Plants have evolved diverse mechanisms to reduce the growth/defense trade-off, involving antagonistic crosstalk among hormones and fine-scale regulation of R genes.
Abstract
Plants have evolved an array of defenses against pathogens. However, mounting a defense response frequently comes with the cost of a reduction in growth and reproduction, carrying critical implications for natural and agricultural populations. This review focuses on how costs are generated and whether and how they can be mitigated. Most well-characterized growth-defense trade-offs stem from antagonistic crosstalk among hormones rather than an identified metabolic expenditure. A primary way plants mitigate such costs is through restricted expression of resistance; this can be achieved through inducible expression of defense genes or by the concentration of defense to particular times or tissues. Defense pathways can be primed for more effective induction, and primed states can be transmitted to offspring. We examine the resistance (R) genes as a case study of how the toll of defense can be generated and ameliorated. The fine-scale regulation of R genes is critical to alleviate the burden of their expression, and the genomic organization of R genes into coregulatory modules reduces costs. Plants can also recruit protection from other species. Exciting new evidence indicates that a plant’s genotype influences the microbiome composition, lending credence to the hypothesis that plants shape their microbiome to enhance defense.
INTRODUCTION
There are spectacular examples of highly defended plants in which a large percentage of biomass is devoted to chemicals that ward off pests (Kempel et al., 2011; Züst et al., 2015). That this metabolic expenditure entails a concomitant decrease in growth and/or reproductive output is hardly surprising. Compensation of costly expenditure is well known in agriculture, where breeders have successfully overcome costs associated with novel resistance traits (Legg et al., 1965; Chaplin and Mann, 1978; Simmonds, 1989; Krattinger and Keller, 2016) and in molecular plant biosciences, where researchers have uncoupled putative growth-resistance trade-offs in the laboratory (Campos et al., 2016). A meta-analysis of the fitness detriment associated with resistance traits in plant populations revealed evidence of a cost of resistance against either herbivores, pathogens, or weeds in only 50% of case studies (Bergelson and Purrington, 1996). How can we reconcile observations of high metabolic costs with the ability to overcome them?
It is often assumed that negative correlations between growth and defense result from pleiotropic effects of a resistance trait. However, a significant fraction of the observations of growth-defense trade-offs may be the consequence of genetic linkage (Bergelson and Purrington, 1996). Indeed, it is well established in agronomy that physiological traits are genetically linked and can be uncoupled with successive rounds of breeding (Bergelson and Purrington, 1996; Monforte and Tanksley, 2000; Brown, 2002; Fu et al., 2013; Hurni et al., 2013). However, when traits are too tightly linked (for example, within a genomic island), breeding strategies may fail to uncouple them. In these cases, it is difficult to distinguish trade-offs due to linkage from those due to pleiotropy.
In this review, we focus on those trade-offs that result from pleiotropy and for which the mechanism underlying the growth-defense trade-off is understood. We begin with the premise that only in rare cases do hosts get something for nothing. Energy diverted toward the production of defense is not available for other needs; thus, trade-offs should typically be inevitable. These costs may be evident in terms of fitness or in terms of success in a disparate ecological setting. The latter are called “ecological costs” (Heil, 2002; Heil and Baldwin, 2002; Strauss et al., 2002) and will be discussed only briefly here. For the purposes of this review, we will consider both biomass accumulation and the production of reproductive tissue to be allocation into “growth.”
We present several strategies that can be used to minimize these costs. These strategies are diverse, reflecting the various mechanisms underlying growth-defense trade-offs. At the same time, we detail obstacles that prevent the full mitigation of costs. The resultant magnitudes of costs have profound effects on selection and, thus, on the evolutionary history of particular loci. Patterns of sequence polymorphism may therefore point toward loci harboring costs, although genetic architecture has also been shown to influence both costs and evolutionary history. We discuss these population level and genomic consequences of costs of immunity.
Finally, we detail those rare instances when hosts co-opt the protection afforded by other species for their own gain. These examples are the closest that one might imagine to cost-free immunity.
ARE TRADE-OFFS INEVITABLE?
It is well appreciated that divergent mechanisms lead to negative correlations between defense and growth. Whether a trade-off can be ameliorated depends on its underlying mechanism, and frequently depends on the environment.
The Role of Nutrient Limitation in Growth-Defense Trade-Offs
Both growth and immunity involve the expression of thousands of genes (Thilmony et al., 2006) and the biosynthesis of myriad compounds (Bennett and Wallsgrove, 1994). If nutrients are limiting, allocation of a nutrient to defense may come with a reduction in allocation to growth.
There is substantial evidence that nutrients influence the growth-defense relationship. For example, Bergelson (1994) grew resistant and susceptible genotypes of a lettuce variety under nutrient-poor and nutrient-rich conditions. As expected with a growth-defense trade-off that is influenced by nutrient allocation, Bergelson found that nutrient-poor conditions reduced the development of reproductive tissue significantly more in the resistant genotype than in the susceptible genotype.
Several nutrients, when limiting, are likely to influence the relationship between growth and defense. For example, defense responses that involve salicylic acid, auxin, glucosinolates, and methyl transferases all rely directly or indirectly on sulfur availability and can involve the upregulation of genes related to sulfur metabolism (Kruse et al., 2007). Király et al. (2012) tested the effect of sulfur addition on the resistance response of Nicotiana tabacum to Tobacco mosaic virus. The presence of sulfur reduced necrotic symptoms associated with the hypersensitive response (HR), thereby likely reducing the cost of mounting a resistance response. Access to nitrogen and phosphorous, two nutrients that are frequently limiting, can similarly influence allocation to defenses (Baldwin et al., 1998; Van Dam and Baldwin, 1998; Zhu et al., 2016). Additional abiotic conditions including drought, temperature, and humidity influence whether a trade-off is observed (Bergelson, 1994; Ohnmeiss and Baldwin, 1994; Heil and Baldwin, 2002; Heidel et al., 2004; Alsdurf et al., 2013). The availability of energy and nutrients is also contingent on interactions with surrounding plants. Cipollini (2002) grew Arabidopsis thaliana alone and in combination with other plant species and then compared the effect of induced defenses on plant growth. The study found significantly reduced growth rates for Arabidopsis plants upregulated for defense, with this effect being greater when the focal plant was competing with other species. These results suggest that competition for resources, or plant-plant signaling, shape the apparent costs of defense induction.
Ecology Influences the Optimal Growth-Defense Relationship
The competing demands imposed by enemies and competitors, as discussed above, illustrate how conditions beyond bioenergetic pulls can dictate the optimal immune response (Kliebenstein, 2016). The age of a plant at the time of pathogenic attack also factors into defining the “optimal” defense response. Imagine a young plant that is energy limited, suffering attack, and finds itself in a highly competitive environment where early growth is essential. The best strategy for this plant may well be to forego defense and grow, at least provided the pathogen is not too virulent. By contrast, that same plant in a sparse field or at an older age may do better to defend itself. In other words, the “optimal” allocation to defense is not static but should be responsive to ecological and phenological conditions (McDowell et al., 2005; Krasileva et al., 2011).
These examples serve to illustrate the contingency of the growth-defense trade-off on the ecology of the plant. It is thus reasonable to expect that a physiological coregulation of growth and immunity that extends beyond energetic pulls could be adaptive.
Coregulation of Growth and Immunity
Only a fraction of trade-offs that are understood mechanistically to date involve nutrient limitation. A major insight from molecular biological studies is that growth and immunity are tightly coregulated, and many negative correlations between growth and defense are the result of regulatory crosstalk (Denancé et al., 2013; Huot et al., 2014).
Evolutionary reasons for the tight coordination between growth and defense remain largely unexplored. It is easy to find anecdotes that imply selection for the coordination of growth and defense. In resistance to blight in rice (Oryza sativa), an intercellular sucrose transporter was recently identified as a major susceptibility locus (Eom et al., 2015). This transporter moves sugars from photosynthetic tissues of a rice plant into the phloem for transport to tissues that require externally supplied sugar for growth and development. A side effect of transporting sugars outside of the plant cell, however, is that the sugars are readily available to endophytes, providing them an abundant food source (Chen et al., 2010; Eom et al., 2015). In this case, plant traits necessary for growth (such as the transport of nutrients) increase susceptibility to microbial invasion. The result is a conflict between the proper distribution of nutrients throughout the plant and the ability to defend against pathogen expansion. In this scenario, limiting growth while infected could reduce the susceptibility to blight.
