The RNA polymerase II transcript elongation complex is an interaction site for transcript elongation factors, facilitating chromatin transcription and its coordination with mRNA processing.
Abstract
Transcript elongation factors (TEFs) are a heterogeneous group of proteins that control the efficiency of transcript elongation of subsets of genes by RNA polymerase II (RNAPII) in the chromatin context. Using reciprocal tagging in combination with affinity purification and mass spectrometry, we demonstrate that in Arabidopsis thaliana, the TEFs SPT4/SPT5, SPT6, FACT, PAF1-C, and TFIIS copurified with each other and with elongating RNAPII, while P-TEFb was not among the interactors. Additionally, NAP1 histone chaperones, ATP-dependent chromatin remodeling factors, and some histone-modifying enzymes including Elongator were repeatedly found associated with TEFs. Analysis of double mutant plants defective in different combinations of TEFs revealed genetic interactions between genes encoding subunits of PAF1-C, FACT, and TFIIS, resulting in synergistic/epistatic effects on plant growth/development. Analysis of subnuclear localization, gene expression, and chromatin association did not provide evidence for an involvement of the TEFs in transcription by RNAPI (or RNAPIII). Proteomics analyses also revealed multiple interactions between the transcript elongation complex and factors involved in mRNA splicing and polyadenylation, including an association of PAF1-C with the polyadenylation factor CstF. Therefore, the RNAPII transcript elongation complex represents a platform for interactions among different TEFs, as well as for coordinating ongoing transcription with mRNA processing.
INTRODUCTION
In eukaryotes, functional mRNA levels in the cell are precisely controlled in a spatially and temporally defined manner. To achieve this goal, transcript synthesis by RNA polymerase II (RNAPII), as well as mRNA processing, are regulated at several stages. Traditionally, the initiation of transcription is considered the crucial step controlling transcript synthesis. In recent years, it has become apparent that the elongation phase of RNAPII transcription is also dynamic and highly regulated. The differential phosphorylation of heptapeptide repeats within the RNAPII C-terminal domain (RNAPII-CTD) characterizes subsequent steps of the elongation phase (Buratowski, 2009; Hajheidari et al., 2013). In addition, a variety of so-called transcript elongation factors (TEFs) have been identified, reflecting the importance of regulating transcript elongation. TEFs facilitate efficient mRNA synthesis in the chromatin context; accordingly, they act as histone chaperones, modify histones within transcribed regions, or modulate the catalytic properties of RNAPII (Jonkers and Lis, 2015; Selth et al., 2010; Sims et al., 2004). Importantly, transcript elongation is also essential for coordinating transcript synthesis and cotranscriptional pre-mRNA processing (Perales and Bentley, 2009).
Genetic and biochemical studies have revealed that various TEFs regulate plant growth and development (Van Lijsebettens and Grasser, 2014). In Arabidopsis thaliana, the histone chaperone complex FACT (facilitates chromatin transcription), consisting of the SSRP1 (structure-specific recognition protein 1) and SPT16 (suppressor of Ty 16) proteins, is involved in the expression of the floral repressor gene FLC and thereby modulates the induction of flowering (Lolas et al., 2010). Likewise, the PAF1 (RNAPII-associated factor 1) complex (PAF1-C), which promotes transcription-coupled histone modifications (i.e., H3K4me, H3K36me, and H2Bubi) (Tomson and Arndt, 2013), was found to regulate flowering by altering the expression of FLC and related MADS box factors (He et al., 2004; Oh et al., 2004). Another TEF that regulates FLC expression is P-TEFb (positive transcript elongation factor b, consisting of CDKC;2 and CYCT1), which mediates the phosphorylation of Ser2 residues within RNAPII-CTD repeats (Wang et al., 2014). The Elongator complex has histone acetyl-transferase activity and modulates developmental and immune response pathways (Woloszynska et al., 2016). TFIIS assists RNAPII progression of transcription through various obstacles and is involved in the expression of DOG1, a key regulator of Arabidopsis seed dormancy (Grasser et al., 2009; Mortensen and Grasser, 2014). Another factor altering the performance of RNAPII is the SPT4/SPT5 heterodimer, which is thought to render the elongation complex stable and processive (Martinez-Rucobo and Cramer, 2013). SPT4/SPT5 is required for the efficient expression of genes involved in auxin signaling and therefore contributes to auxin-related gene expression (Dürr et al., 2014).
In yeast, the assembly of the RNAPII transcript elongation complex (TEC) and the tracking of TEFs with RNAPII along transcribed regions has been examined in quite some detail. Additionally, genetic/biochemical interactions between various TEFs have been studied (Jonkers and Lis, 2015; Selth et al., 2010; Sims et al., 2004). In higher eukaryotes and particularly in plants, considerably less is known about the RNAPII TEC and the collaboration of TEFs. Therefore, in this study, we used a reciprocal tagging approach to affinity purify various TEFs from Arabidopsis cells. Mass spectrometric analyses identified interactions between transcript elongation-related proteins, but they also revealed interplay with mRNA processing factors. Moreover, the examination of plants defective in different combinations of TEFs demonstrated genetic interactions between distinct types of TEFs.
RESULTS
Components of the Arabidopsis RNAPII Elongation Complex
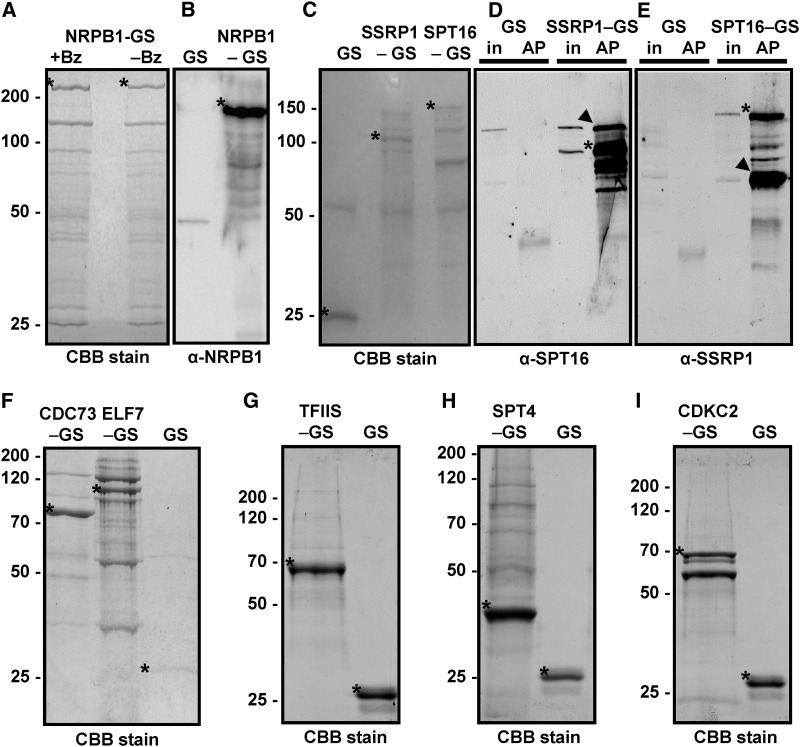
To identify proteins that form the Arabidopsis RNAPII elongation complex, we expressed bait proteins fused to a GS tag (2x protein G domains and streptavidin binding peptide) in PSB-D suspension cultured cells (ecotype Landsberg erecta) (Van Leene et al., 2015), an approach that has been used to characterize other nuclear protein complexes (Dürr et al., 2014; Nelissen et al., 2010; Pauwels et al., 2010). Employing cultured cells enables the production of homogenous biomaterial to characterize the composition of protein complexes involved in basic cellular functions out of a developmental context and in the absence of tissue-specific influences (Van Leene et al., 2015). For GS-tagged bait proteins, we selected the FACT subunits SSRP1 and SPT16, the PAF1-C components ELF7 and CDC73, the SPT4-2 subunit of SPT4/SPT5, TFIIS, and the CDKC;2 component of P-TEFb. For comparison, we analyzed the GS fusion protein harboring the largest subunit (NRPB1) of RNAPII. To identify proteins that interacted with the bait proteins, the GS fusion proteins and the unfused GS control were isolated from cell extracts by IgG affinity purification. In pilot experiments, we performed comparative affinity purifications with cell extracts that were either untreated or treated with the endonuclease benzonase to degrade nucleic acids. According to SDS-PAGE (Figure 1A) and mass spectrometry analyses, we observed only minor changes in protein recovery, with a tendency toward enhanced protein detection upon nuclease treatment, which may be due to an improved solubilization of chromatin-bound proteins (Lambert et al., 2014). All subsequent affinity purifications in this report were performed with nuclease-treated protein extracts; therefore, the observed associations of proteins were likely due to protein interactions rather than mediated by nucleic acids. Proteins in the eluates of the affinity purifications were identified after tryptic digestion by mass spectrometry, and in some cases, the eluates were also examined by immunoblotting. NRPB1 was specifically detected in the NRPB1-GS purification, but not in the control GS purification (Figure 1B). Ten subunits of RNAPII (Ream et al., 2015) were detected by mass spectrometry in the NRPB1-GS eluate (Table 1; Supplemental Data Set 1), but no subunits specific for other RNAPs were observed. Only the two smallest RNAPII subunits (NRPB10/12; 8.3 and 5.9 kD, respectively) were absent, likely due to our gel-based protein separation. Besides the RNAPII subunits, other transcription-related proteins were identified, including the two subunits of the heterodimeric transcription initiation factor TFIIF and the TEFs SPT5, TFIIS, and SPT6, which are known to directly interact with RNAPII. TFIIF and TFIIS are high-affinity interactors of RNAPII (Sopta et al., 1985), and the prominent presence of initiation and elongation factors in the NRPB1-GS sample indicates that functionally different forms of RNAPII were isolated in this experiment.
Figure 1.
Isolation of Components of the Transcript Elongation Complex.
The indicated GS fusion proteins were purified from Arabidopsis cells by IgG affinity chromatography from protein extracts treated with the nuclease benzonase (except right lane in [A]) to degrade DNA and RNA. Isolated proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (CBB) followed by mass spectrometric analyses or used for immunoblotting with the indicated antibodies. The unfused GS tag (GS) served as the control in these experiments.
(A) Eluates of the NRPB1-GS affinity purification with (+Bz) and without (−Bz) benzonase treatment.
(B) Detection of NRPB1 in the NRPB1-GS sample, but not in the GS control, by immunoblotting using an NRPB1 antibody.
(C) to (E) Analysis of the FACT subunits SSRP1-GS and SPT16-GS. Arrowheads indicate the efficient copurification of SPT16 with SSRP1-GS (in [D]) and of SSRP1 with SPT16-GS (in [E]). In addition to the specific bands that react with the respective primary antibodies, the bands of the GS fusion proteins are detected due to the affinity of the antibodies toward the protein G moiety of the tag. The bands are also detected in the input samples (in) but not in the GS control affinity purifications (AP).
(F) to (I) Analysis of PAF1-C (CDC73-GS and ELF7-GS), GS-TFIIS, SPT4-GS, and CDKC;2-GS. Asterisks indicate the different GS fusion proteins (that migrate in the gels according to their expected masses) and the unfused GS.
