Abstract
The PerC protein of enteropathogenic Escherichia coli (EPEC), encoded by the pEAF plasmid, is an activator of the locus of enterocyte effacement (LEE) pathogenicity island via the LEE1 promoter. It has been assumed that the related LEE-containing pathogen enterohemorrhagic E. coli (EHEC) lacks PerC-dependent activation due to utilization of an alternative LEE1 promoter and lack of a perC gene. However, we show here that EPEC PerC can activate both the EPEC and EHEC LEE1 promoters and that the major transcriptional start site is similarly located in both organisms. Moreover, a PerC-like protein family identified from EHEC genome analyses, PerC1 (also termed PchABC), can also activate both promoters in a manner similar to that of EPEC PerC. The perC1 genes are carried by lambdoid prophages, which exist in multiple copies in different EHEC strains, and have a variable flanking region which may affect their expression. Although individual perC1 copies appear to be poorly expressed, the total perC1 expression level from a strain encoding multiple copies approaches that of perC in EPEC and may therefore contribute significantly to LEE1 activation. Alignment of the protein sequences of these PerC homologues allows core regions of the PerC protein to be identified, and we show by site-directed mutagenesis that these core regions are important for function. However, purified PerC protein shows no in vitro binding affinity for the LEE1 promoter, suggesting that other core E. coli proteins may be involved in its mechanism of activation. Our data indicate that the nucleoid-associated protein IHF is one such protein.
Pathogenic bacterial strains such as enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC), which cause disease by forming intestinal attaching-and-effacing (A/E) lesions, possess a chromosomal pathogenicity island (PAI) called the locus of enterocyte effacement (LEE). The EPEC LEE is necessary and sufficient for formation of A/E lesions (21) and encodes a type III secretion system, proteins secreted by this system which form a translocon apparatus, and proteins (intimin and its translocated intimin receptor, Tir) which mediate the intimate bacterial attachment to host cells characteristic of the A/E lesion. Regulatory studies on the LEE show that it is organized into five principal operons: LEE1, LEE2, and LEE3 encode the type III secretion system plus the CesAB chaperone and the EspH effector; LEE4 encodes the translocon proteins; and LEE5 encodes intimin, Tir, and CesT, a chaperone for intimin (22, 35). The promoter of the LEE1 operon is of key importance for regulation of the entire PAI; the first gene of this operon encodes Ler, a regulator related to the H-NS family of nucleoid-associated proteins, which in turn activates the expression of LEE2, LEE3, LEE5, and, at least to some extent in EPEC, LEE4 (10, 11, 22, 38). Given this hierarchical organization of expression, it is apparent that factors and/or environmental conditions which affect LEE1 expression will be able to regulate coordinately the expression of the bulk of the LEE genes and hence the A/E phenotype.
A number of core chromosomal factors and pathogen-specific regulators have been shown to affect LEE1 expression; most of these studies have been performed on EPEC strains. The nucleoid-associated protein H-NS, a common repressor of virulence-related genes in gram-negative bacteria (8), negatively regulates LEE1 expression; its effect is particularly apparent at temperatures below 30°C (40). H-NS is also a temperature-independent repressor of the LEE2 through LEE5 promoters, where its effect is overcome by the related Ler protein (4, 35, 40), and therefore has multiple inputs into LEE regulation. A second nucleoid-associated protein, integration host factor (IHF), is required for efficient expression of LEE1 and interacts with the LEE1 promoter region in vitro; IHF is required for activation of the entire LEE but acts directly only at LEE1 (11). Yet another nucleoid-associated protein, Fis, has been shown to be required for LEE1 expression as well, although surprisingly, this effect did not seem to be transmitted to all of the downstream LEE operons (13). Recent work on mutations in the genes of the LEE from a related A/E pathogen, Citrobacter rodentium, suggests that two open reading frames (ORFs) downstream of LEE1 encode the antagonistic LEE1 regulators GrlA and GrlR, which have an overall positive effect on LEE expression (6); these regulators are conserved in the EPEC and EHEC LEEs and may be assumed to function similarly in those organisms. Thus, pathogen-specific as well as core E. coli regulators can control expression of the LEE.
The principal pathogen-specific LEE regulator which has been studied to date is the per locus from EPEC, which is encoded by the EPEC-specific plasmid pEAF (14, 39). Activation by per is manifested at the LEE1 promoter, thereby activating the Ler regulatory cascade in a manner similar to that for IHF (22). The per locus consists of three ORFs, encoding an AraC-like activator protein, PerA, and PerB and PerC, which do not belong to characterized protein families. Original studies of per suggested that all three proteins might contribute to activation both of the LEE and of the bundle-forming pilus (bfp) operon located on pEAF (14, 39); however, more recently it has become apparent that PerA alone acts directly to autoactivate the per and bfp promoters, while PerC directly or indirectly affects LEE1 expression in a manner which is PerA dependent, but only because expression of PerC requires PerA (4, 20, 29). PerC-dependent LEE1 activation functions in the absence of other EPEC-specific factors in E. coli K-12 (29), contrary to a previous suggestion that the PerC effect might be indirect (4). However, in view of the fact that pEAF is not found in EHEC strains, PerC is apparently EPEC specific, which would confine this activation mechanism to EPEC strains harboring this plasmid. It has been demonstrated that both the EPEC and EHEC LEEs are activated by cell density-dependent (“quorum”) sensing (37), and it has been speculated that activation by Per helps EPEC to compensate for the lower bacterial concentration (and hence reduced quorum-sensing-dependent activation) in its colonization site, the small intestine, compared to that of EHEC, the large intestine. It has also been suggested that the difference in Per dependency between EPEC and EHEC is linked to the use of an alternative LEE1 promoter in EPEC, which is 169 bp upstream of that found in EHEC (10, 37). In EPEC, the downstream promoter mapped in EHEC has a 6-bp duplication overlapping the proposed −10 region and a single-base-pair deletion between the −10 and −35 regions, while in EHEC strains, the upstream promoter mapped in EPEC has a single-base-pair deletion just upstream of the −35 region and a base pair change downstream of the transcriptional start site (10). It is assumed that these nucleotide changes cause only the upstream promoter to be functional in EPEC and only the downstream promoter to function in EHEC, while Per-dependent activation is restricted to the upstream promoter.
Here we have tested the responses of both the EPEC and EHEC LEE1 promoters to activation by EPEC PerC, and we find that both can be activated to similar extents by this EPEC-specific protein. Moreover, the nucleotide differences between the two promoter regions affect the intrinsic activity of the promoter but not PerC-dependent activation, and reexamination of the LEE1 transcriptional start sites from both organisms suggests that both use the upstream promoter as the primary source of LEE1 expression. A subfamily of a group of PerC-like proteins identified from the genome sequences of EHEC strains is also able to activate EPEC and EHEC LEE1 expression, suggesting that activation of the LEE by PerC-like proteins may also function in EHEC. Finally, homology-based mutagenesis of PerC has allowed us to undertake an initial structure-function study on this protein.
MATERIALS AND METHODS
Bacterial strain construction.