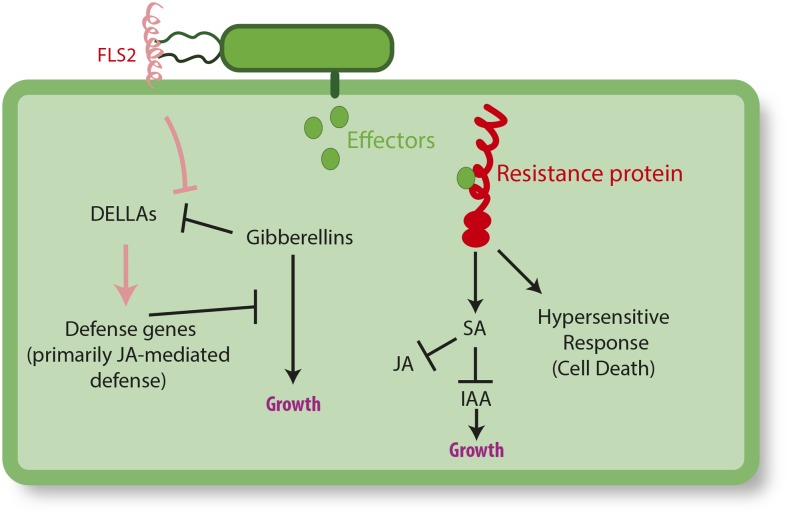
To understand the fine-scale physiological coordination between growth and immunity, consider the activity of gibberellins in the presence versus the absence of microbes. Gibberellins are a class of phytohormones involved in breaking seed dormancy, seed development, and vegetative and floral growth (Davière and Achard, 2013). Gibberellin production destabilizes a class of growth repressing-proteins (DELLAs), thereby promoting growth. When a microbe is detected, however, an immune cascade is initiated that overrides the destabilizing activity of gibberellins, and reestablishes DELLA-mediated suppression of growth (Navarro et al., 2008). The consequence of initiation of defense is the suppression of gibberellin-mediated development (Figure 1).
Figure 1.
Examples of Immune Receptor Crosstalk with Plant Development.
A simplified cartoon illustrating the response of a plant to recognition of pathogen via two different immune proteins: recognition of bacterial flagellin via FLS2 and recognition of a bacterial effector via an R protein. Detection of flagellin by FLS2 causes the stabilization of DELLA proteins, the upregulation of defense (primarily JA-mediated defenses), and the downregulation of gibberellin-mediated growth (Zentella et al., 2007; Navarro et al., 2008). Recognition of bacterial effectors by the corresponding R protein causes upregulation of SA, and downregulation of indole-3-acetic acids (IAAs; involved in growth promotion) and of JA (involved in resistance to necrotrophs and herbivores). The recognition by the R protein also induces HR, a type of programmed cell death in plants. In summary, the recognition of non-self by immune receptors results in both the downregulation of several growth pathways but also in the upregulation of defense genes and cell death. Note, there is additional crosstalk between IAA, SA, JA, and HR not depicted here. The interactions illustrated here are not exhaustive and instead are a small subset of the extensive crosstalk interactions that have been elucidated (for thorough review, see Denancé et al., 2013; Huot et al., 2014).
This type of repressive regulation has been observed for several phytohormones. The two phytohormones most frequently implicated in the immune responses to natural enemies are salicylic acid (SA) and jasmonic acid (JA). SA is a phenolic phytohormone produced in response to challenge by biotrophic and hemibiotrophic pathogens (Glazebrook, 2005). Its production leads to the upregulation of defense pathways, as well as to systemic acquired resistance (Gaffney et al., 1993). The production of SA influences not only resistance to microbes, but also leaf morphology, root development, and floral development (Rivas-San Vicente and Plasencia, 2011). The effect of SA on development is complex. For example, SA treatment was found to both increase and decrease root and vegetative growth depending on plant species and concentration (Rivas-San Vicente and Plasencia, 2011). JA is also a key regulator of defense responses, with its production inhibiting herbivory and colonization by necrotrophic pathogens. Like SA, JA production influences myriad other processes in plant development (Creelman and Mullet, 1995), acting as a promoter of senescence and an inhibitor of root growth (Wasternack, 2007). SA and JA are coregulated within a larger network of phytohormones including auxins, abscisic acid, and gibberellins, all of which are involved in regulating immunity (Denancé et al., 2013) and engage in crosstalk with SA and JA. SA reduces the production of auxins by downregulating pathways involved in auxin generation (Wang et al., 2007). JA alters the distribution of auxins throughout plant tissues by interfering with auxin transport (Huot et al., 2014).
SA and JA antagonize not only stereotypical growth-related phytohormones but also one another. Antagonism between SA and JA production is thought to be a central trade-off in the immune response (Pena-Cortés et al., 1993; Denancé et al., 2013; Huot et al., 2014) albeit dependent both on concentration and genetic background (Thaler et al., 2002; Mur et al., 2006). Given these and many other antagonistic activities (reviewed in Huot et al., 2014), it comes as no surprise that immunity and growth phenotypes are often negatively correlated.
Note that crosstalk between phytohormones, as it is currently understood, is primarily the result of transcriptional and translational coregulation, rather than the limited availability of a common precursor. The suppression of the gibberellin-mediated growth described earlier was the result of repression and induction of regulatory pathways (Navarro et al., 2008). The fact that phytohormonal changes can also alter nutrient metabolism (Gibson, 2004) raises the possibility that phytohormone-induced changes in growth could result (in part) from downstream reallocation of limiting nutrients. Nonetheless, several negative relationships between immunity and growth phytohormones have been successfully uncoupled (Spoel et al., 2007; Campos et al., 2016; Liu et al., 2016). It is questionable whether the uncoupling of this coregulation would benefit the fitness of plants: The tight coregulation of phytohormones, at least in the case of SA and JA, is largely conserved across land plant species, suggesting that their coregulation is important for fitness (Thaler et al., 2012).
Growth-defense coregulation is a pervasive mechanism underlying growth-defense trade-offs (Thaler et al., 2002; Denancé et al., 2013; Huot et al., 2014), having been observed in diverse plant species (Hayat et al., 2010; Rivas-San Vicente and Plasencia, 2011). Going forward, if we wish to determine whether a growth trade-off can be overcome, it will be necessary to determine under which circumstances growth-defense coordination is adaptive. Perhaps insight can be gleaned by exploiting natural variation to compare these trade-offs among accessions adapted to divergent environments.
CAN DEFENSE COSTS BE REGULATED?
Many types of costs simply cannot be overcome by rewiring. However, plants employ strategies that reduce their allocation toward defense. One such strategy is controlling the timing and duration in which the immune response is active. Fine control enables a plant to focus the immune response to times when it is most needed. Here, we detail several mechanisms to regulate defense responses and discuss the conditions that may favor each.
Induced Responses upon Attack
Plants can limit the costs of resistance by expressing some defenses only upon infection. Indeed, two central modes of plant immunity are induced, rather than constitutive. In microbe- or pathogen-associated molecular pattern (MAMP/PAMP)-triggered immunity, plant pattern recognition receptors detect conserved features of many pathogens, such as the bacterial flagellin protein, leading to the induction of defenses that limit further pathogen colonization. Arabidopsis, for instance, upregulates over 1100 defense genes upon treatment with the synthetic flagellin peptide flg22 (Zipfel et al., 2004) (Figure 1). Pathogens that survive or evade this level of defense may inject proteins known as effectors into plant cells; these effectors serve as virulence factors. If plant resistance (R) proteins detect the effectors, effector-triggered immunity results. This form of induced immunity typically leads to targeted plant cell death at the site of infection, known as the HR, to limit the spread of the pathogen (Jones and Dangl, 2006) (Figure 1).
Although PAMP- and effector-triggered immunity can reduce the costs of defense relative to constitutive expression, these forms of immunity take time to deploy and can furthermore be evaded (Pel et al., 2014), thus allowing some pathogenic growth. Plants have evolved additional mechanisms that fine-tune metabolic expenditures on defense. These strategies include defense priming and transgenerational defense induction, which we review below.
Defense Priming
Defense priming is one way in which plants mitigate the costs of defense. Plants exposed to herbivory, pathogen infection, or a chemical elicitor of defense can assume a physiological “state of readiness” that enables a more effective response to subsequent attack relative to individuals that were not previously induced (Pastor et al., 2013). Such primed plants may respond to a subsequent stress by inducing defenses earlier, faster, more strongly, or in response to lower levels of infection (Hilker et al., 2016). Priming thus serves to maintain a heightened defensive state without incurring the costs of sustaining a full-scale defense response.
Defense priming has been identified in multiple plant species in response to diverse herbivores and microbes (reviewed in Pastor et al., 2013). For instance, inoculation of Arabidopsis Col-0 plants with avirulent Pseudomonas syringae (strain PmaDG6/avrRpt2) enabled greater resistance to attack by virulent P. syringae 2 d later (strain PmaDG3; Jung et al., 2009). Note that the cost:benefit ratio of priming is likely to depend on the plant’s physiological state during the priming stimulus, which may be influenced by various factors including abiotic stresses (Hilker et al., 2016). There has been little empirical work on the cost-to-benefit ratio of defense priming itself, but existing evidence suggests that the benefits outweigh the costs. For instance, van Hulten and colleagues (2006) found that priming of Arabidopsis plants with the SA-pathway elicitor β-amino butyric acid incurred only modest costs in the absence of infection, reflected by slight decreases in plant growth without detectable effects on fecundity. The edr1-1 mutant constitutively expressed the priming phenotype and similarly incurred slight fitness costs, but these costs were substantially lower than the fitness costs observed in a mutant (cpr1-1) that constitutively expressed full defenses (see also Wang et al., 2015; Walters et al., 2008). A field study of Arabidopsis found that pretreatment of plants with SA both reduced leaf bacterial population sizes and increased plant fitness compared with controls, indicating that the benefits of these induced defenses outweighed their costs given the natural levels of infection at the time and location studied (Traw et al., 2007).
