The epistatic interaction between two classically defined loci operates via a mutation in an Argonaute protein affecting the small RNA pathway.
Abstract
The soybean (Glycine max) seed coat has distinctive, genetically programmed patterns of pigmentation, and the recessive k1 mutation can epistatically overcome the dominant I and ii alleles, which inhibit seed color by producing small interfering RNAs (siRNAs) targeting chalcone synthase (CHS) mRNAs. Small RNA sequencing of dissected regions of immature seed coats demonstrated that CHS siRNA levels cause the patterns produced by the ii and ik alleles of the I locus, which restrict pigment to the hilum or saddle region of the seed coat, respectively. To identify the K1 locus, we compared RNA-seq data from dissected regions of two Clark isolines having similar saddle phenotypes mediated by CHS siRNAs but different genotypes (homozygous ik K1 versus homozygous ii k1). By examining differentially expressed genes, mapping information, and genome resequencing, we identified a 129-bp deletion in Glyma.11G190900 encoding Argonaute5 (AGO5), a member of the Argonaute family. Amplicon sequencing of several independent saddle pattern mutants from different genetic backgrounds revealed independent lesions affecting AGO5, thus establishing Glyma.11G190900 as the K1 locus. Nonfunctional AGO5 from k1 alleles leads to altered distributions of CHS siRNAs, thus explaining how the k1 mutation reverses the phenotype of the seed coat regions from yellow to pigmented, even in the presence of the normally dominant I or ii alleles.
INTRODUCTION
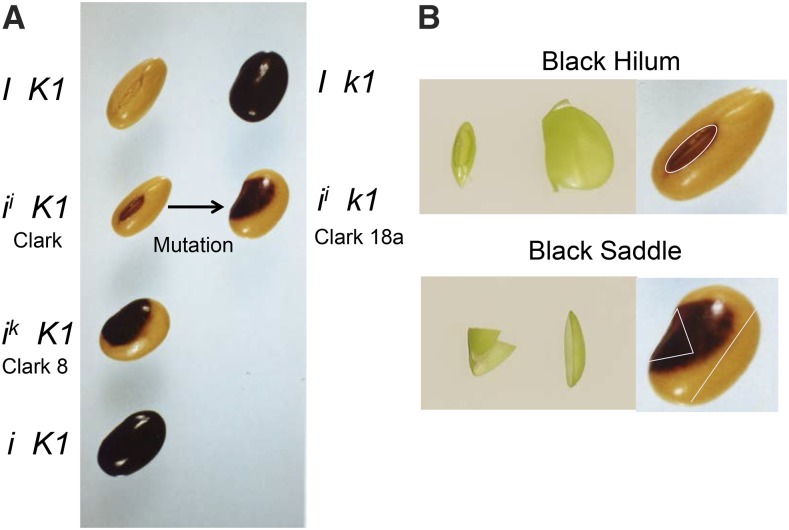
The interaction of the I and K1 loci in soybean (Glycine max) is an intriguing classical example of epistasis in which the phenotype of the dominant I and ii alleles (inhibition of seed coat pigmentation) is not manifested in seed homozygous for the recessive k1 allele. The dominant I and ii alleles are found in most commercial cultivars and in many of the standard varieties used in soybean breeding programs. For example, the Williams, Williams 82 (the cultivar used for the reference soybean genome), and Clark varieties are homozygous for the ii allele, which produces a pigmented hilum (where the seed coat attaches to the pod), but otherwise the majority of the seed coat proper is yellow (nonpigmented). Figure 1 shows the seed coat phenotypes of the four alleles of I in backcrossed or mutant isolines in the background of the Clark variety. Independent, naturally occurring mutations in a number of yellow-seeded cultivars result in completely pigmented (black) seed coats and are homozygous for the recessive i allele. Many of these isogenic recessive mutations result from naturally occurring deletions at the inverted repeat chalcone synthase (CHS) clusters CHS1-3-4 and CHS4-3-1 present in the ii allele (Todd and Vodkin, 1996; Tuteja et al., 2004). This unusual structure of the dominant ii allele spawns tissue-specific primary and secondary CHS short-interfering RNAs (siRNAs) that target at least nine CHS genes. The unlinked CHS7 and CHS8 are the main genes downregulated in immature seed coats (Tuteja et al., 2009; Cho et al., 2013) resulting in yellow rather than pigmented seed coats. The rare ik allele is not used commercially but was introgressed into various lines by backcrossing (see a pedigree summary in Supplemental Figure 1). The ik allele results in a two-colored seed coat, known as the saddle pattern, since the pigment extends from the hilum to occupy a saddle-shaped region on both sides of the seed coat proper.
Figure 1.
Pattern Alleles Exhibited by the Interaction of the I and K1 Loci in Soybean.
Cultivars or isolines are homozygous for the indicated alleles, and, for brevity, only one of the alleles is indicated in the genotypes.
(A) Seeds on the left have the commonly found dominant K1 allele and seeds in the right column are homozygous for the rare, recessive k1 mutation. Dominance relations of the I alleles are in the order I >ii>ik>i, and the dominant I and ii alleles are found in most commercially used cultivars. The Williams and Clark cultivars have the ii allele that limits pigment to the hilum region near the embryonic axis. The saddle pattern isolines used in this study are in the genetic background of the cultivar Clark and are referred to as Clark 8 (PI 547450, ik K1) and Clark 18a (PI 547439, ii k1). The arrow denotes that Clark 18a, which was found in 1956 in Ames, Iowa, was a spontaneous mutation from a Clark parent line. The pigment can be either black or brown of varying intensities depending upon other genes present in the lines (Williams, 1952; Bernard and Weiss, 1973).
(B) An illustration of the dissection of immature green seed coats to obtain sections from seed with the pigmented hilum or saddle pattern phenotypes. Since the immature green seed do not show the pigmented phenotype, the dissections were conducted in a conservative manner to take sections that would avoid tissues with mixed phenotypes at maturity. Left: Actual dissected regions from the immature green seed coats of the 100 to 200 mg seed weight range. Right: An illustration of the regions dissected as outlined on the phenotype of the mature seed.
Most soybean varieties also contain the dominant K1 locus. Interestingly, a recessive k1 spontaneous mutation interacts epistatically to partially overcome the effect of the dominant I and ii alleles and to extend the pigmented region over a larger surface of the seed coat, as shown in Figure 1. Thus, seed with the ii k1 genotype have a saddle pattern that mimics the ik K1 phenotype. The effect of the k1 mutation on the dominant I allele is even more pronounced, and the seed with the I k1 genotype are either completely black or “near black” with a narrow strip of nonpigmented region at the outer edge of the seed coat, which is visible in fully expanded seed before desiccation but often is not apparent on the mature seed. The k1 allele is found in some unadapted germplasm, and new mutations from K1 to k1 have occurred in modern varieties. Except for a brief description of the intriguing phenotypes of this genetic interaction described in a 1958 abstract (Williams, 1958) and in review chapters by Bernard and Weiss (1973), Palmer and Kilen (1987), and Palmer et al. (2004), nothing is known about the nature of the K1 locus other than it segregates independently from the I locus and has a putative map position in the soybean composite reference map at Soybase (Grant et al., 2010).
In this work, our objectives are (1) to determine whether the pattern phenotypes specified by ii K1 (pigmented hilum genotype) or the ik K1 and ii k1 saddle genotypes are conditioned by different levels of CHS siRNAs in the sectors of seed coats with different pigment phenotypes and (2) to use global expression analysis combined with mapping data and structural analysis of whole-genome resequencing data to identify the K1 locus and how it interacts epistatically with alleles of the I locus to reverse the phenotype normally specified by the dominant I and ii alleles. Using small RNA analyses and RNA-seq, we demonstrate that CHS siRNAs are the cause of saddle seed coat patterns produced by the ii and ik alleles of the I locus, which restrict pigment to certain regions of the seed coats. By combining RNA-seq and whole-genome resequencing, we also show that the epistatic K1 locus, which modifies the spatial regulation of CHS siRNA production within the seed coats, encodes Argonaute5 (AGO5), a member of the Argonaute family of proteins. Independently occurring spontaneous mutations having the saddle pattern phenotype also showed different lesions in the AGO5 gene with dramatic effects on its predicted protein structure. The function of AGO5 appears to be integral to the spatial distribution of the CHS siRNAs, thus explaining how the k1 allele reverses the phenotype of the seed coat regions from yellow to pigmented, even in the presence of the normally dominant ii or I alleles, and explains the classical genetic interactions of these two loci.
RESULTS
The Recessive k1 Mutation Modifies the Pigmentation Patterns Specified by Alleles of the I Locus
As shown in Figure 1A, the ii allele produces a pigmented hilum and the ik allele produces a saddle pattern in the presence of the dominant K1 allele. However, in the presence of a homozygous recessive k1 allele, the pigment occupies a larger area, resulting in a saddle pattern on seed with the ii k1 genotype that mimics the ik K1 phenotype. To examine the differential abundance of CHS siRNAs in the two regions, we dissected seed coats and conducted high-throughput sequencing using small RNA libraries, as described in the next section. Since the pigment is not yet visible in the immature green seed stages during which CHS siRNAs are most abundant, we used position to determine how to dissect the two regions. Figure 1B shows some of the actual tissue as well as a schematic of the dissections. While the morphology of the hilum region can be determined visibly, there is no way to differentiate the saddle prior to pigment formation, except by positioning. We dissected the saddle region in a conservative manner including only the central region to ensure that no yellow tissue was likely to be included. Likewise, the yellow tissue was taken far distal from the saddle area. In total, we analyzed data from 16 different small RNA libraries yielding from 22 to 88 million reads per library (Supplemental Data Set 1).
