Abstract
Yersinia spp. inject virulence proteins called Yops into the cytosol of target eukaryotic cells in an effort to evade phagocytic killing via a dedicated protein-sorting pathway termed type III secretion. Previous studies have proposed that, unlike other protein translocation mechanisms, Yops are not recognized as substrates for secretion via a solely proteinaceous signal. Rather, at least some of this information may be encoded within yop mRNA. Herein, we report that the first seven codons of yopE, when fused to the reporter protein neomycin phosphotransferase (Npt), are sufficient for the secretion of YopE1-7-Npt when type III secretion is induced in vitro. Systematic mutagenesis of yopE codons 1 to 7 reveals that, like yopQ, codons 2, 3, 5, and 7 are sensitive to mutagenesis, thereby defining the first empirical similarity between the secretion signals of two type III secreted substrates. Like that of yopQ, the secretion signal of yopE exhibits a bipartite nature. This is manifested by the ability of codons 8 to 15 to suppress point mutations in the minimal secretion signal that change the amino acid specificities of particular codons or that induce alterations in the reading frame. Further, we have identified a single nucleotide position in codon 3 that, when mutated, conserves the predicted amino acid sequence of the YopE1-7-Npt but abrogates secretion of the reporter protein. When introduced into the context of the full-length yopE gene, the single-nucleotide mutation reduces the type III injection of YopE into HeLa cells, even though the predicted amino acid sequence remains the same. Thus, yopE mRNA appears to encode a property that mediates the type III injection of YopE.
The type III mechanism of protein secretion is a common pathogenic strategy employed by many gram-negative bacteria to cause disease in a host animal or plant (14). Yersinia spp. utilize this protein secretion pathway to inject toxic proteins, called Yersinia outer proteins, or Yops, into the cytosol of professional phagocytic cells in an effort to evade phagocytosis (29). This strategy permits bacterial multiplication in lymphoid tissues and the establishment of human disease (10, 26). Along with essential components of the secretion machinery itself, as well as regulatory factors, at least 14 proteins that are substrates for type III secretion by Yersinia enterocolitica are encoded on a 70-kb virulence plasmid (11). In vitro, the secretion of Yops may be induced in the absence of eukaryotic cells by the depletion of Ca2+ ions in rich culture medium (23). Curiously, the concentration of free cytosolic Ca2+ in eukaryotic cells is well below the threshold required to induce type III secretion in vitro (18). It is therefore conceivable that the depletion of Ca2+ in rich medium mimics a situation whereby extracellular yersiniae sense the intracellular environment of target cells via terminal components of the secretion machinery (18).
To date, most if not all protein translocation pathways share a common theme: the information required for translocating a polypeptide across biological membranes is contained solely within the peptide sequence itself (5). It has recently become evident that polypeptides secreted by the type III machinery may represent an exception to this widely held paradigm (2, 9, 32). Early studies indicated that the secretion signal for several Yops resides within the first 15 amino acids (1, 23, 34). A comparison of amino acid sequences of many Yops, however, failed to reveal a property that would suggest a common mode of recognition (1).
Mutational analysis of yopE, yopN, and yopQ revealed that the secretion signal of the first 15 codons, when fused to neomycin phosphotransferase (Npt), a reporter protein, can tolerate a number of frameshift mutations that completely alter its amino acid sequence while its mRNA sequence is largely retained (1, 3). Similar results have also been reported for the secretion signal consisting of the first 28 codons of a plant effector protein from Xanthomonas campestris (AvrBs2) and the secretion signal consisting of codons 4 to 7 of a secretion substrate from Salmonella enterica (InvJ) (24, 30). Further, synonymous substitutions of codons, which change the sequence of the mRNA but leave the amino acid sequence intact, render the minimal secretion signal of yopQ or yopN nonfunctional (27, 28). Taken together, it appears that yop mRNAs may encode information that determines the sorting of Yop proteins.
A recent report has proposed that an N-terminal peptide signal initiates YopE into the type III pathway (21). This hypothesis was borne out of the observation that frameshift mutations introduced into the first 11 codons of yopE abrogate YopE secretion in vitro (21). Further, synonymous mutations that were introduced into yopE codons 1 to 10 failed to abolish the secretion of YopE (21, 22). In this report, we attempt to reconcile these apparently disparate data (20). First, we determined that the first seven codons of yopE, when fused in frame to npt, are sufficient for the secretion of the reporter protein. Similar to yopQ, the ability of the secretion signal to tolerate frameshift mutations depends on the length of the secretion signal, whereby fusions of less than 12 codons harboring a frameshift mutation are unable to promote the secretion of Npt. Like yopQ, codons downstream of the yopE secretion signal encode additional secretion information that is able to suppress mutations within the yopE minimal secretion signal. A systematic mutational analysis of the yopE minimal secretion signal directly reveals, for the first time, nucleotides in the yopE secretion signal that are critical for its function. This analysis also reveals a striking similarity to the yopQ secretion signal: although both signals share no obvious sequence homology, codons 2, 3, 5, and 7 play an important role in the recognition of both proteins by the secretion machinery. Surprisingly, the synonymous substitution of a single nucleotide in codon 3, which encodes the identical amino acid as wild-type yopE at that position, leads to a dramatic reduction in the secretion of YopE1-7-Npt. Introduction of this mutation into the full-length yopE gene has no effect on the secretion of YopE under low-calcium conditions. However, this single-nucleotide substitution leads to a decrease in the type III injection of YopE into HeLa cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Y. enterocolitica W22703 has been described previously (12). Escherichia coli strain DH5α served as a host for DNA manipulations (16).