The bacterial strains used in this study are listed in Table 1, and the oligonucleotide primers used in their construction are shown in Table 2. The single-copy chromosomal fusions of the EHEC LEE1 promoter and mutated EPEC LEE1 promoters to lacZ were constructed by the method previously used to make the EPEC LEE1-lacZ fusion (29). Briefly, an EHEC LEE1 promoter fragment containing 24 codons of the ler gene plus 362 bp of DNA upstream of the ATG was amplified from ZAP198 DNA with primers LEE1A and LEE1TlF, digested with BamHI and KpnI, and cloned into the temperature-sensitive vector pAJR36 (Table 1) to yield pIBlerEHEC-lac, with the LEE1 promoter fused in frame to the lacZ gene. The fusion was then integrated into the chromosome of MG1655 at the lac locus by using a derivative of MG1655 in which the lacZ and lacY genes are replaced by a sacB kan cassette encoding kanamycin resistance and sucrose sensitivity (Table 1). The pIBlerEHEC-lac plasmid was transformed into MG1655 sacB kan at 30°C and then forced to recombine into the chromosome by repeated subculturing in the presence of chloramphenicol at 42°C for 48 h, followed by repeated subculturing at 30°C for 48 h without antibiotic selection to resolve the plasmid cointegrates. Successful recombinants in which the LEE1-lacZ construct had replaced the chromosomal sacB-kan cassette were then selected on agar plates containing 6% sucrose and checked for kanamycin sensitivity, chloramphenicol sensitivity, and the absence of plasmid DNA. Mutant derivatives of the EPEC LEE1-lacZ fusion plasmid pIBler-lac derived by site-directed mutagenesis (see below) were recombined into the MG1655 chromosome similarly. The hns-206::Apr and ihfA82::Tn10 alleles were transduced from the donor strains PD32 and NEC007, respectively, into the MG1655 LEE1-lacZ fusion strain either singly or in combination by using bacteriophage P1cml as described by Silhavy et al. (36).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| BL21(λDE3)/pLysS | Expression strain with IPTG-inducible T7 RNA polymerase | Novagen |
| DH5α | recA cloning strain | Our stocks |
| E2348/69 | Wild-type EPEC O127:H6 | 19 |
| Sakai stx | stx derivative of EHEC O157:H7 strain Sakai | 30 |
| MG1655 LEE1-lacZ | MG1655 with chromosomal LEE1EPEC-lacZ fusion at lac | 29 |
| MG1655 LEE1EHEC-lacZ | MG1655 with chromosomal LEE1EHEC-lacZ fusion at lac | This work |
| MG1655 LEE1Δ6-lacZ | MG1655 LEE1-lacZ with Δ6 mutation in LEE1 promoter | This work |
| MG1655 LEE1I1-lacZ | MG1655 LEE1-lacZ with I1 mutation in LEE1 promoter | This work |
| MG1655 LEE1Δ6I1-lacZ | MG1655 LEE1-lacZ with Δ6 and I1 mutations in LEE1 promoter | This work |
| MG1655 LEE1C→A-lacZ | MG1655 LEE1-lacZ with C→A mutation in LEE1 promoter | This work |
| MG1655 LEE1ΔT-lacZ | MG1655 LEE1-lacZ with ΔT mutation in LEE1 promoter | This work |
| MG1655 LEE1C→A ΔT-lacZ | MG1655 LEE1-lacZ with C→A and ΔT mutations in LEE1 promoter | This work |
| MG1655 LEE1Δ6I1 C→A-lacZ | MG1655 LEE1-lacZ with Δ6, I1, and C→A mutations in LEE1 promoter | This work |
| MG1655 LEE1Δ6I1 ΔT-lacZ | MG1655 LEE1-lacZ with Δ6, I1, and ΔT mutations in LEE1 promoter | This work |
| MG1655 LEE1Δ6I1 C→A ΔT-lacZ | MG1655 LEE1-lacZ with Δ6, I1, C→A, and ΔT mutations in LEE1 promoter | This work |
| MG1655 LEE1-lacZ hns | MG1655 LEE1-lacZ hns-206::Apr | This work |
| MG1655 LEE1-lacZ ihf | MG1655 LEE1-lacZ ihfA82::Tn10 | This work |
| MG1655 LEE1-lacZ hns ihf | MG1655 LEE1-lacZ hns-206::AprihfA82::Tn10 | This work |
| MG1655 sacB kan | MG1655 with lacZY replaced by sacB kan | 29 |
| NEC007 | BL21(λDE3) ihfA82::Tn10 | 12 |
| PD32 | MC4100 hns-206::Apr | 7 |
| ZAP198 | EHEC O157:H7 stx Nalr | 26, 32 |
| Plasmids | ||
| pAC198PerC1-1 | perC1-1 plus flanking DNA from ZAP198 in pACYC184 | This work |
| pAC198PerC1-3 | perC1-3 plus flanking DNA from ZAP198 in pACYC184 | This work |
| pAC198PerC2 | perC2 plus flanking DNA from ZAP198 in pACYC184 | This work |
| pACSakPerC1-2 | perC1-2 plus flanking DNA from Sakai stx in pACYC184 | This work |
| pACSakPerC2 | perC2 plus flanking DNA from Sakai stx in pACYC184 | This work |
| pACYC184 | P15A replicon; Cmr Tcr | New England Biolabs |
| pAJR36 | Temperature-sensitive vector, promoterless lacZ gene flanked by lacIA; Cmr | 29 |
| pBAD198PerC1-1 | Arabinose-inducible PerC1-1 from ZAP198 | This work |
| pBAD198PerC1-3 | Arabinose-inducible PerC1-3 from ZAP198 | This work |
| pBAD198PerC2 | Arabinose-inducible PerC2 from ZAP198 | This work |
| pBAD33 | Arabinose-inducible expression vector; Cmr | 15 |
| pBADSakPerC1-2 | Arabinose-inducible PerC1-2 from Sakai stx | This work |
| pBADSakPerC2 | Arabinose-inducible PerC2 from Sakai stx | This work |
| pBADH6SPerC | Arabinose-inducible His6-S-tagged EPEC PerC | This work |
| pBADPerC | Arabinose-inducible EPEC PerC | 29 |
| pET30b | T7-controlled His6-S-tagged expression vector; Kmr | Novagen |
| pET30PerC | T7-controlled His6-S-tagged EPEC PerC | This work |
| pIBler-lac | EPEC LEE1 promoter fused in frame with lacZ in pAJR36 | 29 |
| pIBlerEHEC-lac | EHEC LEE1 promoter fused in frame with lacZ in pAJR36 | This work |
| pSPT18 | SP6/T7 in vitro transcription vector; Apr | Roche Molecular |
| pSPTperC | Internal fragment of EPEC perC in pSPT18 | 29 |
| pSPTperC1 | Internal fragment of EHEC perC1 in pSPT18 | This work |
| pTH19kr | Low-copy-number cloning vector; Kmr | 16 |
| pTH198PerC1-1 | perC1-1 plus flanking DNA from ZAP198 in pTH19kr | This work |
| pTH198PerC1-3 | perC1-3 plus flanking DNA from ZAP198 in pTH19kr | This work |
| pTH198PerC2 | perC2 plus flanking DNA from ZAP198 in pTH19kr | This work |
| pTHSakPerC1-2 | perC1-2 plus flanking DNA from Sakai stx in pTH19kr | This work |
| pTHSakPerC2 | perC2 plus flanking DNA from Sakai stx in pTH19kr | This work |
| pTHperABC | EPEC perABC plus flanking DNA in pTH19kr | 29 |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a | Use |
|---|---|---|
| LEE1TIF | cgagtggtaccCTCTATAAGCTGAATGTATGG | Cloning of LEE1EHEC-lacZ fusion |
| LEE1A | cgagtggatccGTGAAACGGTTCAGC | Cloning of LEE1EHEC-lacZ fusion/bandshift |
| LerPMut1 | pAGGAAGGACAACAATTAATCA | Δ6 mutation of LEE1EPEC-lacZ fusion |
| LerPMut2 | pGATAAGGTCGCTAATAGCTTT | Δ6 mutation of LEE1EPEC-lacZ fusion |
| IlerPa | CATTTGATTAATTGTTGgTCCTTCCTG | I1 mutation of LEE1EPEC-lacZ fusion |
| IlerPb | CAGGAAGGAcCAACAATTAATCAAATG | I1 mutation of LEE1EPEC-lacZ fusion |
| LEEC-Aa | TTACACATTAGAAAAaAGAGAATAATAACAT | C→A mutation of LEE1EPEC-lacZ fusion |
| LEEC-Ab | ATGTTATTATTCTCTtTTTTCTAATGTGTAA | C→A mutation of LEE1EPEC-lacZ fusion |
| LEEΔTa | GGATTTTAAAAATATATGATTTTTTTGTTGACA | ΔT mutation of LEE1EPEC-lacZ fusion |
| LEEΔTb | TGTCAACAAAAAAATCATATATTTTTAAAATCC | ΔT mutation of LEE1EPEC-lacZ fusion |
| RACE3 | GAGTTCCGGCGAGCGAGTCCATC | RACE cDNA primer |
| RACE4 | CGAGCGAGTCCATCATCAGGCAC | RACE PCR 3′ primer |
| RACE5 | GTATATCCCAGCTCTTGTAAGG | RACE PCR nested 3′ primer |
| LEE1AR | cgagtaagcttGCTTTAATATTTTAAGC | Sequencing of LEE1 5′ RACE products/bandshift |
| LEE1BSc | TTGACATTTAATGATAATG | LEE1 bandshift primer |
| LEE1BSf | CTAATGTGTAAAATACATTATC | LEE1 bandshift primer |
| EHperC1/5 | cgagtggatccTTGGCAGAATAGTTGTTTGG | Cloning of EHEC perC1 plus flanking DNA |
| EHperC1/3 | cgagtaagcttAACACGACCAGAGCACCTGT | Cloning of EHEC perC1 plus flanking DNA |
| EHperC2/5 | cgagtggatccTAACACCGGACAGTCATGCG | Cloning of EHEC perC2-containing ORF |
| EHperC2/3 | cgagtaagcttACGGGCGAGAATACTCATGA | Cloning of EHEC perC2-containing ORF |
| perCEHfara | ctagcaggtaccGCAGTCTGTAGATAAACGGAG | Cloning of arabinose-inducible SakPerC1-2 |
| perC198fara | ctagcaggtaccGCAGTCTGTAGATAATCGGAG | Cloning of arabinose-inducible 198PerC1-1 and -3 |
| perC2fara | ctagcaggtaccGGTAATCAGCCACCAGCGGG | Cloning of arabinose-inducible Sak- and 198PerC2 |
| perC2EHrara | ctagcaaagcttCCTCTGTTGTGTCTGTTTGTTTC | Cloning of arabinose-inducible SakPerC2 |
| perC2198rara | ctagcaaagcttCAACTGGTGCAAAAAAAGCCGG | Cloning of arabinose-inducible 198PerC2 |
| QCC1a | GGCGAAGTACTcGGAAGAAAAAGGG | perC L11S mutagenesis (forward) |
| QCC1b | CCCTTTTTCTTCCgAGTACTTCGCC | perC L11S mutagenesis (reverse) |
| QCC2a | GTACTTGGAAGAAAAAtGGTTTTATAGACGAGC | perC G15W mutagenesis (forward) |
| QCC2b | GCTCGTCTATAAAACCaTTTTTCTTCCAAGTAC | perC G15W mutagenesis (reverse) |
| QCC3a | GAAGAAAAAGGGTTTTAtATACGAGCTGCAG | perC R18I mutagenesis (forward) |
| QCC3b | CTGCAGCTCGTATaTAAAACCCTTTTTCTTC | perC R18I mutagenesis (reverse) |
| QCC4a | GGGTTTTATAGACcAGCTGCAGATC | perC R19P mutagenesis (forward) |
| QCC4b | GATCTGCAGCTgGTCTATAAAACCC | perC R19P mutagenesis (reverse) |
| QCC6a | GCGTGCATTCaGCATTAATAAATCTCTACG | perC C47S mutagenesis (forward) |
| QCC6b | CGTAGAGATTTATTAATGCtGAATGCACGC | perC C47S mutagenesis (reverse) |
Sequences complementary to the target DNA are shown in capital letters; sequences unique to the oligonucleotide primer are lowercased. Restriction sites used for cloning are underlined. p, added 5′ phosphate.