Distribution of CHS siRNAs Results in the Pigmented Hilum Pattern of the Dominant ii Allele
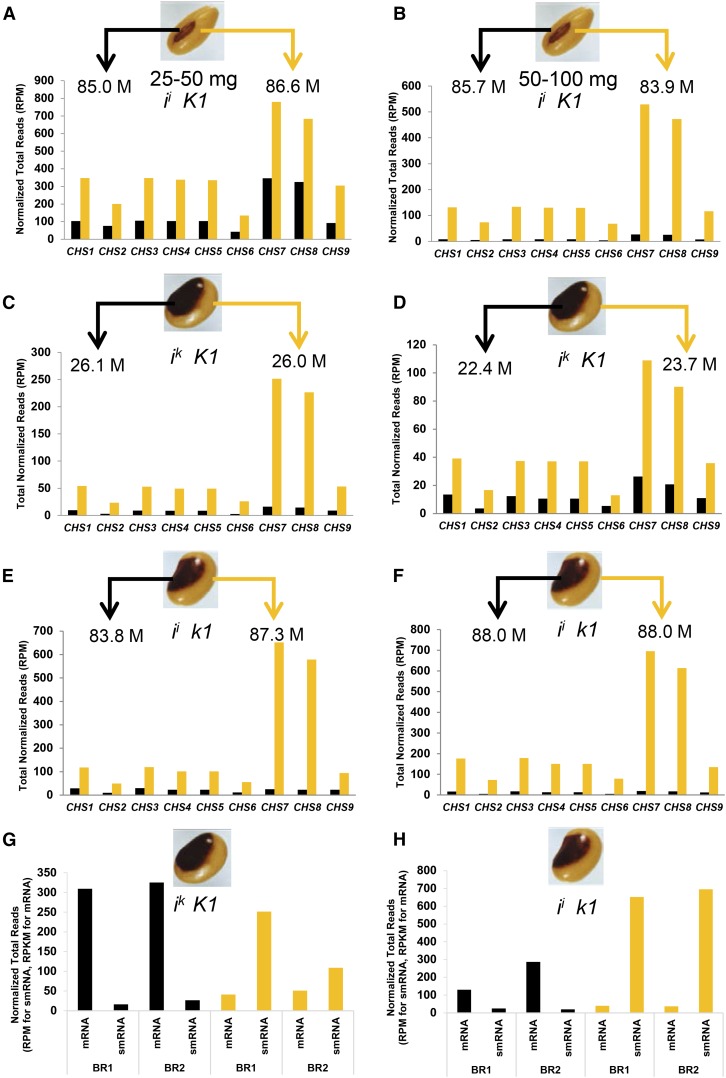
The Williams cultivar is homozygous for the dominant ii allele, which produces a pattern with pigment restricted to the hilum region of the seed coat. To investigate whether the presence of CHS siRNAs is limited to only the nonpigmented region, we dissected the hilum from the seed coat proper from two stages of the immature seed, 25 to 50 mg and 50 to 100 mg (stage demarcations based on the weight of individual seeds). We had previously determined that these two stages contained high levels of small RNAs in the total seed coats of Williams (Cho et al., 2013). We used at least 10 seeds in RNA extractions to even out the biological variation for the dissected hilum and seed coat proper. High-throughput small RNA sequencing was conducted for two biological repeats of each seed coat region and the data are shown in Figures 2A and 2B and in Supplemental Data Set 2. The normalized CHS siRNAs that align to the CHS coding regions were highest in yellow regions of the seed coat and were significantly reduced in the pigmented hilum region in both repeats of both seed weight ranges. The siRNAs representing target genes CHS7 and CHS8 had the highest levels, as expected, each exhibiting an ∼20-fold difference between the pigmented and the nonpigmented regions. The fold increase is not as pronounced in the 25 to 50 mg stage, likely due to the fact that in the earlier stages it is more difficult to obtain tissue dissections of the hilum regions that are free of cells from the seed coat proper. At the immature green stage, the pigment has not formed (Figure 1B); thus, one cannot discern the precise boundaries of the pigmented and nonpigmented tissues. In summary, these data clearly show that the ii allele specifies the high level accumulation of the CHS siRNAs only in the regions of the seed coat that are not pigmented. Thus, not only does the ii allele control highly tissue-specific production of CHS siRNAs (Tuteja et al., 2009; Cho et al., 2013), but it also directs the pattern-specific production of CHS small RNAs within seed coats of the same genotype.
Figure 2.
Distribution of CHS siRNAs or mRNAs within Sectored Regions of Seed Coats of Different Genotypes.
Black bars represent pigmented regions of the seed coats, and yellow bars represent the nonpigmented regions dissected from immature seed coats of the indicated genotypes and seed phenotypes. The number of total small RNA reads in millions (M) derived from each of the tissue sections is indicated below the arrow. RPM, normalized total number of reads per million; BR, biological repeat. The full data for siRNA or mRNA levels for all CHS genes are shown in Supplemental Data Sets 2 and 3.
(A) and (B) CHS siRNAs from two stages of Williams (black hilum; ii K1) immature seed at weight ranges 25 to 50 or 50 to 100 mg. A second biological repeat of each weight range shows the same trend (Supplemental Data Set 1).
(C) and (D) CHS siRNAs from two biological repeats of Clark 8 (black saddle; ik K1) at the 100 to 200 mg seed weight range.
(E) and (F) CHS siRNAs from two biological repeats of Clark 18a (black saddle; ii k1) at the 100 to 200 mg seed weight range.
(G) Inverse relationship of CHS7 mRNA and siRNA (smRNA) levels from the two biological repeats of Clark 8 (black saddle; ik K1).
(H) Inverse relationship of CHS7 mRNA and siRNA levels from the two biological repeats of Clark 18a (black saddle; ii k1).
Quantitative Variation in CHS siRNAs Results in the Saddle Pigment Patterns on Seed Coats
Four small RNA libraries from two biological repeats were constructed from the pigmented saddle region and the nonpigmented seed coat of the Clark 8 isoline, which is homozygous for the ik saddle pattern allele in the presence of the dominant K1 allele (Supplemental Data Set 1). For the saddle pattern genotypes, we used a slightly larger seed weight range to prevent mixing of phenotypes in dissected tissue. Both CHS7 and CHS8 siRNAs were ∼15-fold higher in the nonpigmented region than the pigmented regions dissected from 100 to 200 mg seed (Figure 2C). The biological repeat data also show that expression levels of all CHS siRNAs, especially CHS7 and CHS8 siRNAs, are much higher in the nonpigmented seed coat than in the pigmented saddle (Figure 2D).
We next examined the presence of CHS siRNAs in the pigmented saddle region versus the nonpigmented seed coat region in four small RNA libraries from two biological repeats of the Clark 18a isoline (ii k1). In this isoline, the recessive k1 mutation extends the pigmented region from the hilum to form a saddle pattern that is similar in phenotype to the Clark 8 isoline having ik K1 genotype. Figures 2E and 2F show that the presence of CHS siRNAs was predominantly limited to the nonpigmented yellow regions as in the case of the Clark 8 isoline. CHS7 and CHS8 siRNAs were ∼26-fold higher in the nonpigmented regions in both biological repeats. Supplemental Data Set 2 shows the levels of small RNAs matching each of the annotated CHS Glyma models in the soybean genome within the different regions of the saddle pattern seed coats having the same genotype.
Transcriptome Data Show CHS siRNA and mRNA Levels Have Inverse Patterns of Abundance in the Different Regions of the Seed Coat Saddle Patterns
Figures 2G and 2H clearly show that the expression levels of CHS7 siRNAs and their target CHS7 mRNAs demonstrated inverse patterns of abundance in two biological repeats of small RNA in which the same biological samples were also subjected to RNA-seq. CHS7 siRNAs were more highly expressed in the nonpigmented region where the expression level of CHS7 mRNA was low. However, in the pigmented regions, CHS7 mRNAs were highly expressed and siRNAs were not. The biological repeat shows a similar expression pattern for both CHS7 siRNAs and mRNAs. The results are similar for CHS8 (Supplemental Data Set 3). Taken together, the small RNA and RNA-seq data demonstrate that CHS siRNAs have a critical role in the formation of the pigment pattern on soybean seed coats through the downregulation of their target CHS mRNAs.
Differential Expression of RNA-Seq from ik K1 versus ii k1 Genotypes Revealed a Small Number of Candidate Genes for the K1 Locus
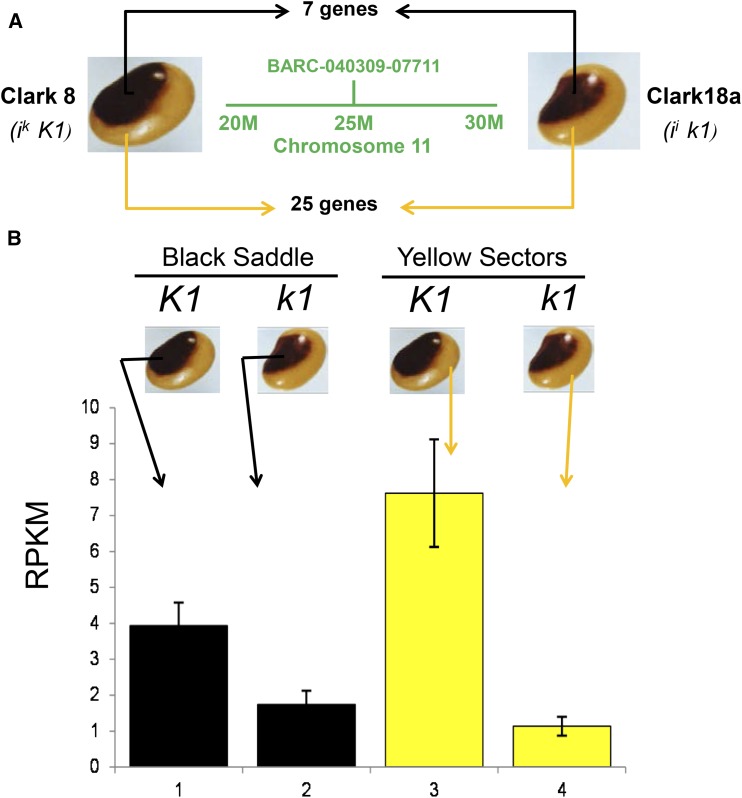
To identify the K1 locus, we compared the transcriptome data from the two saddle pattern Clark isolines, Clark 8 (ik K1) versus Clark 18a (ii k1). In total, we analyzed 12 mRNA libraries containing three biological repeats from the two dissected regions of each genotype, as indicated in Supplemental Data Set 1. The RNA-seq reads (43 to 97 million reads total from each library) were aligned to all 88,647 soybean gene models and splice variants and normalized as reads per kilobase of gene model size per million mapped reads (RPKM) (Mortazavi et al., 2008). We compared the transcriptome data of the pigmented region of Clark 8 (K1 allele) to the pigmented region of Clark 18a (k1 allele) in order to minimize variation due to position on the seed coat while searching for genetic differences specific to the K1 locus. Likewise, the nonpigmented regions were compared between the two varieties. We used both the Cufflinks package (Trapnell et al., 2012) and the DESeq software (Anders and Huber, 2010) for analyses of transcriptome data. Supplemental Figure 2 shows Cufflinks scatterplots and density plots comparing the RNA-seq data. According to soybean public map resources shown in Supplemental Figure 3, the K1 locus has been mapped to Chromosome 11; thus, we concentrated our RNA-seq analyses on that chromosome region near a mapped marker and purposefully took a wide swath of 10 million bases representing ∼30% of the chromosome. Figure 3A illustrates that there were relatively few significantly differentially expressed genes in the DESeq analysis with a Benjamini-Hochberg adjusted P value of <0.05 that were found within a 5-Mb range on either side of the marker used. There were seven differentially expressed genes in the pigmented saddle tissue, and they were also found in the 25 differentially expressed genes in the yellow distal tissue, thus yielding only a small number of potential candidate genes in this region of 10 Mb on chromosome 11, as shown in Supplemental Data Set 4.
Figure 3.
An Argonaute Gene, Glyma.11G190900 (AGO5), Is the Most Likely Candidate for the k1 Mutation Based on Analysis of RNA-Seq Data in the Region of a Mapped Marker.
