The TRAF family proteins MUSE14/TRAF1a and MUSE13/TRAF1b regulate the dynamics of autophagy by facilitating both the proteolysis and stabilization of AUTOPHAGY PROTEIN6.
Abstract
Eukaryotic cells use autophagy to recycle cellular components. During autophagy, autophagosomes deliver cytoplasmic contents to the vacuole or lysosome for breakdown. Mammalian cells regulate the dynamics of autophagy via ubiquitin-mediated proteolysis of autophagy proteins. Here, we show that the Arabidopsis thaliana Tumor necrosis factor Receptor-Associated Factor (TRAF) family proteins TRAF1a and TRAF1b (previously named MUSE14 and MUSE13, respectively) help regulate autophagy via ubiquitination. Upon starvation, cytoplasmic TRAF1a and TRAF1b translocated to autophagosomes. Knockout traf1a/b lines showed reduced tolerance to nutrient deficiency, increased salicylic acid and reactive oxygen species levels, and constitutive cell death in rosettes, resembling the phenotypes of autophagy-defective mutants. Starvation-activated autophagosome accumulation decreased in traf1a/b root cells, indicating that TRAF1a and TRAF1b function redundantly in regulating autophagosome formation. TRAF1a and TRAF1b interacted in planta with ATG6 and the RING finger E3 ligases SINAT1, SINAT2, and SINAT6 (with a truncated RING-finger domain). SINAT1 and SINAT2 require the presence of TRAF1a and TRAF1b to ubiquitinate and destabilize AUTOPHAGY PROTEIN6 (ATG6) in vivo. Conversely, starvation-induced SINAT6 reduced SINAT1- and SINAT2-mediated ubiquitination and degradation of ATG6. Consistently, SINAT1/SINAT2 and SINAT6 knockout mutants exhibited increased tolerance and sensitivity, respectively, to nutrient starvation. Therefore, TRAF1a and TRAF1b function as molecular adaptors that help regulate autophagy by modulating ATG6 stability in Arabidopsis.
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy, or “self-eating”) is a conserved pathway in all eukaryotic cells that is used to recycle nutrients via double-membrane vesicles termed autophagosomes, which target cytoplasmic contents and organelles to the lysosome or vacuole for degradation by resident hydrolases (He and Klionsky, 2009; Liu and Bassham, 2012; Li and Vierstra, 2012; Zhuang et al., 2015; Michaeli et al., 2016). The outer membrane of the autophagosome fuses with the tonoplast and releases an intravacuolar vesicle (autophagic body) containing the inner membrane of the autophagosome and the engulfed components (Liu and Bassham, 2012; Li and Vierstra, 2012). In response to various environmental stresses, such as nutrient starvation, hypoxia, oxidative stress, drought, high salt, and pathogen infection, autophagy serves as a protective mechanism that helps maintain cellular homeostasis and survival (Bassham et al., 2006; Kroemer et al., 2010; Han et al., 2011).
So far, over 36 evolutionarily conserved autophagy-related proteins (ATGs) in the core autophagic machinery have been identified in mammals, yeast, and plants (Liu and Bassham, 2012). In Arabidopsis thaliana, deletion of ATG genes causes hypersensitivity to nutrient deprivation, premature leaf senescence, shortened life span, alteration of the cellular metabolome, activated innate immunity, and impaired biotic and abiotic stress tolerance (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Liu et al., 2005; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Chung et al., 2010; Minina et al., 2013; Li et al., 2014; Chen et al., 2015; Avin-Wittenberg et al., 2015).
Typically, ATG proteins form three different complexes to govern the different steps of autophagosome formation, including initiation, nucleation, expansion, and autophagosome maturation (Yang and Klionsky, 2010; Liu and Bassham, 2012; Li and Vierstra, 2012). In Arabidopsis, the serine/threonine kinase ATG1 interacts with its regulatory components, ATG13, ATG11, and ATG101, forming a kinase complex that stimulates autophagic vesiculation, which is likely controlled by target of rapamycin kinase activity (Liu and Bassham, 2010; Suttangkakul et al., 2011; Li et al., 2014). Surprisingly, the plant ATG1/13 complex likely participates in a later step of autophagosome formation, i.e., autophagosome enclosure (Suttangkakul et al., 2011; Li et al., 2014). Given the evidence that the ATG1/13 kinase complex undergoes rapid autophagy-dependent degradation in the vacuole upon starvation, a novel feedback turnover mechanism that occurs during starvation-induced autophagy has been proposed (Suttangkakul et al., 2011; Li et al., 2014). By contrast, the regulation of nucleation by the ATG6 (Beclin-1 in mammals) complex has not been well defined in plants. In mammals, members of this complex are directly or indirectly regulated by ULK1 (the mammalian homolog of ATG1) through multiple phosphorylations (Yang and Klionsky, 2010). For example, ULK1 phosphorylates Beclin1 and stimulates the activity of the phosphatidylinositol 3-kinase VPS34 and the production of phosphatidylinositol-3-phosphate, which acts as a docking site for the recruitment of other regulatory proteins and as a lipid kinase complex to facilitate the nucleation of autophagic membranes (Yang and Klionsky, 2010). Finally, two ubiquitin-like conjugation pathways, ATG8-PE and ATG12-ATG5, function in mature autophagosome formation and cargo engulfment (Ohsumi, 2001; Geng and Klionsky, 2008; Liu and Bassham, 2012; Li and Vierstra, 2012).
Ubiquitination is an essential mechanism in which ubiquitin molecules are covalently attached to substrate proteins, which are typically targeted to the 26S proteasome for degradation (Kerscher et al., 2006). Recent investigations have highlighted the various roles of ubiquitin modification in regulating ATG protein stability during autophagosome formation (Xie et al., 2015; Popelka and Klionsky, 2015). Upon induction of autophagy, mammalian Beclin-1 undergoes K63-ubiquitination by the E3 ligases Cul4 and TRAF6 (Shi and Kehrl, 2010; Xia et al., 2013). Ubiquitinated Beclin-1 then interacts with VPS34 to increase its activity, thereby promoting autophagy. Conversely, nonubiquitinated Beclin-1 is unable to associate with VPS34, which stays in an inactive state, leading to suppressed autophagosome formation (Shi and Kehrl, 2010; Xia et al., 2013). TRAF6 also mediates K63-linked ubiquitination of ULK1, which subsequently promotes the stabilization and self-association of ULK1 and therefore activates autophagy (Nazio et al., 2013). However, recent studies have revealed that K11- and K48-linked ubiquitination of Beclin-1 by the E3 ligases Nedd4 and RNF216, respectively, have an opposite effect from that of K63-linked ubiquitination, instead leading to degradation of Beclin-1 and the suppression of autophagy (Platta et al., 2012; Xu et al., 2014). Although the regulation of autophagosome formation by ubiquitination of ATG proteins has been extensively studied in mammalian cells, it is still unclear whether plant autophagy involves a similar regulatory mechanism. Here, we report that two Arabidopsis TRAF family proteins, TRAF1a and TRAF1b, play an important role in regulating autophagosome formation by modulating ATG6 stability, a process that is likely mediated by the RING finger E3 ligases SINAT1, SINAT2, and SINAT6.
RESULTS
TRAF1a and TRAF1b Associate with Autophagosomes upon Starvation
The Arabidopsis genome contains more than 70 genes encoding TRAF domain-containing proteins (Oelmüller et al., 2005; Huang et al., 2016), but few of their functions have been characterized to date. To isolate TRAF family proteins with potential roles in autophagy, we cloned half of the TRAF family genes and fused them to GFP for transient expression in Arabidopsis protoplasts. We identified two TRAF proteins (encoded by At5g43560 and At1g04300) that associated with autophagic structures upon carbon starvation (Figure 1). Since these two proteins contain a single TRAF domain and their structures resemble that of mammalian TRAF1, we designated these proteins TRAF1a (At5g43560) and TRAF1b (At1g04300). These proteins were originally named MUSE14 and MUSE13, respectively (MUTANT, SNC1-ENHANCING 14; Huang et al., 2016), but we suggest renaming them to reflect their membership in the TRAF family.
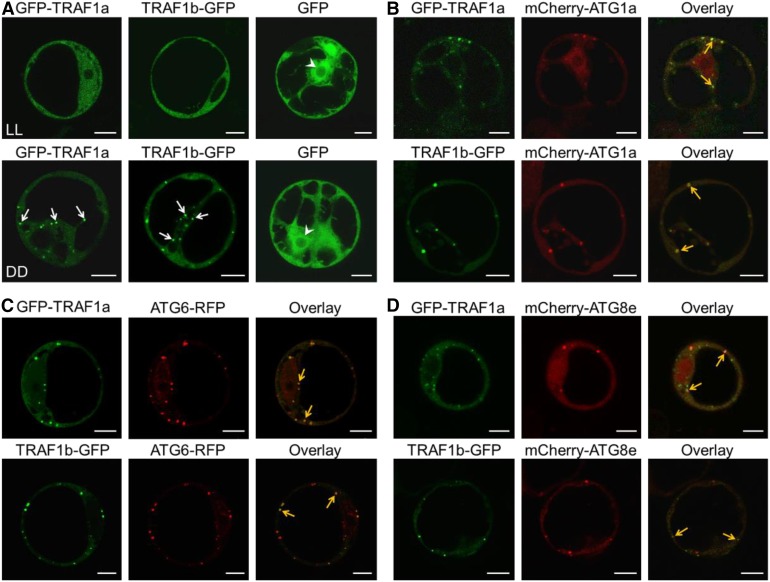
Figure 1.
TRAF1a and TRAF1b Associate with Autophagic Compartments.
(A) Subcellular localization of GFP-TRAF1a and TRAF1b-GFP in Arabidopsis protoplasts under constant light (LL) or dark (DD) conditions. GFP-TRAF1a, TRAF1b-GFP, and the GFP vector control were transiently expressed in Arabidopsis protoplasts, and their localizations were observed 12 h after transformation by confocal microscopy. The arrows indicate the translocation of GFP-TRAF1a and TRAF1b-GFP to the punctate structures upon dark treatment. The arrowheads indicate the nucleus.
(B) to (D) Colocalization of GFP-TRAF1a or TRAF1b-GFP fusions with the autophagy markers mCherry-ATG1a, ATG6-RFP, and mCherry-ATG8e expressed in Arabidopsis protoplasts under dark conditions. GFP-TRAF1a or TRAF1b-GFP was coexpressed with mCherry-ATG1a (B), ATG6-RFP (C), or mCherry-ATG8e (D) in Arabidopsis protoplasts for 12 h, followed by confocal microscopy analysis. Yellow arrows indicate the colocalization of GFP with mCherry/RFP. Bars = 10 μm.
Confocal laser scanning microscopy showed that under constant light conditions, both GFP-TRAF1a and TRAF1b-GFP fusions predominantly localized to the cytoplasm (Figure 1A). However, when the cells were incubated in continuous darkness for 12 h, fluorescent signals from both GFP-TRAF1a and TRAF1b-GFP appeared as punctate dots in the cytoplasm, but not in the vacuole (Figure 1A). As expected, GFP signals expressed from the empty vector control were detected in the cytosol and nuclei under both light and dark conditions (Figure 1A).
The punctate fluorescent signals in the cytoplasm from both GFP-TRAF1a and TRAF1b-GFP resemble signals from autophagosomes (Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010). To further verify the subcellular localization of TRAF1a and TRAF1b, we coexpressed GFP-TRAF1a and TRAF1b-GFP with distinct autophagy markers in protoplasts under constant darkness. As shown in Figure 1, the starvation-inducible punctate dots of GFP-TRAF1a and TRAF1b-GFP clearly colocalized with all of the autophagy markers, including mCherry-ATG1a (Figure 1B), ATG6-RFP (Figure 1C), and mCherry-ATG8e (Figure 1D), suggesting that TRAF1a and TRAF1b associate with starvation-induced autophagosomes in vivo.
Plants Lacking TRAF1a and TRAF1b Are Hypersensitive to Nutrient Deprivation
To further investigate the potential role of TRAF1a and TRAF1b in autophagy, we identified several T-DNA insertional mutants (Supplemental Figure 1) of these two genes. RT-PCR showed that no full-length transcripts were present in the corresponding traf1a or traf1b mutants (Supplemental Figure 1), indicating that all of these lines are knockout mutants. To test their functional redundancy, we crossed traf1a with traf1b to generate three independent double mutants, traf1a/b-1 (traf1a-1 traf1b-2), traf1a/b-2 (traf1a-2 traf1b-1), and traf1a/b-3 (traf1a-2 traf1b-3), for further phenotypic analyses.
Autophagy-defective mutants are characterized by premature leaf senescence and hypersensitivity to nutrient deprivation (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Liu et al., 2005; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Chung et al., 2010). When grown in either nutrient-rich or nutrient-deprived conditions, all of the TRAF1a and TRAF1b homozygous single mutants, including traf1a-1, traf1a-2, traf1b-1, traf1b-2, and traf1b-3, appeared similar to the wild type (Supplemental Figure 2). Also, 2-week-old traf1a/b-1, traf1a/b-2, and traf1a/b-3 double mutants did not display obvious phenotypes under nutrient-rich conditions (Figures 2A, 2B, 2D, and 2E). By contrast, like other autophagy-defective mutants, the traf1a/b-1, traf1a/b-2, and traf1a/b-3 double mutants showed significant hypersensitivity when grown in low-nitrogen Murashige and Skoog (MS) medium or after fixed-carbon starvation (Figure 2; Supplemental Figure 3). Following nitrogen starvation in solid medium for 5 d or in liquid medium for 4 d, all cotyledons of the double mutants exhibited increased yellowing, as calculated by the relative chlorophyll contents of the plants (Figures 2A to 2C; Supplemental Figures 3A and 3B). When 1-week-old wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 plants were transferred to constant darkness and subjected to fixed-carbon starvation for 7 d, the double mutants showed enhanced sensitivity, with yellowing cotyledons and true leaves, compared with the green leaves and significantly higher chlorophyll contents in wild-type plants (Figures 2D to 2G). Following a 7-d recovery under normal light/dark conditions, most of the wild-type plants survived, but >80% of the double mutants died (Figure 2F). The sensitivities of traf1a/b mutants to nutrient deprivation observed in this study were comparable to that observed for the atg10-1 mutant (Figure 2; Phillips et al., 2008).
