Abstract
The replacement of animal use in testing for regulatory classification of skin sensitizers is a priority for U.S. federal agencies that use data from such testing. Machine learning models that classify substances as sensitizers or nonsensitizers without using animal data have been developed and evaluated. Because some regulatory agencies require that sensitizers be further classified into potency categories, we developed statistical models to predict skin sensitization potency for murine local lymph node assay (LLNA) and human outcomes. Input variables for our models included six physicochemical properties and data from three non-animal test methods: the direct peptide reactivity assay, human cell line activation test, and KeratinoSens™ assay. Models were built to predict three potency categories using four machine learning approaches and were validated using external test sets and leave-one-out cross-validation. A one-tiered strategy modeled all three categories of response together while a two-tiered strategy modeled sensitizer/nonsensitizer responses and then classified the sensitizers as strong or weak sensitizers. The two-tiered model using support vector machine with all assay and physicochemical data inputs provided the best performance, yielding accuracy of 88% for prediction of LLNA outcomes (120 substances) and 81% for prediction of human test outcomes (87 substances). The best one-tiered model predicted LLNA outcomes with 78% accuracy and human outcomes with 75% accuracy. By comparison, the LLNA predicts human potency categories with 69% accuracy (60/87 substances correctly categorized). These results suggest that computational models using non-animal methods may provide valuable information for assessing skin sensitization potency.
Keywords: Skin sensitization potency, allergic contact dermatitis (ACD), integrated decision strategy (IDS), machine learning, murine local lymph node assay (LLNA), direct peptide reactivity assay (DPRA), KeratinoSens, h-CLAT (human cell line activation test)
Introduction
Allergic contact dermatitis (ACD) is an adverse health effect that frequently develops in workers and consumers exposed to skin-sensitizing substances and products. ACD can adversely impact quality of life (Brutti et al., 2013; Heisterberg et al., 2011; Kadyk et al., 2003). The prognosis for ACD includes the development of new skin allergies and the persistence of clinical symptoms for approximately 10 years after diagnosis, with occupational cases of ACD generally having poorer prognoses than non-occupational ACD cases (Macan et al., 2013).
Occurrences of ACD can be reduced by minimizing exposure to skin-sensitizing substances. To this end, national and international regulatory authorities require that products be labeled to identify the potential skin sensitization hazards posed by these items. Such hazards have historically been characterized using animal tests that can require large numbers of animals and produce a painful allergic reaction during testing. For example, the guinea pig maximization test and the Buehler test use 20 to 40 animals per substance (OECD 1992). An alternative animal method, the murine local lymph node assay (LLNA), reduces animal use compared to guinea pig tests and causes less pain and distress to test animals. Regardless, even as a reduction alternative, the LLNA requires 20 animals per substance (OECD 2010) and its 72% accuracy for predicting human skin sensitization hazard leaves much room for improvement (ICCVAM 1999).
A number of factors are driving increased international interest in replacement of animal use for chemical safety testing, including ethical concerns about potential pain and distress to test animals, financial concerns about the cost of animal testing, and scientific concerns about the relevance of animal test results to human outcomes. These concerns have resulted in regulatory efforts to limit animal use in testing, including bans on animal use for testing of cosmetics in the European Union and other countries. Prohibitions on animal testing have increased the need for adequately validated tests for skin sensitization and other chemical safety endpoints.
The recent development of a number of non-animal methods for skin sensitization has been aided by the well-developed mechanistic understanding of the process, which has been codified in an adverse outcome pathway for skin sensitization initiated by covalent binding to proteins (OECD 2012a; OECD 2012b). The adverse outcome pathway includes four key events that can be assessed using non-animal test methods: (1) binding of haptens to endogenous proteins in the skin, (2) keratinocyte activation, (3) dendritic cell activation, and (4) proliferation of antigen-specific T cells (OECD 2012b).
A number of in chemico and in vitro tests targeting the different key events for skin sensitization have been developed (reviewed in Mehling et al. (2012) and evaluated in, e.g., Reisinger et al. (2015)). Because skin sensitization is a complex process, it is unlikely that any individual non-animal method will completely replace the current animal tests (Rovida et al., 2015). Thus, a number of approaches have also been developed to integrate relevant information from multiple non-animal methods as a way to overcome the limitations of individual tests and more accurately assess the potential for skin sensitization (Hirota et al., 2015; Jaworska et al., 2015; Natsch et al., 2009; Natsch et al., 2013; Nukada et al., 2013; Strickland et al., 2016a; Strickland et al., 2016b; Urbisch et al., 2015).
For more than a decade, fostering the evaluation and promoting the use of alternative test methods for regulatory use for assessing skin sensitization potential has been a top priority for the U.S. federal agencies participating in the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) (Dean et al., 2001; ICCVAM 1999; NIEHS 2013; Sailstad et al., 2001). Most recently, the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) has supported ICCVAM in building predictive machine learning models for skin sensitization hazard that integrate in chemico, in silico, and in vitro data (Strickland et al., 2016a; Strickland et al., 2016b). Such models could be useful for regulatory applications that require the identification of skin sensitizers for classification and labeling.
In the United States, labeling to identify substances with any potential for skin sensitization hazard is required by the U.S. Environmental Protection Agency (EPA 2011; EPA 2012b; 2012c) and the Occupational Safety and Health Administration (OSHA; OSHA 2012). The European Registration, Evaluation, Authorization and Restriction of Chemicals Regulation (REACH) No. 1907/2008 (EC 2006) mandates similar labeling. However, some regulatory applications require the assessment of skin sensitization potency to distinguish strong sensitizers from weak sensitizers. For example, the U.S. Consumer Product Safety Commission requires labeling for strong sensitizers, and both the European Classification, Labeling, and Packaging Regulation (CLP) No. 1272/2008 (EC 2008) and OSHA require potency classification if the skin sensitization data are adequate to characterize potency (OSHA 2012).
The objective of the project described in this paper was to build predictive models for skin sensitization potency using non-animal data to predict three categories of response for both human and LLNA outcomes based on the Globally Harmonized System of Classification and Labelling of Chemicals (GHS) (UN 2015). The GHS is an internationally harmonized approach for hazard classification and labeling to ensure the safe use, transport, and disposal of chemicals. The GHS criteria for the classification of sensitizers using human or LLNA data are shown in Table 1. OSHA, REACH, and CLP use the GHS for hazard classification and labeling. A previous ICCVAM evaluation of the usefulness of the LLNA for predicting human potency in the GHS categories noted that the LLNA underclassified one-third of strong human sensitizers as weak sensitizers (ICCVAM 2011). The accuracy of the LLNA for predicting human skin sensitization potency in all three categories was 54% (74/136). Our criterion for success was for our models to classify human skin sensitizers more accurately than the LLNA using the GHS potency categories.
Table 1.
GHS potency categories
| GHS Category | LLNA EC3 | Human Threshold |
|---|---|---|
| 1A (strong) | ≤ 2% | ≤ 500 μg/cm2 skin area |
| 1B (other than strong –“weak”a) | > 2% | > 500 μg/cm2 skin area |
| Nonsensitizer | Unclassified | Unclassified |
EC3, estimated test substance concentration that produces a stimulation index of 3, the threshold for a substance to be considered a sensitizer in the LLNA; GHS, United Nations Globally Harmonized System of Classification and Labeling; LLNA, murine local lymph node assay.
For simplicity in this paper, we refer to Category 1B sensitizers as “weak”; this term is not used in the GHS.
Materials and methods
Data collection and substance database
Sources of data used in the study are listed in Supplemental Table S1. We compiled two datasets, one containing LLNA data on 120 substances and the other containing human skin sensitization data on 87 substances. All 87 substances in the human sensitization dataset are represented in the LLNA dataset. Published data for all substances were obtained for three non-animal skin sensitization test methods: direct peptide reactivity assay (DPRA), KeratinoSens™, and human cell line activation test (h-CLAT). DPRA, KeratinoSens, and h-CLAT were selected because the Organisation for Economic Co-operation and Development (OECD) has issued test guidelines for each of these methods (OECD 2015a; 2015b; 2016a).
Most of the LLNA data used in this study were collected previously by NICEATM (http://ntp.niehs.nih.gov/go/40500, NICEATM LLNA Database). LLNA data are expressed as stimulation indices for each concentration tested. For sensitizers, the estimated test substance concentration that produces a stimulation index of three (EC3) represents a measure of skin sensitization potency. LLNA data for five substances that were not in this database were obtained from published literature (Supplemental Table S1).
Most of the human skin sensitization potency data used in this study were previously published either in an ICCVAM test method evaluation report on the usefulness and limitations of the LLNA for human potency categorization (ICCVAM (2011)) or by Basketter et al. (2014), with the exception of chlorobenzene (Basketter and Kimber (2006)). While ICCVAM (2011) compiled sensitization results from human predictive patch tests, the potency assessments listed in Basketter et al. (2014) were developed by a panel of experts that evaluated prevalence from dermatologic clinic data as well as data from predictive patch tests. Conflicts between these references (n = 9) were resolved by choosing the categorizations in Basketter et al. (2014).
We also collected data on six physicochemical properties of these substances relevant to skin exposure and penetration (octanol:water partition coefficient, water solubility, vapor pressure, melting point, boiling point, and molecular weight). These properties have been used in other models or weight-of-evidence assessments for skin sensitization potential (Jaworska et al., 2013; Jaworska et al., 2011; Patlewicz et al., 2014; Strickland et al., 2016a; Strickland et al., 2016b). Data sources for these properties are provided in Supplemental File 1.
Characterization of the substances
LLNA and human sensitizers were categorized using the GHS criteria in Table 1. For sensitizers identified using the LLNA, EC3 values were used to assign 1A and 1B classifications. Classification of 1A is referred to here as a “strong” sensitizer and 1B as a “weak” sensitizer to reflect the relative potency. Negative results are considered “nonsensitizers”. If a substance had multiple sensitizer/nonsensitizer results, the most prevalent result was used. If multiple LLNA EC3 values were available for a particular sensitizer, the geometric mean of the EC3 values for the positive results, regardless of solvent, was calculated and this value used for classification. For sensitizers identified using human patch test data (ICCVAM (2011)), induction doses per unit skin area that produced a 5% response in the test population (DSA05) were used to assign 1A or 1B classifications based on the human threshold criteria in Table 1. The geometric mean DSA05 was used to classify human sensitizers with multiple values. Sensitizers identified using Basketter et al. (2014) were characterized according to the alignment of the six categories used in that publication with the GHS. Basketter et al. (2014) noted that their Categories 1 and 2 corresponded to GHS 1A, Categories 3 and 4 correspond to GHS 1B, and Categories 5 and 6 correspond to nonsensitizers.