Mechanisms of priming are diverse and vary depending on both the type of initial stimulus and the pathogen subsequently encountered. Such mechanisms include accumulation of mitogen-activated protein kinases, transcription (co)factors, hormones, pattern-recognition receptors, and other defense-related proteins and metabolites that can be activated quickly in response to infection (Conrath et al., 2015). Modifications of histone proteins also appear to play a role in defense priming. For instance, treatment of plants with a chemical elicitor of defenses induced histone modifications that are characteristic of expressed genes in the promoter of the defense gene WRKY29 (Jaskiewicz et al., 2011). However, this gene is not expressed until a further stress stimulus is applied. P. syringae infection is also known to induce widespread changes in DNA methylation that correlate with the expression of defense genes (Dowen et al., 2012; Yu et al., 2013). Thus, it appears that a priming stimulus can induce permissive chromatin states that poise defense genes for rapid expression upon subsequent attack. Interestingly, chemical elicitors of defense and pathogen infection may induce the “same” defense phenotype via different molecular and physiological mechanisms (Balmer et al., 2015). This observation raises the question of whether alternative mechanisms of priming differentially influence the costs of immune responses. Such knowledge may be useful for agricultural applications, especially given the interest in using defense priming to enhance disease resistance in crops (Conrath et al., 2015).
Transgenerational Defense Induction
Both ecologists and physiologists have discovered that defenses induced in parents can be inherited by offspring (reviewed in Holeski et al., 2012). Such transgenerational defense induction can take at least two forms. First, primed defensive states induced in parents can be transmitted to offspring, allowing them to deploy stronger, more effective defenses. For instance, treatment of parental Arabidopsis plants with either the bacterial pathogen P. syringae or a chemical elicitor of defense enabled offspring to express stronger immune responses to both P. syringae and the oomycete Hyaloperonospera arabidopsidis (Hpa) (Luna et al., 2012; Slaughter et al., 2012; López Sánchez et al., 2016). Second, transgenerational defense induction can take the form of constitutive expression in offspring. For example, when the leaves of parental Mimulus guttatus plants were damaged by simulated herbivory, offspring increased production of defensive leaf trichomes in the absence of damage to their own leaves (Holeski, 2007; Scoville et al., 2011). Transgenerational defense induction may incur costs if offspring do not experience infection or herbivory (Agrawal, 2001), but any such costs likely depend on whether priming or a constitutively expressed phenotype is involved.
The mechanisms that transmit induced defenses to offspring require further investigation, but existing evidence suggests that induced, heritable epigenetic changes underlie these effects. Mechanistic studies of transgenerational priming reveal that mutations that impair the establishment and maintenance of DNA methylation marks cause global hypomethylation, allowing the mutants to achieve similar levels of disease resistance as transgenerationally primed wild-type plants (Luna et al., 2012; Luna and Ton, 2012; Slaughter et al., 2012; López Sánchez et al., 2016). Such hypomethylation primes the expression of SA pathway genes, such as PATHOGENESIS-RELATED GENE1 (Kathiria et al., 2010), although this increased SA-based resistance trades off with increased susceptibility to necrotrophic pathogens controlled by the JA pathway (López-Sánchez et al., 2016). Transgenerational priming may therefore enhance disease resistance with little to no cost, but only in certain ecological contexts. DNA methylation also mediates the effects of leaf wounding in previous generations on trichome production in M. guttatus (Akkerman et al., 2016). Together, these results suggest that heritable changes to DNA methylation (or to small RNAs that can direct methylation) are involved in the expression of transgenerational priming.
What Is the Optimal Regulation?
The three types of defense regulation described here—directly induced responses, priming, and transgenerational defense induction—occur over different time frames. Directly induced responses act to suppress an ongoing infection. Priming allows for faster induction of future responses following successful resistance to an initial attack. Transgenerational memory has the potential to prevent an infection in a future generation, either as a primed or constitutively expressed phenotype. Thus, the relative value of these strategies will be intertwined with the temporal patterning of enemy attack. Pathogens have a range of specificities, and the frequency of infection differs among pathogen and plant species (Barrett et al., 2009). Direct induction makes most sense when the ability of pathogens to increase in titer is slow relative to the induction of defense.
Priming is likely to be selectively favored when there is some degree of predictability of future enemy attack based on current conditions. Consider the cyclical outbreaks of caterpillars in the northeastern US (Dwyer, 1994). Such outbreaks are recurrent as well as temporally and environmentally predictable (Dwyer and Elkinton, 1993). This recurrence could select for induction mechanisms that allow for more poised responses such as priming. Mathematical models indicate that such adaptive anticipatory effects can evolve even if the degree of predictability is modest (Sultan and Spencer, 2002). Transgenerational memory of pathogen attack makes the most sense when there is generation-to-generation predictability in attack rates. A multitude of ecological factors can influence the predictability of attack rates, including the relation of plant dispersal distances to the spatial heterogeneity of plant enemies and the environmental conditions that influence their presence (Herman et al., 2014). For instance, transgenerational defense induction may be more likely to evolve in species with short dispersal distances, in which offspring are more likely to establish in a microsite that is similar to that of their parents.
Notably, the behavior of priming machinery in field settings is very poorly understood. Studies of mechanisms of immune system regulation have been largely limited to the laboratory (with notable exceptions; see Heil and Baldwin, 2002). Plants in nature are continuously exposed to a diverse and variable array of pathogens and herbivores Consequently, the adaptive value of both within-generation priming and transgenerational defense induction likely depends on the ability to reset these states when necessary (Crisp et al., 2016). For instance, a plant that has been transgenerationally primed for response to biotrophic pathogens may not be able to efficiently respond to necrotrophs due to trade-offs between SA and JA pathways. Resetting the primed state may be adaptive in this scenario, depending on the relative burden imposed by biotrophs versus necrotrophs. These considerations raise the issue of whether we should expect induced defensive states to persist over multiple generations in nature, as has been demonstrated in the laboratory (Luna et al., 2012; Akkerman et al., 2016). For instance, if a specific pathogen or herbivore exposure is consistent, then a primed effect is constantly reprimed. Multigenerational stability of the primed state would be unnecessary and may be maladaptive if ecological conditions change (Herman et al., 2014). However, even nonpathogenic microbes encode molecular patterns that are recognized by plants and that can trigger immune responses (Zipfel et al., 2004; Vetter et al., 2016). Hence, it is possible that some components of priming machinery are triggered in all field settings. An unprimed state could be limited to recently germinated seedlings or to the laboratory. Future work is needed to determine the predictability of enemy attack within and across generations, levels of immune system activation, and effects on fitness in the field.
DEPENDENCE ON A SURVEILLANCE SYSTEM: CASE STUDY OF R GENES
One class of immune genes, R genes, has been repeatedly implicated in the growth-defense trade-off across several plant species (Tian et al., 2003; Yamamoto et al., 2010; Chae et al., 2014; Sicard et al., 2015). R genes mostly encode intracellular immune receptors containing nucleotide binding leucine-rich repeats (NB-LRRs or NLRs). Extensive research on the evolution of these loci and the mechanisms underlying their associated costs provides an insightful case study of the evolution of growth-defense trade-offs.
In this section, we review what is known about the mechanisms underlying the trade-offs associated with R genes, the regulation of these trade-offs, and the evolution of loci to ameliorate these costs. The extensive genetic complexity and variation of these loci complicate their adaptive evolution, illustrating that many costs are likely unavoidable.
R-Gene Expression Can Cause Growth-Defense Trade-offs
R gene products recognize the presence of particular microbes. This recognition induces localized cell death (HR), a type of programmed cell death conserved across plants (Mur et al., 2008) (Figure 1). Although HR plays a critical role in restricting pathogen proliferation at the site of infection, uncontrolled cell death ultimately reduces fitness, as exemplified in a series of lesion mimic mutants and autoimmune syndromes in plants (Bomblies and Weigel, 2007; Bruggeman et al., 2015). HR is often accompanied by elevated levels of the defense hormone SA, which in turn induces systemic acquired resistance at a later phase during infection (Fu and Dong, 2013). Fitness penalties due to SA induction have been revealed by introducing the SA-degrading NahG into the various lesion mimic mutants; these mutants experienced a partial suppression of their lesion phenotype and a concomitant increase in fitness (Aviv et al., 2002; Shirano et al., 2002; Mosher et al., 2010).
Growth-defense trade-offs resulting from R gene expression depend on their level of expression, the effects of genetic background, and environmental conditions. Ectopic expression of several R genes has been shown to trigger HR and to do so in a genetic background-dependent manner (Oldroyd and Staskawicz, 1998; Yang and Hua, 2004; Yi and Richards, 2008; Kim et al., 2010; Palma et al., 2010; Chae et al., 2014), suggesting that plant genotypes fine-tune their basal R gene expression levels. Such ectopic R activity is often temperature dependent, and high or fluctuating temperature suppresses the symptoms most evidently in growth (Yang et al., 2010; Huang et al., 2010; Świadek et al., 2017). Basal R gene expression is presumably regulated at a low level sufficient for monitoring of non-self-mediated changes in the plant cell while minimizing costs of expression. Biotic stresses can induce R gene expression and the upregulation in general correlates with enhanced resistance (Dinesh-Kumar and Baker, 2000; Van Poecke et al., 2007; Zhang and Gassmann, 2007; Tsuchiya and Eulgem, 2013; Boccara et al., 2014).
Fine Regulation of R Genes Ameliorates Costs of Defense
Given the physiological costs of HR and SA induction, an exquisite temporal and spatial control of the many co-occurring R proteins is necessary to avoid compromised growth and development. There has been extensive research on how R activity is controlled both transcriptionally and posttranscriptionally.
