Abstract
HIV-associated neurocognitive disorder (HAND) often complicates HIV infection despite combination antiretroviral therapy (ART) and may be influenced by host genomics. We performed a genome-wide association study (GWAS) of HAND in 1,050 CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) Study participants. All participants underwent standardized, comprehensive neurocognitive and neuromedical assessments to determine if they had cognitive impairment as assessed by the Global Deficit Score (GDS), and individuals with comorbidities that could confound diagnosis of HAND were excluded. Neurocognitive outcomes included GDS-defined neurocognitive impairment (NCI; binary GDS, 366 cases with GDS≥0.5 and 684 controls with GDS<0.5, and GDS as a continuous variable) and Frascati HAND definitions that incorporate assessment of functional impairment by self-report and performance-based criteria. Genotype data was obtained using the Affymetrix Human SNP Array 6.0 platform. Multivariable logistic or linear regression-based association tests were performed for GDS-defined NCI and HAND. GWAS results did not reveal SNPs meeting the genome-wide significance threshold (5.0×10−8) for GDS-defined NCI or HAND. For binary GDS, the most significant SNPs were rs6542826 (p=8.1×10−7) and rs11681615 (1.2×10−6), both located on chromosome 2 in SH3RF3. The most significant SNP for continuous GDS was rs11157436 (p=1.3×10−7) on chromosome 14 in the T-cell-receptor alpha locus; three other SNPs in this gene were also associated with binary GDS (p≤2.9×10−6). This GWAS, conducted among ART-era participants from a single cohort with robust neurological phenotyping, suggests roles for several biologically plausible loci in HAND that deserve further exploration.
Keywords: HIV-Associated Neurocognitive Disorder, neurocognitive impairment, GWAS, genotype, CHARTER study, Global Deficit Score
INTRODUCTION
HIV-associated neurocognitive disorder (HAND) remains a common complication of HIV infection in the combination antiretroviral therapy (ART) era. Although modern ART has substantially improved life expectancy and reduced the incidence of HIV-associated dementia (HAD), milder forms of HAND remain prevalent, including asymptomatic neurocognitive impairment (ANI) and mild neurocognitive disorder (MND). The overall prevalence of HAND is 30 to 50% among unselected HIV-infected persons [Grant 2008; Heaton and others 2010] and is predominantly driven by the ANI category, which predicts decline to symptomatic neurocognitive impairment (NCI) [Grant and others 2014]. The neuropathogenesis of HIV-infection remains incompletely understood, although potential risk factors include persistent low-level central nervous system (CNS) HIV infection, chronic immune activation, and accelerated aging, among others [Fogel and others 2015; Heaton and others 2011; Horvath and Levine 2015].
Host genetics plays a significant role in many neuropsychiatric disorders and has been studied with increasing interest for its potential contribution to HIV disease complications, including HAND [Kallianpur and Levine 2014]. Variants in genes regulating multiple biological processes may impact the risk of NCI, progression of HAND, and response to ART. Candidate-gene association studies [Bol and others 2012; Burt and others 2008] and a single published genome-wide association study (GWAS) [Levine and others 2012] revealed promising genetic variants associated with HAND, including variants in immune-related genes [Pemberton and others 2008], ion channel transporters [Kallianpur and others 2014], and genes related to nuclear and mitochondrial DNA damage [Kallianpur and Levine 2014]. However, most of these associations failed to reach genome-wide statistical significance and have either not been subjected to independent replication attempts, or have not been successfully replicated in subsequent studies, due at least in part to phenotypic and population differences between studies. To be successful, GWAS ideally requires large sample sizes, meticulous and standardized phenotyping, and careful attention to population stratification, and few studies combining neurocognitive and genome-wide genetic data across cohorts for GWAS or meta-analyses have been undertaken to date. In the single published GWAS that was conducted specifically to evaluate HAND [Levine and others 2012], no prior single-nucleotide polymorphism (SNP) association was replicated, and no new SNP association reached genome-wide statistical significance (p=5×10−8).
In this study, we conducted a GWAS of HAND in participants in the CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) Study [Heaton and others 2010], which was specifically designed to study neurological outcomes in HIV infection and is the largest U.S.-based prospective study of neurological complications of HIV/AIDS in the ART era. We performed genome-wide genotyping and comprehensive quality control (QC), resulting in a working dataset comprising 1,050 individuals. For phenotype definition, we employed a commonly used and validated measure of NCI, the global deficit score (GDS). We conducted genome-wide association tests using the GDS score as a dichotomous or continuous variable and Frascati criteria for HAND to define clinical phenotypes. Although we did not discover SNPs meeting the genome-wide (Bonferroni) criterion for statistical significance, we did identify new and promising genes with biological plausibility and p-values approaching this threshold, which warrant further investigation.
METHODS
Study population
DNA samples were collected from 1,082 CHARTER study participants. CHARTER is a prospective, observational study of neurobehavioral outcomes of HIV disease, which enrolled ambulatory, HIV-infected adults from 2003 to 2007 at six U.S. medical centers. Study participants underwent detailed, structured interviews and laboratory assessments to obtain information on HIV disease and treatment-related factors, including nadir CD4+ T-cell counts, ART history, nucleoside reverse-transcriptase inhibitor (NRTI) exposure, history of a major depressive disorder, substance use/dependency, comorbidity, and demographics. Data were also collected on current CD4+ T-cell count, HIV RNA levels in plasma, and hepatitis C virus serology. Details regarding CHARTER study eligibility criteria, enrollment and follow-up procedures have been published previously [Heaton and others 2011; Heaton and others 2015]. CHARTER is approved by the Institutional Review Boards of all participating sites and abides by the Declaration of Helsinki; all study participants provided written informed consent. Comorbid conditions of participants were determined by experienced clinicians to be either minimal (“incidental” to NCI), or mild to moderate severity (“contributing” to NCI) [Heaton and others 2010]. To minimize confounding by conditions other than HIV infection, individuals with severe comorbid conditions that would make it impossible to attribute NCI to HIV infection (e.g., individuals with a history of traumatic brain injury with prolonged loss of consciousness, or developmental learning disorders) were excluded from genetic analyses. Detailed descriptions of minimal, mild/moderate comorbidities, their determinations, and frequency in CHARTER have been presented elsewhere [Antinori and others 2007; Heaton and others 2010; Kallianpur and others 2016].
Neurocognitive phenotypes
All participants were assigned a GDS value at their first or baseline visit [Kallianpur and others 2016]. GDS was analyzed as a continuous measure in HIV-infected persons. Following a conventional threshold [Blackstone and others 2012], we categorized participants as cases and controls according to their GDS values at baseline, i.e., cases with GDS-defined NCI were individuals with GDS≥0.5, while controls were those with GDS<0.5. The binary GDS cutoff of 0.5 is standard in studies of neurocognition in CHARTER and has also been used in other HIV-related studies of neurocognitive function that employ the same test battery [Blackstone and others 2012]. We assigned two GDS-related phenotypes: one quantitative, using continuous GDS values and the other categorical, termed “binary GDS”.