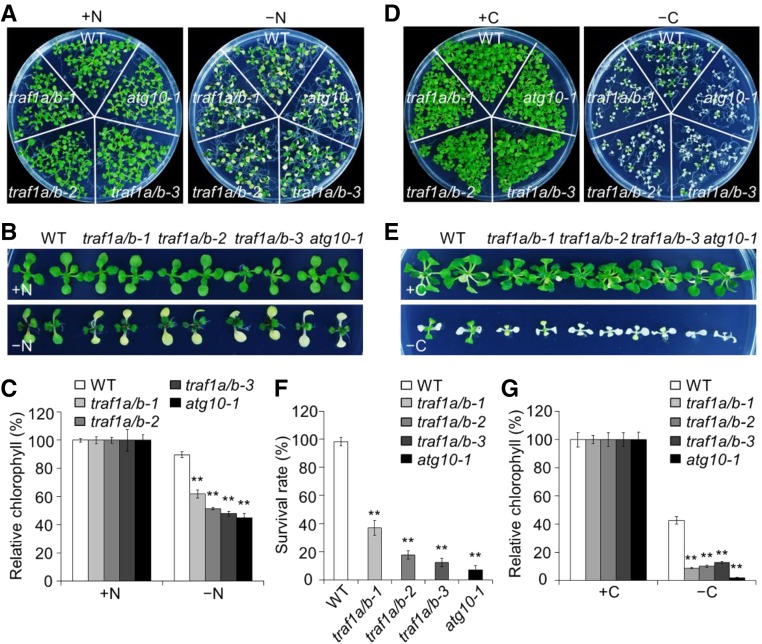
Figure 2.
Deletion of TRAF1a and TRAF1b Confers Hypersensitivity to Nitrogen and Carbon Starvation.
(A) and (B) Phenotypes of traf1a/b double mutants in response to nitrogen starvation. One-week-old wild-type (WT), traf1a/b, and atg10-1 seedlings grown on MS solid medium for 1 week. The seedlings were transferred to N-rich (+N) or N-deficient (–N) medium and photographed at 5 d after treatment.
(C) Relative chlorophyll contents of wild-type, traf1a/b, and atg10-1 seedlings upon 5-d nitrogen starvation. The relative chlorophyll contents were calculated by comparing the values in –N seedlings versus +N seedlings.
(D) and (E) Phenotype of traf1a/b double mutants in response to carbon starvation. One-week-old wild-type, traf1a/b, and atg10-1 were grown on MS agar with sucrose for 1 week. The seedlings were transferred to MS agar with sucrose (+C) or MS agar plates without sucrose followed by constant dark treatment (–C) for 7 d. The images were recorded after a 7-d recovery.
(F) and (G) Survival rates (F) and relative chlorophyll contents (G) of wild-type, traf1a/b, and atg10-1 mutants described in (D) following recovery. The relative chlorophyll contents were calculated by comparing the values of –C seedlings versus +C seedlings.
Relative chlorophyll contents and survival rates are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, five technical replicates pooled with 20 seedlings were used per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
To further investigate the essential role of TRAF1s in plant development and tolerance to nutrient starvation, we performed a complementation test by introducing the TRAF1a-FLAG construct into the traf1a/b-2 mutant to generate the TRAF1a-FLAG traf1a/b-2 line. Phenotypic analyses showed that the dwarfed growth and increased sensitivity to carbon starvation in traf1a/b-2 were completely rescued by the TRAF1a-FLAG transgene (Supplemental Figures 4A and 4B). Moreover, a recent report demonstrated that the growth defects and autoimmunity phenotype in muse13-2 muse14-1 mutants (traf1a/b in this study) were suppressed by the snc1-r1 mutant (Huang et al., 2016). To uncouple the autophagy deficiency-related phenotypes from the autoimmunity phenotypes in these mutants, we analyzed the response of the snc1-r1 muse13-2 muse14-1 triple mutant (Huang et al., 2016) to nutrient starvation. When 3-week-old plants were starved for fixed carbon for 7 d, both the snc1-r1 muse13-2 muse14-1 triple mutant and the muse13-2 muse14-1 double mutant exhibited enhanced sensitivities in comparison with the wild type and the snc1-r1 single mutant (Supplemental Figures 4C and 4D).
During the first 2 weeks of growth, the traf1a/b-1, traf1a/b-2, and traf1a/b-3 double mutants showed few morphological differences compared with the wild type. At 3 weeks old, all three independent traf1a/b lines were extremely dwarfed; however, natural leaf senescence was not detected in wild-type or double mutant plants at this stage (Figure 3). Interestingly, the cotyledons and some true leaves in the 4-week-old traf1a/b lines were yellow, in contrast to the green cotyledons observed in the wild type, indicating the onset of senescence (Figure 3A). However, unlike the classical autophagy-defective mutants, all traf1a/b double mutants showed extended life cycles, with delayed senescence of the remaining true leaves, in 5-week-old and 6-week-old plants compared with the wild type (Figure 3A). We detected significant declines in chlorophyll contents in the leaves of wild-type plants at 5 and 6 weeks old (versus 3 weeks old), but these levels were significantly higher in 5- and 6-week-old traf1a/b leaves than in the wild type (Figure 3B). Statistical analyses showed that the periods of rosette senescence and flowering were significant longer in the traf1a/b mutants than in the wild type, indicative of extended life cycles by knockout of TRAF1a and TRAF1b (Supplemental Figure 5).
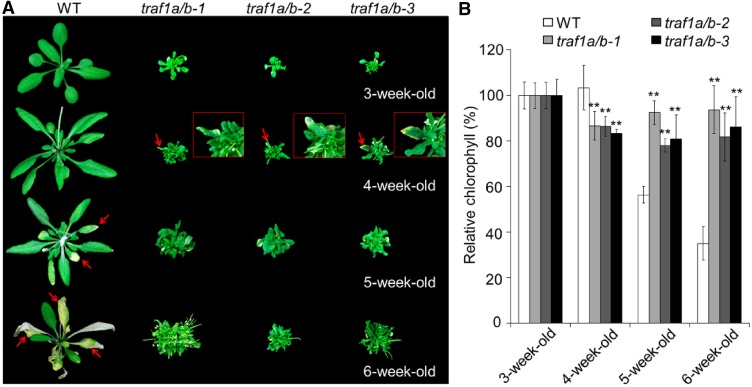
Figure 3.
Age-Dependent Senescence Phenotype of traf1a/b Double Mutants.
(A) Images showing the onset of leaf senescence in wild-type (WT) and traf1a/b lines grown under normal light/dark growth conditions. Photographs were taken at 3, 4, 5, and 6 weeks after germination. Arrows indicate senescent leaves.
(B) Relative chlorophyll content of plants grown under normal light/dark growth conditions for the indicated times. The values of 3-week-old wild-type and traf1a/b plants were set at 100%, and the relative chlorophyll contents of wild-type and traf1a/b leaves at the other stages were calculated accordingly. Data are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, five whole plants (technical replicates) were used per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
The observations of dwarf stature and extended life span in the traf1a/b mutants prompted us to generate TRAF1 knockdown lines. To this end, the TRAF1b-RNAi construct was introduced into the traf1a-1 mutant to obtain two traf1a TRAF1b-RNAi lines (Supplemental Figure 6). Similar to that of traf1a/b double mutants, the traf1a TRAF1b-RNAi lines showed increased sensitivity to both carbon and nitrogen starvation in comparison with the wild type (Supplemental Figures 6A to 6D). At 4 weeks old, the traf1a TRAF1b-RNAi plants displayed smaller sizes than that of the wild type, but they grew normally (Supplemental Figure 6E). In comparison, both traf1a TRAF1b-RNAi lines showed accelerated leaf senescence at 5 and 6 weeks old with similar life cycles to those of wild-type plants (Supplemental Figures 6E and 6F).
Loss of TRAF1a and TRAF1b Constitutively Increases Salicylic Acid Levels, Activates PR Gene Expression, and Enhances Cell Death and Hydrogen Peroxide Production
Autophagy is a protective mechanism that is essential for the disposal of damaged organelles and for the maintenance of cellular reactive oxygen species (ROS) homeostasis in plants under stress conditions, a process likely controlled by the salicylic acid (SA) signaling pathway (Liu and Bassham, 2012; Chen et al., 2015). Given that SA, jasmonates (JAs), and ROS accumulate in Arabidopsis atg mutants (Yoshimoto et al., 2009), we examined the SA, JA, and ROS levels in the TRAF1a- and TRAF1b-knockout lines using liquid chromatography-mass spectrometry. We measured endogenous SA and JA levels in wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 plants under normal growth conditions. As shown in Figure 4A, the SA and JA contents were ∼20-fold higher in the traf1a/b double mutants than in the wild type. A simultaneous increase in SA and JA was also observed in the atg5 mutant (Yoshimoto et al., 2009).
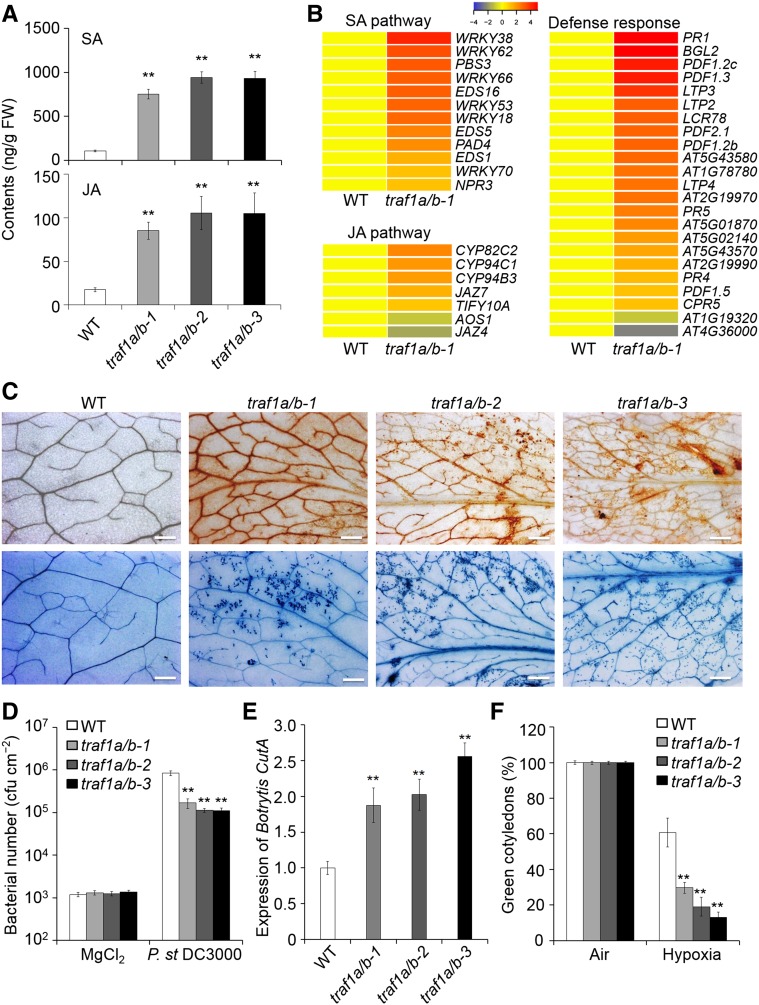
Figure 4.
Loss of TRAF1a and TRAF1b Enhances SA Signaling and Leads to Altered Biotic and Abiotic Stress Tolerance.
(A) Endogenous SA and JA contents in the wild type (WT) and traf1a/b double mutants. Three-week-old wild-type and traf1a/b mutants grown under a normal light/dark cycle were used for plant hormone extraction following liquid chromatography/mass spectrometry analysis. D4-SA and D5-JA were added as internal quantitative standards. The experiment was repeated twice (biological replicates) with similar results. Data are average values ± sd (n = 5) calculated from five independent technical replicates (for each replicate, two to three plants were pooled) per experiment. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test). FW, fresh weight.
(B) Hierarchical cluster analysis applied to the 12 DEGs (more than 2-fold and false discovery rate < 0.05) in the SA pathway, seven DEGs in the JA pathway, and 23 DEGs representing defense response-related genes by comparing the traf1a/b mutant to the wild type. The transcriptional profiles of relative gene expression values were analyzed using the heat map 2.0 command in R. Red and blue represent upregulated and downregulated genes, respectively.
(C) DAB (upper lane) and trypan blue (bottom lane) staining showing ROS accumulation and cell death in the leaves of 3-week-old wild-type and traf1a/b plants under normal growth conditions. Bars = 500 µm.
(D) Number of bacteria in the in planta bacterial growth assay. Plants were inoculated with Pst DC3000 or 10 mM MgCl2, and bacterial growth was evaluated 4 d after inoculation. The experiment was repeated three times (biological replicates). Data are average values ± sd (n = 3) from three independent experiments. For each experiment, five technical replicates (each replicate pooled with three leaves) were used per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test). cfu, colony-forming units.