Modes of posttranscriptional regulation have evolved that coordinately control the expression of clusters of R genes. It appears that microRNAs can target conserved regions of the highly duplicated R genes in a lineage-specific manner, most likely to jointly dampen their immune responses without requiring genic changes in each regulatory element in the cluster (Zhang et al., 2016b). Transposable elements are frequently associated with R gene clusters (Choi et al., 2016) and similarly help control the expression of their members (Tsuchiya and Eulgem, 2013, 2014). For example, the Arabidopsis R gene RPP7 contains an unusually long intron between exon1 and exon2 in which a COPIA-type retrotransposon is inserted (Tsuchiya and Eulgem, 2013, 2014). Tsuchiya and Eulgem (2013, 2014) elegantly demonstrated that a histone methylation mark on the TE influences the ratio of coding versus noncoding alternative RPP7 transcripts by regulating alternative polyadenylation sites. Dynamic changes in this ratio are correlated with RPP7-mediated disease resistance. The presence of alternative splicing variants in other R genes (N and RPS4) and changes in the ratio of transcripts upon pathogenic challenges suggest further mechanisms of fine-scale regulation of R genes (Dinesh-Kumar and Baker, 2000; Zhang and Gassmann, 2007). A recent report from the multi-year study on the epigenetically controlled R gene cluster in rice, the Pigm locus, provides the most compelling evidence for cost reduction via R gene regulation (Deng et al., 2017). In the multi-R gene Pigm cluster that confers broad-spectrum resistance to Magnoporthe oryzae, PigmS and PigmR are antagonistic not only for resistance but also for yield penalty, in which epigenetic expression control of PigmS appears to quantitatively balance the action of PigmR.
More broadly, R gene expression has been found to be induced by several abiotic stresses, which perhaps indicates adaptive regulation. A large-scale experiment probing expression changes of 13 R genes in 12 Arabidopsis accessions upon eight different environmental cues demonstrated that environmental fluctuation per se, either biotic or abiotic, induces expression of these genes, with substantial variation in inducibility among accessions (MacQueen and Bergelson, 2016). A meta-analysis of available genome-wide expression data in Arabidopsis revealed a relatively consistent increase in R expression upon biotic and abiotic challenges, suggesting a general mechanism to initiate R gene expression in response to environmental stress (MacQueen and Bergelson, 2016). As was described for the induction of defenses, this tendency of plants to express R genes at low levels, but induce them upon stimulus, reduces the costs compared with constitutively high expression of the immune system while allowing for a concerted response to challenge. DNA (de)methylation can regulate gene expression in response to both pathogen infection (reviewed by Deleris et al., 2016) and abiotic environmental stresses (e.g., hyperosmotic stress; Wibowo et al., 2016). Furthermore, analysis of met1 and ros1 mutants suggests that the R gene RMG1 is dynamically regulated by RNA-directed DNA methylation of helitron-related repeats in its promoter (Yu et al., 2013), and a recent analysis of over 1100 Arabidopsis transcriptomes and DNA methylomes revealed that R loci harbor extensive variation in TE insertions and associated methylation patterns (Kawakatsu et al., 2016). These results suggest that inducible epigenetic changes may constitute a molecular basis for the shared effects of biotic and abiotic environmental stimuli on R gene expression. Furthermore, the substantial variation among Arabidopsis accessions in the inducibility of R gene expression may derive in part from differences among accessions in the presence/absence of transposable elements near R genes and, hence, the potential for those R genes to be dynamically regulated by DNA methylation.
THE GENETIC LOAD OF THE R GENE SURVEILLANCE SYSTEM
The Cost of the System
In theory, perfectly regulated defense responses would mean that costs are only expressed in the presence of compensating benefits and that there would thus be no reason for hosts to harbor susceptible alleles. That is, the only cost would be the energy/nutrients required to produce minute quantities of recognition proteins that play no active roles in the absence of attack. Since this cost is expected to be small, large costs associated with R genes were not anticipated. Fitness trials testing the consequences of two R genes that exhibit separate presence/absence polymorphisms found that encoding the resistance gene in the absence of infection reduced seed set by 5 to 10% (Tian et al., 2003; Karasov et al., 2014a). In other words, the presence of the resistance gene alone reduced fitness by 5 to 10% in both studies. The high fitness costs associated with single genes suggests that the presence of each of these genes induces overstimulation or misregulation of the immune system. This misregulation was observed for a natural allele of another immune component, ACD6, for which a particular natural allele reduced growth (Todesco et al., 2010).
The high costs associated with the R genes RPM1 and RPS5 have raised questions about the genetic load associated with a large R gene repertoire and the possibility that many R genes are therefore relics that are no longer functional. This is apparently not the case; a series of experiments on rice blast resistance alleles revealed a very high level of functionality among the >300 alleles tested against 12 strains of the pathogen (Yang et al., 2013; Zhang et al., 2015).
Why, then, doesn’t the genetic load associated with a vast repertoire of R genes overwhelm plant fitness? A recent study by MacQueen et al. (2016) suggests that the costs associated with R genes that segregate as presence/absence polymorphisms may not be representative of R genes in general. RPS2 segregates as a single locus with alternative alleles; the R allele recognizes avrRpt2, whereas the specificity of the alternative allele has not been determined. Though these alleles are often called the S class, they retain a sequence that suggests functionality. When isolines varying in their expression of the different alleles are grown, no fitness penalty to resistance was observed. This makes sense in that R and S alleles both retain all functional domains, although likely vary in their specificity. Complete knockouts of the RPS2 alleles were tested for their fitness, and in an unexpected twist, deletion of RPS2 was costly. This result stood in contrast to the large benefits that were apparent for the absence of RPM1 and RPS5. This result may help explain why the RPS2 locus does not harbor true susceptible deletion mutants. In spite of such complexities in the interactions of R alleles, as a generality from numerous studies, the presence and misregulation of R genes typically reduces plant growth and seed production (Yang and Hua, 2004; Yi and Richards, 2007; Kim et al., 2010; Palma et al., 2010), selecting for the tight regulation of surveillance systems.
Immune Gene Interactions Cause/Modulate Costs
Experimental studies to test for costs of R gene resistance, as described above, have focused on loci with very simple genetic architectures. Even still, a profound effect of genetic architecture has already been revealed. R genes show a high degree of copy number variation within and across plant species, and many of them are organized in tandem arrays (Jacob et al., 2013; Zhang et al., 2016b).
A recurring finding in studies of R gene function and evolution is that highly polymorphic R genes interact with one another and that costs from one locus can be amplified or ameliorated by genetic variation at other R gene loci (Bomblies et al., 2007; Chae et al., 2014; Hurni et al., 2014; Stirnweis et al., 2014; Iakovidis et al., 2016), as well as by alleles of the same locus (Todesco et al., 2010, 2014; MacQueen et al., 2016). Lineage-specific expansion and diversification of a subset of immune genes is not unique to plants; in fact, the phenomenon apparently correlates with speciation history in teleost fish (Malmstrøm et al., 2016; Star et al., 2011).
Interactions between R genes can help coordinate immune responses, but misregulation can also lead to overstimulation of the immune system or autoimmunity (Bomblies et al., 2007). The most exhaustive investigation of plant autoimmunity to date revealed deleterious hybrid necrosis in 2% of crosses between Arabidopsis genotypes, suggesting an appreciable frequency of R alleles at risk of inducing autoimmunity (Chae et al., 2014). Intriguingly, the study revealed that most R genes involved in autoimmunity are organized in tandem repeats with high variability both in sequences and copy numbers (Chae et al., 2014). Although multiple evolutionary processes generate and maintain variability in immune genes (Karasov et al., 2014b), the frequency of autoimmune genetic interactions in natural and agricultural plant populations (Hermsen, 1963; Bomblies and Weigel, 2007; Bomblies, 2009; Ispolatov and Doebeli, 2009) suggests that the findings of a genetic load associated with R genes in Arabidopsis may be generalizable.
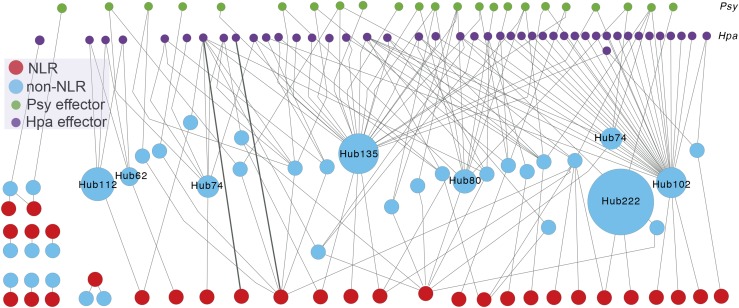
How the activity of one R gene can affect the activity of another becomes clear when one considers the network of protein interactions between R genes (Figure 2). While the majority of functionally annotated R genes were identified for race-specific resistance, most of them turned out to be involved in the surveillance system for host protein homeostasis (Dangl and Jones, 2001). The host proteins that are monitored by the guard R proteins (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008) are not only frequent targets of intruders, but often highly connected to physiological networks that regulate overall plant fitness (Mackey et al., 2002; Mukhtar et al., 2011; Weßling et al., 2014). When we reconstructed the interactome published by Mukhtar et al. (2011) to visualize the complexity that involves R genes, we found numerous connections between R genes and protein-protein interaction hubs.
Figure 2.
The Interconnected Activity of NLRs and Protein Interaction Hubs.
