in soil pots were directly transferred from 22°C to 4°C (rapid), and for the other, plants grown in soil pots were gradually transferred from 22°C to 4°C, with a decrease of 3°C every 10 min (slow). DREB1B expression was significantly induced at 1, 2, and 3 h in response to the rapid cold treatment, and its cold-inducible expression was markedly suppressed in camta123456 and camta35, while DREB1A expression was strongly induced by this treatment in both the wild-type and camta mutant plants (Figure 5B). During the slow cold treatment, expression of DREB1B was markedly decreased in both the wild-type and camta mutant plants, and that of DREB1A was maintained at a high level (Figure 5B). Expression of DREB1B was also decreased in response to the rapid cold treatment in the camta3 and camta5 single mutants, but not in the camta4 mutant, compared with that in the wild type. In contrast, expression of DREB1A was similarly induced by the rapid cold treatment in both the wild type and these three camta single mutants (Supplemental Figure 8). Thus, DREB1B expression was mainly regulated by CAMTA3 and CAMTA5, which were activated by the rapid decrease but not by the slow decrease in temperature, whereas DREB1A expression was not regulated by CAMTA proteins and was induced by both rapid and slow temperature decreases.
Figure 5.
Expression Levels of the DREB1A and DREB1B Genes in Response to Rapid and Slow Temperature Decreases.
(A) A schematic diagram of the temperatures around the plants during the rapid and slow temperature decreases. Three-week-old seedling of the wild-type and camta mutant plants grown in soil pots were immediately cooled from 22°C to 4°C (rapid) or were gradually cooled from 22°C to 4°C, with a decrease of 3°C every 10 min (slow). These cold treatments were started at ZT2.
(B) Expression levels of the DREB1 genes under the two different cold stress treatments. The transcript level of each gene in the plants treated for the indicated duration (0, 1, 2, 3, or 6 h) after the temperature began to decrease was measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The maximum expression level of each gene in the wild-type plants exposed to a rapid temperature decrease was set to 100. The asterisks indicate significant differences (*P < 0.05 and **P < 0.01 according to Student’s t test) in the expression of each gene compared with that in the wild-type plants at each time point in three biological replicates using the plants sampled at different times.
To verify these findings pertaining to DREB1 expression, plants grown on agar plates were transferred from 22°C to 4°C and maintained for specific durations on agar plates without lids, which resulted in strong induction of the cold-inducible expression of DREB1B but slight increase in its expression in plants grown on agar plates with closed lids (Supplemental Figure 9). In addition, DREB1A was expressed in the plants grown on agar plates both with and without lids under cold conditions. Moreover, to examine the temperature dependency of the cold induction of DREB1s, the wild-type and camta sextuple mutant plants grown in soil pots were immediately transferred to various temperatures. DREB1A, DREB1B, and DREB1C expression was first observed at 10°C, and it continuously increased up to 4°C (Supplemental Figure 10). In addition, expression of DREB1B and DREB1C was reduced to low levels in camta123456, but that of DREB1A was not.
Effects of Circadian Rhythm on the Cold-Inducible Expression of DREB1s in Response to a Rapid Temperature Decrease
Then, the effect of circadian rhythm on the cold-inducible expression of DREB1s was assessed in the wild-type and camta mutant plants. Plants grown in soil pots under a 12-h light/12-h dark cycle for 3 weeks were transferred to continuous light conditions and exposed to a rapid temperature decrease beginning at zeitgeber time (ZT) 26 or ZT38 (Supplemental Figure 11A). In both the wild-type and mutant plants, DREB1A expression was strongly induced after initiation of the cold treatment at ZT26 during the subjective day, but its expression was very low after initiation of the cold treatment at ZT38 during the subjective night. In contrast, DREB1B expression was strongly induced after initiation of the cold treatment at both ZT26 and ZT38, especially after 1 h of treatment, but this cold-inducible expression was markedly suppressed in camta35 and camta123456. DREB1C expression exhibited characteristics of both DREB1A and DREB1B expression, with reduced expression in camta35 and camta123456 in both subjective day and night compared with that in the wild-type plants and lower levels of expression in the subjective night compared with those in the subjective day.