We also explored different clinical subcategories of HAND using the well-accepted Frascati criteria [Antinori and others 2007] which incorporate information from self-report regarding functional status. All CHARTER participants were classified into four subgroups, either one of three impaired subgroups (nANI=359, nMND=92, and nHAD=30), or neuropsychiatrically normal (nNCN=568). One individual with a GDS value lacked a HAND assessment. For the purpose of GWAS analyses, we compared different categories of HAND according to the severity of functional impairment and also combined specific groups for comparison, including “all HAND” versus No HAND (NCN), (ANI and MND) versus NCN, MND versus NCN, and ANI versus NCN. The GDS is more conservative for establishing presence of NCI than the Frascati criteria, but a GDS ≥ 0.5 virtually guarantees that Frascati criteria are also met [Blackstone and others 2012]. The GDS as a continuous variable provides additional information regarding the severity of NCI. Therefore, we used GDS for the cognitive impairment definition and HAND categories (Frascati criteria) for establishing symptomatic status and ruling out confounds. To focus the presentation of results, we present the results of GWAS analyses of HAND phenotypes in the Supplementary Results.
Genotyping
Genomic DNA was extracted from peripheral blood mononuclear cells collected at the baseline CHARTER visit using PUREGENE (Gentra Systems, Inc, Minneapolis, MN). All samples were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0TM. Genotyping was conducted by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) at Vanderbilt University in two batches: 576 samples were genotyped before 2009 and 506 (6 were repeats for QC) were genotyped in 2012. Notably, all CHARTER study participants were recruited prior to 2009; however, genotyping was performed in two batches due only to funding availability, rather than to technical or biological concerns. We applied the same QC procedure and analysis pipeline, however, for all samples combined. Nevertheless, batch effect was tested explicitly, and only minor changes were observed (see Results).
Quality control (QC)
We first checked the call rate per individual, which was found to be >95% in all samples. A sex check was performed using PLINK to compute the homozygosity rate of X-chromosome (the F value). F value is close to 1 for male and is less than 0.2 for female; hence, it can be used to predict sex information. Individuals with discrepant sex information were categorized as female or male based on their F values. Samples with misclassified or discrepant sex information were reviewed, double-checked, and revised as necessary. There were 21 samples that were self-reported as female but had an F-value between 0.2 and 0.5. These individuals were assigned as female. Samples with extreme missing rates or heterozygosity rates were also detected using PLINK and removed following the protocol in [Anderson and others 2010].
Unknown relatedness and duplicated individuals were assessed using pairwise identity-by-state (IBS) and identity-by-descent (IBD) estimations. The parameter PI_Hat, which was computed by PLINK for any pair of samples, was used to determine sample correlations. For pairs with high PI_Hat values (i.e., >0.2), the sample with a higher missing rate was removed from the analysis. In total, 28 samples were removed, including technical repeats and controls.
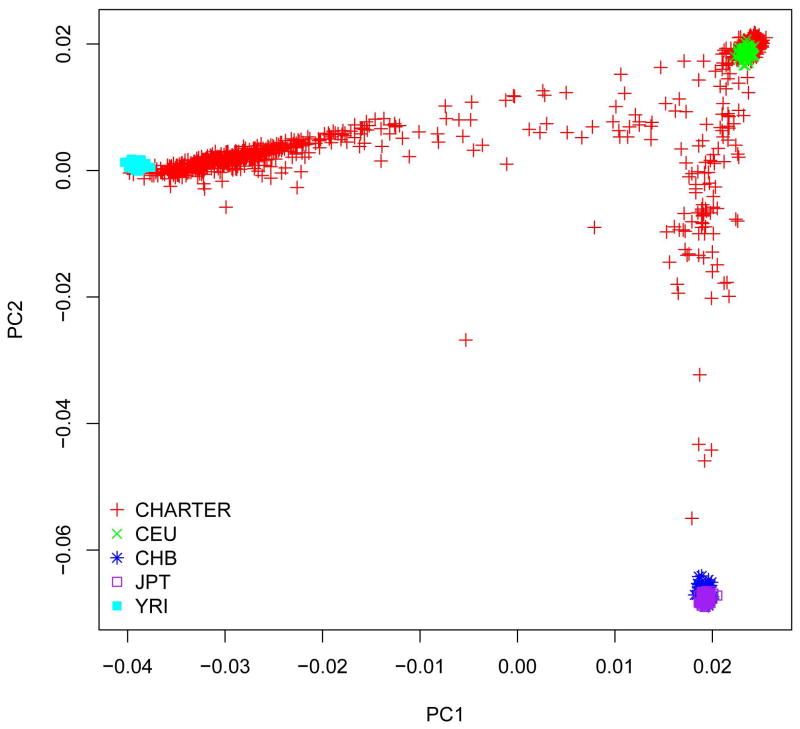
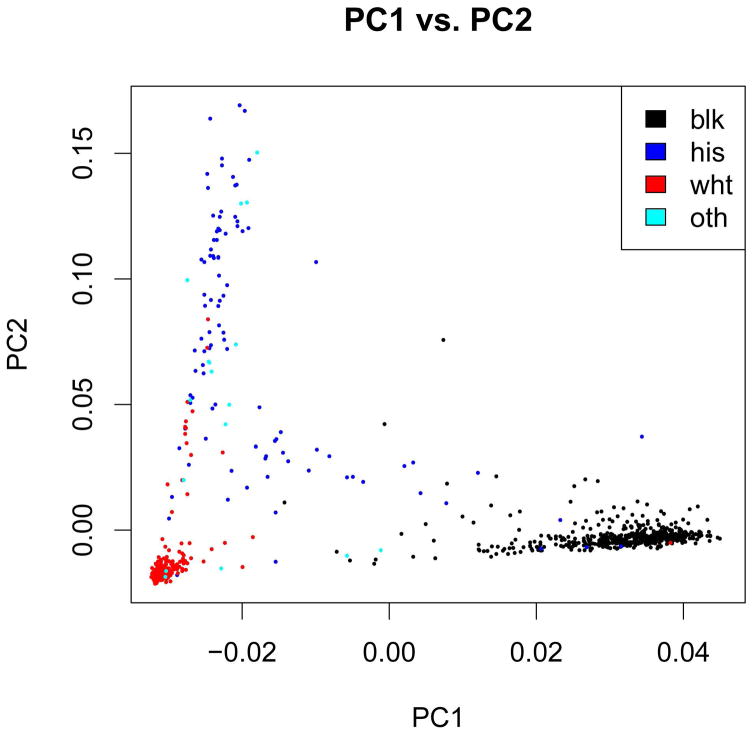
Our samples included self-reported European American (EA), African American (AA), Hispanic (H), and others. We merged CHARTER samples with HapMap samples from 4 populations. We then performed Principal Component (PC) Analysis using EIGENSOFT [Patterson and others 2006] to assess population stratification. To avoid high linkage disequilibrium (LD) between SNPs, we used the r2 parameter, with r2 threshold set to 0.5. This PC analysis process identified two individuals who had greater than six standard deviations (SDs) from the mean for five iterations (default). These two individuals were removed. The PC analysis also identified two samples whose self-reported ethnicity was inconsistent with what the PCs suggested. One sample was self-reported as AA and the other was self-reported as Hispanic but both were predicted to be EA according to the first and second PCs. However, because our association test did not distinguish between these subpopulations, we included the two samples with inconsistent race/ethnicity in the analysis.
