The chloroplast-localized BASS6 transporter works with the PLGG1 protein to balance glycolate export with glycolate import during photorespiration in Arabidopsis thaliana.
Abstract
Photorespiration is an energy-intensive process that recycles 2-phosphoglycolate, a toxic product of the Rubisco oxygenation reaction. The photorespiratory pathway is highly compartmentalized, involving the chloroplast, peroxisome, cytosol, and mitochondria. Though the soluble enzymes involved in photorespiration are well characterized, very few membrane transporters involved in photorespiration have been identified to date. In this work, Arabidopsis thaliana plants containing a T-DNA disruption of the bile acid sodium symporter BASS6 show decreased photosynthesis and slower growth under ambient, but not elevated CO2. Exogenous expression of BASS6 complemented this photorespiration mutant phenotype. In addition, metabolite analysis and genetic complementation of glycolate transport in yeast showed that BASS6 was capable of glycolate transport. This is consistent with its involvement in the photorespiratory export of glycolate from Arabidopsis chloroplasts. An Arabidopsis double knockout line of both BASS6 and the glycolate/glycerate transporter PLGG1 (bass6, plgg1) showed an additive growth defect, an increase in glycolate accumulation, and reductions in photosynthetic rates compared with either single mutant. Our data indicate that BASS6 and PLGG1 partner in glycolate export from the chloroplast, whereas PLGG1 alone accounts for the import of glycerate. BASS6 and PLGG1 therefore balance the export of two glycolate molecules with the import of one glycerate molecule during photorespiration.
INTRODUCTION
Rubisco catalyzes the fixation of CO2 onto the five-carbon ribulose-1,5-bisphosphate (RuBP), generating two molecules of 3-phosphoglycerate (3-PGA). However, at 25°C and our current atmospheric CO2 level of ∼400 ppm (40 Pa), ∼25% of Rubisco catalytic activity in plants with C3 photosynthetic metabolism results in the fixation of the competing substrate O2 instead of CO2. Oxygen fixation by Rubisco results in the conversion of RuBP to one molecule of 3-PGA and one molecule of 2-phosphoglycolate (2-PG) (Bowes et al., 1971; Ogren and Bowes, 1971; Lorimer, 1981; Ogren, 1984; Sharkey, 1988). 2-PG accumulation in the chloroplast stroma inhibits triose phosphate isomerase and phosphofructokinase, thereby decreasing RuBP regeneration capacity (Anderson, 1971; Kelly and Latzko, 1976). Although 2-PG is rapidly dephosphorylated to glycolate by 2-phosphoglycolate phosphatase, the glycolate can inhibit the rate of photosynthesis in the chloroplast and is considered toxic to the cell (Kelly and Latzko, 1976; Gonzalez-Moro et al., 1997).
The inhibition of photosynthesis by 2-PG/glycolate is prevented and partial recovery of the reduced carbon is accomplished via the photorespiratory pathway, an energy-requiring series of reactions involving the chloroplast, peroxisome, mitochondria, and cytosol (Somerville and Ogren, 1979; Ogren, 1984; Eisenhut et al., 2013a). Photorespiration converts two molecules of 2-PG to one molecule of 3-PGA, while releasing one molecule of CO2.Therefore, 75% of the previously fixed carbon that would have been lost as 2-PG is recovered as 3-PGA, but this is not without an energy cost to the cell. The photorespiratory cycle consumes ATP and, as a byproduct of the conversion of glycine to serine, produces ammonia (NH3) in the mitochondria. Plants must then consume reducing equivalents of NAD(P)H to recycle the NH3 and to reduce downstream photorespiratory intermediates. Also, refixing the CO2 molecule released during photorespiration consumes additional ATP and NADPH. As a result, photorespiration under current atmospheric CO2 concentrations results in a significant drag of ∼15 to 50% on C3 photosynthetic efficiency depending upon the regional growing season temperature (Ogren, 1984; Peterhansel et al., 2010; Walker et al., 2016b). Losses in yield due to photorespiration are estimated to total ∼150 trillion calories per year in midwestern US soybean (Glycine max) and wheat (Triticum aestivum) production alone (Walker et al., 2016b). In addition, photorespiration has similar negative impacts on other major C3 crops such as rice (Oryza sativa) and potato (Solanum tuberosum; Sharkey, 1988; Zhu et al., 2010).
The soluble enzymes involved in photorespiration have been well studied over the past four decades, providing much information on the biochemistry and genetics governing photorespiratory metabolism (Peterhansel et al., 2010; Timm and Bauwe, 2013). By contrast, only a small number of transport proteins have been demonstrated to be involved in photorespiration, even though at least 25 transport steps may be involved in the recycling of carbon in photorespiration (Eisenhut et al., 2013a). Importantly, photorespiration is a high-flux pathway that interacts with multiple other metabolic pathways, including the nitrogen cycle and amino acid biosynthesis (Fernie et al., 2013).
The first transporters identified to be involved in photorespiration were the chloroplastic dicarboxylate transporters DiT1 and DiT2 (Somerville and Somerville, 1985; Woo et al., 1987). A single point mutation in DiT2.1 and subsequent biochemical characterization revealed that DiT2 is the glutamate/malate transporter located in the chloroplast inner envelope membrane (Renné et al., 2003). Antisense repression of DiT1 resulted in the classical photorespiratory mutant phenotype of decreased growth under ambient CO2 with complementation by elevated [CO2]. It also resulted in reduced nitrate reassimilation due to a decrease in 2-oxoglutarate transport in the chloroplast (Schneidereit et al., 2006). Together, DiT1 and DiT2 are necessary for the proper refixation of released ammonia from the glycine decarboxylation reaction in photorespiratory metabolism.
More recently, coexpression analysis was used to identify other potential transporters involved in photorespiration (Bordych et al., 2013). Coexpression analysis identified A BOUT DE SOUFFLE (BOU) as a mitochondrial transporter required for normal glycine decarboxylase activity and meristematic growth. Null bou mutants exhibit the photorespiratory mutant phenotype and are complemented by elevated [CO2] (Eisenhut et al., 2013b). Currently, the only known photorespiratory pathway transporter that transports carbon derived directly from glycolate is the plastidic glycolate/glycerate translocator PLGG1 (Pick et al., 2013). PLGG1 is coexpressed with many enzymes involved in photorespiration (Pick et al., 2013). Studies with a PLGG1 T-DNA knockout line in Arabidopsis thaliana (plgg1-1) revealed the role of PLGG1 in both the first and the final transport steps in the photorespiratory pathway: the simultaneous export of glycolate from and import of glycerate into the chloroplast (Pick et al., 2013). Nearly 30 years prior to the molecular identification of PLGG1, the export of glycolate coincident with the import of glycerate import had been demonstrated in purified spinach (Spinacia oleracea) chloroplasts (Howitz and McCarty, 1985, 1986, 1991; Young and McCarty, 1993). Additionally, PLGG1 was identified as a chloroplast protein in proteomic studies. It was originally thought to be involved in programmed cell death, but current evidence now suggests that this phenotype was linked to the accumulation of photorespiratory intermediates (Yang et al., 2012; Pick et al., 2013). However, the Arabidopsis plgg1-1 line showed neither differences in the quantum efficiency of CO2 assimilation nor changes in the photorespiratory CO2 compensation point compared with wild type under low-light conditions (Walker et al., 2016a). Combined, these data not only show that PLGG1 is involved in photorespiratory metabolism, but also suggest an alternative path for glycolate exit from the chloroplast. In addition, they demonstrate the difficulty in phenotypically identifying transporters in the photorespiration pathway (Hodges et al., 2016).
Although both genetic and coexpression approaches have been successful in identifying genes involved in photorespiratory metabolism, many of the transporters involved in the flux of photorespiratory intermediates remain unknown. An alternative approach to coexpression analysis is first to identify candidate chloroplast inner membrane transporters from chloroplast envelope proteomic studies and then to screen T-DNA insertional mutants of the candidate genes for the photorespiratory mutant phenotype of reduced photosystem II efficiency by chlorophyll fluorescence (Badger et al., 2009; Sun et al., 2009). When illuminated in low [CO2], photorespiration-deficient mutants exhibit reduced Fv/Fm chlorophyll fluorescence due to the impaired function of photosystem II (Kozaki and Takeba, 1996; Wingler et al., 2000; Takahashi et al., 2007). This high-throughput fluorescence-based approach, when combined with forward genetics to target putative transporter-like chloroplast inner envelope membrane proteins, can identify additional genes important for photorespiratory metabolite transport.
We used this approach to identify bile acid sodium symporter 6 (BASS6) as a glycolate transporter involved in photorespiration. Bile acid sodium symporters are a family of transport proteins that were first identified as bile acid transporters in the mammalian liver (Alrefai and Gill, 2007). Further analysis showed that members of the BASS family of transporters are present in a wide variety of organisms, including plants, animals, and fungi, and they exhibit a broad range of substrate specificity, including non-bile acid organic compounds such as pyruvate, steroids, and xenobiotics (Furumoto et al., 2011; Claro da Silva et al., 2013). Although bile acids are not produced in plants, BASS family genes are present in both monocots and dicots (Gigolashvili et al., 2009; Sawada et al., 2009; Furumoto et al., 2011). Our analysis of the bass6 T-DNA line in Arabidopsis (bass6-1) revealed that loss of BASS6 resulted in a photorespiratory mutant phenotype and in accumulation of the photorespiratory metabolic intermediates glycine and glycolate. In addition, BASS6 protein was localized to the chloroplast envelope, and its ability to transport glycolate was conclusively demonstrated through combined yeast genetic complementation and transport analysis.