Table 1. Chromatin- and Transcription-Related Proteins Copurifying with RNAPII and TEFs.
| NRPB1a | SSRP1a | SPT16a | ELF7a | CDC73a | TFIISa | SPT4a | CDKC;2a | Interactor | AGI | Complexb |
|---|---|---|---|---|---|---|---|---|---|---|
| 8676/3 | 1040/2 | 1084/2 | 1117/3 | 2177/3 | 1748/3 | NRPB1 | AT4G35800 | RNAPII | ||
| 4992/3 | 911/2 | 1017/3 | 164/3 | 838/3 | 969/3 | NRPB2 | AT4G21710 | RNAPII | ||
| 229/3 | NRPB4 | AT5G09920 | RNAPII | |||||||
| 1441/3 | 364/2 | 489/3 | 365/2 | NRP(A/B/C/D)5 | AT3G22320 | RNAPII | ||||
| 431/3 | 189/3 | NRP(B/C/D/E)6a | AT5G51940 | RNAPII | ||||||
| 1038/3 | 154/2 | 471/3 | 240/2 | NRPB7 | AT5G59180 | RNAPII | ||||
| 525/3 | 160/2 | 94/2 | NRP(A/B/C/D/E)8a | AT1G54250 | RNAPII | |||||
| 874/3 | 305/2 | 165/2 | NRP(A/B/C/D/E)8b | AT3G59600 | RNAPII | |||||
| 142/3 | 102/2 | NRP(B/D/E)9b | AT4G16265 | RNAPII | ||||||
| 663/3 | 128/2 | 207/3 | 122/2 | NRP(B/D/E)11 | AT3G52090 | RNAPII | ||||
| 1074/3 | 224/2 | RAP74 | AT4G12610 | TFIIF | ||||||
| 641/3 | 126/2 | RAP30 | AT1G75510 | TFIIF | ||||||
| 211/3 | SPT4-1 | AT5G08565 | SPT4/SPT5 | |||||||
| 947/3 | SPT4-2 | AT5G63670 | SPT4/SPT5 | |||||||
| 362/2 | 180/3 | 758/3 | 1444/3 | 225/3 | 5714/3 | SPT5-2 | AT4G08350 | SPT4/SPT5 | ||
| 2983/3 | SPT5L | AT5G04290 | RNAPV | |||||||
| 964/3 | AGO4 | AT2G27040 | RNAPV | |||||||
| 258/3 | 541/3 | 2576/3 | 427/2 | TFIIS | AT2G38560 | TFIIS | ||||
| 203/2 | 2892/3 | 4015/3 | 495/3 | CDC73 | AT3G22590 | PAF-C | ||||
| 222/3 | 8944/3 | 5264/3 | 1064/3 | 287/3 | CTR9, ELF8, VIP6 | AT2G06210 | PAF-C | |||
| 859/3 | 3141/3 | 1558/3 | 1062/3 | 150/3 | LEO1, VIP4 | AT5G61150 | PAF-C | |||
| 232/3 | 389/2 | 3196/3 | 1660/3 | 917/3 | 125/2 | PAF1, ELF7 | AT1G79730 | PAF-C | ||
| 1104/2 | 2269/3 | 1241/3 | 603/3 | RTF1, VIP5 | AT1G61040 | PAF-C | ||||
| 542/3 | 1274/3 | 2274/3 | 903/3 | 422/3 | 188/2 | SKI8, VIP3 | AT4G29830 | PAF-C | ||
| 1452/3 | CDKC;2 | AT5G64960 | P-TEFb | |||||||
| 123/3 | CYCT1;3 | AT1G27630 | P-TEFb | |||||||
| 1320/3 | CYCT1;4 | AT4G19600 | P-TEFb | |||||||
| 105/3 | 2804/3 | CYCT1;5 | AT5G45190 | P-TEFb | ||||||
| 2641/3 | 2657/3 | 1696/3 | 320/3 | SSRP1 | AT3G28730 | FACT | ||||
| 2885/3 | 4205/3 | 4220/3 | 764/2 | 155/2 | 858/3 | 107/2 | SPT16 | AT4G10710 | FACT | |
| 1102/3 | 321/3 | 1556/3 | 228/2 | 864/3 | 1174/3 | SPT6L | AT1G65440 | SPT6 | ||
| 510/3 | 297/3 | SPT6 | AT1G63210 | SPT6 | ||||||
| 368/2 | 271/3 | 213/2 | 204/2 | 418/2 | NAP1;2 | AT5G56950 | NAP1 | |||
| 237/2 | 225/3 | 213/2 | 143/2 | NAP1;1 | AT4G26110 | NAP1 | ||||
| 858/3 | 633/3 | 309/3 | 602/2 | 147/3 | NAP1;3 | AT2G19480 | NAP1 | |||
| 233/2 | BSH | AT3G17590 | CRC | |||||||
| 116/2 | MINU1 | AT3G06010 | CRC | |||||||
| 218/3 | SWI3D | AT4g34430 | CRC | |||||||
| 151/2 | 271/3 | SWP73B | AT5G14170 | CRC | ||||||
| 500/3 | 471/3 | BRM | AT2G46020 | CRC | ||||||
| 104/2 | 391/3 | 354/2 | CHR17 | AT5G18620 | CRC | |||||
| 200/2 | INO80 | AT3G57300 | CRC | |||||||
| 625/2 | PKR3 | AT2G13370 | CRC | |||||||
| 229/2 | 182/3 | PKL | At2g25170 | CRC | ||||||
| 263/3 | EAF1B | AT3G24870 | SWR1/NuA4 | |||||||
| 121/2 | SWC4 | AT2G47210 | SWR1/NuA4 | |||||||
| 510/3 | EAF7 | AT1G26470 | SWR1/NuA4 | |||||||
| 109/2 | EPL1B | AT1G79020 | SWR1/NuA4 | |||||||
| 1664/3 | MRG1 | AT4G37280 | SWR1/NuA4 | |||||||
| 99/2 | TRA1B | AT4G36080 | SWR1/NuA4 | |||||||
| 89/2 | HAM1 | AT5G64610 | SWR1/NuA4 | |||||||
| 237/2 | 901/3 | 807/2 | 3018/3 | ELP1, ELO2 | AT5G13680 | Elongator | ||||
| 302/3 | ELP2 | AT1G49540 | Elongator | |||||||
| 794/3 | 747/3 | 222/2 | 2085/3 | ELP3; ELO3 | AT5G50320 | Elongator | ||||
| 126/3 | HDA15 | AT3G18520 | HDAC | |||||||
| 126/2 | HDA19 | AT4G38130 | HDAC | |||||||
| 714/3 | 1178/3 | 539/3 | 1030/3 | HDT3 | AT5G03740 | HDAC | ||||
| 330/3 | 878/3 | 264/2 | 557/2 | HDT4 | AT2G27840 | HDAC | ||||
| 142/2 | HUB1 | AT2G44950 | HUb | |||||||
| 120/2 | 141/2 | WDR5A | AT3G49660 | HMT | ||||||
| 505/3 | SDG4, ASHR3 | AT4G30860 | HMT |
Numbers indicate in which affinity purifications the interactors were identified, and the respective average Mascot scores are given as well the number of times the interactor was detected in three independent affinity purifications; only proteins are listed that were detected at least twice in three experiments.
The protein complex or the protein family to which the interactors belong.
To identify components of the elongation complex, GS-tagged versions of various TEFs were affinity purified. Isolation and immunoblot analysis of the FACT subunits SSRP1 and SPT16 demonstrated that the two proteins robustly were purified together (Figures 1C to 1E), which is in agreement with their occurrence as a heterodimer (Duroux et al., 2004). As expected for a histone chaperone, various core histones (i.e., H2A, H2B, H2A.Z, and H2A.X) copurified with FACT (Supplemental Data Set 2), but since many of them, likely due to their abundance, appear in the list of common contaminants (Van Leene et al., 2015), they are not further discussed here. In addition, various TEFs were found to copurify with FACT, including SPT5-2 and subunits of PAF1-C (Table 1; Supplemental Data Set 2). Members of the NAP1 family of histone chaperones and histone deacetylases (HDACs) were also identified in the SSRP1-GS and SPT16-GS eluates.
Arabidopsis PAF1-C was isolated previously and the subunits VIP3, VIP4, VIP6/ELF8, and CDC73 were identified by immunoblotting (Oh et al., 2004; Park et al., 2010). Affinity purification of ELF7-GS and CDC73-GS (Figure 1F) in combination with mass spectrometry analyses resulted reproducibly in the identification of six predicted subunits of PAF1-C, with high Mascot scores, indicating a high probability of correct identification (Table 1; Supplemental Data Set 3). Therefore, Arabidopsis PAF1-C is likely a hexameric complex consisting of ELF7, VIP6/ELF8, VIP3, VIP4, VIP5, and CDC73, resembling the situation in human and Drosophila melanogaster rather than in yeast, where PAF1-C is composed of five subunits lacking SKI8 (ortholog of Arabidopsis VIP3) (Jaehning, 2010; Tomson and Arndt, 2013). In line with a hexameric PAF1-C, according to publicly available mRNA expression data, the genes encoding the six subunits show a very similar expression profile in Arabidopsis (http://www.arabidopsis.org/). Additionally, other TEFs, including SPT5, TFIIS, FACT, SPT6, and Elongator, copurified with PAF1-C. The SPT6 histone chaperone occurs in two versions in Arabidopsis (Gu et al., 2012), and according to publicly available microarray data, SPT6L appears to be commonly expressed, whereas the SPT6 transcript is barely detectable in most tissues (http://www.arabidopsis.org/). Consistently, only the SPT6L transcript was detected by RT-PCR analysis with seedling RNA, but both genes are expressed in PSB-D cells (Supplemental Figures 1A to 1C). SPT6L, as well as SPT6, were isolated along with PAF1-C from PSB-D cells (Table 1). In addition, subunits of RNAPII, NAP1, and several proteins involved in ATP-dependent chromatin remodeling complexes (CRCs) were identified, primarily in the ELF7-GS eluates.
Along with GS-TFIIS (Figure 1G) diverse subunits of RNAPII and all subunits of PAF1-C were isolated (Table 1; Supplemental Data Set 4). Moreover, other TEFs, including SPT5-2 and SPT6L, as well as NAP1 proteins and HDACs were identified in the GS-TFIIS eluates. We previously investigated SPT4-GS affinity purifications (Dürr et al., 2014). Because the sensitivity of our mass spectrometric analyses has since been markedly improved, the experiment was repeated under comparable conditions (Figure 1H). This experiment confirmed the proteins that were found to copurify with SPT4-GS before (Dürr et al., 2014), but with more robust Mascot scores, and several additional interactors were identified. Thus, in addition to SPT5-2, several subunits of RNAPII, as well as TFIIF and HDACs, were isolated along with SPT4-GS (Table 1; Supplemental Data Set 5). A number of TEFs, including TFIIS, PAF1-C, FACT, SPT6L/SPT6, and Elongator, were also found to copurify. Interestingly, various proteins of ATP-dependent chromatin remodeling complexes (Gentry and Hennig, 2014) were identified in the SPT4-GS eluates. Two components that are involved in RNAPV-mediated RNA-directed DNA methylation, SPT5L and AGO4, were found to specifically associate with SPT4-GS, but with none of the other analyzed GS fusion proteins. The plant-specific protein SPT5L is a direct interactor of SPT4 and AGO4 (Bies-Etheve et al., 2009; Dürr et al., 2014; He et al., 2009), and SPT4 can modulate RNA-directed DNA methylation (Köllen et al., 2015).