(A) An illustration of the small number of significantly differentially expressed genes found in a large region of 10 Mb surrounding a previously mapped marker (BARC 0400309-07711) in the vicinity of the K1 locus on chromosome 11 at ∼25 Mb. Black arrows indicate that only seven genes were significantly differentially expressed in this region when RNA-seq was compared from the black saddle tissue, and yellow arrows indicate only 25 genes were found to be significantly differentially expressed in the yellow seed coat sectors between the two genotypes.
(B) Statistically significant variation in RPKM levels for Glyma.11G190900 (AGO5) between the black saddle comparisons and yellow region comparisons of K1 to k1 genotypes. Data represent the average of three biological repeats of RNA-seq with se bars.
We further determined that Glyma.11G190900 (Chr11:26,361,722..26,367,787), annotated as an Argonaute protein (AGO5), is the closest of these candidate genes to the marker BARC-040309-07711 at position Gm11:25,085,355, which is a closely linked marker to the K1 locus in the current Wm82.a2 reference genome (shown in Supplemental Figure 3D). We also searched for the AGO5 Glyma model in the older version of the soybean reference (Wm82.a1), where it corresponds to Glyma11g19650 (Gm11:16394576-16400897) in the Wm82.a1.v1.1 assembly and is 800 kb from the position of BARC-040309-07711 (Gm11:15,576,898) in Wm82.a1. Thus, Glyma.11G190900 was the strongest candidate gene since others were located much further from the marker position on the genetic map.
Figure 3B shows that the yellow sectors of the K1 genotype had the highest AGO5 transcripts at ∼7.5 RPKM. It also depicts that AGO5 was expressed at significantly higher levels in both the black saddle and yellow sectors of the K1 genotypes compared with seed coats of the k1 genotypes. In summary, Figure 3B shows that AGO5 transcripts were higher by 2.3-fold and 6.2-fold in the black saddle and the yellow regions, respectively, of the K1 compared with k1 seed coats.
Resequencing Reveals a Small Deletion in AGO5 (Glyma.11G190900) in the Saddle Mutant Having the Recessive k1 Allele
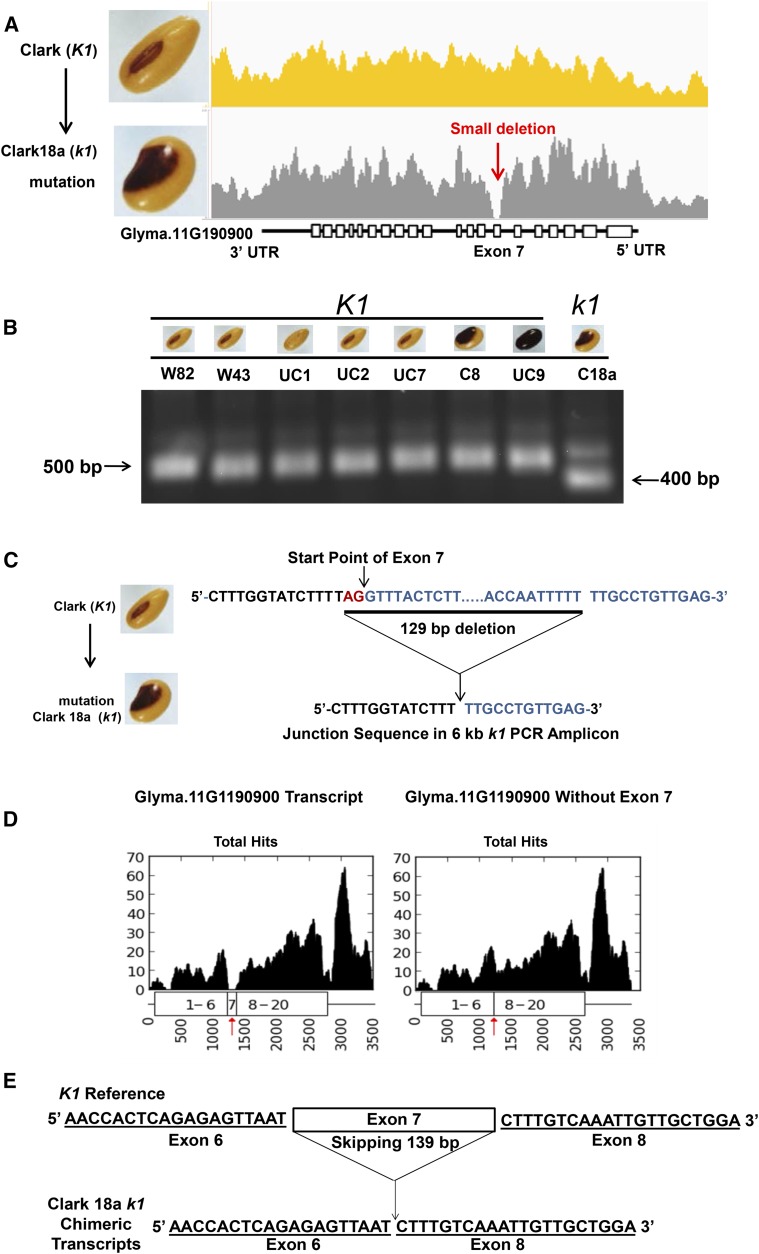
Since the location of genetic map markers is only an approximation, we investigated all candidate genes and searched for structural variants including insertions, deletions, and single-nucleotide polymorphisms by comparing whole-genome resequencing data (Supplemental Data Set 1) between the nonpigmented progenitor Clark (ii K1) and its isogenic saddle mutant Clark 18a (ii k1). Figure 4A illustrates that a small deletion (absence of alignments) was found in the exon 7 region of Glyma.11G190900 in the saddle mutant (k1), while its progenitor (K1) does not have this structural variant. The depth of alignments from the mutant k1 line dropped in the vicinity of the exon 7 region. Bowtie2 allows gaps and partial read alignments at the insertion/deletion breakpoint. Since only partial reads aligned to the K1 reference allele, these delineate the breakpoint. Inspection of the alignments showed there are two extra sequences, 5′-CTTTGGTATCT-3′ and 5′-TTGCCTGTT-3′, flanking the alignments at Gm11:26,365,578 around the 3′ end of intron 6 of Glyma.11G190900 and at Gm11:26,365,450 in exon 7 of the gene. Thus, the chimeric sequence is predicted to be 5′-CTTTGGTATCTTTTTTGCCTGTT-3′. Based on the whole-genome resequencing data showing this chimeric sequence, we project the deletion mutation omits the 129-bp region between Gm11:26,365,450 and Gm11:26,365,578.
Figure 4.
Structural Variation in Glyma.11G190900 and Its Transcripts Encoding AGO5 Found in the Clark 18a k1 Black Saddle Mutation.
(A) The absence of Bowtie2 alignments in genome resequencing reveals a small deletion within exon 7 in the recessive k1 black saddle mutant compared with the dominant K1 allele of yellow seed. The graph represents normalized counts of alignments in 0.25 scale (count at base × 1,000,000/total number of reads as displayed by IGV).
(B) PCR validates that the recessive k1 allele in Clark 18a has a small deletion. All varieties carrying the dominant K1 allele show a band of ∼500 bp in length, while the recessive k1 allele shows 400 bp. Abbreviations for different cultivars are as follows: W82, Williams 82; W, Williams; UC2, Clark; C8, Clark 8; C18a, Clark 18a; UC1, UC7, and UC9 are other Clark isolines with other alleles of the I locus but all are homozygous for K1.
(C) Sequencing the entire 6-kb AGO5 amplicon from the Clark 18a line containing the k1 mutation reveals a 129-bp deletion. The top line is the sequence of Glyma.11G190900 exon 7 and junction regions in Clark K1. The bottom line shows the chimeric sequence covering the deletion point that is found in the Clark 18a line containing the k1 mutation. The 129-bp deletion removes the AG (red) of the 3′ splice site preceding exon 7 and all except 12 bases of exon 7. Exon 7 sequences are shown in blue. See Supplemental Data Set 5 for the complete amplicon sequence of Clark 18a.
(D) Position graphs of RNA-seq alignments of Clark 18a (k1) against the full 3514-bp transcript of Glyma.11G190900 (AGO5) from the reference genome (Williams 82, K1 genotype) or to a modified 3375-bp transcript that lacks the entire 139 bp of exon 7 (right panel). Bowtie1, which does not allow any insertions or deletions, was used with no mismatches allowed. The red arrow marks the position of exon 7 or the fusion position of exon 6 to exon 8 (x axis, position on the transcript; y axis, non-normalized count of the number of times each base occurs within the aligned reads).
(E) Chimeric transcripts validate that the entire 139 bp of exon 7 is spliced out in the recessive k1 saddle mutant. Top: The sequence of the flanking region of exon 7 in the transcript of Glyma.11G190900. Bottom: Chimeric transcripts that were found in the RNA-seq as shown in the alignments in (D) (right) to the modified transcript missing exon 7.
To confirm the deletion detected by genomic resequencing data, we performed PCR amplifying the fragment across the deleted region that spans exon 7. Figure 4B shows that each of the seven soybean lines carrying the dominant K1 allele displayed bands of ∼500 bp in length, while the Clark 18a (ii k1) saddle mutant had a 400-bp fragment. Finally, we subjected the entire 6-kb AGO5 gene amplified from the k1 allele of Clark 18a to next-generation sequencing and found the same chimeric sequence (Figure 4C) that was observed in the whole genomic resequencing data. This chimeric sequence reveals that the 129-bp deletion occurs in a T-rich region and removes the AG at the end of intron 6 and all but 12 bases of the 139 bases of exon 7 of Glyma.11G190900.
The 129-bp Genomic Deletion Results in Loss of the Entire Exon 7 in the k1 Transcript by Exon-Skipping Alternative Splicing
As shown in Figure 3B, the levels of the k1 transcript are apparently affected by the 129-bp genomic deletion since the RPKMs for Glyma.11G190900 were reduced in k1 compared with K1 by 2.3-fold and 6.2-fold in the black saddle and the yellow seed coat regions, respectively. Thus, the deletion in the recessive k1 allele reduces the expression of Glyma.11G190900 rather than abolishing it entirely. We analyzed position graphs of the RNA-seq data alignments in detail to understand which part of the gene is transcribed in the presence of the deletion. Figure 4D shows that the 129-bp genomic deletion appeared to abolish the entire 139 bases of exon 7 from the mRNA transcript even though a partial region of the 3′ end of exon 7 (12 bp) remains in the genome. This was demonstrated by the absence of alignments to the entire exon 7 of Glyma.11G190900 in the recessive k1 saddle mutant (Clark 18a). Since the genomic deletion removes the AG of the 3′ splicing signal immediately prior to exon 7 (Figure 4C), we propose that the loss of the 3′ splice site causes the intron splicing machinery to skip to the next 3′ splicing signal immediately prior to exon 8, thus eliminating the entire exon 7 from the final cytoplasmic transcript.