DNA methods and plasmid construction.
yopE fusions to npt were constructed using a low-copy-number vector backbone containing an engineered NdeI restriction site immediately downstream of the yopE untranslated region described previously (6). Annealed oligonucleotides specifying the desired insertion were synthesized and ligated between the NdeI site and a KpnI site of pDA46 (1), fusing the insertion to npt. Expression of the construct was mediated by a DNA segment comprising 500 bp upstream of the yopE start codon. Point mutations were introduced into full-length yopE by PCR amplification of the yopE coding region with primers with abutted NdeI or BamHI sites (underlined): YopE5′A9CNdeI (5′GGAATTCCATATGAAAATCTCATCATTTATTTCTAC) and YopE3′BamHI (5′AAGGATCCTCACATTCACATCAATGACAGTA), respectively. PCR products were then cloned into 5′ NdeI and 3′ BamHI sites downstream of the yopE promoter. All plasmids encoding yopE secretion signals and their fusions to npt were verified by DNA sequencing.
Protein electrophoresis and immunodetection.
Overnight cultures of Yersinia strains were grown in M9 medium supplemented with 5% Casamino Acids at 26°C and diluted 1:50 into 4 ml of M9-Casamino Acids. A total of 30 μg of chloramphenicol/ml was added to the medium for plasmid maintenance, if necessary. Cultures were grown at 26°C for 2 h and then shifted to 37°C for 3 h. Aliquots of cultures (each, 1.4 ml) were removed and centrifuged at 15,000 × g for 15 min. One milliliter of the culture supernatant was removed and precipitated with 75 μl of 100% trichloroacetic acid (TCA). Cell pellets were suspended in 700 μl of water, and 500 μl of this suspension was precipitated with 500 μl of 10% TCA. Precipitated proteins were washed with acetone, solubilized in sample buffer, and separated by electrophoresis on 15% polyacrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following electrotransfer to a polyvinylidene difluoride (PVDF) membrane, proteins were immunoblotted with α-Npt, α-YopH, and anti-chloramphenicol acetyltransferase (α-CAT) (purified rabbit polyclonal antibodies and α-rabbit horseradish peroxidase conjugates). The chemiluminescent signal was visualized with a Fluorchem 8800 imaging system (Alpha Innotech) and quantified.
Digitonin fractionation of infected HeLa cells.
HeLa cells were grown to 80% confluency in 75-cm2 tissue culture flasks in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, as described previously (17). Overnight cultures of yersiniae grown in tryptic soy broth containing 30 μg of chloramphenicol/ml were diluted 1:20 into 4 ml of tryptic soy broth/cm and grown at 26°C for 2 h. One hour before infection, HeLa cell monolayers were washed twice with phosphate-buffered saline (PBS), and 10 ml of fresh Dulbecco's modified Eagle medium (no fetal bovine serum) was added. One milliliter of yersiniae was harvested, suspended in 1 ml of PBS, added to the monolayer at a multiplicity of infection of 10, and incubated at 37°C and 5% CO2 for 3 h. The medium was removed and centrifuged at 10,000 × g for 15 min. Seven milliliters of the supernatant was removed and retained. The pellet was suspended in 10 ml of PBS-1% SDS, and 7 ml was retained. The monolayer and adherent bacteria were treated with 10 ml (1%) of digitonin in PBS for 20 min. Monolayers were scraped, centrifuged, and treated as above. All fractions were extracted with methanol-chloroform, suspended in 0.5 M Tris-HCl (pH 7.5)-4% SDS, and boiled in sample buffer, and the proteins were separated by SDS-PAGE. Samples were immunoblotted as described above.
Protein purification.
Overnight cultures of Y. enterocolitica strain W22703 carrying pYopE1-7-Npt or pYopE(A9C)1-7-Npt were diluted 1:50 into 2 liter of M9-Casamino Acids and grown for 2 h at 26°C. Cultures were then shifted to 37°C to induce type III secretion for 3 h. Bacteria were sedimented by centrifugation at 8,000 × g, and cells were lysed in a French pressure cell at 10,000 lb/in2. Debris and nonlysed cells were removed by centrifugation, and the resulting supernatant was chromatographed on a MonoQ ion-exchange column and eluted with NaCl buffer (28). Peak fractions were detected by immunoblotting with α-Npt, and corresponding segments of PVDF membrane were analyzed by Edman degradation.
RESULTS
Mapping the minimal secretion signal of yopE.