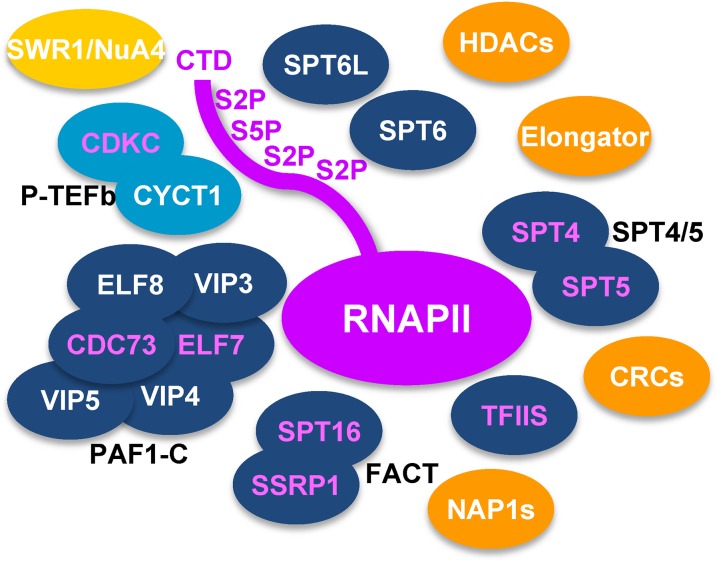
Affinity purification of the P-TEFb component CDKC;2-GS (Figure 1I) demonstrated that it can interact with three different versions of CYCT1 (Table 1; Supplemental Data Set 6), which is consistent with recent results (Cui et al., 2007; Wang et al., 2014). Surprisingly, apart from SPT16, no other TEFs and no RNAPII subunits were found to copurify with CDKC;2-GS. However, various subunits of the NuA4/SWR1 chromatin remodeling complex, with combined histone acetyl-transferase and chromatin remodeling activity (Bieluszewski et al., 2015) (Table 1), as well as several BRD4 (bromodomain-containing protein4)-like proteins (Supplemental Data Set 6) were detected in the CDKC;2-GS eluates. Since BRD4 proteins are involved in recruiting P-TEFb to chromatin containing acetylated histones at target genes in mammalian cells (Bisgrove et al., 2007; Jang et al., 2005), this mechanism may be conserved in plants. In conclusion, our proteomics analyses demonstrate that there is a considerable overlap in the interactions seen with FACT, PAF1-C, TFIIS, and SPT4/SPT5, but the protein interactions of P-TEFb differ markedly from those seen with the other tested TEFs (Figure 2). Additional factors (e.g., NAP1, CRCs, and Elongator) repeatedly copurified with the TEC and may contribute to efficient transcript elongation in Arabidopsis. To examine which form(s) of RNAPII copurified with the TEFs, we analyzed affinity purifications of GS-TFIIS and ELF7-GS by immunoblotting using antibodies directed against the nonphosphorylated RNAPII-CTD and against Ser2-phosphorylated CTD repeats. Relative to the input samples in the affinity-purifications, the Ser2-phosphorylated form of RNAPII was enriched compared with the hypophosphorylated RNAPII (Supplemental Figure 2A). Therefore, the elongating, Ser2-phosphorylated form of RNAPII predominantly copurified with the TEFs.
Figure 2.
Scheme Depicting the Arabidopsis RNAPII Elongation Complex Based on the Targeted Proteomics Results.
The TEFs FACT, TFIIS, SPT4/SPT5, SPT6, SPT6L, and PAF1-C (dark blue) robustly copurified with each other and with RNAPII (magenta), while P-TEFb (light blue) was not enriched in these experiments. However, additional chromatin factors (orange) also repeatedly copurified with the TEFs, except for SWR1/NuA4 (yellow), which was primarily isolated along with P-TEFb. Magenta characters of the oval symbols indicate the proteins that were affinity purified as GS-tagged fusion proteins (for details, see Table 1).
ELF7 and SPT6L Colocalize with RNAPII in Euchromatin
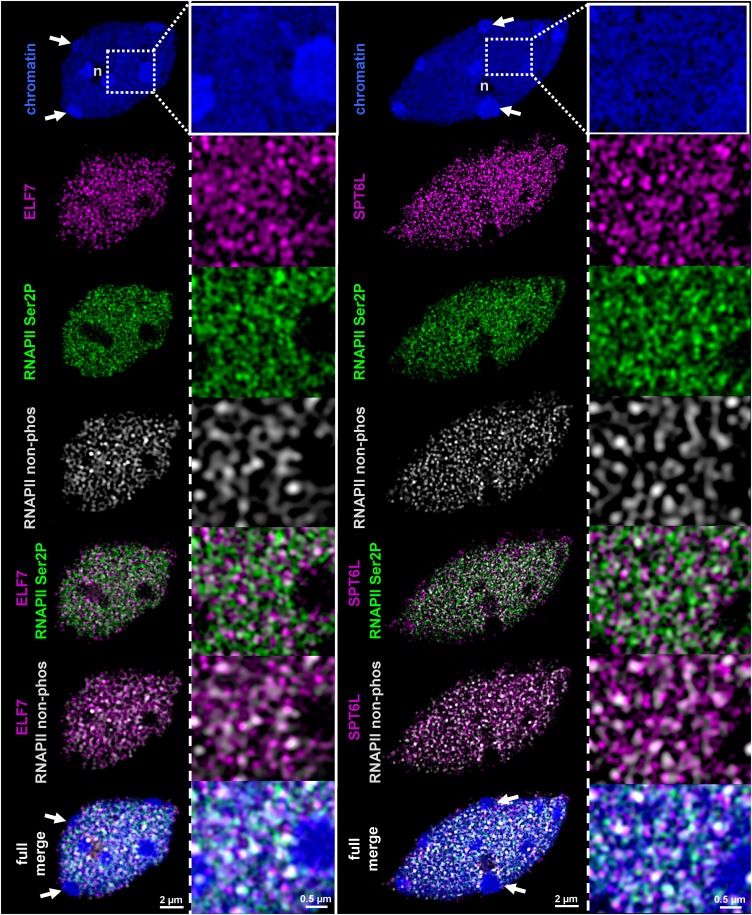
Microscopic immune fluorescence analyses have shown that the TEFs SPT5 and FACT are enriched within transcriptionally active euchromatin in Arabidopsis nuclei and that SPT5 colocalizes with elongating RNAPII (Duroux et al., 2004; Dürr et al., 2014; Lolas et al., 2010). In view of the mutual association of FACT and SPT4/SPT5 with PAF1-C, TFIIS, and SPT6L, we raised antibodies against recombinant ELF7, TFIIS, and SPT6L. While the TFIIS antibodies proved not to be useful, the ELF7 and SPT6L antibodies yielded specific staining patterns in pilot experiments and were used for super-resolution structured illumination microscopy (SIM). ELF7 and SPT6L fluorescent signals were analyzed in flow-sorted 8C nuclei of Arabidopsis leaves (Figure 3). To investigate the subnuclear distribution of ELF7 and SPT6L relative to RNAPII, cells were simultaneously labeled with the respective antibodies and counterstained with 4′,6-diamidino-2-phenylindole. We used antibodies specific for the elongating form (phosphorylated at Ser2 of the CTD repeats) and specific for the nonphosphorylated form of RNAPII. In Arabidopsis interphase nuclei, the majority of RNAPII is globally dispersed in the euchromatic part of the nucleoplasm (Schubert and Weisshart, 2015). Consistently, our SIM analyses revealed that ELF7/SPT6L and both forms of RNAPII comprised separate, dispersed, reticulate structures within the euchromatin, but they were absent from the nucleolus and heterochromatin (Figure 3). Further analysis of the degree of colocalization between ELF7/SPT6L and the RNAPII signals revealed that both TEFs are more often associated with the elongating (CTD-Ser2P) than with the nonphosphorylated form of RNAPII. This tendency is visible in the magnified panels on the right (Figure 3) and in the comparable merged images (Supplemental Figure 3), and it is also in line with the quantification of the colocalizing fluorescence signals (Supplemental Table 1). SPT6 from human and the fungus Candida glabrata binds preferentially to Ser2-phosphorylated RNAPII-CTD repeats (Sun et al., 2010; Yoh et al., 2007), and because of the colocalization of Arabidopsis SPT6L with the elongating RNAPII, we examined its interaction with the RNAPII-CTD. The interaction of the putative CTD-interaction domain SPT6L(Phe1218-Asp1412) with RNAPII-CTD peptides was examined by microscale thermophoresis (MST). The fluorescently labeled peptides were phosphorylated at Ser2 or Ser5 or were unmodified. The MST measurements revealed a clear concentration-dependent interaction with the Ser2-phosphorylated peptides with a Kd of 135 (±26.6) μM, whereas the affinity for the Ser5-phosphorylated and nonphosphorylated peptides was at least 10-fold lower (Supplemental Figure 2B).
Figure 3.
ELF7 and SPT6L Colocalize with RNAPII in Euchromatin.
Colocalization analysis of ELF7 and SPT6L (magenta) with elongating RNAPII (Ser2P, green) and RNAPII (nonphosphorylated, white) within euchromatic regions of flow-sorted 8C nuclei of leaf cells visualized by SIM. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Fluorescent protein signals are not detected in the nucleoli (n) and within heterochromatic chromocenters (arrows). In addition to the individual fluorescent signals, merged images are shown in the three lower panels. The preferential association of ELF7 and SPT6L with RNAPII Ser2P rather than the nonphosphorylated form of RNAPII is visible from the magnified images (on the right of each panel).
Genetic Interactions between Genes Encoding Different TEFs
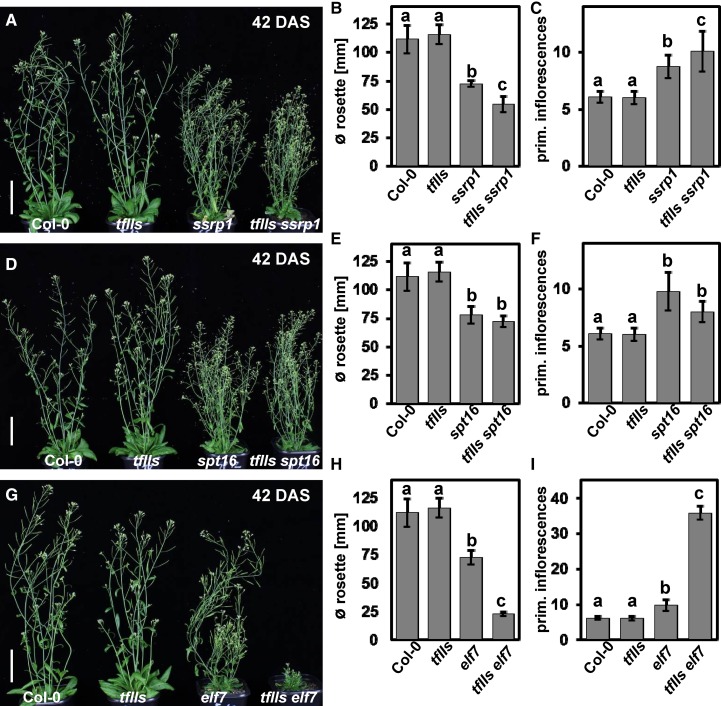
Since several TEFs were found to copurify efficiently and to colocalize with elongating RNAPII, we generated and analyzed various double mutants deficient in different combinations of TEFs. The double mutants, along with the respective single mutants and Col-0 wild-type plants, were primarily examined regarding growth and developmental phenotypes for which differences were observed in the parental lines. We generated plants deficient in TFIIS (which have an essentially wild-type appearance; Grasser et al., 2009) and the FACT subunits (ssrp1 and spt16, which express reduced amounts of the FACT subunits and similarly display various developmental defects; Lolas et al., 2010). While the tfIIs ssrp1 double mutants are synergistically affected regarding rosette diameter and number of primary inflorescences (Figures 4A to 4C; Supplemental Figure 4A), the tfIIs spt16 plants are comparable to the spt16 single mutants (Figures 4D to 4F; Supplemental Figure 4B). The seed set of tfIIs spt16 is clearly reduced relative to Col-0 and tfIIs but comparable to that of spt16 (and ssrp1) (Supplemental Figure 4C), whereas tfIIs ssrp1 plants are sterile. In spt16 plants, the leaf vein patterning is mildly altered and in ssrp1 leaves, the venation is more strongly affected (Lolas et al., 2010), while the leaf venation of tfIIs is comparable to that of Col-0. Examination of the vein pattern in cleared leaves demonstrated that the venation of tfIIs ssrp1 and tfIIs spt16 is similar to that of the respective single mutants defective in the FACT subunits (Supplemental Figure 5). Therefore, concerning different phenotypes, the tfIIs ssrp1 double mutants are synergistically affected, while analysis of the tfIIs spt16 plants indicated that SPT16 acts epistatically to TFIIS. However, regarding bolting time (Supplemental Figures 4D and 4E) and leaf vein patterning, both double mutant combinations behave like the respective single mutant deficient in the FACT subunit. We also generated plants defective in TFIIS and in a PAF1-C subunit (elf7, which show reduced growth and early flowering; He et al., 2004). The tfIIs elf7 double mutants display a synergistically reduced rosette diameter and increased number of primary inflorescences (Figures 4G to 4I; Supplemental Figure 6A). The elf7 single mutant showed reduced leaf venation, lacking most of the tertiary and higher-order veins. In the tfIIs elf7 plants, the defects in vein patterning were severely enhanced (Supplemental Figure 5). Similarly, elf7 plants have a reduced seed set and seed production in tfIIs is comparable to that of Col-0, but the double mutant is sterile. Regarding bolting time, tfIIs elf7 is comparable to elf7, but the number of leaves at bolting is reduced relative to elf7 (Supplemental Figures 6B and 6C). The analysis of double mutants defective in ELF7 and FACT subunits revealed that the elf7 ssrp1 combination is lethal, while elf7 spt16 plants are viable, but their growth is strongly reduced (Supplemental Figure 7). The examination of double mutants defective in different combinations of TEFs demonstrated distinct genetic interactions between the genes encoding FACT, PAF1-C, and TFIIS, yielding synergistic/epistatic effects on the analyzed developmental traits.