RESULTS
Arabidopsis bass6-1 Exhibits a Photorespiratory Mutant Phenotype
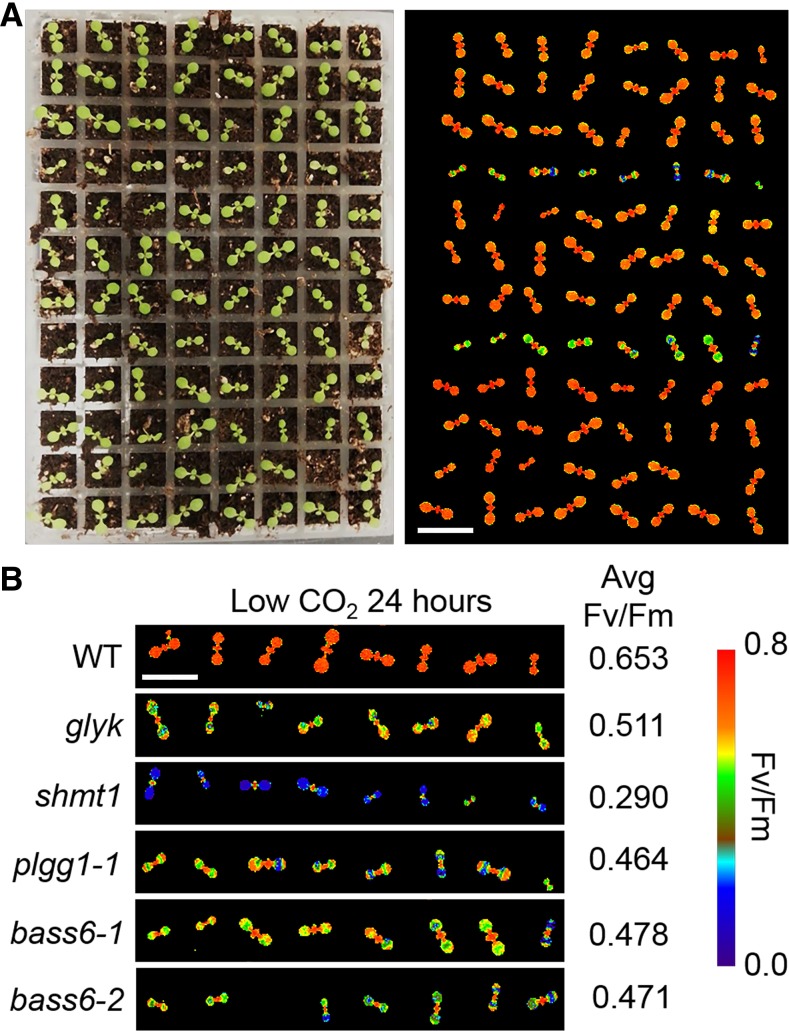
To identify potential transporter proteins involved in photorespiration, we screened a series of known and putative chloroplast inner membrane localized proteins (Supplemental Data Set 1). As controls, T-DNA lines targeting genes known to be involved in photorespiration, glycerate kinase (glyk), serine hydroxymethyltransferase (shm1), and the glycolate/glycerate transporter PLGG1 (plgg1-1), were included in the analysis. To test for mutants involved in photorespiration, we illuminated 9-d-old T-DNA lines under low [CO2]. Under these conditions, photorespiratory mutants show reduced Fv/Fm chlorophyll fluorescence (Badger et al., 2009). From the initial screen of 50 Arabidopsis T-DNA lines, we identified two independent T-DNA insertions targeting the gene At4g22840 (Figure 1; Supplemental Data Set 1). These T-DNA lines lacked expression of the putative chloroplast inner membrane protein BASS6 (Supplemental Data Set 1). To determine whether BASS6 is involved in photorespiration, these two T-DNA lines were grown under ambient CO2 (40 Pa) conditions. Compared with the wild-type control, both bass6 mutant lines had a smaller rosette size, similar to that of plgg1-1, the glycolate/glycerate transporter mutant involved in photorespiration (Figure 2A). To verify that the changes in Fv/Fm observed in the initial screen were also present in more developed plants, 4-week-old T-DNA lines of both plgg1-1 and bass6-1 were analyzed before and after the low [CO2] treatment. Following 24 h of constant illumination at low [CO2], we found a significant reduction in Fv/Fm for both lines when compared with the wild type, even though no reduction in Fv/Fm was observed prior to the low [CO2] treatment (Figure 2B).
Figure 1.
Screen of Arabidopsis T-DNA Lines for a Photorespiration Phenotype.
(A) Representative photos of 9-d-old Arabidopsis seedlings. Seedlings were grown in an 8 × 12 array for screening to identify T-DNA lines defective in Fv/Fm chlorophyll fluorescence due to photorespiration stress. Bar = 1 cm.
(B) Dark-adapted Fv/Fm images of select T-DNA lines. Different colors show changes in Fv/Fm fluorescence after a 24-h illumination in low [CO2]. T-DNA lines are compared with wild-type control, and average Fv/Fm is from the average of eight plants in three replicate experiments. Bar = 1 cm.
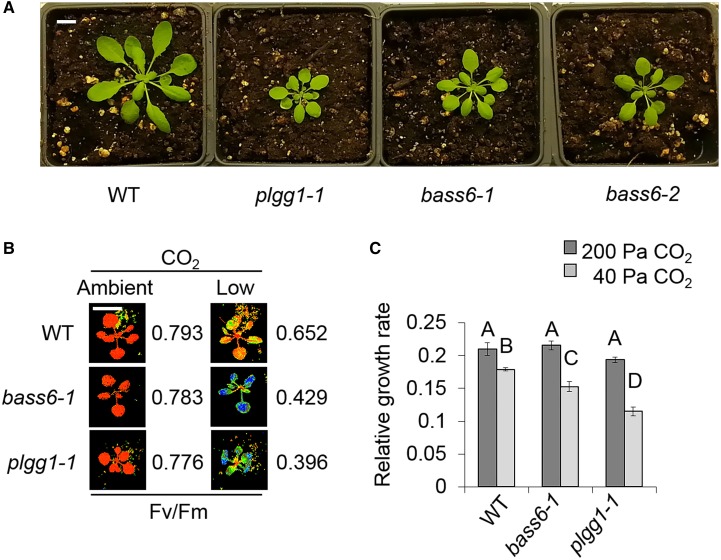
Figure 2.
BASS6 Is Necessary for Growth under Photorespiratory Conditions.
(A) Representative photos of bass6 and plgg1 mutants compared with the wild type. Seedlings were grown under ambient CO2 (8 weeks in 40 Pa CO2 with an 8-h-light/16-h-dark cycle [22°C/18°C] at 250 μmol m−2 s−1 light intensity in growth chambers). Bar = 1 cm.
(B) Changes in Fv/Fm after 24 h of low CO2 and constant illumination. Numbers represent the average of 12 plants from three replicate experiments. Bar = 1 cm.
(C) Relative growth rate of bass6-1 and plgg1-1 compared with the wild type at the indicated CO2 concentration. Error bars indicate sd from five biological replicates, and letters indicate significant difference between genotype and CO2 treatments based on ANOVA analysis (P < 0.05; Supplemental File 1).
To verify the photorespiratory mutant phenotype, we measured growth of the bass6-1 mutant under low, ambient, and elevated [CO2] (12.5, 40, and 200 Pa). Consistent with a classical photorespiratory mutant phenotype, both the bass6-1 and the plgg1-1 mutants failed to grow at 12.5 Pa CO2 (Supplemental Figure 1). Under ambient CO2 conditions, both the bass6-1 and the plgg1-1 T-DNA lines grew more slowly when compared with the wild-type control (Figure 2C; Supplemental Figure 1). Importantly, the wild type, bass6-1, and plgg1-1 grew at the same rate when grown under high [CO2] (Figure 2C; Supplemental Figure 1). Unfortunately, growth characterization of the bass6-2 line showed inconsistent results and its growth was not rescued by exogenous expression of BASS6. Analysis of the T-DNA insertion of bass6-2 revealed that T-DNA insertion is within a shared promoter region with the gene At4g22850. Analysis of gene expression showed reductions in expression of the adjacent gene as well as BASS6 making characterization of BASS6 function in the bass6-2 not feasible (Supplemental Figure 2).
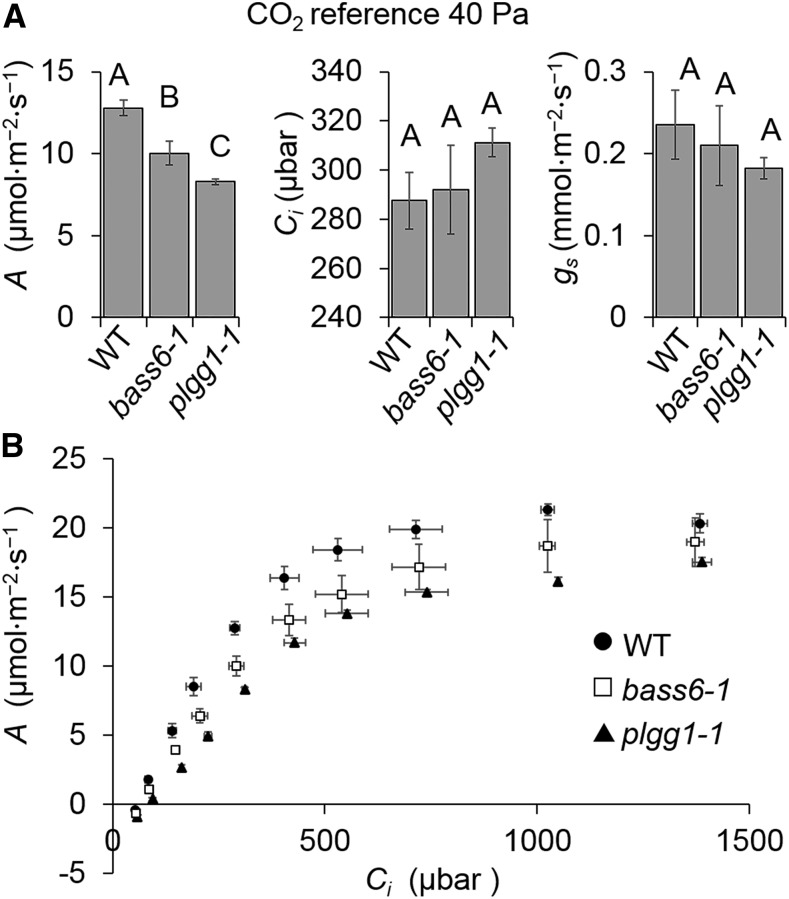
For wild-type or near-wild-type growth, certain classes of mutants in the photorespiratory pathway require high levels of [CO2], in which the Rubisco oxygenation reaction is suppressed to very low levels (Timm and Bauwe, 2013). Under ambient [CO2], photorespiration mutants have reduced photosynthesis that is characterized by reductions in carbon assimilation (A), the maximum rate of Rubisco carboxylation (Vcmax), and the maximum rate of electron transport (Jmax) parameters. Similar to previous reports (Pick et al., 2013; Walker et al., 2016a), plgg1-1 had reduced A compared with the wild type (Figure 3A). The bass6-1 plants also showed a slight reduction in A under ambient [CO2] compared with the wild type and had no detectable changes in internal CO2 concentration (Ci) or stomatal conductance (gs) (Figure 3A). To evaluate the biochemical limitations to photosynthesis of bass6-1 and plgg1-1 lines, the response of A to the intercellular [CO2] (Ci) was used to determine Vcmax and Jmax. We found that both Vcmax and Jmax were reduced in bass6-1 and plgg1-1 compared with the wild type (Figure 3B, Table 1). A was reduced more in plgg1-1 compared with bass6-1, consistent with comparative rosettes sizes and the growth rates of the bass6-1 versus plgg1-1 mutant plants (compare Figures 2A and 2C with Figure 3 and Table 1).
Figure 3.
Loss of BASS6 Impairs Photosynthesis.
(A) Photosynthetic measurements for wild-type and mutant strains. Measurements recorded at 40 Pa CO2 and saturating light (1000 μmol m−2 s−1) for the indicated strains, assimilation (A), internal CO2 concentration (Ci), and stomatal conductance (gs). Letters indicate statistical differences based on ANOVA analysis n = 3 (P < 0.05; Supplemental File 1).
(B) Assimilation (A) based on internal [CO2] (Ci) curves within the leaf of the indicated lines. Error bars indicate sd in (A) and (B), n = 5.
Table 1. Photosynthetic Parameters Based on ACi Data Using the PsFit Model.
| Vcmax 25°C (µmol m−2 s−1) | Jmax 25°C (µmol m−2 s−1) | Rd (µmol m−2 s−1) | gs (mmol m−2 s−1) | |
|---|---|---|---|---|
| Col-0 wild type | 59.19A ± 1.20 | 121.09A ± 3.10 | 1.23A ± 0.16 | 0.20A ± 0.02 |
| bass6-1 | 43.55B ± 3.04 | 104.56B ± 1.29 | 1.25A ± 0.20 | 0.21A ± 0.05 |
| plgg1-1 | 31.47C ± 1.08 | 92.70C ± 1.27 | 1.57B ± 0.17 | 0.16A ± 0.01 |
Superscript letters A, B, and C indicate statistical differences from the wild type based on ANOVA analysis (P ≤ 0.5; Supplemental File 1). Vcmax, maximum carboxylation rate allowed by Rubisco; Jmax, maximum rate of photosynthetic electron transport; Rd, day respiration; gs, stomatal conductance.