(E) Expression of the B. cinerea CutA (Z69264) gene in the wild type and traf1a/b double mutants at 6 d postinoculation. Four-week-old plants were inoculated with B. cinerea or 2% glucose for 6 d, and genomic DNA was extracted from infected plants. Quantification of fungal growth was determined by qPCR analysis using the B. cinerea cutinase-specific primer pairs XS3715/XS3716 (see Supplemental Data Set 2). The experiment was repeated three times (biological replicates). Data are average values ± sd (n = 3) from three independent experiments. For each experiment, three technical replicates (each replicate pooled with three leaves) were used per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
(F) Response of traf1a/b mutants to hypoxia. Wild-type and traf1a/b seeds were germinated on MS with 2% sucrose under normal air or 3% O2 conditions, and the percentage of green cotyledons was calculated after hypoxia treatment for 10 d. The experiment was repeated three times (biological replicates). Data are average values ± sd (n = 3) from three independent experiments. For each experiment, over 20 seedlings were used for calculation per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
To further investigate the genome-wide effects of TRAF1a and TRAF1b depletion, we subjected 3-week-old wild-type and traf1a/b-1 plants to transcriptome sequencing analysis. We detected 2869 genes that were differentially expressed by more than 2.0-fold, including 2523 upregulated and 346 downregulated genes in the traf1a/b-1 mutant compared with the wild type (Supplemental Data Set 1). Further analysis of the differentially expressed genes (DEGs) revealed that the expression of genes involved in cell metabolism, stress responses, plant development, and signaling was significantly altered in the traf1a/b-1 double mutant compared with the wild type (Supplemental Figure 7A). In particular, genes encoding enzymes involved in SA or JA biosynthesis and signaling, including EDS1, PAD4, and AOS1, as well as several WRKY and JAZ genes, were upregulated in traf1a/b-1 (Figure 4B). Due to the accumulation of defense phytohormones in traf1a/b plants, the transcript levels of many SA- and JA-associated defense-responsive genes, such as PR1, PR2, PR5, and PDF1.2, were significantly higher in the double mutants than in the wild type (Figure 4B; Supplemental Figure 7C). Moreover, the transcript levels of senescence-associated genes, including DARK INDUCIBLE2, SENESCENCE1 (SEN1), SENESCENCE-ASSOCIATED GENE13 (SAG13), SAG21, SAG29, SAG101, YELLOW LEAF-SPECIFIC GENE2 (YLS2), and YLS5, were significantly higher in the 3-week-old traf1a/b mutants than in the wild type (Supplemental Figure 7B). Consistent with the extended life cycles in the traf1a/b mutants, we found that the SA and JA levels and SAG12 and SAG101 transcript levels in the 6-week-old traf1a/b mutants were reduced in comparison with the wild type (Supplemental Figure 8).
In addition to increased SA signaling, the autophagy-defective mutants atg2 and atg5 exhibit increased ROS levels and enhanced cell death (Yoshimoto et al., 2009). To investigate whether traf1a/b plants show increased ROS accumulation and constitutive cell death, we examined the rosettes of 3-week-old wild-type and traf1a/b plants using diaminobenzidine (DAB) and trypan blue staining. As shown in Figure 4C, traf1a/b leaves generated high levels of H2O2, as indicated by the brown color upon DAB staining, compared with the wild-type control. When trypan blue staining was used to detect cell death in situ, lesions were apparent in the leaves of traf1a/b plants, but not in the wild type (Figure 4C).
The traf1a/b Mutants Show Altered Sensitivity to Biotic and Abiotic Stress
Given that autophagy contributes to the regulation of plant responses to both biotic (e.g., bacterial and fungal pathogens) and abiotic (e.g., hypoxia) stress in Arabidopsis (Lenz et al., 2011; Chen et al., 2015), we investigated whether the deletion of TRAF1a and TRAF1b would affect plant stress resistance. To this end, we inoculated wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 plants with the virulent bacteria Pseudomonas syringae pv tomato DC3000 and the necrotrophic fungi Botrytis cinerea, which are widely used to evaluate plant defense responses (Lenz et al., 2011; Xiao and Chye, 2011). Bacterial growth, as measured by the number of bacteria before inoculation and 4 d after inoculation, was significantly lower in traf1a/b than in the wild type (Figure 4D). In contrast, the relative transcript levels of the B. cinerea CutA gene, which can be used as a marker for B. cinerea susceptibility (Aubert et al., 2015), were significantly higher in traf1a/b mutants than in the wild type (Figure 4E), indicating that deletion of TRAF1a and TRAF1b resulted in increased resistance to P. syringae pv tomato DC3000 and decreased resistance to B. cinerea.
We recently reported that all of the atg mutants are hypersensitive to hypoxic stress (Chen et al., 2015). When wild-type and traf1a/b-1, traf1a/b-2, and traf1a/b-3 seeds were germinated under low oxygen conditions (3% O2, compared with normal ∼21% atmospheric O2) for 10 d, ∼60% of wild-type seeds germinated normally, as indicated by the percentage of green cotyledons, compared with wild-type seeds under normal air conditions (Figure 4F). By contrast, all three traf1a/b lines showed significantly lower germination rates than the wild type (Figure 4F). These results imply that, like the other atg mutants (Lenz et al., 2011; Chen et al., 2015), the traf1a/b mutants are more resistant to bacterial infection but less tolerant to necrotrophic fungal infection and hypoxic stress than the wild type.
TRAF1a and TRAF1b Are Required for Autophagosome Formation and Autophagy Protein Turnover
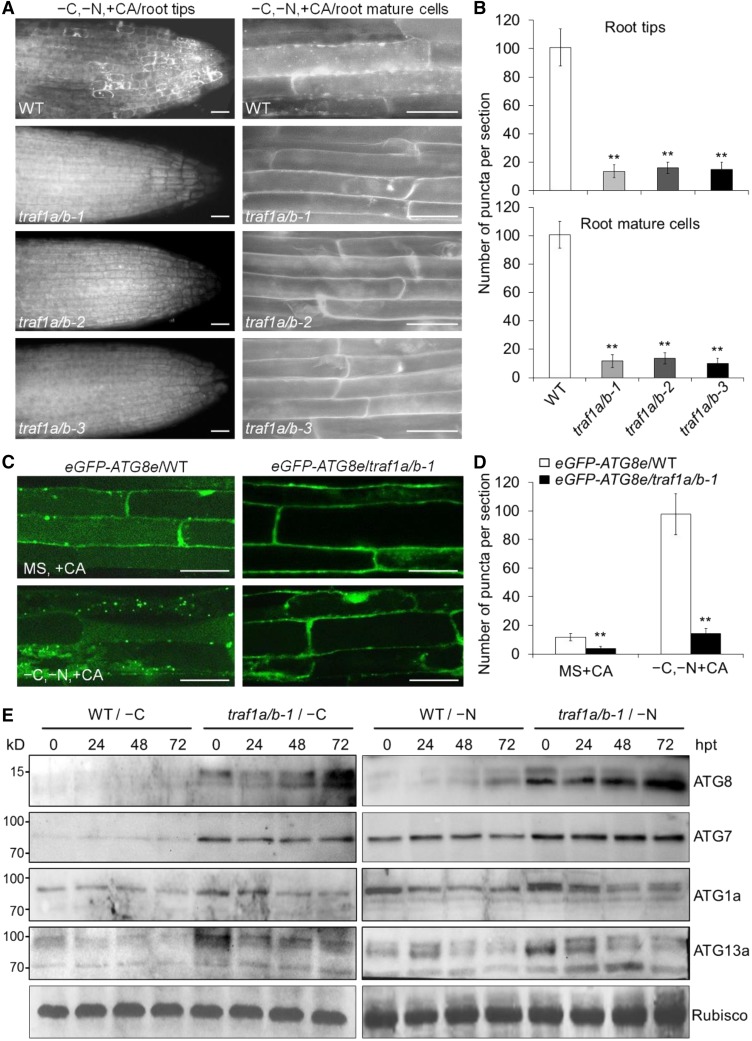
To further assess the role of TRAF1a and TRAF1b in autophagy, we transferred 7-d-old wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 seedlings to nitrogen- and sucrose-deficient MS medium for a 16-h treatment, followed by staining with monodansylcadaverine (MDC) to detect autophagosomes. After the addition of the autophagy inhibitor concanamycin A (CA), MDC-stained vesicles accumulated in wild-type root tips and mature root cells upon starvation (Figure 5A), whereas such accumulation was strongly reduced in the traf1a/b-1, traf1a/b-2, and traf1a/b-3 root cells after nitrogen and sucrose starvation (Figures 5A and 5B). To confirm that the reduction of MDC-labeled dot-like structures in the traf1a/b mutants represented the loss of autophagosomes, we crossed the traf1a/b-1 mutant to the eGFP-ATG8e transgenic line, a well-characterized autophagosome marker line (Xiao et al., 2010), to generate eGFP-ATG8e/traf1a/b-1 plants. Confocal microscopy analysis suggested that after nutrient starvation, the formation of GFP-labeled punctuate structures (autophagosomes or their intermediates) was markedly induced in wild-type root cells (Figures 5C and 5D). However, such accumulation was not evident in the traf1a/b-1 background under either nutrient-rich or starvation conditions (Figures 5C and 5D). We next used the release of free GFP to monitor the autophagic transport and degradation of GFP-ATG8e reporter into the vacuole (Chung et al., 2010; Chen et al., 2015). We found that, consistent with the microscopy results, free GFP levels were substantially reduced in the traf1a/b-1 mutant compared with the wild type in response to carbon starvation (Supplemental Figure 9).
Figure 5.
TRAF1a and TRAF1b Are Required for Autophagosome Formation.
(A) MDC staining of wild-type (WT) and traf1a/b root cells after nitrogen and carbon starvation. One-week-old seedlings were starvation treated (–C and –N) for 16 h with 1 μM CA, followed by staining with MDC. The labeled autophagosomes were visualized by epifluorescence microscopy. Bars = 50 µm.
(B) Numbers of puncta per root section in the root tips (left graph) and mature root cells (right graph) of the wild type and traf1a/b in (A). Data are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, 15 sections were used for the calculation for each genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
(C) Confocal analysis of eGFP-ATG8e/WT and eGFP-ATG8e/traf1a/b-1 lines. One-week-old eGFP-ATG8e/WT and eGFP-ATG8e/traf1a/b-1 lines were exposed to N- and C-sufficient (MS) or N- and C-deficient (–C and –N) conditions with 1 μM CA for 16 h and visualized by fluorescence confocal microscopy. Bars = 50 μm.
(D) Numbers of puncta per root section in the root cells of the wild type and traf1a/b in (C). Data are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, 15 sections were used for the calculation for each genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
(E) ATG (ATG8, ATG7, ATg1a, and ATG13a) protein levels in the wild type and traf1a/b-1 double mutant after carbon starvation (left images) and nitrogen starvation (right images) treatments for the indicated times. Ponceau S-stained membranes are shown below the blots to indicate the amount of protein loaded per lane. The numbers on the left indicate the molecular mass (kD) of the size markers. hpt, hours post-treatment.
Given the evidence that many autophagy proteins such as ATG8, ATG1, and ATG13 are strongly regulated by autophagy and that their levels are elevated in various atg mutant backgrounds (Suttangkakul et al., 2011), we examined the ATG protein levels in the traf1a/b mutants using specific antibodies. Protein gel blot analysis revealed that under either nutrient-rich or starvation conditions, ATG8, ATG7, ATG1a, and ATG13a accumulated to high levels in the traf1a/b-1 double mutant compared with the wild type (Figure 5E). However, qRT-PCR analyses indicated that the traf1a/b-1 double mutant and wild-type plants showed similar starvation-induced transcript levels of the ATG genes (Supplemental Figure 10). We also found that upon carbon or nitrogen starvation treatment for 48 h, the levels of both ATG1a and ATG13a decreased in comparison to that of the untreated control (0 h; Figure 5E), but the proteins were not completely degraded, possibly due to the weaker levels of starvation induced by the separate carbon and nitrogen starvation treatments. Meanwhile, different patterns of ATG8s, as indicated by the different molecular sizes of protein bands, were detected in the traf1a/b mutants upon carbon or nitrogen deprivation (Figure 5E), further indicating that different ATG8s may play distinct roles in response to starvation for various nutrients. Together, these findings suggest that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation and autophagy protein turnover.
