Abstract
A novel staphylolytic enzyme, ALE-1, is a glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. ALE-1 possesses seven histidines. Chemical modification studies using diethylpyrocarbonate and iodoacetic acid suggested that a histidine or tyrosine residue(s) in the molecule is important for the organism's staphylolytic activity. All of the histidine residues, one tyrosine, and one aspartic acid residue in the N-terminally truncated ALE-1 (ΔN-term ALE-1) were systematically altered by site-directed mutagenesis, and the enzyme activities and metal contents of the variants were measured. Our studies indicated that His-150, His-200, His-231, His-233, and Asp-154 are essential for the enzyme activity of ΔN-term ALE-1. Except for His-150 and Asp-154, all of these amino acids were located within the 38-amino-acid region conserved among 11 proteins, including 5 staphylolytic endopeptidases. Inductively coupled plasma-mass spectrometric analysis of ΔN-term ALE-1 revealed that it contains one atom of zinc per molecule. Measurement of the zinc content of the mutant ΔN-term ALE-1 suggested that His-150 and -233 are important for zinc binding; their loss in these variant enzymes coincided with the loss of staphylolytic activity. These results strongly suggest that ALE-1 is a novel member of zinc metalloproteases.
A staphylolytic enzyme, ALE-1, acting on Staphylococcus aureus is a glycylglycine endopeptidase produced by Staphylococcus capitis EPK1 (26). Molecular cloning and subsequent sequencing of the ALE-1 structural gene (ale-1) established that the gene product is composed of 362 amino acid residues and is synthesized as a precursor protein which is cleaved after Ala at position 35, thus producing a mature ALE-1 of 35.6 kDa. The primary structure of mature ALE-1 is very similar to that of the proenzyme form of lysostaphin. Lysostaphin is a well-known glycylglycine endopeptidase produced by Staphylococcus simulans biovar staphylolyticus (5, 24). Like ALE-1, lysostaphin is very effective against S. aureus and also effective against Staphylococcus epidermidis, though it needs a higher concentration of the enzyme (31). It has been extensively used as a tool to lyse staphylococci in the field of basic research. Recently, lysostaphin has gained renewed interest as an antistaphylococcal agent because of the increasing emergence of antibiotic-resistant staphylococci in communities and clinics (7, 13, 19). ALE-1 and prolysostaphin possess similar modular designs of an N-terminal domain with tandem repeats of a 13-amino-acid sequence fused to the active-site-containing domain (26). Prolysostaphin undergoes proteolytic processing of the N-terminal repeat domain when it is secreted in broth culture (9, 17, 21, 27). The N-terminal repeat domain of prolysostaphin was shown not to be essential for staphylolytic activity (27). ALE-1 does not undergo processing in broth culture, unlike lysostaphin, but the N-terminal repeat domain of ALE-1 was not essential for staphylolytic activity either (26). Chemical modification studies using diethylpyrocarbonate and iodoacetic acid suggested that histidine and/or tyrosine, which are potential zinc ligands, are important for the staphylolytic activity of ALE-1 and lysostaphin. ALE-1 possesses seven histidines. A protein homology search suggested that ALE-1 and lysostaphin share a homologous 38-amino-acid-containing motif, Tyr-X-His-X11-Val-X12/20-Gly-X4-5-His, with 18 proteins, including β-metalloproteases, staphylolytic enzymes, such as the newly identified staphylolytic protein LytM, and proteins belonging to the LppB/NlpD lipoprotein family. The histidine residues in the 38-amino-acid region in β-metalloprotease are suggested to correspond to a zinc ligand because of their sequence similarity to the His-X-His motif in carbonic anhydrase (15), although the typical zinc binding motif is His-Glu-X-X-His in metalloproteases (1, 10). Recently, the crystal structure of LytM, with which ALE-1 has high homology in amino acid sequence, was solved at a 1.3- Å resolution (18). In the LytM structure, zinc is tetrahedrally coordinated by the side chains of Asn-117, His-210, Asp-214, and His-293. His-210, His-293, and Asp-214 in LytM correspond to His-150, His-233, and Asp-154 in ALE-1, respectively. Considerable similarity was also found in a sequence of 92 C-terminal amino acid residues with S. aureus LytA and lysostaphin (25, 26, 30). This region is suggested to be a domain that targets staphylococcal cells (3).
The present study was undertaken to identify which histidine(s) is essential for the staphylolytic activity of ALE-1 and to evaluate whether ALE-1 is a metalloprotease. Accordingly, all of the histidine residues in the molecule and one tyrosine residue in the 38-amino-acid domain in ALE-1 were systematically altered by site-directed mutagenesis, and the staphylolytic activities, endopeptidase activities, and metal contents of the wild type and variants were measured.
Our studies indicated that His-150, His-200, His-231, His-233, and Asp-154 are essential for staphylolytic activity. With the exception of His-150 and Asp-154, all of the amino acids were located within the homologous 38-amino-acid domain. Inductively coupled plasma-mass spectrometry (ICP-MS) analysis confirmed that ALE-1 contains one molecule of zinc per molecule. Our data indicate that His-150 and His-233 are important for zinc binding; their loss in these variant enzymes coincided with the loss of staphylolytic activity. These results strongly suggest that ALE-1 is a new member of the bacterial zinc metalloprotease.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotide primers.