To test that all of exon 7 is removed by splicing, we modified the transcript sequence of Glyma.11G190900 by omitting the full 139 bp of exon 7 to use as a reference for the RNA-seq alignments. We used very stringent conditions not allowing any mismatch with the Bowtie1 program, which does not allow any gaps, even a single base deletion or insertion in the alignments. As shown Figure 4D, the alignment gap was found against the original transcript but not the modified transcript (right panel). Figure 4E shows that the chimeric transcripts found in RNA-seq data of the k1 saddle mutant (Clark 18a) cleanly fused exon 6 to exon 8, thus confirming that the 129-bp genomic deletion leads to skipping of the entire 139 bp of exon 7.
The Clark 18a k1 Deletion Results in Early Termination of the AGO5 Protein
Glyma.11G190900 encodes an AGO5 protein, one of the AGO family members. AGO proteins have critical roles in the function of small RNAs in posttranscriptional regulation of target genes due to their ability to bind to nucleic acids (Chapman and Carrington, 2007; Fang and Qi, 2016). The AGO complex recruits single-stranded RNA after Dicer cleaves double-stranded RNA and guides it to the target transcript, which is cleaved. AGO5 (Glyma.11G190900) likely plays a crucial role, either directly or indirectly, in regulating the level of CHS siRNAs in a spatial manner causing the saddle pattern phenotype in the recessive k1 allele saddle mutant (Clark 18a).
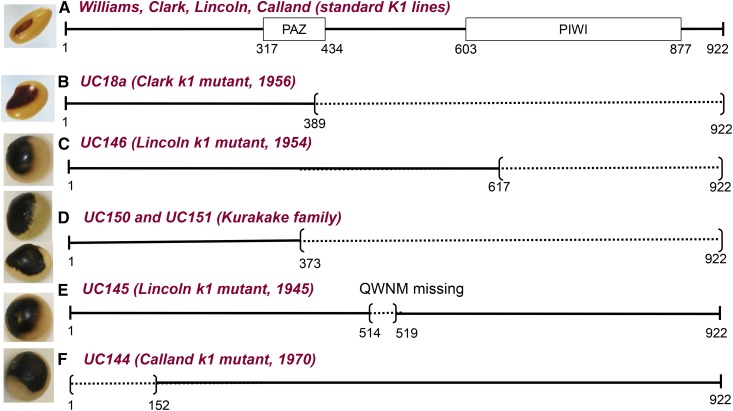
Figure 5A illustrates the amino acid sequence of AGO5 (Glyma.11G190900) in K1 varieties. AGO5 has 922 amino acids and includes the PAZ and PIWI domains, which are reported to be functional domains for interaction with small RNAs. The PAZ domain (amino acids 317–434) is an interface for binding to nucleic acids and the PIWI domain (positions 603–877) is an anchoring site of the 5′ RNA guide strand. Next, we examined the coding potential of AGO5 without exon 7, as found in Clark 18a transcripts. The elimination of exon 7 results in a frame-shifted protein at amino acid 376 and premature termination after 389 amino acids omitting the essential PIWI domain and part of the PAZ domain (Figure 5B). The prematurely terminated k1 protein from the Clark 18a mutation is clearly nonfunctional.
Figure 5.
Schematic of the Predicted Lesions in the AGO5 Protein Found in Independent Saddle Pattern Mutants.
(A) The AGO5 structure found in standard K1 lines and parents including Williams, Clark, Lincoln, and Calland showing the relative positions of PAZ and PIWI domains.
(B) Protein prematurely terminated in the k1 allele of the Clark 18a mutation.
(C) Protein prematurely terminated in the k1 allele of a 1954 saddle mutation in Lincoln.
(D) Proteins prematurely terminated in two different Kurakake lines, large seeded Plant Introduction lines collected in Japan.
(E) Protein missing four amino acids in a 1945 saddle pattern mutant of Lincoln.
(F) Protein of the saddle mutant found in Calland in 1970 and putatively missing a large part of its N terminus due to absence of the initiation codon.
Example seed phenotypes are illustrated. Dark lines represent amino acids of the AGO5 sequence, a tick mark indicates the relative position of altered amino acids introduced after a frameshift mutation, and dotted lines represent missing portions of the protein sequence. The Clark parent line is based on whole-genome resequencing data but the other nine are based on full length sequencing of the AGO5 amplicon. See Supplemental Data Set 5 for a more detailed characterization of the affected positions in the genes and proteins. Amino acid numbers shown underneath are relative to the initiation codon of Glyma.11G190900 in the standard K1 allele. All 10 lines sequenced showed an extra A in exon 20, which would increase the reading frame of the AGO5 protein to 922 amino acids rather than 890 as called by Phytozome for the Glyma.11G190900 predicted protein in the reference genome, representing a correction to the reference.
Multiple Independent Saddle Pattern Mutations Have Lesions in the AGO5 Protein
The recessive k1 allele with the defective AGO5 on chromosome 11 (Clark 18a) arose as a de novo mutation in the variety Clark. Records indicate it was found in Ames, Iowa in 1956. We extended our examination to other black saddle mutations that arose in independent soybean varieties and which have been preserved in the germplasm collection maintained by the USDA (Supplemental Data Set 5). Initially, we surveyed regions of the entire AGO5 gene split into overlapping sections to determine whether any structural changes were visible by gel electrophoresis as was found to be the case for the 129-bp deletion within Clark 18a. However, none were observable. We then designed primers to amplify the AGO5 gene (6067 bases of Glyma.11G190900 + 196 bp upstream of the 5′UTR [untranslated region]); the PCR products were barcoded and subjected to next-generation sequencing and assembly as described in Methods. Supplemental Data Set 5 shows the changes found in the DNA sequence of each amplicon structure. Figures 5B to 5F summarize the predicted effects on the AGO5 protein, and Supplemental Figure 4 shows a Multalin alignment of the amino acid sequences of the nine AGO5 amplicons. Compared with the Lincoln parent line, which produces the full-length protein, a mutant in the variety Lincoln found in 1954 had a single base pair deletion in exon 13, which immediately introduces a stop codon that prematurely terminates the protein at amino acid 617. An independent saddle pattern mutation in Lincoln in 1945 was missing four amino acids due to 12 nucleotides missing within exon 10. Compared with the Calland parent, a saddle mutant found in Calland showed a 15-bp deletion at the junction of the 5′UTR with exon 1, including the initiation methionine codon. The next in-frame methionine is not until position 152 within the protein. Finally, two large-seeded and saddle-patterned varieties, known as Kurakake and Kurakake Daizu, which were collected in Japan, both had the same deletion of 4 bp in exon 6 that results in a frameshift at position 351 and a premature stop codon at position 373 in the protein. These data showing multiple, independent, and large lesions that would inactivate the AGO5 protein confirm Glyma.11G190900 to be the K1 locus and the origin of different k1 alleles that specify a saddle pattern phenotype. The fact that the 1945 Lincoln saddle mutant is missing only four amino acids suggests that these amino acids may be critical to the function of the AGO5 protein.
As shown in the amplicon sequencing results of Supplemental Data Set 5, the number of reads assembled is from 69,609 to 191,356, and the representation of the perfect matches of each base in the sequencing is generally much greater than 1000, allowing very accurate calls of the consensus contig. We found one difference between the Phytozome reference genome Glyma.11G190900 (AGO5 in the Williams 82 cultivar) and all of the other AGO5 genes in the nine soybean varieties and mutants that we subjected to amplicon sequencing, including Williams, the progenitor recurrent parent of Williams 82. There was an extra A in exon 20, which would increase the reading frame of the AGO5 protein to 922 amino acids rather than the 890 called for Glyma.11G190900 in the reference genome. We think this represents a correction to the AGO5 reference sequence at the 3′ end, based on amplicon sequencing of nine different lines. The only other difference relative to the Glyma.11G190900 gene model in the Phytzome call was a single deleted T within intron 6 in both the Calland K1 parent and the Calland k1 mutant line, which is likely a true variant occurring in the parent line relative to the Phytozome sequence.
Epistatic Interaction of the Mutated AGO5 k1 Gene with the Dominant I Allele
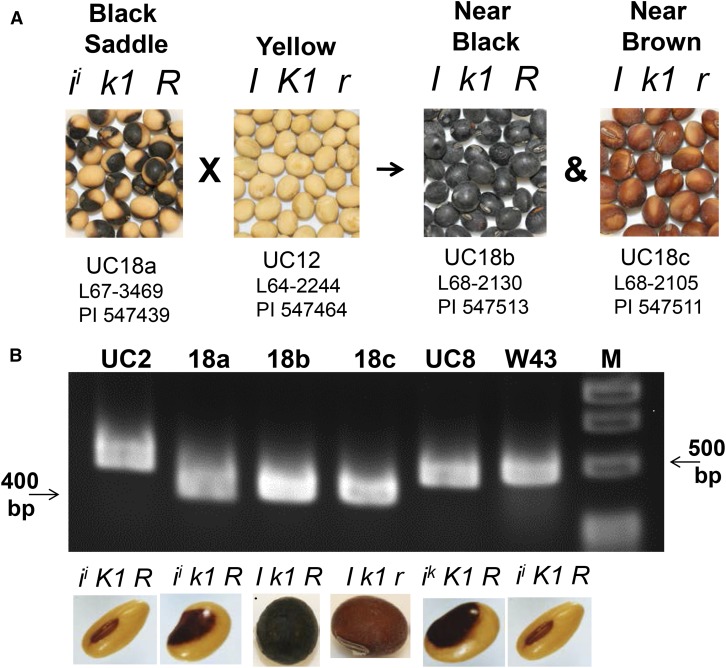
Figure 6A illustrates the nearly complete nullification of dominance of the I allele in the presence of a recessive k1 mutation. This epistatic interaction leads to a near fully pigmented black or near brown seed coat phenotype. While the color of the pigment is dependent on the R locus, known to be a MYB transcription factor regulating enzymes in the later stages of the flavonoid pathway (Gillman et al., 2011; Zabala and Vodkin, 2014), the extent of distribution of the pigment is controlled by the I locus and its interaction with alleles of the K1 locus. Figure 6A illustrates the original line numbers as well as the current Plant Introduction numbers by which these isolines are maintained in the germplasm collection since their release in 1968 by its curator Richard L. Bernard. These are among the relatively few materials and records of the unusual interaction of the two loci (Williams, 1958; Bernard and Weiss, 1973). The source of the k1 allele used in the creation of the near black (Clark 18b) and near brown (Clark 18c) isolines was Clark 18a (L67-3479), the spontaneous mutation that we have demonstrated to be a 129-bp deletion in the AGO5 gene. Thus, we examined these lines by PCR and Figure 6B showed that both contain the smaller PCR fragment that is characteristic of the k1 allele present in Clark 18a, as expected.
Figure 6.
Epistatic Interaction of the Mutated AGO5 k1 Gene with the Dominant I Allele.
(A) Seed of the indicated isolines illustrating the almost fully pigmented phenotypes (known as near black or near brown) that were created by crossing the Clark 18a black saddle k1 mutation with an isoline containing the dominant I allele that specifies all yellow hila and seed coats. The increase in pigmented surface area is an interaction of the dominant I allele in the presence of the k1 mutation, while the color of the pigment is determined by the genotype of the R locus (Williams, 1952; Bernard and Weiss, 1973). The PI and L numbers are official USDA germplasm numbers assigned to these isolines, which were released in 1968 by Richard L. Bernard.