TRAF1a and TRAF1b Physically Interact with ATG6
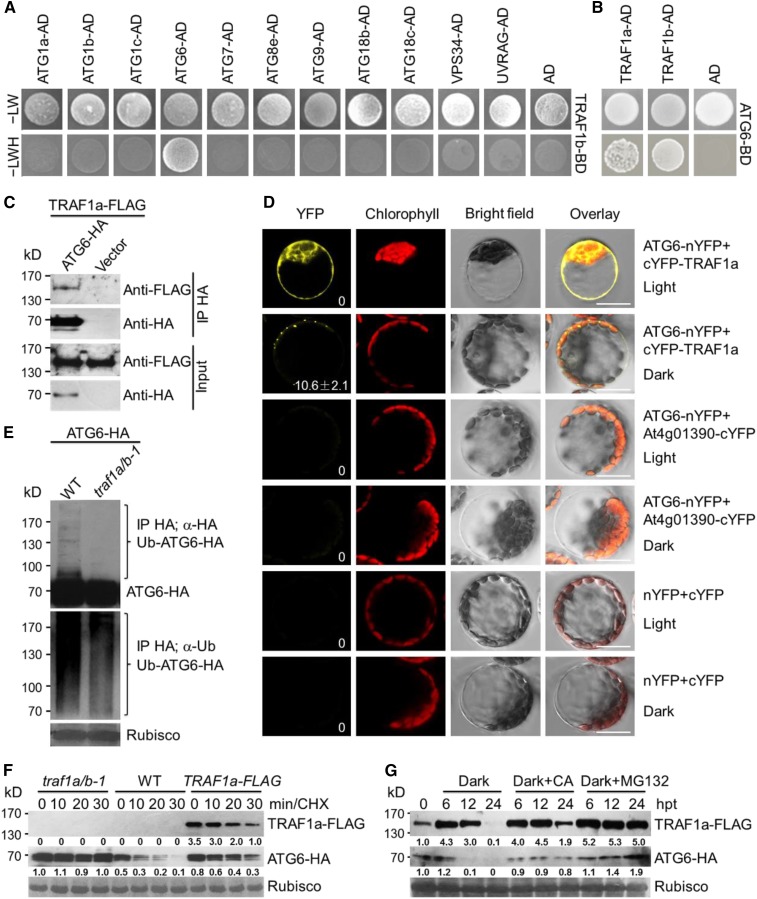
To search for TRAF1a- and TRAF1b-associated autophagy components, we used yeast two-hybrid (Y2H) analysis to screen for interactions of TRAF1a and TRAF1b with known autophagy proteins. Using TRAF1b as bait, we found that TRAF1b specifically interacted with ATG6, but not with other ATG proteins, including ATG1a, ATG1b, ATG1c, ATG7, ATG8e, ATG9, ATG18b, ATG18c, VPS34, and UVRAG (Figure 6A). When ATG6 was used as bait, it interacted with both TRAF1a and TRAF1b (Figure 6B). To further test the interaction between TRAF1a and ATG6 in planta, we generated stable transgenic lines expressing TRAF1a-FLAG. When ATG6-HA was transiently expressed in protoplasts isolated from TRAF1a-FLAG Arabidopsis leaves grown in darkness for 16 h, TRAF1a-FLAG could be immunoprecipitated by ATG6-HA (Figure 6C). The interaction between TRAF1a and ATG6 was further confirmed by bimolecular fluorescence complementation (BiFC) in Arabidopsis protoplasts. When ATG6-nYFP (yellow fluorescent protein) and cYFP-TRAF1a were transiently coexpressed in wild-type protoplasts for 16 h under light or dark conditions, fluorescent BiFC signals were detected in the cytoplasm under light conditions but were observed as punctate structures under dark conditions (Figure 6D). In contrast, coexpression of the negative controls ATG6-nYFP/At4g01390-cYFP and nYFP/cYFP failed to reconstitute an intact YFP in Arabidopsis leaf protoplasts under either light or dark conditions (Figure 6D). These findings imply that in response to starvation, TRAF1s and ATG6 proteins colocalize to autophagosome-related structures.
Figure 6.
TRAF1a and TRAF1b Interact with ATG6 and Modulate Its Stability.
(A) Y2H assay of the interaction between TRAF1b with ATG proteins (ATG1a, ATG1b, ATG1c, ATG6, ATG7, ATG8e, ATG9, ATG18b, ATG18c, VPS34, and UVRAG). Full-length TRAF1b was fused to the BD domain and coexpressed with the indicated ATG-AD proteins in yeast strain YH109. The positive clones were selected on SD medium lacking Trp, Leu, His, and Ade (–LWH). AD indicates empty AD plasmid.
(B) Y2H confirmation of the binary interaction of ATG6 with TRAF1a and TRAF1b. Full-length ATG6 protein was fused to the BD domain and coexpressed with TRAF1a-AD and TRAF1b-AD in yeast. To prevent ATG6 self-activation, 5 mM 3-amino-1,2,4-triazole was added to the selection medium. AD indicates empty AD plasmid.
(C) In vivo CoIP assay of the association between ATG6 and TRAF1a. HA-tagged ATG6 (ATG6-HA) was transiently expressed in protoplasts from transgenic plants expressing TRAF1a-FLAG and immunoprecipitated by HA affinity agarose beads.
(D) BiFC assay of ATG6 and TRAF1a in Arabidopsis. The split nYFP and cYFP fusions ATG6-nYFP and cYFP-TRAF1a were coexpressed in leaf protoplasts for 16 h under light or dark conditions. The ATG6-nYFP/At4g01390-cYFP fusions and nYFP/cYFP vectors were similarly coexpressed as negative controls. Confocal images obtained from YFP, autofluorescent chlorophyll, and bright field are shown. The number in the image of ATG6-nYFP+cYFP-TRAF1a/dark indicates the average autophagosome number ± sd (n = 3) calculated from three independent experiments. For each experiment, 20 cells were used for the calculation per pair of constructs. Bars = 10 μm.
(E) ATG6 ubiquitination is reduced in the traf1a/b-1 mutant. ATG6-HA was transiently expressed in Arabidopsis protoplasts isolated from wild-type (WT) and traf1a/b-1 mutant plants, and its ubiquitination was detected by protein blot analysis. Proteins were extracted at 16 h after expression, and the blots were probed with anti-HA and anti-Ub antibodies.
(F) The degradation of ATG6 is attenuated in both traf1a/b-1 and TRAF1a-FLAG plants. ATG6-HA was transiently expressed in protoplasts from wild-type, traf1a/b-1, and TRAF1a-FLAG plants for 16 h and incubated in the same medium with 50 μM CHX. The samples were collected at 0, 10, 20, and 30 min after treatment. The blots were probed with anti-HA and anti-FLAG antibodies. Relative intensity of TRAF1a-FLAG and ATG6-HA normalized to the loading control is shown below.
(G) TRAF1a-FLAG and ATG6-HA accumulate following treatment with constant darkness, CA, or MG132. One-week-old transgenic lines expressing TRAF1a-FLAG and ATG6-HA were treated with dark, 50 μM MG132, or 1 μM CA for 0, 6, 12, and 24 h. Relative intensity of TRAF1a-FLAG and ATG6-HA normalized to the loading control is shown below. hpt, hours post-treatment.
The numbers on the left indicate molecular masses (kD) of the size markers. Ponceau S-stained membranes are shown below the blots to indicate the equal amounts of protein loaded per lane.
To investigate the relationship between TRAF1a/TRAF1b and ATG6 ubiquitination, we determined the levels of ubiquitinated ATG6 in the absence or presence of TRAF1a and TRAF1b. When ATG6-HA was transiently expressed in protoplasts isolated from wild-type and traf1a/b-1 plants, ATG6-HA ubiquitination was reduced in the traf1a/b-1 mutant compared with the wild type (Figure 6E). To investigate the involvement of TRAF proteins in modulating ATG6 stability, we expressed the ATG6-HA plasmid in protoplasts from wild-type, traf1a/b-1, and TRAF1a-FLAG plants for 16 h and examined the protein stability of ATG6-HA in the presence of cycloheximide (CHX), which blocks new protein biosynthesis. As shown in Figure 6F, the protein level of ATG6-HA without treatment (0 h) was always lower in wild-type cells than in traf1a/b-1 and TRAF1a-FLAG cells. In particular, ATG6-HA was unstable after 10, 20, and 30 min of CHX treatment in wild-type cells. However, the degradation of ATG6-HA was significantly inhibited in traf1a/b-1 cells and in cells expressing TRAF1a-FLAG (Figure 6F). Protein analysis in transgenic lines stably expressing TRAF1a-FLAG and ATG6-HA showed that TRAF1a-FLAG accumulated after exposure to constant darkness for 6 and 12 h but was subsequently degraded at 24 h after carbon starvation. By contrast, ATG6-HA levels increased at 6 h, followed by a rapid decline (Figure 6G). Moreover, when dark-treated seedlings were treated with CA and MG132, the degradation of both TRAF1a-FLAG and ATG6-HA was strongly inhibited by MG132 and CA (Figure 6G), suggesting that TRAF1a and ATG6 are highly regulated by the 26S proteasome and autophagy, the two dominant proteolytic pathways, in Arabidopsis cells. Together, these results suggest that TRAF1a and TRAF1b are likely involved in autophagy by physically interacting with ATG6 in planta.
ATG6 Is a Target of the SINAT1, SINAT2, and SINAT6
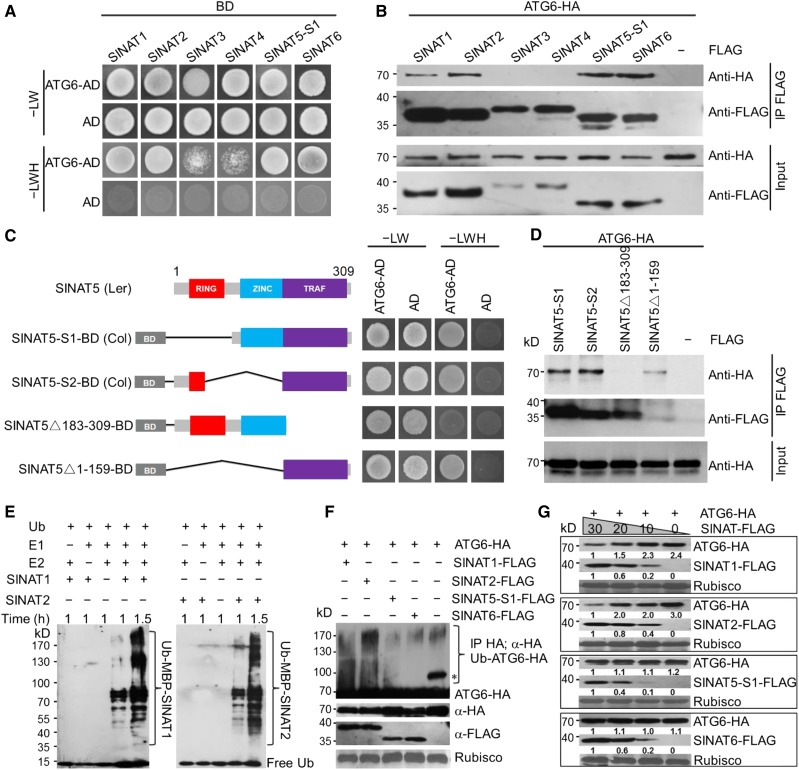
Unlike most mammalian TRAF proteins, which harbor a C-terminal TRAF domain and RING finger domains as well as several zinc finger motifs at their N termini (Arch et al., 1998; Chung et al., 2002), Arabidopsis TRAF1a and TRAF1b contain only an N-terminal TRAF domain and no other conserved domains. The lack of RING finger domains in these two proteins suggests that they recruit other E3 ligases to ubiquitinate ATG6. To further understand the molecular basis of ATG6 processing, we used ATG6 as bait in Y2H experiments to identify the potential interactions of ATG6 with known proteins that contain both TRAF and RING finger domains. One protein that interacted with ATG6 is SINAT2, one of five members of a protein family with diverse functions in Arabidopsis (Xie et al., 2002; Park et al., 2010). We performed Y2H and coimmunoprecipitation (CoIP) assays to investigate the association of ATG6 with all five SINAT proteins, finding that ATG6 interacted strongly with SINAT1 and SINAT2 in both assays (Figures 7A and 7B). The full-length SINAT5 CDS was previously identified from Arabidopsis ecotype Landsberg (Xie et al., 2002). However, we failed to amplify SINAT5 from Arabidopsis ecotype Columbia (Col-0). Instead, we identified two alternatively spliced forms, SINAT5-S1 and SINAT5-S2, lacking the RING finger or zinc finger domain in Col-0 (Figure 7C); Y2H and CoIP assays showed that both SINAT5-S1 and SINAT5-S2 interacted with ATG6 (Figures 7A to 7D). SINA2, which we term SINAT6 for consistency here, is another SINAT homolog with truncated RING finger and zinc finger domains (Bao et al., 2014) and could also interact with ATG6 in both assays (Figures 7A and 7B). To clarify the domains mediating the interaction between SINATs and ATG6, we generated two more truncated SINAT5 mutants containing different domains for analysis (Figure 7C). Y2H and CoIP suggested that the C-terminal TRAF domain in SINAT5 is essential for the interaction between ATG6 and SINAT5 (Figures 7C and 7D).
Figure 7.
ATG6 Is a Target of SINAT1, SINAT2, SINAT5, and SINAT6.
(A) Y2H analysis showing the interaction between ATG6 and SINAT proteins. ATG6-AD and SINAT-BD (SINAT1-BD, SINAT2-BD, SINAT3-BD, SINAT4-BD, SINAT5-S1-BD, and SINAT6-BD) were coexpressed in yeast and selected on SD/-Trp-Leu-His-Ade medium (–LWH) containing 5 mM 3-amino-1,2,4-triazole. AD indicates empty AD plasmid.
(B) In vivo CoIP analysis showing the physical interaction between ATG6 and SINATs. HA-tagged ATG6 (ATG6-HA) was coexpressed with FLAG-tagged SINATs (SINAT1-FLAG, SINAT2-FLAG, SINAT3-FLAG, SINAT4-FLAG, SINAT5-S1-FLAG, and SINAT6-FLAG) in Arabidopsis protoplasts and immunoprecipitated by FLAG affinity magnetic beads.
(C) Truncation analysis of SINAT5 to identify the functional domain mediating the ATG6-SINAT5 association. Full-length SINAT5 amplified from ecotype Landsberg erecta containing a RING finger (RING), a zinc finger (ZINC), and a TRAF domain (TRAF). SINAT5-S1 and SINAT5-S2 are two alternatively spliced products of ecotype Col-0 containing a TRAF domain with impaired RING or ZINC domains. △183-309 and △1-159 are artificial truncated proteins without the TRAF domain or RING/ZINC domains. Truncated SINAT5 was fused to the BD domain and coexpressed with ATG6-AD in yeast. Positive clones were selected on SD medium lacking Trp, Leu, His, and Ade (–LWH). AD indicates empty AD plasmid.
(D) In vivo CoIP assay showing the association between ATG6-HA and FLAG-tagged SINAT5 truncations (SINAT5-S1, SINAT5-S2, SINAT5△183-309, and SINAT5△1-159). The plasmids were coexpressed in Arabidopsis protoplasts and immunoprecipitated by FLAG affinity magnetic beads.
(E) In vitro ubiquitination assay of SINAT1 and SINAT2. Recombinant proteins MBP-SINAT1 and MBP-SINAT2 purified from Escherichia coli were assayed for E3 activity in the presence of wheat E1, human E2, and ubiquitin, as indicated.