Of the 120 substances in the LLNA dataset, 35 (29%) were 1A (strong) sensitizers, 52 (43%) were 1B (weak) sensitizers, and 33 (28%) were nonsensitizers. The 87 substances in the human dataset consisted of 26 (30%) 1A sensitizers, 31 (36%) 1B sensitizers and 30 (34%) nonsensitizers (Fig. 1). For the LLNA substance list, of the 87 sensitizers, three are prehaptens that require oxidation to induce a skin sensitization response, 16 are prohaptens requiring metabolism, and six are pre/prohaptens requiring both oxidation and metabolism to become sensitizers. Out of the 57 human sensitizers, three are prehaptens, 13 are prohaptens, and two are pre/prohaptens (Fig. 2). The substances in both databases represent 14 product categories. The most common product categories were manufacturing use, food additives, pharmaceuticals, chemical synthesis, fragrance agents, personal care products, and pesticides. See Supplemental File 1 for the prehapten/prohapten characterization and product category information on each substance.
Figure 1.
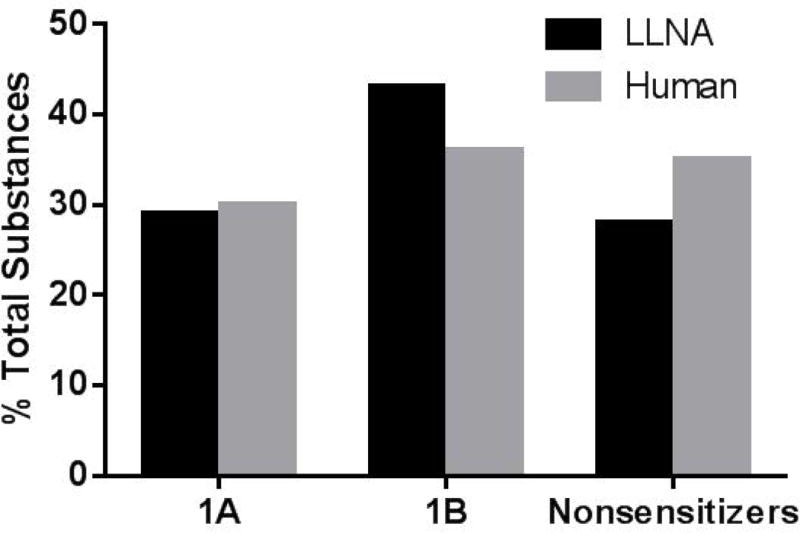
Distribution of substances for the LLNA and human databases across the three GHS categories of skin sensitization. The LLNA database contains 120 substances, including 35 1A (strong) sensitizers and 52 1B (weak) sensitizers. The human database contains 87 substances, including 26 1A sensitizers and 31 1B sensitizers. GHS, United Nations Globally Harmonized System of Classification and Labeling; LLNA, murine local lymph node assay.
Figure 2.
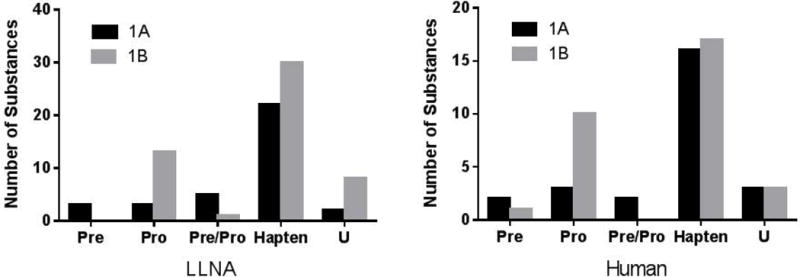
Distribution of prehaptens, prohaptens and pre/prohaptens across the GHS 1A and 1B categories of skin sensitizers. There are 87 LLNA sensitizers (35 1A [strong] and 52 1B [weak]) and 57 human sensitizers (26 1A sensitizers and 31 1B). GHS, United Nations Globally Harmonized System of Classification and Labeling; LLNA, murine local lymph node assay; Pre, prehapten; Pre/Pro, pre- and/or prohapten; Pro, prohapten; U = unknown.
Model variables
The non-animal methods used as input variables in the machine learning approaches align to the adverse outcome pathway (AOP) for skin sensitization initiated by covalent binding to proteins (OECD 2012b).
DPRA
DPRA is an in chemico test that assesses the ability of a substance to form a hapten–protein complex (Gerberick et al., 2004; 2007; OECD 2015a), the molecular initiating event in the skin sensitization AOP as described by OECD (2012b). We used the average percent depletion of cysteine and lysine peptides (Avg.Lys.Cys) as the DPRA input variable.
KeratinoSens
The KeratinoSens test method assesses the ability of a substance to activate cytokines and induce cytoprotective genes in keratinocytes (Emter et al., 2010; OECD 2015b), the second key event in the skin sensitization AOP (OECD 2012b). We used the EC1.5 value, the concentration of test substance that produces 1.5-fold induction of luciferase activity controlled by the antioxidant response element, as the KeratinoSens input variable. KeratinoSens tests that had no significant induction of luciferase activity were assigned a value of 2001 to represent a negative result, since the highest concentration tested was 2000 μM.
h-CLAT
h-CLAT assesses the ability of a substance to activate and mobilize dendritic cells in the skin (Ashikaga et al., 2006; OECD 2016a), the third key event of the skin sensitization AOP (OECD 2012b). Specifically, h-CLAT measures the induction of the CD86 and CD54 cell surface markers, with results expressed as the effective concentration at 150% induction for the CD86 marker (EC150) and the effective concentration at 200% induction for the CD54 marker (EC200). Tests that yielded no significant induction of CD86 or CD54 were arbitrarily assigned 2001 to represent a negative result. We used the minimum induction threshold of the CD54 EC200 and CD86 EC150 as the h-CLAT input variable.
Physicochemical Properties
For each substance in both datasets, we collected experimental data, where available, for octanol:water partition coefficient, water solubility, vapor pressure, molecular weight, melting point, and boiling point. For 10 substances, experimental values for one or more physicochemical properties could not be found in the literature. In these cases, values were imputed via quantitative structure–property relationship models built using binary molecular fingerprints and machine learning approaches (Zang et al., 2016). See Supplemental File 1 for the individual physicochemical properties and data sources.
Data processing and distribution
Table 2 shows the ranges of the nine variables that served as model input data. The distributions of DPRA, h-CLAT and KeratinoSens values in 1A and 1B sensitizer potency classes for both LLNA and human datasets are illustrated in Fig. 3, where h-CLAT and KeratinoSens are log-scaled. The DPRA distribution showed better separation of 1A and 1B classes than the other two non-animal methods, suggesting the DPRA variable can better distinguish between strong and weak sensitizers.
Table 2.
Data ranges of input variables
| Name | Description | Rangea |
|---|---|---|
| DPRA | Average lysine and cysteine peptide depletion measurement (%) | 0 – 95 |
| h-CLAT | Minimum induction threshold [smallest value for CD54 EC200 and CD86 EC150] (μg/mL) | 0.54 – 2001 |
| KeratinoSens | EC1.5 (μM) | 0.50 – 2001 |
| LogP | Octanol:water partition coefficient | −8.28 – 6.46b |
| LogS | Water solubility (mol/L) | −6.39 – 1.92b |
| LogVP | Vapor pressure (mm Hg) | −28.47 – 5.89b |
| MP | Melting point (°C) | −148.50 – 288.00 |
| BP | Boiling point (°C) | −19.10 – 932.20 |
| MW | Molecular weight (g/mol) | 30.03 – 581.57 |
BP, boiling point; DPRA, direct peptide reactivity assay; EC1.5, concentration producing a 1.5-fold induction of luciferase controlled by the antioxidant response element; EC150, estimated concentration inducing a 150% increase for CD86; EC200, estimated concentration inducing a 200% increase for CD54; h-CLAT, human cell line activation test; LogP, log octanol:water partition coefficient; LogS, log water solubility; LogVP, log vapor pressure; MP, melting point; MW, molecular weight.
Human dataset with 87 substances is a subset of LLNA dataset with 120 substances; the two datasets cover the same range.
Range for base 10 logarithm of these measurements.
Figure 3.
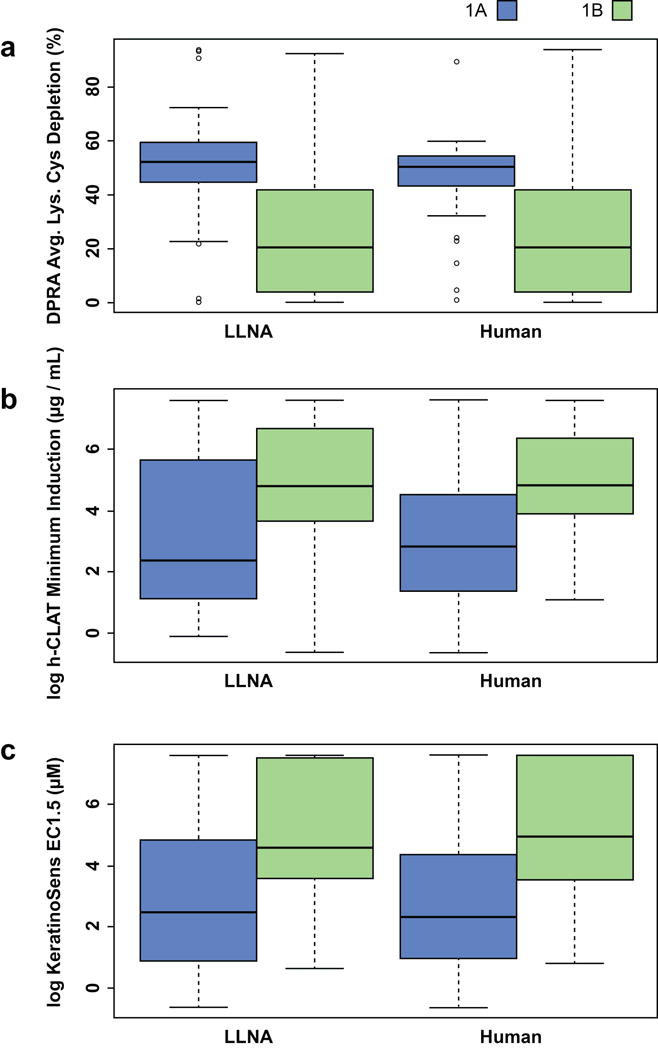
Distributions of (a) DPRA, (b) h-CLAT and (c) KeratinoSens values for substances identified as GHS 1A and 1B sensitizers by LLNA and human data. The boxplot is graphed based on the data quartiles, which divide the distribution into the 25% (Q1), 50% (Q2) and 75% (Q3) percentiles. The height of the box is determined by Q1 and Q3 while the median or Q2 is represented by the dark line inside the box. Avg.Lys.Cys, average depletion of lysine and cysteine peptides; DPRA, direct peptide reactivity assay; EC1.5, concentration producing a 1.5-fold induction of luciferase activity; h-CLAT, human cell line activation test; GHS, Globally Harmonized System of Classification and Labeling of Chemicals (UN 2015); LLNA, murine local lymph node assay.