BASS6 Protein Is Localized in the Chloroplast Envelope
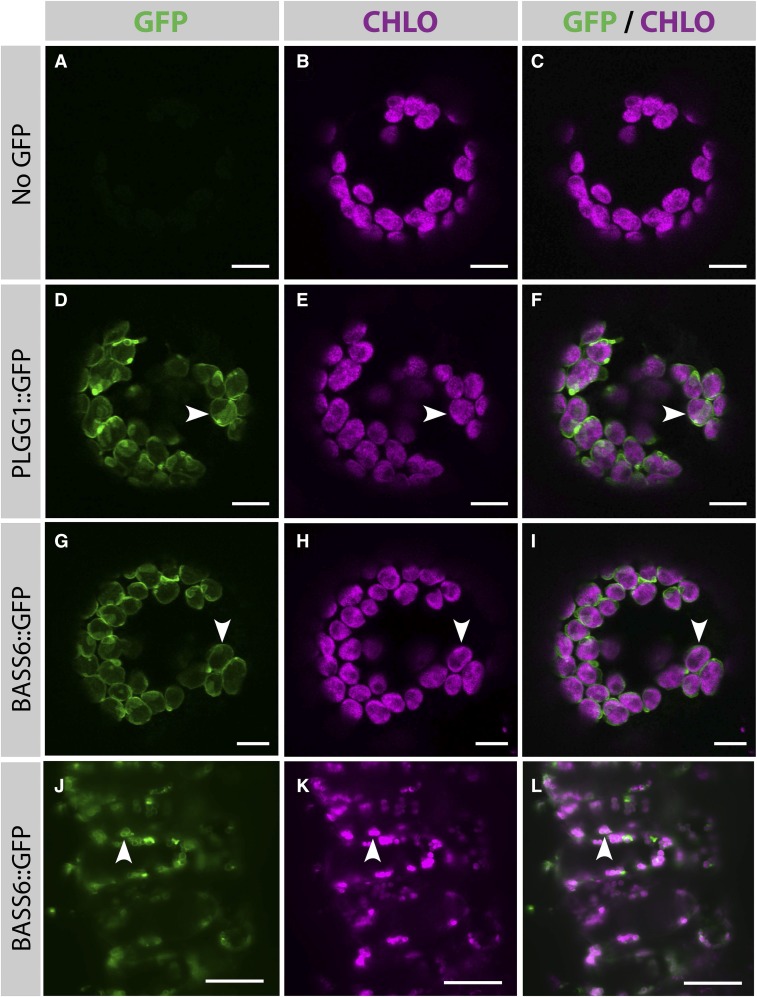
Mass spectrometry proteomic data had previously localized BASS6 protein to the chloroplast inner envelope membrane (Zybailov et al., 2008). To confirm this, we analyzed the transient expression of an Arabidopsis BASS6-GFP fusion protein both in protoplasts and in whole-leaf tissue of Nicotiana benthamiana, using chlorophyll autofluorescence to identify chloroplasts (Figures 4B, 4E, 4H, and 4K). In protoplasts, the BASS6-GFP signal surrounded chloroplast autofluorescence (Figures 4G to 4I), a pattern seen with the Arabidopsis glycolate/glycerate transporter PLGG1 in the chloroplast envelope (Ferro et al., 2003, 2010; Zybailov et al., 2008; Pick et al., 2013; Rolland et al., 2016) (Figures 4D to 4F). This provides evidence that BASS6 is localized in the chloroplast envelope. In these experiments, GFP was not detected outside of the chloroplast compartment, and, notably, no punctate signal suggesting mitochondria or Golgi colocalization was observed. Light-sheet microscopy using whole-leaf tissue further confirmed BASS6-GFP localization to the chloroplast envelope. These whole-tissue experiments further suggested that some of the GFP signal was present outside of chloroplasts in a yet unidentified compartment that did not appear to be cytosolic (Figures 4J to 4L; Supplemental Figure 3A). Whereas this signal may reflect a minor fraction of a BASS6-GFP pool targeted outside the chloroplast, it could also be truncated protein or misfolded protein.
Figure 4.
BASS6 Is Localized to the Chloroplast Envelope.
(A) to (I) Confocal microscopy images of isolated N. benthamiana protoplasts. Protoplasts expressed either no GFP-tagged construct ([A] to [C]), GFP-tagged PLGG1 ([D] to [F]), or GFP-tagged BASS6 ([G] to [I]) at 2 d postinfiltration. These images are maximum projections of four successive planes, and they show that both Arabidopsis PLGG1 and BASS6 localize in the chloroplast envelope (arrowheads), showing that, just like PLGG1, BASS6 is inserted in the chloroplast inner envelope membrane. Bars = 10 μm.
(J) to (L) Light sheet images of N. benthamiana leaf tissue transiently expressing BASS6-GFP expressed from the CaMV 35S promoter. Arrows indicate GFP fluorescence associated with the chlorophyll envelope. Bars = 50 μm.
In all panels, GFP signal is shown in green, and chloroplast autofluorescence is shown in magenta.
To confirm that BASS6 is targeted to chloroplasts, a rabbit polyclonal antibody was raised against a BASS6 peptide (Agrisera) and was used to determine the localization of BASS6 in Arabidopsis (Supplemental Figure 3B). Subcellular fractionation of Arabidopsis Col-0 wild type followed by immunoblot analysis using the BASS6 antiserum revealed that BASS6 was detectable as a band near the predicted ∼44 kD size only in the chloroplast fraction, using RBCL as a marker protein. It was undetectable in the whole-cell soluble fraction, the whole-cell membrane-enriched fraction, and the mitochondrial (COXII marker protein) fraction (Supplemental Figure 3B). The BASS6 band was not detectable either in the whole-cell fraction or in the chloroplast fraction of bass6-1 mutants. The BASS6 antiserum also detected BASS6 in yeast, detecting one band at ∼44 kD along with a larger band at ∼60 kD. The larger band is too small to be a BASS6 dimer and may represent a nonspecific antiserum interaction, different gel migration due to charge, or a possible translation error. BASS6 was not detected in wild-type yeast extracts not expressing BASS6 (Supplemental Figure 3B).
The bass6-1 Mutant Accumulates Photorespiratory Intermediates
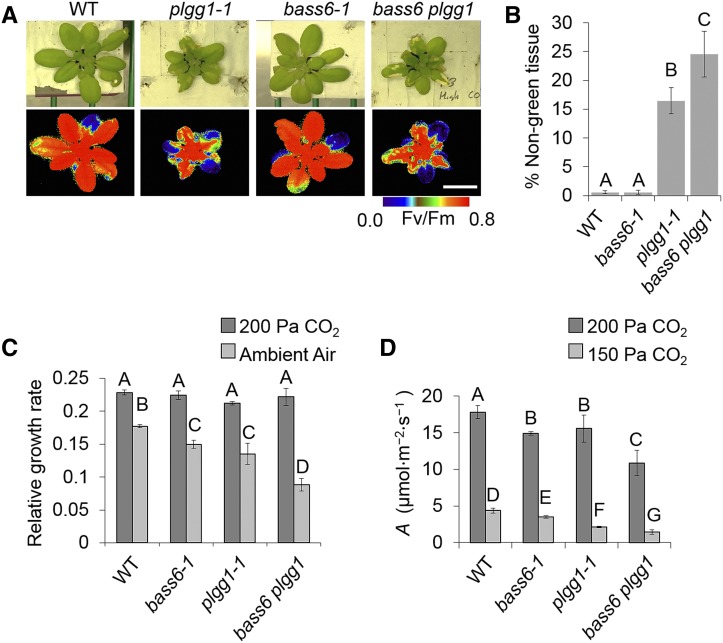
Previous characterization of plgg1-1 mutants demonstrated that they develop chlorotic lesions when grown at high [CO2] and then shifted to ambient air after several days (Pick et al., 2013). The formation of chlorotic lesions is thought to be due to damage from the accumulation of toxic photorespiratory intermediates during the light period. We also observed chlorotic lesions in plgg1-1 mutants 5 d after the shift to ambient air, and these lesions were associated with regions of lowered Fv/Fm as measured using chlorophyll fluorescence imaging (Figure 5A). Whereas the bass6-1 mutant did not develop observable chlorotic lesions on leaves after a shift to ambient [CO2] at 5 d after transfer (Figure 5A), the homozygous F3 generation of the bass6-1 and plgg1-1 cross (bass6 plgg1) developed more expansive chlorotic lesions compared with the plgg1-1 line alone (Figures 5A and 5B). The ∼50% increase in area of chlorotic lesions in the bass6 plgg1 double mutant presaged apparent additive effects of PLGG1 and BASS6 inactivation on growth and photosynthetic rates. The bass6 plgg1 double mutant plants showed a 41% decrease in growth compared plgg1-1 and 31% compared with bass6-1 under ambient [CO2] and also showed lower photosynthetic rates when both PLGG1 and BASS6 functions are missing (Figures 5C and 5D).
Figure 5.
The bass6 plgg1 Double Mutant Has a More Severe Photorespiratory Phenotype.
(A) Images represent indicated plants grown for 4 weeks at 200 Pa CO2 then shifted to ambient CO2 for 5 d. Fv/Fm images show changes in chlorophyll fluorescence due to the formation of chlorotic lesions on leaves. Images are representative of five replicates. Bar = 1 cm.
(B) Extent of chlorotic lesion formation. Percent nongreen tissue based on pixel density measured using photo software (Adobe) comparing nongreen tissue to total rosette tissue.
(C) Relative growth rate of indicated Arabidopsis T-DNA lines under either 200 Pa (dark) or ambient air (light) gray bars. Error bars indicate sd of at least five plants per each of three biological replicates. Letters indicate significant difference between CO2 treatments. Statistical difference based on ANOVA analysis (P < 0.05; Supplemental File 1).
(D) Photosynthetic measurements recorded at indicated CO2 concentration and saturating light (1000 μmol m−2 s−1) for the indicated strains. Letters represent significant difference from ANOVA analysis and Tukey’s post-hoc test. Error bars indicate sd.
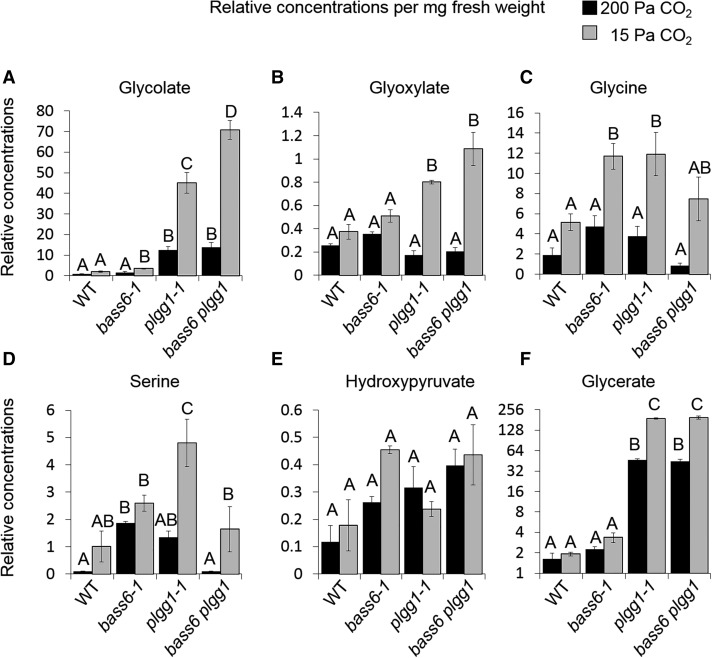
When Rubisco oxygenation rates are high, mutant plants with defects in photorespiration will accumulate various photorespiratory metabolites. To characterize the transport step of BASS6 in the photorespiration pathway, metabolite profiles were performed on bass6-1 leaf tissue from plants exposed either to high (200 Pa) or to low (15 Pa) [CO2]. When leaves were exposed to low [CO2] to increase the Rubisco oxygenation rate, the levels of glycine and glycolate in bass6-1 increased significantly compared with the wild type. By contrast, plgg1-1 plants accumulated glycolate and glycerate at high [CO2] and accumulated glycolate, glycine, glyoxylate, serine, and glycerate at low [CO2] (Figure 6). When leaves were exposed to low [CO2], bass6-1 accumulated levels of glycine similar to levels in the plgg1-1 plants (Figure 6C). The bass6 plgg1 double mutant line had significantly more glycolate than the wild type and the single mutant lines. They also showed increases in other intermediates such as glycerate, similar to what was observed in the plgg1-1 single mutant (Figure 6). Interestingly, serine accumulation also increased in the single mutants but reverted to wild-type levels in the bass6 plgg1 double mutant, suggesting that losing the function of both BASS6 and PLGG1 alters overall photorespiratory metabolism (Figure 6). Importantly, without BASS6 and PLGG1, there is a further increase in glycolate accumulation. The accumulation of photorespiratory metabolites when exposed to low [CO2] conditions indicates a direct role of BASS6 in photorespiratory metabolism.