The bacterial strains used in this study are described in Table 1. Staphylococcus and Escherichia coli were grown in Trypticase soy broth (Becton Dickinson Microbiology Systems, Cockeysville, Md.) and Luria-Bertani broth (5 g of yeast extract, 10 g of polypeptone, 10 g of NaCl per liter [pH 7.2]), respectively. Manipulation of DNA in E. coli XL1-Blue (6) was carried out with the pGEM-T Easy vector system (Promega Co., Madison, Wis.) and pQE30 expression vector (QIAGEN, Tokyo, Japan). When necessary, ampicillin (100 μg/ml) or kanamycin (25 μg/ml) was added for selection or maintenance of plasmid. Oligonucleotides used in this study are described in Table 2.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Vector | Cloning site | Relevant characteristic(s) | Source or reference |
|---|---|---|---|---|
| Strains | ||||
| S. aureus | ||||
| FDA209P | ATCC 6538 | |||
| E. coli | ||||
| XL1-Blue | rec1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZM15 Tn10 (Tet)] | 6 | ||
| M-15 pREP4 | Nals Strs Rifs Thi− Lac− Ara+ Gal+ Mtl− F RecA+ Uvr+ Lon+ | QIAGEN | ||
| TF566 | M-15 pTF561 | This study | ||
| TF578 | M-15 pTF570 | This study | ||
| TF605 | M-15 pTF595 | This study | ||
| TF724 | M-15 pTF721 | This study | ||
| TF606 | M-15 pTF596 | This study | ||
| TF608 | M-15 pTF598 | This study | ||
| TF607 | M-15 pTF597 | This study | ||
| TF583 | M-15 pTF575 | This study | ||
| TF609 | M-15 pTF600 | This study | ||
| TF585 | M-15 pTF577 | This study | ||
| Plasmids | ||||
| pGEM-T Easy | E. coli cloning vector for PCR products | Promega | ||
| pQE-30 | E. coli expression vector | QIAGEN | ||
| pTF561 | pQE-30 | BamHI-SacI | 0.9-kbp BamHI-SacI fragment containing ΔN-term ALE-1 | This study |
| pTF570 | pQE-30 | BamHI-HindIII | 0.8-kbp BamHI-HindIII fragment containing H124A ΔN-term ALE-1 | This study |
| pTF595 | pQE-30 | BamHI-PstI | 0.9-kbp BamHI-PstI fragment containing H150A ΔN-term ALE-1 | This study |
| pTF721 | pQE-30 | BamHI-SacI | 0.9-kbp BamHI-PstI fragment containing D154A ΔN-term ALE-1 | This study |
| pTF596 | pQE-30 | BamHI-PstI | 0.9-kbp BamHI-PstI fragment containing H194A ΔN-term ALE-1 | This study |
| pTF598 | pQE-30 | BamHI-PstI | 0.9-kbp BamHI-PstI fragment containing Y198A ΔN-term ALE-1 | This study |
| pTF597 | pQE-30 | BamHI-PstI | 0.9-kbp BamHI-PstI fragment containing H200A ΔN-term ALE-1 | This study |
| pTF575 | pQE-30 | BamHI-HindIII | 0.8-kbp BamHI-HindIII fragment containing H231A ΔN-term ALE-1 | This study |
| pTF600 | pQE-30 | BamHI-PstI | 0.9-kbp BamHI-PstI fragment containing H233A ΔN-term ALE-1 | This study |
| pTF577 | pQE-30 | BamHI-HindIII | 0.8-kbp BamHI-HindIII fragment containing H327A ΔN-term ALE-1 | This study |
TABLE 2.
Oligonucleotide primers used
| Deletion or mutation | Primer | Sequence (5′ to 3′) | Positions |
|---|---|---|---|
| ALEU | AGGGATCCTTTGTAAGAGAAG | 577-597 | |
| ALER | GGGAATTCATGGGTAGTGATA | 1352-1372 | |
| pQEUV | CGGATAACAATTTCACACAG | ||
| pQERV | GTTCTGAGGTCATTACTGG | ||
| ΔN-term ALE-1 | ALEU4 | CGCCTCGAGTTTGTAAGAGAAGCT | 575-599 |
| ALEL4 | TTCTGCAGTATGGGTAGTGATA | 1452-1474 | |
| H124A | ALEMH1-UV | CTCAATCTAATGCTTCGGCTAGTTGGTTAA | 600-630 |
| ALEMH1-RV | CAACTAGCCGAAGCATTAGATTGAG | 600-625 | |
| H150A | ALEMH2-UV | TGGCGGAAATGCTTATGGCGTTGATTTCTT | 679-709 |
| ALEMH2-RV | ACGCCATAAGCATTTCCGCCATTAA | 676-700 | |
| D154A | ALE-D154A-u | CTATGGCGTTGCTTTCTTTATGAATGTA | 690-719 |
| ALE-D154A-r | CATAAAGAAAGCAACGCCATAGTGATTTC | 684-713 | |
| H194A | ALEMH3-UV | TGATGGTGTTGCTAGACAATGGTATATG | 812-839 |
| ALEMH3-RV | ACCATTGTCTAGCAACACCATCATT | 810-834 | |
| Y198A | ALEMY8-UV | AGACAATGGGCTATGCATTTAAGTAA | 825-850 |
| ALEMY8-RV | ATGCATAGCCCATTGTCTA | 824-842 | |
| H200A | ALEMH4-UV | ATGGTATATGGCTTTAAGTAAATTCAATGT | 830-859 |
| ALEMH4-RV | TACTTAAAGCCATATACCATTGTCT | 825-849 | |
| H231A | ALEMH5-UV | CTACAGCACCGGCTTTACATTTTCAAA | 922-948 |
| ALEMH5-RV | GAAAATGTAAAGCCGGTGCTGTAGC | 921-945 | |
| H233A | ALEMH6-UV | ACCGCATTTAGCTTTTCAAAGAATGACC | 929-956 |
| ALEMH6-RV | TTGAAAAGCTAAATGCGGTGCTGTA | 923-947 | |
| H327A | ALEMH7-UV | AAACAAGATGGTGCTGTATGGGTTGGTTATA | 1209-1239 |
| ALEMH7-RV | ACCCATACAGCACCATCTTGTTTCA | 1207-1231 |
Materials and chemicals.