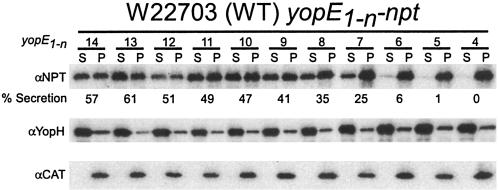
The discovery of the bipartite nature of the secretion signal in the 5′ coding sequence of yopQ was the result of mapping the boundaries of the minimal secretion signal (27, 28). Indeed, our work with the yopQ secretion signal demonstrated that subtleties in type III secretion signals may only be uncovered if mutagenesis is carried out in the context of the minimal type III secretion signal (28). Therefore, we first sought to map the boundaries of the yopE secretion signal. Previous work demonstrated that the first 15 codons of yopE are sufficient for the secretion of a fused reporter protein, Npt (1, 7, 31, 34). Successive single-codon truncations from the 3′ end of yopE codons 1 to 15 were fused to npt, expressed in Y. enterocolitica, and analyzed for the type III secretion of hybrid polypeptides. Briefly, yersiniae were grown in M9 medium supplemented with 5% Casamino Acids at 37°C to induce the type III pathway. Cultures were centrifuged, and the extracellular medium was separated from the cell pellet. Protein in both fractions was precipitated with TCA and analyzed by immunoblotting with α-Npt. As a control for a functional type III pathway, the secretion of YopH was measured. CAT is a cytoplasmic protein; its location was determined as a control for proper fractionation. Figure 1 shows that yopE fusions of more than codons 1 to 9 to npt result in greater than 40% secretion of the reporter. Fusion of the first 7 or 8 codons of yopE to npt resulted in greater than 20% of the reporter protein, which resembled the amount of secretion measured for the minimal yopQ secretion signal (codons 1 to 10). Further deletions of codons from the 3′ end of the secretion signal severely reduced or abolished secretion of hybrid proteins (Fig. 1). In a previous study, Schesser et al. reported that glycine substitutions in the first seven codons of yopE1-11 fused to cya reduced adenylate cyclase activity by whole cells in the presence of calmodulin (31). The authors took this to mean that the secretion of their reporter protein was abrogated. The results reported here are consistent with those reported by Schesser et al. in assigning a critical secretion function to the first seven codons of yopE (31). Thus, although at least the first 10 codons of yopQ are required to direct the secretion of a reporter protein, the minimal secretion signal of yopE comprises just codons 1 to 7, the shortest type III secretion signal we have uncovered thus far in yersiniae (25).
FIG. 1.
Mapping the yopE minimal secretion signal. Specified segments of the yopE coding region are expressed in wild-type Y. enterocolitica strain W22703 as translational fusions to the reporter protein Npt. Type III secretion by yersiniae harboring low-copy-number plasmids expressing npt fusions is measured by separating the medium supernatant (S) and the bacterial pellet (P) of centrifuged cultures that had been induced by temperature shift and low calcium. Proteins in both fractions were separated by SDS-PAGE, electroblotted onto a PVDF membrane, and detected with antisera raised against purified Npt, YopH (a type III secretion substrate), and CAT (a cytoplasmic protein). Chemiluminescent signals were scanned and quantified and are reported as the percentage of signal intensity of protein in the culture supernatant compared to the total amount of protein in both the culture supernatant and the bacterial pellet.
Systematic mutagenesis of the yopE minimal secretion signal.
Our previous mutational analysis of the yopQ minimal secretion signal revealed the contributions of particular nucleotides to the recognition of YopQ as a secretion substrate (27). Such mutations had previously not been uncovered in yop secretion signals when signals longer than the minimal secretion signal had been subjected to mutagenesis (1). We therefore wished to systematically mutagenize the minimal secretion signal of yopE by mutating every single-nucleotide residue individually in an effort to compare and contrast it to the yopQ secretion signal.
Codons 1 to 7 of yopE were fused to npt, and single transversion substitutions were constructed in each of the 18 nucleotides comprising codons 2 to 7, such that each adenyl was changed to thymidyl (uridyl), every guanidyl was changed to cytidyl, and vice versa (Fig. 2). In three cases where such a mutation resulted in a stop codon, an alternate transversion or transition was introduced (positions 4, 11, and 14). Plasmids harboring these individual nucleotide mutations were expressed in wild-type yersiniae and induced for type III secretion. Whereas the wild-type yopE minimal secretion signal routinely allows for over 25% secretion of Npt, mutation of the first two positions of codon 2 resulted in various intermediate phenotypes that slightly reduced Npt secretion. Mutation of position three, however, almost completely abrogated Npt secretion. Surprisingly, like yopQ, mutation of the first two positions of codon 3 led to the dramatic abolishment of Npt secretion, whereas an adenyl-to-uridyl change in position three of codon 3, a synonymous substitution which abrogates the function of the yopQ secretion signal, had no effect on yopE secretion signal function. Nucleotide substitutions at all three positions of codon 4 had either a slight effect or no effect at all on Npt secretion. In codon 5, only the mutation of the second nucleotide position resulted in a significant decrease in the secretion of the reporter protein. Mutation of all three positions of codon 6 led to an overall reduction but not total eradication of Npt secretion. Finally, as the importance of codon 7 was established by the deletion analysis presented in Fig. 1, mutation of the first two nucleotide positions of codon 7 predictably resulted in a dramatic reduction of yopE secretion signal function. To our knowledge, these substitutions represent the first direct measurements of the effects of single-nucleotide or single-amino-acid substitutions that explicitly abrogate the function of the yopE type III secretion signal.