Figure 6.
The bass6-1 Mutant Accumulates Photorespiratory Intermediates.
Levels of photorespiratory intermediates in wild-type, bass6-1, plgg1-1, and homozygous bass6 plgg1 double mutant plants. Plants were grown at elevated CO2 for 6 weeks. Young fully expanded leaves were then acclimated to either elevated (200 Pa) or low (15 Pa) CO2 using a LI-COR 6400 XT gas-exchange system. When photosynthetic rates were stable leaf samples were rapidly frozen and extracted to determine relative concentrations of glycolate (A), glyoxylate (B), glycine (C), serine (D), hydroxypyruvate (E), and glycerate (F). The y axis represents relative differences based on an internal standard (set to 1). Black bars indicate 200 Pa, and gray bars indicate 15 Pa CO2. Error bars indicate se of the mean. Letters indicate statistical differences based on ANOVA analysis (n = 3; Supplemental File 1).
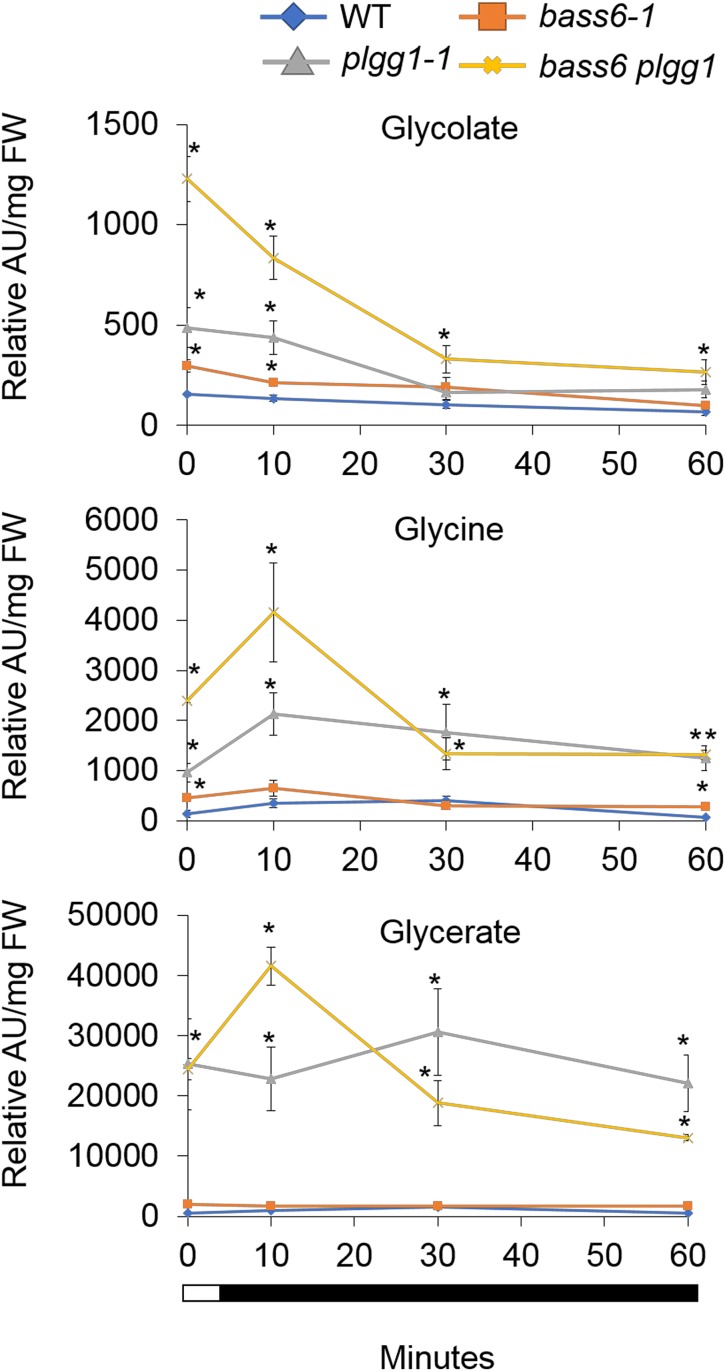
Arabidopsis plgg1-1 shows a light-dependent accumulation of glycolate, glycine, serine, and glycerate (Pick et al., 2013), consistent with impairment of both glycolate export and glycerate import in the chloroplast. Photorespiration is light-dependent, and glycolate and glycine levels in plgg1-1 mutants at night under ambient air returned to wild-type levels accompanied by a significant reduction in glycerate (Pick et al., 2013). To determine changes in glycolate, glycerate, and glycine levels in bass6-1 and the bass6 plgg1 double mutant plants compared with the wild type and plgg1-1, metabolite analysis was performed after a shift from high [CO2] to ambient air and samples were measured at the end of the growth photoperiod and immediately after the light-to-dark transition. Similar to results from Pick et al. (2013), the plgg1-1 mutants showed a reduction in glycolate levels after the end of the light period (Figure 7). At the end of the light period, bass6-1 plants accumulate glycolate and glycine, both of which are significantly reduced within 10 min in the dark (Figure 7). The glycolate level significantly increased in the bass6 plgg1 double mutant compared with wild-type, bass6-1, and plgg1-1 plants, whereas glycine and glycerate levels in the double mutants were similar to those in plgg1-1 mutants.
Figure 7.
BASS6 and PLGG1 Play a Role in Glycolate Metabolism.
Time course of metabolite levels in wild-type and mutant plants. The plant lines indicated were grown in elevated CO2 (200 Pa) for 4 weeks then shifted to ambient air (40 Pa CO2) for 24 h. At the end of the 8-h light cycle, tissue was collected as time 0 min. Each subsequent time point represents sampling during the dark period. The y axis numbers represent relative differences based on an internal standard. Error bars indicate se of the mean. Asterisks indicate statistical differences based on ANOVA analysis comparing wild-type control to T-DNA lines (n = 3; Supplemental File 1).
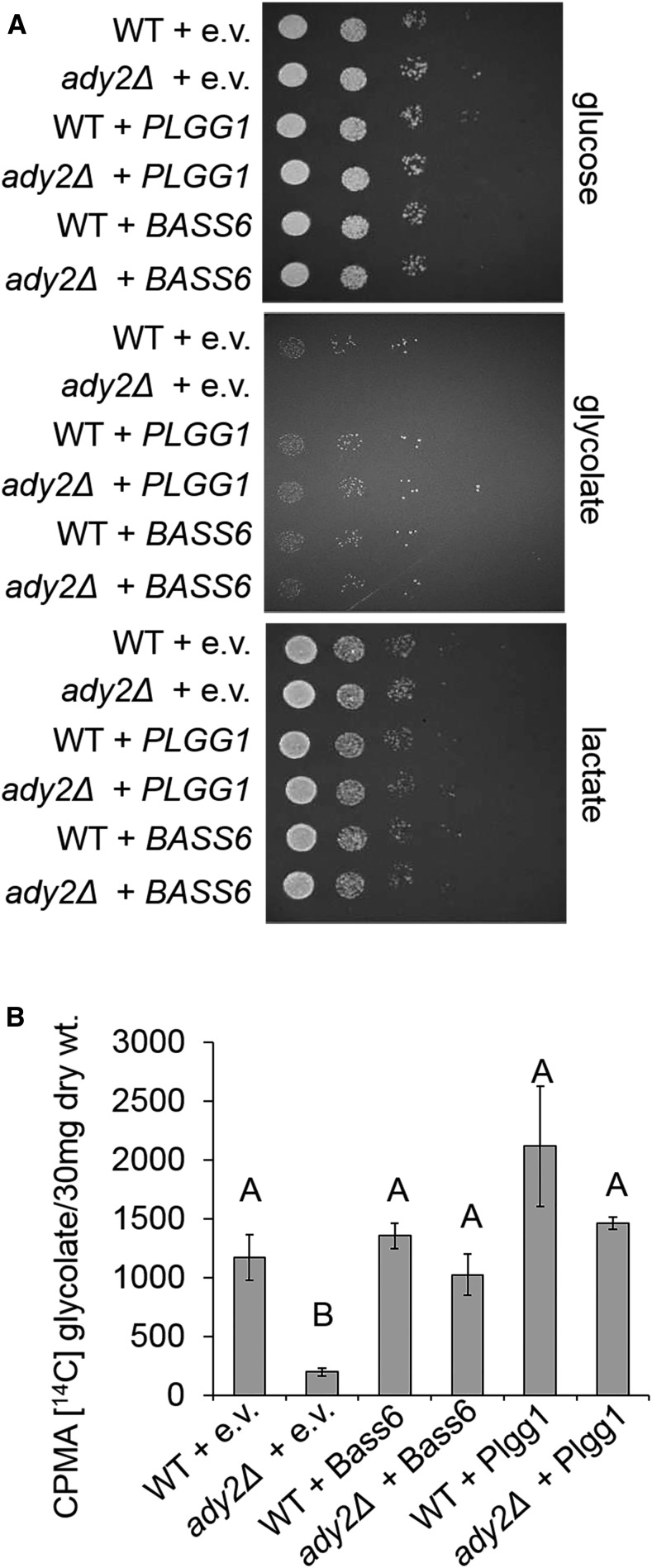
BASS6 and PLGG1 Rescues Growth on Glycolate as a Carbon Source in Yeast
There is no known glycolate transporter in the yeast Saccharomyces cerevisiae, but the plasma membrane-localized acetate transporter ADY2 is homologous to Escherichia coli yjcG, which is known to transport both acetate and glycolate (Gimenez et al., 2003). Expression of chloroplast proteins in yeast often results in localization to the mitochondria (Hurt et al., 1986; Pfaller et al., 1989; Brink et al., 1994). Localization prediction software suggested that BASS6 would localize to the plasma membrane and that PLGG1 would likely localize to the mitochondria membrane. Therefore, we generated yeast vectors expressing both BASS6 and PLGG1 lacking the chloroplast transit peptide (amino acids 1–24) to ensure plasma membrane targeting of both proteins. We then expressed them both in wild-type BY4741 and in the isogenic ady2Δ strain. In spot assays, the ady2Δ strain expressing only empty vector was unable to grow on glycolate as the sole carbon source, demonstrating a loss of glycolate import in the absence of acetate transporter ADY2 function (Figure 8A). The expression of PLGG1 in the ady2Δ yeast strain rescued growth back to the wild-type levels on glycolate as the sole carbon source (Figure 8A). Expression of BASS6 also rescued growth in the ady2Δ strain (Figure 8A). Controls using glucose and lactate as a carbon source showed both that the expression of BASS6 and PLGG1 in yeast does not negatively affect growth and that the ady2Δ strain was able to utilize both sources of carbon (Figure 8A). In addition, the wild type and the ady2Δ strain were both able to utilize glycerate as a carbon source. Unfortunately, this meant we could not determine the role of glycerate-dependent glycolate transport of either transporter because the identity of yeast glycerate transporter(s) is not yet known.