All restriction enzymes and DNA-modifying enzymes were obtained from Roche (Mannheim, Germany) or Takara Shuzo Co. Ltd. (Kyoto, Japan). Other materials and chemicals used were from commercial sources.
DNA manipulations.
Routine DNA manipulations, such as DNA digestion with restriction enzymes, DNA ligations, gel electrophoresis, and DNA sequencing, were performed essentially as described previously (22). The sequences of both DNA strands were determined by the dideoxy chain termination method (23).
Site-directed mutagenesis.
A DNA fragment of the ale-1 gene, encoding the reading frame with multiple truncated N-terminal repeats (ΔN-term ALE-1) was amplified by PCR with the primers ALEU and ALER and cloned into the pGEM-T Easy vector (Promega). A 0.9-kb BamHI-SacI fragment of the recombinant plasmid was cloned in frame downstream from the His tag sequence in the pQE30 expression vector (QIAGEN) and was used as a template for site-directed mutagenesis. PCR-based site-specific mutagenesis was performed to systematically alter each of the histidines or tyrosines by the overlap extension method as described previously (11). Two separate PCRs (PCR1 and PCR2) were performed to construct each mutated fragment. PCR1 involved two separate amplifications, each of which used a unique set of primers. One of the primer sets consisted of pQEUV, which annealed upstream of the His tag sequence in pQE30, and an antisense primer which introduced the desired mutation. The other primer sets consisted of the sense primer, which was complementary to the mutated antisense primer, and pQERV, which annealed downstream of the multicloning site in pQE30. Following PCR1, two partial ale-1 genes with the desired introduced mutation were isolated by agarose gel electrophoresis and were used as a template for the second PCR2. PCR2 was performed in the presence of primers pQEUV and pQERV and these templates. Consequently, the annealed overlapping strands functioned as the initial primers for the strand completion. Then the subsequent amplification of the complete mutant ale-1 gene was carried out with the primers pQEUV and pQERV. The ∼0.8- to ∼0.9-kb BamHI-HindIII, PstI, or SacI mutant fragment was cloned into pQE30. All mutants were confirmed by sequencing with the Thermo sequence fluorescently labeled primer cycle sequencing kit (Amersham Life Science, Little Chalfont, Buckinghamshire, England). For expression of the recombinant protein, the resulting plasmid was transformed into E. coli M-15(pERP4) (QIAGEN).
Expression and purification of the wild-type and variant His-tagged protein.
Transformants carrying the wild-type or mutant ale-1 gene were cultured at 37°C with vigorous shaking until an optical density (OD) value of 0.5 was reached. Then the expression of the His-tagged protein was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 4 h of incubation, cells were harvested by centrifugation, resuspended in lysis buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 8.0]), and incubated for 30 min at room temperature. The cells were then disrupted with an Ultrasonic disruptor (Tomy Seiko, Tokyo, Japan). After centrifugation at 25,000 × g for 40 min at 4°C to remove cellular debris, a Ni-nitrilotriacetic acid (NTA) matrix (QIAGEN) that was equilibrated with lysis buffer B was added to the cleared lysate and mixed gently by shaking for 30 min at room temperature. The lysate-Ni-NTA mixture was then loaded into a column, which was then washed with a 5× bed volume of wash buffer C (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 6.8]). Then the recombinant protein was eluted by eluate buffer E (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 4.5]), and the eluted fraction was dialyzed against the solution containing 4 M urea and 0.1 M phosphate buffer (pH 6.8) and finally against 0.1 M phosphate buffer (pH 6.8).
Assay of enzyme activity.