A network illustrating interactions among pathogen effectors, effector targets, and NLRs reconstructed from the PPIN-1 interactome study by Mukhtar et al. (2011). The reconstruction using Cytoscape visualizes complexity of the interactions that NLR proteins make through other host proteins. Nodes representing effectors are aligned in the top two rows: green nodes for P. syringae (Psy) effectors and purple nodes for effectors from the biotrophic oomycete, Hpa. Red nodes represent 30 NLRs included in the PPIN-1, and turquoise nodes represent Arabidopsis proteins showing interactions with NLRs. Gray edges represent protein-protein interactions assayed by yeast two-hybrid system in PPIN-1. The number after Hub indicates the number of interaction partners of the host protein in the main interactome AI-1 (Mukhtar et al., 2011). A single NLR can be connected with multiple effector targets, while an effector target can be connected to multiple NLRs. Only two NLRs show direct interactions with effector proteins (bold edges), while most other NLRs in the main network make indirect connections to effectors through host proteins. R gene activities are highly interconnected both with one another and with other immune proteins. This interconnectedness makes optimizing the immune response more difficult, as changes in one protein have a high probability of affecting the activity of another immune protein. The interactome network analysis was inferred from yeast two-hybrid associations.
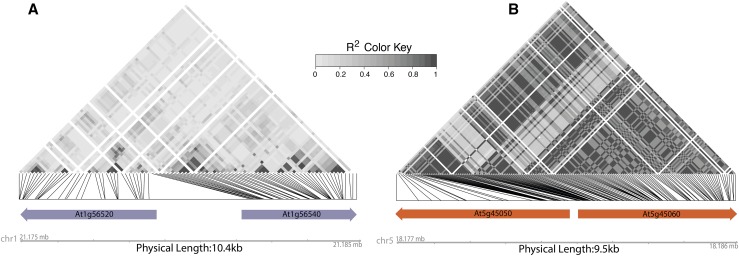
The genomic architecture of R genes in plants suggests that plants have evolved mechanisms that ameliorate the misregulation of R gene interactions. Tight linkage between a pair of R genes in the head-to-head configuration creates a useful coregulatory module, ensuring simultaneous control of an interacting pair due to shared promoter elements. Several such tightly linked pairs have been functionally annotated for their cooperative resistance: Arabidopsis RPP2A/RPP2B for Hpa Cala2, RPS4/RRS1 for bacterial and fungal effectors, and rice RGA4/RGA5 for rice blast (Sinapidou et al., 2004; Narusaka et al., 2009; Eitas and Dangl, 2010; Césari et al., 2014). Their interdependent functional activities require fine control so that the two proteins activate the immune response only with the proper stimulus. Their tight linkage and coregulation of expression reduces the probability of misregulation (Sohn et al., 2014; Saucet et al., 2015; Xu et al., 2015; Chae et al., 2016; Zhang et al., 2016a) (Figure 3). Whether other R genes similarly avoid autoimmune-inducing mispairings is an open question. It is tempting to hypothesize pervasive coselection on immunity genes to reduce misregulation of the immune system. Comparative genomic analyses in combination with genetic analysis can shed light on this possibility (Figure 3).
Figure 3.
Linkage between R Gene Haplotypes May Provide Evidence of Functional Cooperation and Coevolution.
Linkage across 762 Arabidopsis genotypes (Kawakatsu, 2016) is represented as R2 values of SNPs (minor allele frequency > 0.2) within R genes and visualized as a heat map. Only R genes are marked with arrows in these regions. Linkage disequilibrium in the region carrying At1g56520 and At1g56540 (A) is relatively low compared with that in the region carrying At5g45050 (RRS1B) and At5g45060 (RPS4B) (B). The increased linkage (higher R2) across the entire locus in (B) may be the result of the coevolution of alleles of the two R genes for their cooperative function. Functional cooperation of At5g45050 and At5g45060 (B) as a pair was demonstrated for disease resistance specificity as well as for tight cross-regulation against constitutive activity (Narusaka et al., 2009; Saucet et al., 2015). No such interaction was observed for the two R genes depicted in (A). If two R genes nearby or at a distance function together, mismatched allelic combinations could result in fitness costs. It is therefore reasonable to expect such R gene pairs to evolve tight linkage at either short range or long distance. Measuring linkage and population genomic statistics of R gene pairs in large sequencing projects, as we demonstrate here, may allow for the identification of candidate loci for interacting/coevolving pairs and allelic variants.
Insight on Function from Evolutionary History
A central implication of growth-defense trade-offs in plant populations is that plants are exposed to opposing selective pressures. Because these pressures are sensitive to biotic and abiotic factors, the optimal balance between growth and defense is likely a moving target, with the genes that control this balance under strong selection. Genomic studies of plant evolution support this line of logic.
As mentioned above, R genes have among the highest nucleotide diversity of any genes in plant genomes (Cao et al., 2011; Zhang et al., 2016b) as a result of selection and high mutation rates within resistance gene clusters (Dixon et al., 1998; Bergelson et al., 2001; Kuang et al., 2004; Karasov et al., 2014b). Those resistance loci whose alleles exact trade-offs between defense and growth are predicted to undergo balancing selection (Anderson and May, 1982), an evolutionary process that leaves detectable signatures throughout a plant genome (Bakker et al., 2006). These signatures may help pinpoint costly R genes through the analysis of evolutionary history and quantitative genetics.
The ability of selection scans to identify loci that exact fitness costs has been demonstrated several times. Two studies that first identified signatures of balancing selection in the genome were then able to demonstrate a strong fitness cost of resistance alleles (or the absence of resistance alleles) at these same loci (Stahl et al., 1999; Tian et al., 2003; Karasov et al., 2014a). Signatures of selection can also identify those loci with more complex interactions between resistance alleles (MacQueen et al., 2016). The loci identified by Stahl et al. (1999), Tian et al. (2003), and Karasov et al. (2014a) differed from those by MacQueen et al. (2016) not only in the observed cost of resistance but also in their class of sequence polymorphism. As stated above, RPS5 and RPM1 both segregate for presence/absence polymorphisms of the entire gene, while RPS2 exhibits balancing selection on two different allelic classes. An a priori hypothesis is that loci that exhibit presence/absence polymorphisms are most likely to exact large fitness costs, while those loci that exhibit selection on multiple alleles may have negligible costs (MacQueen et al., 2016).
It may also be possible to use linkage information to infer which R genes are coregulated and induce autoimmunity in the absence of coregulation (Narusaka et al., 2009; Chae et al., 2014). A reasonable hypothesis to describe two R genes for which the breakage of linkage is deleterious is that matched alleles of the two loci should be tightly correlated across genotypes. Simple measures of linkage (Figure 3) could reveal such correlations between R gene loci, rapidly identifying a priori candidates for coevolving loci. As the availability of sequencing data for natural plant populations and crop germplasms become increasingly available, it will become more feasible to use signatures of selection and co-occurrence information to identify candidate loci for trade-offs and for crop improvement.
The natural variation of R gene inducibility raises the possibility that the modulation of R gene expression has evolved as an adaptive trait to optimize immune responses in the wild. Given that Arabidopsis habitats range from Mediterranean to northern Scandinavian territories, accumulating transcriptome data sets from natural accessions of this species provide an ideal platform to examine clinal correlations with the R gene expression variation. Initial attempts to exploit previously published worldwide and regional transcriptome data sets (Gan et al., 2011) revealed that the basal R expression levels show clinal correlations, with accessions from high latitudes exhibiting higher basal expression (MacQueen and Bergelson, 2016). It has yet to be investigated how the differences in basal R expression levels contribute to effective immunity in their native and transplanted habitats. With the recently released species-wide transcriptome data set (Kawakatsu et al., 2016), one can begin to test this hypothesis with multiple clinal variables to search for evidence of local adaptation in R gene expression modulation.
Combining comparative genomics with population genetic analysis provides a promising avenue to identify loci in agricultural and nonagricultural plant populations that underlie growth-defense trade-offs. Upcoming advances in sequencing will continue to facilitate such approaches.
GETTING SOMETHING FOR NOTHING
The majority of this review has focused on the costs of resistance and the myriad mechanisms that minimize these costs. However, it is important to recognize an alternative approach that plants adopt: the recruitment of protection from other species. Perhaps the best-known example of this strategy is the ant-plant relationship that provides food and shelter for ants that, in turn, attack the herbivores of their host (reviewed in Mayer et al., 2014). More subtle interactions also exist, including the volatiles that many plants produce when wounded. These volatiles attract parasites and predators that attack the offending herbivore and are also known to have direct antimicrobial properties (Dorman and Deans, 2000; reviewed in Clavijo McCormick et al., 2012).
Recent studies suggest the exciting possibility that plants can protect themselves through the maintenance of an appropriate microbial community. There is still much to be learned about what a protective microbiome would look like. Decades of work in community ecology has revealed a few patterns, albeit with exceptions. First, diverse communities are less likely to be invaded (Levine et al., 2004). From the perspective of host defense, this suggests that hosts harboring rich microbiomes should be less likely to suffer invasion by an infectious agent. For example, a negative relationship between symbiotic bacterial species richness and the density of a pathogenic bacterium, Serratia marsecens, has been found within the gut of the desert locust. Second, communities with a more even distribution of species are less prone to invasion (reviewed in Hillebrand et al., 2008; for a microbial example, see De Roy et al., 2013). Finally, communities that are relatively stable in time are less prone to invasion, most likely due to increased vulnerability while community structures are undergoing change (Robinson et al., 2010).