Figure 8.
BASS6 Can Transport the Photorespiratory Metabolite Glycolate.
(A) Representative serial dilutions of the wild type and ady2Δ yeast with empty vector (e.v.), PLGG1, or BASS6. Five-microliter yeast cultures OD600 = 0.1 in water were spotted on to SC-Leu plates with the indicated carbon source as five 10-fold serial dilutions. Plates were grown for 1 d (glucose), 2 d (lactate), or 7 d (glycolate) at 30°C.
(B) [14C]-glycolate uptake assay. Yeast cultures containing the indicated vector were incubated in SC-Leu glycolate media containing [14C]-glycolate for 10 min before stopping the reaction by the addition of 5 mL of ice-cold water. Error bars indicate sd, and letters indicate statistical differences based on ANOVA analysis (n = 3; Supplemental File 1).
Based on our findings that BASS6 and PLGG1 can complement yeast for growth on glycolate as a carbon source, we tested the transport capabilities of both proteins in uptake experiments in which yeast cells were incubated for 10 min in [14C]-glycolic acid. The expression of PLGG1 increased glycolate uptake both in the wild type and in the ady2Δ strain (Figure 8B), as did the expression of BASS6 (Figure 8B). Our data indicate that both BASS6 and PLGG1 expressed in yeast facilitate transmembrane glycolate transport leading to rescue of the growth in glycolate uptake defective ady2Δ mutant (Figure 8B).
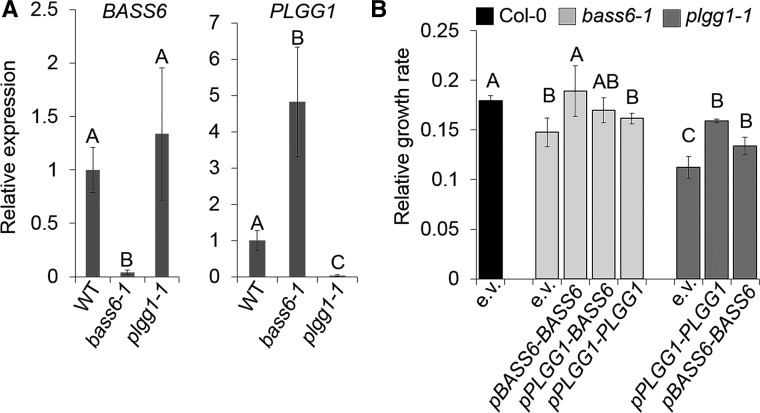
Loss of BASS6 Causes Increased PLGG1 Expression
Although the loss of either BASS6 or PLGG1 resulted in a photorespiratory phenotype, neither T-DNA insertion caused lethality when lines were grown in ambient air, as has been seen with numerous other photorespiratory mutants (Timm and Bauwe, 2013). This leaky photorespiratory phenotype could be due to redundancy in the transport processes and compensation for loss of one gene by the increase in expression of another. To test whether the loss of BASS6 or PLGG1 results in changes in enhanced expression of the other gene, real-time PCR (RT-PCR) experiments were performed. In the plgg1-1 line, there was no detectable difference in BASS6 expression compared with the wild type (Figure 9A). However, the expression of PLGG1 increased 4.8-fold over the wild type in the bass6-1 line, suggesting that PLGG1 expression had markedly increased, possibly to compensate for metabolic changes caused by the loss of BASS6 function (Figure 9A).
Figure 9.
Loss of BASS6 Causes Increased PLGG1 Expression.
(A) Expression of BASS6 and PLGG1 in leaf tissue was determined in the plgg1-1 and the bass6-1 mutants by qRT-PCR analysis. Error bars indicate se of the mean from three biological replicates including three technical replicates each. Letters indicate significant change (P < 0.05).
(B) Relative growth rate of indicated Arabidopsis transgenic lines. Error bars indicate sd of at least five plants per three biological replicates. Letters indicate significant difference between transgenic lines grown under ambient air conditions. Statistical difference based on ANOVA analysis (P < 0.05; Supplemental File 1).
This result prompted us to test whether the expression of either BASS6 or PLGG1 could complement the slow growth phenotype of each of the single mutants. Also, to rule out the possibility that either the plgg1-1 or the bass6-1 line phenotypes were due to another mutation, PLGG1 and BASS6 were also transformed into the plgg1-1 and the bass6-1 lines under the control of their native promoters. Expression of BASS6 under the control of either its own promoter or the PLGG1 promoter rescues the growth rate phenotype in the bass6-1 line, confirming that the loss of BASS6 is the cause of the photorespiratory phenotype (Figure 9B). In addition, as previously reported (Pick et al., 2013), expression of PLGG1 under the control of its own promoter rescues its photorespiratory phenotype (Figure 9B). However, transforming the expression plasmid of PLGG1 into the bass6-1 line showed no significant change in growth rate compared with the bass6-1 mutant (Figure 9B). This could be due to the fact that PLGG1 expression is already substantially upregulated in the bass6-1 background compared with the wild-type background (Figure 9B). Intriguingly, expression of BASS6 under the control of its native promoter in the plgg1-1 line increased its growth as did expression of PLGG1, but neither BASS6 nor PLGG1 fully restored wild-type growth (Figure 9B). Taken together, these results suggest that the expression of both BASS6 and PLGG1 are needed for proper growth at ambient [CO2] and that increasing the expression of one gene can in part compensate for the loss of the other.
DISCUSSION
Our data demonstrate that the bile acid sodium symporter BASS6 is an integral part of photorespiratory metabolism and show that it is required for proper wild-type growth in ambient air. Loss of BASS6 resulted in a slow growth phenotype that was rescued when plants were grown in high [CO2] (Figures 1, 5, and 9; Supplemental Figure 1). Arabidopsis BASS6 protein was localized to the chloroplast in Arabidopsis and shown to be part of the chloroplast envelope membrane by transient expression in N. benthamiana (Figure 4; Supplemental Figure 3). Also, under low [CO2] conditions when photorespiration flux is high, loss of BASS6 caused the accumulation of photorespiratory intermediates including glycine and glycolate (Figures 6 and 7). The combined loss of BASS6 and the glycolate/glycerate translocator PLGG1 resulted in a slower growth phenotype and increased accumulation of glycolate in leaves under low [CO2] compared with either single knockout mutation (Figures 5 to 7). Together with transport complementation in yeast (Figure 8), our study has identified an additional glycolate transporter involved in photorespiratory metabolism, Also, with the observed combined phenotypes, the bass6 mutant shows the class III photorespiratory mutant classification of being still viable in ambient air due to some functional compensation (Timm and Bauwe, 2013). Therefore, BASS6, in addition to PLGG1, contributes to the recycling of glycolate when Rubisco fixes O2 instead of CO2 during photorespiration. Additionally, this study reveals an alternative strategy that is complimentary to coexpression for identifying transport proteins involved in photorespiration.
Genetic screening under high and ambient [CO2], and more recently coexpression analysis, has identified only a few transporters important for photorespiration. Using the same coexpression software used to identify BOU and PLGG1, BASS6 did not show a strong correlation with the expression of other photorespiratory enzymes. Though the Spearman coefficient values were low (≤0.7), BASS6 most strongly associated with the expression of enzymes in the gluconeogenesis and glyoxylate cycles, two pathways that are associated with photorespiration (Supplemental Figure 4). As an alternative approach to discover chloroplast transporters important for photorespiration, we performed targeted fluorescence screening of mutants of known and putative chloroplast inner membrane proteins identified in proteomic studies (Zybailov et al., 2008; Badger et al., 2009). Exposing Arabidopsis T-DNA insertion mutants to 24 h of photorespiratory stress at low [CO2] levels and constant illumination is a very sensitive method for identifying [CO2]-dependent changes in chlorophyll fluorescence (Badger et al., 2009). This study identified bass6 T-DNA insertion mutants in Arabidopsis with reduced chlorophyll fluorescence under photorespiratory stress. Further analysis revealed that bass6-1 mutant plants exhibit a photorespiratory mutant phenotype similar to that of plgg1-1. In addition, our metabolite data and yeast complementation and transport assays demonstrated that BASS6 can transport glycolate (Figures 6 to 8).
Previous biochemical analysis of chloroplasts suggested that a single transporter transports both glycolate and glycerate (Howitz and McCarty, 1985, 1986, 1991). More recent analysis supports this model and identifies the glycolate /glycerate transporter PLGG1 (Pick et al., 2013). However, in each round of the photorespiratory cycle, two molecules of glycolate are transported out of the chloroplast but only one molecule of glycerate is returned (Howitz and McCarty, 1986). Curiously, PLGG1 has been shown to have a 1:1 glycerate:glycolate stoichiometry (Pick et al., 2013). In addition, the plgg1-1 Arabidopsis line under low irradiances exhibits similar photosynthetic rates and quantum efficiencies as the wild type, even under high photorespiratory conditions. This further suggests an alternative transport mechanism for glycolate and glycerate (Walker et al., 2016a).
Alternative transport mechanisms for glycolate and glycerate would also explain why plgg1-1 mutants were not discovered in the initial screens for photorespiratory mutants (Somerville and Ogren, 1981). Our metabolite and transport data support that BASS6, unlike PLGG1, is involved in glycolate but not glycerate transport. BASS6 is a member of the family of sodium-dependent symporters, which are unidirectional in transport, whereas PLGG1 is a glycolate/glycerate antiporter. This is consistent with the metabolite data showing the bass6-1 line does not accumulate glycerate as do plants lacking PLGG1 (Figures 6 and 7). The less severe accumulation of glycolate and the more subtle reduction in photosynthetic and growth rates in the bass6-1 line are also confounded by the increase in expression of PLGG1 (Figures 2, 3, 5, and 9). Together, our data suggest that glycolate flux out of the chloroplast is accomplished by both PLGG1 and BASS6 during normal operation, consistent with the predicted transport stoichiometry of 2:1 glycolate:glycerate of the photorespiratory pathway.
Many transport steps in biological systems have redundancies in which, due to redundant transport proteins, the removal of one transporter does not completely abolish the transport of metabolites across membranes (Vieten et al., 2005; Caffaro et al., 2007; Barberon et al., 2008). In fact, glycolate levels in the plgg1-1 plants reverted to wild-type levels once they were no longer under photorespiratory stress (Figure 7) (Pick et al., 2013). In the bass6 plgg1 double mutant plants, glycolate accumulation was 50 to 200% greater than in the plgg1-1 single mutant (Figures 6 and 7). Even in the bass6 plgg1 double mutant line, when photorespiratory stress was relaxed, there was a reduction in glycolate levels (Figure 7). This may mean that glycolate moved out of the chloroplast or that glycolate was metabolized within the chloroplast (e.g., chloroplastic pyruvate dehydrogenase complex) (Blume et al., 2013).