Finally, SNPs that failed the Hardy-Weinberg exact test were removed (p<10−6). After removing problematic samples, we obtained a cleaned dataset with high-quality SNPs. We then performed PC analysis again using the cleaned dataset and SNPs (minor allele frequency or MAF>0.05) to generate PC covariates.
Association analyses
For dichotomous phenotypes, we used logistic regression, and for continuous phenotypes, linear regression was applied. For all HAND and NCI phenotypes, we adjusted for the following parameters as covariates: age (continuous variable), nadir CD4+ T-cell count (cells/uL, abbreviated as “nadir CD4”, continuous variable), plasma viral load (“plasma VL”, continuous variable), ART use (categorical variable with two levels: current use or non-use), Wide-Range Achievement Test (WRAT) [Casaletto and others 2014; Heaton and others 2015; Olsen and others 2015] (continuous variable), comorbidity status (binary variable: contributing or incidental to NCI), and the first and second PCs, PC1 and PC2. These covariates were selected based on expert review or statistically significant differences in univariate analyses; they were also found to reduce the inflation of test statistics. For the additive effects of SNPs, we report the regression coefficient (β, linear regression) and the odds ratio (OR, logistic regression), which represent the effect of the minor allele denoted by A1 (i.e., a positive regression coefficient indicates that the minor allele increases the risk of NCI) [Purcell and others 2007]. All analyses were conducted using PLINK. Manhattan plots and Quantile-Quantile (QQ) plots were generated using R. Plots of representative SNPs were generated using LocusZoom software.
RESULTS
Sample Characteristics and Genotyping QC
The cleaned dataset included 1,050 individuals (811 male and 239 female) with high-quality genotype data and phenotype information. CHARTER study participants are racially and ethnically diverse. Combined analysis of CHARTER samples and the reference HapMap samples showed that self-reported race information was highly consistent with ancestry predicted by genotype data, with a few outliers (Figures 2 and 3).
Figure 2.
Genetic ancestry (by principal components, PC) in the CHARTER population compared to HapMap samples.
Figure 3.
Population distribution as predicted by genome-wide genotype data versus self-reported race/ethnicity.
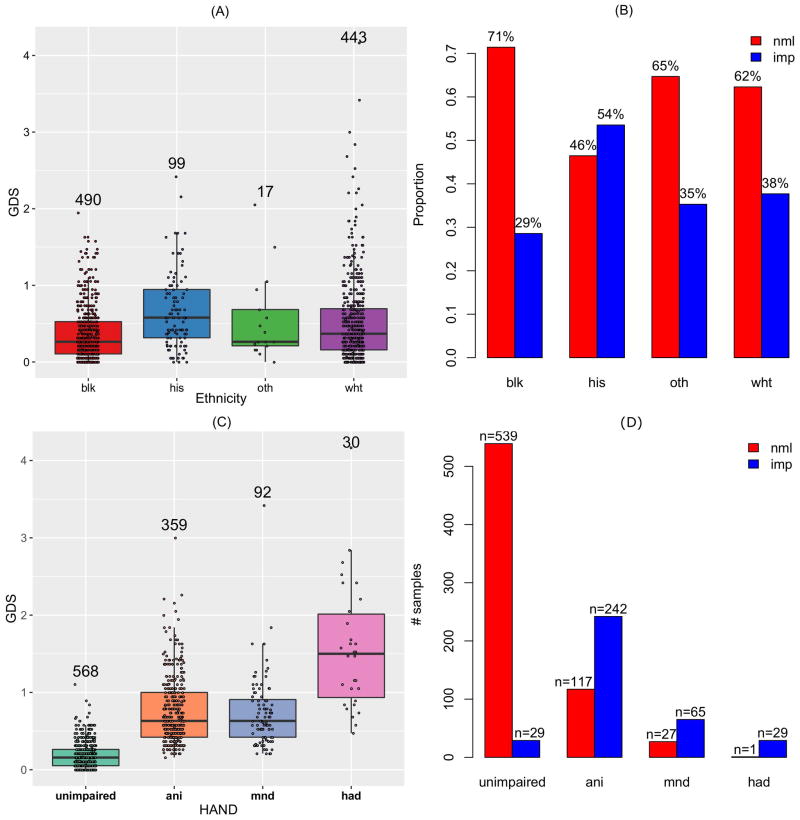
A detailed description of baseline participant characteristics is presented in Table 1. A quantitative phenotype was defined using GDS as a continuous variable, ranging between 0 and 4.17 in this sample (mean=0.46). Individuals were categorized as cases or controls using a GDS threshold of 0.5 (n=366 with GDS≥0.5, referred to as the group with GDS-defined impairment, or imp) and controls (n=684 with GDS<0.5, referred to as unimp) according to the GDS (Table 1). The overall distribution of GDS was presented in Supplemental Figure S1. GDS phenotypes were not significantly associated with age (p=0.72 for continuous GDS and p=0.64 for binary GDS, Table 2). GDS values differed among different ethnicity groups (Figure 1) and were notably higher (worse) among Hispanic participants than in other ethnic groups and lower (better) among self-reported (non-Hispanic) black individuals than in other population subgroups, a finding consistent with prior CHARTER analyses [Heaton and others 2015].
Table 1.
Summary of baseline characteristics by neurocognitive phenotype in the CHARTER genetic study population.
| All (n=1050) | GDS<0.5 (Unimp) (n=684) | GDS≥0.5 (Imp) (n=366) | |
|---|---|---|---|
| Age, median (IQR) | 43 (38, 49) | 43 (38, 48) | 43 (38, 49) |
| Race/ethnicitya | |||
| Black | 490 | 350 | 140 |
| White | 443 | 276 | 167 |
| Hispanic | 99 | 46 | 53 |
| Other | 17 | 11 | 6 |
| Sex | |||
| Female | 239 | 148 | 91 |
| Male | 811 | 536 | 275 |
| CD4+ T-cell nadir, cells/uL, median (IQR) | 180 (53, 308) | 190 (60, 326) | 159.50 (42, 279) |
| Log10 Plasma VL for those on cART, median (IQR) | 2.3 (1.7, 4.0) | 2.4 (1.7, 4.1) | 2.1 (1.7, 3.7) |
| Plasma viral load (n, % undetectable VL of those on ART) | 441, 42.2% | 277, 40.7% | 164, 45.1% |
| HCV serostatus (n, % positive) | 261, 24.9% | 179, 26.2% | 82, 22.4% |
| Current or prior major depression (n,% yes) | 151, 14.5% | 96, 14.1% | 55, 15.2% |
| ART | |||
| On | 749 | 462 | 287 |
| Off | 301 | 222 | 79 |
| WRAT, median (IQR) | 96 (83, 105) | 96 (85, 105) | 92 (79, 102) |
| Comorbidity | |||
| Incidental | 672 | 481 | 191 |
| Contributing | 378 | 203 | 175 |
| HAND groups | |||
| NCN | 568 | 539 | 29 |
| ANI | 359 | 117 | 242 |
| MND | 92 | 27 | 65 |
| HAD | 30 | 1 | 29 |
Abbreviations: GDS, global deficit score; Unimp vs. Imp, neurocognitively unimpaired vs. impaired, respectively; IQR, interquartile range; VL, viral load; HCV hepatitis C virus; ART, combination antiretroviral therapy; WRAT, wide-range achievement test. NCN, neuropsychologically normal; ANI, neurocognitive impairment; MND: mild neurocognitive disorder; HAD, HIV-associated dementia.