(F) In vivo ubiquitination of ATG6 by SINAT1, SINAT2, SINAT5-S1, and SINAT6. ATG6-HA was coexpressed with SINAT1-FLAG, SINAT2-FLAG, SINAT5-S1-FLAG, or SINAT6-FLAG in Arabidopsis protoplasts, and its ubiquitination was detected by protein blot analysis. The asterisk indicates an unidentified nonspecific band in the vector control.
(G) Effects of SINAT1, SINAT2, SINAT5-S1, and SINAT6 on ATG6 protein stability. ATG6-HA was coexpressed with various amounts (0, 10, 20, and 30 μg) of SINAT1-FLAG, SINAT2-FLAG, SINAT5-S1-FLAG, or SINAT6-FLAG for 16 h, and the blots were probed with anti-HA and anti-FLAG antibodies. Relative intensity of each protein normalized to the loading control is shown below. The numbers on the left indicate the molecular mass (kD) of each size marker.
As SINAT5 possesses E3 ligase activity in vitro and in vivo (Xie et al., 2002) and SINAT1, SINAT2, SINAT5-S1, SINAT5-S2, and SINAT6 interact with ATG6 (Figures 7A to 7D), we examined whether SINAT1 and SINAT2 also have E3 ligase activity using an in vitro assay (Zhao et al., 2013). We found that in the presence of ubiquitin, E1 activating enzyme, and E2 conjugating enzyme, both maltose binding protein (MBP)-tagged SINAT1 and SINAT2 recombinant proteins could undergo self-ubiquitination in vitro (Figure 7E).
The interaction between SINATs and ATG6 suggests that SINATs ubiquitinate ATG6 and affect its protein stability. To test this possibility, we coexpressed ATG6-HA with SINAT1-FLAG, SINAT2-FLAG, SINAT5-S1-FLAG, and SINAT6-FLAG fusions in Arabidopsis protoplasts, isolated total proteins, and subjected them to immunoprecipitation and protein gel blot analysis with anti-HA or anti-FLAG antibodies. As shown in Figure 7F, ATG6-HA was strongly ubiquitinated by SINAT1-FLAG and SINAT2-FLAG but was not ubiquitinated by SINAT5-S1-FLAG and SINAT6-FLAG, compared with the vector control. We note that in the absence of SINATs, the immunoblot showed an unspecific band (indicated by the asterisk in Figure 7F), which may represent an unidentified modification of ATG6-HA. The nature of this unspecific band remains to be determined. Analysis of in vivo ATG6 levels in wild-type protoplasts coexpressing SINAT1-FLAG, SINAT2-FLAG, SINAT5-S1-FLAG, or SINAT6-FLAG with ATG6-HA revealed that the expression of SINAT1-FLAG and SINAT2-FLAG increased the degradation of ATG6-HA in a dose-dependent manner, whereas SINAT5-S1-FLAG and SINAT6-FLAG expression had little effect on ATG6-HA stability (Figure 7G). These findings suggest that ATG6 is a direct target of SINAT1, SINAT2, SINAT5, and/or SINAT6 in Arabidopsis cells.
TRAF1a and TRAF1b Are Essential for SINAT1- and SINAT2-Mediated Ubiquitination and Destabilization of ATG6
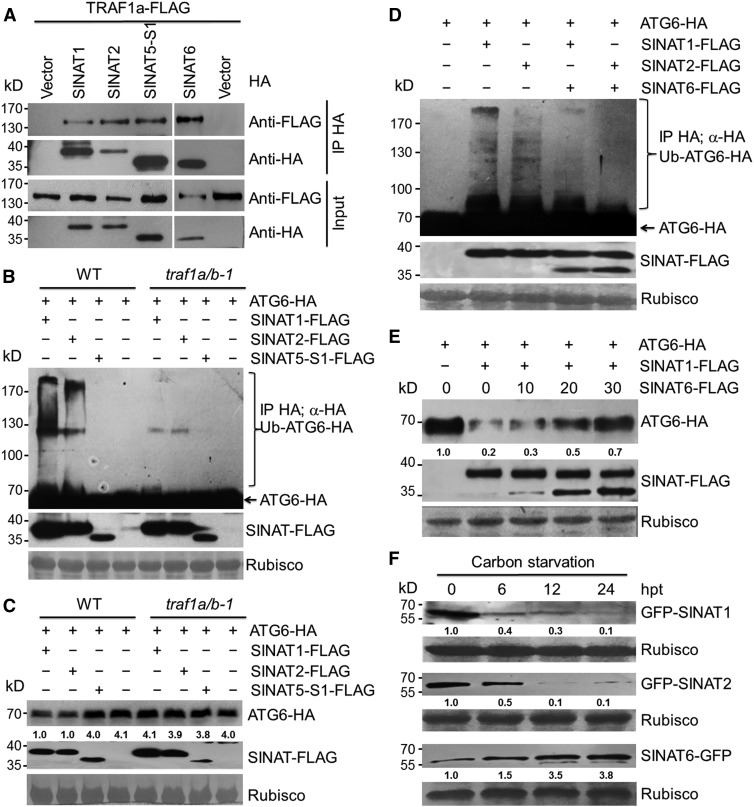
To determine whether TRAFs and SINATs form a common protein complex with ATG6, we examined the interactions between TRAF1a and SINAT proteins by transiently expressing SINAT1-HA, SINAT2-HA, SINAT5-S1-HA, SINAT6-HA, or the empty vector in protoplasts isolated from TRAF1a-FLAG transgenic lines grown in the dark for 16 h. CoIP assays showed that TRAF1a-FLAG was immunoprecipitated by SINAT1-HA, SINAT2-HA, SINAT5-S1-HA, and SINAT6-HA (Figure 8A). Furthermore, when SINAT1-FLAG, SINAT2-FLAG, or SINAT5-S1-FLAG was coexpressed with ATG6-HA in protoplasts prepared from wild-type and traf1a/b-1 leaves, the ubiquitination of ATG6-HA strongly decreased in the traf1a/b-1 mutant compared with the wild type (Figure 8B), suggesting that SINAT-mediated ubiquitination of ATG6 requires TRAF1a and TRAF1b. Analysis of ATG6 protein levels in wild-type and traf1a/b-1 protoplasts coexpressing SINAT1-FLAG, SINAT2-FLAG, or SINAT5-S1-FLAG with ATG6-HA revealed that the degradation of ATG6-HA induced by SINAT1-FLAG and SINAT2-FLAG was impaired in traf1a/b-1 mutant protoplasts (Figure 8C). Together, these findings suggest that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-associated ubiquitination and the degradation of ATG6 in planta.
Figure 8.
TRAF1a and TRAF1b Are Required for SINAT-Mediated Ubiquitination of ATG6.
(A) In vivo CoIP assay showing the interactions between TRAF1a-FLAG and HA-tagged SINAT1, SINAT2, SINAT5-S1, and SINAT6. The SINAT1-HA, SINAT2-HA, SINAT5-S1-HA, and SINAT6-HA plasmids were expressed in Arabidopsis protoplasts prepared from TRAF1a-FLAG transgenic lines and immunoprecipitated by HA affinity magnetic beads.
(B) SINAT-mediated ATG6 ubiquitination is abolished in the traf1a/b-1 mutant. ATG6-HA was transiently expressed in Arabidopsis protoplasts prepared from wild-type (WT) and traf1a/b-1 plants expressing SINAT1-FLAG, SINAT2-FLAG, or SINAT5-S1-FLAG. Ponceau S-stained membranes are shown below the blots to show equal amounts of protein loaded per lane.
(C) SINAT1- and SINAT2-associated degradation of ATG6 is dependent on TRAF1a and TRAF1b. ATG6-HA was transiently expressed in Arabidopsis protoplasts prepared from the wild type and traf1a/b-1 expressing SINAT1-FLAG, SINAT2-FLAG, or SINAT5-S1-FLAG. Ponceau S-stained membranes are shown below the blots to indicate the equal amount of protein loaded per lane.
(D) SINAT1- and SINAT2-mediated ATG6 ubiquitination is impaired by coexpression of SINAT6. ATG6-HA was coexpressed with SINAT1, SINAT2, SINAT1/SINAT6, or SINAT2/SINAT6 in Arabidopsis protoplasts. Ponceau S-stained membranes are shown below the blots to show equal amounts of protein loaded per lane.
(E) SINAT1-associated degradation of ATG6 is inhibited by SINAT6. ATG6-HA was coexpressed with SINAT1-FLAG in the presence of various amounts (0, 10, 20, and 30 μg) of SINAT6-FLAG in Arabidopsis protoplasts. Ponceau S-stained membranes are shown below the blots to show equal amounts of protein loaded per lane. The relative intensity of ATG6-HA normalized to the loading control is shown below.
(F) Stabilities of GFP-SINAT1, GFP-SINAT2, and SINAT6-GFP proteins in response to carbon starvation. One-week-old transgenic lines expressing GFP-SINAT1, GFP-SINAT2, and SINAT6-GFP were transferred to liquid MS medium without sugar under constant darkness for the indicated times. Ponceau S-stained membranes are shown below the blots to indicate the equal amounts of protein loaded per lane. Relative intensity of GFP-SINAT1, GFP-SINAT2, and SINAT6-GFP normalized to loading control is shown below. The numbers on the left indicate the molecular mass (kD) of each size marker. hpt, hours post-treatment.
Compared with SINAT1 and SINAT2, SINAT6 contains only a short, truncated RING finger domain (Bao et al., 2014) and has little effect on the ubiquitination and destabilization of ATG6 (Figures 7F and 7G). Therefore, we hypothesize that SINAT6 may be involved in maintaining ATG6 homeostasis by competitively associating with ATG6 under certain growth conditions. To test this possibility, we transiently coexpressed SINAT1-FLAG or SINAT2-FLAG with SINAT6-FLAG in Arabidopsis protoplasts and detected the ubiquitination and degradation of ATG6 by protein blot analyses. As shown in Figure 8D, both SINAT1- and SINAT2-mediated ubiquitination ATG6 were strongly reduced by coexpression with SINAT6. Competition assays further revealed that the addition of SINAT6 protein prevented the SINAT1-mediated degradation of ATG6 in a dose-dependent manner (Figure 8E), implying that SINAT1/2 and SINAT6 may function to regulate ATG6 stability under nutrient-rich and nutrient starvation conditions, respectively. To further confirm this, we generated transgenic plants expressing GFP-SINAT1, GFP-SINAT2, and SINAT6-GFP fusions and monitored their protein levels in response to starvation. The results revealed that after carbon starvation for 6 h, GFP-SINAT1 and GFP-SINAT2 levels decreased significantly and were almost undetectable at 24 h after treatment (Figure 8F). By contrast, SINAT6-GFP was present at low levels under normal growth conditions and gradually accumulated to ∼3.8-fold higher than initial levels from 6 to 24 h of carbon starvation (Figure 8F).
SINAT1/SINAT2 and SINAT6 Play Opposing Role in Regulating Autophagy
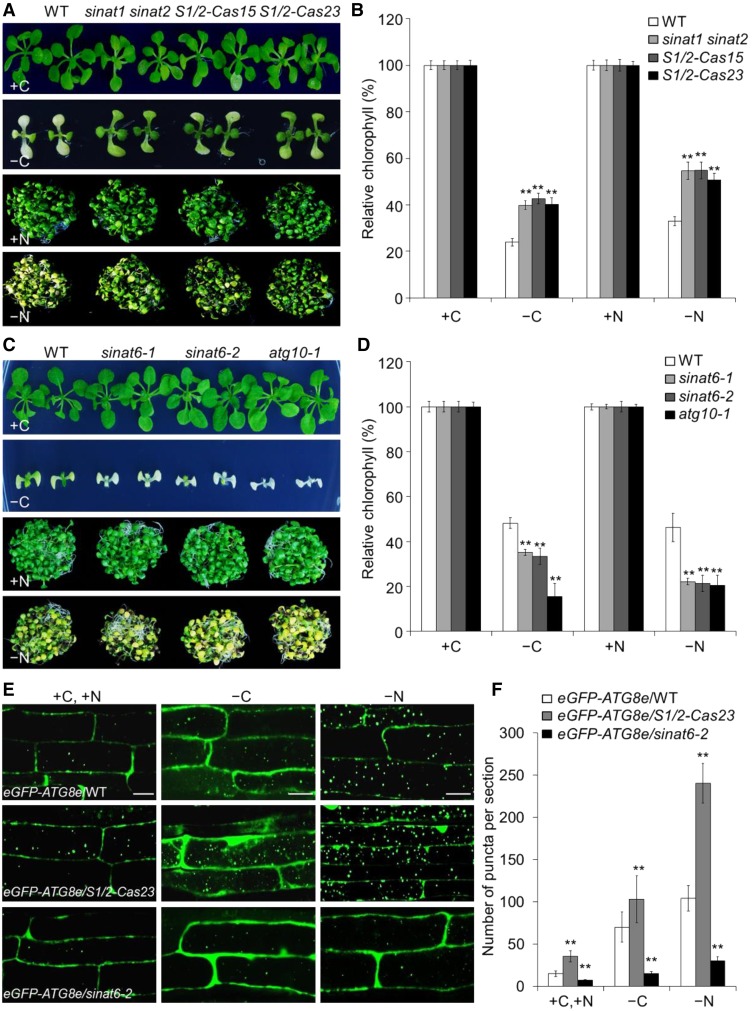
To obtain genetic evidence for the involvement of SINAT1 and SINAT2 in autophagy, we identified two single knockout mutants, sinat1 and sinat2 (Supplemental Figure 11A), and crossed them to obtain the sinat1 sinat2 double mutant. We also used CRISPR-Cas9 to generate two transgenic lines with knockout mutations of SINAT1 and SINAT2 (designated S1/2-Cas15 and S1/2-Cas23; Supplemental Figure 12). Phenotypic analyses showed that upon carbon and nitrogen starvation, the responses of the sinat1 and sinat2 single mutants were similar to those of the wild type (Supplemental Figure 11B). By contrast, the sinat1 sinat2 double mutant and S1/2-Cas lines (S1/2-Cas15 and S1/2-Cas23) displayed more tolerance than the wild type under both carbon and nitrogen starvation, as reflected by their delayed yellowing phenotypes (Figure 9A) and increased chlorophyll contents (Figure 9B), suggesting that SINAT1 and SINAT2 act redundantly to negatively regulate the plant response to autophagy-associated nutrient starvation in Arabidopsis. To further validate whether SINAT6 regulates plant responses to nutrient starvation, we isolated two independent T-DNA knockout lines, sinat6-1 and sinat6-2 (Supplemental Figure 11C). The results showed that, like the atg10-1 mutant, the sinat6-1 and sinat6-2 mutants were more sensitive to carbon and nitrogen starvation than the wild type (Figures 9C and 9D).