(B) PCR shows that the small deletion in the AGO5 gene of Clark 18a (black saddle, ii k1 R) is also found in the Clark 18b (near black, I k1 R) and Clark 18c (near brown, I k1 r) isolines.
Multiple AGOs Are Expressed in the Immature Seed Coats
Alignments to the soybean genome with Glyma.11G190900 revealed high similarity to Glyma.12G083500, which is also annotated as AGO5. Although pairwise alignments of the two transcripts showed 90% nucleotide similarity overall (Supplemental Figure 5), the polymorphisms are distributed throughout the coding regions. Therefore, alignments by Bowtie1, which allow up to only three mismatches and no gaps, will distinguish between these two paralogous transcripts. The chromosome 12 AGO5 with <0.6 RPKM in the yellow sectors was expressed much less than the chromosome 11 AGO5 at 8 RPKM. The AGO5 on chromosome 12 does not have the 129-bp deletion observed in the k1 mutant and has intact PAZ and PIWI domains. Although its transcripts are much less abundant, it is possible that it could perform some of the function of AGO5 in the k1 mutant line.
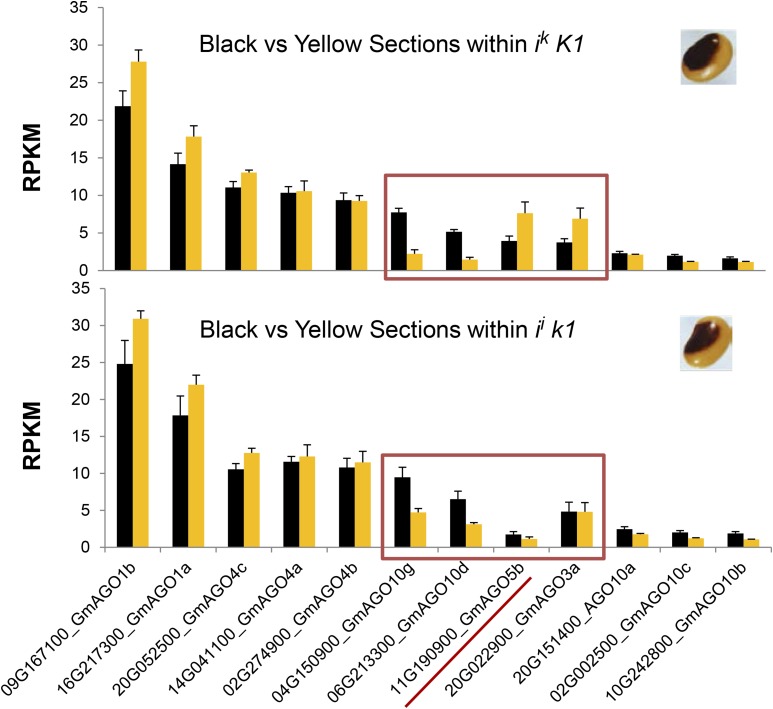
In total, there are 20 genes annotated as AGO in the soybean genome that have been examined by homology to known Arabidopsis thaliana AGO genes (Liu et al., 2014). A phylogenetic tree of these 20 proteins is shown in Supplemental Figure 6, and the RNA-seq levels found for 12 AGO family members with the highest expression are shown in Figure 7. The highest level of transcripts in the seed coats was observed for two AGO1 genes (Glyma.16G217300.1 and Glyma.09G167100.1) at ∼22 and 28 RPKM, respectively. The full table of RPKM levels of all tissues and genotypes is shown in Supplemental Data Set 6. Several of the AGOs, including AGO5b (Glyma.11G190900), AGO10g, and AGO10d, differed in transcript levels within the black and yellow sectors of the same genotype. Of these 20 AGO genes, only AGO5b (Glyma.11G190900) showed statistically significant expression differences between K1 and k1 genotypes. The other AGO5-related sequence, Glyma.12G083500, was not among the 12 most highly expressed AGOs.
Figure 7.
Comparison of the Expression Levels of 12 Argonaute Genes within Different Tissues of the Same Genotypes.
Top: RNA-seq levels within the black versus yellow tissue sectors of the ik K1 saddle pattern genotype. Bottom: RNA-seq levels within the black versus yellow tissue sectors of the ii k1 saddle pattern genotype. Data represent the average of three biological repeats of RNA-seq data normalized as RPKM with se bars. Black bars are from the black saddle region and yellow are from the yellow tissue. The 12 Argonaute genes with highest expression are shown, and the complete data set for all 20 soybean Argonaute genes is in Supplemental Data Set 6. The subclassification of the AGOs by a, b, c, etc., follows the nomenclature used by Liu et al. (2014). The red box denotes several Argonautes with variable expression between the sectors. The red underline denotes the K1 locus, Glyma.11G190900, which is AGO5b, the more highly expressed of the two AGO5 soybean genes.
In summary, RNA-seq analyses of isolines with patterned tissues contrasting black saddle genotypes (ik K1 versus ii k1) combined with genome resequencing of isogenic mutant lines enabled a focus on a reduced number of candidate genes in the vicinity of the legacy map position of the K1 gene including Glyma.11G190900 encoding AGO5. The presence of a 129-base deletion in the k1 mutation that arose in 1956 in the variety Clark was a strong indication that AGO5 is the k1 allele producing a prematurely terminated protein. Amplicon sequencing of several independent saddle pattern mutations from different genetic backgrounds revealed independent lesions affecting production of the AGO5 protein, thus establishing Glyma.11G190900 as the K1 locus and implicating a role for AGO5 in controlling the spatial patterning of the CHS siRNAs generated at the I locus.
DISCUSSION
Based on knowledge of the small RNA pathway in other organisms, we previously presented a model for the naturally occurring dominant I and ii alleles in preventing pigment formation in soybean seed coats (Tuteja et al., 2009). The long inverted repeat of the ii allele contains two inverted repeat clusters, cluster A (CHS1-3-4) and cluster B (CHS4-3-1), each of which also have inverted repeats of CHS genes within them. A precursor CHS double-stranded RNA (dsRNA) forms at some point within the region. The cleavage of the progenitor CHS dsRNA from the CHS1-3-4 cluster regions by Dicer-Like proteins (DCL) generates primary CHS siRNAs, some of which have similarity to the more distantly related CHS7 and CHS8 transcripts that are expressed in seed coat development. After cleavage at the CHS7/8 sites targeted by the primary CHS siRNAs, RNA-dependent RNA polymerase synthesizes dsRNA from the cleaved CHS7 and CHS8 mRNAs, which are then diced to generate secondary CHS siRNAs that also target CHS7 and CHS8. Thus, the action of RNA-dependent RNA polymerase amplifies the silencing response as well as spreading it over a larger region of the target. Sequence polymorphisms between the CHS1-3-4 genes at the origin of the dsRNA and the CHS7/8 gene targets allowed us to differentiate the primary CHS1-3-4 siRNAs that were present in the very small seed at 12 to 14 d after flowering and the transition to the secondary CHS7/8 siRNAs beginning at the 5 to 6 mg seed weight range through their maximal expression at the 50 to 75 mg seed weight (Cho et al., 2013). The primary CHS siRNAs were lower in abundance but composed of a higher proportion of 22-nucleotide small RNAs, whereas the more abundant secondary CHS siRNAs were primarily 21 nucleotides.
C2-Idf, a dominant allele of the C2 locus, which encodes chalcone synthase in maize (Zea mays), also has repeated copies of the CHS genes and operates via endogenous silencing through siRNAs (Della Vedova et al., 2005), though silencing by this locus is not tissue specific or pattern specific as is the case for the alleles of the soybean I locus. Small RNA sequencing of cosuppressed, nonpigmented transgenic petunia (Petunia hybrida) flowers with introduced CHS genes has shown that CHS siRNAs are the causative factor of silencing the pigment pathway in flowers (De Paoli et al., 2009). There are many commonalities between the naturally occurring soybean seed coat and the transgenic petunia systems (Eckardt, 2009). The first observations of silencing of chalcone synthase by cosuppression in transgenic petunia (Napoli et al., 1990; van der Krol et al., 1990) were also associated with variegated or patterned flowers as well as white flowers. More recently, it has been shown that the naturally occurring color patterns in petunia are the result of tandem arrangements of CHS genes and that CHS siRNAs are present in the nonpigmented sectors (Morita et al., 2012). They propose that other loci are likely involved in the patterning phenomena in petunia, but none have been identified to date. Here, we investigated the soybean system to determine whether the patterns in soybean are due to the differential levels of CHS siRNAs and whether we could identify the K1 locus that might be interacting with the small RNA pathway since it so dramatically changes the phenotype mediated by the I and ii alleles.
Even more intriguing than the tissue specificity exemplified by the I locus are the pattern-specific alleles that result in the two color regions of yellow and pigmented tissue on the same seed coats (Figure 1A). Using small RNA sequencing and RNA-seq of dissected regions, we demonstrated definitively that these patterns are due to the distribution of CHS siRNAs in the sectors destined to be yellow and not in the regions destined to be pigmented (Figure 2). These results clearly show that the levels of the CHS siRNAs follow a developmental program that leads to their increased expression within certain subregions of the seed coat that is mediated in genotypes with the ii and ik alleles.
The K1 Locus, Identified as AGO5, Influences the Spatial Distribution of CHS siRNAs
The RNAi pathway in plants evolved as a defense mechanism against RNA viruses and endogenous transposable elements (Baulcombe, 2004; Matzke and Birchler, 2005; Fusaro et al., 2006; Chapman and Carrington, 2007; Borges and Martienssen, 2015). Certain viral proteins can interfere with the pathway and suppress posttranscriptional silencing by siRNAs (Mallory et al., 2002; Diaz-Pendon et al., 2007). The posttranscriptional downregulation of CHS mRNAs can be interrupted by viral proteins that are suppressors of silencing (Senda et al., 2004; Nagamatsu et al., 2007) leading to pigment mottling on the seed coat in plants infected with soybean mosaic virus. Cold temperature also interferes with the production of small RNAs and is associated with partial discoloration of the yellow seed with pigment (Kasai et al., 2009).
In addition to the environmental effects of viruses and temperature, the seed color is influenced genetically by the K1 locus, which interacts with the I locus to control the pigment distribution in the seed coat. The k1 mutant allele extends the pigmented region of the ii allele to form a saddle pattern that mimics the phenotype of the ik allele (Figure 1A). Thus, it appears to overcome the silencing of the I locus. Here, we asked whether the phenotype was mediated also by the distribution of CHS siRNAs within the different color regions of the seed coat. Similar to patterned seed coats with genotype ik K1, those with genotype ii k1 also displayed quantitative variation and inverse correlation of the levels of CHS siRNAs and CHS7/8 mRNAs (Figures 3C to 3H). Thus, we conclude that the k1 allele results in altered spatial distribution of the CHS siRNAs. Interestingly, the effect of the k1 mutation on the dominant I allele is even more pronounced with the inhibition of silencing over most of the seed coat surface (Figures 1 and 7).