FIG. 2.
Mutational analysis of the yopE minimal secretion signal. Single transversion mutations (from purines to pyrimidines and vice versa) were engineered into the first seven codons of yopE and fused to npt. A transversion mutation at nucleotide position 4, 11, or 14 would have resulted in a stop codon, so an alternate mutation was introduced. Fusions to Npt were analyzed for type III secretion as described in the legend to Fig. 1.
Synonymous codon substitutions in the yopE minimal secretion signal.
In a previous report that investigated the yopE secretion signal, the simultaneous synonymous substitution of codons 2 to 10 of yopE, which comprised a change of 17 nucleotides, did not adversely affect YopE secretion when introduced into the context of the full-length protein (21). Indeed, we observed that introducing this mutation into the context of just the yopE minimal secretion signal also failed to abolish its function (data not shown.) The mRNA signal hypothesis would have predicted that an RNA-encoded secretion signal could not tolerate such a dramatic change. Although such dramatic mutations do, in fact, abrogate the function of the yopQ minimal secretion signal, we did not uncover any synonymous codon substitutions that abolished the function of the yopE minimal secretion signal in the course of our systematic mutagenesis (Fig. 2). Do yopQ and yopE represent drastically different modes of substrate recognition, or might there be an undiscovered silent mutation that abrogates yopE signal function? To distinguish between these possibilities, we sought to test every possible individual synonymous substitution in the first seven codons of yopE that were not yet tested in an effort to uncover any silent mutations that might abrogate the function of the yopE minimal secretion signal.
Codons 1 to 7 of yopE were fused to npt, and every possible codon specifying each amino acid at positions 2 to 7 was substituted individually (Fig. 3). While the replacement of codon 4 (UCA) by an AGU, codon 5 (UCA) by AGC, and codon 6 (UUU) by UUC result in a moderate reduction in the secretion of Npt, most of the substitutions we tested did not appreciably abrogate yopE secretion signal function. A glaring exception was the substitution of codon 3, an isoleucine normally encoded by an AUA. Although mutation of the third position of codon 3 to uridyl does not affect the function of the secretion signal (Fig. 3), mutating codon 3 to AUC, a synonymous isoleucine substitution, almost completely abolishes the secretion of the reporter protein. Comparatively, codon 3 of yopQ also encodes isoleucine but uses AUU. However, while the yopQ signal does not tolerate an AUA substitution at codon 3, it does tolerate an AUC codon. In any case, the third codon seems to play a critical role in the recognition of both YopQ and YopE as secretion substrates.
FIG. 3.
Synonymous codon substitutions in the yopE minimal secretion signal. All possible synonymous substitutions of codons (which do not alter the amino acid specificity) that were not tested in the experiments described in the legend to Fig. 2 were individually introduced at each position from codon 2 to 7 of the yopE minimal secretion signal and fused to npt. Fusions to Npt were analyzed for type III secretion as described in the legend to Fig. 1.
We sought to ascertain that yopE1-7 and yopE1-7 (A9C) indeed generated the same polypeptide sequence (Fig. 4). Y. enterocolitica strain W22703 cells expressing plasmid-encoded yopE1-7-npt hybrids, were lysed in a French pressure cell, and cytoplasmic protein was prepurified by ion-exchange chromatography on a MonoQ column (Fig. 4A and C). Following separation by SDS-PAGE, proteins were electrotransferred to a PVDF membrane and stained with Coomassie brilliant blue, and amino acid sequences were determined by Edman degradation (Fig. 4B and D). The chromatography data in Fig. 4B and D quantify phenylthiohydantoin amino acyl residues during each of the seven sequencing cycles. The eluted compound of YopE1-7-Npt following the first Edman degradation cycle was the initiator methionine. Every phenylthiohydantoin amino acyl eluting after the initial cycle matched the residues predicted by the sequence of mRNA codons (Fig. 3B and D). Thus, these results suggest that yopE1-7 and yopE1-7 (A9C) indeed generated the identical polypeptide sequence.
FIG. 4.
Edman degradation analysis of the N-termini of YopE1-7-Npt and YopE(A9C)1-7-Npt. (A and C) Purification of YopE1-7-Npt and YopE(A9C)1-7-Npt, respectively. Npt fusions were purified from Y. enterocolitica cell extracts after induction of type III secretion in low-Ca2+ medium by ion-exchange chromatography (MonoQ). Purification was followed by immunoblotting with antisera raised against Npt. Immunoblots and Coomassie-stained polyacrylamide gels separating peak fractions are shown. The stained band subjected to Edman degradation is outlined, and the N-terminal amino acid sequence assigned to each terminus is indicated. (B and D) Edman degradation of YopE1-7-Npt and YopE(A9C)1-7-Npt, respectively. The first seven cycles of analysis are shown, and peak residues are highlighted and listed on the right.
yopE codons 8 to 15 contain additional secretion information.