Of course, in order for a plant to utilize its microbiome for protection, it must be able to control it. We now have ample evidence that a plant’s microbiome can be heritable (Peiffer et al., 2013; Horton et al., 2014), although there is also an environmental component to the microbial community structure (Lundberg et al., 2012; Agler et al., 2016; Wagner et al., 2016). This heritability at least opens the door to host control of beneficial microbes as a defense mechanism. We also know that the presence within leaves of key microbial species can provide protection. For example, the introduction of five native bacterial species protected Nicotiana attenuata from fungal disease in the field (Santhanam et al., 2015). The key then will be deciphering the extent and ability of plant hosts to structure their microbial community. Significant headway has been made by Agler et al.’s (2016) demonstration that microbial hub species strongly influence microbial communities and are themselves influenced by host genotype. The challenges now are to understand which community structures are most protective to plants, to determine the host factors shaping the microbiome, and to determine the extent to which host variability in associated microbiomes is adaptive. Then, one might envision agricultural breeding strategies to enhance resistance through the co-option of microbial symbionts.
Finally, ecologists have long recognized the importance of escaping in time. Winter annuals, like Arabidopsis, bolt and reproduce sufficiently early in the spring that most herbivores are not yet active (Crawley, 1997). While useful as an escape from herbivory, this phenology is unlikely to provide an advantage in terms of reduced pathogen pressures. By contrast, species that restrict their growth to the hot, dry summer months should suffer less attack by pathogens that favor cool, moist climates (Holub et al., 1995). It would be useful to examine differentiation in the defense strategy of cohorts that are segregated in time; it is possible that host species reduce costs by focusing defense on the classes of enemy most likely to attack during that window of time.
An extension of the idea that the modulation of plant defenses matches risks of exposure was revealed by the demonstrations that defenses follow a circadian rhythm. This is a general phenomenon, affecting both constitutive defenses and JA- and SA-regulated responses, although there are idiosyncrasies associated with the response to particular enemies and of particular defenses (reviewed in Baldwin and Meldau, 2013). Indeed, even indirect defenses such as extrafloral nectars can show diurnal patterns (Radhika et al., 2008). While it is tempting to speculate that these diurnal cycles are adaptive, the evidence is unclear. Zhou et al. (2015) showed that the major immune regulator NPR1 senses the redox state of Arabidopsis plants and regulates transcription of core circadian clock genes. They went on to unravel how SA/NPR1-mediated induction of the circadian clock helps control stronger resistance responses in the morning, when conditions are more favorable for pathogen growth. The adaptive value of circadian allocation to defense has not been tested in nature, where plants are exposed to the natural rhythm of attack, nor has it been tested with natural enemies under controlled conditions.
These studies, in addition to numerous others, reveal the probable importance of tritrophic species interactions in plant defense against pathogens. Given the interconnectedness of the plant immune system that we discuss above, perfectly balancing the defense response by tweaking expression and surveillance within the immune system seems to be infeasible. The ability of other organisms to protect the plant could reduce the reliance of a plant on its immune system. Studying the immune responses of plants exposed to pathogen in the presence or absence of protective microbes or commensal invertebrates could provide evidence of how plants use their immune systems in the presence of other beneficial organisms.
CONCLUSION
Recent work has led to a paradigm shift from viewing costs of immunity as an inescapable consequence of metabolic expenditure to a trade-off that is carefully regulated by the plant (Huot et al., 2014; Kliebenstein, 2016). The implications of this new perspective are profound in raising questions about how plants shift the balance between growth and defense in response to environmental change. For example, the transport of sugars during plant growth makes plants vulnerable to invasion by pathogen (Eom et al., 2015). A reduction in growth (concomitant with the mounting of an immune response) can therefore reduce the spread of infection. What is the optimal balance between reducing pathogen expansion by reducing growth or enduring pathogen expansion while growing? Fine-tuning of these trade-offs should underlie adaptation to enemies in a natural context. Unfortunately, we currently know little about which induced shifts in growth-defense trade-offs are adaptive. Field trials that uncouple components of growth and defense and then measure the outcomes of challenge by pathogen and abiotic stresses could begin to address these issues.
Of course, plants also adopt strategies to reduce the cost of mounting an immune response. These strategies largely involve restriction of defenses to particular tissues (Velasco et al., 2008), developmental stages, and/or windows of time. While there is an obvious risk to not investing in constitutive, ubiquitous defense, there can be tremendous cost savings if only the most vulnerable tissues are protected. For example, the circadian regulation of defenses (Zhou et al., 2015) could limit the highest expression of defense to times with greatest pathogen pressure. Still, nature is heterogeneous and there could be information to be gained from an attack, provided that attacks are correlated. It is perhaps in this spirit that we should consider the value of priming and transgenerational defense induction. We know little of spatial and temporal predictability in the abundance of various natural enemies and as a consequence cannot assess the adaptive value of these responses. This area seems an especially ripe for rigorous, long-term field investigations.
Perhaps the least explored but among the most promising of the mechanisms of cost amelioration is that of defense via tritrophic interactions. These interactions involve the activity of another species such as a protective microbiome, in which the other species prevents the invasion of pathogen. This type of interaction could involve reduced reliance on the immune system and its complexities.
In this review, we described several mechanisms by which plants reduce the magnitude of the growth-defense trade-off. However, it is important to reiterate that reducing the trade-off is not always advantageous to the fitness of a plant and that the mechanisms of amelioration may be advantageous only under specific conditions. Ultimately, if we wish to draw conclusions about the generalizability and basis of trade-offs, it will be necessary to assess trade-offs and mechanisms of their amelioration in a range of conditions, and in environments with other species.
Supplementary Material
Acknowledgments
We thank Ignacio Rubio-Somoza and Michael Werner for thoughtful discussion and comments on the manuscript and Moises Exposito-Alonso for providing genomic data.
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
References
- Agler M.T., Ruhe J., Kroll S., Morhenn C., Kim S.-T., Weigel D., Kemen E.M. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14: e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A.A. (2001). Transgenerational consequences of plant responses to herbivory: an adaptive maternal effect? Am. Nat. 157: 555–569. [DOI] [PubMed] [Google Scholar]
- Akkerman K.C., Sattarin A., Kelly J.K., Scoville A.G.(2016). Transgenerational plasticity is sex-dependent and persistent in yellow monkeyflower (Mimulus guttatus). Environ. Epigenet. 2: dvw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsdurf J.D., Ripley T.J., Matzner S.L., Siemens D.H. (2013). Drought-induced trans-generational tradeoff between stress tolerance and defence: consequences for range limits? AoB Plants 5: plt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., May R.M. (1982). Coevolution of hosts and parasites. Parasitology 85: 411–426. [DOI] [PubMed] [Google Scholar]
- Aviv D.H., Rustérucci C., Holt B.F. III, Dietrich R.A., Parker J.E., Dangl J.L. (2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29: 381–391. [DOI] [PubMed] [Google Scholar]
- Bakker E.G., Toomajian C., Kreitman M., Bergelson J. (2006). A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18: 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin I.T., Gorham D., Schmelz E.A., Lewandowski C.A., Lynds G.Y. (1998). Allocation of nitrogen to an inducible defense and seed production in Nicotiana attenuata. Oecologia 115: 541–552. [DOI] [PubMed] [Google Scholar]
- Baldwin I.T., Meldau S. (2013). Just in time: circadian defense patterns and the optimal defense hypothesis. Plant Signal. Behav. 8: e24410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer A., Pastor V., Gamir J., Flors V., Mauch-Mani B. (2015). The ‘prime-ome’: towards a holistic approach to priming. Trends Plant Sci. 20: 443–452. [DOI] [PubMed] [Google Scholar]
- Barrett L.G., Kniskern J.M., Bodenhausen N., Zhang W., Bergelson J. (2009). Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytol. 183: 513–529. [DOI] [PubMed] [Google Scholar]
- Bennett R.N., Wallsgrove R.M. (1994). Secondary metabolites in plant defence mechanisms. New Phytol. 127: 617–633. [DOI] [PubMed] [Google Scholar]
- Bergelson J. (1994). The effects of genotype and the environment on costs of resistance in lettuce. Am. Nat. 143: 349–359. [Google Scholar]
- Bergelson J., Kreitman M., Stahl E.A., Tian D. (2001). Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Bergelson J., Purrington C.B. (1996). Surveying patterns in the cost of resistance in plants. Am. Nat. 148: 536–558. [Google Scholar]
- Boccara M., Sarazin A., Thiébeauld O., Jay F., Voinnet O., Navarro L., Colot V. (2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10: e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K. (2009). Too much of a good thing? Hybrid necrosis as a by-product of plant immune system diversification. Botany 87: 1013–1022. [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J.L., Weigel D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. (2007). Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393. [DOI] [PubMed] [Google Scholar]
- Brown J.K.M. (2002). Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5: 339–344. [DOI] [PubMed] [Google Scholar]