Using a combination of RNA-seq and genomic resequencing coupled with the known chromosome location of the K1 locus, we identified the K1 locus as Glyma.11G190900 encoding an AGO5 protein (Figures 3 and 4). The chromosome 11 AGO5 in the Clark 18a k1 line appears to be a nonfunctional protein. More evidence that Glyma.11G190900 is the K1 locus is the finding of different lesions in the AGO5 protein in independent mutant pairs in Lincoln and Calland as identified by amplicon sequencing (Figure 5). We conclude that AGO5 at the K1 locus participates in the spatial patterning of the CHS siRNAs produced from the CHS1-3-4 clusters of the I locus.
How Could a Deficit of AGO5 Affect Spatial Biogenesis of CHS siRNAs?
A complete lack of AGO family proteins would be expected to have highly detrimental effects, since the small RNA pathway is critical to many cellular functions through the action of microRNAs (miRNAs) that often regulate transcription factors as well as through siRNAs that are involved in methylation. Glyma.11G190900 is one of at least 20 different gene models annotated as encoding Argonaute proteins in the soybean genome. There is an AGO5 paralog, Glyma.12G083500, which has 90% nucleotide similarity to Glyma.11G190900 that encodes the K1 locus. The AGO5a (Glyma.12G083500) on chromosome 12 was also expressed in seed coats, though at a much lower level than AGO5b (Glyma.11G190900) on chromosome 11. In addition, there are many AGOs more highly expressed in the seed coats (Figure 7) that may partially compensate for the loss of function of one of the AGO5 genes in processing essential siRNAs and miRNAs. Multiple genes for Dicer proteins also exist in soybean. Mutant plants constructed with zinc finger nuclease technology disrupting either DCL1a or DCL1b homologs did not show a pronounced morphological or molecular phenotype, but double mutant plants were severely affected in both (Curtin et al., 2015).
The quantities of CHS siRNAs were very different in the pigmented and nonpigmented regions of seed coats with the saddle phenotype, implying that the production of CHS siRNAs is affected. Reduced or nonfunctional AGO5 could lead to altered efficiency of particular miRNAs or other siRNAs to downregulate their targets. One of these targets might be a transcription factor affecting a suite of genes or other miRNAs that regulate spatial patterning in the seed coats. In Arabidopsis, for example, a transcription factor complex that generates the precursor of miR166 defines cell fate in roots (Carlsbecker et al., 2010).
The AGOs have been grouped into three major phylogenetic clades (Fang and Qi, 2016) based on nomenclature for the 10 AGO genes in Arabidopsis: AGO1/5/10, AGO2/3/7, and AGO4/6/8/9. AGO5 belongs to the same group as the founding Argonaute member AGO1, whose function is to load miRNAs and trans-acting siRNAs that are involved in many developmental processes. Sorting of small RNAs into Argonaute complexes can be directed by the 5′ terminal nucleotide of the small RNA with different AGOs having preferences for different 5′ nucleotides. Although in the same group as AGO1, AGO5 has been shown to preferentially recruit sequences with a 5′ C-terminal nucleotide which are derived from intergenic sequences (Mi et al., 2008). AGO5 has been implicated to be important in male germline development in Arabidopsis (Borges et al., 2011) and to participate in female gametogenesis in the ovules (Tucker et al., 2012). A protein complex MEL1/AGO5c is associated with 21-nucleotide phased siRNAs (phasiRNAs), which are mostly generated from long noncoding intergenic regions in Arabidopsis that are initially cleaved by the 22-nucleotide miR2118 (Komiya et al., 2014). The closest homolog in maize, AGO5c, is also potentially the binding partner in anther development of premeiotic 21-nucleotide phasiRNAs, which are initially cleaved by the 22-nucleotide miR2118 (Zhai et al., 2015).
PhasiRNAs are particularly abundant in soybean with over 500 loci having been found of which 483 overlapped protein coding genes and 41% of those corresponded to the abundant NB-LRR class of proteins that are often involved in disease resistance (Arikit et al., 2014). To date, we have not observed any small RNAs mapping to the intergenic regions of the I locus clusters on chromosome 8, rather only small RNAs mapping to the CHS transcript regions. Since the CHS siRNAs are not formed from an initial cleavage step by a miRNA, but rather by dicing of a dsRNA originating from the unique structure of the dominant ii allele, they are not classified as phasiRNAs. The secondary amplification that produces phasiRNAs can be triggered by 22-nucleotide miRNAs (Chen et al., 2010; Cuperus et al., 2010; Fei et al., 2013). Interestingly, the primary CHS siRNAs representing the CHS1-3-4 origin region are higher in 22-nucleotide small RNAs compared with the more abundant 21-nucleotide secondary CHS siRNAs that map to the target CHS7 and CHS8 genes (Cho et al., 2013). It is possible that the soybean AGO5 homolog might participate as a binding partner for some of the CHS siRNAs as it does for phasiRNAs in Arabidopsis (Komiya et al., 2014) and likely also in maize (Zhai et al., 2015). This could explain the lack of silencing in the black saddle region but cannot readily explain the effective silencing of the target CHS7 and CHS8 mRNAs in the yellow sectors of the ii k1 seed coats where the AGO5 protein is terminated prematurely. More likely, the need for functional AGO5 in mediating patterning may be manifested only very early in development of the seed coat either directly in the function of the low abundance primary CHS siRNAs or indirectly by altering pattern development through the action of other miRNAs or siRNAs.
The I and K1 Alleles Reflect an Epistatic Interaction Involving the Small RNA Pathway
The I locus is one of the examples showing how small RNAs determine the dominance of alleles by their trans-acting influence on gene expression. We think understanding the interaction between the ii and k1 alleles, producing the saddle pattern phenotype of soybean seed, can shed light on the molecular mechanism of epistatic interactions through the small RNA pathway. Various molecular mechanisms of epistatic interactions have been discovered in diverse organisms (reviewed in Lehner, 2011). However, few reports have demonstrated that small RNAs are involved in the epistatic interactions between classical gene loci.
How is the expression of CHS siRNAs so precisely regulated within the same seed coat tissue with the same genotype (ii k1) to lead to a clear phenotypic boundary of the saddle color pattern? It is very possible that the boundary represents a larger suite of genes and small RNAs that are undergoing developmental shifts as the cells of the seed coat expand and differentiate, but only the change in CHS mRNAs and siRNAs is reflected as an easily visible phenotype. To address this question, we are currently correlating the global view of mRNA, siRNA, and miRNA populations from the black versus yellow sectors within each genotype as well as across genotypes. In this manner, we may find miRNAs and non-CHS siRNAs that show significant differential expression which would give us an insight to the molecular mechanism of whether other small RNAs are affected that could lead to developmental timing of the sharp boundary between the two interacting ii and k1 alleles. While many of the AGO family members appear to have near equal expression in the black versus yellow regions of seed coats with the same genotype, some of them, including AGO5b, AGO10g, and AGO10d, showed differential expression within the different regions of the seed coat (Figure 7). Differential expression of AGOs between tissue types or in response to stress or pathogens is one of the many factors that influence miRNA biogenesis and function (Jeong et al., 2013).
The mechanism of the dominant alleles of the I locus (I and ii) is due to the unusual structure that generates CHS siRNAs. The sequence of the ii allele is well defined through BAC sequencing (Clough et al., 2004; Tuteja and Vodkin, 2008). Restriction fragment length polymorphism and PCR mapping previously showed that several of the independent, naturally occurring, recessive i mutations that result in complete seed coat pigmentation were deletions in the CHS1-3-4 cluster regions (Todd and Vodkin, 1996; Tuteja et al., 2004). In this article, we demonstrated that the first step in the mechanism of the epistatic k1 mutation is mediated through deficient AGO5, which effectively reduces the quantities of CHS siRNAs in a pattern-specific manner when k1 is in the presence of the dominant ii or I alleles.
The recessive k1 allele with the defective AGO5b on chromosome 11 (Clark 18a) arose as a de novo mutation in the variety Clark. Records indicate it was found in Ames, Iowa in 1956. Since all commercially developed soybean varieties carry the yellow seed coat combination of either I K1 or ii K1, any rare seed arising with fully pigmented or saddle pattern forms are easily visible in the field harvests of inbred lines (Williams, 1945; Wilcox, 1988). The USDA germplasm collection maintains at least 60 of these pigmented lines that were initially observed as de novo mutations in field plots of named varieties dating back to 1945 through the present. The lines breed true for the pigmented phenotype and are often assumed to be mutations of either the classical I or K1 loci, although traditional tests of allelism have not been conducted for most of them. However, the basic genetics of interaction of the dominant I alleles with a k1 mutation was delineated by Leonard F. Williams (Williams, 1958) and Richard L. Bernard (Bernard and Weiss, 1973) who served among the earliest soybean geneticists, breeders, and curators of the USDA germplasm collection located at the University of Illinois.
Using these seed resources from the 1960s, we found that two isolines created to demonstrate the epistatic interaction of the k1 allele with the dominant I allele also carry the small AGO5 deletion (Figure 6). This is as expected, since the source of the k1 allele was derived from the Clark 18a mutation. Even more powerful evidence than cosegregation of the molecular variant with the seed phenotype is our finding that three independent black saddle mutations also display lesions in the AGO5 gene (Figure 5) relative to their progenitor parent lines (two from the variety Lincoln and one from Calland). The types of lesions displayed in the AGO5 gene are primarily from small deletions occurring in exonic regions (ranging from 1–129 nucleotides in size) that generally affect the reading frame and induce premature termination of the protein. Exon skipping was also demonstrated for the Clark 18a variant, deletion of the initiation codon for the Calland mutation, and deletion of only four amino acids for one of the Lincoln mutations. Kurakake varieties are large pod, flat-seeded, and black saddle pattern varieties used in Japan for vegetable (edamame) soybeans. The earliest report of a genetic factor named k was for the black saddle trait in Kurakake that segregated as recessive to a non-saddle variety (Takagi, 1929). Crosses with two of the early spontaneous saddle pattern mutations in named varieties developed in the United States revealed allelism with the Kurakake saddle pattern gene (reviewed in Bernard and Weiss, 1973). As we showed in Figure 5, two Kurakake black saddle varieties also have lesions in the AGO5 gene.
While mutations arising in the ii K1 varieties are easy to spot as they produce either the black or brown saddle pattern, those that occur in a I K1 parent line are near black or brown, which are difficult to distinguish from recessive i mutations that are larger deletions within the CHS1-3-4 regions of the I locus. Thus, we initially concentrated on the saddle pattern mutations rather than those that are possibly near black or near brown. Now that K1 is known to be AGO5, all of the lines could be surveyed by amplicon sequencing to verify de novo k1 mutations in the AGO5 locus. It is also possible that some de novo saddle pattern phenotypes that revert from yellow seed to saddle or completely pigmented may be controlled by loci other than K1 or I. Thus, they would reflect a release of the posttranscriptional silencing imposed by the dominant I or ii alleles. For example, classical tests of allelism have shown that a black saddle mutant derived from an X-rayed Clark line is a different gene, and it was named k3 (Bernard and Weiss, 1973). It will be of interest to determine whether examination of the k3 line or other de novo mutations with saddle phenotypes lead to discovery of different genes involved in controlling the pattern distributions of the CHS siRNAs. The distribution of the CHS siRNAs is likely to represent a larger developmental program than that reflected in the highly visible phenotype of seed color.