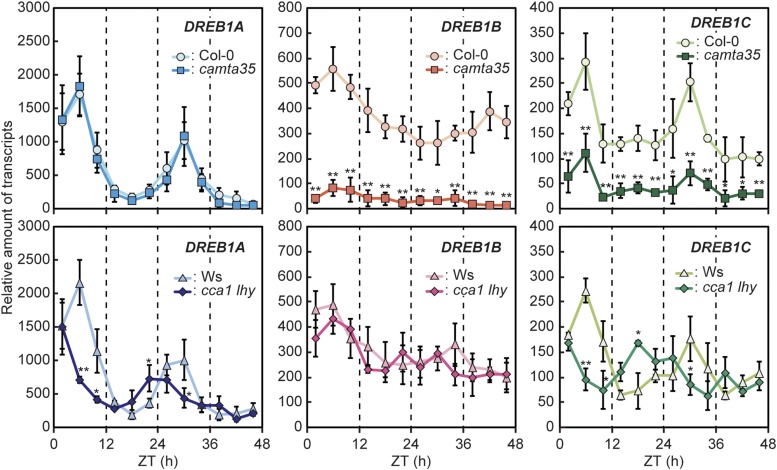
Furthermore, cold-inducible DREB1 expression was assessed at several time points in camta35 and wild-type (Col-0) plants following transfer to free-running conditions (Figure 6). Three-week-old plants grown in soil pots under a 12-h-light/12-h-dark cycle were transferred to continuous light conditions, and cold treatments were initiated every 4 h from ZT2 to ZT46 by immediate cooling to 4°C and incubation for 1 h at each time point. RT-qPCR analyses indicated that DREB1A expression in response to the rapid temperature decrease was highly induced during the subjective day but that it decreased to low levels during the subjective night. However, its cold-inducible expression during the subjective day was not significantly reduced in camta35. In contrast, DREB1B was highly expressed in response to the cold treatment during both the subjective day and night, and its expression was clearly decreased in camta35 at all time points. DREB1C was expressed in a circadian manner, but its expression was higher than that of DREB1A during the subjective night. Moreover, DREB1C expression was significantly decreased in camta35 at all time points. As the cold-inducible expression of DREB1s has been reported to be reduced in cca1 lhy double mutant plants (Dong et al., 2011), we further analyzed the expression of DREB1s in cca1 lhy (Figure 6). The cold-inducible expression of DREB1A and DREB1C at ZT6, ZT10, and ZT30 was lower in the mutant than in the wild type (Wassilewskija), but obvious expression of both genes was still detected in the mutant, suggesting that other factors are also involved in their expression. However, DREB1B expression was not significantly altered in cca1 lhy, in contrast to the findings of a previous study (Dong et al., 2011). The same treatments were performed after the diurnal cycle on plants grown under a 12-h-light/12-h-dark photoperiod from ZT2 to ZT22, and similar results were obtained (Supplemental Figure 11B). These data indicate that CAMTA3 and CAMTA5 function in the cold-responsive expression of DREB1B and DREB1C during the day and night in response to a rapid temperature decrease and that CCA1/LHY and other factors are involved in the cold-responsive expression of DREB1A and DREB1C during the day.
Figure 6.
Effects of Circadian Clock on Expression of the DREB1 Genes in Response to a Rapid Temperature Decrease.
Three-week-old plants grown in soil pots under a 12-h-light/12-h-dark cycle were transferred to free-running conditions under continuous light, and cold treatments were initiated every 4 h from ZT2 to ZT46 by immediate cooling to 4°C and incubation for 1 h at each time point. Several plants were treated and harvested at each time point. The transcript level of each gene in the plants treated at 4°C was measured by RT-qPCR. The values represent the average of three technical replicates, and the error bars indicate the sd. The transcript levels in the wild type plants (Col-0 or Wassilewskija) before the cold treatment at ZT2 were set to 1. The asterisks indicate significant differences (*P < 0.05 and **P < 0.01 according to Student’s t test) in the expression of each gene compared with that in the wild-type plants at each time point in three biological replicates using the plants sampled at different times.
