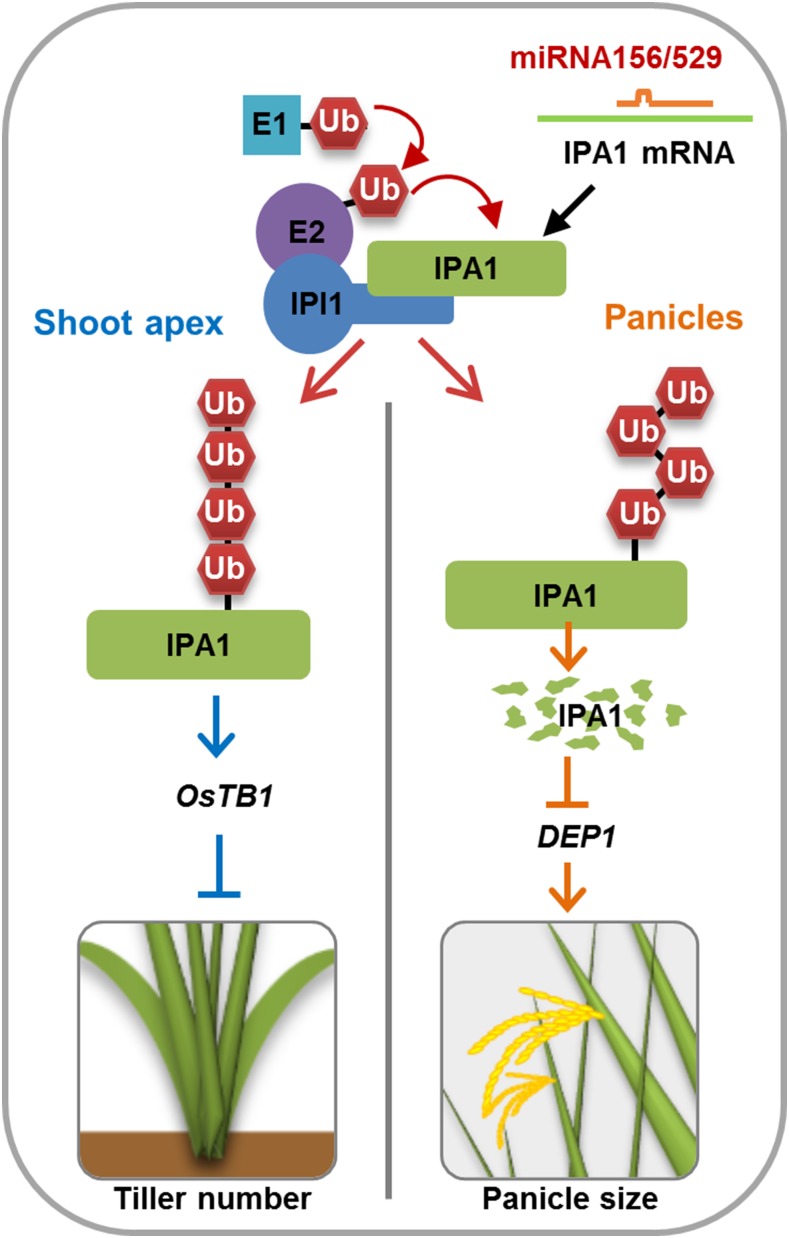
IPA1 INTERACTING PROTEIN1 functions as a RING-finger E3 ligase and adds different types of polyubiquitination to IPA1 in a tissue-specific manner to regulate plant architecture.
Abstract
Plant architecture, a collection of genetically controlled agronomic traits, is one of the decisive factors that determine grain production. IDEAL PLANT ARCHITECTURE1 (IPA1) encodes a key transcription factor with pleiotropic effects on regulating plant architecture in rice (Oryza sativa), and IPA1 expression is controlled at the posttranscriptional level by microRNA156 and microRNA529. Here, we report the identification and characterization of IPA1 INTERACTING PROTEIN1 (IPI1), a RING-finger E3 ligase that can interact with IPA1 in the nucleus. IPI1 promotes the degradation of IPA1 in panicles, while it stabilizes IPA1 in shoot apexes. Consistent with these findings, the ipi1 loss-of-function mutants showed markedly altered plant architecture, including more tillers, enlarged panicles, and increased yield per plant. Moreover, IPI1 could ubiquitinate the IPA1-mediated complex with different polyubiquitin chains, adding K48-linked polyubiquitin chains in panicles and K63-linked polyubiquitin chains in the shoot apex. These results demonstrate that IPI1 affects plant architecture through precisely tuning IPA1 protein levels in different tissues in rice and provide new insight into the tissue-specific regulation of plant architecture and important genetic resources for molecular breeding.
INTRODUCTION
Rice (Oryza sativa) plant architecture, including plant height, tiller number, and panicle morphology, is a key agronomic trait that determines rice grain yield (Wang and Li, 2008). IDEAL PLANT ARCHITECTURE1 (IPA1) encodes a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family transcription factors, SPL14 (Jiao et al., 2010; Miura et al., 2010). Increased expression of IPA1 repressed shoot branching but promoted panicle branching, resulting in an ideal plant architecture with reduced tiller number, increased panicle size, and enhanced lodging resistance. To understand the organ-specific functions of this transcription factor, a genome-wide study on IPA1 binding sites was performed and a complex network orchestrated by IPA1 in regulating plant architecture was revealed (Lu et al., 2013). IPA1 could directly bind to the “GTAC” motif in the promoters of TEOSINTE BRANCHED1 (OsTB1) and DENSE AND ERECT PANICLE1 (DEP1), which in turn regulate rice tillering and panicle morphology (Lu et al., 2013). Posttranscriptional regulation of SPL transcription factors is an important and conserved mechanism for plant development (Wang et al., 2009). The IPA1 mRNA contains a target site for microRNA156 (miR156) and a mutation in this target sequence perturbs miR156-mediated transcript cleavage in the ipa1 (renamed ipa1-1D to indicate its dominance nature) mutant, which results in high accumulation of IPA1 (Jiao et al., 2010). Massive analysis of rice small RNAs revealed that IPA1 could also be targeted by miR529, but this relationship is absent in IPA1 homologs in Arabidopsis thaliana (Jeong et al., 2011). Previous work showed that miR156 is preferentially expressed in seedlings and miR529 is mainly expressed in panicles, showing the complex spatiotemporal regulation of IPA1 by small RNAs. In addition, the change of DNA methylation was also found to affect the IPA1 expression in its allelic mutant, wealthy farmer’s panicle (Miura et al., 2010). These findings suggested that IPA1 is subjected to multiple types of posttranscriptional regulation by miR156, miR529, and epigenetic modifications. However, whether and how IPA1 is regulated at the protein level still need to be elucidated.
Ubiquitination is a posttranslational modification that modulates protein activities and plays important roles in various aspects of plant growth and development, including embryogenesis, floral development, plant senescence, and disease resistance (Vierstra, 2003; Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Lee et al., 2009; Park et al., 2012). Ubiquitination of a substrate requires a cascade of enzymatic reactions: activating ubiquitin by a ubiquitin activation enzyme E1, transferring ubiquitin to a ubiquitin conjugating enzyme E2, and then transferring the ubiquitin to a substrate catalyzed by a ubiquitin ligase E3 (Ciechanover and Schwartz, 1998). E3 ligases, the key enzymes that determine substrate specificities, can be classified into four groups, including Really Interesting New Gene (RING)/U-box, Anaphase Promoting Complex, Homology to E6-AP C terminus, and SKP1-CULLIN-F-box (Vierstra, 2003). Protein substrates can be modified with a single ubiquitin protein (monoubiquitination) or a chain of ubiquitins (polyubiquitination) (Ikeda and Dikic, 2008). Ubiquitin is a highly conserved protein consisting of 76 amino acid residues with seven lysines. Polyubiquitin chains can be divided into eight types, including ub-ub chains, K6, K11, K27, K29, K33, K48, or K63 chains. In most instances, the ubiquitinated proteins are destined for degradation by the 26S proteasome system, but sometimes the proteins go to various nonproteolytic pathways, which are determined by different types of ubiquitin chains (Smalle and Vierstra, 2004; Chen and Sun, 2009; Lim and Lim, 2011). Proteins modified with the K48-linked polyubiquitin chain are more likely to be degraded by the 26S proteasome (Pickart and Fushman, 2004), but proteins modified with the K63-linked polyubiquitin chain are mostly involved in proteasome-independent pathways such as endocytosis and signal transduction (Mukhopadhyay and Riezman, 2007).