The lytic activity of wild-type or variant ΔN-term ALE-1 was assayed by zymography or turbidimetry. Separately, endopeptidase activity was measured by quantitation of increased free amino groups by using S. aureus peptidoglycan as the substrate. Zymography was performed as described previously (26). Briefly, purified recombinant proteins were electrophoresed in 12% polyacrylamide gel containing heat-killed S. aureus FDA209P cells (0.5 mg [dry weight]/ml) under denaturing condition in the presence of sodium dodecyl sulfate (SDS). After electrophoresis, gels were washed with distilled water and incubated with 0.1 M phosphate buffer (pH 6.8) at 37°C. Staphylolytic activity was visualized as a clear lytic zone with Immunoviewer MU (Jookoo Sangyo Co. Ltd., Tokyo, Japan). Turbidimetry was performed using SDS-treated and heat-killed S. aureus FDA209P cells. Cells were suspended in 0.1 M Tris-HCl (pH 8.5) (1 mg [dry weight] of cells/ml). Recombinant protein was added to 2 ml of the cell suspension and incubated at 37°C. The rate of decrease in turbidity was measured at an OD at 595 nm (OD595) in a spectrometer. To measure endopeptidase activity, purified peptidoglycan of S. aureus FDA209P was used. Peptidoglycan was prepared as described by Maidhof et al. (16). A sample of the appropriate dilution of the test specimen was mixed with 200 μl of the peptidoglycan suspension in 0.1 M phosphate buffer (pH 6.8) (OD595 = 0.8). After incubation at 37°C for 90 min, the concentration of free amino groups (fragments of enzymatically hydrolyzed peptidoglycan), which appeared in the soluble fraction, was determined by a Ghuysen procedure using 1-fluoro-2,4-dinitrobenzene. The supernatant was incubated with 133.6 μl of 2% sodium borate and 3.3 μl of 0.1 M 1-fluoro-2,4-dinitrobenzene at 60°C for 30 min in the dark. Then the dinitrophenyl derivative was hydrolyzed by adding 733.3 μl of 6 N HCl at 100°C for 20 min. The hydrolyzed sample was diluted with 560 μl of distilled water, and the concentration of the free amino group was estimated by measuring the OD420 and using 2,4-dinitrophenyl-alanine as a reference.
Binding assay.
S. aureus FDA209P cells grown in Trypticase soy broth to mid-log phase were suspended with 0.1 M phosphate buffer (pH 6.8) containing 4% SDS, incubated in boiling water for 1 h, and then washed until the SDS was removed. The obtained SDS-treated, heat-killed cells were suspended with 0.1 M phosphate buffer (pH 6.8) containing the endopeptidase inhibitor iodoacetic acid at 0.1 M (OD595 = 1.0). A 100-μl mixture of cell suspension and recombinant protein was incubated for 1 h at 4°C. After the cells were washed three times with 0.1 M phosphate buffer (pH 6.8) containing 0.1 M iodoacetic acid, the bound recombinant protein was eluted with 4% SDS. The eluted recombinant protein was separated by SDS-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide gel) and stained with Coomassie brilliant blue. The amount of bound recombinant protein was estimated by NIH-Image version 1.52 with the scanned protein band.
Susceptibility to trypsin digestion.
Wild-type or variant ΔN-term ALE-1 was incubated with 0.05 μg of trypsin (Nakalai Tesque, Kyoto, Japan) in 20 mM Tris-HCl (pH 8.0) for 30 min at 37°C. The reaction mixture was separated by SDS-PAGE (12% polyacrylamide gel) and stained with Coomassie brilliant blue.
Circular-dichroism spectra.
Circular-dichroism spectra were recorded in a J-600 spectropolarimeter (JASCO, Tokyo, Japan) fitted with a thermostatted cell holder and with a thermostatic bath. Far-UV spectra were recorded in 0.1-cm-path-length quartz cells at a protein concentration of 0.2 mg/ml.
Screening for the presence of metals and determination of zinc content in wild-type and variant ΔN-term ALE-1.
The presence of metals and the amounts of zinc in the recombinant protein were determined by using ICP-MS (model 4500 spectrometer; Agilent). The purified recombinant protein was dialyzed against 0.1 M phosphate buffer (pH 6.8) and was decomposed by heating it with nitric acid and sulfuric acid, resolved in diluted nitric acid, and used for analysis.
Other procedures.
SDS-PAGE and zymography for bacteriolytic enzymes were carried out as described previously (26). Protein concentrations were determined with the Bio-Rad (Richmond, Calf.) protein assay reagent, with bovine serum albumin used as the standard.
RESULTS
Purification of wild-type and variant ΔN-term ALE-1.
ALE-1 contains seven histidine residues at positions 124, 150, 194, 200, 231, 233, and 327, all of which are conserved in lysostaphin (26) (Fig. 1A). Moreover, a protein alignment study revealed that His-200, His-233, and Tyr-198 are thoroughly conserved among 18 proteins sharing a homologous 38-amino-acid region and that His-150, His-200, His-231, His-233, and Asp-154 are also conserved in LytM (Fig. 1). To examine which amino acid is important for staphylolytic activity, we altered ΔN-term ALE-1 at His-124, His-150, His-194, His-200, His-231, His-233, His-327, and the conserved Asp-154 and Tyr-198 to Ala by site-directed mutagenesis. These His-tagged recombinant proteins were purified with a Ni-NTA matrix from the lysate of E. coli transformants carrying the wild-type ΔN-term ALE-1 gene or the variant ΔN-term ALE-1 genes. Purified mutant ΔN-term ALE-1 with the Ala replacement indicated one band on SDS-PAGE, and molecular masses of the purified variant ΔN-term ALE-1 were approximately 30 kDa.
FIG. 1.