Figure 4.
Analysis of Double Mutants Defective in Different Combinations of TEFs.
tfIIs ssrp1([A] to [C]), tfIIs spt16 ([D] to [F]), and tfIIs elf7 ([G] to [I]) mutants were examined relative to the respective single mutants and the Col-0 wild type. Representative images of the plants at 42 DAS under long-day conditions are shown ([A], [D], and [G]) with bars representing 5 cm. The rosette diameter (∅ in [B], [E], and [H]) and the number of primary inflorescences ([C], [F], and [I]) were determined (n = 13). Data were analyzed by two-way ANOVA, and error bars indicate sd calculated from the measurements of 13 individual plants for each line. The letters above the histogram bars indicate the outcome of a multi comparisons Tukey’s test (P < 0.05).
Interaction of the RNAPII TEC with mRNA Processing Factors
In view of the central role of the RNAPII TEC in coordinating the synthesis and processing of transcripts, as established in yeast and metazoa (Perales and Bentley, 2009), we examined our proteomics data obtained from the affinity purification of various TEFs for the occurrence of factors involved in pre-mRNA processing. Since almost no interactions with mRNA processing factors were observed in the data obtained with P-TEFb, we did not include these in our analysis (Table 2). However, we performed additional affinity purifications with the polyadenylation factors CstF64-GS and CstF77-GS (Supplemental Figure 8) to explore possible interactions among the mRNA processing machineries and with the TEC. Many splicing factors were found to copurify with SPT4, SPT16, and ELF7 and to a lesser extent with TFIIS, SSRP1, and CDC73 (Table 2; Supplemental Data Sets 2 to 5). Various spliceosomal complexes, including U1, U2, U5, Sm, and NTC, were identified, suggesting that spliceosomes of different assembly stages associate with the TEC. A few polyadenylation factors were also identified in the SPT4-GS and ELF7-GS eluates. The robust copurification of CstF50, CstF64, and CstF77 in the CstF64-GS and CstF77-GS samples (Table 2; Supplemental Data Set 7) indicates that the composition of the Arabidopsis CstF complex resembles the situation in metazoa rather than in yeast (Hunt et al., 2008; Yang and Doublié, 2011). In mammals, CstF is a trimeric complex consisting of CstF50, CstF64, and CstF77, and it is believed to dimerize, forming a functional hexameric complex that is critically involved in pre-mRNA 3′end processing (Shi and Manley, 2015). Other polyadenylation factors were also identified in the CstF77-GS eluates. In addition, multiple splicing factors (e.g., Sm and NTC) copurified with CstF77-GS (Table 2), which is in agreement with recent studies reporting a close cooperation between mRNA splicing and polyadenylation (Kaida, 2016; Misra and Green, 2016).
Table 2. mRNA Processing Factors Copurifying with TEFs and CstF.
| SSRP1a | SPT16a | SPT4a | TFIISa | ELF7a | CDC73a | CstF77a | CstF64a | Interactor | AGI | Complexb | Funct.c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 116/3 | Luc7B | AT5G17440 | U1 | Splice | |||||||
| 125/2 | Luc7-RL | AT5G51410 | U1 | Splice | |||||||
| 215/3 | PRP39A | AT1G04080 | U1 | Splice | |||||||
| 702/3 | PRP40A | AT1G44910 | U1 | Splice | |||||||
| 233/2 | PRP40B | AT3G19670 | U1 | Splice | |||||||
| 557/3 | SAP94 | AT1G60200 | U1 | Splice | |||||||
| 107/2 | SAP61 | AT5G06160 | U2 | Splice | |||||||
| 171/2 | U2A' | AT1G09760 | U2 | Splice | |||||||
| 282/2 | U2B”B | AT2G30260 | U2 | Splice | |||||||
| 204/2 | 449/3 | 192/3 | SAP130A | AT3G55200 | U2 | Splice | |||||
| 107/3 | 233/2 | SAP145 | AT4G21660 | U2 | Splice | ||||||
| 212/2 | 137/2 | 125/2 | P14-1 | AT5G12190 | U2 | Splice | |||||
| 140/2 | 1297/3 | 390/3 | 330/2 | SAP155 | AT5G64270 | U2 | Splice | ||||
| 96/2 | 206/2 | 114/2 | 118/2 | 230/3 | SAP49A | AT2G18510 | U2 | Splice | |||
| 222/2 | 106/2 | SR140-1 | AT5G25060 | U2 | Splice | ||||||
| 138/2 | PRP24 | AT4G24270 | U4/U6 | Splice | |||||||
| 166/2 | PRP31 | AT1G60170 | U4/U6 | Splice | |||||||
| 1894/3 | U5-200-1 | AT5G61140 | U5 | Splice | |||||||
| 135/2 | U5-52 | AT5G09390 | U5 | Splice | |||||||
| 556/3 | 165/3 | U5-102 | AT4G03430 | U5 | Splice | ||||||
| 191/2 | 281/2 | 172/2 | 283/3 | U5-40 | AT2G43770 | U5 | Splice | ||||
| 241/2 | 1061/3 | 1998/2 | 529/2 | 1478/2 | U5-200-2A | AT1G20960 | U5 | Splice | |||
| 94/2 | 201/2 | SmE-B | AT2G18740 | Sm | Splice | ||||||
| 302/2 | SmB-B | AT4G20440 | Sm | Splice | |||||||
| 168/2 | SmD1-B | AT4G02840 | Sm | Splice | |||||||
| 81/2 | 89/2 | SmD3-A | AT1G76300 | Sm | Splice | ||||||
| 120/2 | 156/2 | SmD2-B | AT3G62840 | Sm | Splice | ||||||
| 167/2 | 164/2 | 143/2 | SmD1-A | AT3G07590 | Sm | Splice | |||||
| 170/2 | SYF3 | AT5G41770 | NTC | Splice | |||||||
| 137/2 | SPF27 | AT3G18165 | NTC | Splice | |||||||
| 97/2 | 238/2 | SYF1 | AT5G28740 | NTC | Splice | ||||||
| 297/2 | PRL1 | AT4G15900 | NTC | Splice | |||||||
| 103/2 | 189/2 | 357/3 | ISY1 | AT3G18790 | NTC | Splice | |||||
| 141/2 | 184/2 | 163/2 | PRP19A | At1G04510 | NTC | Splice | |||||
| 154/3 | 236/3 | 532/2 | CDC5 | AT1G09770 | NTC | Splice | |||||
| 242/2 | 360/2 | 383/2 | PRP19B | AT2G33340 | NTC | Splice | |||||
| 168/3 | 197/2 | 334/2 | SKIP | AT1G77180 | NTC | Splice | |||||
| 130/2 | BUD31 | AT4G21110 | NTC | Splice | |||||||
| 230/3 | ECM2-1B | AT2G29580 | NTC | Splice | |||||||
| 105/2 | Prp17-1 | AT1G10580 | NTC | Splice | |||||||
| 338/3 | CWFJ-L | AT1G56290 | NTC | Splice | |||||||
| 146/2 | CCCH64 | AT5G56900 | NTC | Splice | |||||||
| 89/2 | 141/2 | CWC22 | AT1G80930 | NTC | Splice | ||||||
| 443/3 | 148/3 | 171/2 | ECM2-1A | AT1G07360 | NTC | Splice | |||||
| 141/2 | 997/3 | 646/2 | 124/2 | 782/2 | Aquarius | AT2G38770 | NTC | Splice | |||
| 92/2 | 214/2 | ESP4 | AT5G01400 | CPSF | PA | ||||||
| 105/2 | 354/2 | FIPS5 | AT5G58040 | CPSF | PA | ||||||
| 111/2 | CPSF100 | AT5G23880 | CPSF | PA | |||||||
| 255/2 | CPSF160 | AT5G51660 | CPSF | PA | |||||||
| 222/2 | CPSF30 | AT1G30460 | CPSF | PA | |||||||
| 181/2 | CFIS2 | AT4G25550 | CFI | PA | |||||||
| 1474/3 | 807/3 | CSTF50 | AT5G60940 | CstF | PA | ||||||
| 899/3 | 1639/3 | CSTF64 | AT1G71800 | CstF | PA | ||||||
| 3278/3 | 2058/3 | CSTF77 | AT1G17760 | CstF | PA |
Numbers indicate in which affinity purifications the interactors were identified, and the respective average Mascot scores are given as well the number of times the interactor was detected in three independent affinity purifications; only proteins are listed that were detected at least twice in three experiments.
The protein complex or the protein family to which interactors belong.
The process, splicing or polyadenylation (PA), in which the interactors are primarily involved, according to the literature.