- Bruggeman Q., Raynaud C., Benhamed M., Delarue M. (2015). To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Césari S., Kanzaki H., Fujiwara T., Bernoux M., Chalvon V., Kawano Y., Shimamoto K., Dodds P., Terauchi R., Kroj T. (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33: 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., et al. (2014). Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., Tran D.T.N., Weigel D. (2016). Cooperation and conflict in the plant immune system. PLoS Pathog. 12: e1005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin J.F., Mann T.J. (1978). Evaluation of tobacco mosaic resistance factor transferred from burley to flue-cured tobacco. J. Hered. 69: 175–178. [Google Scholar]
- Chen L.-Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., et al. (2016). Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 12: e1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini D.F. (2002). Does competition magnify the fitness costs of induced responses in Arabidopsis thaliana? A manipulative approach. Oecologia 131: 514–520. [DOI] [PubMed] [Google Scholar]
- Clavijo McCormick A., Unsicker S.B., Gershenzon J. (2012). The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci. 17: 303–310. [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G.J.M., Langenbach C.J.G., Jaskiewicz M.R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53: 97–119. [DOI] [PubMed] [Google Scholar]
- Crawley M.J. (1997). Life history and environment. In Plant Ecology, 2nd ed, M.J. Crawley, ed (Oxford, UK: Blackwell Scientific Press), pp. 73–131. [Google Scholar]
- Creelman R.A., Mullet J.E. (1995). Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 92: 4114–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp P.A., Ganguly D., Eichten S.R., Borevitz J.O., Pogson B.J. (2016). Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2: e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Deleris A., Halter T., Navarro L. (2016). DNA methylation and demethylation in plant immunity. Annu. Rev. Phytopathol. 54: 579–603. [DOI] [PubMed] [Google Scholar]
- Denancé N., Sánchez-Vallet A., Goffner D., Molina A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355: 962–965. [DOI] [PubMed] [Google Scholar]
- De Roy K., Marzorati M., Negroni A., Thas O., Balloi A., Fava F., Verstraete W., Daffonchio D., Boon N. (2013). Environmental conditions and community evenness determine the outcome of biological invasion. Nat. Commun. 4: 1383. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Baker B.J. (2000). Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97: 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M.S., Hatzixanthis K., Jones D.A., Harrison K., Jones J.D. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10: 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman H.J., Deans S.G. (2000). Antimicrobial agents from plants: antibacterial activity of plant volatile oils. J. Appl. Microbiol. 88: 308–316. [DOI] [PubMed] [Google Scholar]
- Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer G. (1994). Density dependence and spatial structure in the dynamics of insect pathogens. Am. Nat. 143: 533–562. [Google Scholar]
- Dwyer G., Elkinton J.S. (1993). Using simple models to predict virus epizootics in gypsy moth populations. J. Anim. Ecol. 62: 1–11. [Google Scholar]
- Eitas T.K., Dangl J.L. (2010). NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J.-S., Chen L.-Q., Sosso D., Julius B.T., Lin I.W., Qu X.-Q., Braun D.M., Frommer W.B. (2015). SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25: 53–62. [DOI] [PubMed] [Google Scholar]
- Fu C.-Y., Wang F., Sun B.-R., Liu W.-G., Li J.-H., Deng R.-F., Liu D.-L., Liu Z.-R., Zhu M.-S., Liao Y.-L., Chen J.-W. (2013). Genetic and cytological analysis of a novel type of low temperature-dependent intrasubspecific hybrid weakness in rice. PLoS One 8: e73886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Gaffney T., Friedrich L., Vernooij B., Negrotto D., Nye G., Uknes S., Ward E., Kessmann H., Ryals J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756. [DOI] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S.I. (2004). Sugar and phytohormone response pathways: navigating a signalling network. J. Exp. Bot. 55: 253–264. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Hayat Q., Hayat S., Irfan M., Ahmad A. (2010). Effect of exogenous salicylic acid under changing environment: A review. Environ. Exp. Bot. 68: 14–25. [Google Scholar]
- Heidel A.J., Clarke J.D., Antonovics J., Dong X. (2004). Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M. (2002). Ecological costs of induced resistance. Curr. Opin. Plant Biol. 5: 345–350. [DOI] [PubMed] [Google Scholar]
- Heil M., Baldwin I.T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7: 61–67. [DOI] [PubMed] [Google Scholar]
- Herman J.J., Spencer H.G., Donohue K., Sultan S.E. (2014). How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68: 632–643. [DOI] [PubMed] [Google Scholar]
- Hermsen J.G.T. (1963). Hybrid necrosis as a problem for the wheat breeder. Euphytica 12: 1–16. [Google Scholar]
- Hilker M., et al. (2016). Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. Camb. Philos. Soc. 91: 1118–1133. [DOI] [PubMed] [Google Scholar]
- Hillebrand H., Bennett D.M., Cadotte M.W. (2008). Consequences of dominance: a review of evenness effects on local and regional ecosystem processes. Ecology 89: 1510–1520. [DOI] [PubMed] [Google Scholar]
- Holeski L.M. (2007). Within and between generation phenotypic plasticity in trichome density of Mimulus guttatus. J. Evol. Biol. 20: 2092–2100. [DOI] [PubMed] [Google Scholar]
- Holeski L.M., Jander G., Agrawal A.A. (2012). Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. (Amst.) 27: 618–626. [DOI] [PubMed] [Google Scholar]
- Holub E.B., Brose E., Tör M., Clay C., Crute I.R., Beynon J.L. (1995). Phenotypic and genotypic variation in the interaction between Arabidopsis thaliana and Albugo candida. Mol. Plant Microbe Interact. 8: 916–928. [DOI] [PubMed] [Google Scholar]
- Horton M.W., Bodenhausen N., Beilsmith K., Meng D., Muegge B.D., Subramanian S., Vetter M.M., Vilhjálmsson B.J., Nordborg M., Gordon J.I., Bergelson J. (2014). Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 5: 5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li J., Bao F., Zhang X., Yang S. (2010). A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B., Yao J., Montgomery B.L., He S.Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7: 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S., Brunner S., Buchmann G., Herren G., Jordan T., Krukowski P., Wicker T., Yahiaoui N., Mago R., Keller B. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76: 957–969. [DOI] [PubMed] [Google Scholar]
- Hurni S., Brunner S., Stirnweis D., Herren G., Peditto D., McIntosh R.A., Keller B. (2014). The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 79: 904–913. [DOI] [PubMed] [Google Scholar]
- Iakovidis M., Teixeira P.J.P.L., Exposito-Alonso M., Cowper M.G., Law T.F., Liu Q., Vu M.C., Dang T.M., Corwin J.A., Weigel D., Dangl J.L., Grant S.R. (2016). Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204: 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ispolatov I., Doebeli M. (2009). Speciation due to hybrid necrosis in plant-pathogen models. Evolution 63: 3076–3084. [DOI] [PubMed] [Google Scholar]
- Jacob F., Vernaldi S., Maekawa T. (2013). Evolution and conservation of plant NLR functions. Front. Immunol. 4: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhänsel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. (2009). Priming in systemic plant immunity. Science 324: 89–91. [DOI] [PubMed] [Google Scholar]
- Karasov T.L., et al. (2014a). The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., Horton M.W., Bergelson J. (2014b). Genomic variability as a driver of plant-pathogen coevolution? Curr. Opin. Plant Biol. 18: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiria P., Sidler C., Golubov A., Kalischuk M., Kawchuk L.M., Kovalchuk I. (2010). Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 153: 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., et al.; 1001 Genomes Consortium (2016). Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempel A., Schädler M., Chrobock T., Fischer M., van Kleunen M. (2011). Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc. Natl. Acad. Sci. USA 108: 5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Gao F., Bhattacharjee S., Adiasor J.A., Nam J.C., Gassmann W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király L., Künstler A., Höller K., Fattinger M., Juhász C., Müller M., Gullner G., Zechmann B. (2012). Sulfate supply influences compartment specific glutathione metabolism and confers enhanced resistance to Tobacco mosaic virus during a hypersensitive response. Plant Physiol. Biochem. 59: 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J. (2016). False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol. Mol. Plant Pathol. 95: 55–59. [Google Scholar]
- Krasileva K.V., Zheng C., Leonelli L., Goritschnig S., Dahlbeck D., Staskawicz B.J. (2011). Global analysis of Arabidopsis/downy mildew interactions reveals prevalence of incomplete resistance and rapid evolution of pathogen recognition. PLoS One 6: e28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger S.G., Keller B. (2016). Molecular genetics and evolution of disease resistance in cereals. New Phytol. 212: 320–332. [DOI] [PubMed] [Google Scholar]
- Kruse C., Jost R., Lipschis M., Kopp B., Hartmann M., Hell R. (2007). Sulfur-enhanced defence: effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol (Stuttg.) 9: 608–619. [DOI] [PubMed] [Google Scholar]
- Kuang H., Woo S.-S., Meyers B.C., Nevo E., Michelmore R.W. (2004). Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16: 2870–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg P.D., Matzinger D.F., Mann T.J. (1965). Genetic variation and covariation in a Nicotiana tabacum L. synthetic two generations after synthesis. Crop Sci. 5: 30. [Google Scholar]
- Levine J.M., Adler P.B., Yelenik S.G. (2004). A meta-analysis of biotic resistance to exotic plant invasions. Ecol. Lett. 7: 975–989. [Google Scholar]
- Liu L., Sonbol F.-M., Huot B., Gu Y., Withers J., Mwimba M., Yao J., He S.Y., Dong X. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7: 13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Sánchez A., Stassen J.H.M., Furci L., Smith L.M., Ton J. (2016). The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 88: 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T.J.A., Roberts M.R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Ton J. (2012). The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 7: 615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D.S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Holt B.F. III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- MacQueen A., Bergelson J. (2016). Modulation of R-gene expression across environments. J. Exp. Bot. 67: 2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A., Sun X., Bergelson J. (2016). Genetic architecture and pleiotropy shape costs of Rps2-mediated resistance in Arabidopsis thaliana. Nat. Plants 2: 16110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M., et al. (2016). Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 48: 1204–1210. [DOI] [PubMed] [Google Scholar]