RING proteins form an abundant E3 ubiquitin ligase family and can directly catalyze the transfer of ubiquitin from E2s to substrate proteins (Deshaies and Joazeiro, 2009). The RING domain contains four pairs of zinc ligands formed by cysteine and histidine residues with two zinc ions, which are essential for E3 ubiquitin ligase activity, and the mutations in zinc binding residues could perturb the domain structure and abolish ligase activity (Deshaies and Joazeiro, 2009). The roles of some RING domain-containing E3 ligases have been implicated in plant hormone signaling and defense responses (Bu et al., 2009; Ryu et al., 2010; Cho et al., 2011; Li et al., 2011; Cheng et al., 2012; Park et al., 2012; Kim and Kim, 2013; Mural et al., 2013). However, whether RING domain-containing E3 ligases participate in regulating rice plant architecture is still unclear.
Here, we report that a RING-containing E3 ligase, IPA1 INTERACTING PROTEIN1 (IPI1), regulates rice plant architecture by ubiquitinating and specifically controlling IPA1 protein levels in different tissues. Overexpression of IPI1 results in a decreased IPA1 protein levels in the panicles but increased levels in the shoot apexes and thus alters plant architecture accordingly. The ipi1 mutant shows significant increases in tiller number, panicle size, and yield per plant. We also find that the IPA1 complex could be targeted with different polyubiquitin chains in different tissues promoted by IPI1. These discoveries demonstrate that a context-dependent mechanism modulates the stability of IPA1, which provides insight into the tissue-specific posttranslational regulation of IPA1 abundance and a valuable resource for breeding high-yield elite rice varieties.
RESULTS
Identification of IPI1
IPA1 functions as a key transcription factor in forming ideal plant architecture (Jiao et al., 2010; Miura et al., 2010; Lu et al., 2013). To understand whether and how IPA1 is regulated at the protein level, we screened a rice cDNA library for IPA1 interacting proteins by yeast two-hybrid assay. Several positive clones were obtained and sequence analysis revealed that these putative IPA1 interactors included an RNA binding protein, a protein kinase, and a transcriptional regulator (Supplemental Table 1). Among them, LOC_Os01g24880, a C3H2C3-type RING finger-containing protein, is particularly interesting for its potential in regulating the ubiquitination and abundance of the IPA1 protein; it was named IPI1 and used for in-depth investigation.
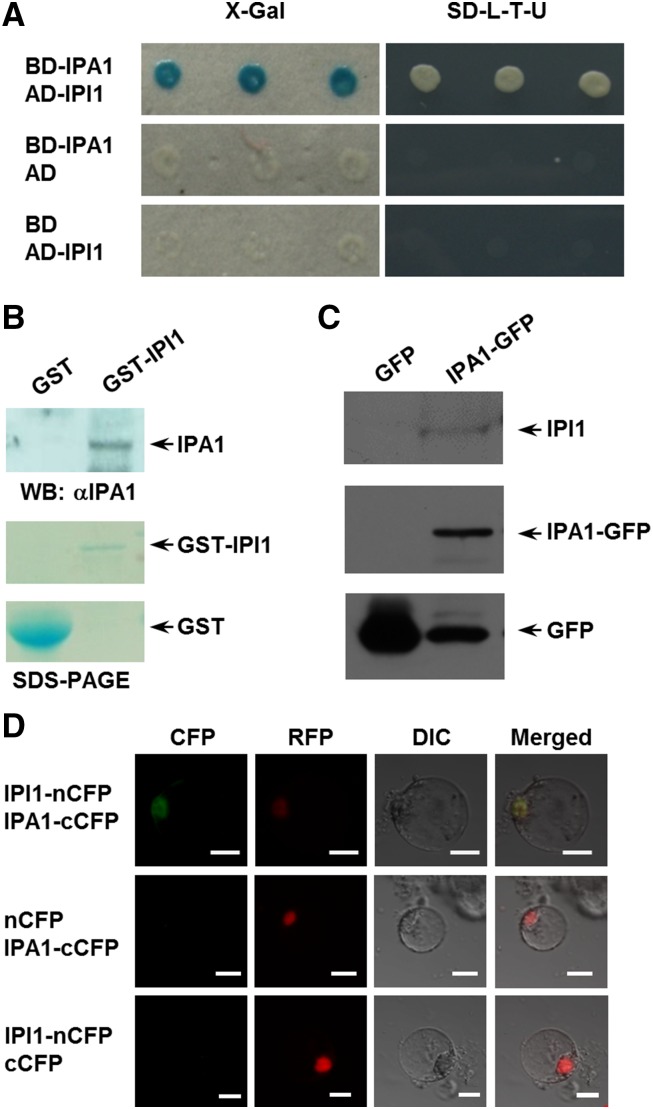
To further examine the interaction between IPA1 and IPI1, we fused the full-length IPA1 and IPI1 to the GAL4 DNA binding and activation domains, respectively. The cotransformed yeast cells that expressed both IPA1 and IPI1 could activate the expression of URA3 and the LacZ reporter gene, suggesting the existence of an interaction between IPI1 and IPA1 (Figure 1A). We further found that the SBP (SQUAMOSA PROMOTER BINDING PROTEIN) domain of IPA1 was essential for its interaction with IPI1 in the yeast two-hybrid assay (Supplemental Figure 1). The interaction between IPI1 and IPA1 was confirmed by a GST pull-down assay using purified GST-IPI1 incubated with the total proteins extracted from Nipponbare calli (Figure 1B), the coimmunoprecipitation experiment (Figure 1C) using the IPA1-GFP fusion protein extracted from IPA1:7mIPA1-GFP plants (Jiao et al., 2010; Lu et al., 2013), and a bimolecular fluorescence complementation (BiFC) assay (Figure 1D) using rice protoplasts. These results demonstrated that IPI1 could interact with IPA1 in the nucleus.
Figure 1.
Identification of IPI1.
(A) Interaction between IPI1 and IPA1 in the yeast two-hybrid assay. The IPA1 protein was fused with the GAL4 binding domain to generate DB-IPA1 and IPI1 with the GAL4 activation domain to form AD-IPI1. Blue clones in an X-Gal assay or the clones grown on the medium SD-L-T-U indicate the protein interaction in yeast cells.
(B) The GST pull-down assay, showing the interaction between IPI1 and IPA1. IPI1 was fused to a GST tag, and IPA1 was detected with anti-IPA1 polyclonal antibodies.
(C) The coimmunoprecipitation assay, indicating the in vivo interaction between IPI1 and IPA1. Immunoprecipitation was performed with an anti-GFP mAb-agarose, and immunoblotting was conducted with an anti-GFP monoclonal antibody and anti-IPI1 polyclonal antibodies, respectively. Transgenic plants expressing GFP were used as a negative control.
(D) The BiFC assay, showing the interaction between IPI1 and IPA1 in rice protoplasts. IPA1 was fused with cCFP (C terminus of CFP) and IPI1 with nCFP (N terminus of CFP). The visible light indicates the interaction between IPI1 and IPA1 in the nucleus. The NLS-RFP was cotransformed as the nucleus marker. Bars = 10 μm.
IPI1 Functions as an E3 Ligase and Promotes Polyubiquitination of IPA1
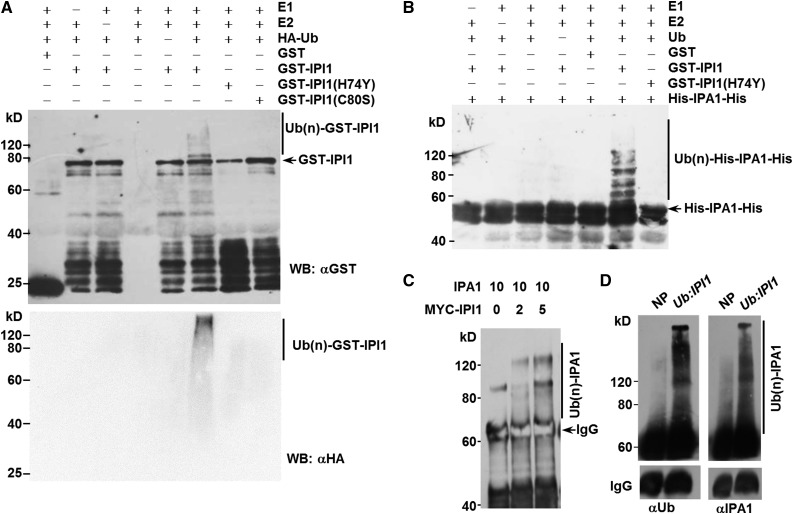
Since IPI1 is a putative C3H2C3-type RING finger protein, we first tested whether IPI1 functions as an E3 ligase using an in vitro self-ubiquitination assay with purified recombinant GST-IPI1, ubiquitin, E1, and E2 proteins. As shown in Figure 2A, a high molecular weight smear ladder was detected with GST-IPI1 incubated with E1, E2, and ubiquitin, suggesting that IPI1 is a functional E3 ligase. To test whether the RING finger domain of IPI1 is essential for its E3 ligase activity, we mutated the conserved histidine at 74 to tyrosine to form GST-IPI1(H74Y) and cysteine at 80 to serine to form GST-IPI1(C80S) in the RING finger domain (Supplemental Figure 2) and then assayed the enzyme activities of the mutant proteins. As expected, the high molecular weight smear ladder could not be detected with purified GST-IPI1(H74Y) or GST-IPI1(C80S) recombinant proteins in the presence of ubiquitin, E1, and E2 (Figure 2A), indicating that the intact RING finger domain of IPI1 is essential for its E3 ligase activity.