DISCUSSION
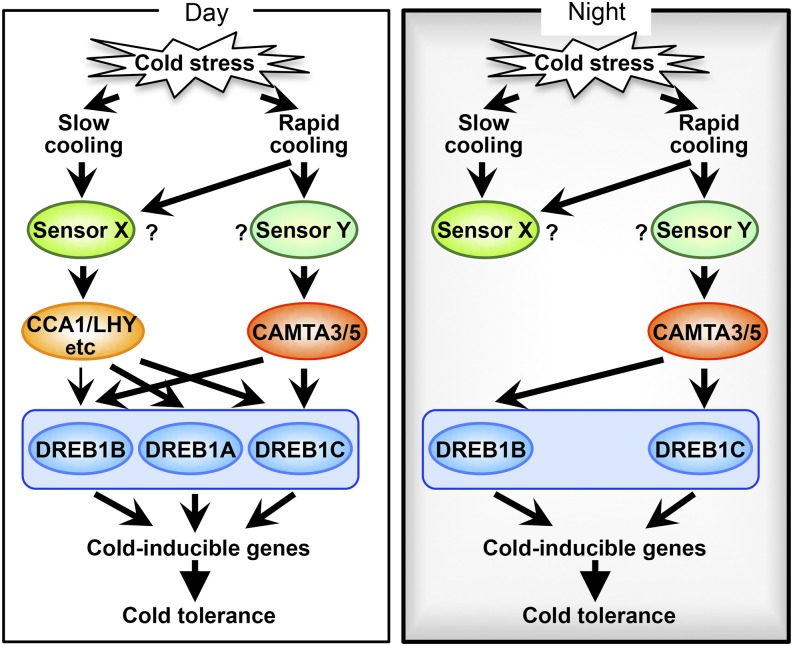
In this study, we analyzed the cold-responsive expression of DREB1s using various CAMTA mutants, including a camta123456 sextuple mutant lacking all CAMTA family genes, and found that each DREB1 gene is differentially controlled by CAMTA proteins under cold stress conditions (Figures 3 and 5). The cold-inducible expression of DREB1B was mainly regulated by CAMTA3 and CAMTA5, but DREB1A expression was regulated by clock factors, such as CCA1/LHY. Importantly, comparison of the results of expression analyses of DREB1A and DREB1B in plants grown in soil pots and on agar plates demonstrated that the plants recognized the changes in the temperature decrease rates (Figures 5 and 6). DREB1B expression was mainly induced by a rapid temperature decrease, i.e., a direct change from 22°C to 4°C, but not by a slow decrease, i.e., a gradual change from 22°C to 4°C, with a 3°C decrease every 10 min. These findings suggest that the CAMTA proteins are activated by a rapid temperature decrease but not by a slow decrease. In contrast, DREB1A expression was induced in response to both rapid and slow temperature decreases. Thus, this research has highlighted that plants respond differently to rapid and gradual temperature decreases, with induction of the expression of different DREB1 genes encoding main switches of the cold stress response. Plants recognize cold stress as two different signals, a low temperature of less than 10°C and a sudden drop in temperature to less than 10°C, and two different signaling pathways function in response to these gradual and rapid temperature decreases (Figure 7). It is possible that two different sensing systems may function in the recognition of cold stress in plants. A number of factors that function in the cold-responsive expression of DREB1s have been identified (Chinnusamy et al., 2003; Doherty et al., 2009; Kidokoro et al., 2009; Nakamichi et al., 2009; Dong et al., 2011; Jeon and Kim, 2013), making it difficult to elucidate the molecular mechanisms underlying the regulation of cold-responsive gene expression (Zhao et al., 2015). These complex mechanisms may be further clarified by distinguishing between these two types of cold stress signals.
Figure 7.
Model of the Cold Stress Response in Arabidopsis during the Day and Night.