(A) Schematic representation of ALE-1, ΔN-term ALE-1, and LytM, indicating the location of the conserved 38-amino-acid region and relevant amino acids. (B) Amino acid sequence alignments of 18 proteins sharing the homologous 38-amino-acid region. Conserved or semiconserved residues which have been mutated in ΔN-term ALE-1 by site-directed mutagenesis are in boxes. A bar indicates a His-X-His-like motif. Abbreviations (GenBank accession numbers in parentheses): A. l. protease, beta-lytic metalloproteases from Achromobacter lyticus (P27458); L. e. metalloprotease, beta-lytic metalloprotease from Lysbacter enzymogenes (P00801); LasA, LasA from Pseudomonas aeruginosa (A33661); Preprolysostaphin, lysostaphin precursor (X06121, M15686); PreALE-1, ALE-1 precursor (D86328); LytM, S. aureus autolysin (L77184); S. z. ZooA, ZooA from Streptococcus zooepidemicus (U50357); E. c. OrfU, Escherichia coli OrfU (P24204); H10409, Haemophilus influenzae hypothetical protein (P44693); TagE, Vibrio cholerae ToxR-activated protein TagE (JC2569); S. e. Orf1, Synechococcus elongates Orf1 (D13173); H. i. NlpD, NlpD of H. influenzae (P44833); E. c. NlpD, NlpD of E. coli (P33648); S. t. NlpD, NlpD of Salmonella enterica serovar Typhi (X81641); H. s. Lppb, LppB of Haemophilus somnus (P36685); Y. e. NlpD, NlpD of Yersinia enterocolitica (U16312); P. a. NlpD, NlpD of Pseudomonas aeruginosa (P45682); P. p. NlpD, NlpD of Pseudomonas putida (X91546).
Limited digestion by trypsin of wild-type and variant ΔN-term ALE-1.
To test whether replacement of histidine or tyrosine residues of the wild-type ΔN-term ALE-1 causes conformational changes, wild-type and variant ΔN-term ALE-1 were analyzed by digesting them with trypsin for 30 min at 37°C. Similar digestion patterns were observed with the wild-type and variant ΔN-term ALE-1 (data not shown).
Circular-dichroism spectroscopy of wild-type and variant ΔN-term ALE-1.
To further assess the effect of the replacement of the histidine or tyrosine residues on the secondary structure of wild-type ΔN-term ALE-1, the far-UV spectra of the wild-type and variant proteins were recorded. The far-UV spectrum of wild-type ΔN-term ALE-1 is characterized by a large positive band at 224 nm, which is attributable to the chiral contribution of aromatic side chains (2, 4, 8), and a minor positive shoulder at 209 nm. The far-UV spectra of variant enzymes were very similar to that of the wild type, although the minor induction of a positive band at 204 nm was noted in the Ala mutant at His-150, His-200, His-231, and His-233. These results suggested that there were no significant changes in secondary structure in these variant enzymes studied.
Staphylolytic and endopeptidase activities of the wild-type and the variant ΔN-term ALE-1.
To determine the consequences of ALE-1 mutation, the staphylolytic activities of wild-type and variant ΔN-term ALE-1 were assayed by zymography and by turbidimetry. In both assays, the complete loss of staphylolytic activity was observed in variant enzymes with His-150, His-200, His-231, His-233, and Asp-154 replaced with Ala (Fig. 2A and B). On the other hand, mutations at His-124, His-194, His-327, and Tyr-198 had no effect on these staphylolytic activities. To study whether this loss of staphylolytic activity corresponded to the loss of endopeptidase activity, the endopeptidase activities of wild-type and variant ΔN-term ALE-1 were characterized by measuring the increased amount of free N-terminal amino groups induced by hydrolysis of the purified peptidoglycan by these enzymes. As shown in Fig. 2C, the loss of staphylolytic activities in variant enzymes with a mutation at His-150, His-200, His-231, His-233, and Asp-154 coincided well with the loss of endopeptidase activities in these enzymes. These results suggested that mutation at His-150, His-200, His-231, His-233, and Asp-154 to Ala resulted in the loss of endopeptidase activity in these variant enzymes.
FIG. 2.
Enzyme activities of ΔN-term ALE-1 variants. Staphylolytic activity was measured by either zymography (A) or turbidimetry (B). (A) Wild-type ΔN-term ALE-1 or variant enzymes (0.135 μg) were electrophoresed in 12% polyacrylamide gel with (upper panel) or without (lower panel) S. aureus FDA209P cells in the presence of SDS. After electrophoresis, the gel without bacteria was subjected to Coomassie blue staining and the gel with bacteria was washed in distilled water and further incubated in 0.1 M phosphate buffer (pH 6.8) and lytic bands were photographed. (B) Wild-type ΔN-term ALE-1 orvariant enzyme (1.35 μg) was incubated with a 2-ml cell suspension of heat-killed S. aureus FDA209P in 0.1 M Tris-HCl (pH 8.5) for 4 h at 37°C. The decrease in turbidity was measured at 595 nm in a spectrometer. The initial OD595 of the cell suspension was 0.55, and the OD595 decreased to 0.15 after the incubation with wild-type ΔN-term ALE-1. Activity was expressed as the relative percentage of the decrease in the turbidity of the cell suspension incubated with wild-type ΔN-term ALE-1. Data are representative of three independent experiments. Endopeptidase activity (C) was determined by measuring the amount of free N-terminal amino groups that was increased by hydrolyzing the S. aureus FDA209P peptidoglycan with the wild-type ΔN-term ALE-1 or variant enzymes. A 200-μl mixture of peptidoglycan suspension (OD595 = 0.8) and recombinant protein (17.4 μg) in 0.1 M phosphate buffer (pH 6.8) was incubated for 1.5 h at 37°C. After incubation, the amount of free N-terminal amino group was measured at 420 nm in a spectrometer. Activity was expressed as the percentage relative to the amount of the free N-terminal amino group from peptidoglycan by incubation with wild-type ΔN-term ALE-1. Data are representative of three independent experiments.