Plasmid construction.
Plasmids used or constructed in this study are listed in Table 1, and again the oligonucleotide primers employed in construction are shown in Table 2. The EHEC perC1 and perC2 genes were amplified from Sakai stx or ZAP198 chromosomal DNA by using the primer pairs EHperC1/5-EHperC1/3 and EHperC2/5-EHperC2/3, respectively, digested with BamHI and HindIII, cloned into pACYC184, subsequently amplified from the pACYC184 clones with the same primers, and subcloned into pTH19kr. For construction of arabinose-inducible derivatives of the same genes, the perC1 or perC2 ORF was amplified from the pACYC184 clones by using primers perCEHfara and EHperC1/3 (for SakPerC1-2), perC198fara and EHperC1/3 (for 198PerC1-1 and 198 PerC1-3), perC2fara and perC2EHrara (for SakPerC2), or perC2fara and perC2198rara (for 198PerC2), digested with KpnI and HindIII, and cloned downstream of the araBAD promoter in similarly digested pBAD33. A tagged PerC derivative for expression and purification (pET30PerC) was made by amplifying the perC ORF from the low-copy-number perABC clone pTHperABC by using perCex1 and perCex2, digesting with NcoI and BamHI, and cloning into pET30b; this entire tagged ORF was also amplified with PETBADF and PETBADR and cloned into KpnI-HindIII-digested pBAD33 to produce an arabinose-inducible construct (pBADH6SPerC) to test the functionality of the tagged protein.
Growth conditions and enzyme assays.
For analysis of LEE1-lacZ transcription, strains were grown at 37°C overnight in Luria-Bertani (LB) medium and then subinoculated 1:100 into high-glucose Dulbecco's modified Eagle medium (DMEM) containing HEPES and lacking phenol red (catalog no. 21063-029; Gibco-Invitrogen Corporation). The DMEM cultures were grown to mid-logarithmic phase (optical density at 600 nm, ∼0.6) and assayed for β-galactosidase activity as described by Miller (23). Cultures were assayed in duplicate, and the assays were repeated at least twice. Standard deviations were less than 10%. Antibiotic selection with chloramphenicol (20 μg/ml) or kanamycin (25 μg/ml) was carried out as appropriate. When arabinose-inducible derivatives of PerC or PerC-like proteins were used, arabinose was added at a concentration of 1.0% to overcome catabolite repression of the araBAD promoter by glucose in the medium (15).
Site-directed mutagenesis of PerC and the EPEC LEE1 promoter.
Site-directed mutagenesis of the perC gene was performed using the pTHperABC template, oligonucleotide primers as listed in Table 2, and a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Single-base-pair insertions, deletions, and substitutions in the EPEC LEE1 promoter were also engineered by the QuikChange method using the fusion plasmid pIBler-lac as a template and specific primers as listed in Table 2. The Δ6 mutation in the EPEC LEE1 promoter was constructed by outward PCR from the pIBler-lac template using the 5′-phosphate-containing primers LerPMut1 and LerPMut2, followed by digestion of template DNA with DpnI and religation.
RNA extraction, probe synthesis, and Northern blotting.
Total cellular RNA was extracted from cultures grown to mid-log phase by cell lysis in boiling REB buffer (20 mM sodium acetate [pH 5.2], 2% sodium dodecyl sulfate, 0.3 M sucrose) followed by phenol extraction and DNase I treatment as described previously (28). A 143-bp internal HincII-PvuII fragment of the EHEC perC1 gene was cloned into the in vitro transcription vector pSPT18 to generate pSPTperC1. This plasmid and the previously described plasmid pSPTperC (Table 1) were linearized with HindIII and transcribed in vitro with SP6 RNA polymerase to generate digoxigenin-UTP-labeled RNA probes by using a DIG-RNA labeling kit supplied by Roche Molecular according to the manufacturer's instructions. Samples of total cellular RNA (20 μg) were electrophoresed on morpholinepropanesulfonic acid (MOPS)-formaldehyde-agarose gels, transferred to positively charged Hybond-N+ membranes (Amersham), and hybridized with the labeled RNA probes overnight. Following stringency washes, bound probes were detected with alkaline phosphatase-conjugated anti-digoxigenin and the chemiluminescent substrate CSPD (Roche Molecular) as described previously (28). Transcript bands were quantified by using ImageQuant software.
5′ RACE analysis of LEE1 transcripts.
Total cellular RNA preparations from EPEC strain E2348/69 and EHEC strain ZAP198 grown to mid-logarithmic phase in DMEM or LB medium (see above) were used as templates for first-strand cDNA synthesis employing a SMART rapid amplification of cDNA ends (RACE) kit (BD Biosciences) as described by the manufacturer, except that the oligo(dT) primer supplied with the kit was replaced with the LEE1-specific primer RACE3, which primes from 229 bp downstream of the ler initiation codon. RACE PCR was then performed with the kit-specific universal primer mix and the LEE1-specific RACE4 primer, and nested PCR on these products was carried out using nested universal primer A from the kit and the RACE5 primer, which primes from 194 bp downstream of the ler initiation codon. These steps were performed as described by the kit manufacturer, except that annealing temperatures of 60 and 50°C, respectively, were used. Products were analyzed on 3% Agarose-MS (molecular screening agarose) gels (Roche Molecular), and individual bands were purified from the gels and sequenced directly by using the LEE1AR primer.
Protein purification and mobility shift assays.
Five-hundred-milliliter cultures of BL21(λDE3)/pLysS transformed with pET30PerC were grown to mid-log phase (optical density at 600 nm, ∼0.6) in LB medium under kanamycin selection (50 μg/ml) and then induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a concentration of 1 mM for 3 h. Cells were pelleted by centrifugation, washed in 40 ml of 50 mM HEPES-0.1 M NaCl, and resuspended in 15 ml of His-Tag binding buffer (5 mM imidazole, 0.25 M NaCl, 20 mM Tris [pH 7.9], 10% glycerol, 0.1% Triton X-100, 10 mM β-mercaptoethanol). Following cell lysis by sonication (MSE Soniprep 150; 3 to 6 pulses of 1 min at an amplitude of 5 μm, interspersed with cooling on ice), cell debris was pelleted by centrifugation at 30,000 × g for 20 min, and the cleared supernatants were loaded onto columns of Ni2+-nitrilotriacetic acid agarose (QIAGEN) equilibrated with His-Tag binding buffer. The columns were washed twice with 15 ml of His-Tag binding buffer supplemented with 30 mM imidazole to remove weakly bound proteins, and the specifically bound proteins were then eluted with His-Tag binding buffer supplemented with 0.5 M imidazole. Eluted proteins were dialyzed against 20 mM Tris (pH 7.9)-50 mM KCl-5 mM NaCl-1 mM EDTA-1 mM dithiothreitol-20% glycerol-0.1% NP-40 overnight and then stored in aliquots at −80°C. EPEC LEE1 promoter DNA fragments for use in mobility shift assays were amplified with primer pairs LEE1A-LEE1BSf (−368 to −157 with respect to the ler start codon), LEE1BSc-LEE1AR (−191 to −1), or LEE1A-LEE1AR (−368 to −1) and end labeled with digoxigenin-ddUTP by using an end-labeling kit (Roche Molecular) according to the manufacturer's instructions. Approximately 1.2 ng of labeled probe was mixed with purified protein in binding buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 5 mM NaCl, 50 mM KCl, 8% glycerol, 0.05 mg of bovine serum albumin/ml, 1 mM dithiothreitol) in the presence of a ∼1,000-fold excess (1 μg) of the nonspecific competitor poly(dI-dC) and was incubated at room temperature for 20 min. Protein-DNA complexes were electrophoresed on native 4% polyacrylamide gels at 100 V and 4°C for 5 h and were then transferred to Hybond-N+ membranes (Amersham). The labeled DNA probe was then detected by using the alkaline phosphatase-CSPD-based chemiluminescence system (Roche Molecular) as described for Northern blotting.
Computational analysis.
Protein secondary-structure predictions were performed at http://www.embl-heidelberg.de/predictprotein/predictprotein.html by using the PHD algorithm (33, 34). Protein and DNA sequences were aligned by using the BLASTP, BLASTN, and TBLASTN programs (2) at http://www.ncbi.nlm.nih.gov/BLAST/.
RESULTS
The LEE1 promoters from both EPEC and EHEC can be activated by PerC in the absence of other pathogen-specific factors.