Plants recognize cold stress as two different signals and induce the three DREB1 genes encoding main switches of the cold stress response. The DREB1 genes are tandemly arrayed in the Arabidopsis genome in the order of DREB1B, DREB1A, and DREB1C. One of the signals is caused by a rapid drop in temperature. In this signaling pathway, CAMTA3 and CAMTA5 activate expression of DREB1B and DREB1C. The other signal is caused by low temperatures in general and includes both rapid and slow decreases in temperature. This signal activates expression of DREB1A and DREB1C strongly and that of DREB1B weakly. Activation of this signaling pathway may be controlled by the central oscillator of the circadian clock, involving CCA1 and LHY. During the day, both signaling systems efficiently function in response to slow and rapid temperature decreases. During the evening, only the signaling system involving CAMTAs is activated in response to a rapid temperature decrease. The arrows indicate positive regulatory pathways, and the question marks denote factors that remain to be confirmed.
During the season change from autumn to winter, the temperature gradually drops, with repeated rises and falls. Eventually, when it falls under 10°C, DREB1A is expressed and induces downstream target genes to enhance freezing stress tolerance. However, DREB1A is repressed during the night (Figures 6 and 7), despite the lower temperature compared with during the daytime, and its expression may be mainly regulated by central oscillators of the circadian clock, such as CCA1 and LHY, which are expressed in the morning and repressed in the evening (Nagel et al., 2015). In contrast, DREB1B and DREB1C expression is regulated by the CAMTA transcription factors and is induced by rapid temperature decreases during both the day and night (Figures 6; Supplemental Figure 6). Thus, plants are thought to use at least two different signaling pathways to respond to the sudden drop in temperature that occurs during the night and under abnormal weather conditions and to the gradual temperature decrease that occurs with the change in seasons from autumn to winter. The circadian clock is likely involved in the regulation of cold-responsive gene expression during the season change because it can recognize the photoperiod. From autumn to winter, the daylength gradually decreases, but during the spring, it progressively increases. Expression of DREB1s in short-day (8-h photoperiod) plants has been shown to be 3- to 5-fold higher compared with that in long-day (16-h photoperiod) plants (Lee and Thomashow, 2012). Therefore, plants may express cold-inducible genes at higher levels to enhance freezing stress tolerance during the winter.
The CAMTA proteins are considered to be involved in Ca2+ signaling because they contain conserved calmodulin binding sites (Bouché et al., 2002). AtSR1/CAMTA3 has been shown to act as a repressor of SA signaling (Du et al., 2009). Ca2+/calmodulin binding to AtSR1/CAMTA3 has been reported to be required for the suppression of SA biosynthesis-related genes. In addition, CAMTA1 and CAMTA2 have been demonstrated to function together with CAMTA3 at warm temperatures to repress the expression of genes related to SA biosynthesis in a study comparing double and triple mutants of CAMTA1-3 (Kim et al., 2013). In this study, we showed that CAMTA3 plays an important role and that CAMTA2 has a minor role in the suppression of SA biosynthesis using CAMTA quintuple mutants (Figure 3). Although CAMTA1 may also function in this process, its effect is likely weaker compared with CAMTA2 and CAMTA3. On the other hand, we have revealed that CAMTA3 and CAMTA5 are important transcriptional activators that regulate the cold-inducible expression of DREB1B and DREB1C. CAMTA3 functions in the two key regulatory systems, but CAMTA2 and CAMTA5 function in only one system each, namely, SA signaling and the cold stress response, respectively. As expression of the CAMTA3 and CAMTA5 genes is not induced by cold stress, it is important to analyze their activation mechanisms in response to cold stress. Requirement of Ca2+ signaling for the regulation of CAMTA activity has not been observed in the cold stress response, but because the calmodulin binding site is also conserved in CAMTA5 and involvement of Ca2+ signaling has been reported in the cold stress response, Ca2+ signaling may also be important for the regulation of cold-responsive CAMTA activity (Townley and Knight, 2002; Liu et al., 2015). However, it remains unclear how Ca2+ signaling affects CAMTA protein activity in relation to cold stress-responsive gene expression. Further analysis is required to verify the roles of Ca2+ signaling in the cold stress response.