Figure 2.
IPI1 Is a Functional E3 Ligase and Promotes Polyubiquitination of IPA1.
(A) The E3 ubiquitin ligase activity of IPI1, revealed by an in vitro assay with purified GST-IPI1, GST-IPI1(H74Y), and GST-IPI1(C80S) proteins. Immunoblotting was performed with an anti-GST monoclonal antibody. The presence (+) or absence (−) of components in the reaction mixture is indicated.
(B) In vitro polyubiquitination of IPA1 by IPI1. GST-IPI1(H74Y) and GST were used as negative controls. Immunoblotting was performed with an anti-His or anti-HA monoclonal antibody. The presence (+) or absence (−) of components in the reaction mixture is indicated.
(C) Enhanced polyubiquitination of IPA1 with increased MYC-IPI1 in tobacco. Immunoprecipitation and immunoblotting were performed with anti-IPA1 polyclonal antibodies. Numbers indicate the ratio of the concentration of agrobacteria used in coinfiltration.
(D) IPI1 promotes the polyubiquitination of IPA1 in vivo. The assay was performed with Nipponbare (NP) and transgenic plants overexpressing IPI1 driven by the rice Ubiquitin promoter (Ub:IPI1). Immunoprecipitation was performed with anti-IPA1 polyclonal antibodies, and immunoblotting was performed with an anti-Ub monoclonal antibody and anti-IPA1 polyclonal antibodies. IgG was added as an internal control.
Based on the facts that IPI1 can physically interact with IPA1 and that IPI1 functions as an E3 ligase, we tested whether IPA1 is a substrate of IPI1 via an in vitro ubiquitination assay and found that His-IPA1-His was indeed polyubiquitinated by IPI1 in the presence of E1, E2, and ubiquitin proteins, but not by the mutated GST-IPI1(H74Y) (Figure 2B), suggesting that IPA1 was targeted by IPI1 for polyubiquitination.
To further understand the relationship between IPA1 and IPI1, we coexpressed IPA1 and MYC-IPI1 in tobacco (Nicotiana benthamiana) leaves and found that stronger smear bands were detected on IPA1 when MYC-IPI1 protein levels were increased (Figure 2C). Furthermore, we generated the Ubiquitin promoter-driven IPI1-overexpressing (Ub:IPI1) transgenic plants in rice cultivar Nipponbare and detected IPA1 ubiquitination in vivo using 4-week-old seedlings to test whether ubiquitination of IPA1 was enhanced in Ub:IPI1 transgenic plants. As shown in Figure 2D, more abundant ubiquitinated IPA1 was detected by both anti-IPA1 and anti-Ub antibodies in Ub:IPI1 transgenic plants than in the control plant. Taken together, these results clearly demonstrated that IPI1 functions as an E3 ligase to polyubiquitinate IPA1.
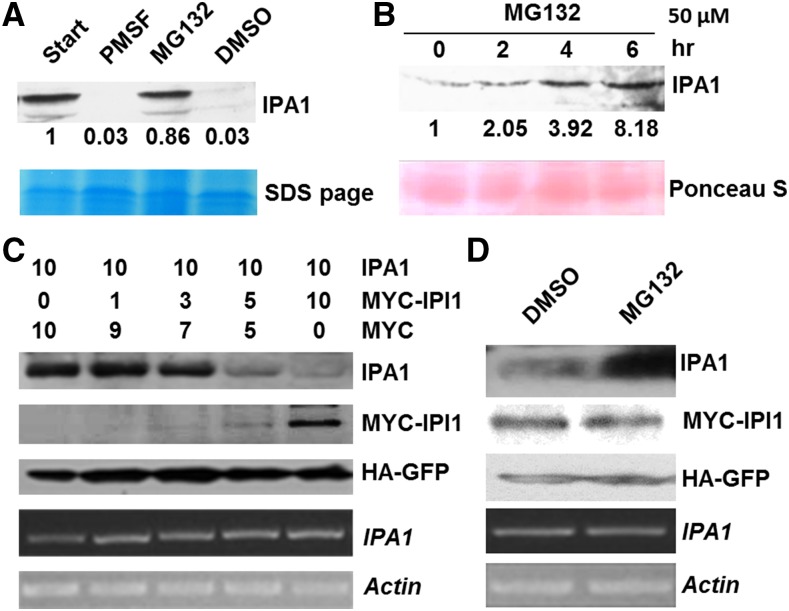
IPI1 Promotes Degradation of IPA1 through the Ubquitin-26S Proteasome System
To test whether IPA1 is degraded through the ubquitin-26S proteasome system, we performed a cell-free degradation assay in the presence or absence of MG132, a 26S proteasome inhibitor. As shown in Figure 3A, the degradation of IPA1 could be strongly inhibited by MG132, but not by the protease inhibitor PMSF or the organic solvent DMSO. Similar results were also obtained in the 4-week-old seedlings treated with MG132 (Figure 3B), indicating that IPA1 could be degraded via the 26S proteasome system. To further explore the effect of IPI1 on the degradation of IPA1, we coinfiltrated Agrobacterium tumefaciens EHA105 cells that express IPA1 and MYC-IPI1 into tobacco leaves and found that the protein level of IPA1 decreased correspondingly with increased MYC-IPI1 proteins, but we observed no obvious changes at the transcriptional level (Figure 3C). Furthermore, the degradation of IPA1 promoted by IPI1 could also be inhibited by MG132 (Figure 3D). These results strongly suggest that IPA1 is polyubiquitinated by IPI1 and then degraded via the ubquitin-26S proteasome pathway.
Figure 3.
IPI1 Promotes Degradation of IPA1.
(A) In vitro degradation assay of IPA1. DMSO, MG132 (40 μM), or PMSF (4 mM) was added in the cell lysate, respectively. Immunoblotting was performed with anti-IPA1 polyclonal antibodies. Relative amounts of proteins were determined by densitometry and normalized to loadings determined by Coomassie blue staining (blue) and expressed relative to the value at zero time.
(B) The in vivo stability of IPA1 enhanced by MG132. Seedlings were treated with MG132 (50 μM) and then collected at the time points as indicated. Protein detected as in (A), and relative amounts of proteins were normalized to loadings determined by Ponceau staining (red).
(C) Effect of IPI1 levels on the degradation of IPA1. Numbers indicate the ratio of the concentration of agrobacteria used in coinfiltration. HA-GFP was used as an internal control for protein synthesis. The transcript levels of IPA1 and Actin were analyzed by RT-PCR.
(D) Inhibition of IPA1 degradation by MG132 in tobacco leaves. MG132 (50 μM) or DMSO was infiltrated into tobacco leaves 12 h before samples were collected. The transcripts of IPA1 and Actin were analyzed by RT-PCR.