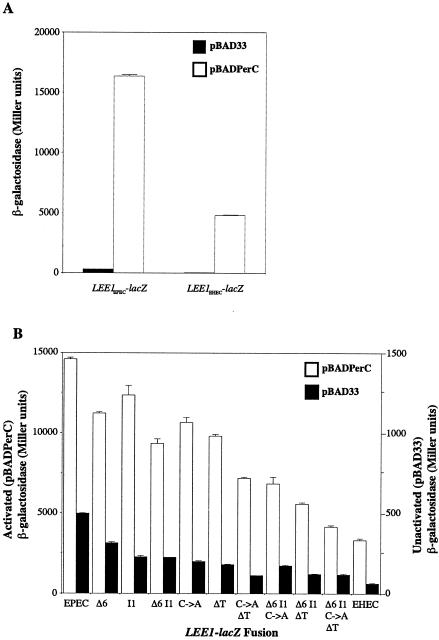
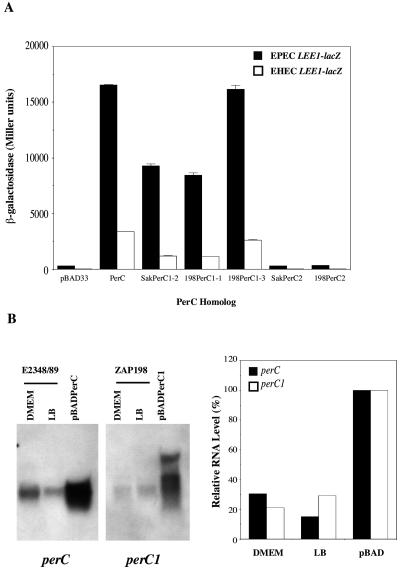
In order to compare the abilities of PerC from EPEC strain E2348/69 to activate the LEE1 promoters from EPEC and EHEC, we used single-copy chromosomal fusions of the two promoters to lacZ integrated into the genome of the E. coli K-12 strain MG1655 at the native lac locus. The LEE1EPEC-lacZ fusion has been described previously (29), and the LEE1EHEC-lacZ fusion was constructed by similar methods (see Materials and Methods). The EPEC fusion contains 368 bp of DNA upstream of the ATG of the ler gene, extending as far as the enterobacterial repetitive intergenic consensus sequence found upstream of LEE1 in this organism (9); because of differences in the promoter sequence, this corresponds to 362 bp in the EHEC fusion (Fig. 1). Each fusion also contains the first 24 codons of ler, which are fused in frame to lacZ. Into these two fusion strains we introduced plasmid pBADPerC, in which the PerC protein is expressed from the arabinose-inducible araBAD promoter without a requirement for its natural activator, the AraC-like PerA protein. Compared to the vector control, pBAD33, pBADPerC mediated ∼40-fold activation of the EPEC LEE1 promoter when induced with 1.0% arabinose in DMEM at mid-logarithmic phase (Fig. 2A). Unexpectedly, a similar fold activation by PerC was also seen at the EHEC LEE1 promoter, although the actual level of expression, both unactivated and activated, from this promoter was three- to fourfold lower than that from the EPEC promoter.
FIG. 1.
Promoter regions of the LEE1 operon from EPEC O127:H6 and EHEC O157:H7 cloned as chromosomal LEE1-lacZ fusion constructs in this study. The −35 and −10 regions associated with previously mapped transcriptional start sites (10, 22, 37) are boxed, and these start sites are indicated by angled arrows. Mutations in the EPEC promoter sequence engineered in this study are indicated above the EPEC sequence. Nucleotides identified as RNA 5′ ends by 5′ RACE analysis are boldfaced. The first 24 amino acids of the EPEC and EHEC Ler protein sequences, which were encoded by the fusion constructs, are indicated above and below the respective DNA sequences.
FIG. 2.
(A) Expression of the LEE1EPEC-lacZ and LEE1EHEC-lacZ fusion constructs in the presence of the arabinose-inducible EPEC PerC construct pBADPerC (open bars) or the vector control pBAD33 (solid bars). Strains were grown in DMEM with 1.0% arabinose to mid-logarithmic phase and assayed. (B) Effects of EHEC-like promoter mutations on expression of the LEE1EPEC-lacZ fusion construct. Fusion strains containing the mutations indicated (see also Fig. 1) were assayed in the presence of pBADPerC (open bars, left-hand scale) or pBAD33 (solid bars, right-hand scale) as described for panel A. The unmutated LEE1EPEC-lacZ and LEE1EHEC-lacZ fusion constructs were assayed as controls. The activated and unactivated expression levels are plotted on different scales to enable both to be seen clearly.
Point mutations in the EPEC LEE1 promoter that make it more like the EHEC promoter reduce its activity but do not affect activation by PerC.
EHEC LEE1 transcription has been postulated to be Per independent due to the use of a promoter different from that in EPEC as a result of base changes in the DNA upstream of the ler ORF. Around the downstream LEE1 promoter previously mapped in EHEC (37), EPEC contains two sets of mutations, a duplication of a 6-bp sequence overlapping the putative −10 region and a deletion of a G nucleotide between the −10 and −35 regions (10) (Fig. 1). Conversely, around the upstream LEE1 promoter mapped in EPEC (22), EHEC contains a C-to-A transversion 5 bp downstream of the transcriptional start site and a deletion of a T nucleotide 2 bp upstream of the −35 region (Fig. 1). We therefore introduced these changes individually and in combination into the LEE1EPEC-lacZ fusion construct on a plasmid and recombined all the constructs into the MG1655 chromosome as described above. The mutated constructs were then assayed in comparison to the native EPEC and EHEC constructs when the strains were grown to mid-log phase in DMEM with or without PerC-dependent activation (Fig. 2B). The LEE1EPEC-Δ6 and LEE1EPEC-I1 constructs with EHEC-like mutations around the downstream promoter sequences had slightly reduced expression, which was reduced further to around two-thirds that of the native EPEC promoter when the two mutations were combined (LEE1EPEC-Δ6I1) but was still significantly higher than that obtained from the EHEC promoter. In comparison, single-base-pair mutations around the upstream promoter sequences (LEE1EPEC-C→A or LEE1EPEC-ΔT) had slightly greater effects on expression than the Δ6 or I1 mutation, and when the two were combined (LEE1EPEC-C→AΔT), β-galactosidase activity was reduced by ∼50%. This result suggests that the upstream promoter sequences are more important for expression than the downstream sequences, as expected in EPEC. When these upstream mutations were combined with those around the downstream promoter sequences, EPEC promoter constructs with activity reduced almost to that of the EHEC construct were obtained (Fig. 2B). The effect of combining the single mutations in the EPEC promoter is therefore a stepwise reduction in activity toward that of the EHEC promoter. The slightly elevated activity of the quadruple-mutant construct (LEE1EPEC-Δ6I1 C→AΔT) relative to that of LEE1EHEC presumably reflects the effect of the final base difference between the two promoters, a G-to-A transition 89 bp upstream of the EPEC transcriptional start site (Fig. 1). It is also possible that the change of Thr to Asn at codon 11 of the EHEC Ler protein may affect expression of this translational fusion. Importantly, however, all of the fusion constructs, like their EPEC and EHEC parents, were activated similarly (around 40-fold) by expression of PerC (Fig. 2B). This suggests either that the LEE1 promoter region in general is activated similarly by PerC irrespective of which promoter is functional or that the location of the LEE1 transcriptional start sites in EPEC and EHEC strains requires reexamination.
The major transcriptional start sites of the EPEC and EHEC LEE1 promoters map within a few base pairs of each other.
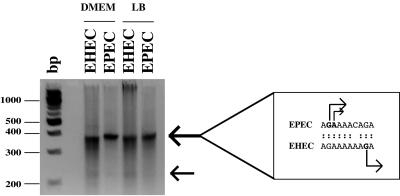
In order to verify which transcriptional start sites operate in vivo at the EPEC and EHEC LEE1 operons, we performed 5′-RACE analysis of LEE1 transcripts in total cellular RNA from EPEC strain E2348/69 and EHEC strain ZAP198 grown to mid-log phase in DMEM or LB medium (see Materials and Methods for details). This methodology allows the detection of RNA end points from within the entire LEE1 promoter region with a single set of primers, thus eliminating inconsistencies due to variations in primer binding. Using a 3′ primer amplifying from 194 bp downstream of the Ler initiation codon, we obtained major products of 350 to 400 bp from both strains in either growth medium, with the EPEC product appearing slightly larger than the EHEC product (Fig. 3). These sizes are consistent with the upstream start site previously mapped in EPEC (168 bp upstream of the ATG in EPEC; 163 bp in EHEC [22]). Purification and sequencing of these RACE products confirmed that the EPEC transcript started at the previously identified G nucleotide or at the A immediately downstream, while the EHEC transcript started at a G nucleotide 7 bp downstream of the major EPEC start (Fig. 1 and 3). Thus, it seems that essentially the same LEE1 promoter is used by both EPEC and EHEC in the strains tested here; the apparent 7-bp difference in the initiating nucleotide could be due to the EHEC-specific C-to-A transversion downstream of the EPEC start site. Much weaker RACE products of ∼220 bp were also obtained from both EPEC and EHEC RNA; the abundance of the EHEC product was slightly greater (Fig. 3). Purification and sequencing of these products revealed in EHEC a variety of 5′ ends clustered around (but not including) the previously identified EHEC start site, and in EPEC dual 5′ ends at equivalent G residues within the 6-bp EPEC-specific duplication (Fig. 1). This heterogeneity suggests that these 5′ ends may represent processing sites within the LEE1 transcript rather than genuine transcriptional starts and that the effect of the 6-bp duplication within this region of the EPEC 5′ untranslated sequence is to affect this processing rather than to abolish an alternative promoter. Whatever the true nature of the downstream transcript ends, it is clear that the upstream LEE1 promoter is dominant in both organisms, and this finding correlates with the ability of PerC to activate both EPEC and EHEC LEE1-lacZ fusions.
FIG. 3.
5′ RACE analysis of LEE1 transcripts from EPEC strain E2348/69 and EHEC strain ZAP198 grown to mid-logarithmic phase in DMEM or LB medium. The products were analyzed on 3% Agarose-MS gels, and the positions of molecular weight markers (in base pairs) are shown. Major and minor RACE products are indicated by large and small arrows, respectively. The initiating nucleotides corresponding to the major products are indicated in the box at the right of the figure; those corresponding to the minor products are shown in Fig. 1.
PerC-homologous proteins exist in a variety of pathogenic E. coli strains with different copy numbers.