- Mayer V.E., Frederickson M.E., McKey D., Blatrix R. (2014). Current issues in the evolutionary ecology of ant-plant symbioses. New Phytol. 202: 749–764. [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Williams S.G., Funderburg N.T., Eulgem T., Dangl J.L. (2005). Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol. Plant Microbe Interact. 18: 1226–1234. [DOI] [PubMed] [Google Scholar]
- Monforte A.J., Tanksley S.D. (2000). Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor. Appl. Genet. 100: 471–479. [Google Scholar]
- Mosher S., Moeder W., Nishimura N., Jikumaru Y., Joo S.-H., Urquhart W., Klessig D.F., Kim S.-K., Nambara E., Yoshioka K. (2010). The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 152: 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M.S., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Kenton P., Lloyd A.J., Ougham H., Prats E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59: 501–520. [DOI] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., Narusaka Y. (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60: 218–226. [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D.G. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18: 650–655. [DOI] [PubMed] [Google Scholar]
- Ohnmeiss T.E., Baldwin I.T. (1994). The allometry of nitrogen to growth and an inducible defense under nitrogen-limited growth. Ecology 75: 995–1002. [Google Scholar]
- Oldroyd G.E., Staskawicz B.J. (1998). Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci. USA 95: 10300–10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor V., Luna E., Ton J., Cerezo M., García-Agustín P., Flors V. (2013). Fine tuning of reactive oxygen species homeostasis regulates primed immune responses in Arabidopsis. Mol. Plant Microbe Interact. 26: 1334–1344. [DOI] [PubMed] [Google Scholar]
- Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L., Buckler E.S., Ley R.E. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel M.J.C., van Dijken A.J.H., Bardoel B.W., Seidl M.F., van der Ent S., van Strijp J.A.G., Pieterse C.M.J. (2014). Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol. Plant Microbe Interact. 27: 603–610. [DOI] [PubMed] [Google Scholar]
- Pena-Cortés H., Albrecht T., Prat S., Weiler E.W., Willmitzer L. (1993). Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191: 123–128. [Google Scholar]
- Radhika V., Kost C., Bartram S., Heil M., Boland W. (2008). Testing the optimal defence hypothesis for two indirect defences: extrafloral nectar and volatile organic compounds. Planta 228: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Robinson C.J., Bohannan B.J.M., Young V.B. (2010). From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74: 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam R., Luu V.T., Weinhold A., Goldberg J., Oh Y., Baldwin I.T. (2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. USA 112: E5013–E5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. (2015). Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 6: 6338. [DOI] [PubMed] [Google Scholar]
- Scoville A.G., Barnett L.L., Bodbyl-Roels S., Kelly J.K., Hileman L.C. (2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano Y., Kachroo P., Shah J., Klessig D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Kappel C., Josephs E.B., Lee Y.W., Marona C., Stinchcombe J.R., Wright S.I., Lenhard M. (2015). Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat. Commun. 6: 7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds N.W. (1989). How frequent are superior genotypes in plant breeding populations? Biol. Rev. Camb. Philos. Soc. 64: 341–365. [Google Scholar]
- Sinapidou E., Williams K., Nott L., Bahkt S., Tör M., Crute I., Bittner-Eddy P., Beynon J. (2004). Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38: 898–909. [DOI] [PubMed] [Google Scholar]
- Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.H., Segonzac C., Rallapalli G., Sarris P.F., Woo J.Y., Williams S.J., Newman T.E., Paek K.H., Kobe B., Jones J.D.G. (2014). The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10: e1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E.A., Dwyer G., Mauricio R., Kreitman M., Bergelson J. (1999). Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Star B., et al. (2011). The genome sequence of Atlantic cod reveals a unique immune system. Nature 477: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweis D., Milani S.D., Brunner S., Herren G., Buchmann G., Peditto D., Jordan T., Keller B. (2014). Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J. 79: 893–903. [DOI] [PubMed] [Google Scholar]
- Strauss S.Y., Rudgers J.A., Lau J.A., Irwin R.E. (2002). Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 17: 278–285. [Google Scholar]
- Sultan S.E., Spencer H.G. (2002). Metapopulation structure favors plasticity over local adaptation. Am. Nat. 160: 271–283. [DOI] [PubMed] [Google Scholar]
- Świadek M., et al. (2017). Novel allelic variants in ACD6 cause hybrid necrosis in local collection of Arabidopsis thaliana. New Phytol. 213: 900–915. [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Fidantsef A.L., Bostock R.M. (2002). Antagonism between jasmonate- and salicylate-mediated induced plant resistance: effects of concentration and timing of elicitation on defense-related proteins, herbivore, and pathogen performance in tomato. J. Chem. Ecol. 28: 1131–1159. [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T., Whiteman N.K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270. [DOI] [PubMed] [Google Scholar]
- Thilmony R., Underwood W., He S.Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46: 34–53. [DOI] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77. [DOI] [PubMed] [Google Scholar]
- Todesco M., et al. (2010). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Kim S.-T., Chae E., Bomblies K., Zaidem M., Smith L.M., Weigel D., Laitinen R.A.E. (2014). Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet. 10: e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traw M.B., Kniskern J.M., Bergelson J. (2007). SAR increases fitness of Arabidopsis thaliana in the presence of natural bacterial pathogens. Evolution 61: 2444–2449. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 110: E3535–E3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2014). The PHD-finger module of the Arabidopsis thaliana defense regulator EDM2 can recognize triply modified histone H3 peptides. Plant Signal. Behav. 9: e29202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam N.M., Baldwin I.T. (1998). Costs of jasmonate-induced responses in plants competing for limited resources. Ecol. Lett. 1: 30–33. [Google Scholar]
- van der Hoorn R.A.L., Kamoun S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulten M., Pelser M., van Loon L.C., Pieterse C.M.J., Ton J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poecke R.M.P., Sato M., Lenarz-Wyatt L., Weisberg S., Katagiri F. (2007). Natural variation in RPS2-mediated resistance among Arabidopsis accessions: correlation between gene expression profiles and phenotypic responses. Plant Cell 19: 4046–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco P., Soengas P., Vilar M., Cartea M.E., del Rio M. (2008). Comparison of glucosinolate profiles in leaf and seed tissues of different Brassica napus crops. J. Am. Soc. Hortic. Sci. 133: 551–558. [Google Scholar]
- Vetter M., Karasov T.L., Bergelson J. (2016). Differentiation between MAMP triggered defenses in Arabidopsis thaliana. PLoS Genet. 12: e1006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M.R., Lundberg D.S., Del Rio T.G., Tringe S.G., Dangl J.L., Mitchell-Olds T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7: 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.R., Paterson L., Walsh D.J., Havis N.D. (2008). Priming for plant defense in barley provides benefits only under high disease pressure. Physiol. Mol. Plant Pathol. 73: 95–100. [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17: 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wang L., Jin P., Wang J., Jiang L., Shan T., Zheng Y. (2015). Methyl jasmonate primed defense responses against Penicillium expansum in sweet cherry fruit. Plant Mol. Biol. Report. 33: 1464–1471. [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R., et al. (2014). Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A., et al. (2016). Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5: e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhu C., Cevik V., Johnson K., Liu Y., Sohn K.-H., Jones J.D.G., Holub E.B., Li X. (2015). Autoimmunity conferred by chs3-2D relies on CSA1, its adjacent TIR-NB-LRR encoding neighbour. Sci. Rep. 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Takashi T., Morinaka Y., Lin S., Wu J., Matsumoto T., Kitano H., Matsuoka M., Ashikari M. (2010). Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol. Genet. Genomics 283: 305–315. [DOI] [PubMed] [Google Scholar]
- Yang H., Shi Y., Liu J., Guo L., Zhang X., Yang S. (2010). A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J. 63: 283–296. [DOI] [PubMed] [Google Scholar]
- Yang S., Hua J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li J., Zhang X., Zhang Q., Huang J., Chen J.-Q., Hartl D.L., Tian D. (2013). Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. USA 110: 18572–18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2007). A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19: 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2008). Phenotypic instability of Arabidopsis alleles affecting a disease Resistance gene cluster. BMC Plant Biol. 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., Abraham A.-L., Penterman J., Fischer R.L., Voinnet O., Navarro L. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 110: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., et al. (2015). A genome-wide survey reveals abundant rice blast R genes in resistant cultivars. Plant J. 84: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-C., Gassmann W. (2007). Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145: 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Liu J., Ding Y., Wang S., Zhang X., Liu Y., Yang S. (2016a). Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol. 213: 1330–1345. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xia R., Kuang H., Meyers B.C. (2016b). The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 33: 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wang W., Karapetyan S., Mwimba M., Marqués J., Buchler N.E., Dong X. (2015). Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang J., Chen Q., Chen G., Huang Y., Yang Z. (2016). Costs and trade-offs of grazer-induced defenses in Scenedesmus under deficient resource. Sci. Rep. 6: 22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Züst T., Rasmann S., Agrawal A.A. (2015). Growth–defense tradeoffs for two major anti-herbivore traits of the common milkweed Asclepias syriaca. Oikos 124: 1404–1415. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.