Self-reported race/ethnicity was not available for one participant.
Table 2.
Effects of covariates in univariate linear (using GDS as a continuous variable) or logistic (using GDS as a binary variable) regression analysis of neurocognitive phenotypes.
| GDS (continuous) | GDS (binary) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Estimate | z value | Pr(>|z|) | Estimate | z value | Pr(>|z|) | |
| (Intercept) | 1.49 | 10.504 | < 2×10−16 | 2.84 | 4.261 | 2.03×10−5 |
| Age | 6.15×10−4 | 0.358 | 0.7201 | −4.04×10−3 | −0.474 | 0.6356 |
| Nadir CD4 | −1.65×10−4 | −1.894 | 0.0585 | −3.63×10−4 | −0.833 | 0.4049 |
| Plasma VL | 7.56×10−3 | 0.528 | 0.5975 | 6.07×10−2 | 0.865 | 0.3869 |
| ART usea | −7.52×10−2 | −1.636 | 0.1021 | −0.585 | −2.532 | 0.0114 |
| WRAT | −6.80×10−3 | −6.361 | 3.01×10−10 | −3.04×10−2 | −5.728 | 1.02×10−8 |
| PC1 | −4.15 | −7.967 | 4.28×10−15 | −15.8 | −5.913 | 3.35×10−9 |
| PC2 | 0.298 | 0.627 | 0.5306 | 2.02 | 0.913 | 0.3614 |
| comorbidityb | −0.188 | −6.24 | 6.38×10−10 | −0.692 | −4.85 | 1.23×10−6 |
categorical parameters (i.e., “on” versus “off” ART).
categorized as “minimal” or “not contributing” to NCI versus “contributing” to NCI.
Abbreviations: nadir CD4, nadir CD4+ T-cell count; plasma VL, log10(plasma HIV RNA copies/mL); ART use, on vs. off antiretroviral therapy at the time of neurocognitive assessment; WRAT, wide-range achievement test (estimate of reading comprehension and IQ); PC, principal component.
Figure 1.
Distribution of Global Deficit Score (GDS) in the CHARTER study population. GDS as a continuous (quantitative) variable (A) or as a binary variable (B) in different population subgroups. GDS as a continuous variable (C) or as a binary variable (D) in different HAND sub-categories.
A related categorical definition of HAND was also evaluated, based on self-report of functional status and published Frascati criteria [Antinori and others 2007]. Comparisons of the GDS between HAND subcategories are shown in Figure 1. Individuals with HAD, as anticipated, had significantly higher GDS than individuals with other forms of HAND.
Covariate effects
We performed linear or logistic regression analysis of each covariate with each phenotype to determine the effect of covariates. Statistical results are presented in Table 2 (GDS-related phenotypes) and in Supplemental Table S1 (HAND-related phenotypes). According to these results, nadir CD4, plasma VL, WRAT score, comorbidity status (contributing vs. incidental to NCI), and PC1 were significantly (p<0.05), or marginally (p<0.1), associated with either or both GDS phenotypes, supporting the selection of these covariates [Hulgan and others 2015; Kallianpur and others 2016] for inclusion in subsequent multivariable analyses of genotype associations with GDS-defined NCI or HAND.
Association results
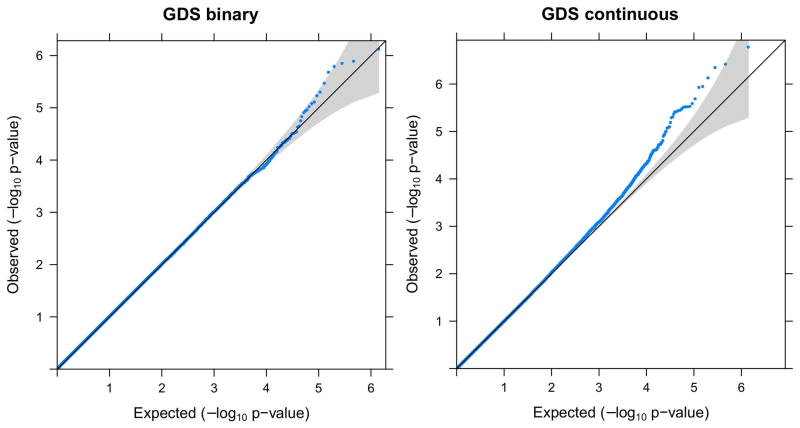
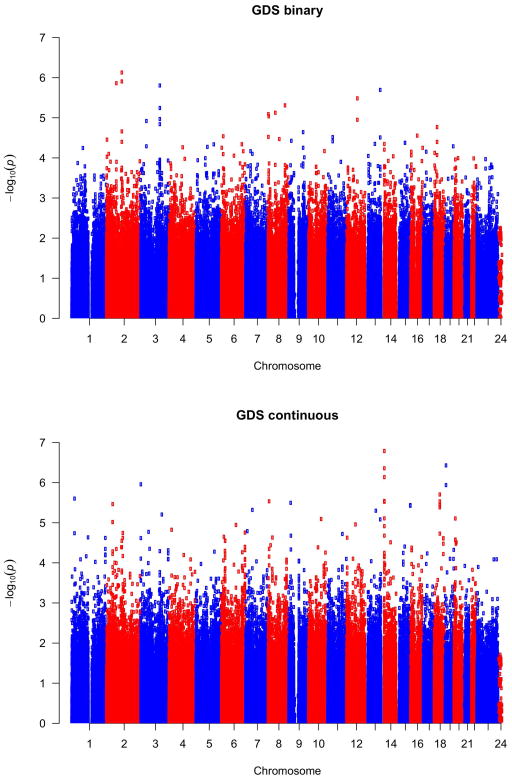
By fitting linear regression models (for continuous GDS) or logistic regression models (for binary GDS), we conducted association tests for all evaluable SNPs genome-wide. We calculated the genomic inflation factor for each result, which was found to range between 0.99 and 1.02 for all phenotypes, indicating that our QC process was highly effective, with well-controlled genomic inflation. Visualization of the QQ plots of p-values also showed that the majority of SNPs had expected p-values following approximately the line y=x, and the most significant SNPs departed from the expectation dramatically (Figure 4 and Supplemental Figure S2).
Figure 4.
Q-Q plot of the genome-wide association study (GWAS) results for GDS as a binary phenotype (left) or GDS as a quantitative phenotype (right). The grey area indicates 95% confidence interval (CI).
Notably, we examined additional PCs (i.e., PC3 and PC4) for their potential association with these phenotypes. We compared p-values obtained by including either 2 PCs plus other biological covariates in regression models, or 4 PCs plus other covariates. As shown in Supplemental Figure S3, we observed highly consistent results. Thus, we determined that the 3rd and 4th PCs had minor impact on our results. In addition, neither PC3 nor PC4 was significantly associated with GDS phenotypes. Taken together, the inclusion of PC1 and PC2 was sufficient to control for population stratification in multivariable analyses.