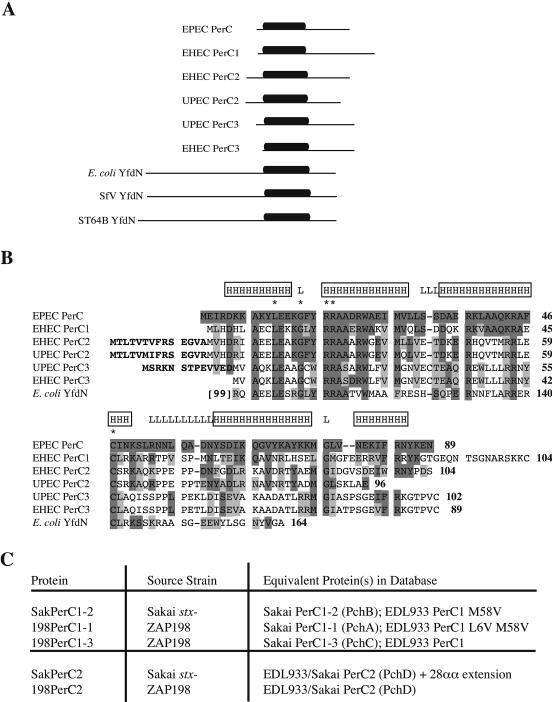
When it was first characterized, PerC from EPEC was considered a unique protein with no known homologues in other organisms (14, 39). Based on this assumption, the PerC-dependent activation of the EHEC LEE1 promoter demonstrated above would be an artifactual effect never seen in EHEC strains. However, the complete genome sequencing of the EHEC strains EDL933 and Sakai (17, 27), of UPEC strain CFT073 (41), and even of the E. coli K-12 laboratory strain MG1655 (3) reveals that all of these organisms encode proteins with differing degrees of similarity to PerC. The chromosome of the EHEC strain Sakai encodes a total of five PerC-like proteins, all located within prophages or prophage-like regions scattered throughout the chromosome (Table 3 and Fig. 4). The closest relative of PerC, which we term EHEC PerC1, is 47% identical and 67% similar to PerC, has a 14-amino-acid nonconserved C-terminal extension, and exists in three copies encoded by the lambda-like prophages Sp4 (PerC1-1), Sp11 (PerC1-2), and Sp14 (PerC1-3). PerC1-1 has conservative amino acid substitutions at two nonconserved amino acids (L6V and M58V) compared to PerC1-3, while PerC1-2 has only the M58V substitution; the 200 bp of DNA upstream of the perC1 ORFs are also highly divergent among the three copies, which may affect expression. EHEC PerC2, which is 39% identical and 65% similar to PerC and has a putative 14-amino-acid N-terminal extension which overlaps an upstream ORF, is encoded by the nonlambdoid Sp7 prophage. Finally, EHEC PerC3, 25% identical and 54% similar to PerC, is found in the CP4 phage-like element SpLE1 (Table 3; Fig. 4). Since completion of this work, characterization of the PerC1-1, -2, and -3, PerC2, and PerC3 proteins from the Sakai strain has also been reported by Iyoda and Watanabe (18), who termed them PchA through PchE, respectively; these alternative designations are also shown in Table 3 and Fig. 4C. The genome sequence of EHEC strain EDL933 reveals only a single PerC1 protein, equivalent to PerC1-3 from Sakai and encoded by the CP-933R lambdoid prophage, as well as a PerC2 protein identical to that in Sakai and encoded by the CP-933C prophage (which is equivalent to Sp7 and is inserted at the same location) and two identical copies of the PerC3 protein due to the duplication of the SpLE1 phage-like element in this strain (24, 27) (Table 3). However, Iyoda and Watanabe (18) report that EDL933 also contains PerC1-1 (PchA) and PerC1-2 (PchB) proteins, although they are not present in the database sequence.
TABLE 3.
PerC-homologous proteins identified from completed bacterial and phage genomes
| Organism | Protein | Length (aa) | Database reference | Location (bp) | % PerC identity/ similarity (no. of residues) |
|---|---|---|---|---|---|
| EHEC O157:H7 Sakai (NC_002695.1) | EHEC PerC1-1 (PchA) | 104 | NP_309118.1 | 1183678-1183364 | 47/67 (87) |
| EHEC PerC1-2 (PchB) | 104 | NP_310209.1 | 2183078-2182764 | 47/67 (87) | |
| EHEC PerC1-3 (PchC) | 104 | NP_310764.1 | 2690650-2690336 | 47/67 (87) | |
| EHEC PerC2 (PchD) | 104a | NP_309615.1 | 1601592-1601906 | 39/65 (89) | |
| EHEC PerC3 (PchE) | 89 | NP_309415.1 | 1439736-1440005 | 25/54 (81) | |
| EHEC O157:H7 EDL933 (NC_002655.2) | EHEC PerC1 | 104 | NAc | 2139131-2138823 | 47/67 (87) |
| EHEC PerC2 | 104a | NA | 1685565-1685879 | 39/65 (89) | |
| EHEC PerC3-1 | 89 | NA | 1127902-1128171 | 25/54 (81) | |
| EHEC PerC3-2 | 89 | NA | 1523508-1523777 | 25/54 (81) | |
| UPEC CFT073 (NC_004431.1) | UPEC PerC2 | 96a | NA | 1384917-1385207 | 42/59 (76) |
| UPEC PerC3 | 102b | NP_754054.1 | 4962162-4961854 | 25/53 (86) | |
| UPEC YfdN | 164 | NP_755073.1 | 3057063-3056569 | 30/48 (56) | |
| E. coli K-12 MG1655 (NC_000913.1) | YfdN | 164 | NP_416858.1 | 2470901-2470407 | 36/50 (46) |
| S. flexneri SfV phage (NC_003444.1) | YfdN | 162 | NP_599072.1 | 29126-29614 | 32/46 (56) |
| Salmonella serovar Typhimurium ST64B phage (NC_004313.1) | YfdN | 174 | NP_700416.1 | 31803-32327 | 32/51 (49) |
EHEC and UPEC PerC2 proteins have a putative N-terminal extension of 14 amino acids compared to database entry NP_309615.1(shown boldfaced in Fig. 4) which overlaps with a putative upstream open reading frame.
UPEC PerC3 has a putative N-terminal extension of 13 amino acids compared to EHEC PerC3 proteins (shown boldfaced in Fig. 4) due to the presence of an alternative upstream start codon.
NA, not annotated.
FIG. 4.
(A) Schematic representation of PerC-like proteins identified in EHEC and UPEC strains, E. coli K-12, and bacteriophages SfV and ST64B by genome studies (see Table 3 for details). The extent of the protein sequence is indicated by the horizontal line, while the more-conserved core region corresponding to amino acids 5 to 50 of EPEC PerC is indicated by a solid box. (B) BLAST alignment of EPEC PerC with the EHEC PerC1, PerC2, and PerC3 proteins, PerC2 from UPEC strain CFT073, and YfdN from E. coli K-12 (shown from amino acid 100 only). The PerC1 sequence from strain EDL933 (corresponding to PerC1-3 from the Sakai strain [Table 3]) is shown. Putative N-terminal extensions of EHEC PerC2, UPEC PerC2, and UPEC PerC3 (see the text) are boldfaced. Residues identical to those in EPEC PerC are boxed in dark gray, and conservative substitutions are boxed in light gray. Predicted secondary structure motifs are boxed above the EPEC PerC sequence (H, α-helix; S, β-strand); L, predicted loop regions. Residues of EPEC PerC mutated by site-directed mutagenesis in this work are indicated by asterisks above the EPEC PerC sequence. (C) Summary of the various EHEC PerC-like proteins cloned in this study and their source strains. Identical proteins (with amino acid substitutions where appropriate) encoded by sequenced EHEC strains are listed.
PerC-like proteins are not restricted to EHEC strains, either. UPEC strain CFT073 encodes single copies of PerC2 and PerC3 within prophage 3 at phoQ and a CP4-like element at pheU, respectively (41); the UPEC PerC3 protein has a potential N-terminal extension of 13 amino acids relative to the EHEC PerC3s due to the presence of an alternative upstream start codon, while UPEC PerC2 is truncated at the C terminus with respect to all the other PerC-like proteins (Fig. 4; Table 3). Both CFT073 and E. coli K-12 (but not the EHEC strains) also encode YfdN, a protein with a heterologous N-terminal domain but with significant C-terminal homology to the first 70 amino acids of PerC (Fig. 4). CFT073 YfdN is encoded within a lambdoid prophage inserted at ssrA, while in MG1655 the yfdN gene is contained within the phage-like element KpLE1 inserted at argW (17). YfdN-like proteins are also encoded by the Shigella flexneri bacteriophage SfV (1) and the ST64B bacteriophage from Salmonella enterica serovar Typhimurium (Table 3). Alignments of the amino acid sequences of these PerC-like proteins suggest that they have a core homologous region extending from approximately amino acid 5 to amino acid 50 of PerC (Fig. 4A and B), although PerC1 and PerC2 in particular have more extensive homology over the full length of PerC.
Comparison of PerC homologues from EHEC O157:H7 strains ZAP198 and Sakai.