A previous report had opined that mutational analyses of type III secretion signals that are not performed in the context of a full-length protein are of limited value because they do not represent a physiologically relevant substrate (21). However, since yopQ mRNA encodes secretion information downstream of the minimal secretion signal which suppresses mutations introduced elsewhere, we felt the need to thoroughly examine minimal sections of type III secretion signals before testing their effects in the context of full-length proteins. Accordingly, yop signals longer than the minimal secretion signal required drastic alterations to produce loss of function phenotypes. We therefore wanted to test if the addition of codons downstream of the yopE minimal secretion signal could suppress mutations introduced into codons 2 to 7.
yopE codons 1 to 7 harboring a codon 3 mutation at nucleotide 8, when fused to npt, resulted in the quantitative block of Npt secretion (Fig. 2). When introduced into the context of the first 8, 9, or 10 codons of yopE, the mutation was still unable to promote secretion of the reporter protein (Fig. 5). However, the sequential addition of codons downstream of codon 10 resulted in the steady increase of Npt secretion. When introduced in the context of the first 15 codons, the nucleotide 8 mutation no longer significantly affected the secretion of the reporter protein, as more than 25% of Npt was secreted. Thus, the addition of codons 8 to 15 can suppress minor mutations introduced into the yopE minimal secretion signal. The suppression of this point mutation is reminiscent of the yopQ “suppressor region,” where a nucleotide 8 transversion mutation in the context of 13 to 15 codons is also increasingly suppressed.
FIG. 5.
yopE codons 8 to 15 contain additional secretion information. (A) Suppression of a yopE codon 3 mutation by the addition of downstream codons was measured by introducing the U8A transversion into successively longer yopE secretion signals and fusion to npt. (B) The minimum number of yopE codons required to tolerate a frameshift mutation. One nucleotide immediately following the AUG start codon was deleted to introduce a −1 frameshift in the yopE reading frame. The reading frame was restored by addition of an adenyl immediately after the 3′ end of the yopE sequence before fusion to npt. Fusions to Npt were analyzed for type III secretion as described in the legend to Fig. 1.
Our analysis of the yopQ secretion signal indicated that the suppressor region (yopQ codons 11 to 15) can also suppress frameshift mutations introduced into the minimal secretion signal (27). Furthermore, this suppressor ability is maintained despite the introduction of frameshift mutations to the suppressor region itself (27). This suppression phenomenon is manifested by the inability of shorter secretion signals to tolerate frameshift mutations. Indeed, Wolf-Watz and colleagues have reported that full-length YopE is not secreted when frameshift mutations are introduced into codons 2 to 11 (see Discussion, below) (21). Additionally, we have demonstrated that, while frameshift mutations introduced into the first 15, 14, or 13 codons of yopQ do not abrogate the secretion of a reporter protein, secretion signals comprised of the first 12, 11, or 10 codons of yopQ are unable to tolerate frameshift mutations (27). Therefore, we wished to determine the minimum number of codons that the yopE secretion signal requires to tolerate frameshift mutations.
Codons 1 to 15 of yopE were fused to npt, and the adenyl nucleotide at position 4 was removed to change the reading frame of the secretion signal. The reading frame was restored at the junction to the reporter protein by the addition of an adenyl at position 45. Despite resulting in an entirely new amino acid sequence for the secretion signal, the frameshift did not abolish secretion of the reporter protein (Fig. 5B). Subsequent deletion of codons from the 3′ end of this secretion signal harboring a frameshift mutation resulted in the steady lowering of Npt secretion. Fusion of 12 or fewer codons harboring a −1 frameshift to Npt resulted in barely detectable amounts of the reporter in the culture supernatant, consistent with the observations of Lloyd et al. (21). Thus, like yopQ, codon 13 marks the boundary of tolerance to frameshift mutations in yopE.
Synonymous substitution of yopE codon 3 reduces YopE injection into HeLa cells.
Prior investigations have reported that substitution of codons 2 to 15 of yopE with codons 2 to 15 of nonsecreted proteins such as CAT or β-galactosidase abolish the type III injection of YopE, demonstrating that the first 15 codons of yopE are necessary for the type III injection of YopE (17, 34, 38). We therefore wondered if the introduction of the A9C synonymous codon 3 substitution would hinder the translocation YopE into eukaryotic cells.
We eschewed the introduction of a restriction enzyme site into codons 12 and 13 (which lies within the suppressor region of both yopE and yopQ) and simply PCR amplified the yopE encoding region and introduced the A9C mutation into the 5′ amplifying primer. The result, as confirmed by DNA sequence, was a yopE coding region that is identical to that of wild-type yopE, except at nucleotide position 9. By extension, the predicted amino acid sequence of the mutant YopE was identical to the wild-type sequence. The mutant sequence was cloned downstream of the yopE promoter and expressed from a low-copy-number plasmid in Y. enterocolitica LC1 (ΔyopE). Compared to the secretion of wild-type plasmid-encoded YopE, the secretion of YopE (A9C) was unaffected when the yersiniae were induced for type III secretion in low-calcium medium (Fig. 6A). This is expected, since the presence of downstream sequences suppresses mutations made in the minimal secretion signal (7, 8).
FIG. 6.