Figure 9.
SINAT1/SINAT2 and SINAT6 Play Opposing Roles in Autophagy.
(A) Phenotype of wild-type (WT), sinat1 sinat2 double mutant, and S1/2-Cas (S1/2-Cas15 and S1/2-Cas23) plants upon carbon or nitrogen starvation. One-week-old wild-type, sinat1 sinat2 double mutant, and S1/2-Cas plants were grown on MS agar with sucrose for 1 week. The seedlings were transferred to MS agar with sucrose (+C) or MS liquid medium (+N) or MS agar plates without sucrose followed by constant dark treatment (−C) for 10 d or MS liquid medium without nitrogen under normal light/dark growth conditions (−N) for 6 d.
(B) Relative chlorophyll contents in wild-type, sinat1 sinat2 double mutant, and S1/2-Cas (S1/2-Cas15 and S1/2-Cas23) plants upon carbon or nitrogen starvation. The relative chlorophyll contents were calculated by comparing the values of –C or –N seedlings to those of +C or +N seedlings.
(C) Phenotype of wild-type, sinat6 mutants (sinat6-1 and sinat6-2), and atg10-1 plants upon carbon or nitrogen starvation. One-week-old wild-type, sinat6 mutants, and atg10-1 plants were grown on MS agar with sucrose for 1 week. The seedlings were transferred to MS agar with sucrose (+C) or MS liquid medium (+N) or MS agar plates without sucrose followed by constant dark treatment (–C) for 7 d or MS liquid medium without nitrogen under normal light/dark growth conditions (–N) for 4 d.
(D) Relative chlorophyll contents in wild-type, sinat6 mutants (sinat6-1 and sinat6-2), and atg10-1 plants upon carbon or nitrogen starvation. The relative chlorophyll contents were calculated by comparing the values of –C or –N seedlings to those of +C or +N seedlings. Relative chlorophyll contents are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, five technical replicates pooled with 20 seedlings were used per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
(E) Confocal analysis of eGFP-ATG8e/WT, eGFP-ATG8e/S1/2-Cas23, and eGFP-ATG8e/sinat6-2. One-week-old eGFP-ATG8e/WT, eGFP-ATG8e/S1/2-Cas23, and eGFP-ATG8e/sinat6-2 seedlings were exposed to N- and C-sufficient (MS) or C-deficient (–C) and N-deficient (–N) conditions with 1 µM CA for 16 h and visualized by confocal microscopy. Bars = 50 µm.
(F) Numbers of puncta per root section in the root cells of eGFP-ATG8e/WT, eGFP-ATG8e/S1/2-Cas23, and eGFP-ATG8e/sinat6-2 in (E). Data are average values ± sd (n = 3) calculated from three independent experiments. For each experiment, 15 sections were used for calculation per genotype. Asterisks indicate significant differences from the wild type (**P < 0.01 by Student’s t test).
To investigate the role of SINAT1/SINAT2 and SINAT6 in autophagosome formation, we crossed S1/2-Cas23 and sinat6-2 to a transgenic line expressing eGFP-ATG8e to obtain eGFP-ATG8e/S1/2-Cas23 and eGFP-ATG8e/sinat6-2. Confocal microscopy showed that under nutrient-rich (+C or +N) or nutrient starvation (−C or −N) conditions, the numbers of autophagic puncta in the root cells of eGFP-ATG8e/S1/2-Cas23 and eGFP-ATG8e/sinat6-2 were significantly increased and reduced, respectively, compared with the eGFP-ATG8e line in the wild-type background (Figures 9E and 9F). Consistent with this finding, the carbon starvation-inducible release of free GFP was significantly enhanced in eGFP-ATG8e/S1/2-Cas23 seedlings and inhibited in eGFP-ATG8e/sinat6-2 compared with eGFP-ATG8e/WT (Supplemental Figure 13A). Furthermore, the different functions of SINAT1/2 and SINAT6 in autophagosome formation were confirmed by MDC staining of the root cells of wild-type, sinat1 sinat2, S1/2-Cas23, sinat6-1, and sinat6-2 plants (Supplemental Figures 13B and 13C). Taken together, these findings indicate that SINAT1/SINAT2 and SINAT6 play opposing role in the regulation of autophagy dynamics in Arabidopsis.
DISCUSSION
Autophagy is a conserved biological process that maintains cellular homeostasis in either a housekeeping capacity or in response to environmental stress (Michaeli et al., 2016). Upon induction, autophagy occurs via phagophore (isolation membrane) initiation, nucleation, expansion, and maturation, leading to the formation of autophagosomes, which subsequently fuse with the vacuole and deliver the engulfed materials for degradation (Yang and Klionsky, 2010; Liu and Bassham, 2012; Li and Vierstra, 2012). To date, several ATGs and their regulatory factors have been shown to function in autophagy in plants (Michaeli et al., 2016). Among these, ATG6 is a key component of the nucleation complex that functions during autophagy by interacting with the phosphatidylinositol-3-kinases PI3K and VPS15. In Arabidopsis, ATG6 knockout mutants are male gametophytic lethal (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008), suggesting that ATG6 is essential for plant development. Analysis of ATG6 antisense transgenic lines demonstrated the vital role of ATG6 in autophagosome formation (Patel and Dinesh-Kumar, 2008). Here, we discovered that TRAF1a and TRAF1b play a role in autophagosome formation by regulating ATG6 ubiquitination and stability in Arabidopsis. During this process, the SINAT1- and SINAT2-based E3 ligases have critical functions in ubiquitinating ATG6 and promoting its degradation under nutrient-rich conditions. By contrast, the starvation-induced accumulation of SINAT6 is likely involved in maintaining ATG6 stability and promoting autophagy in response to starvation.
In mammals, TRAF family proteins were originally identified as adaptors that help transduce upstream signals from cell surface receptors, such as the TNFs and Toll/interleukin-1 receptors, to downstream effectors (Inoue et al., 2000; Xie, 2013). There are seven known TRAF members, TRAF1 to TRAF7, in mammalian cells, which play key roles in diverse signaling cascades, including innate and adaptive immunity, the survival, proliferation, and differentiation of cells, and abiotic stress responses. All of these proteins contain a conserved C-terminal TRAF domain that mediates their self-association and interactions with various protein partners (Inoue et al., 2000; Xie, 2013). All TRAF proteins except TRAF1 have an N-terminal RING finger domain and various numbers of zinc finger motifs that provide them with E3 ubiquitin ligase catalytic activity during protein ubiquitination and degradation (Pineda et al., 2007). Thus, these TRAF proteins function as both adaptors and E3 ubiquitin ligases in controlling various cellular signaling pathways.
Similar to TRAF1, which is unique in mammals, the two Arabidopsis TRAF proteins identified in the study, TRAF1a and TRAF1b, contain a TRAF domain at their N termini but no other conserved domains, suggesting that they likely serve as molecular adaptors rather than E3 protein ligases to regulate signaling. Despite the similarities in the domain structures between mammalian TRAF1 and Arabidopsis TRAF1a/TRAF1b, these proteins appear to perform different functions in mammals and plants. TRAF1 functions as a positive or negative regulator of TNFR2 signaling and neuron cell death by directly interacting with TRAF2 (Xie, 2013). By contrast, Arabidopsis TRAF1a and TRAF1b appear to play an important role in maintaining autophagy dynamics by interacting with ATG6, the RING finger E3 ligases SINAT1 and SINAT2, and SINAT6 (this study). The involvement of mammalian TRAF1 in autophagy signaling is still unknown. Increasing evidence suggests that upon autophagy induction, another TRAF member, TRAF6, interacts with Beclin-1, the mammalian homolog of ATG6, to modulate its protein stability, which subsequently promotes autophagy (Shi and Kehrl, 2010; Nazio et al., 2013). These findings confirm that although the working mechanisms vary, the regulation of TRAF-mediated autophagy is evolutionarily conserved between mammals and plants. Consistent with this notion, a recent report revealed that similar to mammalian TRAFs, Arabidopsis TRAF1a and TRAF1b (termed MUSE14 and MUSE13, respectively, in that study) are involved in regulating plant immunity and pathogen resistance by modulating the homeostasis of NLR immune sensors SNC1 and RPS2 (Huang et al., 2016).
Beclin-1/ATG6, a core component of the Beclin-1/ATG6-PI3K complex during autophagosome formation, is regulated in a sophisticated manner by posttranslational ubiquitination to determine the proper levels of autophagy in eukaryotic cells. In addition to TRAF6, several other regulatory cofactors, such as BCL-2, DAPK, Cul4, Nedd5, and RNF216, also associate with Beclin-1 to regulate its activity (He and Levine, 2010; Wirawan et al., 2012; Mei et al., 2016). Of these, TRAF6/Cul4 and Nedd4/RNF216 are two types of E3 ligases that promote the stabilization and degradation, respectively, of Beclin-1 and subsequently regulate autophagy dynamics in mammals (Shi and Kehrl, 2010; Platta et al., 2012; Xia et al., 2013; Xu et al., 2014). Upon induction of autophagy, TRAF6 and Cul4 ubiquitinate Beclin-1 at the K63 residue, which allows it to interact with VPS34 and then activate autophagy (Shi and Kehrl, 2010; Xia et al., 2013). However, the Nedd4/RNF216 ligases mediate K11- and K48-linked ubiquitination of Beclin-1, which leads to the degradation of Beclin-1 and thereby the suppression of autophagy (Platta et al., 2012; Xu et al., 2014).
We found that upon short-term (6 h) carbon starvation, Arabidopsis ATG6 accumulated (Figure 6G), which is indicative of induction of autophagy. However, when starvation treatment lasted for more than 12 h, ATG6 levels decreased; MG132 treatment strongly inhibited this reduction in ATG6 levels (Figure 6G). These results imply that like mammalian Beclin-1, Arabidopsis ATG6 is also controlled by ubiquitination-mediated proteolysis to regulate autophagy dynamics. Because ATG6 degradation was inhibited in both the traf1a/b double mutant and TRAF1a-FLAG-overexpressing lines (Figure 6F), it is conceivable that during autophagy induction, TRAF1a and TRAF1b serve as both positive and negative regulators by recruiting different E3 ligases to maintain the ATG6 pool in plant cells. Consistent with this notion, phenotypic analysis of the traf1a/b double knockout mutants revealed that although all three independent mutants were more sensitive to carbon and nitrogen starvation than the wild type, they displayed premature senescence only at 4 weeks old (Figures 2 and 3). By contrast, all of the double mutants showed delayed leaf senescence at 5 and 6 weeks old compared with the visible leaf senescence in wild-type plants (Figure 3). Indeed, the opposing age-dependent senescence phenotypes in the traf1a/b double mutants may be explained by the potential existence of E3 ligases involved in promoting ATG6 stabilization during senescence in Arabidopsis. It is conceivable that natural senescence signals may trigger the association of the unknown positive E3 ligases with TRAF1a/TRAF1b and ATG6, which contributes to the ubiquitination and stabilization of ATG6. In contrast to their accumulation at 6 and 12 h upon starvation, the TRAF1 proteins were degraded at 24 h after treatment (Figure 6G), suggesting that TRAF1 protein levels are highly regulated in different nutrient conditions and at different stages of starvation. However, we observed that, unlike the pattern of TRAF1s, the SINAT6 protein was continually activated from 6 to 24 h after starvation (Figure 8F). It is possible that SINAT6 may perform other functions, rather than autophagy, during nutrient starvation.
Here, we obtained several lines of evidence suggesting that two RING finger domain-containing E3 ligases, SINAT1 and SINAT2, together with their truncated homologous protein SINAT6, act as positive and negative regulators, respectively, of TRAF1a/TRAF1b-regulated ATG6 degradation. First, Y2H and CoIP assays showed that SINAT1, SINAT2, and SINAT6 physically interact with ATG6 in vivo (Figures 7A to 7D). Second, SINAT1 and SINAT2 directly ubiquitinate ATG6 and promote its degradation in protoplasts (Figures 7F and 7G). Third, SINAT1 and SINAT2 associate with TRAF1 in planta (Figure 8A). Fourth, TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated ubiquitination and degradation of ATG6, which are abolished in the presence of SINAT6 (Figures 8B to 8F). Fifth, the sinat1 sinat2 double mutant and S1/2-Cas mutants show enhanced tolerance to autophagy-associated starvation and increased autophagosome formation, while the sinat6 knockout mutants display deficiencies in nutrient starvation tolerance and starvation-induced autophagosome formation (Figure 9). Together, these observations provide strong evidence supporting the idea that under normal growth conditions, SINAT1 and SINAT2 are key E3 ligases that regulate the interaction, ubiquitination, and degradation of ATG6, thereby suppressing autophagy. By contrast, nutrient starvation triggers significant declines in the levels of SINAT1 and SINAT2 proteins through a yet unknown mechanism but induces the accumulation of SINAT6 protein (Figure 8F), which directly or indirectly promotes autophagy in Arabidopsis. It is likely that TRAF1a and TRAF1b function as molecular adaptors that are essential to facilitate the effect of SINAT1/SINAT2 and SINAT6 on ATG6 upon autophagy induction.