Interplay between PAF1-C and CstF
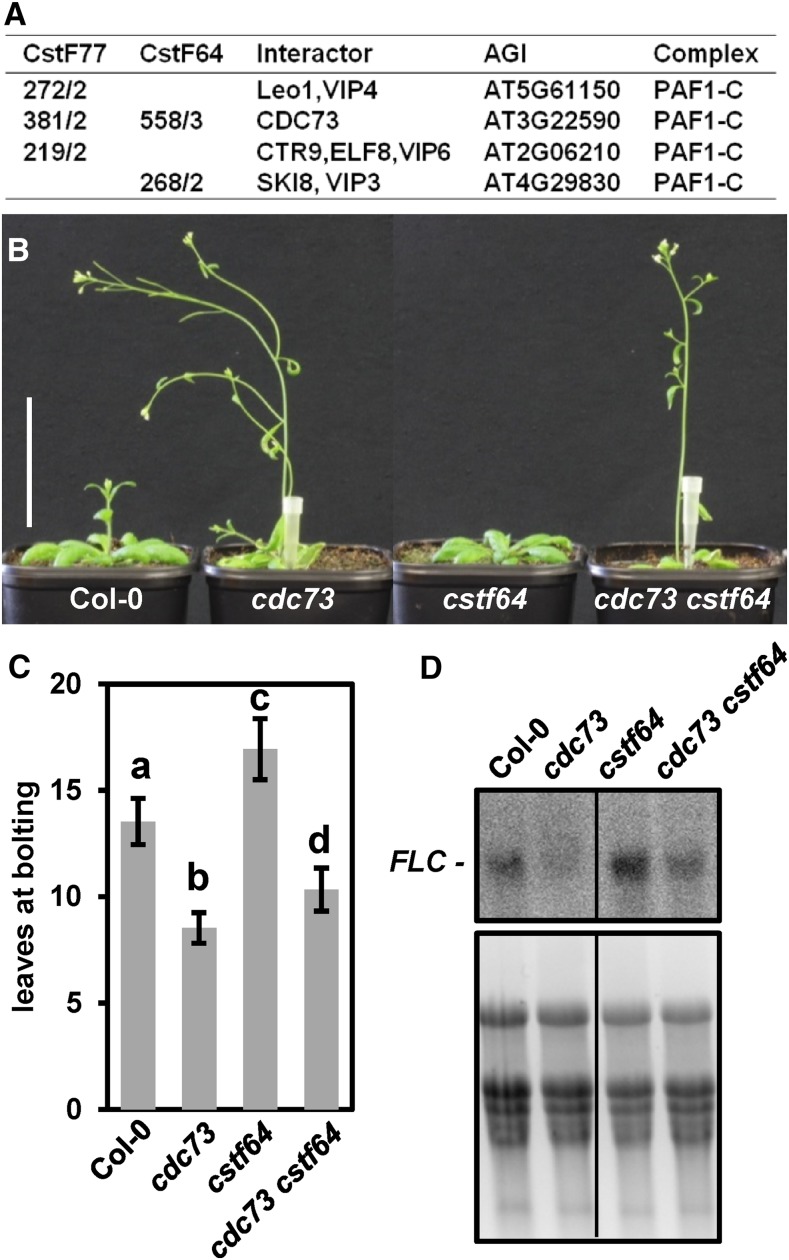
In yeast and metazoa, PAF1-C has various functions related to transcription and mRNA processing, including interactions with the polyadenylation machinery (Jaehning, 2010; Tomson and Arndt, 2013). Thus, PAF1-C is involved in recruiting certain polyadenylation factors to transcribed regions and can modulate the 3′end processing of mRNAs (Nagaike et al., 2011; Nordick et al., 2008). Interestingly, several subunits of PAF1-C, including CDC73, were identified in the CstF77-GS and CstF64-GS eluates (Figure 5A; Supplemental Data Set 7), albeit for some reason, CstF was not observed in the affinity purification of ELF7/CDC73. The possible interaction of Arabidopsis PAF1-C and CstF is in accordance with the association of human CstF with CDC73 (Rozenblatt-Rosen et al., 2009). Arabidopsis mutants deficient in CDC73 are early flowing, whereas mutants lacking CstF64 are late flowering, which is ultimately mediated by altered transcript levels of the floral repressor FLC (Liu et al., 2010; Park et al., 2010; Yu and Michaels, 2010). We generated double mutants deficient in CDC73 and CstF64 to examine the induction of flowering in the cdc73 cstf64 combination. The cdc73 cstf64 plants bolted earlier than Col-0, but not as early as the cdc73 single-mutant (Figures 5B and 5C; Supplemental Figure 9A). Consistent with the early flowering phenotype relative to Col-0, reduced FLC transcript levels were detected by RNA gel blot analysis and qRT-PCR in the double mutant, but the amount was slightly higher than in cdc73 (Figure 5D; Supplemental Figure 9B), indicating that in the double mutant, the influence of cdc73 on flowering and FLC expression is more pronounced than that of cstf64.
Figure 5.
Interaction of PAF1-C and the CstF Complex.
(A) Copurification of PAF1-C subunits with CstF77-GS and CstF64-GS. In the first two columns, the average Mascot scores are given as well as the number of times the interactor was detected in three independent affinity purifications.
(B) Representative images of the cdc73 cstf64 double mutant as well as the respective single mutants and Col-0 at 35 DAS under long-day conditions are shown. Bar = 5 cm.
(C) Quantification of the number of leaves at bolting (n = 15). Data were analyzed by two-way ANOVA (significance level P < 0.05) and error bars depict sd calculated from the measurements of 15 individual plants for each line.
(D) RNA gel blot analysis of RNA isolated from the different genotypes using an antisense probe that detects the FLC sense transcript (top panel). For comparison, an ethidium bromide stain of the RNA in the agarose gel prior to membrane transfer is shown (bottom panel).
Specificity of TEFs for RNAPII-Mediated Transcription
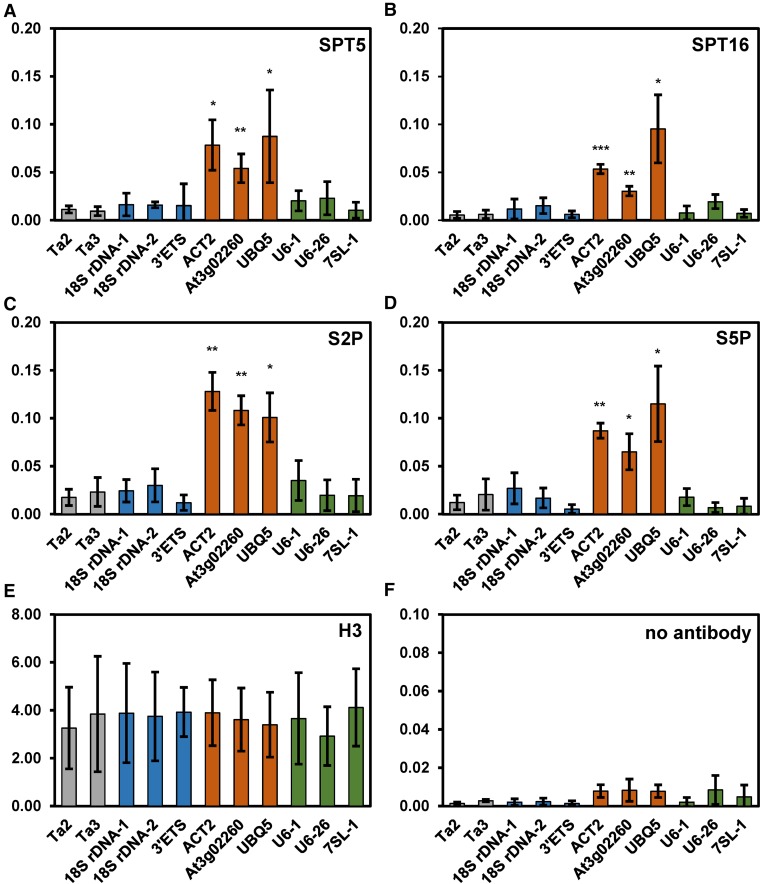
Certain TEFs, including SPT5 and SPT6, were reported to play a role in RNAPI-catalyzed transcription in yeast (Anderson et al., 2011; Engel et al., 2015). Additionally, there is evidence that mammalian FACT is involved in chromatin transcription by RNAPI and RNAPIII (Birch et al., 2009). Characteristic of TEFs, SPT5, and FACT (SPT16 and SSRP1) were found to associate with transcribed regions of various genes actively transcribed by RNAPII in Arabidopsis (Duroux et al., 2004; Dürr et al., 2014; Perales and Más, 2007). Here, using chromatin immunoprecipitation with antibodies directed against SPT5 and SPT16, we investigated whether the two TEFs also associate with genomic regions transcribed by RNAPI and RNAPIII. As expected, SPT5 and SPT16 were detected at ACT2, UBQ5, and At3g02260, which are transcribed by RNAPII (Figures 6A and 6B), whereas only background levels were observed at the transcriptionally inactive retrotransposons TA2 and TA3. For both SPT5 and SPT16, background levels were also detected at three regions transcribed by RNAPI (18S rDNA and ETS). Similarly, no enrichment of the two TEFs was seen at three loci transcribed by RNAPIII (U6-1, U6-26, and 7SL-1). For comparison, we analyzed the association of RNAPII with these loci using antibodies directed against RNAPII-CTD phosphorylated at Ser2 and Ser5 as well as an antibody against histone H3. Comparable to SPT5 and SPT16, both forms of transcribing RNAPII associated with the RNAPII-transcribed loci, but not with the other analyzed regions (Figures 6C and 6D). As expected, H3 was detected at all tested genomic loci (Figure 6E), whereas no enrichment of these loci was observed in the controls without the addition of antibodies (Figure 6F). Therefore, according to our chromatin immunoprecipitation (ChIP) experiments, SPT5 and SPT16 do not associate with Arabidopsis loci transcribed by RNAPI and RNAPIII. In addition, we determined the transcript levels of the 35S pre-rRNA that is synthesized by RNAPI in mutants deficient in different TEFs relative to Col-0. Using RNA gel blot analyses, no significant differences in the amounts of 35S pre-rRNA were detected in the mutant lines compared with Col-0 (Supplemental Figure 10). Therefore, our data do not indicate that the analyzed TEFs collaborate with RNAPI (and RNAPIII) in Arabidopsis.
Figure 6.
SPT5 and SPT16 Associate with RNAPII-Transcribed Loci, but Not with Loci Transcribed by RNAPI and RNAPIII.
SPT5 and SPT16 are enriched at genes transcribed by RNAPII (orange), but not at loci transcribed by RNAPI (blue) and RNAPIII (green), or at nontranscribed regions (gray). ChIP analyses of various loci transcribed by different RNA polymerases. The antibodies used are directed against SPT5 (A), SPT16 (B), RNAPII-CTD-S2P/S5P ([C] and [D]), histone H3 (E), or without antibody (F). For the ChIP experiments, percentage input was determined by qPCR, and P values for all qPCR data were calculated using two-sample t tests (unpaired and two-tailed). Error bars indicate sd of at least three biological (independently pooled collections of tissue) and three technical replicates (independent qPCR measurements each). Data sets marked with asterisks are significantly different from TA3 (gray): *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
Affinity purification of various TEFs from yeast cells has revealed that several TEFs copurified with each other and with RNAPII (Krogan et al., 2002; Lindstrom et al., 2003; Squazzo et al., 2002), and various TEFs were found to associate with the transcribed regions of all genes in the yeast genome that are actively transcribed by RNAPII (Mayer et al., 2010). Together, these experiments indicate that the assembly of certain TEFs with RNAPII promotes efficient chromatin transcription. Our reciprocal tagging approach using various TEFs demonstrated that elongating Arabidopsis RNAPII reproducibly copurified with transcript elongation-related proteins. The set of TEFs that frequently copurified comprised TFIIS, SPT4/SPT5, PAF1-C, and FACT, and to a lesser extent Elongator, which are all implicated in the regulation of Arabidopsis transcript elongation (Dürr et al., 2014; Grasser et al., 2009; He et al., 2004; Lolas et al., 2010; Nelissen et al., 2010; Oh et al., 2008). SPT6L and SPT6 were also identified in several TEF eluates, which is consistent with the colocalization of SPT6L with elongating RNAPII and its preferential binding to Ser2-phosphorylated CTD repeats. Therefore, it is likely that SPT6L and SPT6 also act as TEFs in plants. In contrast, the elongation factor P-TEFb (Quaresma et al., 2016) was not enriched in the affinity purifications of the other TEFs and may interact with the TEC more dynamically, or its association with the complex may not be stable under our purification conditions. Other proteins that repeatedly coeluted with TEFs include variants of the histone chaperone NAP1, suggesting that NAP1 may collaborate with other histone chaperones such as FACT and SPT6 to facilitate transcript elongation (Zhou et al., 2015). Interestingly, a variety of proteins involved in ATP-dependent chromatin remodeling (Gentry and Hennig, 2014) were isolated along with ELF7 and SPT4. So far, there is no evidence from plants, but in other organisms, CRCs are associated with the modulation of transcript elongation, probably by altering the structure of nucleosomes in transcribed regions (Murawska and Brehm, 2011; Subtil-Rodríguez and Reyes, 2011). Regarding histone modifying enzymes, only a few proteins were coisolated with the TEFs. These include the histone methyltransferases SDG4 and WDR5A, which copurified with CDKC;2 and FACT, respectively, as well as the histone ubiquitinase HUB1, which copurified with CDC73 (Table 1). Compared with these proteins, enzymes involved in histone acetylation (i.e., Elongator, SWR1/NuA4, and HDACs) were somewhat overrepresented. In other organisms, histone acetyltransferases and histone deacetylases are enriched along RNAPII-transcribed regions, although only relatively low levels of histone acetylation are observed in the transcribed regions of active genes, suggesting that dynamic histone acetylation is required for efficient elongation (Selth et al., 2010). The integration of our proteomics analyses shows that similar to yeast (Krogan et al., 2002), in Arabidopsis, TFIIS, SPT4/SPT5, SPT6, PAF1-C, and FACT associate with elongating RNAPII (Figure 2). Furthermore, additional chromatin factors, such as NAP1, CRCs, and enzymes involved in histone acetylation (i.e., Elongator and HDACs), interacted with the complex in Arabidopsis cells and may contribute to productive transcript elongation.