Binding of variant ΔN-term ALE-1 to S. aureus cells.
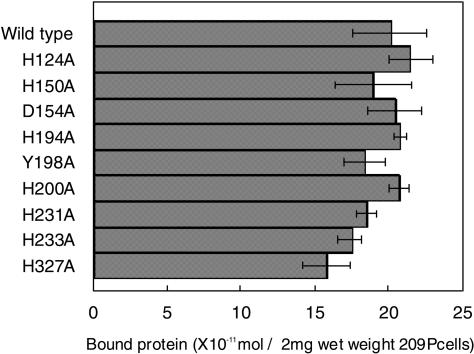
The variant ΔN-term ALE-1 was also tested to determine whether the alteration of histidine, tyrosine, or aspartic acid residues had any effect on enzyme affinity to S. aureus cells. After incubation of S. aureus 209P SDS-treated, heat-killed cells with wild-type or variant ΔN-term ALE-1 for 1 h at 4°C, cells were washed, and bound protein was eluted with 4% SDS and analyzed by SDS-PAGE. Under the experimental conditions, wild-type ΔN-term ALE-1 did not show any staphylolytic activity in cell suspension. When wild-type ΔN-term ALE-1 was incubated with 1 mg (dry weight) of heat-killed S. aureus cells, dose-dependent binding of the enzyme to the cells was observed. The amount of maximum binding of wild-type ΔN-term ALE-1 to S. aureus was 25 × 10−11 mol/mg (dry weight) of cells. Therefore, 12.5 ×10−11 mol of each variant ΔN-term ALE-1 was used for binding to 1 mg (dry weight) of heat-killed S. aureus cells. All variant ΔN-term ALE-1s showed levels of binding to heat-killed S. aureus cells almost identical to those of wild-type ΔN-term ALE-1, suggesting that the mutation of histidine, tyrosine, and aspartic acid had no effect on the binding of the variant enzyme to the target cells (Fig. 3).
FIG. 3.
Binding activity of variant ΔN-term ALE-1 to S. aureus cells. Wild-type ΔN-term ALE-1 or variant enzyme was incubated with SDS-treated, heat-killed S. aureus FDA209P cells (0.5 mg [dry weight]) suspended in 100 μl of 0.1 M phosphate buffer (pH 6.8) containing 0.1 M iodoacetic acid for 1 h at 4°C. After several washes with 0.1 M phosphate buffer (pH 6.8), the cells were harvested and suspended with 4% SDS. The supernatant was separated by SDS-PAGE (12% polyacrylamide gel) and stained with Coomassie brilliant blue. The amount of bound recombinant protein was measured with an image scanner connected to the computer and estimated by NIH-Image version 1. Data are representative of three independent experiments.
Zinc content in wild-type and variant ΔN-term ALE-1.
Lysostaphin has been reported to be a zinc enzyme which contains one zinc atom per molecule (28). We previously demonstrated that ALE-1 contains one zinc atom per molecule (26). Although both enzymes do not possess the primary sequence motif for bacterial zinc metalloproteases, HEXXH, it has been pointed out that both possess His-Leu-His (His is at positions 231 and 233 in ALE-1), which is similar to the His-X-His sequence serving as the zinc ligand in carbonic anhydrase. Furthermore, this sequence is well conserved in several staphylolytic endopeptidases, including Lysbacter enzymogenes and Achromobacter lyticus β-lytic proteases, Pseudomonas aeruginosa LasA, and S. aureus LytM (Fig. 1). Inhibitor experiments suggested that β-lytic proteases and LasA are zinc metalloproteases. However, there is no systemic study to assess the correlation between the metal content and staphylolytic activities of these staphylolytic enzymes. Therefore, we reevaluated the heavy metal of wild-type ΔN-term ALE-1 by ICP-MS and assessed the metal contents of the variant enzymes. As shown in Table 3, ICP-MS revealed that wild-type ΔN-term ALE-1 contains one zinc atom per molecule. To determine whether the loss of staphylolytic activity of some variant ΔN-term ALE-1s was due to the loss of the zinc molecule, the zinc contents of variant ΔN-term ALE-1s were measured. The zinc contents of variant ΔN-term ALE-1s were significantly lower in those variants with mutations at His-150 and His-233 (Table 3). On the other hand, mutation at His-200, His-231, or Asp-154 did not cause a complete loss of the zinc content of the variant enzyme, although the enzymes lost staphylolytic as well as endopeptidase activity (Table 3 and Fig. 2).
TABLE 3.
Zinc contents of wild-type and variant enzymes
| Mutation | Enzymatic activity | Zn content (mol/mol of protein)a |
|---|---|---|
| Wild type | + | 1.0 |
| H124A | + | 0.9 |
| H150A | − | 0.1 |
| D154A | − | 0.6 |
| H194A | + | 0.5 |
| Y198A | + | 1.2 |
| H200A | − | 0.6 |
| H231A | − | 0.7 |
| H233A | − | 0.1 |
| H327A | + | 0.5 |
The amounts of zinc in wild-type or variant enzymes were determined by ICP-atomic emission spectrometry as described in Materials and Methods.