More recently, TRAF1a and TRAF1b were shown to form a plant-type TRAFasome by interacting with the NLR sensors SNC1 and RPS1 and with SCFCPR1 E3 ubiquitin ligase, which negatively regulates plant immune responses (Huang et al., 2016). Although the snc1 mutation can suppress the dwarf phenotype of traf1a/b in the snc1-r1 muse13-2 muse 14-1 triple mutant, it showed similar hypersensitivity to carbon starvation to that of the traf1a/b mutant (Supplemental Figure 4), indicating that the autophagy deficiency in the traf1a/b does not rely on the NB-LRR immune receptors. Therefore, it is also conceivable that two protein complexes consisting of TRAF1a/1b-SINAT1/SINAT2-ATG6 and TRAF1a/1b-SINAT6-ATG6 function as a plant-specific TRAFasome and are essential for the negative or positive regulation of autophagosome formation by modulating ATG6 stability. Our results indicate that rather than SINAT6, other potential E3 protein ligase(s) might also be involved in TRAF1a/1b-mediated positive regulation of the autophagic process by ubiquitinating and stabilizing ATG6. Therefore, these findings increase our understanding of the mechanism used by plants to assemble specific cofactors to maintain autophagy dynamics and cellular homeostasis upon nutrient starvation. Studies aimed at identifying additional TRAF1a/1b interactors and investigating the functional links of these proteins in regulating autophagosome formation are needed to better understand how TRAF proteins dynamically mediate autophagy signaling in Arabidopsis.
METHODS
Plant Materials, Growth Conditions, and Treatments
All wild-type, mutant, and transgenic Arabidopsis thaliana plants used in this study are in the Columbia ecotype (Col-0) background. The T-DNA insertional mutants described in this study were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org) with the locus names traf1a-1 (SALK_059309C), traf1a-2 (SALK_120387), traf1b-1 (SALK_146938C), traf1b-2 (SALK_035658), traf1b-3 (CS821400), sinat1 (SALK_010417C), sinat2 (SALK_002174C), sinat6-1 (SALK_077413), and sinat6-2 (SALK_129594). The T-DNA insertion in each mutant was identified by PCR using a gene-specific primer paired with a T-DNA-specific primer. The traf1a and traf1b mutants were crossed to generate the traf1a/1b-1 (traf1a-1 traf1b-2), traf1a/1b-2 (traf1a-2 traf1b-1), and traf1a/1b-3 (traf1a-2 traf1b-3) double mutants. The sinat1 and sinat2 mutants were crossed to generate the sinat1 sinat2 double mutants. The primers used to genotype the single and double mutants are listed in Supplemental Data Set 2. All Arabidopsis seeds were surface sterilized with 20% bleach containing 0.1% Tween 20 for 20 min, washed with sterilized water, and sown on MS medium (Sigma-Aldrich) containing 2% sucrose (w/v) and 0.8% agar (w/v). Following cold treatment for 3 d, the plates were incubated at 22°C under a 16-h-light/8-h-dark cycle with a light intensity of 170 µmol/m2/s using bulbs (Philips F17T8/ TL841 17W). After 1 week, the seedlings were transferred to soil for further growth.
For nitrogen starvation, 1-week-old seedlings grown on MS medium were transferred to MS or nitrogen-deficient MS medium (solid or liquid) and grown under normal growth conditions for the indicated times. For carbon starvation, 1-week-old MS-grown seedlings or 3-week-old soil-grown plants were transferred to continuous darkness for the indicated duration following recovery under normal growth conditions for 7 d. Samples were collected or photographed at the indicated time points. To calculate the survival rate after carbon starvation, over 10 plants per genotype were dark-treated followed by a 7-d recovery. The number of surviving plants, as indicated by the ability to produce new leaves, was recorded.
Pathogen infection was performed as described previously (Xiao and Chye, 2011). The pathogen strains Pseudomonas syringae pv tomato DC3000 and Botrytis cinerea were obtained from the American Type Culture Collection. For hypoxia treatment, the seeds of various genotypes were germinated on MS medium under normal air or hypoxia (3% oxygen) conditions for 10 d. The number of seedlings with green cotyledons was recorded and the relative percentages were calculated by comparing the cotyledon greening rates for each genotype under hypoxia to their respective survival rates under normal air conditions.
Plasmid Construction
Most plasmids used in this study were generated using an In-Fusion HD Cloning Kit (Takara) according to the manufacturer’s instructions. The gene-specific primers with 15-bp extensions homologous to the corresponding vectors are listed in Supplemental Data Set 2. To construct the mCherry-ATG8e and mCherry-ATG1a expression vectors, mCherry was inserted into BamHI/SpeI-digested pBI221-UBQ-YFP-ATG8f to create pBI221-UBQ-mCherry-ATG8f (Zhuang et al., 2013), and the full-length cDNA fragments of ATG8e (digested with XbaI and XhoI) and ATG1a (digested with SpeI and XhoI) were inserted into pBI221-UBQ-mCherry-ATG8f to replace ATG8f. The ATG6 fragment from pBI221-UBQ-ATG6-YFP was digested with BamHI and KpnI and inserted into pBI221-UBQ-SH3P2-RFP (Zhuang et al., 2013) to create ATG6-RFP. Plasmids for the remaining transient expression analyses were derived from vector pUC119 or pHBT (Li et al., 2013), which contains an eGFP fragment followed by 2×HA or 2×FLAG tags. Briefly, full-length coding sequences of TRAF1a and TRAF1b were inserted into StuI- and BamHI-digested pUC119, respectively, and fused with the C or N terminus of eGFP. For the TRAF1a-HA, TRAF1b-HA, and TRAF1a-FLAG constructs, the pUC119 plasmids were digested with StuI and BamHI to remove the eGFP sequence, and the TRAF1a and TRAF1b fragments were then inserted into the plasmids and fused with the 2×HA or 2×FLAG tag. SINAT1, SINAT2, SINAT3, SINAT4, SINAT5, and SINAT6, SINAT5 deletion fragments, and ATG6 were cloned into StuI- and BamHI-digested pHBT to generate FLAG- or HA-tagged SINAT or ATG6 constructs.
To generate stable transgenic lines expressing TRAF1a-FLAG, ATG6-HA, SINAT1-GFP, SINAT2-GFP, and GFP-SINAT6, the UBQ10pro:TRAF1a-FLAG, 35SPPDKpro:ATG6-HA, 35SPPDKpro:GFP-SINAT1, 35SPPDKpro:GFP-SINAT2, and 35SPPDKpro:SINAT6-GFP fragments derived from AscI-digested pUC119 and pHBT constructs were cloned into binary vector pFGC-RCS (Li et al., 2013). To generate SINAT1/2-Cas transgenic lines, the 19- or 20-bp target sites of SINAT1 and SINAT2 were assembled with specific promoters and gRNAs to form expression cassettes by overlap PCR. Cassettes of U3d:TSINAT1-gRNA and U3b:TSINAT2-gRNA were then ligated to pYLCRISPR-Cas9Pubi-B (Ma et al., 2015). The genome editing efficiency of the transgenic lines was tested by sequencing of PCR products. The TRAF1b-RNAi construct was generated by PCR amplification of a 0.5-kb sense TRAF1b CDS fragment using the primer pair XS3053 and XS3054 (Supplemental Data Set 2) and its corresponding 0.5-kb antisense fragment using the primer pair XS3055 and XS3056 (Supplemental Data Set 2). The sense and antisense fragments were respectively cloned into the XhoI-EcoRI and HindIII-XbaI sites of binary vector pFGC-pHANNIBAL modified from pHANNIBAL (Xiao et al., 2010). The expression cassettes were subsequently introduced into wild-type Arabidopsis (Col-0) or traf1a-1 mutants (for traf1a TRAF1b-RNAi) by Agrobacterium tumefaciens-medium transformation via the floral dip method (Clough and Bent, 1998).
To generate plasmids for the Y2H assay, full-length coding sequence fragments of TRAF1a, TRAF1b, ATG1s, ATG6, ATG7, ATG8e, ATG9, ATG18b, ATG18c, VPS34, UVRAG, SINAT1, SINAT2, SINAT3, SINAT4, SINAT5, SINAT6, and SINAT5 deletions were amplified and inserted into EcoRI-digested vectors pGADT7 and pGBKT7 (Clontech). To generate plasmids for BiFC, the split fragments of nYFP and cYFP were inserted into StuI- and BamHI-digested pHBT to replace eGFP and generate vectors pHBT-YN and pHBT-YC, respectively. Full-length coding sequence fragments of TRAF1a and ATG6 were then inserted into StuI-digested pHBT-YC and BamHI-digested pHBT-YN, respectively, to generate a fusion with the C terminus of nYFP or N terminus of cYFP. As a control, the full-length cDNA sequence of another TRAF-domain containing protein, At4g01390, was also inserted into BamHI-digested pHBT-YC to generate At4g01390-cYFP. For protein expression, MBP-SINAT1 and MBP-SINAT2 plasmids were constructed by cloning SINAT1 and SINAT2 cDNA fragments into the PstI-digested vector pMAL-c5x (New England Biolabs).
Transient Expression in Arabidopsis Protoplasts and Microscopy Analyses
Transient expression assays in Arabidopsis protoplasts were conducted essentially as described by Miao and Jiang (2007) and Zhuang et al. (2013). To determine the effect of TRAF1a and TRAF1b deletion on autophagosome formation, plants from the stable eGFP-ATG8e line (Xiao et al., 2010) were crossed with the traf1a/1b-1 mutant to generate the eGFP-ATG8e/traf1a/1b-1 double combination. Seven-day-old eGFP-ATG8e/Col-0 and eGFP-ATG8e/traf1a/1b-1 seedlings grown on MS medium were transferred to MS medium with or without sucrose and nitrogen containing 1 µM CA (Sigma-Aldrich) as described by Spitzer et al. (2015). Primary root cells were observed under an LSM 780 NLO laser scanning confocal microscope (Carl Zeiss).
MDC staining was performed as described previously (Contento et al., 2005; Chen et al., 2015). Briefly, the roots of 7-d-old seedlings were stained with 0.05 mM MDC (Sigma-Aldrich) in PBS for 10 min. Following two washes with PBS buffer, the root cells were observed under an Axio Observer Z1 inverted microscope (Carl Zeiss) with a 4′,6-diamidino-2-phenylindole-specific filter.
Measurement of Chlorophyll Contents
Chlorophyll contents were measured following Xiao et al. (2010). Arabidopsis leaves were extracted by immersion in 1 mL of N,N-dimethylformamide for 48 h in the dark at 4°C. Absorbance was determined at 664 and 647 nm, and total chlorophyll content was measured and normalized to gram fresh weight per sample.
SA and JA Measurements
SA and JA were extracted and measured as described previously (Pan et al., 2010; Chen et al., 2015). Powdered tissue was weighed in a 2-mL centrifuge tube and extracted with 1 mL of extraction buffer (2-propanol/water/concentrated HCl [2:1:0.005, v/v/v]) with internal standards (10 ng D4-SA and 10 ng D5-JA; Sigma-Aldrich). The mixtures were shaken at 100g for 30 min at 4°C, followed by the addition of 1 mL dichloromethane and shaking for an additional 30 min at 4°C. The samples were then centrifuged at 13,000g for 10 min. Solvent (900 µL) from the lower phase was collected and concentrated using a nitrogen evaporator with nitrogen flow. The samples were dissolved in a 100-μL mixture of 60 μL methanol and 40 μL distilled water. Quantitative determination of SA and JA was performed using a Triple TOF 5600 (AB SCIEX) system according to Chen et al. (2015).
RNA Extraction, RNA-Seq, and qRT-PCR Analysis
Total RNA was extracted from Arabidopsis rosettes using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Illumina library construction and sequencing were performed as described by Yu et al. (2012).
The isolated RNA was reverse transcribed using a PrimeScript RT reagent kit with gDNA Eraser (Takara). The qRT-PCR was performed using SYBR Green master mix (Takara) on a StepOne Plus real-time PCR system (Applied Biosystems). The conditions for qRT-PCR were initial denaturation at 95°C for 5 min, followed by 40 cycles of PCR (denaturing, 95°C for 10 s; annealing, 55°C for 15 s; extension, 72°C for 30 s). Three experimental replicates were performed per reaction. ACTIN2 was used as a reference gene. The gene-specific primers used for qRT-PCR are listed in Supplemental Data Set 2.
Trypan Blue and DAB Staining
Trypan blue staining and DAB staining were performed according to Xiao and Chye (2011). For trypan blue staining, leaves of 3-week-old wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 double mutants plants were collected, boiled for 1 min in trypan blue staining buffer (12.5% phenol, 12.5% glycerol, 12.5% lactic acid, 48% ethanol, and 0.025% trypan blue), and incubated for 10 min at room temperature, followed by destaining five times in 70% chloral hydrate. For DAB staining, leaves of 3-week-old wild-type, traf1a/b-1, traf1a/b-2, and traf1a/b-3 double mutant plants were collected and incubated in 1 mg mL−1 DAB solution (pH 3.8) for 2 h at room temperature in darkness and subsequently cleared in 95% boiling ethanol for 10 min.
Protein Isolation and Immunoblot Analysis
For total protein extraction, Arabidopsis samples grown on solid or liquid medium were ground in liquid nitrogen and homogenized in ice-cold extraction buffer (50 mM sodium phosphate, pH 7.0, 200 mM NaCl, 10 mM MgCl2, 0.2% β-mercaptoethanol, and 10% glycerol) supplemented with protease inhibitor cocktail (Roche). The samples were incubated on ice for 30 min and centrifuged at 4°C for 30 min at 12,000g. The supernatant was transferred to a new microfuge tube prior to electrophoresis.