The bass6-1 mutant also demonstrated an increase in glycine when exposed to low [CO2] (Figures 6 and 7). Increase in glycine accumulation in Arabidopsis T-DNA insertion mutants of serine hydroxymethyltransferase (shm1), hydroxypyruvate reductase (hpr1), and glycerate kinase (glyk1), which are all genes encoding enzymes involved in reactions occurring after the conversion of glyoxylate to glycine (Timm et al., 2012). Other mutants also show an accumulation in glycine that is not directly linked to photorespiration. T-DNA bou-2 mutants in Arabidopsis and those with antisense targeting of TCA cycle enzymes in tomato (Solanum lycopersicum) accumulate glycine, showing a close relationship between respiration and photorespiration (Carrari et al., 2003; Sienkiewicz-Porzucek et al., 2008; Eisenhut et al., 2013b). Unexpectedly, 2PG-phosphatase (pglp1) T-DNA lines also demonstrate an increase in glycine levels even though PGLP acts before glycine formation (Timm et al., 2012). Together, disruption of photorespiration and closely related pathways can have unexpected effects on metabolite levels as appears to be the case for bass6-1 mutants (Figure 6).
In Arabidopsis, there are five loci annotated as BASS family proteins, including BASS1, BASS2, BASS4, BASS5, and BASS6, and all are either predicted or have been shown to be localized to the chloroplast envelope based on data from The Arabidopsis Information Resource (TAIR; Sawada et al., 2009). BASS2 was identified as highly expressed in C4 species and was shown to be a sodium-dependent pyruvate transporter located in the chloroplast envelope (Furumoto et al., 2011). In addition, BASS5 was recently shown to be localized to the Arabidopsis chloroplast envelope and to play a role in the sodium dependent transport of 2-keto acids involved in glucosinolate biosynthesis (Gigolashvili et al., 2009; Sawada et al., 2009). In fact, the characterization of BASS5 by Gigolashvili et al. (2009) is the only previously published study on BASS6 function concluding that, despite some similarity of BASS6 to BASS5 (69% identical), neither BASS6 nor BASS4 was involved in aliphatic glucosinolate biosynthesis. Thus, current work on BASS family proteins in plants suggests that they function in the transport of various organic acids involved in diverse metabolic pathways.
In this study, microscopy analysis revealed that Arabidopsis BASS6-GFP fusion proteins localize to the chloroplast envelope of N. benthamiana protoplasts in a similar fashion to that observed for PLGG1. Our light-sheet microscopy images and our immunoblot data also confirmed that Arabidopsis BASS6 proteins localize to the chloroplast in whole N. benthamiana leaf tissue and in Arabidopsis leaf tissue expressing BASS6 from its endogenous locus. Tobacco (Nicotiana tabacum) contains two copies of the BASS6 homolog, which is not surprising given that tobacco has an allotetraploid genome. Interestingly the BASS6 proteins in tobacco show 91% identity to each other but only 63% identity with Arabidopsis BASS6, with major differences occurring primarily in the N terminus (Supplemental Figure 5). One of the BASS6 proteins in tobacco contains an N-terminal extension that predicts localization in the mitochondria (NtBass6M), and the other is predicted to be chloroplastic (NtBass6C) (ARAMEMMNON) (Supplemental Figure 5). BASS6 does not appear to be dual-localized in Arabidopsis. Whether the tobacco proteins are present in different organelles remains to be determined.
In summary, BASS6 has been shown to be localized to the chloroplast envelope where it functions along with PLGG1 to export glycolate from chloroplast during photorespiratory metabolism. Localization of the BASS6 protein to the chloroplast envelope is supported by mass spectrometry proteomic analysis of the chloroplast inner envelope (Zybailov et al., 2008), by Arabidopsis BASS6-GFP localization in N. benthamiana (Figure 4), and by immunoblot analysis of Arabidopsis leaf tissue extracts (Supplemental Figure 3B). BASS6 glycolate transport function was demonstrated by genetic complementation of growth on glycolate (Figure 8A) and uptake of [14C] glycolate (Figure 8B) in yeast. In addition, the overexpression of BASS6 complemented the slow-growth phenotype of plgg1-1 (Figure 9B), and the loss of BASS6 resulted in the accumulation of glycolate, which was even higher in the bass6 plgg1 double mutant. Whereas BASS6 and PLGG1 work together in glycolate export, PLGG1 alone accounts for the import of the glycerate that balances the export of two glycolate molecules during photorespiration. Evidence that BASS6 does not transport glycerate includes the fact that BASS family proteins are unidirectional transporters and metabolite data showing the bass6-1 line does not accumulate glycerate as do plants lacking PLGG1 (Figures 6 and 7). The direct role of BASS6 in photorespiratory metabolism is shown by rescue of the slow growth phenotype of bass6-1 by growth at high [CO2] and by genetic complementation with BASS6 (Figure 2; Supplemental Figure 1). Further evidence is the additive effect of the bass6 plgg1 double mutant on the high [CO2] reversible suppression of growth (Figure 5C) and photosynthesis (Figure 5D) and the accumulation of photorespiratory metabolites when exposed to low [CO2] (Figure 6).
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) was used as the wild type reference. Salk lines used in this study are listed in Supplemental Data Set 1. Salk T-DNA lines were obtained from the Arabidopsis Biological Resource Center, and homozygous mutations were verified using oligonucleotide primers described in Supplemental Table 1. Plants were grown in elevated (200 Pa), ambient (40 Pa), or low (12.5 Pa) CO2 in an 8-h-light/16-h-dark cycle (22°C/18°C) at 250 µmol m−2 s−1 PAR and 65% relative humidity in growth chambers (Conviron) using LC1 Sunshine Mix (Sun Gro).
Determination of Growth
Growth was monitored by recording images using the CF Imager Technologica (http://www.technologica.co.uk/). The Fluorimager software determined the rosette size in mm2. The leaf area for each individual plant was determined on days 7, 11, 14, 18, and 21. For each individual plant, the growth rate was modeled with an exponential function [A(t) = A0ert] where A = area, A0 = initial size, r = growth rate, and t = time (El-Lithy et al., 2004).
Chlorophyll Fluorescence Measurements
Arabidopsis plants were grown under ambient air or elevated CO2 conditions and moved to a sealed clear plastic container at low CO2 conditions under constant illumination for 24 h before chlorophyll fluorescence measurements. Chlorophyll fluorescence measurements were done as previously described (Oxborough and Baker, 1997; Badger et al., 2009). Briefly, Fv/Fm images were taken after 15 min dark adaptation of either 9-d-old seedlings or 4-week-old plants using the CF Imager Technologica (http://www.technologica.co.uk/). Maximum flash intensity was 6800 µmol m−2 s−1 for 800 ms. Image values were obtained for each individual plant using the fluorimager software to define each position.
Photosynthesis Measurements
The youngest fully expanded leaves of 30- to 40-d-old Arabidopsis plants grown at elevated [CO2] were used for analysis of CO2 assimilation by gas exchange. Gas exchange measurements were performed using a LI-COR 6400XT with a 2-cm2 fluorescence measuring head with gasket leaks corrected for as outlined in the manual (LI-COR Biosciences). A, gs, and Ci measurements were obtained at indicated reference CO2 concentrations at a leaf temperature of 25°C and saturating light (1000 µmol m−2 s−1) from the full A-Ci data set. The A-Ci curves were measured in a range of [CO2] (50–2000 µbar) under the temperature and light conditions stated above after acclimation under ambient CO2. Vcmax, Jmax, Rd, and gs were determined using A-Ci data and the PsFit fitting routine (Bernacchi et al., 2003).
Cloning Procedures
All expression vectors are described in Supplemental Table 2. Promoters and open reading frames were synthesized based on sequences obtained from TAIR.
For the transient expression work on isolated protoplasts, the BASS6-GFP construct was cloned as follows. The coding sequence of Arabidopsis BASS6 (At4g22840) was synthesized by GENEWIZ with a C-terminal tag containing mGFP6 (Haseloff, 1999), 6xHIS, and MYC and was then cloned into a modified Gateway-compatible pUC57 plasmid from which it was recombined in pMDC32. The Arabidopsis PLGG1-GFP and P19 constructs have been described (Rolland et al., 2016).
For the remaining experiments, restriction sites and 4-bp regions of homology were designed according to common syntax in plant synthetic biology (Patron et al., 2015). Constructs were then assembled using the Golden Gate cloning protocol then subcloned into a binary vector (EC50505) (Werner et al., 2012; Engler et al., 2014; Marillonnet and Werner, 2015; Patron et al., 2015). For stable transformation, the binary vectors were transformed into Agrobacterium tumefaciens C58C1 by electroporation and then transformed into the Col-0 wild-type strain, plgg1-1, or bass6-1 T-DNA insertion lines using the floral dip method (Clough and Bent, 1998). Transformed lines were selected based on BASTA resistance, and gene insertion was verified by PCR analysis. For transient expression, constructs were designed as above driven by the CaMV35S promoter with a C-terminal GFP fusion.
Agroinfiltration and Microscopy of Nicotiana benthamiana
For the transient expression work on isolated protoplasts, growth and infiltration experiments were as described by Rolland et al. (2016). Briefly, Agrobacterium GV3101(pMP90) cells were transformed with plasmids of interest and grown in LB media containing rifampicin (50 µg/mL) and kanamycin (30 µg/mL). Cultures were grown for ∼24 h in a 28 to 30°C incubator and used for transformation of N. benthamiana leaves. Agrobacteria containing the P19 plasmid described above (OD600 = 0.3) were mixed with bacteria containing the plasmid of interest and/or the endoplasmic reticulum compartment marker (OD600 = 0.5) (plasmid CD3-959 from the Arabidopsis Biological Resource Center; http://abrc.osu.edu; Nelson et al., 2007). Cells were centrifuged for 8 min at 2150g and resuspended in 10 mM MES, pH 5.6, 10 mM MgCl2, and 150 µM acetosyringone. The cells were incubated for 2 h at room temperature and infiltrated into 3- to 4-week-old N. benthamiana leaves.
Protoplasts were prepared as described by Rolland et al. (2016). Two days after infiltration, a 4-cm2 area of infiltrated leaf was cut with a scalpel and transferred to a 5-mL syringe in which 2 mL of digestion solution (1.5% [w/v] Cellulase R-10, 0.4% [w/v] macerozyme R-10, 0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.6, 10 mM CaCl2, and 0.1% [w/v] BSA) was added and a gentle vacuum was manually applied. The digestion solution and the leaf tissue were transferred to a 2-mL Eppendorf tube and incubated for 1 h at room temperature. Leaf debris was removed using a forceps, and protoplasts were allowed to sediment before the solution was replaced with imaging solution (0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.6, 10 mM CaCl2, and 0.1% [w/v] BSA).
Protoplasts were observed and imaged using an upright Zeiss LSM780 confocal laser scanning microscope (Carl Zeiss), a 40× water immersion objective (NA = 1.1), and the Zen 2011 software package (Carl Zeiss). GFP and chlorophyll were excited at 488 nm and recorded at 499 to 534 nm and 630 to 735 nm, respectively.
For the transient expression work on whole tissue, sliced whole-leaf tissue was visualized using a Zeiss Lightsheet Z.1 microscope (Carl Zeiss) as described, using a 40× objective (NA = 1.0) and the Zen Lightsheet software package (Carl Zeiss). GFP and chlorophyll were excited at 488 nm, and emission selection was recorded at 505 to 545 nm and 660 to 735 nm, respectively.