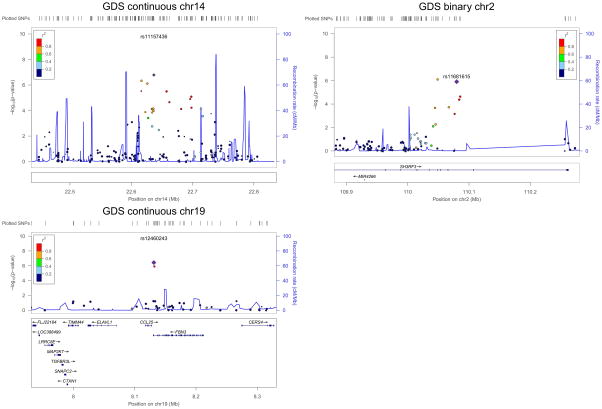
No SNP met the genome-wide significance (5×10−8) criterion for association with either GDS-related phenotype (Figure 5, Table 3), or with any of the four Frascati-defined HAND phenotypes (Supplemental Figure S4). We list the top 10 most significant SNPs for each phenotype in Table 4 and Supplemental Tables S2–S5. For continuous GDS, the most significant SNP was rs11157436 (p=1.26×10−7) located on chromosome 14 and in the gene region of T-cell receptor alpha locus (TRAα). Three other SNPs, located nearby, were also found among the top 10 most significant genes: rs12437004 (p=4.41×10−7), rs1076546 (p=7.31×10−7), and rs2293731 (p=2.88×10−6). All four SNPs were highly correlated with one another and in LD (Figure 6). The second most significant SNP associated with continuous GDS was located on chromosome 19, rs12460243 (p=3.73×10−7), which was located in the intergenic region between the genes CCL25 and FBN3. For binary GDS, the most significant SNPs were rs6542826 (p=8.06×10−7) and rs11681615 (1.22×10−6) located on chromosome 2 and in the gene SH3RF3. The next most significant SNP was located on chromosome 3, rs9814567 (p=1.38×10−6), located in the gene CEP63.
Figure 5.
Manhattan plot of GWAS results for GDS as a binary phenotype (top) or GDS as a quantitative phenotype (bottom).
Table 3.
Summary of association results.
| # SNPs | GDS (binary) | GDS (continuous) |
|---|---|---|
| p<×10−6 | 1 | 4 |
| p<×10−5 | 12 | 27 |
| p<×10−4 | 62 | 123 |
| p<×10−3 | 734 | 855 |
| Minimum p-value | 7.465×10−7 | 1.635×10−7 |
| Lambdaa | 1.014 | 0.982 |
Lambda: genomic inflation factor
Table 4.
Most significant SNPs associated with GDS.
| GDS continuous
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | SNP ID | BP | Genes | A1 | A2 | OR | STAT | P | MAF | ||
|
| |||||||||||
| White | Black | All | |||||||||
| 14 | rs11157436 | 21706714 | TRAα | T | C | 0.155 | 5.274 | 1.64×10−7 | 0.212 | 0.083 | 0.155 |
| 19 | rs12460243 | 8037240 | CCL25, FBN3 | A | G | 0.166 | 5.115 | 3.73×10−7 | 0.164 | 0.046 | 0.105 |
| 14 | rs12437004 | 21687255 | TRAα | A | C | 0.12 | 5.083 | 4.41×10−7 | 0.254 | 0.176 | 0.22 |
| 14 | rs1076546 | 21696788 | TRAα | T | A | 0.12 | 4.984 | 7.31×10−7 | 0.257 | 0.162 | 0.215 |
| 3 | rs17038463 | 1400168 | CNTN6 | G | T | 0.173 | 4.901 | 1.11×10−6 | 0.058 | 0.114 | 0.085 |
| 19 | rs17160128 | 8038698 | CCL25, FBN3 | G | A | 0.162 | 4.892 | 1.16×10−6 | 0.162 | 0.036 | 0.1 |
| 18 | rs978490 | 40178035 | - | A | G | 0.168 | 4.782 | 1.99×10−6 | 0.148 | 0.044 | 0.094 |
| 1 | rs829418 | 21816091 | RAP1GAP | A | G | 0.159 | 4.734 | 2.51×10−6 | 0.106 | 0.083 | 0.093 |
| 14 | rs2293731 | 21686674 | TRAα | G | C | 0.139 | 4.706 | 2.88×10−6 | 0.21 | 0.082 | 0.153 |
| 8 | rs17154702 | 8647290 | - | A | G | −0.118 | −4.701 | 2.94×10−6 | 0.26 | 0.169 | 0.2 |
|
| |||||||||||
| GDS binary | |||||||||||
|
| |||||||||||
| Chr. | SNP ID | BP | Genes | A1 | A2 | OR | STAT | P | MAF | ||
|
| |||||||||||
| White | Black | All | |||||||||
|
| |||||||||||
| 2 | rs6542826 | 109416151 | SH3RF3 | G | A | 1.703 | 4.949 | 7.47×10−7 | 0.632 | 0.278 | 0.468 |
| 2 | rs11681615 | 109446413 | SH3RF3 | C | T | 1.659 | 4.848 | 1.25×10−6 | 0.576 | 0.256 | 0.43 |
| 2 | rs6723162 | 70955794 | - | A | T | 0.605 | −4.828 | 1.38×10−6 | 0.411 | 0.268 | 0.338 |
| 3 | rs9814567 | 135701247 | ANAPC13, CEP63 | T | C | 0.592 | −4.802 | 1.57×10−6 | 0.339 | 0.34 | 0.327 |
| 13 | rs4772857 | 106615670 | FAM155A | G | A | 1.604 | 4.751 | 2.03×10−6 | 0.45 | 0.503 | 0.476 |
| 12 | rs795943 | 77187598 | - | A | G | 0.589 | −4.651 | 3.30×10−6 | 0.194 | 0.609 | 0.398 |
| 8 | rs876084 | 121170703 | - | C | T | 0.635 | −4.568 | 4.93×10−6 | 0.611 | 0.361 | 0.493 |
| 3 | rs11915964 | 135831620 | KY | G | T | 0.621 | −4.536 | 5.73×10−6 | 0.351 | 0.387 | 0.355 |
| 8 | rs2915495 | 52442292 | PXDNL | A | G | 1.698 | 4.478 | 7.52×10−6 | 0.157 | 0.354 | 0.254 |
| 8 | rs7840128 | 3665395 | CSMD1 | A | T | 2.17 | 4.461 | 8.15×10−6 | 0.028 | 0.146 | 0.084 |
Abbreviations: Chr., chromosome; BP, physical position (base-pair); genes, lists of genes located within 20kb upstream or downstream of the SNP (if there were multiple genes, they are all listed and separated by a comma; if there was no gene available within 20kb of the SNP, we used “-” to indicate such cases); A1, Allele 1 code (minor allele); A2, Allele 2 code (major allele); BETA/OR, regression coefficient (linear) or odds ratio (logistic); STAT, coefficient t-statistic; MAF, minor allele frequency.
Figure 6.
Examples of top SNPs associated with GDS as a quantitative phenotype or binary phenotype in the CHARTER Study. Note in the region on chromosome 14, the gene TRAα had many transcripts and was not shown in the figure.
Considering that our study sample comprises at least three populations (EA, AA, and, Hispanic), we also reported allele frequencies for the top SNPs in each population (Table 4).