The following conventions were adopted for data processing:
KeratinoSens and h-CLAT values that were provided as less than a specified value were replaced with the specified value. For example, a CD86 EC150 for the h-CLAT expressed as <9 μg/mL was replaced with 9 μg/mL.
Substances with multiple test results for the same assay were represented by the median response value. If a substance had multiple Avg.Lys.Cys values for the DPRA, the median of those results was calculated after negative peptide depletion values were set to zero.
Values for octanol:water partition coefficient were provided as log values.
We converted water solubility and vapor pressure to log values because the range of values covered several orders of magnitude.
Selection of training and test sets
For each dataset, the substances were divided into training and test sets in approximate proportions of 75% to 25% for building and evaluating the predictive models, respectively. Substances were placed in the training and test sets so that the sets would have similar ranges of activity and structural characteristics, such as the distributions of potency, product use categories, diversity of chemical structures, prehaptens/prohaptens and mechanistic protein binding domains. For predictive modeling of LLNA results, this procedure placed 94 and 26 substances into the training set and test set, respectively. For predictive modeling of human results, this yielded a training set of 63 substances and a test set of 24 substances. Table 3 summarizes the number and percentage of substances in each potency category in the training and test sets.
Table 3.
| a. Distributions of training and test substances for LLNA models | ||||||
|---|---|---|---|---|---|---|
| Potency categorya | Entire set (n = 120) | Training set (n = 94) | Test set (n = 26) | |||
| Number | % | Number | % | Number | % | |
| 1A | 35 | 29 | 28 | 30 | 7 | 27 |
| 1B | 52 | 43 | 40 | 42 | 12 | 46 |
| Neg | 33 | 28 | 26 | 28 | 7 | 27 |
| b. Distributions of training and test substances for human models | ||||||
|---|---|---|---|---|---|---|
| Potency categorya | Entire set (n = 87) | Training set (n = 63) | Test set (n = 24) | |||
| Number | % | Number | % | Number | % | |
| 1A | 26 | 30 | 19 | 30 | 7 | 29 |
| 1B | 31 | 36 | 22 | 35 | 9 | 38 |
| Neg | 30 | 34 | 22 | 35 | 8 | 33 |
LLNA, murine local lymph node assay.
The 1A category consists of strong sensitizers and the 1B category consists of weak sensitizers based on the Globally Harmonized System of Classification and Labeling of Chemicals (UN 2015); Neg, nonsensitizer.
Random forest for variable importance ranking
A random forest (RF) analysis was conducted to assess the relative importance of the three non-animal methods and physicochemical properties for discriminating between 1A and 1B sensitizer potency categories. RF is a consensus algorithm that constructs an ensemble of decision trees via bootstrap sampling, conducted by random selection with replacement from the substances in the training data, where substances not used for tree growth are referred to as out-of-bag (OOB) substances. Each tree provides a prediction for the OOB substance set, and the average of these predictions over all trees produces an overall OOB validation (Diaz-Uriarte 2007; Hao et al., 2011). RF can assess the importance of variables to the model based on the deterioration of classification performance via random permutation of the variables. After all the variables have been estimated, RF returns a list of the variables ranked according to their importance (Zang et al., 2013).
In general, the RF results ranked the three non-animal methods higher in importance when distinguishing between strong and weak sensitizers, indicating that they were more discriminative as individual variables than any of the physicochemical properties (Fig. 4). LogP was ranked as having greater importance than the other physicochemical properties for both LLNA and human data and ranked higher than h-CLAT and KeratinoSens when classifying 1A and 1B sensitizers in the LLNA data set (Fig. 4a). For the human data set, the most important variable was KeratinoSens followed by DPRA and h-CLAT, and the least important variables were the physicochemical properties (Fig. 4b). For classification modeling, the variables were used in four sets as defined in Table 4, with Variable Set IV composed of the four most important variables identified in the RF analysis.
Figure 4.
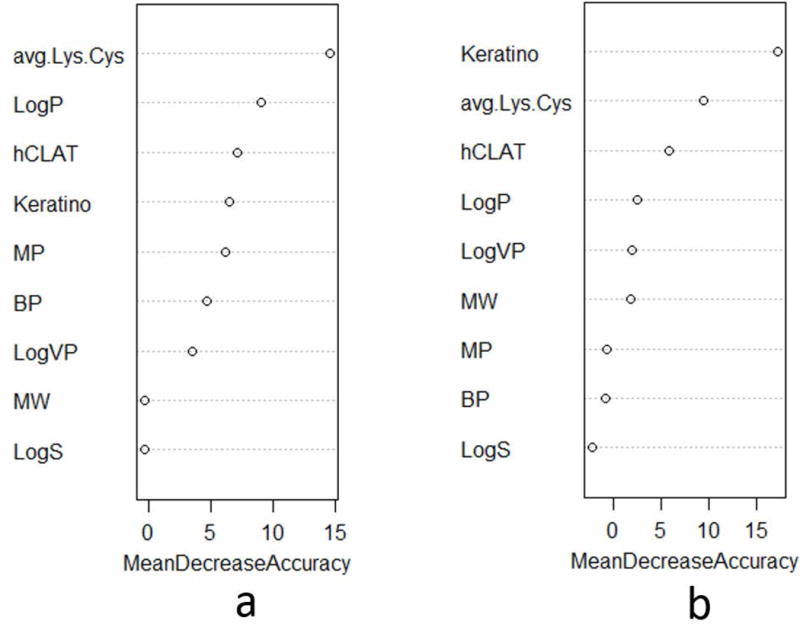
Ranking of variable importance by random forest algorithm for (a) LLNA data set and (b) human data set for distinguishing between GHS 1A and 1B sensitizers. Avg.Lys.Cys, average depletion of lysine and cysteine peptides; BP, boiling point; GHS, Globally Harmonized System of Classification and Labeling of Chemicals (UN 2015); hCLAT, minimum induction values of the CD86 EC150 and the CD54 EC200; Keratino, EC1.5 for the induction of luciferase activity controlled by the antioxidant response element; LLNA, murine local lymph node assay; LogP, log octanol:water partition coefficient; LogS, log water solubility; LogVP, log vapor pressure; MP, melting point; MW, molecular weight.
Table 4.
Variable sets for model building
| Variable set | Combination of input variables |
|---|---|
| I | DPRA + h-CLAT + KeratinoSens |
| II | LogP + LogS + LogVP + MP + BP + MW |
| III | DPRA + h-CLAT + KeratinoSens + LogP + LogS + LogVP + MP + BP + MW |
| IV | DPRA + h-CLAT + KeratinoSens + LogP |
BP, boiling point; DPRA, direct peptide reactivity assay; h-CLAT, human cell line activation test; LogP, log octanol:water partition coefficient; LogS, log water solubility; LogVP, log vapor pressure; MP, melting point; MW, molecular weight.
Machine learning approaches and modeling strategies
Predictive models were developed using four machine learning approaches with different algorithm principles. These included classification and regression tree (CART) (Deconinck et al., 2005; Questier et al., 2005; Zang et al., 2011a), linear discriminant analysis (LDA) (Luan et al., 2005; Zang et al., 2011b), logistic regression (LR) (Varmuza and Peter 2009), and support vector machine (SVM) (Shen et al., 2011). The models were trained to predict potency classifications for LLNA and human data using the training sets described above.
Two strategies were applied to model strong potency, weak potency, and nonsensitizers. Strategy A, a one-tiered strategy, was a multiple-category classification that simultaneously modeled all three classes of substances (Fig. 5a). Strategy B shows a two-tiered approach (Fig. 5b), in which binary models were used for each tier. Tier One categorized substances into sensitizers and nonsensitizers based on models from Strickland et al. (2016a, 2016b), and Tier Two categorized sensitizers into 1A (strong) and 1B (weak) subcategories based on the Globally Harmonized System of Classification and Labeling of Chemicals (UN 2015).
Figure 5.
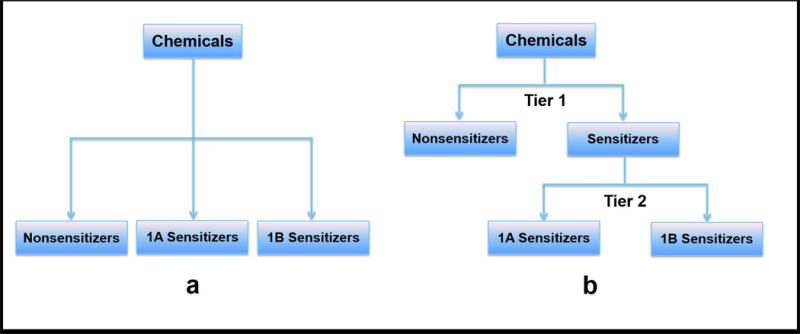
Two classification strategies for modeling three categories of sensitization potency.
OECD Toolbox (OECD 2016b) can classify substances as sensitizers or nonsensitizers, but cannot predict if a substance belongs to 1A or 1B category. Thus, the read-across prediction from OECD Toolbox can be used only as a variable for sensitizer/nonsensitizer modeling (Tier One in Strategy B), and cannot be applied to Strategy A and Tier Two in Strategy B. Therefore the two-tiered strategy uses the variables described in this work with the addition of read-across predictions generated using the OECD Toolbox.
Evaluation of model performance
Once the predictive models developed using the various machine learning approaches were trained using the training sets, the test sets were used to evaluate model performance. We also applied a leave-one-out cross-validation (LOOCV) procedure on each complete dataset to assess model robustness and reliability. In LOOCV, n - 1 substances from the complete set of n substances were used as the training data for building the model and the remaining substance was used for testing the model. The cross-validation process was repeated n times with each of the substances used exactly once as the test set. The predictive accuracy was calculated by averaging individual values over the n runs.
Model performance was measured using accuracies of individual classes and overall accuracies for the three-category classification. For binary models, the performance was examined in terms of sensitivity, specificity, and overall accuracy given the assumption that the sensitizers and nonsensitizers were positive and negative classes, respectively, for the sensitizer-nonsensitizer model, while strong sensitizers and weak sensitizers represented positive and negative classes, respectively, for strong-weak potency models. These metrics were calculated by the following formulae:
We calculated 95% confidence limits of proportions for correct classification rate, overclassification rate, and underclassification rate using the formula (Brown et al., 2001):
with p being the rate and N the number of chemicals.
Statistical software
All data processing, variable ranking, and model building operations were conducted using the following packages in the R statistical analysis software for Windows (v3.2.1) (The R Core Team 2013): package randomForest for random forest, package MASS for linear discriminant analysis and logistic regression, package rpart for classification and regression tree, and package e1071 for support vector machine.