Given the existence of this variety of PerC homologues in bacterial pathogens other than EPEC, we wished to investigate the possibility that some or all of them might mediate activation of the LEE1 promoter as seen for EPEC PerC. Initial investigations with the PerC2 and PerC3 proteins encoded by UPEC strain CFT073 indicated that they could not activate the EPEC LEE1-lacZ chromosomal fusion in place of EPEC PerC (data not shown). However, considering that the UPEC PerC2 protein is truncated with respect to EHEC PerC2 and PerC itself, it was possible that the truncation, which removes a group of amino acids conserved between EHEC PerC2 and PerC, might have affected function. We therefore focused our attention on the EHEC PerC1 and (full-length) EHEC PerC2 proteins. The chromosomal DNAs of two different EHEC O157:H7 strains—ZAP198, a stx Nalr derivative of a strain isolated from an outbreak in Washington State (26, 32) (Table 1), and a stx derivative of the sequenced Sakai strain (30) (Table 1)—were used as templates for PCR amplification with primers binding to the flanking regions of PerC1 and PerC2 from Sakai (see Materials and Methods for details). PerC1-specific PCR products of the expected size (650 bp) were cloned into the multicopy vector pACYC184 and sequenced. The potential PerC1 clones revealed three different sequences: two independent clones from ZAP198 corresponding to the Sakai PerC1-3 (PchC) and Sakai PerC1-1 (PchA) proteins, which we termed 198PerC1-3 and 198PerC1-1, respectively, and a clone from the Sakai stx strain corresponding to Sakai PerC1-2 (PchB), which we termed SakPerC1-2 (Fig. 4C). The upstream flanking DNA of the 198PerC1-1 and SakPerC1-2 clones was identical to the Sakai database entries, except that SakPerC1-2 had a T-to-A transversion 205 bp upstream of the PerC1-2 start codon and a T-to-G change 105 bp downstream of the stop codon (data not shown). As well as these perC1 genes, we were also able to amplify a perC2 gene from both ZAP198 and the Sakai stx strain by using flanking primers. The gene encoding the ZAP198 protein (198PerC2) was identical to the Sakai PerC2 (PchD) genome sequence, while the gene encoding the Sakai stx strain protein (SakPerC2) had a T-to-C transition at the first base of the PerC2 stop codon, resulting in a protein with a 28-amino-acid extension at the C terminus (Fig. 4C). These studies suggest that ZAP198 encodes at least PerC1-1 and PerC1-3 proteins as in the Sakai strain, while strain-to-strain point mutations can also occur, as evidenced by the stop codon mutation in PerC2 and flanking mutations in PerC1-2 present in the Sakai stx strain. Following sequencing, the various perC1- and perC2-containing inserts were also subcloned into the low-copy-number vector pTH19kr.
EHEC PerC1 proteins, but not PerC2 proteins, can mediate activation of the EPEC and EHEC LEE1 promoters.
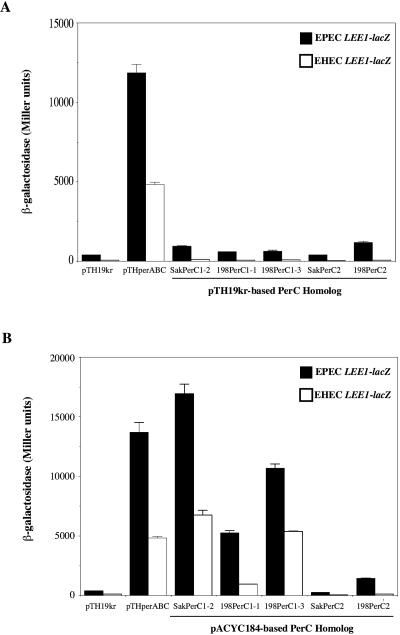
The effects of the cloned EHEC PerC-like proteins on the chromosomal LEE1EPEC-lacZ and LEE1EHEC-lacZ fusions in E. coli K-12 were then tested. Compared to a low-copy-number clone of the EPEC per locus (pTHperABC), low-copy-number pTH19kr-based clones of EDLPerC1-2, 198PerC1-1, 198PerC1-3, SakPerC2, and 198PerC2 mediated little or no activation of LEE1 (Fig. 5A). This may be because unlike EPEC perC, which is transcribed from the strong, autoactivated perA promoter (20, 29), the EHEC genes are individually poorly expressed. Provision of PerA (and PerB) in trans does not mediate any further activation of LEE1 by these low-copy-number perC1 and perC2 clones (our unpublished data). However, by increasing SakPerC1-2, 198PerC1-1, and 198PerC1-3 protein expression by using pACYC184-based multicopy clones, we did obtain 20 to 125% of the activation achieved by pTHperABC, with SakPerC1-2 having the greatest effect while 198PerC1-1 was the least active (Fig. 5B). In contrast, the pACYC184-based PerC2 clones gave no more activation than their low-copy-number counterparts. These data suggest that the expression levels of the PerC1 proteins are critical to their effects on LEE1 expression and may differ for the perC1-1, perC1-2, and perC1-3 genes due to their variable upstream flanking regions. To eliminate this variability, the SakPerC1-2, 198PerC1-1, and 198PerC1-3 proteins, as well as SakPerC2 and 198PerC2, were cloned downstream of the inducible araBAD promoter in the pBAD33 vector (Table 1). For the PerC2 constructs, both of the two potential initiator methionines (Fig. 4B) were included, as was the 28-amino-acid C-terminal extension of SakPerC2. The abilities of these constructs to activate LEE1-lacZ expression following induction with 1.0% arabinose in DMEM were then compared to that of the EPEC pBADPerC construct and the vector control pBAD33 (Fig. 6A). While 198PerC1-3 gave activation equivalent to that of PerC, SakPerC1-2 and 198PerC1-1 gave 50 to 60% activation. This finding contrasts with the situation observed with the multicopy clones, where SakPerC1-2 was most active, and presumably reflects differences in expression due to changes in the flanking DNA. Since 198PerC1-3 and SakPerC1-2 are identical apart from an M58V change, with an additional L6V change in 198PerC1-1, this line of reasoning suggests that the substitution of valine for methionine at position 58 adversely affects function, despite the fact that this residue is not conserved with respect to EPEC PerC (Fig. 4B), while the L6V change has little effect. In agreement with the results seen with the pTH19kr- and pACYC184-based clones, neither of the PerC2 proteins was able to activate LEE1-lacZ expression when expressed from the araBAD promoter (Fig. 6A).
FIG. 5.
Activation of EPEC (solid bars) and EHEC (open bars) LEE1-lacZ fusion constructs by low-copy-number (A) or medium-copy-number (B) clones of the SakPerC1-2, 198PerC1-1, 198PerC1-3, SakPerC2, and 198PerC2 proteins plus flanking DNA. The PerC2 clones contain a putative operon of four genes, of which perC2 is the third. A low-copy-number clone of the EPEC perABC locus was used as a control. Strains were grown to mid-logarithmic phase in DMEM and assayed.
FIG. 6.
(A) Activation of EPEC (solid bars) and EHEC (open bars) LEE1-lacZ fusion constructs by arabinose-inducible clones of the SakPerC1-2, 198PerC1-1, 198PerC1-3, SakPerC2, and 198PerC2 minimal open reading frames. The vector control pBAD33 and the arabinose-inducible EPEC PerC construct pBADPerC are shown as controls. Strains were grown to mid-logarithmic phase in DMEM plus 1.0% arabinose and assayed. (B) Northern blot analysis of perC transcription in EPEC strain E2348/69 (left panel) and perC1 transcription in EHEC strain ZAP198 (center panel) grown to mid-logarithmic phase in DMEM or LB medium, with transcription from E. coli MG1655 strains harboring the arabinose-inducible pBADPerC or pBADPerC1 plasmid grown in DMEM as a control. The amount of total RNA loaded per lane was 20 μg. The RNA transcripts are quantified in the bar graph on the right (solid bars, perC; open bars, perC1).
Total perC1 transcription in EHEC strain ZAP198 is only slightly lower than that of perC in EPEC strain E2348/69.
Based on the results described above, LEE1 activation by PerC-like proteins in EHEC appears to depend on the PerC1 proteins, but not on PerC2 (or PerC3), of the particular EHEC strain concerned; its extent is likely to vary depending on the precise variants of PerC1 present, the number of copies, and their expression dependent on changes in the upstream flanking sequence. To assess how the total level of PerC1 expression in an EHEC strain compares to that of PerC in EPEC, we compared total perC1 transcripts in EHEC strain ZAP198 grown to mid-log phase in DMEM or LB medium with those of perC in EPEC strain E2348/69 grown under identical conditions by using probes specific for perC1 and perC, respectively; to allow comparison of the levels of perC and perC1 expression, the respective gene transcribed from the araBAD promoter was used as a control and its expression level was set to 100% (Fig. 6B). As expected given previous knowledge of the effect of the growth medium on the expression of EPEC virulence genes, perC was expressed better in DMEM than in LB medium, although not to as high a level as expression from the araBAD promoter. In contrast, total perC1 expression in ZAP198 (which corresponds to at least the two genes that we have cloned) was no higher in DMEM than in LB medium. Significantly, though, it was only slightly lower relative to expression from the araBAD promoter than was that of perC. Therefore, although EHEC strains may lack an efficient transcriptional activation mechanism for their perC1-like genes equivalent to the PerA-dependent activation of perC in EPEC, the existence of multiple copies of perC1 in EHEC strains can partially overcome this deficiency. Thus, although activation of LEE1 by a PerC-like protein in EHEC may be less strong than that in EPEC even when multiple perC1 genes are present in the same strain, it could make a significant contribution toward total LEE1 expression.
Homology- and structure-based mutagenesis of EPEC PerC reveals five single-amino-acid changes which eliminate activity.