Arabidopsis mutants defective in TEFs exhibit a great variety of alterations in growth/development, ranging from mildly affected, for instance, in the case of tfIIs and cdc73 (Grasser et al., 2009; Park et al., 2010; Yu and Michaels, 2010), to severe/lethal phenotypes, for instance, in the case of spt5-2 and spt6l (Dürr et al., 2014; Gu et al., 2012). We examined double mutants deficient in different combinations of TEFs (Supplemental Table 2) to elucidate the consequences on plant growth and development as well as possible interactions between the genes encoding the TEFs. Mutant plants that express reduced amounts of the FACT subunits SSRP1 and SPT16 are phenotypically similar (Lolas et al., 2010), but when these mutations are combined with tfIIs or elf7, regarding most phenotypes, the ssrp1 version of the double mutant is clearly more severely affected than the spt16 version, suggesting that under these conditions, the SSRP1 subunit is more critical for plant growth and development. Consistently, in mammals, SSRP1 has additional, SPT16-independent roles in RNAPII transcription (Antosch et al., 2012). Thus, SSRP1 can act as a coactivator of sequence-specific transcription factors (Spencer et al., 1999; Zeng et al., 2002), and transcript profiling revealed SSRP1-specific targets (Li et al., 2007). Moreover, Arabidopsis SSRP1, but not SPT16, is required for the action of the DNA demethylase DME in the central cell of the female gametophyte (Ikeda et al., 2011). FACT and TFIIS can help RNAPII transcribe through nucleosomes (Belotserkovskaya et al., 2003; Bondarenko et al., 2006; Nock et al., 2012), and they might collaborate regarding this function, which may explain the impaired growth of the tfIIs ssrp1 mutants relative to the respective single mutants. In metazoa, PAF1-C and FACT can apparently recruit each other to sites of transcription (Adelman et al., 2006; Pavri et al., 2006), and yeast cells lacking both PAF1 and SPT16 exhibit synthetic growth defects (Squazzo et al., 2002). In a highly reconstituted mammalian chromatin transcription system, the establishment of H2B monoubiquitination, which is associated with transcriptional activity, is dependent on FACT and PAF1-C (Pavri et al., 2006). Genetic interactions were observed between Arabidopsis SSRP1/SPT16 and the HUB1 gene (encoding an H2B monoubiquitinase) (Lolas et al., 2010). In view of these findings, the severe growth defect of the elf7 spt16 plants and the lethality of elf7 ssrp1 suggest that there is also a close cooperation of FACT and PAF1-C in plants, possibly involving transcription-related histone ubiquitination. Human PAF1-C and TFIIS physically interact and cooperatively bind to RNAPII, promoting activated chromatin transcription (Kim et al., 2010), and yeast cells lacking PAF1 and TFIIS exhibit severe synthetic growth defects (Squazzo et al., 2002). tfIIs elf7 double mutant plants show strong synergistic defects in growth and development (e.g., number of primary inflorescences and leaf vein pattern) relative to the parental lines. The increased number of inflorescences, which may be due to reduced apical dominance and decreased leaf venation, could be associated with defects in auxin signaling (Benjamins and Scheres, 2008; Chapman and Estelle, 2009). Several TEFs, including SPT4/SPT5 and Elongator, are involved in auxin-mediated gene expression (Dürr et al., 2014; Nelissen et al., 2010), but to date, PAF1-C and TFIIS have not been linked to auxin signaling. Our study of selected Arabidopsis double mutants defective in different combinations of TEFs contributes to an emerging interaction network among factors involved in transcript elongation. The type of genetic interactions (epistatic and synergistic) detected in our comparative analysis of the single/double mutants depends on the respective trait and is likely determined by distinct changes in gene expression brought about by the deficiencies in the different TEF(s). Generally, in plants (and other organisms) lacking individual TEFs, a relatively small number of genes is differentially expressed, and currently, it is still unclear what determines the requirement of subsets of genes for TEF action to achieve proper expression (Van Lijsebettens and Grasser, 2014). Our experiments also illustrate that the examination of Arabidopsis double mutants is a promising approach to elucidating the complex interplay of TEFs in higher eukaryotes, although further analyses are required to fully exploit the opportunities that the generated plant lines present.
Originally, TEFs were identified as factors modulating transcript elongation by RNAPII (Jonkers and Lis, 2015; Selth et al., 2010; Sims et al., 2004). A few studies yielded the finding that TEFs (i.e., SPT5, SPT6, and FACT) known to cooperate with RNAPII also play a role in RNAPI (and RNAPIII)-mediated transcription (Anderson et al., 2011; Birch et al., 2009; Engel et al., 2015). Our proteomics experiments did not provide conclusive evidence for the interaction of the seven analyzed TEFs with RNAPI/RNAPIII, as no RNAPIII-specific subunits copurified with the TEFs and three RNAPI subunits were only isolated along with SPT4 (Supplemental Data Set 5). In view of the structural differences between RNAPI and RNAPII (Engel et al., 2013), it is unlikely that the TEFs that directly bind RNAPII (i.e., SPT5 and SPT6) interact in a similar manner with RNAPI. Consistently, subnuclear localization analyses demonstrated that several TEFs, including SPT5 (Dürr et al., 2014) as well as SPT6L and ELF7 (this work), are excluded from nucleoli in Arabidopsis. Furthermore, using ChIP, SPT5 and SPT16 were found to associate with RNAPII-transcribed genes, but only background levels were detected at sites transcribed by RNAPI and RNAPIII. In agreement with these findings, relative to wild-type plants, no altered levels of RNAPI-transcribed 35S pre-rRNA were detected in mutants deficient in different TEFs. Therefore, our combined results do not support a role for the analyzed TEFs in RNAPI (and RNAPIII)-mediated transcription in Arabidopsis, which does not exclude the possibility that certain TEFs may collaborate with RNAPs other than RNAPII under specific conditions.
In addition to revealing interplay between various TEFs, our proteomics analyses demonstrated interactions between the RNAPII TEC and mRNA processing factors. Many splicing factors (e.g., components of U1, U2, U5 RNPs, Sm, and NTC) and some polyadenylation-related proteins copurified with the TEFs. It is likely that a portion of the mRNA processing factors that is enriched in the affinity purifications of the TEFs physically interacts with the elongating RNAPII (Elkon et al., 2013; Perales and Bentley, 2009; Saldi et al., 2016). Several subunits of PAF1-C also copurified with the polyadenylation factor CstF. This is in line with the finding that mammalian TEFs including the PAF1-C component CDC73 contribute to the recruitment of CstF to transcribed genes (Martincic et al., 2009; Rozenblatt-Rosen et al., 2009) and that PAF1-C can modulate mRNA 3′end processing events (Nagaike et al., 2011; Nordick et al., 2008). The analysis of the cdc73 cstf64 double mutant revealed that the bolting time and the transcript level of the floral repressor FLC are in between the values of Col-0 and the cdc73 single mutant, indicating that the effect of cdc73 clearly exceeds that of cstf64. CDC73 regulates FLC expression by modulating histone H3 methylation (H3K4me3 and H3K27me3) (Park et al., 2010; Yu and Michaels, 2010), while CstF64 is required for 3′processing of FLC antisense transcripts, which influences the level of the FLC sense transcript (Liu et al., 2010). In the cdc73 cstf64 double mutant, both effects are potentially combined and, in view of the observed interaction between CDC73 (PAF1-C) and the CstF complex, the recruitment of CstF to the FLC locus might be affected. In recent years, it turned out that both splicing and polyadenylation are linked to transcript elongation and that there is a close interrelationship between ongoing transcription and mRNA processing (Perales and Bentley, 2009). For example, the RNAPII elongation rate, which is under control of TEFs, influences the efficiency of splicing and polyadenylation events (Elkon et al., 2013; Saldi et al., 2016). In plants, the interplay between transcript elongation and mRNA processing has only recently become apparent, as exemplified by the connection of TFIIS and PAF1-C with splicing (Dolata et al., 2015; Li et al., 2016). We also observed that a range of splicing factors copurified with CstF77. This is in accordance with recent studies showing a close cooperation of mRNA splicing and polyadenylation, particularly at the last exons (Kaida, 2016; Misra and Green, 2016). In conclusion, our proteomic and genetic experiments underscore the important role of the plant RNAPII elongation complex in the production of mRNAs, representing an interaction site for different TEFs and mRNA processing factors.
METHODS
Plasmid Construction
The required gene or cDNA sequences were amplified by PCR with KAPA DNA polymerase (PeqLab) using Arabidopsis thaliana genomic DNA or cDNA as template and the primers (also providing the required restriction enzyme cleavage sites) listed in Supplemental Table 3. The PCR fragments were inserted into suitable plasmids using standard methods. For affinity purification, the coding sequences were fused to a C-terminal GS tag under the control of the 35S promoter (except for TFIIS, where for steric reasons an N-terminal fusion was generated and for technical reasons it was expressed under the control of its native promoter). All plasmid constructions were checked by DNA sequencing. Details about the plasmids generated in this work are summarized in Supplemental Table 3.
Plant Material and Documentation
Arabidopsis (Col-0) was grown at 21°C and 60% relative humidity in a growth chamber under long-day conditions (16-h photoperiod per day, 120 μmol m−2 s−1 using a combination of Osram Lumilux cool-white and Sylvania Lumiline plus warm white fluorescent tubes) on soil, while for segregation analyses, plants were grown on Murashige and Skoog medium (Launholt et al., 2006; Lolas et al., 2010). Seeds of the T-DNA insertion lines elf7-2 and elf7-3 (He et al., 2004), cdc73-2 (Yu and Michaels, 2010), and cstf64-3 (Liu et al., 2010) were obtained from the European Arabidopsis Stock Center (http://www.arabidopsis.info/), and we previously reported those of ssrp1-2, spt16-1, and tfIIs-1 (Grasser et al., 2009; Lolas et al., 2010). By crossing the parental lines as previously described (Lolas et al., 2010), the following double mutant lines were generated: tfIIs-1 elf7-3, tfIIs-1 ssrp1-2, tfIIs-1 spt16-1, ssrp1-2 elf7-2, spt16-1 elf7-2, and cdc73-2 cstf64-3. After sowing, the seeds were stratified in darkness for 48 h at 4°C prior to incubation in the plant growth chamber. All phenotypic analyses were independently performed at least twice and representative examples of the reproduced experiments are shown. Plant phenotypes including leaf vein patterning were documented as previously described (Dürr et al., 2014; Lolas et al., 2010).