In summary, we have shown that spatial regulation of the production of soybean CHS siRNAs in the seed coat is influenced by two genetically defined variants of the I locus (ii and ik) as well as the independent k1 allele using quantitative data from small RNA and mRNA populations of these variant lines. These results illustrate that not only are the CHS siRNAs in the soybean system tissue specific as shown in a large data set of RNA-seq results (Cho et al., 2013), but also that they are exquisitely regulated in a pattern-specific manner during development. The nature of an epistatic interaction between the I locus alleles and the k1 mutation has been shown to operate via chromosome 11 AGO5, which has a role in pattern distribution of CHS siRNAs generated by the I locus on chromosome 8 to downregulate the target mRNAs from the CHS7 and CHS8 genes that reside on chromosomes 1 and 11, respectively. The k1 mutation should provide a way to assess other small RNAs and their target genes and pathways affected by AGO5 malfunction in the k1 allele. Molecular and genomic investigation of additional multiple independent mutations producing saddle pattern phenotypes may reveal additional genes interacting with the small RNA pathway in soybean.
METHODS
Plant Materials and Tissue Collection
The soybean (Glycine max) lines used in this study are inbred and homozygous for the indicated loci. All lines were developed by soybean geneticists and breeders during the 1960s and 1970s and are available from the USDA germplasm collection through GRIN (Germplasm Resources Information Network). The variety Williams (PI 548631), maturity group III, has genotype ii K1 with a black hilum on an otherwise yellow seed coat. The genome of the closely related Williams 82 isoline was recently sequenced (Schmutz et al., 2010) and is used as a reference genome standard in soybean. The saddle pattern isolines are in the background of the cultivar Clark (ii K1), maturity group IV, and have internal lab numbers of Clark 8 and Clark 18a. Clark 8 (PI 547450, L70-4204) has the saddle pattern with genotype ik K1 and results from repetitive backcrossing and selection of the ik allele from Black Eyebrow into Clark. Clark 18a (PI 547439, L67-3479) has the saddle pattern specified by interaction of the ii allele and the k1 mutation originally found in Clark. The PI number is an accession number by which information on the cultivar is searchable in the GRIN database. The L number is a breeding line number and is also searchable. Supplemental Figure 1 is a pedigree summary. Other lines with the saddle pattern mutations that were investigated include Lincoln (UC80, PI548362) and two Lincoln black saddle mutations arising independently in 1945 (UC145, L88-5422, PI634895) and 1954 (UC146, L88-5424, PI634896); Calland (UC82, PI548527), and a Calland black saddle mutant arising in 1970 (UC144, L88-5344, PI634873); and two large-seeded Japanese varieties Kurakake (UC150, PI506948) and Kurakake Daizu (UC151, PI506949), which both have the black saddle phenotype.
Soybean plants were grown in the greenhouse and immature seeds were harvested over the course of several weeks. The seeds were shelled, pooled together, and then sorted by weight range. The seeds were first dissected to separate the seed coat and the cotyledon. To obtain the regions within the seed coat, they then were dissected again to separate the pigmented and the nonpigmented regions of the seed coat with each part placed in different 15-mL tubes. Figure 1B shows an illustration of the dissection method since the pigmentation cannot be observed in the green stages at which the CHS siRNA levels are maximal. The seed coats were frozen in liquid nitrogen for 10 min and stored in the freezer (−80°C) until they were lyophilized. At least 10 samples were used for each extraction in order to minimize biological variation.
RNA and Small RNA Isolation
Total RNA was isolated by standard methods using phenol and chloroform extractions (Wang and Vodkin, 1994) and precipitated with ethanol but without lithium chloride in order to preserve small RNAs. For mRNA isolation, the sample was further purified by lithium chloride precipitation. In the case of the seed coats with pigments, the pigmented regions contain proanthocyanidins that bind to the RNA in the regions of the seed coat that will eventually contain the anthocyanin pigments (Todd and Vodkin, 1993). The modified RNA isolation method that was designed specifically to overcome this problem uses hydrated polyvinylpolypyrrolidone, polyproline, and BSA as competitors for the proanthocyanidin compounds in order to extract high-quality RNA (Wang and Vodkin, 1994; Wang et al., 1994) throughout seed coat development with pigmented genotypes.
Small RNA-Seq and Data Analysis
Small RNA libraries and high-throughput sequencing were performed using the TruSeq Small RNA kit according to the manufacturer’s instructions (Illumina) with the Genome Analyzer-II and HiSeq 2000 (Illumina) by the Keck Center (University of Illinois, Urbana, IL) using standard Illumina protocols (https://support.illumina.com). Some of the sequences were barcoded for sequencing within a single lane. Generally, a total of 20 to 80 million reads were obtained from these deep sequencing libraries. Adapter trimming was performed using Illumina’s Flicker pipeline, which finds the presence of the adapter in each read by finding the best alignment of the adapter to the read, and then removing it. The sizes of the small RNAs after adapter trimming ranged from 14 to 33 nucleotides with the majority in the range of 18 to 25 nucleotides. Adapter trimmed sequences were compared with obtain the number and occurrences of unique sequences. Alignments of siRNA sequences to CHS Glyma models (Phytozome, Joint Genome Institute) from the current assembly of the Williams 82 reference genome (Wm82.a2) of G. max (Schmutz et al., 2010) and to the known CHS genes from BACs from Williams 82 including BAC77G7a, accession EF623858, containing the I locus clusters (Tuteja and Vodkin, 2008; Clough et al., 2004) were performed using the Bowtie v.1 program (Langmead et al., 2009). Supplemental Data Set 2 shows the correspondence of CHS gene names to Glyma models. Alignments were made to individual CHS sequences with no mismatches allowed. Small RNA sequencing data were normalized in reads per million.
mRNA Sequencing Data Analysis
Transcriptome libraries and high-throughput sequencing (RNA-seq) were performed using the TruSeq RNA sample kit or Stranded TruSeq RNA Sample Kit (Supplemental Data Set 1) per the manufacturer’s instructions (Illumina) and sequenced with the HiSeq 2000 (Illumina) by the Keck Center using the standard Illumina protocol (https://support.illumina.com). A total of 43 to 97 million reads from each library of either 75 or 100 nucleotides were obtained from these deep sequencing libraries. Alignments of mRNA sequences to all 88,647 Glyma models, including splice variants, from the Williams 82 reference genome of G. max (Phytozome, Joint Genome Institutes) were performed using the Bowtie program v.1 (Langmead et al., 2009). The DESeq package (Anders and Huber, 2010) was used for statistical testing, and significantly expressed genes were selected based on an adjusted P value <0.05 controlled for false discovery (Benjamini and Hochberg, 1995). Independently, the Cufflinks package (Trapnell et al., 2012) was also used to compare differential gene expression. Transcriptome data were also normalized in RPKM as the transcriptome depends on the transcript length (Mortazavi et al., 2008).
DNA Isolation and PCR Amplifications
Soybean genomic DNA was isolated from freeze-dried shoot tips by standard methods using phenol-chloroform precipitation. PCR experiments were performed in a 50-μL reaction mixture containing 1 µg of genomic DNA in 5 μL of PCR buffer, 2.5 mM Mg2+, 0.4 mM deoxynucleotide triphosphate, 2.5 units of Takara EX Taq polymerase, and 0.2 µM forward and reverse primers. The following primers were used to span exon 7 of Glyma.11G190900 with a predicted fragment of 583 bp: BC41F, 5′-GTGAGCAAGTTATGCTATTGTTTCCCTTC-3′, and BC41R, 5′-TAGAGTTTTCTCTATCATGAGGACGCTGA-3′. PCR amplifications were conducted in a PTC-200 programmable thermocycler (MJ Research) via an initial denaturation step at 94°C for 1 min followed by 30 cycles of denaturing at 94°C for 10 s, annealing at 55°C for 1 min, and elongation at 72°C for 1 min, to end with a 2-min extension at 72°C. The amplified reactions were separated on a 0.7% agarose gel and bands stained via Gel Green (Biotium).
Whole-Genome Resequencing and Data Analysis
Whole-genome sequencing library construction and high-throughput sequencing were performed by the Keck Center. The libraries were prepared with Kapa library construction kits (https://www.kapabiosystems.com) and quantitated by qPCR and sequenced on one lane for 100 cycles from each end of the fragments on a HiSeq 2000 (Illumina) using a TruSeq SBS sequencing kit version 3 and analyzed with Casava1.8 (pipeline 1.9). The average size of the DNA fragments was around 500 nucleotides; insert size is 300 nucleotides. Quality check for sequencing data was done by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Alignments were performed using Bowtie2 (Langmead and Salzberg, 2012). We used default alignment and reporting options. Bowtie2 output files, in SAM format, were converted to the BAM format and sorted by Samtools (Li et al., 2009). The Integrative Genomic Viewer (IGV) was used to convert from BAM to TDF format and visualize normalized [count at base × one million/total number of reads] data (Robinson et al., 2011). To call single-nucleotide polymorphisms, we used the Samtools command (mpileup -uf) and converted BCF to VCF format to visualize in the IGV.
Amplicon Sequencing and Analyses
A 6.2-kb amplicon containing the entire region of Glyma.11G190900 plus 196 bases of the region upstream of the 5′UTR was amplified from nine different soybean parent lines or mutations containing either K1 or k1 genotypes using the following primers: forward primer, 5′-TTCACCACACTACTCAAGAT-3′ and reverse primer 5′-ATTAATATAACAAGCTGACG-3′. PCR amplifications were performed as above in Corning Axygen thin walled tubes in a PTC-200 DNA Engine from MJ Research using modified reaction times as follows: initial denaturation step at 96°C for 2 min followed by 39 cycles of denaturing at 96°C for 20 s, annealing at 55°C for 1 min, and elongation at 72°C for 4 min, to end with a 7-min extension at 72°C. For each soybean genotype, four reactions were pooled and purified with a Zymo DNA Clean and Concentrator kit and the concentration adjusted to between 40 and 64 ng/μL. A total of 35 μL was submitted for barcoding of PCR reactions, amplicon sequencing with the Illumina MySeq, and automated assembly by the Center for Computational and Integrative Biology DNA Core Facility at Massachusetts General Hospital (Cambridge, MA). Coverage of each amplicon ranged from 69,609 to 191,356 reads, and representation of each base in an assembled contig sequence was generally >1000 with the great majority being perfect matches to the consensus contig. Amplicon sequences were aligned to the gene, transcript, or protein sequences of Glyma.11G190900 AGO5 using Multalin (Corpet, 1988).