The discovery of a family of related PerC-like proteins with differential abilities to activate LEE1 expression enables the structure-function of EPEC PerC to be probed systematically for the first time. Previously, randomly occurring missense mutations of PerC (I27V, L30H, R44C, D63V, K70H, and N85S) have been observed either singly or in combination in different EPEC strains (25), but the effects of these mutations were not clear, since all of these strains also contain PerA mutations and PerC function was not tested in isolation. Secondary-structure predictions based on the PerC sequence suggest that the core conserved region (amino acids 5 to 50) of the PerC family is composed of three α-helices separated by short loops and is separated from a further helix-loop-helix at the C terminus by a long loop or linker region which is proline rich in many of the homologues but not in EPEC PerC (Fig. 4B). Based on this predicted structure and the homology within the family, we targeted five residues within the core conserved region by site-directed mutagenesis. A conserved leucine within the first predicted helix, L11, was mutated to serine, while the glycine in the loop between helices 1 and 2, G15, was mutated to tryptophan in an attempt to disrupt this putative loop. Two basic residues at the start of helix 2, R18 and R19, were mutated to the hydrophobic residue isoleucine or the helix-disrupting residue proline, respectively. Finally, the sulfhydryl group of the conserved cysteine 47 in helix 3 was removed by mutating it to serine (all the mutations are indicated in Fig. 4B). The effects of the mutations were tested in the context of a low-copy-number perABC clone activating the EPEC LEE1 promoter. All the mutated proteins were observed to have lost the ability to activate LEE1-lacZ expression, yielding 300 to 650 U of β-galactosidase activity (the vector control yielded 400 U) compared to 12,500 U for the wild-type clone. This finding suggests that these conserved residues are essential for the stability and/or function of PerC. Particularly interesting was the effect of the relatively conservative C47S mutation. It is not clear whether this residue would actually form disulfide bonds with another PerC monomer or a different protein in vivo, but it is apparent that replacing the sulfhydryl group in the side chain with a hydroxyl affects function significantly. Likewise, the importance of the putative helix 1-loop-helix 2 structure (the G15W and R19P mutations), a conserved hydrophobic residue in helix 1 (the L11S mutation), and a basic residue in helix 2 (the R18I mutation) is established.
LEE1 promoter DNA-PerC complexes cannot be detected in vitro but may depend on other cellular factors in vivo.
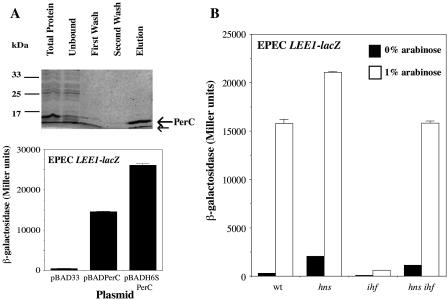
The discovery that members of a family of PerC-related proteins can activate LEE1 expression in both EPEC and EHEC raises the question by what mechanism this activation occurs. The simplest explanation is that these proteins constitute a novel family of DNA-binding proteins that bind the LEE1 promoter DNA and activate transcription directly. To test this possibility, we purified EPEC PerC as a His-S peptide-tagged fusion protein in E. coli (Fig. 7A). The tagged protein was pure but showed a tendency to break down despite the presence of protease inhibitors throughout the procedure; the observed smaller product was demonstrated by Western blotting with labeled S protein to be a breakdown product of the intact protein (data not shown). Based on the size of the breakdown product and the predicted structure of PerC, the most likely site of cleavage is the extended loop or linker region following helix 3 (Fig. 4B). The functionality of the N-terminal tagged protein was checked by expressing it from the araBAD promoter in the LEE1EPEC-lacZ fusion strain (Fig. 7A); in fact, tagged PerC mediated more activation of the fusion construct than the untagged control. This enhanced activity might reflect stabilization of the tagged protein compared to its untagged parent. Despite this activity in vivo, His-S-tagged PerC was unable to bind an EPEC LEE1 promoter fragment encompassing the mapped transcriptional initiation site and upstream regions in mobility shift assays in vitro, even at a PerC concentration as high as 1 μM (data not shown). It was also unable to bind either to the downstream DNA region between the promoter and the start of the ler gene or to a larger DNA probe comprising both the upstream and downstream fragments. This result implies that either PerC does not act as a DNA-binding protein or it binds DNA only in the presence of other cellular factors.
FIG. 7.
(A) Purification and activity of His6-S-tagged EPEC PerC. (Upper panel) A Coomassie-stained gel of samples of total soluble proteins, the unbound Ni2+-agarose column fraction in the presence of 5 mM imidazole, wash fractions in the presence of 35 mM imidazole, and the fraction eluted with 0.5 M imidazole are shown. Intact tagged PerC (15.2 kDa) and its breakdown product are indicated by arrows. Positions of molecular size markers are given on the left. (Lower panel) Activation of the EPEC LEE1-lacZ fusion construct by the His6-S-tagged PerC protein expressed from the pBADH6SPerC plasmid in the presence of 1.0% arabinose, compared to activation by untagged PerC expressed from pBADPerC and the vector control, is shown. (B) Activation of the EPEC LEE1-lacZ fusion construct by pBADPerC in the absence of induction (solid bars) or after induction with 1.0% arabinose (open bars) in E. coli K-12 wild-type (wt), hns-206::Apr, ihfA82::Tn10, and hns-206::Apr ihfA82::Tn10 strains. Strains were grown to mid-logarithmic phase in DMEM and assayed.
To investigate the latter possibility, we examined the role of known regulators of LEE1 expression in PerC-dependent activation. The nucleoid-associated proteins H-NS and IHF are known to repress and activate the EPEC LEE1 promoter, respectively (11, 40). An hns-null mutation was seen to lead to slight derepression of EPEC LEE1 in the absence of PerC (Fig. 7B), a result consistent with the weak repressive effect of H-NS at 37°C (40); this derepression was still detectable but weaker in the presence of PerC-dependent activation. In contrast, a mutation in the ihfA gene encoding the α-subunit of IHF weakened basal expression of LEE1-lacZ and abolished most of the PerC-dependent activation, showing not only that IHF acts as a positive regulator of the fusion construct, as expected (11), but also that it is required for correct activation of the promoter by PerC. When the hns and ihf mutations were combined, the double-mutant strain exhibited derepressed basal LEE1 expression similar to that of the hns single mutant but PerC-activated expression similar to that of the wild type (Fig. 7B). Therefore, H-NS and IHF act antagonistically at the EPEC LEE1 promoter, and PerC can activate the promoter normally when neither nucleoid-associated protein is present. However, activation by PerC in an hns wild-type strain exhibits a strong dependency on IHF, a finding that suggests that IHF may mediate a promoter conformation which either stimulates PerC binding or brings a PerC-DNA complex into contact with the transcriptional machinery (see Discussion). Despite this possibility, we were still unable to detect a PerC-dependent mobility shift of the LEE1 promoter in the presence of purified IHF (data not shown). It is possible that additional cellular proteins are also required for PerC to bind DNA efficiently.
DISCUSSION
The work presented here has demonstrated that, rather than being a specific regulatory mechanism confined to EPEC strains, PerC-dependent activation of LEE expression via the LEE1 promoter is functional in EHEC strains as well and may therefore have more general importance for control of the A/E phenotype than has previously been appreciated. This conclusion is based on two principal sets of observations: (i) that the upstream, PerC-activatable LEE1 promoter is the primary source of transcription in both EPEC and EHEC and (ii) that EHEC strains encode PerC-related proteins which are able to activate this promoter as PerC itself does in EPEC. It is clear from our 5′ RACE experiments that the majority of the LEE1-specific transcripts isolated from EPEC strain E2348/69 and EHEC strain ZAP198 are initiating in the region previously suggested to contain the LEE1 promoter in EPEC (22), irrespective of the growth medium. Interestingly, the actual initiating nucleotides in the two organisms seem to be slightly different, with those in EHEC being 6 or 7 bp, respectively, downstream of the two positions identified in EPEC here. In fact, the mapped initiating nucleotides in EPEC are unusually close to the proposed −10 region of this promoter (10) (Fig. 1), and it is possible that the downstream initiating nucleotide used in EHEC is a true alternative initiation site for RNA polymerase binding to this promoter sequence. Alternatively, the difference may be due to a processing event; either mechanism may be affected by the single base change located within this transcript initiation region in EHEC compared to EPEC. However, it is apparent that the same core promoter is being utilized in both organisms.
We also detected less-abundant transcripts, with 5′ ends in the downstream region previously proposed to be the LEE1 promoter in EHEC (10, 37), in RNA samples from both organisms. The heterogeneity of these transcript ends and the fact that none of them correlates with the initiating nucleotide suggested previously from primer extension studies leads us to conclude that they are more likely to represent downstream processing products of a primary transcript from the upstream promoter. It is also noteworthy that the core promoter sequences proposed for the putative downstream promoter (10) have a very long (19-bp) spacer between the −35 and −10 regions, and the ribosome binding site proposed to be associated with this transcript actually lies downstream of the ATG codon that is normally assumed to be the initiation codon for the Ler protein. Our observation that point mutations around the upstream promoter affect the activity of an LEE1-lacZ fusion more than those around the downstream promoter also suggests that it is the upstream promoter that is the primary determinant of LEE1 expression; the downstream mutations found in EHEC may affect RNA processing rather than transcriptional initiation and could account for the increased abundance of shorter transcripts with heterogeneous 5′ ends in EHEC.