The results of transcriptome analysis using the camta123456 mutant revealed the upregulation of many SA-inducible genes (Supplemental Figure 6). Because CAMTA2 and CAMTA3 repress the expression of SA synthesis-related genes, such as EDS1 and ICS1 (Du et al., 2009; Kim et al., 2013), SA was likely synthesized in the sextuple mutant that lacked these CAMTA genes, which resulted in the upregulation of SA-inducible genes. In fact, the W-box, the binding site of WRKY transcription factors involved in SA-related gene expression, was detected among the top 10 overrepresented hexamer sequences in the promoters of the upregulated genes (Supplemental Table 2). In contrast, the DRE, the binding site of DREB1s, was not observed among the overrepresented hexamers in these promoters. Because transcriptional activators other than the CAMTA proteins, such as CCA1/LHY, function in the induction of DREB1s, the lack of CAMTAs was compensated for by these transcription factors; thus, it did not affect the expression of cold-inducible DREB1 target genes in camta123456.
CCA1/LHY have been shown to regulate gene expression only during the day, as demonstrated by their expression patterns (Dong et al., 2011; Nagel et al., 2015; Kamioka et al., 2016). A previous transcriptome analysis using the prr5/7/9 triple mutant has revealed that expression of the DREB1A, DREB1B, and DREB1C genes is upregulated under cold conditions, especially during the night (Nakamichi et al., 2009), because PRR5/7/9 are expressed during the evening and suppress the expression of CCA1/LHY. DREB1A and DREB1C contain an evening element (AAATATCT), which is a CCA1/LHY binding site, near the TATA boxes in their promoters, and DREB1B contains an evening element at ∼350 bp upstream of the TATA box in its promoter. Therefore, it seems that DREB1A and DREB1C are strongly and DREB1B is weakly regulated by CCA1/LHY. We detected their certain involvement in the cold-responsive expression of DREB1A and DREB1C using cca1 lhy (Figure 6). However, these genes were still expressed in cca1 lhy, although their expression was significantly reduced. Moreover, the cold-responsive expression of DREB1B was not clearly altered in cca1 lhy, in contrast to a previous study (Dong et al., 2011) that showed that the expression of DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 is nearly absent in cca1 lhy. These discrepancies between this previous study and our findings might be attributable to differences in experimental conditions. Because the CCA1/LHY proteins are expressed during the daytime, even at normal temperatures, a mechanism should exist that suppresses the expression of DREB1s at normal temperatures. This negative regulatory system might be essential for their expression, activated by not only CCA1/LHY but also CAMTAs, to precisely regulate cold-specific gene expression. It is necessary to analyze proteins that interact with CCA1/LHY and CAMTAs and their modifications, such as phosphorylation, to elucidate the mechanism underlying suppression of the expression of DREB1 genes at normal temperatures.
In summary, we have revealed that plants recognize cold stress as two different signals, rapid and gradual temperature decreases, and induce the DREB1 genes encoding main switches of the cold stress response (Figure 7). One signal is caused by a rapid temperature drop to less than 10°C. In this signaling pathway, CAMTA3 and CAMTA5 mainly function as transcriptional activators in the expression of DREB1B and DREB1C. The other signal is caused by both rapid and gradual temperature decreases to less than 10°C. The central oscillator of the circadian clock, involving CCA1/LHY, may play a role in this signaling pathway to strongly induce the expression of DREB1A and DREB1C and weakly induce the expression of DREB1B. The presence of these two signaling pathways suggests that two different sensor systems might function in the recognition of cold stress signals in plants. During the day, both signaling pathways efficiently function in response to gradual and rapid temperature decreases. During the evening, CAMTAs are activated in response to a rapid temperature decrease. The clock factors could control the expression of DREB1 genes in a photoperiod-dependent manner, resulting in high expression during the winter and low expression during the spring (Lee and Thomashow, 2012). Thus, plants efficiently acquire freezing tolerance through two signaling pathways in response to a gradual temperature decrease during season changes and a sudden temperature drop during the night.
METHODS
Sequence Alignment and Phylogenetic Analysis
The peptide sequences of the CAMTA family proteins were obtained from Phytozome v11.0 (https://phytozome.jgi.doe.gov/pz/portal.html). Alignments were performed and a phylogenetic tree was constructed using a bootstrap test of neighbor-joining method with 1000 bootstrap trials (Mizoi et al., 2013). A text file of the alignment is provided as Supplemental File 1.