IPI1 Regulates Rice Architecture by Modulating IPA1 Abundance
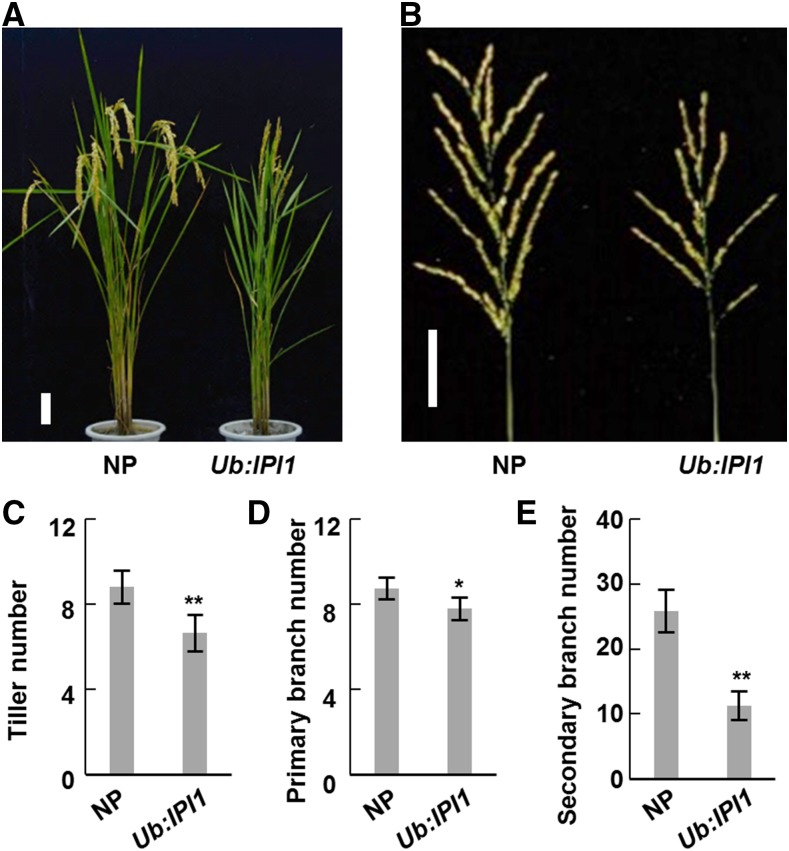
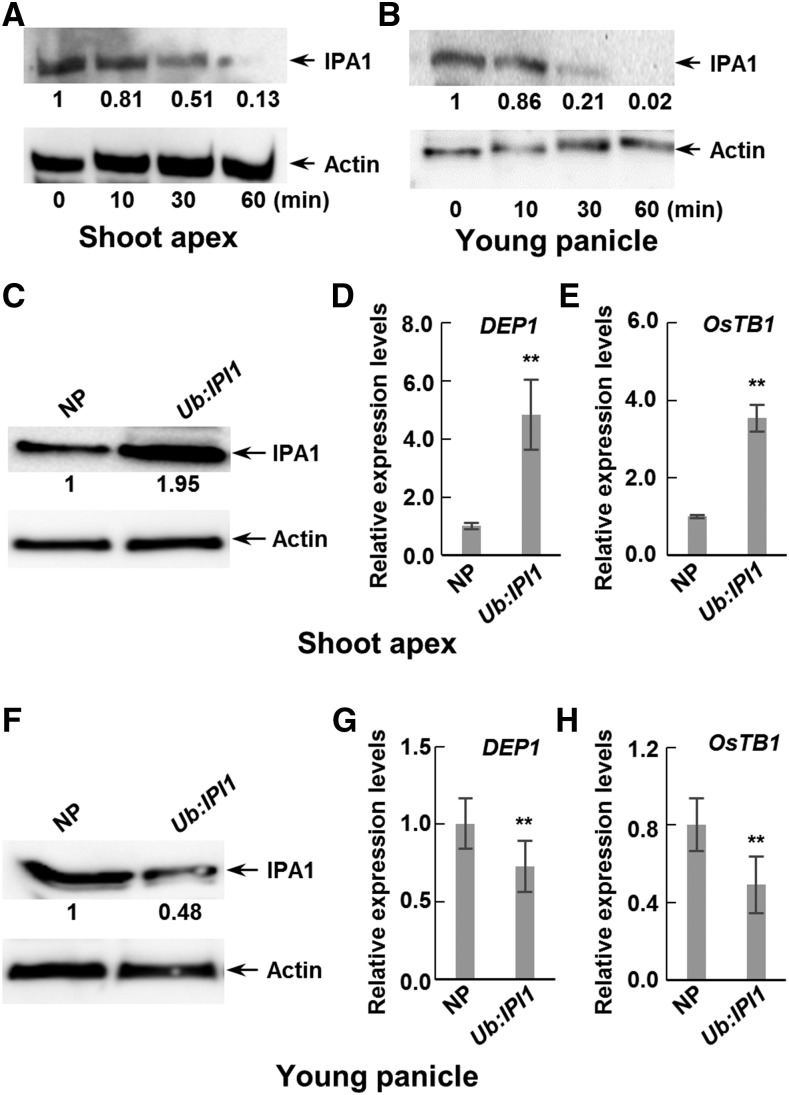
To understand whether IPI1-mediated polyubiquitination and degradation of IPA1 are involved in the regulation of rice plant architecture, we examined the phenotypes of IPI1-overexpressing transgenic plants by overexpressing IPI1 driven by the rice Ubiquitin promoter (Ub:IPI1 and Ub:HA-IPI1) in cultivar Nipponbare (Supplemental Figures 3A, 4C, and 5C). Compared with the control Nipponbare, IPI1-overexpressing transgenic plants exhibited obvious alterations in plant architecture, including fewer tillers (Figures 4A and 4C; Supplemental Figures 4A and 5A) and smaller panicles (Figures 4B, 4D, and 4E; Supplemental Figures 3B, 4B, and 5B). We should point out that the change of tiller number is surprisingly similar to that of IPA1-overexpressing transgenic plant lines, but the other morphological changes are opposite to those of IPA1-overexpressing transgenic plants (Jiao et al., 2010). Furthermore, we found that IPA1 protein is less stable in young panicles than in shoot apexes (Figures 5A and 5B). Considering that the IPA1 abundance is negatively related to tiller number, we therefore detected the protein levels of IPI1 and IPA1 in shoot apexes and young panicles (<3 cm) of the IPI1-overexpressing transgenic plants and found that the IPI1 protein level increased in both tissues but the IPA1 protein level unexpectedly increased in the shoot apexes but decreased in the young panicles, which is consistent with their morphological phenotypes (Figures 5C and 5F; Supplemental Figures 5C and 6). To further confirm the effect of different IPA1 abundance in two tissues of IPI1-overexpressing transgenic plants on the downstream genes, we examined the expression level of two reported IPA1 target genes, DEP1 and OsTB1, which were identified as the direct targets of IPA1 via ChIP-seq (Lu et al., 2013). Consistent with the protein level of IPA1, the transcript levels of DEP1 and OsTB1 were significantly increased in shoot apexes but decreased in young panicles (Figures 5D, 5E, 5G, and 5H) in IPI1-overexpressing transgenic plants. Expression level analysis revealed that IPI1 transcript abundances were high in leaves but low in shoot apex and young panicles (Supplemental Figure 7), obviously opposite to the IPA1 expression pattern. Therefore, to rule out the possible effect of the strong promoter, we generated IPI1 native promoter-driven IPI1-overexpressing transgenic plants (IPI1:IPI1-GUS) and found a similar relationship between the phenotype and IPA1 protein abundance to that in Ub:IPI1 plants (Supplemental Figure 8).
Figure 4.
Overexpression of IPI1 Affects Plant Architecture.
(A) Morphological phenotypes of Nipponbare (NP) and IPI1-overexpressing transgenic plants (Ub:IPI1). Bar = 10 cm.
(B) Morphologies of main panicles of NP and Ub:IPI1. Bar = 5 cm.
(C) to (E) Statistical analysis of tiller numbers (C) and the primary (D) and secondary (E) branch numbers of NP and Ub:IPI1. Double asterisk indicates P < 0.01 and single asterisk indicates 0.01 < P < 0.05 (Student’s t test, n = 10 independent plants). Bars indicate sd.
Figure 5.
Abundance of IPA1 and Expression Levels of Downstream Genes Are Modulated by Overexpressing IPI1.
(A) and (B) An in vitro degradation assay was performed in shoot apexes (A) and young panicles (B) to detect the IPA1 protein. The samples were collected as indicated and relative amounts of proteins were determined by densitometry normalized to actin.
(C) IPA1 abundance in shoot apexes in Nipponbare (NP) and IPI1-overexpressing transgenic (Ub:IPI1) plants. Relative amounts of IPA1 protein were determined as (A) and expressed relative to the value of Nipponbare.
(D) and (E) Quantitative real-time PCR was performed to detect the transcript levels of DEP1 (D) and OsTB1 (E) in shoot apexes in NP and Ub:IPI1. Double asterisk indicates P < 0.01 (Student’s t test, n = 3), and bars indicate sd.
(F) IPA1 abundance in young panicles in NP and Ub:IPI1. The proteins were detected as in (C).
(G) and (H) Quantitative real-time PCR was performed to detect the transcript levels of DEP1 (G) and OsTB1 (H) in young panicles in NP and Ub:IPI1. Double asterisk indicates P < 0.01 (Student’s t test, n = 3), and bars indicate sd.