We also tested the effect of batch by including it as a covariate. As shown in Supplemental Table S6, the p-values obtained with or without the batch effect showed minor changes. The top SNPs remained nearly the same with or without inclusion of batch in the models.
Examination of SNPs implicated in previous studies
Iron-related genes have been previously implicated in neurological complications of HIV infection in our studies [Kallianpur and others 2014]. We also evaluated 20 iron-related candidate genes noted to be of interest previously [Kallianpur and others 2014] as part of a nested candidate-gene analysis, based on their direct or indirect roles in iron metabolism, transport, storage, and/or regulation, and on the existence of prevalent SNPs in these genes. We examined the association of these iron-metabolism-related SNPs with GDS (Table 5). A total of 391 SNPs located within or around (20 kb upstream or downstream) these iron-related genes were genotyped in our dataset. The most significant SNP in these genes associated with GDS as a quantitative phenotype was rs7753111 (p=1.91×10−3) in BMP6, and another SNP associated with binary GDS was rs1690313 (p=3.73×10−3) in B2M. In our prior study [Kallianpur and others 2014], both genes were found to harbor SNPs that were associated with neuropathic pain, but the SNPs identified were not identical to those observed here.
Table 5.
Significant SNPs in iron genes associated with GDS.
| Gene | SNP | Chr. | BP | P | Distance |
|---|---|---|---|---|---|
| GDS continuous | |||||
| B2M | rs1690313 | 15 | 42792952 | 0.029 | 0 |
| BMP6 | rs7753111 | 6 | 7675943 | 1.83×10−3 | 0 |
| BMP6 | rs1358893 | 6 | 7731412 | 6.56×10−3 | 0 |
| BMP6 | rs9379137 | 6 | 7733601 | 3.93×10−3 | 0 |
| BMP6 | rs911750 | 6 | 7738181 | 0.045 | 0 |
| BMP6 | rs9392924 | 6 | 7740099 | 0.030 | 0 |
| BMP6 | rs927406 | 6 | 7752303 | 0.028 | 0 |
| BMP6 | rs267180 | 6 | 7775140 | 0.024 | 0 |
| CP | rs13075921 | 3 | 150398318 | 0.040 | 0 |
| HEPH | rs6624875 | 23 | 65325170 | 7.85×10−3 | 0 |
| HEPH | rs5965110 | 23 | 65333737 | 0.040 | 0 |
| HEPH | rs1264212 | 23 | 65341266 | 4.03×10−3 | 0 |
| HEPH | rs809363 | 23 | 65343849 | 0.041 | 0 |
| HEPH | rs806608 | 23 | 65345254 | 0.048 | 0 |
| HEPH | rs806610 | 23 | 65350264 | 0.045 | 0 |
| HEPH | rs1091486 | 23 | 65356189 | 0.035 | 0 |
| HEPH | rs1598697 | 23 | 65417224 | 0.030 | 13.3kb |
| SLC40A1 | rs6434347 | 2 | 190122538 | 0.042 | Upstream 11.02kb |
| GDS binary | |||||
| B2M | rs1690313 | 15 | 42792952 | 4.18×10−3 | 0 |
| BMP6 | rs267180 | 6 | 7775140 | 8.34×10−3 | 0 |
| FTH1 | rs2028062 | 11 | 61502529 | 0.041 | 10.8kb |
| FTH1 | rs10792320 | 11 | 61502867 | 0.019 | 11.16kb |
| HEPH | rs806668 | 23 | 65284240 | 0.016 | Upstream 15.15kb |
| HEPH | rs6624875 | 23 | 65325170 | 1.86×10−4 | 0 |
| HEPH | rs5965110 | 23 | 65333737 | 9.27×10−3 | 0 |
| HEPH | rs5964501 | 23 | 65341013 | 0.041 | 0 |
| HEPH | rs1264212 | 23 | 65341266 | 1.08×10−4 | 0 |
| HEPH | rs809363 | 23 | 65343849 | 7.37×10−3 | 0 |
| HEPH | rs806608 | 23 | 65345254 | 5.90×10−3 | 0 |
| HEPH | rs806610 | 23 | 65350264 | 5.45×10−3 | 0 |
| HEPH | rs1091486 | 23 | 65356189 | 3.96×10−3 | 0 |
| HEPH | rs708968 | 23 | 65386371 | 0.014 | 0 |
| HEPH | rs708969 | 23 | 65386620 | 0.012 | 0 |
| HEPH | rs1598697 | 23 | 65417224 | 0.039 | 13.3kb |
| SLC11A2 | rs17125137 | 12 | 49651273 | 0.042 | Upstream 14.77kb |
Abbreviations: Chr., chromosome; BP, physical position (base-pair); Dist, distance between the SNP and the start or end position of the gene. A distance of 0 indicates the SNP is located within the gene body.
We also evaluated 19 SNPs that were implicated in previous GWAS or association studies of HAND, including rs1130371 (MIP1-α), rs1800629 (TNF-α), rs1024611 (MCP-1), rs1801157 (CXCL12), rs2839619 (PKNOX1), rs1800450 (MBL2), rs1800451 (MBL2), and rs5030737 (MBL2) [Levine and others 2012]. Only five SNPs were represented in our data, and only one SNP, rs1024611 in the gene MCP1 (monocyte chemoattractant protein-1), was found to be nominally significant in association with binary GDS (p=0.023) (Table 6). In CHARTER, APOE-e4 genotype has not been associated with HAND or with HIV neuroimaging outcomes in older HIV-infected persons [Cooley and others 2016; Morgan and others 2013], and it did not emerge in this GWAS.
Table 6.
Association results for SNPs identified in previous studies.
| Chr. | SNP | BP | A1 | A2 | BETA/OR | STAT | P |
|---|---|---|---|---|---|---|---|
| GDS continuous | |||||||
| 2 | rs404005 | 40249582 | C | T | −0.030 | −1.498 | 0.134 |
| 10 | rs1801157 | 44188263 | T | C | −0.027 | −0.880 | 0.379 |
| 17 | rs1130371 | 31440650 | A | G | −0.008 | −0.322 | 0.747 |
| 17 | rs1024611 | 29603901 | G | A | −0.027 | −1.160 | 0.246 |
| 21 | rs2839619 | 43309246 | G | A | −0.016 | −0.792 | 0.429 |
| GDS binary | |||||||
| 2 | rs404005 | 40249582 | C | T | 0.890 | −1.182 | 0.237 |
| 10 | rs1801157 | 44188263 | T | C | 0.932 | −0.472 | 0.637 |
| 17 | rs1130371 | 31440650 | A | G | 0.915 | −0.724 | 0.469 |
| 17 | rs1024611 | 29603901 | G | A | 0.773 | −2.277 | 0.023 |
| 21 | rs2839619 | 43309246 | G | A | 0.914 | −0.921 | 0.357 |
Abbreviations: Chr., chromosome; BP, physical position (base-pair); A1, Allele 1 code (minor allele); A2, Allele 2 code (major allele); BETA/OR, regression coefficient (linear) or odds ratio (logistic); STAT: coefficient t-statistic.