Results
Strategy A: one-tier approach
To predict classification of test substances as strong sensitizers, weak sensitizers, or nonsensitizers, we developed a series of models for both LLNA and human potency outcomes using each of the four machine learning approaches and each of the four variable sets described in Table 4, yielding a total of 32 models. When all three potency categories were predicted simultaneously using Strategy A, models using the SVM approach provided superior predictions to models using other machine learning approaches across all four variable sets in Table 4 for both LLNA and human endpoint data. We report the detailed classification results from the SVM models in Table 5 and accuracy for all models in Supplemental Table S2.
Table 5.
Accuracy of individual category and overall classification predictions using Strategy Aa and SVM
| LLNA/Humanb | Variable Set | Data Set | 1Ac (%) | 1Bc (%) | Neg (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| LLNA | I | Training | 79 ± 15 | 63 ± 15 | 85 ± 14 | 73 ± 9 |
| Test | 86 ± 26 | 50 ± 28 | 71 ± 34 | 65 ± 18 | ||
| LOOCV | 83 ± 12 | 62 ± 13 | 73 ± 15 | 71 ± 8 | ||
| II | Training | 82 ± 14 | 75 ± 13 | 65 ± 18 | 75 ± 9 | |
| Test | 71 ± 34 | 58 ± 28 | 43 ± 37 | 58 ± 19 | ||
| LOOCV | 68 ± 15 | 60 ± 13 | 55 ± 17 | 61 ± 9 | ||
| III | Training | 96 ± 7 | 80 ± 12 | 96 ± 8 | 89 ± 6 | |
| Test | 86 ± 26 | 67 ± 27 | 86 ± 26 | 77 ± 16 | ||
| LOOCV | 83 ± 12 | 69 ± 12 | 85 ± 12 | 78 ± 7 | ||
| IV | Training | 96 ± 7 | 73 ± 14 | 96 ± 8 | 86 ± 7 | |
| Test | 71 ± 34 | 58 ± 28 | 86 ± 26 | 69 ± 18 | ||
| LOOCV | 83 ± 12 | 60 ± 13 | 85 ± 12 | 73 ± 8 | ||
| Human | I | Training | 79 ± 18 | 55 ± 21 | 95 ± 9 | 76 ± 11 |
| Test | 71 ± 34 | 56 ± 32 | 75 ± 30 | 67 ± 19 | ||
| LOOCV | 77 ± 16 | 58 ± 17 | 80 ± 14 | 71 ± 10 | ||
| II | Training | 58 ± 22 | 64 ± 20 | 77 ± 18 | 67 ± 12 | |
| Test | 43 ± 37 | 44 ± 32 | 50 ± 35 | 46 ± 20 | ||
| LOOCV | 58 ± 19 | 65 ± 17 | 57 ± 18 | 60 ± 10 | ||
| III | Training | 89 ± 13 | 59 ± 21 | 82 ± 16 | 76 ± 11 | |
| Test | 86 ± 26 | 56 ± 32 | 75 ± 30 | 71 ± 18 | ||
| LOOCV | 85 ± 14 | 61 ± 17 | 80 ± 14 | 75 ± 9 | ||
| IV | Training | 84 ± 16 | 64 ± 20 | 82 ± 16 | 76 ± 11 | |
| Test | 86 ± 26 | 56 ± 32 | 75 ± 30 | 71 ± 18 | ||
| LOOCV | 88 ± 13 | 58 ± 17 | 80 ± 14 | 75 ± 9 |
LLNA, murine local lymph node assay; LOOCV, leave-one-out cross-validation; SVM, support vector machine.
The values after ± indicate 95% confidence limits of proportion for correct classification rate.
Strategy A modeled all three categories of response simultaneously.
The LLNA data set contained 120 substances: 35 strong sensitizers, 52 weak sensitizers, and 33 nonsensitizers. The human data set contained 87 substances: 26 strong sensitizers, 31 weak sensitizers, and 30 nonsensitizers.
1A (strong) and 1B (weak) are subcategories for sensitizers in the Globally Harmonized System of Classification and Labeling of Chemicals (UN 2015).
The SVM LLNA models using Variable Set III input data, which included data from the three non-animal methods and the six physicochemical properties, achieved the best predictive performance for the three potency classes with a test set accuracy of 77%. This was followed by the models using Variable Set IV input data (three non-animal methods and LogP) with a test set accuracy of 69%. Models using Variable Set II input data, consisting of the six physicochemical properties only, had the poorest accuracy of 58% for the test set. However, it should be noted that these differences in performance are magnified by the small number of substances in the test set, which included only seven substances each in class Neg and class 1A. Misclassification of one or two substances resulted in highly variable classification accuracies. Using LOOCV to evaluate the classification models using the entire dataset (35 strong sensitizers, 52 weak sensitizers, and 33 nonsensitizers), Variable Set III achieved the highest accuracy of 78%, with 83%, 69% and 85% accuracy for classifying strong, weak, and nonsensitizers, respectively. This was superior to results from models using only input data from the three non-animal methods (Variable Set I) or physicochemical properties (Variable Set II), with overall LOOCV accuracies of 71% and 61%, respectively. These results suggest that, using a one-tiered classification approach, both non-animal assay data and physicochemical properties significantly contribute to the LLNA modeling, and the assays play more important roles than the properties.
Contrary to the LLNA predictions, the one-tiered human potency models using input data from Variable Sets III (all variables) and IV (three non-animal methods and LogP) achieved comparable predictive performance when considering the overall accuracy, although there were differences in the ability of these models to differentiate between 1A and 1B sensitizers. The model using input data from Variable Set III produced slightly better accuracy for identification of strong sensitizers than the model using input data from Variable Set I and hence had better overall accuracy of 71% vs 67% on the test set and 75% vs 71% on the LOOCV. Similar to LLNA modeling, the human models using Variable Set II input data (only physicochemical properties) produced the lowest accuracy of 46% on the test set and 60% on the LOOCV. Therefore, using a one-tiered classification approach, data from the three non-animal methods appear to make more significant contributions to accurate human classifications than the physicochemical properties.
Strategy B: two-tier approach
Strategy B used a two-tiered approach for potency classification. Substances were first classified as either sensitizers or nonsensitizers. Sensitizers were then further classified as 1A or 1B sensitizers.
Tier One: Sensitizers vs Nonsensitizers
For the Tier One sensitizer vs. nonsensitizer classification, we used previously published integrated decision strategies using non-animal in chemico and in vitro data, in silico predictions, and the six physicochemical properties to predict classification of test substances as sensitizers or nonsensitizers based on LLNA or human results (Strickland et al., 2016a; Strickland et al., 2016b). The prior studies used the same chemical set as the current study, and therefore the results from those studies are being used in the current study for the Tier One results for Strategy B.
Tier Two: Strong vs. Weak Potency of Sensitizers
Substances predicted to be sensitizers in Tier One were further classified in Tier Two as strong or weak sensitizers. We developed a series of binary models for both LLNA and human outcomes to distinguish strong from weak sensitizers. For each machine learning approach, the variable sets that produced the best performance are shown in Table 6. Models using SVM performed best in modeling LLNA outcomes using input data from Variable Set III (the three non-animal methods and six physicochemical properties) with a balanced sensitivity and specificity of 86% and 92% and an overall accuracy of 89% for the test set. For human potency prediction, classification models using SVM and input data from Variable Sets III and IV produced the highest test set accuracy of 81% (sensitivity of 86% and specificity of 78%). It is worth noting that although the human model using the CART machine learning approach achieved a specificity of 100%, sensitivity was low (43%) due to a bias towards the 1B class, leading to an overall accuracy of only 75%.
Table 6.
Individual category accuracy and overall accuracy of sensitizer classificationa from Tier Two of Strategy B using four machine learning approaches
| LLNA/Humana | Approach | Variable setb | Data set | Sensitivity (1A %) | Specificity (1B %) | Accuracy (%) |
|---|---|---|---|---|---|---|
| LLNA | CART | III/IV | Training | 86 ± 13 | 88 ± 10 | 87 ± 8 |
| Test | 57 ± 37 | 75 ± 25 | 68 ± 21 | |||
| LDA | I | Training | 71 ± 17 | 75 ± 13 | 74 ± 10 | |
| Test | 71 ± 34 | 83 ± 21 | 79 ± 18 | |||
| LR | I | Training | 82 ± 14 | 73 ± 14 | 77 ± 10 | |
| Test | 86 ± 26 | 67 ± 27 | 74 ± 20 | |||
| SVM | III | Training | 89 ± 12 | 88 ± 10 | 88 ± 8 | |
| Test | 86 ± 26 | 92 ± 15 | 89 ± 14 | |||
| Human | CART | I/III/IV | Training | 68 ± 21 | 86 ± 15 | 78 ± 13 |
| Test | 43 ± 37 | 100 ± 0 | 75 ± 21 | |||
| LDA | I | Training | 79 ± 18 | 73 ± 19 | 76 ± 13 | |
| Test | 71 ± 34 | 78 ± 27 | 75 ± 21 | |||
| LR | III | Training | 84 ± 17 | 64 ± 20 | 73 ± 14 | |
| Test | 57 ± 37 | 78 ± 27 | 69 ± 23 | |||
| SVM | III/IV | Training | 90 ± 14 | 77 ± 18 | 83 ± 11 | |
| Test | 86 ± 26 | 78 ± 27 | 81 ± 19 |
CART, classification and regression tree; LDA, linear discriminant analysis; LR, logistic regression; LLNA, murine local lymph node assay; SVM, support vector machine.
The values after ± indicate 95% confidence limits of proportion for correct classification rate.
Chemicals predicted to be sensitizers using the Strickland et al., 2016, models were used in Tier Two. The LLNA and human datasets respectively included 84 (34 1A and 50 1B) and 53 (26 1A and 27 1B) chemicals predicted to be sensitizers.
Variable sets are defined in Table 4.
As described previously (Fig. 4), RF ranked the three non-animal methods and LogP as the top four variables for discriminating between strong and weak potency. To determine whether all of the physicochemical properties were necessary for the modeling, we compared the models using input data from Variable Set III (all nine input features) with those using input data from Variable Set IV (three assays and LogP). For this comparison we used the highest performing machine learning approach (SVM); the results are summarized in Supplemental Table S3.
For the LLNA data, models using input data from Variable Set III predicted test set classifications with a higher accuracy (89%) than models using input data from Variable Set IV (accuracy of 79%). The sensitivity for these two models was the same (86%), but the specificity decreased when LogP was used as the only physicochemical property input (92% for Variable Set III vs. 75% for Variable Set IV). For the LOOCV, models using input data from Variable Set III had the same sensitivity as those using input data from Variable Set IV (91%), but higher specificity (85% vs. 79%). Hence, using Variable Set III produced a marginal increase in overall accuracy (87% vs. 84%). Models using only LogP instead of all six physicochemical properties did not predict LLNA potency as well, implying that the other physicochemical properties play important roles in determining sensitization potency.