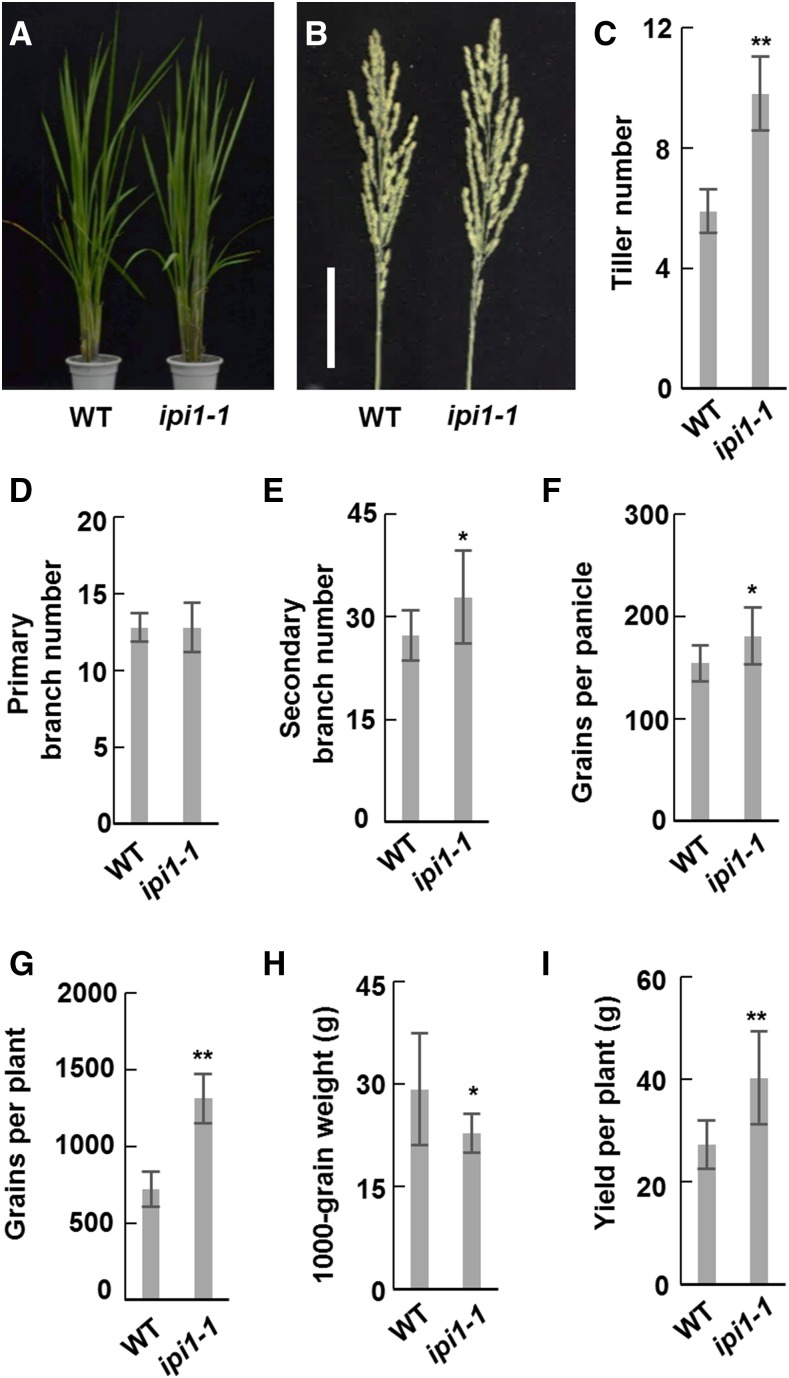
As tiller number and panicle size decreased in IPI1-overexpressing lines, we wondered whether both traits and plant yield could be increased by loss of IPI1 function. Using CRISPR/Cas9 technology, we generated ipi1-1 mutant plants in the TEIPEI 309 (TP309) background. Sequence analysis revealed that a G was inserted in the fourth exon of IPI1, which results in a truncated IPI1 lacking 283 amino acids at the C terminus (Supplemental Figures 9A and 9B). We found the 152 amino acids on the C terminus of IPI1 is essential for its nucleus localization and interaction with IPA1 (Supplemental Figures 9C and 9D), suggesting that ipi1-1 should be a loss-of-function mutant for regulation of IPA1’s stability. Furthermore, the ipi1-1 plants showed significantly altered plant architecture, including increased tiller number and grain number per panicle, which is contrast to IPI1-overexpressing transgenic plants (Figures 6A to 6F). These phenotypes led to an obvious increase in grain yield per plant in the ipi1-1 line compared with TP309 (Figures 6G to 6I). To further confirm these results, we also isolated an ipi1-2 mutant, which lost 7 bp in the IPI1 cDNA sequence and showed similarly altered plant architecture (Supplemental Figure 10).
Figure 6.
The ipi1-1 Mutant Showed Altered Plant Architecture.
(A) Morphological phenotypes of the wild type and ipi1-1 mutant. Bar = 5 cm.
(B) Morphologies of main panicles of the wild type and ipi1-1.
(C) to (H) Statistical analysis of tiller numbers (C), the primary (D) and secondary (E)branch numbers, grains per panicle (F), grains per plant (G), 1000-grain weight (H), and yield per plant (I) of the wild type and ipi1-1. Double asterisk indicates P < 0.01 and single asterisk indicates 0.01 < P < 0.05 (Student’s t test, n = 10 independent plants). Bars indicate sd.
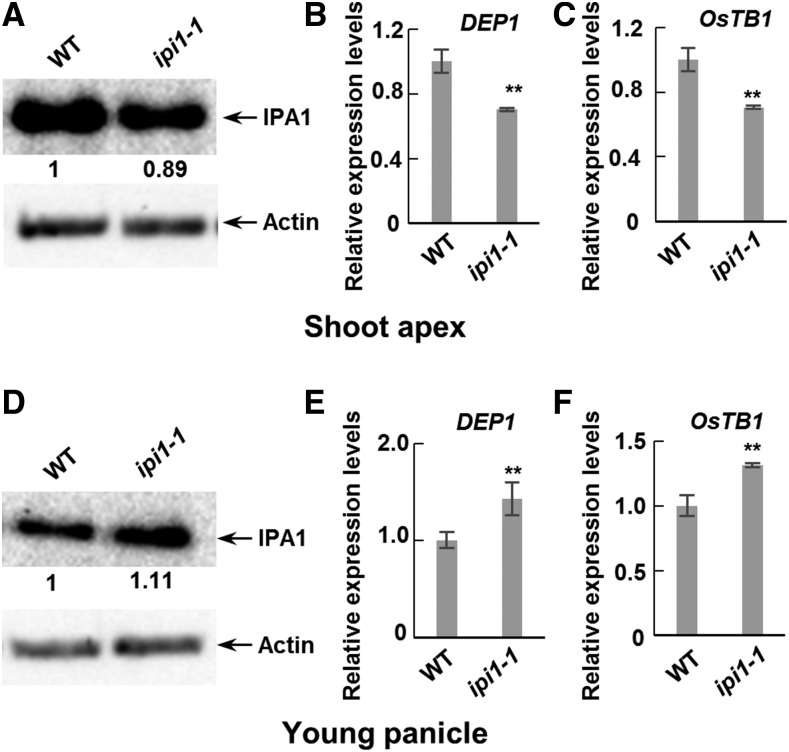
We then examined the IPA1 protein level and downstream gene expression levels in ipi1-1 plants and found that the IPA1 protein level is decreased in the shoot apexes while increased in the young panicles and that the expression levels of OsTB1 and DEP1 were altered correspondingly, which are all consistent with the morphological changes (Figure 7). Taken together, these results suggest that IPI1 promotes the degradation of IPA1 in young panicles but enhances the stability of IPA1 in shoot apexes to regulate downstream genes in determining rice architecture; therefore, loss of function of IPI1 could increase rice tiller number, panicle size, and yield per plant.
Figure 7.
Abundance of IPA1 and Expression Levels of Downstream Genes Are Modulated in ipi1-1 Mutants.
(A) IPA1 abundance in shoot apexes in the wild type and ipi1-1 mutant (ipi1-1). Relative amounts of IPA1 protein were determined by densitometry and normalized to loadings determined by the actin protein level and expressed relative to the value of the wild type.
(B) and (C) Quantitative real-time PCR was performed to detect the transcript levels of DEP1 (B) and OsTB1 (C) in shoot apexes in the wild type and ipi1-1. Double asterisk indicates P < 0.01 (Student’s t test, n = 3), and bars indicate sd.
(D) IPA1 abundance in young panicles in the wild type and ipi1-1. Proteins were detected as in (A).