Metabolic Profiling
For metabolite analysis, ∼40 mg of fresh leaf tissue was frozen in liquid nitrogen, crushed, and extracted with 500 μL of 100% methanol. Samples were then submitted to the Metabolomics Center, Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, where two additional extractions were performed: isopropanol:acetonitrile:water (3:3:2 v/v) and chloroform:methanol (2:2 v/v). Metabolites were analyzed using a GC-MS system (Agilent) consisting of an Agilent 7890 gas chromatograph, an Agilent 5975 mass selective detector, and a HP 7683B autosampler. Gas chromatography was performed on a ZB-5MS capillary column (60-m × 0.32-mm i.d. and 0.25-µm film thickness; Phenomenex). The inlet and MS interface temperatures were 250°C, and the ion source temperature was adjusted to 230°C. An aliquot of 1 μL was injected with the split ratio of 10:1. The helium carrier gas was kept at a constant flow rate of 2 mL/min. The temperature program was a 5-min isothermal heating at 70°C, followed by an oven temperature increase of 5°C min−1 to 310°C and a final 10 min at 310°C. The mass spectrometer was operated in positive electron impact mode at 69.9 eV ionization energy at m/z 30 to 800 scan range. The spectra of all chromatogram peaks were compared with electron impact mass spectrum libraries NIST08 (NIST), W8N08 (Palisade), and a custom-built database (464 unique metabolites). All known artificial peaks were identified and removed. To allow comparison among samples, all data were normalized to the internal standard in each chromatogram and the sample wet weight. The spectra of all chromatogram peaks were evaluated using the AMDIS 2.71 program (NIST). Metabolite concentrations were reported as concentrations relative to the internal standard (i.e., target compound peak area divided by peak area of hentriacontanoic acid): Ni (relative concentration) = Xi (target compound peak area) * X−1IS (peak area of hentriacontanoic acid) per gram wet weight. The instrument variability was within the standard acceptance limit of 5%.
Yeast Complementation and Glycolate Uptake Assays
All yeast strains are described in Supplemental Table 3. Yeast plasmids are described in Supplemental Table 2 and were assembled as previously described (South et al., 2010). Briefly, RNA was obtained from Col-0 wild-type Arabidopsis and converted to cDNA using an RNeasy extraction kit and a Quantitec reverse transcription kit (Qiagen). The coding sequences of full-length BASS6 and PLGG1 minus the chloroplast localization signal (1−24) were amplified by PCR using primers described in Supplemental Table 1. The PCR product was cloned into the pRS415-ADH1 vector (ATCC) using BamHI and XhoI restriction sites. Yeast transformations were performed as previously described with BY4741 mat a wild-type and ady2Δ strains (GE Dharmacon) (South et al., 2010, 2013).
Complementation of the ady2Δ strain was performed by comparing growth analysis using glycolate as a carbon source. Wild-type and ady2Δ strains transformed with BASS6, PLGG1, or empty vector expression plasmids were grown in 50 mL cultures in synthetic complete media lacking leucine (SC-Leu yeast nitrogen base without amino acids, 20% glucose, and synthetic complete dropout mix lacking leucine; Sunrise Biosciences) until reaching an optical density of OD600 between 0.6 and 0.8. Cells were then washed in water twice and resuspended in water. A spot assay was performed on SC-Leu plates containing 2% glucose, lactate, or glycolate as a carbon source. Five-microliter spots were dropped onto plates with five 10-fold serial dilutions starting with an OD600 of 0.1. Plates were then incubated at 30°C. Photographs of plates were taken at 1 d (glucose), 2 d (lactate), and 7 d (glycolate).
Glycolate uptake measurements were performed similarly to glycerol uptake assays described previously (Oliveira et al., 1996). Yeast strains were grown in 50-mL cultures in SC-Leu at 30°C to an optical density of OD600 between 0.6 and 0.8. Cells were harvested and washed twice with water and resuspended in 100 mM Tris/citrate buffer, pH 5.0, at a concentration of 30 mg/mL dry weight. After a 2-min incubation at 25°C, the reaction was started by adding 150 μL of SC-Leu/glycolate containing 1 μL of an aqueous solution of 50 mCi/ mmol (3.7*103 [Bq] total) [14C]-glycolic acid (American Radiolabeled Chemicals). After 10 min, the reaction was stopped by the addition of 5 mL of ice cold water. The reaction mixtures were then filtered on glass fiber filters (Fisher Scientific) and washed three times with 5 mL of ice-cold water. [14C]-glycolate uptake was measured by scintillation counting using the filters plus 4 mL of scintillation fluid (RPI Bio-safe II) using a Packard Tri-Carb liquid scintillation counter (Perkin-Elmer).
RT-PCR Analysis
cDNA was generated from RNA extracted using the plant RNeasy extraction kit and Quantitec reverse transcription kit (Qiagen) from 4-week-old Arabidopsis plants grown under an 8-h/16-h day/night cycle at 180 µmol m−2 s−1 PAR and 22°C/18°C temperature regime at 65% relative humidity. Three biological replicates including three technical replicates each were used for all samples. Samples were analyzed using a Bio-Rad CFX connect real-time PCR system (Bio-Rad Laboratories), and relative changes in transcript abundance were determined using the ΔΔCt method with primers directed toward violaxanthin deepoxidase, PLGG1, and BASS6 transcripts. cDNA was amplified using a SSO advanced SYBR green master mix (Bio-Rad), and primer sequences are described in Supplemental Table 1.
Immunoblot Analysis
Leaf tissue was collected from wild-type (Col-0) and bass6-1 plants grown at elevated CO2, and organelles were isolated following Lang et al. (2011), making adaptations for Arabidopsis organelle isolation as described by Seigneurin-Berny et al. (2008) and Klein et al. (1998). Unless otherwise noted, all steps were performed on ice or at 4°C. Leaves were briefly homogenized in extraction buffer (50 mM HEPES-NaOH, pH 6.9, 0.33 M sorbitol, 2 mM EDTA, 2 mM MgCl2, 1 mM MnCl2, and 0.1% [w/v] BSA) and filtered through three layers of Miracloth (Calbiochem). A fraction of filtrate from this soluble leaf tissue was centrifuged for 10 min at 16,000g to enrich for total organelle membrane fraction. The filtrate was centrifuged for 10 min at 1500g to pellet chloroplasts, and the remaining supernatant was decanted into a new tube to use for the isolation of mitochondria (see below). Pelleted chloroplasts were resuspended in 3 mL of buffer (50 mM HEPES-NaOH, pH 7.6, 0.33 M sorbitol, 2 mM EDTA, 2 mM MgCl2, 1 mM MnCl2, 10 mM KCl, and 1 mM NaCl) using a fine paintbrush, applied to a 15-mL Percoll density gradient (top to bottom: 10% [v/v], 40% [v/v], and 80% [v/v] Percoll in resuspension buffer), and centrifuged for 20 min at 16,000g. Intact chloroplasts accumulated at the 40 to 80% interface and were removed by aspiration, washed twice in 10 volumes of washing buffer (50 mM HEPES-NaOH, pH 7.6, 0.33 M sorbitol, 2 mM EDTA, and 2 mM MgCl2), and centrifuged for 10 min at 1500g.
Plastid proteins were extracted by lysing the chloroplasts in a hypotonic buffer (10 mM Tricine-NaOH, pH 7.6, and 5 mM DTT) and briefly sonicating the sample on ice. Proteins were precipitated with 4 times the sample volume of ice-cold acetone, and the final protein pellet was dissolved in SDS sample buffer (60 mM Tris-HCl, pH 6.8, 10% glycerol [v/v], and 2% [w/v] SDS). Using the supernatant of the original filtrate, mitochondria were enriched by a three-step centrifugation: two 5-min centrifugations at 3000g and 6000g (organelles in supernatant) and one centrifugation at 18,000g for 20 min (organelles in pellet). The crude mitochondrial fraction was resuspended in 5 mL of wash buffer containing 20%[v/v] Percoll and overlaid on a two-step density gradient (top to bottom: sample in 20% [v/v] Percoll, 5 mL of 33% [v/v], and 5 mL 80% [v/v] Percoll). After centrifugation for 60 min at 18,000g, mitochondria were isolated by aspiration. Percoll was removed by two successive washes with 10 volumes washing buffer followed by centrifugation at 16,000g for 10 min. Mitochondria were suspended in 10 mM Tricine-NaOH (pH 7.6), homogenized in a glass tissue homogenizer (Tenbroeck), and briefly sonicated on ice.
Proteins were precipitated with acetone and the final protein pellet was dissolved in SDS sample buffer. The purity of the isolated organelles and the cellular localization of BASS6 were determined by immunoblotting. Protein from whole-cell lysates (soluble and membrane enriched), chloroplasts, and mitochondria were loaded on a total protein concentration basis, as determined by a Bradford assay (Bio-Rad), and separated via SDS-PAGE. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane (PVDF-F Millipore) using the Trans-Blot electrophoresis transfer cell (Bio-Rad). Membranes were incubated in Odyssey blocking buffer (LI-COR) or 5% nonfat milk in TBS-T (20 mM Tris-HCl, 150 mM NaCl, and 0.1% [v/v] Tween 20), prior to incubation with either anti-RBCL antibody (1:5000; Agrisera AS03 037), anti-COXII antibody (1:1000; Agrisera AS04 053A), or custom rabbit polyclonal anti-BASS6 serum raised against a peptide from AtBASS6 (Agrisera). Signals were developed with either an alkaline phosphate-conjugated secondary antibody and colorimetric reagent (1:2500; Bio-Rad) or a fluorescent goat anti-rabbit IgG (H+L) 800 CW secondary antibody and visualized with the LI-COR Odyssey scanner and software (LI-COR Biosciences).
Statistical Analysis
All experiments had at least three biological replicates and data indicate the average values. Relative growth analysis and relative changes in mRNA levels include sd. Relative growth, relative changes in mRNA levels, metabolites, and photosynthetic measurements were analyzed either by a one-way ANOVA (genotype) or two-way ANOVA (genotype by CO2 treatment) with a significance threshold of P < 0.05 (Supplemental File 1). All ANOVA analyses were followed with a Tukey’s post-hoc test and determined using statistical software (OriginPro 9.1; OriginLab).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: BASS6 (At4g22840), GLYK (At1g80380), PLGG1 (At1g32080), and SHM1 (At4g37930). Germplasm included bass6-1 (CS859747), bass6-2 (SALK_052903C), glyk (SALK_036371), plgg1-1 (SALK_053469C), shm1-1 (a gift from Andreas P.M. Weber, originally called stm in Somerville and Ogren, 1981).
Supplemental Data
Supplemental Figure 1. BASS6 Is Necessary for Growth at Low CO2.
Supplemental Figure 2. Characterization of BASS6 T-DNA Insertions.
Supplemental Figure 3. BASS6 Is Chloroplast-Localized.
Supplemental Figure 4. Coexpression Analysis of BASS6 in Arabidopsis.
Supplemental Figure 5. BASS6 Amino Acid Sequence Alignments.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Table 2. Plasmids Used in This Study.
Supplemental Table 3. Yeast Strains Used in This Study.
Supplemental Data Set 1. T-DNA lines Included in the Chlorophyll Fluorescence Screen.
Supplemental File 1. ANOVA Tables.
Supplementary Material
Acknowledgments
We thank Richard D. Field, Caroline Keller, and Helen W. Liu for assistance with experiments. We also thank the Core Facilities at the Carl R. Woese Institute for Genomic Biology, University of Illinois, and the Center of Advanced Microscopy of the Australian National University for light-sheet and confocal microscope access, respectively. Funding was provided via a subcontract by the Bill and Melinda Gates Foundation (OPP1060461) titled ‘RIPE-Realizing Increased Photosynthetic Efficiency for Sustainable Increases in Crop Yield’. Gene synthesis and assembly were supported by the Crop Engineering Consortium sponsored by the project Engineering Nitrogen Symbiosis for Africa through a grant to the John Innes Centre by the Bill and Melinda Gates Foundation (OPP1028264).