Type III injection of YopE into HeLa cells is impaired if a synonymous substitution is introduced into codon 3. (A) Y. enterocolitica LC1 harboring either the plasmid-encoded wild type or the A9C mutant yopE was induced for type III secretion in low-Ca2+ medium. Secretion of YopE was measured as described in the legend to Fig. 1. (B) HeLa cell monolayers were infected with LC1 harboring either the plasmid-encoded wild type or the A9C mutant yopE. After 3 h, the medium was removed and separated by centrifugation to yield supernatant (S) and pellet (P) fractions. The monolayer with adherent bacteria was treated with 1% digitonin and separated by centrifugation to yield supernatant (S) and pellet (P) fractions. Protein samples were treated and analyzed as described in the legend to Fig. 1.
We then infected monolayers of HeLa cells with Yersinia strains harboring either wild-type or the A9C yopE construct for 3 h. The infection media were removed and centrifuged to separate nonadherent bacteria from secreted proteins. The monolayer was treated with 1% digitonin, a nonionic detergent that preferentially disrupts cholesterol-containing membranes (17). The treated monolayers were then centrifuged to separate intact adherent bacteria and cellular debris from liberated eukaryotic cytosolic proteins. The four fractions were separated by SDS-PAGE and analyzed by immunoblotting for various proteins, including those that control for proper fractionation (Fig. 6B).
CAT, a marker for the bacterial cytosol, was found only in the pellet fractions of the infection media and digitonin-solubilized samples (Fig. 6B). This is consistent with the separation of nonadherent bacteria by centrifugation and demonstrates that the digitonin treatment did not lyse adherent bacteria. YopR, a type III secreted protein exclusively sorted to the extracellular milieu, either is found in the supernatant of the infection medium or remains associated with adherent bacteria (17). IκB, a marker for the eukaryotic cytosol, is recovered only in the supernatant of digitonin solubilized material, indicating the quantitative lysis of the HeLa cells. YopH, a positive control for the fitness of the type III injection machinery, is extractable by digitonin. In three trials, the ratio of YopH injection by strain LC1 pYopE(A9C) to LC1 pYopE was 1.03 ± 0.17, suggesting that YopH injection was not affected by the mutation introduced into the plasmid-carried yopE. In contrast, the ratio of YopE injection in three trials was reduced to 0.67 ± 0.13 compared to wild-type YopE (Fig. 6B). In our studies, treatment of the infected monolayer with trypsin protease prior to digitonin extraction did not affect the digitonin solubility of either YopE or YopH (data not shown), suggesting that digitonin-soluble YopE and YopH are found solely in the eukaryotic cytosol. Thus, the secretion of YopE in low-calcium medium was not affected by the introduction of a synonymous mutation at codon 3. However, the effect of a single-nucleotide mutation caused a subtle reduction in the type III injection of YopE during the course of infection.
DISCUSSION
To date, it is clear that yersiniae secrete 14 polypeptides in response to low-calcium conditions in vitro and that a subset of these proteins is translocated into target eukaryotic cells in vivo (10). However, the nagging question of how these proteins are recognized as secretion substrates by the type III secretion machinery has required far more analysis than originally perceived. In accordance with canonical views of protein sorting, the type III secretion signal was mapped to the N-terminal approximately 15 amino acids of various Yops (34). In the absence of a predictable motif encoded in the primary sequence of amino acids, it was assumed that perhaps a structural aspect of Yop N-termini could direct the secretion of Yops (20). Subsequent reports of solution and crystal structures of type III secreted proteins indicated that their N termini usually assume an unordered motif, although exceptions arose where clearly defined folds were apparent (4, 13, 33, 35-37). Thus, in the absence of a conserved identity in either the primary or tertiary structure of Yop amino acids, no model emerged that explained how a polypeptide sequence could direct Yop secretion.
Two lines of evidence intimated that the secretion signal may not be entirely encoded within Yop proteins. First, the secretion signals of many Yops, comprising the first 15 codons, tolerate drastic changes to their reading frame and therefore their amino acid sequence by the simple insertion of nucleotides (1). These results demonstrated that polypeptide sequences of Yop secretion signals could be drastically altered, provided that the mRNA sequence specifying those amino acids remained relatively unchanged. Second, data are now accumulating that demonstrate that the minimal secretion signals of various Yops do not tolerate the synonymous substitutions of codons, even though the codons specify the identical amino acid sequence as the wild-type secretion signal (15, 28). Thus, changes introduced in the mRNA sequence alone can abrogate the function of yop minimal secretion signals.