Comparison of significant SNPs across phenotypes
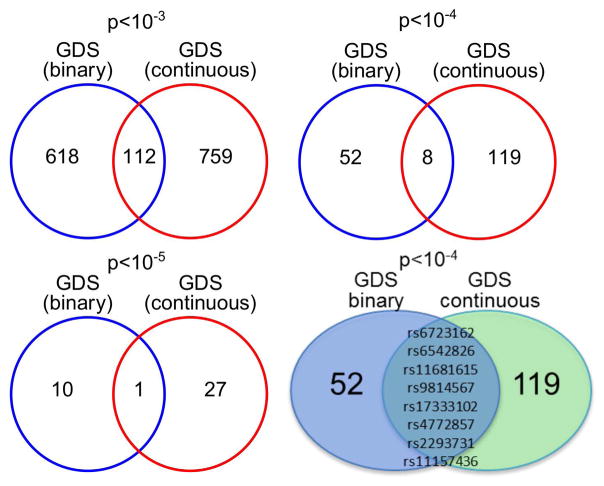
We compared the top SNPs associated with GDS as a continuous variable and as a categorical variable (Figure 7). We evaluated the top SNPs with p<10−3, p<10−4, and p<10−5. Although binary and continuous GDS phenotypes are expected to be correlated, we observed relatively few SNPs common to both phenotypes at each p-value cut-off. A single SNP, rs4772857 in gene FAM155A, overlapped all of these phenotypes at p<10−5 (Figure 7). Eight SNPs were associated with both binary GDS and continuous GDS at p<10−4, and 112 SNPs overlapped these phenotypes at p<10−3 (13% of continuous GDS SNPs and 15% of binary GDS SNPs). We found no overlapping SNPs among the 10 most significant SNPs identified for each phenotype (see above, Table 4). While both GDS outcomes represent related measures, binary GDS may be a more clinically relevant measure of NCI, while GDS as a continuous measure may be more powerful for detecting genetic effects among those participants who are only mildly impaired.
Figure 7.
Comparison of top SNPs associated with GDS as a quantitative phenotype and GDS as a binary phenotype at different cut-off values and overlap of associations between phenotypes.
Additionally, we compared the top SNPs among all six phenotypes. We defined the top SNPs with different cutoff values as p<10−3, p<10−4, p<10−5, and p<10−6 (Supplemental Figure S5).
DISCUSSION
We conducted a GWAS among CHARTER study participants with or without cognitive impairment phenotypes, defined by either the GDS or Frascati (HAND) criteria at enrollment. While no SNPs meeting stringent, genome-wide (Bonferroni) significance criteria were associated with any of the neurocognitive phenotypes, several biologically plausible associations approaching the GWAS level of significance were identified in multivariable analyses, adjusting for numerous host- and HIV-related potential confounders. Our results therefore provide candidate SNPs and variants for future validation.
Several associations with biological potential include 4 SNPs (rs11157436, rs12437004, rs1076546, and rs2293731) in the T-cell receptor-alpha (TCRα) locus, 2 SNPs in FBN3 and CCL25 (rs12460243 and rs17154702) and 2 SNPs in the gene SH3RF3. Studies in knock-out mouse models have established that the gene TCRα has an important role in adult neurogenesis [Huang and others 2010], particularly in the hippocampus, an area of the brain involved in the HAND phenotype. The SH3RF3 gene (and the SH3 domain) has been associated with age of onset in familial Alzheimer’s disease [Lee and others 2015]. At a type I error cut-off of p<10−4, 8 SNPs were associated with both binary GDS and continuous GDS. Several intronic variants, as well as SNPs in CNTN6, RAP1GAP, FAM155A, KY, PXDNL, and CSMD1, were also among the top SNPs identified. Interestingly, ANAPC13, CEP63, and KY were also among genes whose predicted RNA expression values based on genomic sequence were associated with GDS in CHARTER (p=3×10−7 and 1×10−6, respectively, Kallianpur, Hulgan, and Bush et al, unpublished data). ANAPC13 functions in the ubiquitin-mediated proteolysis pathway. CNTN6 encodes a neural adhesion molecule which has been associated with intellectual disability [Kashevarova and others 2014] in children, and copy number variants in this gene have recently been identified it as a candidate for a wide spectrum of neurobehavioral disorders [Hu and others 2015]; this gene may also function in a large number of protein interaction networks important for normal brain development [Zuko and others 2016]. The gene CSMD1 has been associated with sporadic Alzheimer’s disease [Parcerisas and others 2014], schizophrenia [Koiliari and others 2014], cannabis dependence [Koiliari and others 2014], and other neuropsychiatric disorders. FBN3 encodes an extracellular matrix molecule, which has been shown to be regulated by iron in animal studies [Hill and others 2007]. Fibrillins have emerged as critical mediators of the immune response and, like other extracellular matrix proteins, may have a particular role in regulating neuroinflammation [Summers and others 2013; Zeyer and Reinhardt 2015]. The chemokine CCL25 is important for T-cell homing and antigen-specific mucosal immunity, both of which are defective in HIV and simian immunodeficiency virus (SIV) infection [Mavigner and others 2012; Qin and others 2008]. Microbial translocation across the gut mucosa is postulated to contribute to HIV-associated persistent immune activation and neurocognitive dysfunction [Hoenigl and others 2016].
A number of iron-regulatory gene variants, many of which were within the same gene or genes known to be closely related (HEPH and CP) were also nominally associated with binary GDS or continuous GDS outcomes, 3 of which were shared between these phenotypes (B2M, BMP6 and HEPH). Genes CP, SLC40A1, FTH1, and SLC11A2 were also nominally associated with GDS-defined HAND in this study and have been associated (after adjustment for multiple testing) with HAND-related neuroimaging traits in CHARTER (e.g., subcortical gray matter volume, abnormal white matter volume, and basal ganglia choline (unpublished data [Thornton-Wells and others 2015]). HEPH encodes the membrane-based homolog of ceruloplasmin (encoded by CP). The gene FTH1 has been associated with HIV-mediated dysregulation of CXCL12/CXCR4 signaling in HIV-positive persons with a history of opioid abuse [Bandaru and others 2011]. Finally, SNP rs1024611 in MCP1, which is one of very few genes that have been replicated in previous candidate-gene association studies of HAND, was also associated with HAND at a nominal level of significance in our study. MCP-1 is an important chemokine with postulated roles in immune activation and monocyte-mediated neuro-inflammation in HIV infection [Bethel-Brown and others 2012; Kelder and others 1998; Marcotte and others 2013]. In this CHARTER study, the MCP-1 G allele was linked nominally to reduced risk of HAND, whereas prior studies have suggested increased or no change in risk [Bol and others 2012; Levine and others 2012]. Prior associations of rs1024611 in the MCP1 gene (G allele) with HAND have been similarly inconsistent, possibly due to interacting polymorphisms in the PREP1 gene or significant differences in population stratification [Kallianpur and Levine 2014]. Notably, the SNPs in iron-related genes were only nominally significant, and these associations would not survive multiple-testing correction. Caution is therefore needed when interpreting these results. Future studies with a significantly larger sample size are necessary to potentially validate these results.