Recombinant Proteins, Peptides, and Antibodies
For the MST experiments, SPT6L(Phe1218-Asp1412) fused to GST was expressed in Escherichia coli and purified by glutathione-sepharose affinity chromatography as previously described (Kammel et al., 2013; Krohn et al., 2002). Using the pQE plasmids described in Supplemental Table 3, full-length TFIIS, ELF7(D401-E589), SPT6L(V1121-M1430), and NRPB1(L1746-P1839) were expressed as 6xHis-tagged proteins in E. coli and purified by metal-chelate chromatography as previously described (Kammel et al., 2013). Purified recombinant proteins were analyzed by SDS-PAGE and mass spectrometry and used for commercial immunization (Eurogentec), and the obtained antisera were tested as previously described (Kammel et al., 2013; Launholt et al., 2006). Antibodies against SPT5, SPT16, and SSRP1 were previously described (Duroux et al., 2004; Dürr et al., 2014). For ChIP, the following commercial primary antibodies were used: RNAPII-CTD-S2P (rabbit, ab5095, lot GR124547-2, Abcam, diluted 1:100), RNAPII-CTD-S5P (rabbit, ab1791, lot GR193750-4, Abcam, diluted 1:100), and H3 C-terminal region (rabbit, ab1791, lot GR176735-2, Abcam, diluted 1:250), as well as the previously described antibodies against Arabidopsis SPT16 (Duroux et al., 2004) and SPT5 (Dürr et al., 2014). For immunostaining, we used primary antibodies against nonphosphorylated RNAPII-CTD (mouse, ab817, GR81285-5, Abcam, diluted 1:200), and RNAPII-CTD-S2P (rat, 04-1571, lot 2377414, Millipore, diluted 1:100), as well as the secondary antibodies anti-rabbit-rhodamine (cat. no. 111-295-144, lot 81183, Jackson ImmunoResearch, diluted 1:300), anti-rat-Alexa488 (cat. no. 112-545-167, lot 103333, Jackson ImmunoResearch, diluted 1:200), and anti-mouse-Cy5 (cat. no. 715-175-151, lot 109542, Jackson ImmunoResearch, diluted 1:50). N-terminally FITC-labeled peptides corresponding to RNAPII-CTD repeats were commercially synthesized (Biomatik): CTD-noP (PSYSPTSPSYSP), CTD-Ser2P [TSPSY(pS)PTSPSY], and CTD-Ser5P [SYSPT(pS)PSYSPT].
Affinity Purification and Characterization of GS-Tagged Proteins from Arabidopsis Cells
Arabidopsis suspension cultured PSB-D cells were maintained and transformed as previously described (Van Leene et al., 2015). Protein isolation, purification, and mass spectrometric analyses were essentially performed as previously described (Dürr et al., 2014). In brief, after sonicating 15 g cells in extraction buffer (25 mM HEPES-KOH, pH 7.4, 0.05% IGEPAL CA-630, 1 mM DTT, 100 mM NaCl, 2 mM MgCl2, 5 mM EGTA, 10% glycerol, proteinase inhibitor cocktail, and 1 mM PMSF), the nonspecific endonuclease Benzonase (50 units/mL extract) was added to degrade DNA and RNA. GS-tagged proteins were affinity purified using IgG-coupled magnetic beads (Hamperl et al., 2014), and eluted proteins were analyzed by SDS-PAGE and digested with trypsin. Peptides were separated by reverse-phase chromatography on an UltiMate 3000 RSLCnano System (Thermo Scientific) using a Reprosil-Pur Basic C18 nano column (75 µm i.d. × 250 mm; Dr. Maisch) and applying a linear 90-min gradient of 4 to 40% acetonitrile in 0.1% formic acid. The LC system was coupled online to a maXis plus UHR-QTOF system (Bruker Daltonics) via a nanoflow electrospray source (Bruker Daltonics). Data-dependent acquisition of tandem mass spectrometry (MS/MS) spectra by CID fragmentation was performed using a dynamic method with a fixed cycle time of 3 s (Compass 1.7; Bruker Daltonics). Protein Scape 3.1.3 (Bruker Daltonics) in connection with Mascot 2.5.1 (Matrix Science) facilitated database searching of the NCBInr database. Search parameters were as follows: trypsin, one missed cleavage allowed; deamidation (N,Q), oxidation (M), carbamidomethyl (C), and propionamid (C) as variable modifications; precursor tolerance, 5 ppm; MS/MS tolerance, 0.04 D; significance threshold, P < 0.05. Mascot peptide ion score cutoff was set to 25. A protein score of minimum 80 and at least two peptides found with an individual ion score of ≥25 were considered as criteria for reliable protein identification. The experimental background of contaminating proteins that were isolated with the unfused GS tag or that copurify nonspecifically independent of the used bait protein was subtracted. The list of 760 known nonspecific Arabidopsis proteins is based on 543 affinity purifications with 115 bait proteins, mainly from PSB-D cells (Van Leene et al., 2015).
Immunostaining and Super-Resolution Microscopy
Nuclei of differentiated rosette leaves were fixed in 4% paraformaldehyde in Tris buffer and flow sorted as described (Weisshart et al., 2016). Immunostaining with different antibodies was performed as previously described (Weisshart et al., 2016). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (2 µg/mL) in Vectashield (Vector Laboratories). To analyze the substructures and spatial arrangement of immunosignals and chromatin beyond the classical Abbe/Raleigh limit (super-resolution), spatial SIM (3D-SIM) was applied using a Plan-Apochromat 63×/1.4 oil objective of an Elyra PS.1 microscope system and the software ZEN (Zeiss). The images were captured using 405-, 488-, 561-, and 642-nm laser lines for excitation and the appropriate emission filters and merged using ZEN software (Weisshart et al., 2016). Imaris 8.0 (Bitplane) software was used to measure the degree of colocalization between RNAPII and TEFs. Briefly, after loading SIM image stacks, the Imaris colocalization tool was applied. An automatic threshold defined by the point spread function was calculated and used to establish a new colocalization channel originating from the ELF7/SPT6 and RNAPII channels. This resulting channel contains the channel statistics including the degree of colocalization (in %) and the Pearson’s and Mander’s coefficients. Imaris 8.0 was also applied to measure the amount of ELF7, SPT6, and RNAPII in flow-sorted 8C rosette leaf nuclei via voxel intensities and to produce 3D movies.
PCR-Based Genotyping and qRT-PCR
To distinguish between plants that were wild type, heterozygous, or homozygous for the T-DNA insertions, genomic DNA was isolated from leaves. The genomic DNA was used for PCR analysis with Taq DNA polymerase (PeqLab) and primers specific for DNA insertions and the target genes (Supplemental Table 3). For RT-PCR, total RNA was extracted from ∼100 mg of frozen plant tissue using the TRIzol method (Invitrogen) before the RNA samples were treated with DNase. Reverse transcription was performed using 2 μg of RNA and Revert Aid H minus M-MuLV reverse transcriptase (Thermo Scientific). The obtained cDNA was amplified by PCR using Taq DNA polymerase (PeqLab) or qPCR with KAPA SYBR FAST Universal reagents (PeqLab) and a Mastercycler ep realplex2 (Eppendorf), as previously described (Dürr et al., 2014).
RNA Gel Blot Analysis
Total RNA (20 μg) isolated from plants at 14 d after stratification (DAS) was separated by agarose gel electrophoresis for RNA gel blot analysis as previously described (Grasser et al., 2009). Hybridization probes were generated by PCR (Supplemental Table 3) and 32P-labeled, including the antisense probe that was used to detect the FLC sense transcript.
Fluorescent MST Binding Assay
MST binding experiments were performed with 40 nM FITC-labeled peptide in 10 mM sodium phosphate (pH 7.0), 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF with various concentrations of GST-SPT6L(Phe1218-Asp1412) at 40% MST power and 80% LED power in premium capillaries on a Monolith NT.115 device at 25°C (NanoTemper Technologies). Thermoporesis + TJump was used to analyze the data. The recorded fluorescence was normalized to fraction bound (0 = unbound, 1 = bound), processed using the software KaleidaGraph 4.5, and fitted using the Kd fit formula derived from the law of mass action. Technical duplicates were performed for each experimental setup.
ChIP
ChIP assays using 21 DAS Col-0 plants and different antibodies were performed as previously described in detail (Dürr et al., 2014; Fiil et al., 2008). Five microliters of precipitated DNA (diluted 1:200 for input and 1:10 for H3, SPT5, SPT16, RNAPII-CTD-S2P, RNAPII-CTD-S5P, and the control without antibody) was analyzed by qPCR with locus-specific primers (Supplemental Table 3). Data were normalized to the input, and P values were calculated with the software R version 3.2.3.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: NRPB1, AT4G35800; SPT4-2, AT5G63670; SPT5-2, AT4G08350; TFIIS, AT2G38560; CDC73, AT3G22590; ELF7, AT1G79730; SSRP1, AT3G28730; SPT16, AT4G10710; SPT6L, AT1G65440; SPT6, AT1G63210; and CDKC;2, AT5G64960.
Supplemental Data
Supplemental Figure 1. Expression of SPT6L and SPT6.
Supplemental Figure 2. Enrichment of Ser2-phosphorylated RNAPII in TEF affinity purifications and interaction of SPT6L with Ser2-phosphorylated RNAPII-CTD repeats.
Supplemental Figure 3. ELF7 and SPT6L colocalize with RNAPII in euchromatin.
Supplemental Figure 4. Analysis of tfIIs ssrp1 and tfIIs spt16 double mutants in comparison to the respective single mutants and the Col-0 wild type.
Supplemental Figure 5. Leaf vein patterning of Col-0, single, and double mutants.
Supplemental Figure 6. Analysis of tfIIs elf7 double mutants in comparison to the respective single mutants and the Col-0 wild type.
Supplemental Figure 7. Phenotype of spt16 elf7 double mutant plants in comparison to the respective single mutants and Col-0.
Supplemental Figure 8. Isolation of the CstF complex.
Supplemental Figure 9. Early bolting phenotype of cdc73 cstf64 double mutant plants.
Supplemental Figure 10. 35S pre-rRNA levels in mutants deficient in different TEFs.
Supplemental Table 1. Degree of colocalization of ELF7 and SPT6L with RNAPII (phosphorylated at Ser2 or nonphosphorylated).
Supplemental Table 2. Overview of phenotypes observed for single and double mutants defective in various TEFs.
Supplemental Table 3. Oligonucleotide primers used in this study and construction of plasmids.
Supplemental Data Set 1. Proteins that copurified with NRPB1-GS identified by mass spectrometry.
Supplemental Data Set 2. Proteins that copurified with SPT16-GS and/or SSRP1-GS identified by mass spectrometry.
Supplemental Data Set 3. Proteins that copurified with ELF7-GS and/or CDC73-SGS identified by mass spectrometry.
Supplemental Data Set 4. Proteins that copurified with TFIIS-GS identified by mass spectrometry.
Supplemental Data Set 5. Proteins that copurified with SPT4-GS identified by mass spectrometry.
Supplemental Data Set 6. Proteins that copurified with CDKC2-GS identified by mass spectrometry.
Supplemental Data Set 7. Proteins that copurified with CstF77-GS and/or CstF64-GS identified by mass spectrometry.