Accession Numbers
The data have been entered into Gene Expression Omnibus under superseries GSE43347 for the 16 small RNA samples comparing the patterned seed coats and series GSE89126 for the 12 RNA-seq samples of PI547450 (Clark 8 ik K1) and PI547439 (Clark 18a ii k1), respectively, and SRP092073 and SRP092133 for the paired-end genomic sequencing of PI548533 Clark (ii K1) and PI547439 (Clark 18a, ii k1), respectively. Amplicon sequences were entered in GenBank as KY621334 for Clark 18a (ii k1) and KY631911 to KY631917 and KY785328 for other K1 and k1 lines as shown in Supplemental Data Set 5. Accession numbers for CHS genes can be found in Supplemental Data Set 2. Sequences for Glyma.11G190900 (AGO5b) and Glyma.12G083500 (AGO5a) can be found in Phytozome at https://phytozome.jgi.doe.gov.
Supplemental Data
Supplemental Figure 1. The Pedigree of Clark Isolines That Show the Saddle Pattern Phenotype.
Supplemental Figure 2. Cufflinks Plots Comparing RNA-Seq Data from Clark (ii K1) and Clark 18a (ii k1).
Supplemental Figure 3. Mapping Information Resources at Soybase for the K1 Locus.
Supplemental Figure 4. Alignment of Nine AGO5 Proteins Derived from Amplicon Sequences Comparing K1 and k1 Lines.
Supplemental Figure 5. Alignment of Two Closely Related Soybean AGO5 Coding Sequence Transcripts.
Supplemental Figure 6. Phylogenetic Tree for Soybean Argonaute Genes.
Supplemental Data Set 1. Summary of Sequencing Data Generated for Small RNAs, mRNAs, and Genomic DNAs.
Supplemental Data Set 2. Small RNA Levels of 21 CHS Glyma Models Comparing Tissues from Pattern Genotypes as Shown in Figures 3 and 4.
Supplemental Data Set 3. RNA-Seq Levels of 21 CHS Glyma Models Comparing Tissues from the Pattern Genotypes of Clark 8 (ik K1) and Clark 18a (ii k1).
Supplemental Data Set 4. Glyma Models with Differential RNA-Seq Expression on Chromosome 11 in the Region of a Putative Marker near the K1 Locus.
Supplemental Data Set 5. Summary of AGO Amplicon Sequencing Results for Standard K1 and Mutant k1 Lines and Example Output.
Supplemental Data Set 6. Expression from RNA-Seq Data for 20 AGO Genes within Different Regions of the Saddle Pattern Seed Coats Having the Same Genotypes.
Supplemental File 1. Fasta File of the Alignment Used to Generate the Phylogenetic Tree in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank Alvaro Hernandez and staff of the University of Illinois High-Throughput Sequencing Unit of the Biotechnology Center for the small RNA, RNA-seq, and genome resequencing services. We thank Nicole Stang-Thomann and the staff of the Center for Computational and Integrative Biology (CCIB) at Massachusetts General Hospital for the use of the CCIB DNA Core Facility (Cambridge, MA), which provided amplicon sequencing services. We thank undergraduate and academic assistants Drew Metz, Achira Kulasekara, and Berwin Xie for help with data handling and python scripting. We acknowledge seed obtained from the USDA Soybean Germplasm Collections (Urbana, IL) available from the Germplasm Information Resources Network (GRIN). The research was supported by grants from the United Soybean Board, the USDA, and the Illinois Soybean Association.
AUTHOR CONTRIBUTIONS
Y.B.C. and S.I.J. collected tissue samples, performed experiments, analyzed and interpreted data, and drafted results. L.O.V. designed initial approach, led and coordinated the project, interpreted data, drafted sections, and edited the manuscript. All authors read and approved the manuscript.
Glossary
- siRNA
short-interfering RNA
- RPKM
reads per kilobase of gene model size per million mapped reads
- UTR
untranslated region
- dsRNA
double-stranded RNA
- miRNA
microRNA
- phasiRNAs
phased siRNAs
- IGV
Integrative Genomic Viewer
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S., Xia R., Kakrana A., Huang K., Zhai J., Yan Z., Valdés-López O., Prince S., Musket T.A., Nguyen H.T., Stacey G., Meyers B.C. (2014). An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to statistical testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bernard R.L., Weiss M.G. (1973). Qualitative genetics. In Soybeans: Improvement, Production, and Uses, 1st ed, Caldwell B.E., ed (Madison, WI: American Society of Agronomy; ), pp. 117–149. [Google Scholar]
- Borges F., Pereira P.A., Slotkin R.K., Martienssen R.A., Becker J.D. (2011). MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 62: 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8: 884–896. [DOI] [PubMed] [Google Scholar]
- Chen H.-M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.B., Jones S.I., Vodkin L. (2013). The transition from primary siRNAs to amplified secondary siRNAs that regulate chalcone synthase during development of Glycine max seed coats. PLoS One 8: e76954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Tuteja J.H., Li M., Marek L.F., Shoemaker R.C., Vodkin L.O. (2004). Features of a 103-kb gene-rich region in soybean include an inverted perfect repeat cluster of CHS genes comprising the I locus. Genome 47: 819–831. [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S.J., et al. (2015). MicroRNA maturation and microRNA target gene expression regulation are severely disrupted in soybean dicer-like1 double mutants. G3 (Bethesda) 6: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Vedova C.B., Lorbiecke R., Kirsch H., Schulte M.B., Scheets K., Borchert L.M., Scheffler B.E., Wienand U., Cone K.C., Birchler J.A. (2005). The dominant inhibitory chalcone synthase allele C2-Idf (inhibitor diffuse) from Zea mays (L.) acts via an endogenous RNA silencing mechanism. Genetics 170: 1989–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paoli E., Dorantes-Acosta A., Zhai J., Accerbi M., Jeong D.-H., Park S., Meyers B.C., Jorgensen R.A., Green P.J. (2009). Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA 15: 1965–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon J.A., Li F., Li W.X., Ding S.W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt N.A. (2009). Tissue-specific siRNAs that silence CHS genes in soybean. Plant Cell 21: 2983–2984. [Google Scholar]
- Fang X., Qi Y. (2016). RNAi in plants: an Argonaute-centered view. Plant Cell 28: 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A.F., Matthew L., Smith N.A., Curtin S.J., Dedic-Hagan J., Ellacott G.A., Watson J.M., Wang M.B., Brosnan C., Carroll B.J., Waterhouse P.M. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7: 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman J.D., Tetlow A., Lee J.-D., Shannon J.G., Bilyeu K. (2011). Loss-of-function mutations affecting a specific Glycine max R2R3 MYB transcription factor result in brown hilum and brown seed coats. BMC Plant Biol. 11: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D., Nelson R.T., Cannon S.B., Shoemaker R.C. (2010). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38 (suppl. 1): D843–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.-H., Thatcher S.R., Brown R.S., Zhai J., Park S., Rymarquis L.A., Meyers B.C., Green P.J. (2013). Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol. 162: 1225–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai A., Ohnishi S., Yamazaki H., Funatsuki H., Kurauchi T., Matsumoto T., Yumoto S., Senda M. (2009). Molecular mechanism of seed coat discoloration induced by low temperature in yellow soybean. Plant Cell Physiol. 50: 1090–1098. [DOI] [PubMed] [Google Scholar]
- Komiya R., Ohyanagi H., Niihama M., Watanabe T., Nakano M., Kurata N., Nonomura K. (2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78: 385–397. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. (2011). Molecular mechanisms of epistasis within and between genes. Trends Genet. 27: 323–331. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lu T., Dou Y., Yu B., Zhang C. (2014). Identification of RNA silencing components in soybean and sorghum. BMC Bioinformatics 15: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Bartel D., Vance V.B., Bowman L.H. (2002). A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99: 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Birchler J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6: 24–35. [DOI] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Saito R., Ban Y., Tanikawa N., Kuchitsu K., Ando T., Yoshikawa M., Habu Y., Ozeki Y., Nakayama M. (2012). Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 70: 739–749. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nagamatsu A., Masuta C., Senda M., Matsuura H., Kasai A., Hong J.-S., Kitamura K., Abe J., Kanazawa A. (2007). Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol. J. 5: 778–790. [DOI] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R.G., Kilen T.C. (1987). Qualitative genetics and cytogenetics. In Soybeans: Improvement, Production and Uses, 2nd ed, Wilcox J.R., ed (Madison, WI: American Society of Agronomy; ), pp. 135–209. [Google Scholar]
- Palmer R.G., Pfeiffer T.W., Buss G.R., Kilen T.C. (2004). Qualitative genetics. In Soybeans: Improvement, Production and Uses, 3rd ed, H.G. Boerma and J.E. Specht, eds (Madison, WI: American Society of Agronomy; ), pp. 137–233. [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Senda M., Masuta C., Ohnishi S., Goto K., Kasai A., Sano T., Hong J.S., MacFarlane S. (2004). Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16: 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, F. (1929). On the inheritance of some characters in Glycine soja (Bentham) (soybean). Sci. Rep. Tohuku Univ. Ser. 4: 577–589. [Google Scholar]
- Todd J.J., Vodkin L.O. (1993). Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiol. 102: 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J.J., Vodkin L.O. (1996). Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Okada T., Hu Y., Scholefield A., Taylor J.M., Koltunow A.M. (2012). Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139: 1399–1404. [DOI] [PubMed] [Google Scholar]
- Tuteja J.H., Clough S.J., Chan W.C., Vodkin L.O. (2004). Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja J.H., Vodkin L.O. (2008). Structural features of the endogenous CHS silencing and target loci in the soybean genome. Crop Sci. 48: 49–69. [Google Scholar]
- Tuteja J.H., Zabala G., Varala K., Hudson M., Vodkin L.O. (2009). Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in glycine max seed coats. Plant Cell 21: 3063–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol A.R., Mur L.A., Beld M., Mol J.N.M., Stuitje A.R. (1990). Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.S., Vodkin L.O. (1994). Extraction of RNA from tissues that contain high levels of procyanidins that bind RNA. Plant Mol. Biol. Report. 12: 132–145. [Google Scholar]
- Wang C.S., Todd J.J., Vodkin L.O. (1994). Chalcone synthase mRNA and activity are reduced in yellow soybean seed coats with dominant I alleles. Plant Physiol. 105: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox J.R. (1988). Performance and use of seed coat mutants in soybean. Crop Sci. 28: 30–32. [Google Scholar]
- Williams L.F. (1945). Off-colored seeds in the Lincoln soybean. Soybean Digest 5: 50–61. [Google Scholar]
- Williams L.F. (1952). The inheritance of certain black and brown pigments in the soybean. Genetics 37: 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.F. (1958). Alteration of dominance and apparent change in direction of gene action by a mutation at another locus affecting the pigmentation of the seed coat of the soybean. Proc. Intl. Natl. Cong. Genet. 10: 315–316. [Google Scholar]
- Zabala G., Vodkin L.O. (2014). Methylation affects transposition and splicing of a large CACTA transposon from a MYB transcription factor regulating anthocyanin synthase genes in soybean seed coats. PLoS One 9: e111959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Zhang H., Arikit S., Huang K., Nan G.-L., Walbot V., Meyers B.C. (2015). Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 112: 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.