DISCUSSION
Here we investigated the role of histidine, tyrosine, and aspartic acid residues in the staphylolytic activity of ALE-1 by site-directed mutagenesis. The change of His-150, His-200, His-231, His-233, and Asp-154 to Ala resulted in an almost complete loss of enzymatic activity, although their binding activities to the substrate cells were almost identical to that of wild-type ΔN-term ALE-1 (Fig. 2 and 3). ICP-MS analysis indicated that a mutation at His-150 or His-233 resulted in the loss of a zinc molecule. His-200, His-231, and His-233 are part of a homologous 38-amino-acid region shared by 18 proteins (Fig. 1B). Among the 18 proteins, LasA, lysostaphin, ALE-1, and two β-metalloproteases are endopeptidases (12, 26, 29). Also, LasA, lysostaphin, and ALE-1 exhibited staphylolytic activity. LytM is a newly identified staphylolytic protein with a 38-amino-acid region (20). The two β-metalloproteases have bacteriolytic activities (15). On the other hand, seven of the proteins belong to the NlpD/LppB lipoprotein family. NlpD was suggested to have cell wall lytic activity (14), but there is no evidence that it has staphylolytic activity. We constructed the recombinant plasmid, which encodes His-tagged NlpD from E. coli and purified the recombinant His-tagged NlpD as a protein of 41 kDa. His-tagged NlpD was assayed for staphylolytic activity by using turbidimetry or zymography. The results showed that NlpD had no staphylolytic activity (data not shown). This result suggested that proteins belonging to the NlpD/LppB family sharing a 38-amino-acid region have amino acid sequences similar to those of staphylolytic enzymes, but they have distinct functions.
Recently, the crystal structure of LytM was solved at a 1.3-Å resolution (18). In the LytM structure, zinc is tetrahedrally coordinated by the side chains of Asn-117, His-210, Asp-214, and His-293. Interestingly, full-length LytM is a latent enzyme. Deprivation of Asn-117 coordination by site-directed mutagenesis of Asn-117 or by deletion of the N-terminal segment containing Asn-117 resulted in activation of LytM. Among the 18 homologous proteins, the amino acid corresponding to Asn-117 in LytM is not conserved at all. For instance, the amino acid corresponding to Asn-117 in LytM is Ser in lysostaphin and Pro in ALE-1, both of which are located in the N-terminal tandem repeat region, which is not essential for staphylolytic activity. His-210, His-293, and Asp-214 in LytM correspond to His-150, His-233, and Asp-154 in ALE-1, respectively (Fig. 1). This result is in part in agreement with our observation that the mutation of either His-150 or His-233 to Ala resulted in loss of zinc together with a loss of endopeptidase activity, suggesting that these histidines are zinc ligands in ALE-1. On the other hand, mutation at Asp-154 did not result in a complete loss of zinc, although the variant enzyme completely abolished the endopeptidase activity. This result is apparently inconsistent with the data for the LytM crystal structure. Several interpretations of this result are possible. Since His-260 and His-291 in LytM are spatially proximate to the zinc ion in LytM, it is possible that His-200 or His-231 (equivalent to His-260 or His-291 in LytM) may compensate for the zinc coordination of Asp-154 in ALE-1. Another possibility is that Asp-154 is involved in the activation of zinc-bound H2O so that the mutation may abolish the enzyme activity but the zinc remains intact. Or Asp-154 is simply not a ligand for zinc and another amino acid acts as the ligand for the zinc. Previously, the histidines in the His-X-His sequence found in staphylolytic enzymes sharing the 38-amino-acid region (for instance, His-231-Leu-232-His-233 in ALE-1) have been suggested to be the unique zinc ligands of novel bacterial metalloproteases (26). However, our results have contradicted the notion and suggest that His-231 may not be a ligand for zinc. Also, the structure of LytM clearly indicated that His-291 (equivalent to His-231 in ALE-1) is not a ligand for zinc (18). Nevertheless, the site-directed mutagenesis assay demonstrated that His-291 is essential for the staphylolytic and endopeptidase activities of the N-terminally deleted active form of LytM. Consistently, our results indicated that His-231 is essential for the staphylolytic and endopeptidase activities of ALE-1. Odintsov et al. (18) suggested that His-291 in LytM might play a role as a functional equivalent of glutamate in HEXXH metalloproteases, thus activating incoming water molecules, or play a role in stabilizing a reaction intermediate. They noted that His-260, which is also close to the metal center, might be another candidate for such a function, although they did not show any direct evidence that mutation at His-260 affects LytM activity. We demonstrated that His-200 in ALE-1 (equivalent to His-260 in LytM) is essential for staphylolytic as well as endopeptidase activities. Taken together, these results strongly suggest that ALE-1 is a zinc metalloprotease and that His-150 and His-233 are the possible ligands for zinc. Clustering of the functionally important His in the previously documented 38-amino-acid region further suggested that this region is catalytically important for bacteriolytic enzymes sharing the homologous 38-amino-acid region. Most of the catalytic sites of zinc enzymes are tetrahedrally, trigonally, or bipyramidally coordinated by 4 or 5 amino acid residues, typically His, Glu, Asp, and Lys (1). Identification of other amino acids for zinc ligands and further characterizations of functionally important histidines and aspartic acid at position 154 require the crystal structural analysis of ALE-1.