Given these observations on the concurrency of LEE1 transcriptional initiation in EPEC and EHEC, it is not surprising that the EPEC PerC protein can mediate a similar fold activation of both promoters. Indeed, the only effect of the nucleotide differences between the two upstream regions seems to be to make the expression of EPEC LEE1 threefold higher than that of EHEC. The observation that both promoters can be activated by PerC is relevant given the additional finding that EHEC encodes PerC-like proteins which can also activate LEE1 expression. The EHEC PerC-like protein family is obviously heterogeneous, given both the information obtained from complete genome sequencing (17, 27) and our own findings here. The genome sequences reveal that the EHEC strain Sakai encodes five PerC-like proteins of three different families (PerC1, PerC2, and PerC3) while strain EDL933 has one extra copy of PerC3. Although only one copy of PerC1 is present in the EDL933 database entry (compared to three in Sakai), it is reported that the other two PerC1 proteins are encoded by EDL933 genomic DNA (18). Our cloning studies on the perC1 gene family show that at least two of the PerC1 variants are present in the independently isolated strain ZAP198, while a stx derivative of Sakai contains a PerC1-2 protein with flanking nucleotide substitutions relative to the sequenced parental isolate. All of these proteins are chromosomally encoded in prophage-like sequences, giving the potential for strain-to-strain variability in their sequence and copy number. There is also variability in the PerC2 and PerC3 proteins, as illustrated by our cloning of a PerC2 protein with a novel C-terminal extension from the Sakai stx strain and by the duplication of the prophage-like sequence encoding PerC3 in EDL933. These results emphasize the importance of prophages in strain-to-strain variation between bacterial pathogens (5, 24) and provide a link between this variation and the regulation of virulence phenotypes.
We demonstrate here that PerC1 proteins are able to mediate activation of the EPEC and EHEC LEE1 promoters in a PerC-like manner, while PerC2 (and PerC3) seems to be inactive. This finding correlates with the fact that PerC1 is more closely related to PerC than is PerC2, and much more so than PerC3 (Table 3; Fig. 4). The activating ability of an individual PerC1 protein is dependent on both its amino acid sequence (despite the fact that the variation in amino acid sequence among different PerC1s is restricted to positions not conserved in PerC) and its expression level, as shown by the different relative activation levels obtained when the PerC1 proteins were expressed from their native upstream regions or from the araBAD promoter. PerC1 exists in multiple copies in EHEC strains, and the level of total perC1 expression as assayed in strain ZAP198, which encodes at least two copies of the protein, is only slightly lower than that of perC in EPEC strain E2348/69. Therefore, PerC1-dependent activation of LEE1 in EHEC has the potential to be an important effect, and indeed Iyoda and Watanabe (18) have recently shown that individual deletions of the pchA (perC1-1) and pchB (perC1-2) genes, or double deletions of pchA and pchB or of pchA and pchC (perC1-3), in the Sakai strain reduce Esp protein secretion and adherence to HEp-2 cells via an effect on LEE1 transcription. Interestingly, PerC1-3, deletion of which alone has no effect on Esp secretion in the latter study, is shown here to be the most active PerC1 protein when expressed from the araBAD promoter. This finding confirms that the contribution of each PerC1 protein to LEE1 activation in EHEC is dependent both on the amino acid sequence of the protein and on the level of its expression from its native promoter.
The apparently weak expression of individual perC1 genes and the lower intrinsic activity of the EHEC LEE1 promoter that we have demonstrated here mean that, despite the presence of multiple copies of perC1, EHEC strains may still require additional transcriptional activation mechanisms to achieve the same degree of LEE expression as EPEC. A candidate for such an additional activating mechanism is the potentially greater quorum-sensing-dependent activation of LEE expression experienced by EHEC at its natural site of colonization (37). It is noteworthy that EPEC is apparently unique in having acquired a PerC protein that is encoded not in a chromosomal phage-related element but on a virulence-associated plasmid downstream of a strong, autoactivated promoter (the perABC promoter). This recruitment and upregulation of what was presumably originally a phage-encoded protein may have allowed EPEC to activate its LEE expression in host environments where quorum-sensing-dependent activation is weak. Interestingly, although PerC is an efficient activator of LEE1 fusions in E. coli K-12 (29; this work) and can complement a polar perA mutation in EPEC (4), loss of the pEAF plasmid does not reduce LEE2 and LEE3 expression as would be expected (4). One possible explanation might be the existence of additional perC homologues in EPEC as well as EHEC. The incomplete genome sequence of EPEC strain E2348/69 (http://www.sanger.ac.uk/Projects/Escherichia_Shigella/) reveals no PerC1 proteins but a much more distantly related PerC-like protein with a long N-terminal extension which is also found in certain bacteriophage; however, this protein does not activate LEE1 expression (our unpublished data). We have also been unable to amplify a perC1 gene from E2348/69 genomic DNA by using specific primers. It is to be hoped that completion of the EPEC genome sequence will reveal whether this organism also encodes other PerC-like proteins that can contribute to LEE1 activation.
The discovery that PerC-dependent activation is not confined to EPEC strains containing pEAF makes the requirement for some understanding of how PerC acts as an activator more pressing. Alignments of the known PerC-related proteins afford the opportunity to probe the structure-function characteristics of these proteins in a systematic way. Our secondary-structure predictions suggest that the well-conserved core region of the family (amino acids 5 to 50 of EPEC PerC) is likely to adopt a 3-helix structure with intervening loop regions, and the disruption of the first loop and second helix structure by G15W and R19P mutations, respectively, indicates that these structural elements are critical for functionality. Likewise, a conserved hydrophobic side chain within the (predominantly hydrophilic) first helix and a second conserved arginine in helix 2 (one of three in that helical element) are shown by our mutagenesis studies to be essential. The absolutely conserved cysteine within helix 3 is also important, as shown by the inactivity of the C47S mutant, and the rest of this helix may also be significant for LEE1 activation, given that this is the major region in which EPEC PerC resembles EHEC PerC1 (which can activate LEE1) more closely than PerC2 (which cannot). The C-terminal region of PerC is also predicted to be predominantly helical, although it is less well conserved across the family and, as suggested by our studies with purified PerC, may be subject to proteolytic cleavage. Although none of the helices composing PerC are predicted to form classical DNA-binding motifs such as the helix-turn-helix, the simplest explanation for the ability of PerC to activate LEE1 expression is that it acts directly as a DNA-binding protein which interacts with the LEE1 promoter. We know from our expression studies that no other pathogen-specific proteins are required to mediate activation by PerC, but we were unable to demonstrate any binding of purified PerC to a LEE1 promoter fragment in vitro. This could be because PerC requires additional cellular factors present in E. coli K-12 (and presumably also in EPEC and EHEC) to form complexes on the promoter and/or because its complexes are not stable under the conditions of the mobility shift assay. It is interesting in this context that efficient activation by PerC is shown here to require the accessory protein IHF. IHF binds upstream of the −35 region of the EPEC LEE1 promoter (11) and is known to be able to introduce a sharp bend of 160° or more into DNA (31). It could, therefore, either interact with PerC itself or mediate an interaction between an upstream-bound PerC or PerC-dependent complex and the core transcription machinery. We have also shown that when the H-NS protein is absent, IHF is no longer required for full activation by PerC; this could be because when it is no longer constrained by bound H-NS protein, the promoter DNA can adopt such a looped structure without the need for the DNA-bending activity of IHF. However, inclusion of purified IHF in a bandshift reaction with PerC on the LEE1 promoter does not permit formation of a PerC-dependent complex on this promoter (our unpublished data). It is apparent that further studies on the mechanism of PerC-dependent activation, particularly on whether it can interact with DNA either on its own or as part of a multiprotein complex, are urgently required.
Acknowledgments
We thank the staff of the ICMB DNA Sequencing Service for DNA sequencing; Mark Pallen for the Sakai stx strain; Padraig Deighan for the hns-206::Apr mutant; Seiichi Yasuda of the Cloning Vector Collection, National Institute of Genetics, Mishima, Japan, for providing the pBAD33 and pTH19kr vectors; and members of the ZAP Laboratory for useful discussions.
This work was supported by grants 066381/Z/01/Z and 065574/Z/01/Z from the Wellcome Trust.
REFERENCES
- 1.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 5.Canchaya, C., G. Fournous, and H. Brüssow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 6.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vázquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 8.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, S., L. A. Wainwright, T. McDaniel, B. MacNamara, L.-C. Lai, M. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. P. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 12.Gally, D. L., J. Leathart, and I. C. Blomfield. 1996. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 21:725-738. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, M. D., M. Johnson, J. C. D. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 14.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto-Gotoh, T., M. Yamaguchi, K. Yasojima, A. Tsujimura, Y. Wakabayashi, and Y. Watanabe. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185-191. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 18.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to Hep-2 cells. Microbiology 150:2357-2371. [DOI] [PubMed] [Google Scholar]
- 19.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 21.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 22.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Ohnishi, M., K. Kurokawa, and T. Kayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 25.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostroff, S. M., P. M. Griffin, R. V. Tauxe, L. D. Shipman, K. D. Greene, J. G. Wells, J. H. Lewis, P. A. Blake, and J. M. Kobayashi. 1990. A statewide outbreak of Escherichia coli O157:H7 infections in Washington State. Am. J. Epidemiol. 132:239-247. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Porter, M. E., and C. J. Dorman. 1997. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 256:93-103. [DOI] [PubMed] [Google Scholar]
- 29.Porter, M. E., P. Mitchell, A. J. Roe, A. Free, D. G. E. Smith, and D. L. Gally. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54:1117-1133. [DOI] [PubMed] [Google Scholar]
- 30.Ren, C.-P., R. R. Chaudhuri, A. Fivian, C. M. Bailey, M. Antonio, W. M. Barnes, and M. J. Pallen. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186:3547-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice, P. A., S. Yang, K. Mizuuchi, and H. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced U-turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 32.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. E. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71:5900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rost, B., and C. Sander. 1993. Prediction of protein structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 34.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez-SanMartín, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 39.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 40.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 41.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Science 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]