For the human data, models using input data from Variable Set IV produced similar results to those using data from Variable Set III. For predicting classification of test set substances, the models both achieved sensitivity of 86%, specificity of 78%, and overall accuracy of 81%. Models using input data from Variable Set IV performed almost as well as those using input data from Variable Set III on the LOOCV with an overall accuracy of 79% vs 81%. These results show that, unlike for the LLNA predictions, physicochemical properties other than LogP do not appear to add value in predicting human test outcomes.
For the LLNA potency models, overall accuracy assessed by LOOCV of the two-tiered Strategy B for the highest performing SVM model (using input data from Variable Set III) was 88% (Table 7). This accuracy is much higher than the overall accuracy assessed by LOOCV for the best performing SVM model for the one-tiered Strategy A (which also used input data from Variable Set III), which was 78% (Tables 5 and 7). Individual accuracies of 91%, 81%, and 97% for classification of strong, weak, and nonsensitizers using Strategy B were higher than the corresponding accuracies of 83%, 69%, and 85% from Strategy A.
Table 7.
Performance of one-tiered (A) and two-tiered (B) classification strategies using SVMa
| Model | Strategy | Classification Rate | Overall Accuracy | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1A (Strong) | 1B (Weak) | Neg (Nonsensitizers) | |||||||
| Correct | Under | Over | Correct | Under | Correct | Over | |||
| LLNA | A | 83 ± 12% (29/35) |
17 ± 12% (6/35) |
17 ± 10% (9/52) |
69 ± 12% (36/52) |
14 ± 9% (7/52) |
85 ± 12% (28/33) |
15 ± 12% (5/33) |
78 ± 7% (93/120) |
| B | 91 ± 9% (32/35) |
9 ± 9% (3/35) |
15 ± 10% (8/52) |
81 ± 11% (42/52) |
4 ± 5% (2/52) |
97 ± 6% (32/33) |
3 ± 6% (1/33) |
88 ± 6% (106/120) |
|
| Human | A | 85 ± 14% (22/26) |
15 ± 14% (4/26) |
26 ± 15% (8/31) |
61 ± 17% (19/31) |
13 ± 12% (4/31) |
80 ± 14% (24/30) |
20 ± 14% (6/30) |
75 ± 9% (65/87) |
| B | 85 ± 14% (22/26) |
15 ± 14% (4/26) |
22 ± 15% (7/31) |
65 ± 17% (20/31) |
13 ± 12% (4/31) |
93 ± 9% (28/30) |
7 ± 9% (2/30) |
81 ± 8% (70/87) |
|
LLNA, murine local lymph node assay; SVM, support vector machine.
The values after ± indicate 95% confidence limits of proportion for correct classification rate.
Leave-one-out cross-validation results.
For the human potency models, overall accuracy assessed by LOOCV of the two-tiered Strategy B for the highest performing SVM model (using input data from Variable Set III) was 81% (Table 7). This accuracy is higher than the overall accuracy assessed by LOOCV for the best performing SVM model for the one-tiered Strategy A (which also used input data from Variable Set III), which was 75% (Tables 5 and 7). While the classification accuracy for strong sensitizers (85%) was the same and the classification accuracy for weak sensitizers was essentially the same (61% vs 65%) for Strategy A and Strategy B, individual accuracy of 93% for classification of nonsensitizers via Strategy B was substantially higher than the corresponding accuracy of 80% from Strategy A. It is important to note that, regardless of whether Strategy A or Strategy B was used, the highest performing model predicted human potency categories better than the LLNA. For the entire human data set the accuracy of the LLNA was 65% (17/26), 74% (23/31), and 67% (20/30) for classification of strong, weak, and nonsensitizers, respectively, and the overall accuracy was 69%.
Discussion
The adverse outcome pathway for skin sensitization involves multiple steps linking the structure and properties of a chemical to allergic contact dermatitis. These steps are not likely to be successfully modeled by a single non-animal method. Accordingly, the use of integrated decision strategies bringing together data from several non-animal methods is needed for reliable prediction of this adverse outcome (OECD 2012b). Previously published integrated decision strategies to predict skin sensitization hazard without the use of animals have principally focused on differentiation between sensitizers and nonsensitizers. However, potency data are necessary for some regulatory authorities and for use in risk assessment to identify the threshold level of exposure to a substance below which it is unlikely to produce skin sensitization. Some studies have been published describing approaches to predict LLNA potency categories using simple test batteries (Natsch et al., 2009; Nukada et al., 2013), testing strategies (Roberts and Patlewicz 2014; Takenouchi et al., 2015), or machine learning approaches (Jaworska et al., 2013; Jaworska et al., 2011; Jaworska et al., 2015; Luechtefeld et al., 2015; Tsujita-Inoue et al., 2014). In this study, we developed a novel machine learning approach capable of predicting GHS LLNA and human sensitizer potency categories using data from non-animal alternative methods.
To achieve the best results, we compared two strategies; Strategy A modeled all GHS potency categories simultaneously, while Strategy B first made sensitizer vs. nonsensitizer determinations and then performed a potency classification of predicted sensitizers. Comparing the two strategies demonstrated that the two-tiered approach outperformed the one-tiered approach for both LLNA and human potency predictions.
Consistent with our previous work (Strickland et al., 2016a; Strickland et al., 2016b), of the four machine learning approaches tested, SVM performed the best. For prediction of LLNA potency categories, SVM models using input data from Variable Set III (three non-animal methods plus six physicochemical properties) performed best for all individual GHS classes with an overall accuracy of 78% for the LOOCV when using the one-tiered strategy. While including the six physicochemical properties increased the accuracy of potency prediction, models relying solely on the physicochemical properties performed poorly. This suggests that data from both non-animal assays and physicochemical properties contribute to successful LLNA modeling. Greater predictive performance was achieved using the two-tiered strategy (overall accuracy for the LOOCV was 88%).
The performance of our approach was on par with the aforementioned approaches to categorizing substances into LLNA potency categories. While the Bayesian network approach by Jaworska et al. (2015) predicted the correct GHS LLNA potency category 96% of the time in a test set of 60 chemicals, our approach uses fewer inputs and requires no unit conversions of test data. Our approach also utilizes open-source tools, which we believe makes it more accessible to the research community.
This work is one of only two studies to use an integrated decision strategy approach to predict human skin sensitizer potency. Natsch et al. (2015) used a regression approach that incorporated in vitro data to predict human skin sensitizer potency. However, this approach used only data from the KeratinoSens assay and a non-OECD validated kinetic peptide binding assay. Converting the results of this analysis to percent accuracy produced accuracy of 55% and 70% for classification of weak and strong sensitizers, respectively. Importantly, 36% of the weak sensitizers and 11% of the strong sensitizers were misclassified as nonsensitizers. In our models, use of input data from Variable Sets III (three non-animal methods and six physicochemical properties) and IV (three non-animal methods and LogP) achieved similar predictive performance (75% accuracy) for human potency categories when using the one-tiered strategy. Predictive performance was improved using the two-tiered strategy, producing accuracy assessed by LOOCV of 81%.
While the predictive capacity of our models may have been impacted by the small size of the human dataset, the disparity between LLNA and human potency prediction may be due to other factors. For instance, the in chemico and in vitro tests employed for this study were calibrated against the LLNA during assay development and, for this reason, the relatively high performance for predicting LLNA potency categories is not surprising. Additionally, it has been demonstrated that chemicals penetrate rodent skin more readily than that of humans (Garnett et al., 1994; Mint et al., 1994). An added complication is that the purity of the test substances was not necessarily the same for all tests or over time. Sensitization potency can be influenced by impurities and degradants, as well the actual amount of the parent chemical.
Furthermore, in the case of nickel allergy, it has been previously shown that species differences between rodents and humans at the molecular level can profoundly alter susceptibility to skin sensitization (Kimber et al., 2011). For example, mice lack the TLR4 pathway required for the dermal sensitization effects induced by cobalt and nickel in humans (Schmidt et al, 2010). It is conceivable that other interspecies differences exist and have yet to be discovered. These points of divergence may bias in vitro model development towards prediction of rodent outcomes as long as the metric for success during development of alternative methods is the ability to predict rodent outcomes. Keeping in mind that in chemico and in vitro assays are often designed to model one specific event in the multi-step process leading to skin sensitization, these points further support the concept that reliable prediction of skin sensitization requires the integration of data from many sources.
In our study, the two-tiered strategy classified weak and strong sensitizers with 65% and 85% accuracy, respectively (Table 7). Supplemental Tables S4a and S4b list the substances misclassified by the LLNA and human models that are summarized in Table 7. Seventeen substances were misclassified by the human model using Strategy B. In Tier 1, four weak sensitizers were misclassified as nonsensitizers and two nonsensitizers were misclassified as sensitizers. No strong sensitizers were misclassified as nonsensitizers. In Tier 2, four strong sensitizers were misclassified as weak sensitizers and seven weak sensitizers were misclassified as strong sensitizers. The three sensitizers misclassified by both the LLNA and human prediction models were formaldehyde, isoeugenol and 2-mercaptobenzothiazole. Isoeugenol is a prehapten that requires oxidation to induce a skin sensitization reaction and 2-mercaptobenzothiazole is a prohapten that requires metabolic activation to produce skin sensitization. Coumarin was the only nonsensitizer that was misclassified as a sensitizer in both the LLNA and human prediction models. While our models appear to be somewhat more predictive than the approach developed by Natsch et al. (2015), the performance of our models cannot be directly compared with theirs because they were not tested using the same substance set and they did not evaluate performance using an external set. Consequently, additional studies using more chemicals are warranted to more accurately gauge performance of the models.
The biological mechanisms underlying sensitizer potency are not fully understood (Natsch et al., 2015). Thus, the reasons for the potency misclassifications we observed are not entirely clear. The strength of peptide reactivity contributes to skin sensitization potency. However, the DPRA was not designed to quantify reactivity; the stated goal of the test is to determine if “a chemical is reactive enough to be a sensitizer (Roberts and Patlewicz 2014). Recently, Jaworska et al. (2015) reported that cytotoxicity contributes more to the prediction of potency categories than Cys and Lys reactivity. Thus, inclusion of other assays in models similar to ours may reduce potency misclassifications.