AUTHOR CONTRIBUTIONS
P.F.S., A.P.C., and D.R.O. designed the experiments. P.F.S., B.J.W., A.P.C., and V.R. performed experiments. P.F.S., D.R.O., B.J.W., A.P.C., V.R., and M.B. analyzed data. P.F.S. and D.R.O. wrote the article.
Glossary
- 3-PGA
3-phosphoglycerate
- 2-PG
2-phosphoglycolate
Footnotes
Articles can be viewed without a subscription.
References
- Alrefai W.A., Gill R.K. (2007). Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm. Res. 24: 1803–1823. [DOI] [PubMed] [Google Scholar]
- Anderson L.E. (1971). Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235: 237–244. [DOI] [PubMed] [Google Scholar]
- Badger M.R., Fallahi H., Kaines S., Takahashi S. (2009). Chlorophyll fluorescence screening of Arabidopsis thaliana for CO2 sensitive photorespiration and photoinhibition mutants. Funct. Plant Biol. 36: 867–873. [DOI] [PubMed] [Google Scholar]
- Barberon M., Berthomieu P., Clairotte M., Shibagaki N., Davidian J.-C., Gosti F. (2008). Unequal functional redundancy between the two Arabidopsis thaliana high-affinity sulphate transporters SULTR1;1 and SULTR1;2. New Phytol. 180: 608–619. [DOI] [PubMed] [Google Scholar]
- Bernacchi C.J., Pimentel C., Long S.P. (2003). In vivo temperature response functions of parameters required to model RuBP-limited photosynthesis. Plant Cell Environ. 26: 1419–1430. [Google Scholar]
- Blume C., Behrens C., Eubel H., Braun H.P., Peterhansel C. (2013). A possible role for the chloroplast pyruvate dehydrogenase complex in plant glycolate and glyoxylate metabolism. Phytochemistry 95: 168–176. [DOI] [PubMed] [Google Scholar]
- Bordych C., Eisenhut M., Pick T.R., Kuelahoglu C., Weber A.P.M. (2013). Co-expression analysis as tool for the discovery of transport proteins in photorespiration. Plant Biol (Stuttg.) 15: 686–693. [DOI] [PubMed] [Google Scholar]
- Bowes G., Ogren W.L., Hageman R.H. (1971). Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 45: 716–722. [DOI] [PubMed] [Google Scholar]
- Brink S., Flügge U.I., Chaumont F., Boutry M., Emmermann M., Schmitz U., Becker K., Pfanner N. (1994). Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. J. Biol. Chem. 269: 16478–16485. [PubMed] [Google Scholar]
- Caffaro C.E., Hirschberg C.B., Berninsone P.M. (2007). Functional redundancy between two Caenorhabditis elegans nucleotide sugar transporters with a novel transport mechanism. J. Biol. Chem. 282: 27970–27975. [DOI] [PubMed] [Google Scholar]
- Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M.E., Fernie A.R. (2003). Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133: 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claro da Silva T., Polli J.E., Swaan P.W. (2013). The solute carrier family 10 (SLC10): beyond bile acid transport. Mol. Aspects Med. 34: 252–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Eisenhut M., Pick T.R., Bordych C., Weber A.P.M. (2013a). Towards closing the remaining gaps in photorespiration--the essential but unexplored role of transport proteins. Plant Biol (Stuttg.) 15: 676–685. [DOI] [PubMed] [Google Scholar]
- Eisenhut M., et al. (2013b). Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J. 73: 836–849. [DOI] [PubMed] [Google Scholar]
- El-Lithy M.E., Clerkx E.J.M., Ruys G.J., Koornneef M., Vreugdenhil D. (2004). Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol. 135: 444–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Youles M., Gruetzner R., Ehnert T.-M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S. (2014). A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Fernie A.R., et al. (2013). Perspectives on plant photorespiratory metabolism. Plant Biol (Stuttg.) 15: 748–753. [DOI] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2: 325–345. [DOI] [PubMed] [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T., et al. (2011). A plastidial sodium-dependent pyruvate transporter. Nature 476: 472–475. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U.-I. (2009). The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21: 1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez R., Nuñez M.F., Badia J., Aguilar J., Baldoma L. (2003). The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 185: 6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Moro B., Lacuesta M., Becerril J.M., Gonzalez-Murua C., Munoz-Rueda A. (1997). Glycolate accumulation causes a decrease of photosynthesis by inhibiting rubisco activity in maize. J. Plant Physiol. 150: 388–394. [Google Scholar]
- Haseloff J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58: 139–151. [DOI] [PubMed] [Google Scholar]
- Hodges M., Dellero Y., Keech O., Betti M., Raghavendra A.S., Sage R., Zhu X.-G., Allen D.K., Weber A.P.M. (2016). Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. J. Exp. Bot. 67: 3015–3026. [DOI] [PubMed] [Google Scholar]
- Howitz K.T., McCarty R.E. (1985). Substrate-specificity of the pea chloroplast glycolate transporter. Biochemistry 24: 3645–3650. [Google Scholar]
- Howitz K.T., McCarty R.E. (1986). D-glycerate transport by the pea chloroplast glycolate carrier - studies on [1-C-14] d-glycerate uptake and d-glycerate dependent O-2 evolution. Plant Physiol. 80: 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz K.T., McCarty R.E. (1991). Solubilization, partial purification, and reconstitution of the glycolate/glycerate transporter from chloroplast inner envelope membranes. Plant Physiol. 96: 1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E.C., Soltanifar N., Goldschmidt-Clermont M., Rochaix J.D., Schatz G. (1986). The cleavable pre-sequence of an imported chloroplast protein directs attached polypeptides into yeast mitochondria. EMBO J. 5: 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G.J., Latzko E. (1976). Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 68: 55–58. [DOI] [PubMed] [Google Scholar]
- Klein M., Binder S., Brennicke A. (1998). Purification of mitochondria from Arabidopsis. In Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 49–53. [DOI] [PubMed]
- Kozaki A., Takeba G. (1996). Photorespiration protects C3 plants from photooxidation. Nature 384: 557–560. [Google Scholar]
- Lang E.G., Mueller S.J., Hoernstein S.N., Porankiewicz-Asplund J., Vervliet-Scheebaum M., Reski R. (2011). Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 30: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer G.H. (1981). The carboxylation and oxygenation of ribulose 1,5-bisphosphate - the primary events in photosynthesis and photo-respiration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 32: 349–383. [Google Scholar]
- Marillonnet S., Werner S. (2015). Assembly of multigene constructs using Golden Gate cloning. In Glyco-Engineering: Methods and Protocols, A. Castilho, ed (New York: Springer), pp. 269–284. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ogren W.L. (1984). Photorespiration - pathways, regulation, and modification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35: 415–442. [Google Scholar]
- Ogren W.L., Bowes G. (1971). Ribulose diphosphate carboxylase regulates soybean photorespiration. Nat. New Biol. 230: 159–160. [DOI] [PubMed] [Google Scholar]
- Oliveira R.P., Lages F., Lucas C. (1996). Isolation and characterisation of mutants from the halotolerant yeast Pichia sorbitophila defective in H+/glycerol symport activity. FEMS Microbiol. Lett. 142: 147–153. [DOI] [PubMed] [Google Scholar]
- Oxborough K., Baker N.R. (1997). An instrument capable of imaging chlorophyll a fluorescence from intact leaves at very low irradiance and at cellular and subcellular levels of organization. Plant Cell Environ. 20: 1473–1483. [Google Scholar]
- Patron N.J., et al. (2015). Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol. 208: 13–19. [DOI] [PubMed] [Google Scholar]
- Peterhansel C., Horst I., Niessen M., Blume C., Kebeish R., Kürkcüoglu S., Kreuzaler F. (2010). Photorespiration. The Arabidopsis Book 8: e0130, doi/10.1199/tab.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller R., Pfanner N., Neupert W. (1989). Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem. 264: 34–39. [PubMed] [Google Scholar]
- Pick T.R., Bräutigam A., Schulz M.A., Obata T., Fernie A.R., Weber A.P.M. (2013). PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc. Natl. Acad. Sci. USA 110: 3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renné P., Dressen U., Hebbeker U., Hille D., Flügge U.I., Westhoff P., Weber A.P.M. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35: 316–331. [DOI] [PubMed] [Google Scholar]
- Rolland V., Badger M.R., Price G.D. (2016). Redirecting the cyanobacterial bicarbonate transporters BicA and SbtA to the chloroplast envelope: Soluble and membrane cargos need different chloroplast targeting signals in plants. Front. Plant Sci. 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Toyooka K., Kuwahara A., Sakata A., Nagano M., Saito K., Hirai M.Y. (2009). Arabidopsis bile acid:sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 50: 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit J., Häusler R.E., Fiene G., Kaiser W.M., Weber A.P.M. (2006). Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J. 45: 206–224. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Salvi D., Joyard J., Rolland N. (2008). Purification of intact chloroplasts from Arabidopsis and spinach leaves by isopycnic centrifugation. Curr. Protoc. Cell Biol. 3: 30. [DOI] [PubMed] [Google Scholar]
- Sharkey T.D. (1988). Estimating the rate of photorespiration in leaves. Physiol. Plant. 73: 147–152. [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D.C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R., Ogren W.L. (1979). Gas-exchange analysis of a photosynthesis-photorespiration mutant of Arabidopsis thaliana. Plant Physiol. 63: 152–152.16660670 [Google Scholar]
- Somerville C.R., Ogren W.L. (1981). Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 67: 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville S.C., Somerville C.R. (1985). A mutant of arabidopsis deficient in chloroplast dicarboxylate transport is missing an envelope protein. Plant Sci. Lett. 37: 217–220. [Google Scholar]
- South P.F., Harmeyer K.M., Serratore N.D., Briggs S.D. (2013). H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc. Natl. Acad. Sci. USA 110: E1016–E1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South P.F., Fingerman I.M., Mersman D.P., Du H.N., Briggs S.D. (2010). A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J. Biol. Chem. 285: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D.B., van Wijk K.J. (2009). PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 37: D969–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Bauwe H., Badger M. (2007). Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 144: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Bauwe H. (2013). The variety of photorespiratory phenotypes - employing the current status for future research directions on photorespiration. Plant Biol (Stuttg.) 15: 737–747. [DOI] [PubMed] [Google Scholar]
- Timm S., Mielewczik M., Florian A., Frankenbach S., Dreissen A., Hocken N., Fernie A.R., Walter A., Bauwe H. (2012). High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS One 7: e42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wiśniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531. [DOI] [PubMed] [Google Scholar]
- Walker B.J., South P.F., Ort D.R. (2016a). Physiological evidence for plasticity in glycolate/glycerate transport during photorespiration. Photosynth. Res. 129: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., VanLoocke A., Bernacchi C.J., Ort D.R. (2016b). The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 67: 107–129. [DOI] [PubMed] [Google Scholar]
- Werner S., Engler C., Weber E., Gruetzner R., Marillonnet S. (2012). Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3: 38–43. [DOI] [PubMed] [Google Scholar]
- Wingler A., Lea P.J., Quick W.P., Leegood R.C. (2000). Photorespiration: metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K.C., Flügge U.I., Heldt H.W. (1987). A two-translocator model for the transport of 2-oxoglutarate and glutamate in chloroplasts during ammonia assimilation in the light. Plant Physiol. 84: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Jin H., Chen Y., Lin W., Wang C., Chen Z., Han N., Bian H., Zhu M., Wang J. (2012). A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 193: 81–95. [DOI] [PubMed] [Google Scholar]
- Young X.K., McCarty R.E. (1993). Assay of proton-coupled glycolate and d-glycerate transport into chloroplast inner envelope membrane-vesicles by stopped-flow fluorescence. Plant Physiol. 101: 793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61: 235–261. [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.