(E) and (F) Quantitative real-time PCR was performed to detect the transcript levels of DEP1 (E) and OsTB1 (F) in young panicles in the wild type and ipi1-1. Double asterisk indicates P < 0.01 (Student’s t test, n = 3), and bars indicate sd.
Mutation in the miR156 Target Site in ipa1-1D Has No Effect on Polyubiquitination Mediated by IPI1
In the Ri22 background, the ipa1-1D allele that contains a point mutation in the miR156 and miR529 target sites confers a gain-of-function phenotype with thick culm, large panicle, and reduced tillers (Jiao et al., 2010). We found that this point mutation in ipa1-1D does not affect its association with IPI1 (Supplemental Figure 11A) nor the polyubiquitination and degradation mediated by IPI1 (Supplemental Figures 11B and 11C).
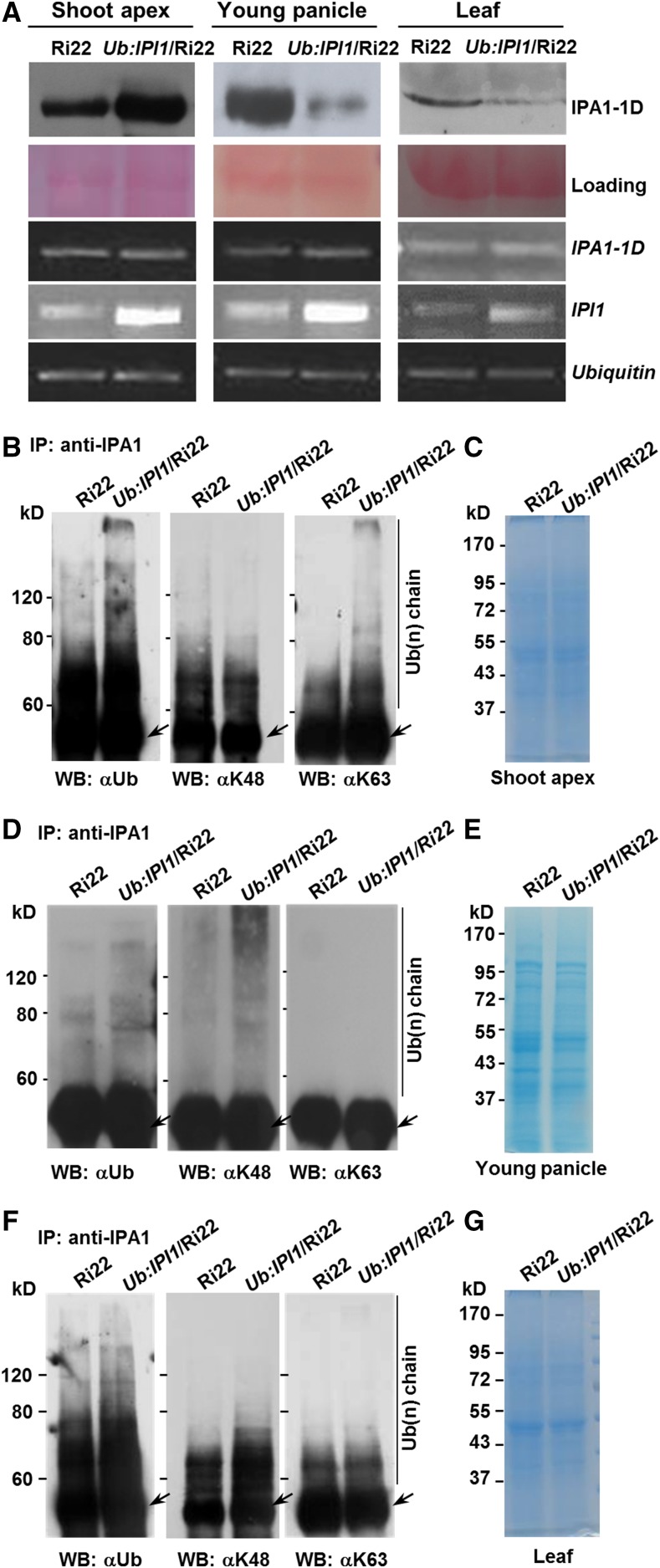
We further generated IPI1-overexpressing transgenic plants in the Ri22 background and found that most of the ipa1-1D gain-of-function phenotypes, such as thick culm and large panicle, were reverted, but the tiller number of IPI1-overexpressing transgenic plants was unchanged compared with that of Ri22 (Supplemental Figures 12A to 12C), which may result from the high protein level of ipa1-1D in Ri22. To confirm this, we examined the ipa1-1D protein levels in Ub:IPI1/Ri22 and found that it was decreased in young panicles and leaves, but increased in the shoot apexes (Figure 8A). These results provide further evidence that a complex tissue-specific regulation mechanism underling the IPI1-mediating polyubiquitination of IPA1 proteins and the miRNA regulated IPA1 transcripts determines rice architecture.
Figure 8.
Differential Ubiquitination Modes of IPA1-Mediated Complex Promoted by IPI1 in Different Tissues.
(A) Protein and transcript levels of ipa1-1D in Ri22 and Ub:IPI1/Ri22 transgenic plants. The Ponceau S staining was used as a protein loading control and the Ubiquitin as a transcript level control.
(B) to (G) IPI1 promotes the ubiquitination of IPA1-mediated complex with different polyubiquitin chains in shoot apexes ([B] and [C]), young panicles ([D] and [E]), and leaf ([F] and [G]). The total protein extracted from different tissues was immunoprecipitated with anti-IPA1 polyclonal antibodies and then detected by immunoblotting with different specific polyclonal antibodies for Ub (left), K48 ubiquitin chain (middle), or K63 ubiquitin chain (right). Total protein for immunoblot was shown in the Coomassie blue-stained SDS-PAGE gels. Arrow indicates IgG.
IPI1 Promotes Differential Ubiquitination of the IPA1 Complex in Shoot Apexes and Panicles
Proteins modified with different ubiquitin chains can undergo different cellular processes, which may differently determine the stability of target proteins (Kulathu and Komander, 2012). We then tested whether IPI1 could promote the modification of proteins targets with different types of ubiquitin chains. The transgenic plant Ub:IPI1/Ri22 was used to assay the polyubiquitination status of IPA1 due to a high level of IPA1 in Ri22 for better detection (Jiao et al., 2010). Proteins immunoprecipitated with anti-IPA1 polyclonal antibodies were immunoblotted with two specific ubiquitin antibodies, K48- and K63-Ub chain antibodies (Supplemental Figure 13). We found that in Ri22 plants, the proteins immunoprecipitated with anti-IPA1 antibodies could be tagged with the K48- and K63-polyubiquitin chains in shoot apexes and leaves (Figures 8B, 8C, 8F, and 8G), but mainly with the K48-polyubiquitin chain in young panicles (Figures 8D and 8E), suggesting that IPA1-mediated complex could be targeted with different polyubiquitin chains in vivo. Furthermore, in Ub:IPI1/Ri22 plants, the K63-polyubiquitination of IPA1-mediated complex was enhanced only in shoot apexes, while its K48-polyubiquitination was enhanced in young panicles and leaves. These results suggested that IPI1 could promote targeting of the IPA1 complex with different polyubiquitin chains, and the diverse fate of IPA1 proteins could in turn regulate downstream genes and determine plant architecture in different tissues (Figure 9).
Figure 9.
A Proposed Model of IPI1 Mediated Posttranslational Modification of IPA1.
IPI1 modulates IPA1 at the posttranscriptional level, which leads to different fates of IPA1 and thus alters rice plant architecture.
DISCUSSION
IPA1 acts as a pivotal regulator for plant architecture, and its overexpression could profoundly change rice plant architecture with reduced tiller number, enlarged panicle size, and enhanced grain yield (Jiao et al., 2010; Miura et al., 2010; Lu et al., 2013). Therefore, elucidating the molecular mechanism of IPA1 regulation is of great value for understanding the mechanisms underlying plant architecture formation. Previous studies have demonstrated that the expression level of IPA1 is regulated by miR156- and miR529-directed cleavage and epigenetic modifications (Jiao et al., 2010; Miura et al., 2010). In this study, we showed that the E3 ligase IPI1 could interact with IPA1, promote the differential polyubiquitination and degradation of IPA1 in different tissues and consequently alter plant architecture (Figures 1, 4, 5, and 8). These results provide new insight into the posttranslational regulation of IPA1 in the establishment of plant architecture in rice (Figure 9).