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on peat moss in plastic pots or on germination medium agar plates at 22 ± 2°C under a under a 12-h-light/12-h-dark cycle at a photon flux density of 50 ± 10 μmol m−2 s−1 of white light. Three-week-old seedlings grown under these conditions were used for the stress treatments. The cold stress treatment was performed in an LH-1-120S incubator (Nippon Medical and Chemical Instruments). For a rapid cooling treatment, plants were directly transferred to the incubator set at 4°C, and for a slow cooling treatment, plants were transferred to the incubator set at 22°C, and the temperature was decreased by 3°C every 10 min. The temperature was monitored using a MCR-4TC thermocouple logger (T&D). The T-DNA insertion lines for the CAMTA family genes, including SALK_108806 (camta1), SALK_139868 (camta2), SALK_001152 (camta3), SALK_087870 (camta4), SALK_134491 (camta5), SALK_078900 (camta6), and cca1-11 lhy-21 (cca1 lhy; CS9380), were obtained from the ABRC (Alonso et al., 2003). T-DNA insertion was confirmed by amplification of the left border flanking region of the genomic DNA using the primer set listed in Supplemental Table 3.
Plasmid Construction
For histochemical GUS staining, 4-kb CAMTA3 and 1.5-kb CAMTA1, CAMTA2, CAMTA4, CAMTA5, and CAMTA6 promoter fragments upstream of the translational start site were amplified from Col-0 plants. These fragments were cloned into a pGK-GUS vector (Qin et al., 2008). For GFP fluorescence observation, the coding sequences of six CAMTA cDNAs were inserted into a pGKX-NsGFP or pGH-35Spro:sGFP vector (Qin et al., 2008; Fujita et al., 2009), and for their expression, each CAMTA promoter fragment was inserted into a pGH-35Spro:CAMTA-sGFP vector, from which the CaMV 35S promoter had been removed. For the effector plasmids in transactivation assay, the coding sequences of the CAMTA proteins were PCR amplified from Arabidopsis cDNA clones using the primers indicated in Supplemental Table 3 and cloned into the site of each restriction enzyme of a pGKX vector (Qin et al., 2008). For gel mobility shift assay, fragments of CAMTA5 (amino acids 1–160) and CAMTA3 (amino acids 1–150) were each inserted into a pGEX-4T-1 vector (GE Healthcare).
RNA Preparation, RT-PCR, and RT-qPCR
Total RNA was isolated from Arabidopsis plants using RNAiso plus (Takara Bio) according to the manufacturer’s instructions. cDNA was synthesized with a High-Capacity cDNA reverse transcription kit (Applied Biosystems) and RT-qPCR was performed using Power SYBR Green Master Mix (Applied Biosystems) (Tanaka et al., 2012). RT-PCR was performed using KOD FX (TOYOBO). The primers used for RT-PCR and RT-qPCR are listed in Supplemental Table 3. In RT-qPCR analyses, triplicate measurements were performed for each cDNA sample, and the obtained values with the standard curve methods were normalized to those of 18S rRNA. Samples and their technical replicates for the same time point were run on the same plates. All experiments were performed three times, using plants sampled at different times. Average and sd of the three biological replicates are shown in the figures. The significance of expression changes was evaluated according to the expression levels of the genes calculated from the biological replicates, using Student’s t test for comparisons between two groups and the Tukey-Kramer method for comparisons among multiple groups.
Histochemical GUS Staining and GFP Fluorescence Observation
Histochemical GUS staining was performed as previously described (Qin et al., 2008). GUS staining was observed using an M205C stereomicroscope and a DFC490 digital color camera (Leica Microsystems). Seedlings that expressed fluorescent proteins were observed using a confocal laser scanning microscope (LSM5 PASCAL; Zeiss). For 4′,6-diamidino-2-phenylindole staining, 2-week-old seedlings were soaked in a 4′,6-diamidino-2-phenylindole solution (2 μg/mL) for 15 to 30 min.
Transient Expression Assays with Arabidopsis Mesophyll Protoplasts
Transient transformation of Arabidopsis mesophyll protoplasts was performed as described (Kidokoro et al., 2009). Plasmids expressing Pyrearinus termitilluminans luciferase (Emerald Luc; TOYOBO) under the control of four tandem repeats of the promoter fragments of DREB1A (−139 to −74), DREB1B (−126 to −62), and DREB1C (−113 to −47) and a pBI221 (35S-GUS; Takara Bio) vector were used as reporters and an internal control, respectively.