The three non-animal methods used in our models assess three of the four key events in the AOP for skin sensitization initiated by covalent binding to proteins. All three methods are described in internationally accepted test guidelines adopted by OECD. In isolation, each of these test methods has documented limitations, including incorrect identification of pre- and prohaptens, that hinder the identification of potential sensitizers (OECD 2015a; OECD 2015b; OECD 2016a). DPRA has consistently classified prehaptens correctly for skin sensitization hazard (OECD 2015a), but KeratinoSens (OECD 2015b) and h-CLAT (OECD 2016a) have not. Although DPRA has no metabolic capacity and is not be expected to correctly classify prohaptens as sensitizers (OECD 2015a), Patlewicz et al. (2016) notes that it has correctly identified some prohaptens and pre/prohaptens. KeratinoSens has also correctly identified some prohaptens and pre/prohaptens (OECD 2015b; Patlewicz et al. 2016) and h-CLAT has correctly classified some (OECD 2016a) or all of the evaluated (Patlewicz et al. 2016) prohaptens and pre/prohaptens. The individual non-animal methods used in this study only predicted human skin sensitization hazard with 63–79% accuracy for the test set when used in isolation; however, integrating the data from these assays increased predictive capacity to 92% (Strickland et al., 2016b). Potency prediction adds a new layer of complexity to development of an integrated decision strategy to predict skin sensitization, and the ability to properly classify prehaptens and prohaptens is an important consideration for the prediction of skin sensitization classifications. Of the 25 pre- and prohaptens included in this study, one prohapten (2-mercaptobenzothiazole) was overclassified as an LLNA strong sensitizer and one prehapten (isoeugenol) was underclassified using Strategy B as an LLNA weak sensitizer. In the case of human sensitizer categories, one prehapten (isoeugenol) and one prohapten (3-dimethylaminopropylamine) were underclassified as weak sensitizers, while one prehapten (1,4-dihydroquinone) and one prohapten (2-mercaptobenzothiazole) were overclassified as strong sensitizers. Given the known limitations of the individual assays and the relatively small dataset, additional studies using more chemicals are warranted to more accurately gauge performance of the models and bolster confidence in this approach.
Allergic contact dermatitis is the second most commonly reported occupational illness (Anderson et al., 2011) imparting a significant economic burden to industry (NIOSH 2012). Current hazard classification labeling schemes are based largely on animal test results (EPA 2012a; UN 2015) and the LLNA has been the preferred method for this purpose for a number of years (Basketter et al., 2009; ECHA 2015; EPA 2011). A wealth of data has been generated using the LLNA and regulators are accustomed to evaluating these data. The human predictive patch test data are sparse (Api et al., 2015) and, for cosmetic, are often negative because the goal of a human study in this case is usually to confirm that a particular dose determined not to cause an adverse reaction in an animal study will in fact not cause an adverse reaction in humans (Politano and Api 2008). These limitations make it challenging to model human outcomes. Given these points, it is not surprising that many efforts to develop an integrated decision strategy to predict skin sensitization emphasize LLNA outcomes (Bauch et al., 2012; Hirota et al., 2015; Jaworska et al., 2013; Jaworska et al., 2011; Luechtefeld et al., 2015; Natsch et al., 2009; Natsch et al., 2013; Nukada et al., 2013; Pirone et al., 2014; Strickland et al., 2016a; Takenouchi et al., 2015; Tsujita-Inoue et al., 2014; Urbisch et al., 2015; van der Veen et al., 2014). This approach relies on the assumption that there is a clear and reliable linkage between LLNA and human outcomes. However, previous evaluations of LLNA performance report accuracies as low as 72% for LLNA predictions of human sensitization hazard (ICCVAM 1999). The utility of LLNA potency classification for human risk assessment is also questionable. An ICCVAM evaluation (ICCVAM (2011)) revealed that the LLNA underclassified one-third of strong human sensitizers as weaker sensitizers. In that one-tiered analysis of 136 substances, the overall accuracy of LLNA for predicting human potency categories was only 54%. LLNA EC3 values were deemed suitable for classification of 1A sensitizers (71% accuracy), but not 1B sensitizers (52% accuracy). As a result, ICCVAM does not recommend the use of the LLNA as a stand-alone method to predict skin sensitization potency, but instead recommends including other types of supporting data (e.g. in chemico data and in vitro data) into an integrated decision strategy for categorization.
U.S. federal agencies represented on ICCVAM are committed to identifying and implementing reliable non-animal approaches to predicting human skin sensitization potency. The models developed here are an important step towards that goal. Future work will focus on testing the models with an expanded set of substances, including chemical formulations such as pesticides and a greater range of chemical structures. We will also investigate the impact of including additional data inputs such as cytotoxicity in predictive models for human sensitizer potency classification. Ultimately, we hope to advance a non-animal approach that is considered sufficient for routine regulatory use and end the use of animals for identifying skin sensitization hazards.
Supplementary Material
Acknowledgments
For contributions of DPRA, h-CLAT, and KeratinoSens data, the authors thank Drs. Hitoshi Sakaguchi and Masaaki Miyazawa of Kao Corporation, Dr.Takao Ashikaga of SHISEIDO Co., Ltd., Dr. Andreas Natsch of Givaudan Schweiz AG, and Dr. Daniel Urbisch of BASF. The authors thank Drs. D. Germolec, R. Luebke, M. Ward, and B.A. Merrick for their thoughtful critical review of this manuscript, and Ms. C. Sprankle for editorial review. This project was funded in whole or in part with federal funds from the National Institute of Environmental Sciences (NIEHS), National Institutes of Health (NIH) under contract HHSN273201500010C to ILS in support of NICEATM.
Footnotes
Disclaimer
This article may be the work product of an employee or group of employees of the NIEHS, NIH, U.S. Environmental Protection Agency, U.S. Food and Drug Administration, U.S. Consumer Product Safety Commission, or other organizations. However, the statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of the NIEHS, NIH, U.S. Environmental Protection Agency, U.S. Food and Drug Administration, U.S. Consumer Product Safety Commission, the United States government, or other organizations. ILS staff provide technical support for NICEATM, but do not represent NIEHS, NTP, or the official positions of any federal agency. The use of commercial product names is for comparative purposes only and does not constitute endorsement by any of the authors, organizations, or agencies.
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
- Anderson SE, Siegel PD, Meade BJ. The LLNA: A brief review of recent advances and limitations. J Allergy. 2011;2011:424203. doi: 10.1155/2011/424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Api AM, Basketter D, Lalko J. Correlation between experimental human and murine skin sensitization induction thresholds. Cutan Ocul Toxicol. 2015;34:298–302. doi: 10.3109/15569527.2014.979425. [DOI] [PubMed] [Google Scholar]
- Ashikaga T, Sakaguchi H, Sono S, Kosaka N, Ishikawa M, Nukada Y, Miyazawa M, Ito Y, Nishiyama N, Itagaki H. A comparative evaluation of in vitro skin sensitisation tests: the human cell-line activation test (h-CLAT) versus the local lymph node assay (LLNA) Altern Lab Anim. 2010;38:275–284. doi: 10.1177/026119291003800403. [DOI] [PubMed] [Google Scholar]
- Ashikaga T, Yoshida Y, Hirota M, Yoneyama K, Itagaki H, Sakaguchi H, Miyazawa M, Ito Y, Suzuki H, Toyoda H. Development of an in vitro skin sensitization test using human cell lines: the human Cell Line Activation Test (h-CLAT). I. Optimization of the h-CLAT protocol. Toxicol In Vitro. 2006;20:767–773. doi: 10.1016/j.tiv.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Ball N, Cagen S, Carrillo JC, Certa H, Eigler D, Emter R, Faulhammer F, Garcia C, Graham C, Haux C, Kolle SN, Kreiling R, Natsch A, Mehling A. Evaluating the sensitization potential of surfactants: Integrating data from the local lymph node assay, guinea pig maximization test, and in vitro methods in a weight-of-evidence approach. Regul Toxicol Pharmacol. 2011;60:389–400. doi: 10.1016/j.yrtph.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Alepee N, Ashikaga T, Barroso J, Gilmour N, Goebel C, Hibatallah J, Hoffmann S, Kern P, Martinozzi-Teissier S, Maxwell G, Reisinger K, Sakaguchi H, Schepky A, Tailhardat M, Templier M. Categorization of chemicals according to their relative human skin sensitizing potency. Dermatitis. 2014;25:11–21. doi: 10.1097/DER.0000000000000003. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Gerberick GF, Kimber I, Loveless SE. The local lymph node assay: a viable alternative to currently accepted skin sensitization tests. Food Chem Toxicol. 1996;34:985–997. doi: 10.1016/s0278-6915(96)00059-2. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Kimber I. Predictive tests for irritants and allergens and their use in quantitative risk assessment. In: Frosch P, Menné T, Lepoittevin J-P, editors. Contact Dermatitis. Springer Verlag; Heidelberg: 2006. pp. 179–188. [Google Scholar]
- Basketter DA, McFadden JF, Gerberick F, Cockshott A, Kimber I. Nothing is perfect, not even the local lymph node assay: A commentary and the implications for REACH. Contact Dermatitis. 2009;60:65–69. doi: 10.1111/j.1600-0536.2008.01444.x. [DOI] [PubMed] [Google Scholar]
- Bauch C, Kolle SN, Fabian E, Pachel C, Ramirez T, Wiench B, Wruck CJ, Ravenzwaay BV, Landsiedel R. Intralaboratory validation of four in vitro assays for the prediction of the skin sensitizing potential of chemicals. Toxicol In Vitro. 2011;25:1162–1168. doi: 10.1016/j.tiv.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Bauch C, Kolle SN, Ramirez T, Eltze T, Fabian E, Mehling A, Teubner W, van Ravenzwaay B, Landsiedel R. Putting the parts together: combining in vitro methods to test for skin sensitizing potentials. Regul Toxicol Pharmacol. 2012;63:489–504. doi: 10.1016/j.yrtph.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Brown LD, Cai TT, DasGupta A. Interval estimation for a binomial proportion. Statistical Science. 2001;16:101–133. [Google Scholar]