In panicles, overexpression of IPI1 promotes the polyubiquitination and degradation of IPA1, resulting in a similar phenotype as IPA1 RNAi transgenic plants. However, in shoot apexes, overexpression of IPI1 could promote the accumulation of IPA1 and repress rice tillering (Figures 4 and 5). Recent studies showed that different types of polyubiquitination could determine the variant destinations of target proteins, which enable more flexible and complex regulation to the stabilities of E3 ligase substrates (Chen and Sun, 2009; Kulathu and Komander, 2012). Proteins modified with K48-linked polyubiquitin chain usually go to the 26S proteasome pathway for degradation, while the K63-linked polyubiquitin chain labeled proteins may enter proteasome-independent pathways for signal transduction (Mukhopadhyay and Riezman, 2007). Our results show that for the posttranslational regulation by IPI1, the IPA1 complex in panicles is tagged with a K48-linked polyubiquitin chain, but in shoot apexes is tagged with a K63-linked polyubiquitin chain (Figure 8). Therefore, it appears that the differential polyubiquitination of K48 or K63 promoted by IPI1 may lead to different fates of IPA1 in different tissues, resulting in fine and precise regulation of rice plant architecture.
However, the mechanisms that modify IPA1-mediated complex with different polyubiquitin chains by IPI1 remain unclear. In the ethylene signaling pathway, the MKK9 cascade phosphorylates EIN3 on T174 to enhance its stability, whereas the MAPK pathway mediated by CTR1 phosphorylates T592 to promote the EIN3’s degradation via the proteasome (Yoo et al., 2008), suggesting that different phosphorylation forms will lead to different protein fates. Robust and comprehensive motif search algorithms have identified several putative phosphorylation sites for IPA1. The phosphorylation status might be an important event to determine the special ubiquitin chain ligated to IPA1 in different tissues. On the other hand, it has been noticed that an E3 ubiquitin ligase could recognize multiple E2s, and their interactions with different E2s may allow the synthesis of different types of ubiquitin conjugates.
In addition to the protein level modifications, the posttranscriptional regulation of IPA1 also shows tissue specificity. Both miR156 and miR529 could target the mRNA of IPA1 and trigger miRNA-mediated cleavage, while miR156 is preferentially expressed in seedlings while miR529 preferentially in panicles. This tissue-specific regulation of IPA1 at both the RNA and protein levels may indicate the importance and diverse functions of IPA1 in different tissues and organs. Therefore, a deep study of the specific modifications that can change the various fates of IPA1 in different tissues and organs will provide many benefits. Besides, overexpression of IPA1 will lead to an increase in panicle size but a decrease in rice tiller number, which limits the application of IPA1 in rice breeding. In this study, the elucidation of the different roles of IPI1 in regulating IPA protein abundance in different tissues provides a novel and realistic application for IPA1 in molecular breeding, by which the knocking out of IPI1 could increase both tiller number and panicle size and thus lead to high yield potential. This will greatly benefit the improvement of rice architecture in different varieties by applying different strategies of IPA1 utilization. Together, the IPI1-mediating ubiquitination of IPA1 and the miRNA-mediating posttranscriptional cleavage form a complex and precise regulatory network in fine-tuning plant development in rice and provide an important genetic resource for manipulating rice tillering and panicle size in molecular breeding.
METHODS
Plant Materials
Rice (Oryza sativa) ssp japonica variety Nipponbare, Ri22, and Ub:IPI1 transgenic lines were grown in either the greenhouse or experimental field of Institute of Genetics and Developmental Biology. The Ubi-IPI1 plasmid was introduced into Agrobacterium tumefaciens strain EHA105 and transformed into Nipponbare and Ri22 as previously reported (Hiei et al., 1994). Two independent Ub:IPI1 lines with increased expression of IPI1 were obtained and used in the further investigation.
The ipi1-1 and ipi1-2 plants were generated using CRISPR/Cas9 by Biogle Co. To minimize the potential off-target effects induced by CRISPR/Cas9, we performed two distinct transformations, using two sgRNAs targeting IPI1 at different locations: sg1516, 5′-GGCAGCCATTCCGCTTCCAA-3′, and sg1517, 5′-GAGAACTTACAGGTTACGGG-3′. The single sgRNA was created in the BGK03 vector containing Cas9, which was introduced into Agrobacterium strain EHA105 and transformed into TEIPEI309. Eight independent lines of sg1516 and sg1517 were obtained. To examine the function of CRISPR/Cas9 in vivo, genomic DNA was extracted from transgenic plants and primer pairs flanking the designed target site were used for PCR amplification (Supplemental Table 2). Sequence alignment revealed that two independent mutants, ipi1-1 and ipi1-2, were obtained (Supplemental Figures 9 and 10).
Constructs
In brief, target fragments were generated by PCR amplification using primers listed in Supplemental Table 2. The PCR products were digested with appropriate restriction enzymes and ligated into desired vectors. The vectors used in the yeast two-hybrid assay were pDBLeu for bait and pPC86 for prey (Clontech). Kanll-SCYCE and Hygll-SCYNE vectors were used for a BiFC assay. The coding regions of targets were inserted into the PGEX-6P-1 (GE Healthcare) vector or into PET-28a (Merck). Point mutation constructs were generated with the QuikChange site-directed mutagenesis kit (Stratagene).
Yeast Two-Hybrid Screen
The ProQuest System (Invitrogen) was used to screen for the interaction proteins of IPA1 following the manufacturer’s instructions. Plasmids were cotransferred into the yeast strain MAV203, and transformed cells were grown on SD medium without Leu, Trp, and His and containing 40 mM 3-amino-1,2,4-triazole. The positive clones were then confirmed by growing on SD medium without Leu, Trp, and Ura for 2 d.
Proteins were extracted from yeast cells according to the ProQuest System hand book (Invitrogen) using the cracking buffer containing 8 M Urea, 5% SDS, 40 mM Tris-HCl (pH 6.8), 0.1 mM EDTA, 0.4 mg/mL Bromophenol blue, and cocktail protease inhibitors (Roche; 11873580001) and then immunoblotted with anti-IPA1 and anti-IPI1 polyclonal antibodies.
GST Pull-Down Assay
Total proteins were extracted from rice suspension culture cells by a pull-down buffer containing 40 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 0.4 M sucrose, 1 mM EDTA, 1 mM DTT, 0.2% Triton-X100, and cocktail protease inhibitors (Roche; 11873580001). The pull-down assay was performed according to a previous method (Nakashima et al., 2008) with some modifications. Briefly, the homogenate was centrifuged at 13,000g for 30 min, and the supernatant was collected for affinity chromatography. The purified GST-IPI1 or GST protein was bound to glutathione sepharose 4FF beads (GE Healthcare; 17-5132-01). The total protein was precleared with the glutathione Sepharose 4FF beads and then loaded on a GST-IPI1 or GST affinity column. After washing with the pull-down buffer three times, an equal volume of the 2× SDS buffer was added and samples were boiled at 100°C for 10 min. Then the immunoblotting analysis was performed with anti-IPA1 polyclonal antibodies.
Transient Expression in the Leaf Protoplasts
For transient expression, plasmids were introduced into rice protoplasts prepared from leaves by polyethylene glycol-mediated transformation (Bart et al., 2006). After incubation in the dark for 16 h, the visible signals were examined under a confocal microscope (Nikon A1).
In Vitro Ubiquitination Assay
The coding sequence of IPI1 was cloned into the vector PGEX-6P-1 and then transferred into Escherichia coli stain BL21 (DE3) cells. The fusion proteins were purified based on the manufacturer’s instructions. The IPI1(H74Y) and IPI1(C80S) were prepared with the QuikChange site-directed mutagenesis kit (Stratagene) according to the protocol. The ubiquitination assay was performed as described previously (Qin et al., 2008) with some modification: 1 μg GST-IPI1, GST-IPI(H74Y), GST-IPI1(C80S), or GST protein were incubated in a 20-μL reaction mixture containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 2 mM DTT, 5 mM ATP, 3 mM creatine phosphate, 1 unit creatine kinase, 100 ng E1 (Boston Biochem; E-305), 100 ng E2 (Boston Biochem; E2-607), and 4 μg ubiquitin. To confirm IPI1-mediated ubiquitination of IPA1, the IPA1 coding sequence was constructed into the vector PET28a (Novagen), and expressed His-IPA1-His was purified with Ni Sepharose 6 Fast Flow (GE Healthcare; 17-5318-06). Purified His-IPA1-His protein (2 μg) was added to the reaction mixture and incubated at 30°C for 1.5 h, stopped with adding of 5× SDS sample buffer and boiling at 100°C for 10 min. The mixtures were then subjected to the 10% (w/v) SDS-PAGE gel and immunoblotting analysis.