Fusion Protein Preparation and Purification and Gel Mobility Shift Assay
Expression and purification of the GST fusion proteins, probe labeling, and gel mobility shift assays were performed as previously described (Kidokoro et al., 2009) with minor modifications. The probe oligonucleotides and mutation fragments were PCR amplified and digested by XbaI. A mixture of 2000 dpm of 32P-labeled probe and 100 ng of the fusion proteins was incubated for 15 min at 4°C. The reaction mixtures were resolved by electrophoresis through a 6% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer at 150 V for 90 min. For the competition experiments, the competitors were incubated with the fusion proteins for 5 min at room temperature prior to the addition of labeled probes.
RNA-Seq Analysis
Total RNA was isolated from Arabidopsis plants as described above. A RiboMinus Plant Kit for RNA-seq and Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) were used for the removal of rRNAs from the total RNA samples and construction of a cDNA library, respectively. The qualities of the total RNA and cDNA library were monitored with an Agilent Tape Station (Agilent Technologies). The cDNA libraries were pooled for emulsion PCR using an Ion PI Template OT2 200 Kit v3 (Thermo Fisher Scientific). The samples were loaded onto an Ion Proton PI chip v2 and sequenced with an Ion Proton instrument using an Ion PI sequencing 200 kit v3 (Thermo Fisher Scientific). The raw reads from the libraries were filtered to remove adaptor sequences and low-quality bases, and the clean reads from each library were mapped to the reference Arabidopsis genome (TAIR 10) using CLC Genomics Workbench (version 8.0; CLC Bio). Gene expression levels were measured by RPKM (reads per kilobase of the transcript per million mapped reads) values for assessment. Two biological replicates of the wild type and camta sextuple mutants were used for cDNA library construction and sequencing. To obtain statistical confirmation of the differences in gene expression, we compared the RPKM values using Pearson’s χ2 test. The raw sequence reads were deposited into National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) under accession number PRJNA326982. Overrepresentation analysis of the hexamer sequences in the promoters of the up- and downregulated genes was performed as described previously (Maruyama et al., 2012).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CAMTA1 (At5g09410), CAMTA2, (At5g64220), CAMTA3 (At2g22300), CAMTA4 (At1g67310), CAMTA5 (At4g16150), CAMTA6 (At3g16940), DREB1A (At4g25480), DREB1B (At4g25490), DREB1C (At4g25470), EDS1 (At3g48090), ICS1 (At1g74710), PR1 (At2g14610), XTH31 (At3g44990), COR15A (At2g42540), COR15B (At2g42530), GolS3 (At1g09350), and KIN2 (At5g15970).
Supplemental Data
Supplemental Figure 1. Phylogenetic tree of CAMTA family proteins.
Supplemental Figure 2. Subcellular localization of CAMTA proteins in transgenic plants.
Supplemental Figure 3. Transactivation analyses of the CAMTA family proteins using each 1-kb DREB1 promoter.
Supplemental Figure 4. Electrophoresis mobility shift assay of binding of the recombinant CAMTA3BD protein to the DREB1 promoters.
Supplemental Figure 5. Construction of mutants of the CAMTA family genes.
Supplemental Figure 6. Transcriptome analysis of sextuple mutants of the CAMTA family genes grown in soil pots.
Supplemental Figure 7. Expression levels of the CAMTA family genes in Arabidopsis grown in soil pots or on agar plates.
Supplemental Figure 8. Effects of cooling rates and growth conditions on cold-inducible expression of the DREB1A and DREB1B genes in various camta mutants.
Supplemental Figure 9. Cold-inducible expression of the DREB1A and DREB1B genes in plants grown on agar plates with or without lids.
Supplemental Figure 10. Cold-inducible expression of the DREB1 genes in plants grown in soil pots at various temperatures.
Supplemental Figure 11. Effects of the diurnal cycle and circadian clock on cold-inducible expression of the DREB1 genes.
Supplemental Table 1. The results of yeast one-hybrid screens.