Acknowledgments
We thank Junichi Yamagishi, Dainippon Pharmaceutical Co., Ltd., for technical advice and protein sequencing.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Aoki, S., and E. Kimura. 2004.. Zinc hydrolases. Comp. II. Coordinat. Chem. 8:601-604.
- 2.Auer, H. E. 1973. Far-ultraviolet absorption and circular dichroism spectra of l-tryptophan and some derivatives. J. Am. Chem. Soc. 95:3003-3011. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 4.Brahms, S., and J. Brahms. 1980. Determination of protein structure in solution by vacuum ultraviolet circular dichroism. J. Mol. Biol. 138:149-178. [DOI] [PubMed] [Google Scholar]
- 5.Browder, H. P., W. A. Zygmunt, J. R. Young, and P. A. Tavormina. 1965. Lysostaphin: enzymatic mode of action. Biochem. Biophys. Res. Commun. 19:383-389. [DOI] [PubMed] [Google Scholar]
- 6.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 7.Dajcs, J., E. Hume, J. Moreau, A. Caballero, B. Cannon, and R. O'Callaghan. 2000. Lysostaphin treatment of methicillin-resistant Staphylococcus aureus keratitis in rabbits. Investig. Ophthalmol. Vis. Sci. 41:1432-1437. [PubMed] [Google Scholar]
- 8.Day, L. A. 1973. Circular dichroism and ultraviolet absorption of a deoxyribonucleic acid binding proteins of filamentous bacteriophage. Biochemistry 12:5329-5339. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich, P., R. Rosenstein, M. Böhmer, P. Sonner, and F. Götz. 1987. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol. Gen. Genet. 209:563-569. [DOI] [PubMed] [Google Scholar]
- 10.Häse, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, W., H. Ishiguro, and Y. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102:67-70. [DOI] [PubMed] [Google Scholar]
- 12.Iversen, O., and A. Grov. 1973. Studies on lysostaphin. Separation and characterization of three enzymes. Eur. J. Biochem. 38:293-300. [DOI] [PubMed] [Google Scholar]
- 13.Kerr, D., K. Plaut, A. Bramley, C. Williamson, A. Lax, K. Moore, K. Wells, and R. Wall. 2001. Lysostaphin expression in mammary glands confers protection against staphylococcal infection in transgenic mice. Nat. Biotechnol. 19:66-70. [DOI] [PubMed] [Google Scholar]
- 14.Lange, R., and R. Hengge-Aronis. 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13:733-743. [DOI] [PubMed] [Google Scholar]
- 15.Li, S. L., S. Norioka, and F. Sakiyama. 1990. Molecular cloning and nucleotide sequence of the β-lytic protease gene from Achromobacter liticus. J. Bacteriol. 172:6506-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maidhof, H., B. Reinicke, P. Blümel, B. Berger-Bächi, and H. Labischinski. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuman, V. C., H. E. Heath, P. A. LeBlanc, and G. L. Sloan. 1993. Extracellular proteolytic activation of bacteriolytic peptidoglycan hydrolases of Staphylococcus simulans biovar staphylolyticus. FEMS Microbiol. Lett. 110:205-212. [DOI] [PubMed] [Google Scholar]
- 18.Odintsov, S. G., I. Sabala, M. Marcyjaniak, and M. Bochtler. 2004. Latent LytM at 1.3 Å resolution. J. Mol. Biol. 335:775-785. [DOI] [PubMed] [Google Scholar]
- 19.Patron, R. L., M. W. Climo, B. P. Goldstein, and G. L. Archer. 1999. Lysostaphin treatment of experimental aortic valve endocarditis caused by a Staphylococcus aureus isolate with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 43:1754-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramadurai, L., and R. K. Jayaswal. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179:3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Recsei, P. A., A. D. Gruss, and R. P. Novick. 1987. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc. Natl. Acad. Sci. USA 84:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 23.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler, C. A., and V. T. Schuhardt. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 51:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shad, P. A., and B. H. Iglewski. 1988. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J. Bacteriol. 170:2784-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugai, M., T. Fujiwara, T. Akiyama, M. Ohara, H. Komatsuzawa, S. Inoue, and H. Suginaka. 1997. Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. J. Bacteriol. 179:1193-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thumm, G., and G. Götz. 1997. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 23:1251-1265. [DOI] [PubMed] [Google Scholar]
- 28.Trayer, H. R., and C. E. Buckley III. 1970. Molecular properties of lysostaphin, a bacteriolytic agent specific for Staphylococcus aureus. J. Biol. Chem. 245:4842-4846. [PubMed] [Google Scholar]
- 29.Wadström, T., and O. Vesterberg. 1971. Studies on endo-beta-N-acetylglucosaminidase, staphylolytic peptidase, and N-acetylmuramyl-l-alanine amidase in lysostaphin and from Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B 79:248-264. [DOI] [PubMed] [Google Scholar]
- 30.Wang, X., B. J. Wilkinson, and R. K. Jayaswal. 1991. Sequence analysis of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. Gene 102:105-109. [DOI] [PubMed] [Google Scholar]
- 31.Zygmunt, W., and P. A. Tavormina. 1972. Lysostaphin: model for a specific enzymatic approach to infectious disease. Prog. Drug Res. 16:309-333. [DOI] [PubMed] [Google Scholar]