In this study, we begin to demonstrate the generality of at least three observations that we have made for the yopQ secretion signal. First, yopE mRNA appears to encode information that contributes to the sorting of YopE protein. Consistent with a nonproteinaceous entity, the minimal secretion signal of yopE (codons 1 to 7) does not tolerate the synonymous substitution of at least one codon (codon 3). Further, the yopE secretion signal consisting of at least 13 codons tolerates frameshift mutations that completely alter the amino acid profile of the YopE N terminus. Second, the yopE secretion signal exhibits a bipartite nature, similar to that of the yopQ signal. Accordingly, mutations introduced into the minimal secretion signal may be suppressed by the addition of downstream codons that do not, by themselves, represent an independent secretion signal. In the case of yopE, point mutations introduced into codons 2 to 7 may abrogate the secretion of YopE1-7-Npt. These point mutations may either change the amino acid specificity of a particular codon or alter the reading frame of the entire minimal secretion signal. However, these same mutations do not destroy the function of the signal if approximately 7 to 8 downstream codons are present. Thus, yopE codons 1 to 7 define a minimal secretion signal, whereas codons 8 to 15 comprise a suppressor region that masks mutations introduced into codons 1 to 7. Third, the systematic mutagenesis of yopE codons 2 to 7 revealed nucleotides that contribute to the function of the minimal secretion signal. When aligned with sensitive nucleotides in the yopQ minimal secretion signal, an astonishing pattern emerges (Fig. 6). Codons 2, 3, 5, and 7, but not codons 4 and 6, contribute to the secretion of both YopE and YopQ. To date, these empirical analyses have revealed the most striking similarity between the secretion signals of two type III secreted substrates. The accumulation of mutagenesis data of other yop secretion signals will test whether or not this pattern of sensitive codons is universal. Third, the secretion signals of both yopE and yopQ require at least the first 13 codons to tolerate frameshift mutations. The characteristics of the yopE and yopQ secretion signals are compared and summarized in Fig. 7.
FIG. 7.
Comparison of the yopQ and yopE mRNA type III secretion signals. Nucleotides that comprise the minimal secretion signals of yopQ and yopE are shown in red, and those comprising the suppressor region are shown in blue. Amino acids encoded by the each codon are indicated above the codon using single-letter abbreviations. Nucleotides that are sensitive to mutagenesis are underlined, and codons that contain critical nucleotides are highlighted in yellow. The boundary beyond which frameshift mutations are tolerated for yopQ and yopE secretion signals, codon 13, is also shown.
The ambiguity associated with the final destination of the YopQ polypeptide during the course of infection (19) limited our ability to analyze the effect that mRNA mutations have on the in vivo sorting of full-length YopQ. Thus, we redirected our efforts at analyzing a bona fide injection substrate, YopE. At the onset, it was clear that the yopE secretion signal was much more resilient to silent mutations in the mRNA sequence. We therefore systematically analyzed all possible synonymous substitutions within yopE codons 1 to 7 and found that the replacement of codon 3 (AUA) by AUC destroyed the function of the minimal secretion signal. We then introduced this mutation by PCR amplification into the full-length yopE gene, resulting in an otherwise-unaltered full-length yopE. As expected, this mutation had no discernible effect on YopE secretion in low-Ca2+ medium, since the presence of downstream codons suppressed point mutations introduced into the minimal secretion signal. However, while the low-calcium induction of Yop secretion is an invaluable tool in dissecting type III secretion, it ultimately represents an exaggerated scenario where subtleties may be overlooked. Therefore, we tested the effect of the yopE codon 3 substitution in the type III injection of YopE into HeLa cells. In this situation, which more closely approximates an in vivo infection, the mutation of a single-nucleotide residue that results in the insertion of the wild-type amino acid at position three reduced the translocation of YopE by about 30%. Since the indispensability of yopE codons 1 to 15 in the injection of YopE has been established, further investigations may uncover mutations downstream of codon 7, in the suppressor region, which completely abolish YopE injection.
Our studies were initiated with small segments of yop coding regions fused to reporter proteins to demonstrate the sufficiency of particular secretion signals. In the course of these studies, we uncovered redundancies and subtleties existing within the mechanism whereby Yops are recognized as secretion substrates. The insights gained from these reductionist approaches were then applied to the study of full-length Yops during the course of their translocation into target eukaryotic cells. In this manner, we have begun to explain the discrepant conclusions that have been reached by various investigators in the past. Studies by Lloyd et al. suggested that the mRNA signal hypothesis was an artifactual result stemming from the use of reporter proteins, rather than performing mutagenesis on full-length Yop proteins (21). Specifically, Lloyd et al. demonstrated that introducing a −1 frameshift mutation to the first 11 codons of yopE did not promote secretion of YopE (21). We have, to date, demonstrated that the secretion signals of both yopE and yopQ do not tolerate frameshift mutations introduced into less than the first 13 codons. Why, then, was the frameshift mutation introduced in the study by Lloyd et al. not suppressed by the presence of sequences downstream of codon 11? One possible explanation may involve the construction of the full-length yopE in that study (22). Presumably to aid in cloning, the authors introduced two nucleotide mutations in codons 12 and 13 in order to accommodate the overhangs of an NdeI restriction site. It is conceivable that these substitutions effectively abolished the ability of codons 8 to 15 to suppress the point mutation in codons 2 to 11. Lloyd et al. also clearly demonstrated that the yopE secretion signal is astonishingly resistant to the simultaneous silent change of codons 2 to 10 within the context of the full-length protein, a phenomenon that we, too, have observed within the context of just codons 1 to 7 (22). For reasons that are not immediately obvious, we have observed that just the mutation of yopE nucleotide 9 (normally adenyl) to a cytidyl dramatically affects the function of the yopE minimal signal. As a comparison, the study by Lloyd et al. mutated this residue to uridyl, a substitution which we also demonstrated has no phenotype in the yopE minimal secretion signal (Fig. 2).