- Brutti CS, Bonamigo RR, Cappelletti T, Martins-Costa GM, Menegat AP. Occupational and non-occupational allergic contact dermatitis and quality of life: a prospective study. An Bras Dermatol. 2013;88:670–671. doi: 10.1590/abd1806-4841.20131950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JH, Twerdok LE, Tice RR, Sailstad DM, Hattan DG, Stokes WS. ICCVAM evaluation of the murine local lymph node assay: II. Conclusions and recommendations of an independent scientific peer review panel. Regul Toxicol Pharmacol. 2001;34:258–273. doi: 10.1006/rtph.2001.1497. [DOI] [PubMed] [Google Scholar]
- Deconinck E, Hancock T, Coomans D, Massart DL, Heyden YV. Classification of drugs in absorption classes using the classification and regression trees (CART) methodology. J Pharm Biomed Anal. 2005;39:91–103. doi: 10.1016/j.jpba.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Diaz-Uriarte R. GeneSrF and varSelRF: A web-based tool and R package for gene selection and classification using random forest. BMC Bioinformatics. 2007;8:328. doi: 10.1186/1471-2105-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EC. Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 2006 [Google Scholar]
- EC. Regulation (EC) No 1272/2008 of the European Parliament and of the Council of 16 December 2008 2008 [Google Scholar]
- ECHA. Chapter R.7a: Endpoint Specific Guidance (Version 4.0) European Chemicals Agency (ECHA); Helsinki: 2015. Guidance on Information Requirements and Chemical Safety Assessment. [Google Scholar]
- Emter R, Ellis G, Natsch A. Performance of a novel keratinocyte-based reporter cell line to screen skin sensitizers in vitro. Toxicol Appl Pharmacol. 2010;245:281–290. doi: 10.1016/j.taap.2010.03.009. [DOI] [PubMed] [Google Scholar]
- EPA. Expansion of the Traditional Local Lymph Node Assay for the Assessment of Dermal Sensitization Potential of End Use Pesticide Products; and Adoption of a “Reduced” Protocol for the Traditional LLNA (Limit Dose) Office of Pesticide Programs; Washington, DC: 2011. [Google Scholar]
- EPA. Label Review Manual [Internet] U.S. Environmental Protection Agency; Washington, DC: 2012a. [Google Scholar]
- EPA. Toxicology Data Requirement Table, 40 CFR 158.500. EPA; 2012b. Available: http://www.gpo.gov/fdsys/pkg/CFR-2012-title40-vol25/xml/CFR-2012-title40-vol25-sec158-500.xml. [Google Scholar]
- EPA. Toxicology Data Requirements, 40 CFR 161.340. EPA; 2012c. Available: http://www.gpo.gov/fdsys/pkg/CFR-2012-title40-vol25/xml/CFR-2012-title40-vol25-sec161-340.xml. [Google Scholar]
- Estrada E, Patlewicz G, Chamberlain M, Basketter D, Larbey S. Computer-aided knowledge generation for understanding skin sensitization mechanisms: the TOPS-MODE approach. Chem Res Toxicol. 2003;16:1226–1235. doi: 10.1021/tx034093k. [DOI] [PubMed] [Google Scholar]
- Garnett A, Hotchkiss SA, Caldwell J. Percutaneous absorption of benzyl acetate through rat skin in vitro. 3. A comparison with human skin. Food Chem Toxicol. 1994;32:1061–1065. doi: 10.1016/0278-6915(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Bailey RE, Chaney JG, Morrall SW, Lepoittevin JP. Development of a peptide reactivity assay for screening contact allergens. Toxicol Sci. 2004;81:332–343. doi: 10.1093/toxsci/kfh213. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Foertsch LM, Price BB, Chaney JG, Lepoittevin JP. Quantification of chemical peptide reactivity for screening contact allergens: a classification tree model approach. Toxicol Sci. 2007;97:417–427. doi: 10.1093/toxsci/kfm064. [DOI] [PubMed] [Google Scholar]
- Hao M, Li Y, Wang Y, Zhang S. A classification study of respiratory syncytial virus (RSV) inhibitors by variable selection with random forest. Int J Mol Sci. 2011;12:1259–1280. doi: 10.3390/ijms12021259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterberg MV, Menné T, Johansen JD. Contact allergy to the 26 specific fragrance ingredients to be declared on cosmetic products in accordance with the EU cosmetics directive. Contact Dermatitis. 2011;65:266–275. doi: 10.1111/j.1600-0536.2011.01962.x. [DOI] [PubMed] [Google Scholar]
- Hirota M, Fukui S, Okamoto K, Kurotani S, Imai N, Fujishiro M, Kyotani D, Kato Y, Kasahara T, Fujita M, Toyoda A, Sekiya D, Watanabe S, Seto H, Takenouchi O, Ashikaga T, Miyazawa M. Evaluation of combinations of in vitro sensitization test descriptors for the artificial neural network-based risk assessment model of skin sensitization. J Appl Toxicol. 2015;35:1333–1347. doi: 10.1002/jat.3105. [DOI] [PubMed] [Google Scholar]
- ICCVAM. The Murine Local Lymph Node Assay: A Test Method for Assessing the Allergic Contact Dermatitis Potential of Chemicals/Compounds. The Results of an Independent Peer Review Evaluation Coordinated by the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) and the National Toxicology Program Center for the Evaluation of Alternative Toxicological Methods (NICEATM). National Institute of Environmental Health Sciences; Research Triangle Park, NC: 1999. [Google Scholar]
- ICCVAM. ICCVAM Test Method Evaluation Report: Usefulness and Limitations of the Murine Local Lymph Node Assay for Potency Categorization of Chemicals Causing Allergic Contact Dermatitis in Humans. National Institute of Environmental Health Sciences; Research Triangle Park, NC: 2011. [Google Scholar]
- Jaworska J, Dancik Y, Kern P, Gerberick F, Natsch A. Bayesian integrated testing strategy to assess skin sensitization potency: From theory to practice. J Appl Toxicol. 2013;33:1353–1364. doi: 10.1002/jat.2869. [DOI] [PubMed] [Google Scholar]
- Jaworska J, Harol A, Kern PS, Gerberick GF. Integrating non-animal test information into an adaptive testing strategy - Skin sensitization proof of concept case. ALTEX. 2011;28:211–225. doi: 10.14573/altex.2011.3.211. [DOI] [PubMed] [Google Scholar]
- Jaworska JS, Natsch A, Ryan C, Strickland J, Ashikaga T, Miyazawa M. Bayesian integrated testing strategy (ITS) for skin sensitization potency assessment: a decision support system for quantitative weight of evidence and adaptive testing strategy. Arch Toxicol. 2015;89:2355–2383. doi: 10.1007/s00204-015-1634-2. [DOI] [PubMed] [Google Scholar]
- Joint Research Centre of the European Union. EURL ECVAM Recommendation on the Direct Peptide Reactivity Assay (DPRA) for Skin Sensitisation Testing. Publications Office of the European Union; Luxembourg: 2013. [Google Scholar]
- Joint Research Centre of the European Union. EURL ECVAM Recommendation on the KeratinoSens™ assay for skin sensitisation testing. Publications Office of the European Union; Luxembourg: 2014. [Google Scholar]
- Kadyk DL, McCarter K, Achen F, Belsito DV. Quality of life in patients with allergic contact dermatitis. J Am Acad Dermatol. 2003;49:1037–1048. doi: 10.1016/s0190-9622(03)02112-1. [DOI] [PubMed] [Google Scholar]
- Kimber I, Basketter DA, Gerberick GF, Ryan CA, Dearman RJ. Chemical allergy: Translating biology into hazard characterization. Toxicol Sci. 2011;120:S238–S268. doi: 10.1093/toxsci/kfq346. [DOI] [PubMed] [Google Scholar]
- Luan F, Zhang R, Zhao C, Yao X, Liu M, Hu Z, Fan B. Classification of the carcinogenicity of N-nitroso compounds based on support vector machines and linear discriminant analysis. Chem Res Toxicol. 2005;18:198–203. doi: 10.1021/tx049782q. [DOI] [PubMed] [Google Scholar]
- Luechtefeld T, Maertens A, McKim JM, Hartung T, Kleensang A, Sa-Rocha V. Probabilistic hazard assessment for skin sensitization potency by dose-response modeling using feature elimination instead of quantitative structure-activity relationships. J Appl Toxicol. 2015;35:1361–1371. doi: 10.1002/jat.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macan J, Rimac D, Kezic S, Varnai VM. Occupational and non-occupational allergic contact dermatitis: A follow-up study. Dermatology. 2013;227:321–329. doi: 10.1159/000354763. [DOI] [PubMed] [Google Scholar]
- Mehling A, Eriksson T, Eltze T, Kolle S, Ramirez T, Teubner W, van Ravenzwaay B, Landsiedel R. Non-animal test methods for predicting skin sensitization potentials. Arch Toxicol. 2012;86:1273–1295. doi: 10.1007/s00204-012-0867-6. [DOI] [PubMed] [Google Scholar]
- Mint A, Hotchkiss SA, Caldwell J. Percutaneous absorption of diethyl phthalate through rat and human skin in vitro. Toxicol In Vitro. 1994;8:251–256. doi: 10.1016/0887-2333(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Montelius J, Wahlkvist H, Boman A, Wahlberg JE. Murine local lymph node assay for predictive testing of allergenicity: Two irritants caused significant proliferation. Acta Derm Venereol. 1998;78:433–437. doi: 10.1080/000155598442728. [DOI] [PubMed] [Google Scholar]
- Natsch A, Emter R, Ellis G. Filling the concept with data: Integrating data from different in vitro and in silico assays on skin sensitizers to explore the battery approach for animal-free skin sensitization testing. Toxicol Sci. 2009;107:106–121. doi: 10.1093/toxsci/kfn204. [DOI] [PubMed] [Google Scholar]
- Natsch A, Emter R, Gfeller H, Haupt T, Ellis G. Predicting skin sensitizer potency based on in vitro data from keratinosens and kinetic peptide binding: Global versus domain-based assessment. Toxicol Sci. 2015;143:319–332. doi: 10.1093/toxsci/kfu229. [DOI] [PubMed] [Google Scholar]
- Natsch A, Ryan CA, Foertsch L, Emter R, Jaworska J, Gerberick F, Kern P. A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J Appl Toxicol. 2013;33:1337–1352. doi: 10.1002/jat.2868. [DOI] [PubMed] [Google Scholar]
- NIEHS. Request for Information on Alternative Skin Sensitization Test Methods and Testing Strategies and for Comment on ICCVAM’s Proposed Activities. Fed Regist. 2013;78:68076–68077. [Google Scholar]
- NIOSH. Skin Exposures and Effects. Workplace Safety and Health. 2012 http://www.cdc.gov/niosh/topics/skin/ [April 3, 2013]
- Nukada Y, Ashikaga T, Miyazawa M, Hirota M, Sakaguchi H, Sasa H, Nishiyama N. Prediction of skin sensitization potency of chemicals by human cell line activation Test (h-CLAT) and an attempt at classifying skin sensitization potency. Toxicol In Vitro. 2012;26:1150–1160. doi: 10.1016/j.tiv.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Nukada Y, Ashikaga T, Sakaguchi H, Sono S, Mugita N, Hirota M, Miyazawa M, Ito Y, Sasa H, Nishiyama N. Predictive performance for human skin sensitizing potential of the human cell line activation test (h-CLAT) Contact Dermatitis. 2011;65:343–353. doi: 10.1111/j.1600-0536.2011.01952.x. [DOI] [PubMed] [Google Scholar]