Supplemental Table 2. Enriched hexamers in the promoters of the genes downstream of the CAMTA family genes.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. The CAMTA-downstream, cold-inducible, and/or DREB1-downstream genes.
Supplemental File 1. Text file of the alignment used for phylogenetic analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Yuriko Tanaka for providing excellent technical assistance and Etsuko Toma for providing skillful editorial assistance. We also thank Motomu Endo (Kyoto University) for the fruitful discussions and valuable suggestions on the experimental design concerning circadian rhythm. This work was financially supported by grants from a Grant-in-Aid for Scientific Research on Innovative Areas (no. 15H05960 to K.Y.-S.) and for Scientific Research (A) (no. 25251031 to K.Y.-S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Program for the Promotion of Basic Research Activities for Innovative Biosciences (BRAIN) of Japan (to K.S. and K.Y.-S.).
AUTHOR CONTRIBUTIONS
S.K. and K.Y.-S. designed the study. S.K., K.Y., H.T., and F.T. performed the experiments and analyzed the data. S.K., K.S., and K.Y.-S. wrote the article. All of the authors discussed the results and commented on the manuscript.
Glossary
- DRE
dehydration-responsive element
- SA
salicylic acid
References
- Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281: 37636–37645. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Baker S.S., Wilhelm K.S., Thomashow M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24: 701–713. [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277: 21851–21861. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330. [DOI] [PubMed] [Google Scholar]
- Choi M.S., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280: 40820–40831. [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G.S., Simons K.A., Hou J., Yang T., Reddy A.S., Poovaiah B.W. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Whitelam G.C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39: 1410–1413. [DOI] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2013). Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 161: 408–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Kopka J., Sung D.Y., Zhao W., Popp M., Porat R., Guy C.L. (2007). Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50: 967–981. [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291. [DOI] [PubMed] [Google Scholar]
- Kidokoro S., Maruyama K., Nakashima K., Imura Y., Narusaka Y., Shinwari Z.K., Osakabe Y., Fujita Y., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151: 2046–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S.J., Thomashow M.F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75: 364–376. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Lee M., Lee J.H., Lee H.J., Park C.M. (2015). The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 89: 187–201. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Thomashow M.F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Whalley H.J., Knight M.R. (2015). Combining modelling and experimental approaches to explain how calcium signatures are decoded by calmodulin-binding transcription activators (CAMTAs) to produce specific gene expression responses. New Phytol. 208: 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.L., Newton L., Liu M.J., Shiu S.H., Farré E.M. (2016). A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in arabidopsis. Plant Physiol. 170: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38: 982–993. [DOI] [PubMed] [Google Scholar]
- Maruyama K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150: 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Thomashow M.F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60: 328–339. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., Fujita M., Shinozaki K., Matsui M., Ohme-Takagi M. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51: 2145–2151. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Isono T., Sato M.H. (2003). Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 44: 975–981. [DOI] [PubMed] [Google Scholar]
- Miura K., Lee J., Miura T., Hasegawa P.M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51: 103–113. [DOI] [PubMed] [Google Scholar]
- Mizoi J., Ohori T., Moriwaki T., Kidokoro S., Todaka D., Maruyama K., Kusakabe K., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2013). GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161: 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 109: 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462. [DOI] [PubMed] [Google Scholar]
- Park S., Lee C.M., Doherty C.J., Gilmour S.J., Kim Y., Thomashow M.F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82: 193–207. [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., Yang J., Xu Y.P., Munyampundu J.P., Cai X.Z. (2016). Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 7: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., Hayashizaki Y., Shinozaki K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinwari Z.K., Nakashima K., Miura S., Kasuga M., Seki M., Yamaguchi-Shinozaki K., Shinozaki K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250: 161–170. [DOI] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Osakabe Y., Katsura S., Mizuno S., Maruyama K., Kusakabe K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70: 599–613. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Townley H.E., Knight M.R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128: 1169–1172. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277: 45049–45058. [DOI] [PubMed] [Google Scholar]
- Zarka D.G., Vogel J.T., Cook D., Thomashow M.F. (2003). Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 133: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Lang Z., Zhu J.K. (2015). Cold responsive gene transcription becomes more complex. Trends Plant Sci. 20: 466–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.