Detection of IPA1 Ubiquitination in Vivo
In vivo ubiquitination of IPA1 proteins was assayed as described previously (Lee et al., 2009) with some modifications. Briefly, samples were ground into powder in liquid nitrogen and then homogenized with protein extraction buffer NB1 containing 50 mM Tris-MES (pH 8.0), 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, and cocktail protease inhibitor (Roche; 11873580001) (Liu et al., 2010). The crude extracts containing 500 μg proteins were coincubated with anti-IPA1 polyclonal antibodies and 50 μM MG132. After gentle shaking for 1 h, 30 μL protein agarose beads (GE Healthcare) was added into the mixtures and incubated for another 1 h with gentle shaking, and then the agarose beads were washed with NB1 buffer three times. After an equal volume of 2× SDS buffer was added and boiled at 100°C for 10 min, the sample was loaded on a 10% (w/v) SDS-PAGE and visualized by staining with Coomassie Brilliant Blue or immunoblotted with anti-Ub (Abcam; ab7254), anti-K48 ubiquitin chain, or anti-k63 ubiquitin chain polyclonal antibodies (Millipore; 05-1307 and 05-1308).
In Vitro Degradation Assay
A cell-free protein degradation assay was performed as previously described (Spoel et al., 2009). Briefly, the total protein was extracted from cultured cells with proteolysis buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 10 mM ATP, 5 mM DTT, and 0.05 mg/mL cycloheximide), and the cell lysate was incubated with the inhibitors PMSF (4 μM), MG132 (40 μM), or organic solvent DMSO at 30°C for 2 h. Finally, after the 5× SDS buffer was added and boiled at 100°C for 10 min, the sample was loaded on 10% (w/v) SDS-PAGE for immunoblotting with anti-IPA1 antibody (Jiao et al., 2010).
To detect the stability of IPA1 in shoot apex and young panicles, total proteins were extracted from the tissues as indicated with the proteolysis buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 10 mM ATP, 5 mM DTT, and 0.05 mg/mL cycloheximide). The cell lysate was incubated at 30°C for different times as indicated. Finally, the 5× SDS buffer was added and boiled at 100°C for 10 min, and then the sample was loaded on a 10% (w/v) SDS-PAGE for immunoblotting with anti-IPA1 antibody.
To examine the stability of IPA1 in vivo, 4-week-old seedlings treated with 50 μM MG132 at different time points as indicated. The samples were ground into powder in liquid nitrogen and then homogenized with the DB protein extraction buffer (4 M urea, 0.1% Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 1 mM PMSF [optional]) (Liu et al., 2010). The supernatant was boiled with 5× SDS buffer at 100°C for 10 min. Immunoblotting assay was performed with anti-IPA1 polyclonal antibodies.
In Vivo Degradation Assay
An in vivo protein degradation assay was performed as previously described (Liu et al., 2010). Agrobacterium cells carrying the MYC-IPI1 and IPA1 plasmids were coinfiltrated into the same area of Nicotiana benthamiana leaves with the MYC plasmid as a control and the HA-GFP plasmid as an internal control. Two days after infiltration, samples were harvested for analysis. Concentration of IPA1 was detected with anti-IPA1 polyclonal antibodies (Jiao et al., 2010) and anti-GFP and anti-MYC monoclonal antibodies (Roche; 11814460001 and 11667149001). The transcripts of ACTIN were analyzed as control by RT-PCR as previously described (Liu et al., 2010).
RNA Extraction, RT-PCR, and Quantitative Real-Time PCR
Total RNA was prepared with a TRIzol Kit (Life Technology; 15596-018) according to the user’s manual. RNA samples (2 μg each) were treated with DNase I (Toyobo) and then subjected to the SuperScript III first-strand synthesis system (Life Technology; 18080-051). The primers used for RT-PCR and quantitative real-time PCR are listed in Supplemental Table 2. UBIQUITIN (LOC_Os03g13170) was used as an internal control. The quantitative real-time PCR was detected with SYBR Green Kit (Qiagen; 208054) in a real-time PCR apparatus (Bio-Rad CFX96).
Protein Extraction and Immunoblotting Analysis
Samples were collected and grounded into powder in liquid nitrogen and resuspended in DB buffer on ice. Extracts were centrifuged at 13,000g at 4°C for 10 min, and the supernatant was boiled with 5× SDS buffer at 100°C for 10 min. Immunoblotting assay was performed according to the protocol of GE. The protein level of Actin (Affinity; T0022 β-Actin) was used as an internal control. Two independent replicates were performed.
Preparation of Polyclonal Antibodies
The IPI1 polyclonal antibodies were produced by Kangweishiji Co., and the quality was tested with expressed Myc-IPI1 and Myc-LOC_Os05g06270 (another RING finger E3 ligase) in tobacco leaves. The immune serum was purified using Protein A/G agarose (Thermo; 20421) following the instructions and used for immunoblotting analysis (Supplemental Figure 13).
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project or GenBank/EMBL databases under accession numbers LOC_Os08g39890 (IPA1) and LOC_Os01g24880 (IPI1).
Supplemental Data
Supplemental Figure 1. The SBP Domain of IPA1 Is Essential for the Interaction between IPA1 and IPI1.
Supplemental Figure 2. Scheme of the IPI1 RING Finger Composition.
Supplemental Figure 3. Expression and Phenotype of Ub:IPI1 Transgenic Plants.
Supplemental Figure 4. Overexpression of IPI1 Affects Plant Architecture.
Supplemental Figure 5. Overexpression of IPI1 Affects Plant Architecture via Modulating IPA1 Abundance.
Supplemental Figure 6. IPA1 Negatively Regulates Tiller Numbers.
Supplemental Figure 7. The IPI1 Expression Pattern.
Supplemental Figure 8. Overexpression of IPI1 Driven by its Native Promoter Affects Plant Architecture via Modulating IPA1 Abundance.
Supplemental Figure 9. Identification of ipi1-1 Mutant Plants.
Supplemental Figure 10. ipi1-2 Mutant Shows Altered Plant Architecture as ipi1-1.
Supplemental Figure 11. Mutation in ipa1-1D Has No Effect on Polyubiquitination Mediated by IPI1.
Supplemental Figure 12. Overexpression of IPI1 in the ipa1-1D Background Affects Plant Architecture.
Supplemental Figure 13. Specificity of K48- or K63-Polyubiquitin Antibodies.
Supplemental Figure 14. Specificity of IPI1 Polyclonal Antibodies.
Supplemental Table 1. List of Putative Proteins Interacting with IPA1.
Supplemental Table 2. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the assistance in the assays in tobacco. This work was supported by the National Key Research and Development Program of China (Grant 2016YFD0101800), the National Natural Science Foundation of China (Grant 91335204), and the Chinese Academy of Sciences (Grant XDA08030101).
AUTHOR CONTRIBUTIONS
J.W. and H.Y. designed experiments and analyzed data. J.W., Z.L., Y.J., X.M., G.L., and X.C. performed the experiments. G.X., H.Y., Y.W., and J.L. analyzed the data and wrote the article. J.L. designed the research and conceived and supervised the project.
Glossary
- BiFC
bimolecular fluorescence complementation
Footnotes
Articles can be viewed without a subscription.
References
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Li H., Zhao Q., Jiang H., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Wang D., Li C. (2009). The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 150: 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33: 275–286. [DOI] [PubMed] [Google Scholar]
- Cheng M.C., Hsieh E.J., Chen J.H., Chen H.Y., Lin T.P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Seo D.H., Kang B.G., Kim W.T. (2011). The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 157: 2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A.L. (1998). The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 95: 2727–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Ikeda F., Dikic I. (2008). Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 9: 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.-H., Park S., Zhai J., Gurazada S.G.R., De Paoli E., Meyers B.C., Green P.J. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kim W.T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162: 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y., Komander D. (2012). Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13: 508–523. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhong S., Li G., Li Q., Mao B., Deng Y., Zhang H., Zeng L., Song F., He Z. (2011). Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res. 21: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.L., Lim G.G. (2011). K63-linked ubiquitination and neurodegeneration. Neurobiol. Dis. 43: 9–16. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Lu Z., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205. [DOI] [PubMed] [Google Scholar]
- Mural R.V., Liu Y., Rosebrock T.R., Brady J.J., Hamera S., Connor R.A., Martin G.B., Zeng L. (2013). The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25: 3615–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Chen S., Shirsekar G., Zhou B., Khang C.H., Songkumarn P., Afzal A.J., Ning Y., Wang R., Bellizzi M., Valent B., Wang G.L. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24: 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M., Fushman D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8: 610–616. [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M.Y., Cho S.K., Kim W.T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154: 1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.