- Nukada Y, Miyazawa M, Kazutoshi S, Sakaguchi H, Nishiyama N. Data integration of non-animal tests for the development of a test battery to predict the skin sensitizing potential and potency of chemicals. Toxicol In Vitro. 2013;27:609–618. doi: 10.1016/j.tiv.2012.11.006. [DOI] [PubMed] [Google Scholar]
- OECD. OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects. OECD Publishing; Paris: 1992. Test No 406. Skin Sensitisation. [Google Scholar]
- OECD. LLNA Test Guideline 429 2010 [Google Scholar]
- OECD. Part 2: Use of the AOP to Develop Chemical Categories and Integrated Assessment and Testing Approaches. OECD Publishing; Paris: 2012a. OECD Series on Testing and Assessment No 168. The Adverse Outcome Pathway for Skin Sensitisation Initated by Covalent Binding to Proteins. [Google Scholar]
- OECD. Part 1: Scientific Assessment. OECD Publishing; Paris: 2012b. OECD Series on Testing and Assessment No 168. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins. [Google Scholar]
- OECD. OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects. OECD Publsihing; Paris: 2015a. Test No. 442C. In Chemico Skin Sensitization: Direct Peptide Reactivity Assay (DPRA) [Google Scholar]
- OECD. OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects. OECD Publishing; Paris: 2015b. Test No. 442D. In Vitro Skin Sensitisation: ARE-Nrf2 Luciferase Test Method. [Google Scholar]
- OECD. OECD guideline for the testing of chemicals: human cell line activation test (h-CLAT) 2016a http://www.oecd-ilibrary.org/docserver/download/9716121e.pdf?expires=1472755387&id=id&accname=guest&checksum=094DA96195CA77C00F5FA1C0790057C3 [September 1, 2016]
- OECD. The OECD QSAR Toolbox. 2016b http://www.oecd.org/chemicalsafety/risk-assessment/theoecdqsartoolbox.htm [August 31, 2016]
- OSHA. Washington DC. Occupational Safety and Health Standards, 29 CFR 1910.1200. OSHA; 2012. Available: http://www.osha.gov/pls/oshaweb/owadisp.show_document?p_table=STANDARDS&p_id=10099. [Google Scholar]
- Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts DW, Mekenyan O. Towards AOP application–implementation of an integrated approach to testing and assessment (IATA) into a pipeline tool for skin sensitization. Regul Toxicol Pharmacol. 2014;69:529–545. doi: 10.1016/j.yrtph.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Casati S, Basketter DA, Asturiol D, Roberts DW, Lepoittevin JP, Worth AP, Aschberger K. Can currently available non-animal methods detect pre and pro-haptens relevant for skin sensitization? Regul Toxicol Pharmacol. 2016 Aug 26; doi: 10.1016/j.yrtph.2016.08.007. pii: S0273-2300(16)30228-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Pirone JR, Smith M, Kleinstreuer NC, Burns TA, Strickland J, Dancik Y, Morris R, Rinckel LA, Casey W, Jaworska JS. Open source software implementation of an integrated testing strategy for skin sensitization potency based on a Bayesian network. ALTEX. 2014;31:336–340. doi: 10.14573/altex.1310151. [DOI] [PubMed] [Google Scholar]
- Politano VT, Api AM. The Research Institute for Fragrance Materials’ human repeated insult patch test protocol. Regul Toxicol Pharmacol. 2008;52:35–38. doi: 10.1016/j.yrtph.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Questier F, Put R, Coomans D, Walczak B, Heyden YV. The use of CART and multivariate regression trees for supervised and unsupervised feature selection. Chemometr Intell Lab. 2005;76:45–54. [Google Scholar]
- Reisinger K, Hoffmann S, Alepee N, Ashikaga T, Barroso J, Elcombe C, Gellatly N, Galbiati V, Gibbs S, Groux H, Hibatallah J, Keller D, Kern P, Klaric M, Kolle S, Kuehnl J, Lambrechts N, Lindstedt M, Millet M, Martinozzi-Teissier S, Natsch A, Petersohn D, Pike I, Sakaguchi H, Schepky A, Tailhardat M, Templier M, van Vliet E, Maxwell G. Systematic evaluation of non-animal test methods for skin sensitisation safety assessment. Toxicol In Vitro. 2015;29:259–270. doi: 10.1016/j.tiv.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Roberts DW, Patlewicz GY. Integrated testing and assessment approaches for skin sensitization: a commentary. J Appl Toxicol. 2014;34:436–440. doi: 10.1002/jat.2943. [DOI] [PubMed] [Google Scholar]
- Rovida C, Alepee N, Api AM, Basketter DA, Bois FY, Caloni F, Corsini E, Daneshian M, Eskes C, Ezendam J, Fuchs H, Hayden P, Hegele-Hartung C, Hoffmann S, Hubesch B, Jacobs MN, Jaworska J, Kleensang A, Kleinstreuer N, Lalko J, Landsiedel R, Lebreux F, Luechtefeld T, Locatelli M, Mehling A, Natsch A, Pitchford JW, Prater D, Prieto P, Schepky A, Schuurmann G, Smirnova L, Toole C, van Vliet E, Weisensee D, Hartung T. Integrated Testing Strategies (ITS) for safety assessment. ALTEX. 2015;32:25–40. doi: 10.14573/altex.1411011. [DOI] [PubMed] [Google Scholar]
- Sailstad DM, Hattan D, Hill RN, Stokes WS. ICCVAM evaluation of the murine local lymph node assay: I. The ICCVAM review process. Regul Toxicol Pharmacol. 2001;34:249–257. doi: 10.1006/rtph.2001.1496. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Ryan C, Ovigne JM, Schroeder KR, Ashikaga T. Predicting skin sensitization potential and inter-laboratory reproducibility of a human cell line activation test (h-CLAT) in the European Cosmetics Association (COLIPA) ring trials. Toxicol In Vitro. 2010;24:1810–1820. doi: 10.1016/j.tiv.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Raghavan B, Müller V, Vogl T, Feier G, Tchaptchet S, Keck S, Kalis C, Nielsen PJ, Galanos C, Roth J, Skerra A, Martin SF, Freudenberg MA, Goebeler M. Crucial role for human toll-like receptor 4 in the development of contact allergy to nickel. Nat Immunol. 2010;11(9):814–819. doi: 10.1038/ni.1919. [DOI] [PubMed] [Google Scholar]
- Shen MY, Su BH, Esposito EX, Hopfinger AJ, Tseng YJ. A comprehensive support vector machine binary hERG classification model based on extensive but biased end point hERG data sets. Chem Res Toxicol. 2011;24:934–949. doi: 10.1021/tx200099j. [DOI] [PubMed] [Google Scholar]
- Smith CK, Hotchkiss SAM. Allergic Contact Dermatitis Chemical and Metabolic Mechanisms. Taylor and Francis; London and New York: 2001. [Google Scholar]
- Strickland J, Zang Q, Kleinstreuer N, Paris M, Lehmann DM, Choksi N, Matheson J, Jacobs A, Lowit A, Allen D, Casey W. Integrated decision strategies for skin sensitization hazard. J Appl Toxicol. 2016a;35(9):1150–1162. doi: 10.1002/jat.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland J, Zang Q, Paris M, Lehmann DM, Kleinstreuer N, Allen D, Choksi N, Matheson J, Jacobs A, Casey W. Multivariate models for prediction of human skin sensitization hazard. J Appl Toxicol. 2016b Aug 2; doi: 10.1002/jat.3366. 2016. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi O, Fukui S, Okamoto K, Kurotani S, Imai N, Fujishiro M, Kyotani D, Kato Y, Kasahara T, Fujita M, Toyoda A, Sekiya D, Watanabe S, Seto H, Hirota M, Ashikaga T, Miyazawa M. Test battery with the human cell line activation test, direct peptide reactivity assay and DEREK based on a 139 chemical data set for predicting skin sensitizing potential and potency of chemicals. J Appl Toxicol. 2015;35(11):1318–1332. doi: 10.1002/jat.3127. [DOI] [PubMed] [Google Scholar]
- Takenouchi O, Miyazawa M, Saito K, Ashikaga T, Sakaguchi H. Predictive performance of the human cell line activation Test (h-CLAT) for lipophilic chemicals with high octanol-water partition coefficients. J Toxicol Sci. 2013;38:599–609. doi: 10.2131/jts.38.599. [DOI] [PubMed] [Google Scholar]
- The R Core Team. R: A Language and Environment for Statistical Computing Reference Index. R Foundation for Statistical Computing; 2013. [Google Scholar]
- Tsujita-Inoue K, Hirota M, Ashikaga T, Atobe T, Kouzuki H, Aiba S. Skin sensitization risk assessment model using artificial neural network analysis of data from multiple in vitro assays. Toxicol In Vitro. 2014;28:626–639. doi: 10.1016/j.tiv.2014.01.003. [DOI] [PubMed] [Google Scholar]
- UN. Globally Harmonized System for Classification and Labelling of Chemicals. United Nations; New York: 2015. [Google Scholar]
- Urbisch D, Mehling A, Guth K, Ramirez T, Honarvar N, Kolle S, Landsiedel R, Jaworska J, Kern PS, Gerberick F, Natsch A, Emter R, Ashikaga T, Miyazawa M, Sakaguchi H. Assessing skin sensitization hazard in mice and men using non-animal test methods. Regul Toxicol Pharmacol. 2015;71:337–351. doi: 10.1016/j.yrtph.2014.12.008. [DOI] [PubMed] [Google Scholar]
- van der Veen JW, Rorije E, Emter R, Natsch A, van Loveren H, Ezendam J. Evaluating the performance of integrated approaches for hazard identification of skin sensitizing chemicals. Regul Toxicol Pharmacol. 2014;69:371–379. doi: 10.1016/j.yrtph.2014.04.018. [DOI] [PubMed] [Google Scholar]
- Van Och FMM, Slob W, De Jong WH, Vandebriel RJ, Van Loveren H. A quantitative method for assessing the sensitizing potency of low molecular weight chemicals using a local lymph node assay: employment of a regression method that includes determination of the uncertainty margins. Toxicology. 2000;146:49–59. doi: 10.1016/s0300-483x(00)00165-7. [DOI] [PubMed] [Google Scholar]
- Varmuza K, Peter F. Introduction to Multivariate Statistical Analysis in Chemometrics. CRC Press; Boca Raton, Florida: 2009. [Google Scholar]
- Zang Q, Keire DA, Buhse LF, Wood RD, Mital DP, Haque S, Srinivasan S, Moore CM, Nasr M, Al-Hakim A, Trehy ML, Welsh WJ. Identification of heparin samples that contain impurities or contaminants by chemometric pattern recognition analysis of proton NMR spectral data. Anal Bioanal Chem. 2011a;401:939–955. doi: 10.1007/s00216-011-5155-4. [DOI] [PubMed] [Google Scholar]
- Zang Q, Keire DA, Wood RD, Buhse LF, Moore CM, Nasr M, Al-Hakim A, Trehy ML, Welsh WJ. Combining (1)H NMR spectroscopy and chemometrics to identify heparin samples that may possess dermatan sulfate (DS) impurities or oversulfated chondroitin sulfate (OSCS) contaminants. J Pharm Biomed Anal. 2011b;54:1020–1029. doi: 10.1016/j.jpba.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Zang Q, Rotroff DM, Judson RS. Binary classification of a large collection of environmental chemicals from estrogen receptor assays by quantitative structure-activity relationship and machine learning methods. J Chem Inf Model. 2013;53:3244–3261. doi: 10.1021/ci400527b. [DOI] [PubMed] [Google Scholar]
- Zang Q, Mansouri K, Williams AJ, Judson RS, Allen DG, Casey WM, Kleinstreuer NC. In Silico Prediction of Physicochemical Properties of Environmental Chemicals Using Molecular Fingerprints and Machine Learning. Submitted to Journal of Chemical Information and Modeling. doi: 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