For immunoblot analysis, total proteins were subjected to SDS-PAGE and electrophoretically transferred to a Hybond-C membrane (Amersham). Specific anti-ATG1a, anti-ATG13a (Suttangkakul et al., 2011; 1:1000), anti-ATG7 (Abcam, cat. no. ab9901, 1:2000), anti-ATG8a (Abcam, cat. no. ab77003, 1:1500), anti-HA (Sigma-Aldrich, cat. no. H6533, 1:5000), anti-FLAG (Sigma-Aldrich, cat. no. A8592, 1:5000), anti-Ub (Proteintech, cat. no. 10201-2-AP, 1:2000), and anti-GFP (Cell Signaling Technology, cat. no. 2955, 1:1000) antibodies were used in the protein blotting analysis. Quantification of the protein signal was done using Image J software.
Y2H, CoIP, and BiFC Assays
Preparation of yeast competent cells and yeast transformation were performed as described by Chen et al. (1992). To avoid self-activation of the transformants, 5 mM 3-amino-1,2,4-triazole was added to the medium.
Plasmids for transient expression analysis were extracted using a Maxi Kit (Omega; D6922-02). Arabidopsis mesophyll protoplast preparation and transfection were performed according to Yoo et al. (2007). Protoplasts isolated from 4-week-old rosettes were transfected with the indicated plasmids and cultured for 16 h for protein expression. For the CoIP assays, the cells were then collected and lysed in immunoprecipitation (IP) buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 10% glycerol) with 0.3% Triton. A portion of the total lysis (10%) was reserved for input, and the remainder was incubated with HA or FLAG affinity beads (Sigma-Aldrich) for 4 h at 4°C. The beads were then collected and washed five times with IP buffer containing 0.1% Triton, followed by sample buffer elution at 95°C for 5 min. For the BiFC assay, the split nYFP and cYFP plasmids ATG6-nYFP/cYFP-TRAF1a, ATG6-nYFP/At4g01390-cYFP, or nYFP/cYFP were coexpressed in leaf protoplasts for 16 h under light or dark conditions, and the YFP signal was detected by confocal microscopy.
In Vitro and in Vivo Ubiquitination Assays
The full-length cDNAs of SINAT1 and SINAT2 were cloned into the vector pMAL-c5x (New England Biolabs) to generate MBP-SINAT1 and MBP-SINAT2 fusions. For protein expression, IPTG at a final concentration of 300 μM and 2% glucose were added to a 250-mL culture when its OD600 value reached 0.6. After an additional 4 h of culturing at 30°C, the cells were harvested and purified using a pMAL Protein Fusion and Purification System (New England Biolabs) following the manufacturer’s instructions. Fusion proteins were eluted, followed by desalting and concentration, using Amicon Ultra-4 centrifugal filter devices (Millipore). The in vitro ubiquitination assay was performed as described by Zhao et al. (2013). The ubiquitination reaction mixture (total volume of 30 μL) contained 40 ng His-tagged wheat E1 (GI:136632), 200 ng His-tagged human E2 (UBCH5B), 1 μg MBP-SINAT1 or MBP-SINAT2, and 2 μg His-Ub (AtUBQ14) in reaction buffer (50 mM Tris HCl, pH 7.5, 2 mM ATP, 5 mM MgCl2, and 2 mM DTT). The reaction mixture was incubated at 30°C for 1 to 1.5 h and terminated by the addition of SDS sample buffer.
For the in vivo ubiquitination assays, the ATG6-HA plasmid was either expressed in Arabidopsis mesophyll protoplasts isolated from wild-type or traf1a/b-1 plants or cotransfected with TRAF1a/TRAF1b-HA or SINAT1/SINAT2/SINAT5-S1/SINAT6-FLAG plasmids into wild-type protoplasts for overnight expression. The cells were then collected and lysed in IP buffer containing 1% Triton with vigorous vortexing. The ubiquitination pattern of ATG6-HA was detected by HA affinity immunoprecipitation and immunoblot analyses.
Statistical Analysis
Data reported in this study are means ± sd of three independent experiments unless otherwise indicated. The significance of the differences between groups was determined by a two-tailed Student’s t test. The P values < 0.05 or < 0.01 were considered significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TRAF1a (At5g43560), TRAF1b (At1g04300), SINAT1 (At2g41980), SINAT2 (At3g58040), SINAT3 (At3g61790), SINAT4 (At4g27880), SINAT5 (At5g53360), SINAT6 (At3g13672), ATG6 (At3g61710), PR1 (At2g14610), PR2 (At3g57260), PR5 (At1g75040), PDF1.2a (At5g44420), SEN1 (At4g35770), SAG12 (At5g45890), SAG101 (At5g14930), ATG1a (At3g61960), ATG1b (At3g53930), ATG1c (At2g37840), ATG2 (At3g19190), ATG5 (At5g17290), ATG7 (At5g45900), ATG8a (At4g21980), ATG8e (At2g45170), ATG9 (At2g31260), ATG10 (At3g07525), ATG13a (At3g49590), ATG18b (At4g30510), ATG18c (At2g40810), VPS34 (At1g60490), and UVRAG (AT2g32760).
Supplemental Data
Supplemental Figure 1. Molecular Identification of TRAF1a and TRAF1b Knockout Mutants.
Supplemental Figure 2. Phenotypic Characterization of traf1a and traf1b Single Mutants.
Supplemental Figure 3. Phenotypic Analysis of traf1a/b Double Mutants.
Supplemental Figure 4. Phenotypic Analyses of TRAF1a traf1a/b-2 and snc1-r1 muse13-2 muse14-1 Mutants.
Supplemental Figure 5. Lifespan of the Wild Type and traf1a/b Mutants.
Supplemental Figure 6. Phenotypic Analyses of traf1a TRAF1b-RNAi Transgenic Plants.
Supplemental Figure 7. Gene Expression Profiling of traf1a/b by RNA-Seq Analysis.
Supplemental Figure 8. Endogenous SA and JA contents and Senescence-Associated Gene Expression in the 6-Week-Old Wild Type and traf1a/b Double Mutants.
Supplemental Figure 9. Immunoblot Analysis Showing the Processing of GFP-ATG8e in Wild-Type and traf1a/b-1 Plants in Response to Carbon Starvation.
Supplemental Figure 10. ATG Gene Expression in the Wild Type and traf1a/b by qRT-PCR Analysis.
Supplemental Figure 11. Analysis of sinat1, sinat2, and sinat6 Single Mutants.
Supplemental Figure 12. Generation of SINAT1/2-Cas Transgenic Lines.
Supplemental Figure 13. Autophagosome Formation in the sinat Mutants.
Supplemental Data Set 1. Differentially Expressed Genes in the traf1a/b-1 Mutant Compared with the Wild Type.
Supplemental Data Set 2. Sequences of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC for providing all of the T-DNA seed pools, X. Li (University of British Columbia) for the muse13-2 muse14-1, snc1-r1 muse13-2 muse14-1, and snc1-r1 lines, M.L. Chye (University of Hong Kong) for the eGFP-ATG8e line, and J.F. Li (Sun Yat-sen University) for pUC119, pHBT, and pFGC-RCS. This work was supported by the National Natural Science Foundation of China (Projects 31370298, 31461143001, and 31670276), by the Program for New Century Excellent Talents in University (Project NCET-13-0614), by Sun Yat-sen University (start-up fund to S.X.), by the Foundation of Guangzhou Science and Technology (Project 201504010021), as well as by RGC (Projects C4011-14R and AoE/M-05/12 to L.J.).
AUTHOR CONTRIBUTIONS
S.X. designed the research. H.Q., F.N.X., L.J.X., L.J.Y., Q.F.C., X.H.Z., and Q.W. carried out the experiments. S.X., H.Q., F.N.X., F.L., L.J., and Q.X. analyzed the data. S.X., H.Q., F.N.X., and L.J.X. wrote the manuscript.
Glossary
- MS
Murashige and Skoog
- ROS
reactive oxygen species
- SA
salicylic acid
- JA
jasmonate
- DEGs
differentially expressed genes
- DAB
diaminobenzidine
- MDC
monodansylcadaverine
- CA
concanamycin A
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- CHX
cycloheximide
- CoIP
coimmunoprecipitation
- IP
immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Arch R.H., Gedrich R.W., Thompson C.B. (1998). Tumor necrosis factor receptor-associated factors (TRAFs)--a family of adapter proteins that regulates life and death. Genes Dev. 12: 2821–2830. [DOI] [PubMed] [Google Scholar]
- Aubert Y., Widemann E., Miesch L., Pinot F., Heitz T. (2015). CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 66: 3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T., Bajdzienko K., Wittenberg G., Alseekh S., Tohge T., Bock R., Giavalisco P., Fernie A.R. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27: 306–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Wang C., Jiang C., Pan J., Zhang G., Liu H., Zhang H. (2014). The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 202: 174–187. [DOI] [PubMed] [Google Scholar]
- Bassham D.C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L.J., Yoshimoto K. (2006). Autophagy in development and stress responses of plants. Autophagy 2: 2–11. [DOI] [PubMed] [Google Scholar]
- Chen D.C., Yang B.C., Kuo T.T. (1992). One-step transformation of yeast in stationary phase. Curr. Genet. 21: 83–84. [DOI] [PubMed] [Google Scholar]
- Chen L., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11: 2233–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.Y., Park Y.C., Ye H., Wu H. (2002). All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115: 679–688. [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y. (2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143: 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Klionsky D.J. (2008). The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Yu B., Wang Y., Liu Y. (2011). Role of plant autophagy in stress response. Protein Cell 2: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe N.J., Olsen L.J. (2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4: 339–348. [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D.J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Levine B. (2010). The Beclin 1 interactome. Curr. Opin. Cell Biol. 22: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chen X., Zhong X., Li M., Ao K., Huang J., Li X. (2016). Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 19: 204–215. [DOI] [PubMed] [Google Scholar]
- Inoue Ji., Ishida T., Tsukamoto N., Kobayashi N., Naito A., Azuma S., Yamamoto T. (2000). Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp. Cell Res. 254: 14–24. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22: 159–180. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. (2010). Autophagy and the integrated stress response. Mol. Cell 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830. [DOI] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Li J.F., Chung H.S., Niu Y., Bush J., McCormack M., Sheen J. (2013). Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Ma X., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Miao Y., Jiang L. (2007). Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2: 2348–2353. [DOI] [PubMed] [Google Scholar]
- Michaeli S., Galili G., Genschik P., Fernie A.R., Avin-Wittenberg T. (2016). Autophagy in Plants--What’s New on the Menu? Trends Plant Sci. 21: 134–144. [DOI] [PubMed] [Google Scholar]
- Mei Y., Glover K., Su M., Sinha S.C. (2016). Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond. Protein Sci. 25: 1767–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Sanchez-Vera V., Moschou P.N., Suarez M.F., Sundberg E., Weih M., Bozhkov P.V. (2013). Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell 12: 327–329. [DOI] [PubMed] [Google Scholar]
- Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., Gretzmeier C., Dengjel J., Piacentini M., Fimia G.M., Cecconi F. (2013). mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15: 406–416. [DOI] [PubMed] [Google Scholar]
- Oelmüller R., Peškan-Berghöfer T., Shahollari B., Trebicka A., Sherameti I., Varma A. (2005). MATH domain proteins represent a novel protein family in Arabidopsis thaliana, and at least one member is modified in roots during the course of a plant-microbe interaction. Physiol. Plant. 124: 152–166. [Google Scholar]
- Ohsumi Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Park B.S., Eo H.J., Jang I.C., Kang H.G., Song J.T., Seo H.S. (2010). Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 398: 242–246. [DOI] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S.P. (2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27. [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda G., Ea C.K., Chen Z.J. (2007). Ubiquitination and TRAF signaling. Adv. Exp. Med. Biol. 597: 80–92. [DOI] [PubMed] [Google Scholar]
- Platta H.W., Abrahamsen H., Thoresen S.B., Stenmark H. (2012). Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 441: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popelka H., Klionsky D.J. (2015). Post-translationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J. 282: 3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.J., Gu H. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17: 249–263. [DOI] [PubMed] [Google Scholar]
- Shi C.S., Kehrl J.H. (2010). TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Li F., Buono R., Roschzttardtz H., Chung T., Zhang M., Osteryoung K.W., Vierstra R.D., Otegui M.S. (2015). The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E., Lippens S., Vanden Berghe T., Romagnoli A., Fimia G.M., Piacentini M., Vandenabeele P. (2012). Beclin1: a role in membrane dynamics and beyond. Autophagy 8: 6–17. [DOI] [PubMed] [Google Scholar]
- Xia P., et al. (2013). WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32: 2685–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Chye M.L. (2011). Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 156: 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q.F., Chan S.W., Zheng S.X., Ma J., Wang M., Welti R., Chye M.L. (2010). Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P. (2013). TRAF molecules in cell signaling and in human diseases. J. Mol. Signal. 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Xie Y., Kang R., Sun X., Zhong M., Huang J., Klionsky D.J., Tang D. (2015). Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11: 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546. [DOI] [PubMed] [Google Scholar]
- Xu C., Feng K., Zhao X., Huang S., Cheng Y., Qian L., Wang Y., Sun H., Jin M., Chuang T.H., Zhang Y. (2014). Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy 10: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.J., Luo Y.F., Liao B., Xie L.J., Chen L., Xiao S., Li J.T., Hu S.N., Shu W.S. (2012). Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 195: 97–112. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Tian M., Li Q., Cui F., Liu L., Yin B., Xie Q. (2013). A plant-specific in vitro ubiquitination analysis system. Plant J. 74: 524–533. [DOI] [PubMed] [Google Scholar]
- Zhuang X., Wang H., Lam S.K., Gao C., Wang X., Cai Y., Jiang L. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Cui Y., Gao C., Jiang L. (2015). Endocytic and autophagic pathways crosstalk in plants. Curr. Opin. Plant Biol. 28: 39–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.