Supplementary Material
Acknowledgments
We thank Lena Dietz, Stefan Faderl, and Irene Fuhrmann for contributions during early stages of the project, Antje Böttinger for technical assistance, Jelle Van Leene and Geert De Jaeger for advice regarding cell culture transformation and for providing Arabidopsis PSB-D cells, Eduard Hochmuth for recording mass spectra, and the Nottingham Arabidopsis Stock Centre for providing Arabidopsis T-DNA insertion lines. This work was supported by the German Research Foundation (DFG) through Grant SFB960 to M.G. and K.D.G. and through Grant GR1159/14-1 to K.D.G., as well as the EC Marie Curie Research Training Network “Chromatin in Plants–European Training and Mobility” (CHIP-ET, FP7-PEOPLE-2013-ITN607880) to K.D.G.
AUTHOR CONTRIBUTIONS
M.G. and K.D.G. designed the research and wrote the manuscript. W.A., A.P., H.F.E., P.H., K.K., S.A.M., T.S., V.S., and M.G. performed the research. W.A., A.P., H.F.E., P.H., K.K., S.A.M., A.B., T.S., G.L., J.G., V.S., M.G., and K.D.G. analyzed the data. A.B., G.L., J.G., V.S., M.G., and K.D.G. provided the reagents and the tools for the analysis. All authors commented on the manuscript and contributed to the writing.
Glossary
- TEF
transcript elongation factor
- TEC
transcript elongation complex
- HDAC
histone deacetylase
- CRC
chromatin remodeling complex
- SIM
structured illumination microscopy
- MST
microscale thermophoresis
- ChIP
chromatin immunoprecipitation
- DAS
days after stratification
Footnotes
Articles can be viewed without a subscription.
References
- Adelman K., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T. (2006). Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 26: 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.J., Sikes M.L., Zhang Y., French S.L., Salgia S., Beyer A.L., Nomura M., Schneider D.A. (2011). The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J. Biol. Chem. 286: 18816–18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosch M., Mortensen S.A., Grasser K.D. (2012). Plant proteins containing high mobility group box DNA-binding domains modulate different nuclear processes. Plant Physiol. 159: 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D. (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Bieluszewski T., Galganski L., Sura W., Bieluszewska A., Abram M., Ludwikow A., Ziolkowski P.A., Sadowski J. (2015). AtEAF1 is a potential platform protein for Arabidopsis NuA4 acetyltransferase complex. BMC Plant Biol. 15: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T. (2009). RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 10: 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch J.L., Tan B.C., Panov K.I., Panova T.B., Andersen J.S., Owen-Hughes T.A., Russell J., Lee S.C., Zomerdijk J.C. (2009). FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 28: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove D.A., Mahmoudi T., Henklein P., Verdin E. (2007). Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA 104: 13690–13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko V.A., Steele L.M., Ujvári A., Gaykalova D.A., Kulaeva O.I., Polikanov Y.S., Luse D.S., Studitsky V.M. (2006). Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24: 469–479. [DOI] [PubMed] [Google Scholar]
- Buratowski S. (2009). Progression through the RNA polymerase II CTD cycle. Mol. Cell 36: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Cui X., Fan B., Scholz J., Chen Z. (2007). Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19: 1388–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolata J., Guo Y., Kołowerzo A., Smoliński D., Brzyżek G., Jarmołowski A., Świeżewski S. (2015). NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 34: 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroux M., Houben A., Růzicka K., Friml J., Grasser K.D. (2004). The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 40: 660–671. [DOI] [PubMed] [Google Scholar]
- Dürr J., Lolas I.B., Sørensen B.B., Schubert V., Houben A., Melzer M., Deutzmann R., Grasser M., Grasser K.D. (2014). The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 42: 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Ugalde A.P., Agami R. (2013). Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P. (2013). RNA polymerase I structure and transcription regulation. Nature 502: 650–655. [DOI] [PubMed] [Google Scholar]
- Engel K.L., French S.L., Viktorovskaya O.V., Beyer A.L., Schneider D.A. (2015). Spt6 is essential for rRNA synthesis by RNA Polymerase I. Mol. Cell. Biol. 35: 2321–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil B.K., Qiu J.L., Petersen K., Petersen M., Mundy J. (2008). Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc. 3: prot5049. [DOI] [PubMed] [Google Scholar]
- Gentry M., Hennig L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321: 40–46. [DOI] [PubMed] [Google Scholar]
- Grasser M., Kane C.M., Merkle T., Melzer M., Emmersen J., Grasser K.D. (2009). Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J. Mol. Biol. 386: 598–611. [DOI] [PubMed] [Google Scholar]
- Gu X.L., Wang H., Huang H., Cui X.F. (2012). SPT6L encoding a putative WG/GW-repeat protein regulates apical-basal polarity of embryo in Arabidopsis. Mol. Plant 5: 249–259. [DOI] [PubMed] [Google Scholar]
- Hajheidari M., Koncz C., Eick D. (2013). Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 18: 633–643. [DOI] [PubMed] [Google Scholar]
- Hamperl S., Brown C.R., Perez-Fernandez J., Huber K., Wittner M., Babl V., Stöckl U., Boeger H., Tschochner H., Milkereit P., Griesenbeck J. (2014). Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. Methods Mol. Biol. 1094: 329–341. [DOI] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.-S., Jin H., Zhu J.K. (2009). An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Doyle M.R., Amasino R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.G., et al. (2008). Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genom. 9: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Kinoshita Y., Susaki D., Ikeda Y., Iwano M., Takayama S., Higashiyama T., Kakutani T., Kinoshita T. (2011). HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev. Cell 21: 589–596. [DOI] [PubMed] [Google Scholar]
- Jaehning J.A. (2010). The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K. (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19: 523–534. [DOI] [PubMed] [Google Scholar]
- Jonkers I., Lis J.T. (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D. (2016). The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip. Rev. RNA 7: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammel C., Thomaier M., Sørensen B.B., Schubert T., Längst G., Grasser M., Grasser K.D. (2013). Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS One 8: e60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Guermah M., Roeder R.G. (2010). The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140: 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllen K., Dietz L., Bies-Etheve N., Lagrange T., Grasser M., Grasser K.D. (2015). The zinc-finger protein SPT4 interacts with SPT5L/KTF1 and modulates transcriptional silencing in Arabidopsis. FEBS Lett. 589: 3254–3257. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22: 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn N.M., Yanagisawa S., Grasser K.D. (2002). Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. J. Biol. Chem. 277: 32438–32444. [DOI] [PubMed] [Google Scholar]
- Lambert J.P., Tucholska M., Pawson T., Gingras A.C. (2014). Incorporating DNA shearing in standard affinity purification allows simultaneous identification of both soluble and chromatin-bound interaction partners. J. Proteomics 100: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launholt D., Merkle T., Houben A., Schulz A., Grasser K.D. (2006). Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18: 2904–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xia C., Feng J., Yang D., Wu F., Cao Y., Li L., Ma L. (2016). The SNW domain of SKIP Is required for Its integration into the spliceosome and its interaction with the Paf1 complex in Arabidopsis. Mol. Plant 9: 1040–1050. [DOI] [PubMed] [Google Scholar]
- Li Y., Zeng S.X., Landais I., Lu H. (2007). Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J. Biol. Chem. 282: 6936–6945. [DOI] [PubMed] [Google Scholar]
- Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R. III, Hartzog G.A. (2003). Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23: 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97. [DOI] [PubMed] [Google Scholar]
- Lolas I.B., Himanen K., Grønlund J.T., Lynggaard C., Houben A., Melzer M., Van Lijsebettens M., Grasser K.D. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61: 686–697. [DOI] [PubMed] [Google Scholar]
- Martincic K., Alkan S.A., Cheatle A., Borghesi L., Milcarek C. (2009). Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat. Immunol. 10: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo F.W., Cramer P. (2013). Structural basis of transcription elongation. Biochim. Biophys. Acta 1829: 9–19. [DOI] [PubMed] [Google Scholar]
- Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010). Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17: 1272–1278. [DOI] [PubMed] [Google Scholar]
- Misra A., Green M.R. (2016). From polyadenylation to splicing: Dual role for mRNA 3′ end formation factors. RNA Biol. 13: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen S.A., Grasser K.D. (2014). The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Lett. 588: 47–51. [DOI] [PubMed] [Google Scholar]
- Murawska M., Brehm A. (2011). CHD chromatin remodelers and the transcription cycle. Transcription 2: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T., Logan C., Hotta I., Rozenblatt-Rosen O., Meyerson M., Manley J.L. (2011). Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell 41: 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., et al. (2010). Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 107: 1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock A., Ascano J.M., Barrero M.J., Malik S. (2012). Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordick K., Hoffman M.G., Betz J.L., Jaehning J.A. (2008). Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Park S., van Nocker S. (2008). Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 4: e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Zhang H., Ludwig P., van Nocker S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Oh S., Ek-Ramos J., van Nocker S. (2010). PLANT HOMOLOGOUS TO PARAFIBROMIN is a component of the PAF1 complex and assists in regulating expression of genes within H3K27ME3-enriched chromatin. Plant Physiol. 153: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717. [DOI] [PubMed] [Google Scholar]
- Perales M., Más P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R., Bentley D. (2009). “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaresma A.J., Bugai A., Barboric M. (2016). Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 44: 7527–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Pontvianne F., Nicora C.D., Norbeck A.D., Paša-Tolić L., Pikaard C.S. (2015). Subunit compositions of Arabidopsis RNA polymerases I and III reveal Pol I- and Pol III-specific forms of the AC40 subunit and alternative forms of the C53 subunit. Nucleic Acids Res. 43: 4163–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Nagaike T., Francis J.M., Kaneko S., Glatt K.A., Hughes C.M., LaFramboise T., Manley J.L., Meyerson M. (2009). The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. USA 106: 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L. (2016). Coupling of RNA Polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 428: 2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V., Weisshart K. (2015). Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei. J. Exp. Bot. 66: 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L.A., Sigurdsson S., Svejstrup J.Q. (2010). Transcript elongation by RNA Polymerase II. Annu. Rev. Biochem. 79: 271–293. [DOI] [PubMed] [Google Scholar]
- Shi Y., Manley J.L. (2015). The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 29: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J. III, Belotserkovskaya R., Reinberg D. (2004). Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Sopta M., Carthew R.W., Greenblatt J. (1985). Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260: 10353–10360. [PubMed] [Google Scholar]
- Spencer J.A., Baron M.H., Olson E.N. (1999). Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J. Biol. Chem. 274: 15686–15693. [DOI] [PubMed] [Google Scholar]
- Squazzo S.L., Costa P.J., Lindstrom D.L., Kumer K.E., Simic R., Jennings J.L., Link A.J., Arndt K.M., Hartzog G.A. (2002). The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil-Rodríguez A., Reyes J.C. (2011). To cross or not to cross the nucleosome, that is the elongation question.... RNA Biol. 8: 389–393. [DOI] [PubMed] [Google Scholar]
- Sun M., Larivière L., Dengl S., Mayer A., Cramer P. (2010). A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J. Biol. Chem. 285: 41597–41603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson B.N., Arndt K.M. (2013). The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10: 169–187. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Grasser K.D. (2014). Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 19: 717–726. [DOI] [PubMed] [Google Scholar]
- Wang Z.W., Wu Z., Raitskin O., Sun Q., Dean C. (2014). Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. USA 111: 7468–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K., Fuchs J., Schubert V. (2016). Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio Protoc. 6: e1725. [Google Scholar]
- Woloszynska M., Le Gall S., Van Lijsebettens M. (2016). Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim. Biophys. Acta 1859: 1025–1033. [DOI] [PubMed] [Google Scholar]
- Yang Q., Doublié S. (2011). Structural biology of poly(A) site definition. Wiley Interdiscip. Rev. RNA 2: 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh S.M., Cho H., Pickle L., Evans R.M., Jones K.A. (2007). The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Michaels S.D. (2010). The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol. 153: 1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S.X., Dai M.-S., Keller D.M., Lu H. (2002). SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 21: 5487–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Zhu Y., Dong A., Shen W.-H. (2015). Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J. 83: 78–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.