Is it conceivable that the use of a specific, modified tRNA molecule during the course of translating yop messages may mediate the export of Yops? In this model, nucleotide sequences downstream of the minimal secretion signal may recruit such tRNA and promote its preferential usage at particular codons within the minimal signal, for example, the isoleucine codon 3 of yopE or yopQ (25). If so, codon usage by specific tRNAs could function as a targeting mechanism that directs ribosomes to the type III machinery. Consistent with this hypothesis, a comparison between the yopQ and yopE minimal secretion signal reveals an obvious importance of certain codons in the recognition of both proteins as substrates for secretion. Codon 3, encoding isoleucine, represents an especially critical codon as it is sensitive to synonymous substitutions in both yopE and yopQ. This model also accounts for the increase in efficiency of secretion of reporter proteins when they are fused to yop segments longer than the minimal signal and also provides a testable hypothesis for the mechanism by which point mutations in the minimal signal are suppressed. The discovery of extragenic factors required for this suppression may eventually explain how an aspect of mRNAs can mediate the secretion of proteins.
Acknowledgments
We thank Ratul Raychauduri for technical assistance, and Joe Sorg and Debra Anderson for critical review of the manuscript.
Protein sequence analysis was provided by The Rockefeller University Protein/DNA Technology Center, which is supported in part by NIH shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment. The work on Yersinia type III secretion is supported by U.S. Public Health Service grant AI42797 to O.S.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., and O. Schneewind. 1999. Type III machines of gram-negative pathogens: injecting virulence factors into host cells and more. Curr. Opin. Microbiol. 2:18-24. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 4.Birtalan, S. C., R. M. Phillips, and P. Gosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 5.Blobel, G. 1980. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 77:1496-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambronne, E. D., and O. Schneewind. 2002. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J. Bacteriol. 184:5880-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: on the role of SycE in targeting YopE into HeLa cells. J. Biol. Chem. 274:22102-22108. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G. R. 2003. How Yop proteins find their way out of Yersinia. Mol. Microbiol. 50:1091-1094. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, G. R., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by the recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87:285-291. [DOI] [PubMed] [Google Scholar]
- 13.Evdokimov, A. G., J. E. Tropea, K. M. Routzahn, T. D. Copeland, and D. S. Waugh. 2001. Structure of the N-terminal domain of Yersinia pestis YopH at 2.0 A resolution. Acta Crystallogr. D Biol. Crystallogr. 57:793-799. [DOI] [PubMed] [Google Scholar]
- 14.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1333. [DOI] [PubMed] [Google Scholar]
- 15.Goss, J. W., J. A. Sorg, K. S. Ramamurthi, H. Ton-That, and O. Schneewind. 2004. The secretion signal of YopN, a regulatory protein of the Yersinia enterocolitica type III secretion pathway. J. Bacteriol. 186:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-572. [DOI] [PubMed] [Google Scholar]
- 17.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 18.Lee, V. T., S. K. Mazmanian, and O. Schneewind. 2001. A program of Yersinia enterocolitica type III secretion reactions is triggered by specific host signals. J. Bacteriol. 183:4970-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, V. T., and O. Schneewind. 1999. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31:1619-1629. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of the type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 23.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mudgett, M. B., O. Chesnokova, D. Dahlbeck, E. T. Clark, O. Rossier, U. Bonas, and B. J. Staskawicz. 2000. Molecular signal required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. USA 97:13324-13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramamurthi, K. S., and O. Schneewind. 2003. Substrate recognition by the Yersinia type III protein secretion machinery. Mol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 26.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed] [Google Scholar]
- 27.Ramamurthi, K. S., and O. Schneewind. 2002. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J. Bacteriol. 184:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramamurthi, K. S., and O. Schneewind. 2003. Yersinia yopQ mRNA encodes a bipartite type III secretion signal in the first fifteen codons. Mol. Microbiol. 50:1189-1198. [DOI] [PubMed] [Google Scholar]
- 29.Rosqvist, R., K.-E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russmann, H., T. Kubori, J. Sauer, and J. E. Galan. 2002. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46:769-779. [DOI] [PubMed] [Google Scholar]
- 31.Schesser, K., E. Fritzh-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhavy, T. J. 1997. Death by lethal injection. Science 278:1085-1086. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. L., P. Khandelwal, K. Keliikuli, E. R. Zuiderweg, and M. A. Saper. 2001. Structure of the type III secretion and substrate binding domain of Yersinia YopH phosphatase. Mol. Microbiol. 42:967-979. [DOI] [PubMed] [Google Scholar]
- 34.Sory, M.-P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA. 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stebbins, C. E., and J. E. Galan. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 36.Vonderviszt, F., R. Ishima, K. Akasaka, and S. Aizawa. 1992. Terminal disorder: a common structural feature of axial proteins in the bacterial flagellum? J. Mol. Biol. 226:575-579. [DOI] [PubMed] [Google Scholar]
- 37.Vonderviszt, F., M. Sonoyama, M. Tasumi, and K. Namba. 1992. Conformational adaptability of the terminal regions of flagellin. Biophys. J. 63:1672-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woestyn, S., M.-P. Sory, A. Boland, O. Lequenne, and G. R. Cornelis. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20:1261-1271. [DOI] [PubMed] [Google Scholar]