Only one prior GWAS of HAND has been published to date (see [Levine and others 2012]). In that study of 1,287 HIV-infected men from the Multicenter AIDS Cohort Study (MACS), no novel genetic susceptibility loci were found to be associated at genome-wide levels of significance with HAD, neurocognitive decline over time, or either information processing speed or executive function domains, based on a combination of structured clinical interviews and neuropsychological tests. That study combined three different clinical datasets, and several genotyping platforms were used to generate genomic data, introducing additional heterogeneity into analyses. Particular strengths of the present CHARTER analysis include the racial/ethnic diversity of the population we studied (N=1,050, both men and women), and the uniformity and detail with which comprehensive phenotypic assessments were conducted in CHARTER, a cohort designed specifically to study neurological outcomes of HIV infection. Genetic ancestry, a critical covariate in any genetic model of HAND, was also determined for all participants in CHARTER using identical methods to generate the PCs, and it was adjusted for accordingly in all models. Since all genetic data were available within one cohort and were obtained using a single (Affymetrix) platform, no imputation of SNP genotypes was conducted, and multiple (albeit related) measures of NCI of varying severity were used as outcomes for analysis, facilitating evaluation of robustness of any observed SNP associations across these phenotypes. This has not been possible in the field until now. However, this study did not include genome-wide analysis of longitudinal data or individual neurocognitive domains as in the study by Levine et al; these analyses are ongoing. Results of ongoing longitudinal genome-wide analyses of neurocognitive decline will be reported in future. Results of imputation analyses, which have the advantage of revealing genotype calls for millions of additional SNPs beyond those included on the current platform, will also be reported in future work. Finally, this GWAS was conducted in a population that included women, and may be more representative of the modern ART era, since the distribution of HAND phenotypes more closely resembles the epidemiology of HAND in the ART era, with only a small minority of individuals carrying the diagnosis of HAD.
As has been noted previously, HAND is a challenging clinical phenotype, akin to other neuropsychiatric disorders, and its investigation is complex, due in part to fluctuations in the results of neurocognitive assessments within individuals over time [Kallianpur and Levine 2014]. This study, performed using cross-sectional neurocognitive outcomes data obtained at baseline in CHARTER participants, is therefore subject to random misclassification of individuals as impaired or unimpaired, which would be expected to bias our results toward the null [Hennekens and Buring 1987]. As others have noted, detection of additive SNP associations with 80% power using an alpha (type I error) level of 5×10−8 requires that the SNP account for at least 3.5% of the variability in the phenotype [Levine and others 2012]. Despite being relatively common, the genetic variants we evaluated might not account for a sufficient proportion of the variability in the HAND phenotype for detection with 80% power in our study population, which also comprised several different ancestral subgroups. In addition to phenotypic complexity and population heterogeneity, the modest size of our sample, while not small, is still likely the major reason why more significant associations were not detected. Finally, since etiologic factors in HAND are varied, with involvement of immune/inflammatory, lifestyle, and concomitant comorbid and aging-related conditions, residual confounding of genetic effects by these other factors remains a consideration [Clifford and Ances 2013]. However, the associations between neurocognitive outcomes and covariates other than age that have been linked to these outcomes in prior CHARTER studies and other HIV cohorts are reassuring in this regard. Due to the important need for replication, we have initiated collaborations with other large HIV cohorts with neurocognitive assessment data, to in order to significantly augment the sample size for genetic studies (N approximately 7000). The planned replication attempts will require genomic as well as outcome data harmonization beforehand, since neurocognitive assessment methods and genotyping platforms differed across studies.
In conclusion, this GWAS conducted among CHARTER study participants identified several new genes with possible roles in HAND, some of which approach genome-wide levels of significance. This study did not replicate SNPs previously observed to be associated with HAND, with the exception of a nominal association for MCP1. These new associations with HAND, although biologically plausible, remain to be replicated in larger studies of HIV-infected persons with longitudinal NC assessments.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contributions of all participants in CHARTER. S. L. L. has received support for research projects from Gilead Sciences, Merck & Co., and ViiV Healthcare. He has also been an advisor for Cipla, CytoDyn, Merck & Co., and ViiV Healthcare. R. J. E. gave sponsored talks and received honoraria for serving on the scientific advisory board of GlaxoSmithKline. A. C. C. has received past research support from Schering-Pough, Merck, and Roche Molecular Systems and has served as a data, safety, and monitoring board member for Merck-sponsored studies; she previously owned stock in Abbott Laboratories, Bristol-Myers Squibb, Johnson & Johnson, and Pfizer. C. M. M receives royalties from Lippincott Williams and Wilkins and from UptoDate. D. B. C. has received support for consulting/advisory boards from Sanofi, Genentech, Quintiles, Inhibikase, Bristol-Myers Squibb, GlaxoSmithKline, Millennium, Biogen Idec, Amgen, Pfizer, AstraZeneca, Cytheris and Merck and receives research support from Lilly, Roche, Bavarian Nordic, Gilead, the Alzheimer Association and the National Institutes of Health (National Institute of Allergy and Infectious Diseases, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute on Aging, and National Institute of Nursing Research). J. A. M. authors chapters on HIV for the Merck Manual. D. M. S. has provided consultancy to Astellas, Acorda, Allergan, Merz, and Ipsen; has received speaking honoraria from Allergan and Acorda; and received research grants from Merz, Allergan, Ipsen, Acorda, and Astellas. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
FUNDING SUPPORT
Financial support for this CHARTER study was provided by the National Institutes of Health (contracts N01 MH22005 and HHSN271201000036C to I. G., R01 MH095621 to A. R. K. and T. H., and R01 LM011177 (to Z. Z.).
References
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nature protocols. 2010;5(9):1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69(18):1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru VV, Patel N, Ewaleifoh O, Haughey NJ. A failure to normalize biochemical and metabolic insults during morphine withdrawal disrupts synaptic repair in mice transgenic for HIV-gp120. J Neuroimmune Pharmacol. 2011;6(4):640–649. doi: 10.1007/s11481-011-9289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethel-Brown C, Yao H, Hu G, Buch S. Platelet-derived growth factor (PDGF)-BB-mediated induction of monocyte chemoattractant protein 1 in human astrocytes: implications for HIV-associated neuroinflammation. Journal of neuroinflammation. 2012;9:262. doi: 10.1186/1742-2094-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone K, Moore DJ, Franklin DR, Clifford DB, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, Ellis RJ, Atkinson JH, Grant I, Heaton RK. Defining neurocognitive impairment in HIV: deficit scores versus clinical ratings. Clin Neuropsychol. 2012;26(6):894–908. doi: 10.1080/13854046.2012.694479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol SM, Booiman T, van Manen D, Bunnik EM, van Sighem AI, Sieberer M, Boeser-Nunnink B, de Wolf F, Schuitemaker H, Portegies P, Kootstra NA, van ‘t Wout AB. Single nucleotide polymorphism in gene encoding transcription factor Prep1 is associated with HIV-1-associated dementia. PloS one. 2012;7(2):e30990. doi: 10.1371/journal.pone.0030990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt TD, Agan BK, Marconi VC, He W, Kulkarni H, Mold JE, Cavrois M, Huang Y, Mahley RW, Dolan MJ, McCune JM, Ahuja SK. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(25):8718–8723. doi: 10.1073/pnas.0803526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto KB, Cattie J, Franklin DR, Moore DJ, Woods SP, Grant I, Heaton RK, Group H. The Wide Range Achievement Test-4 Reading subtest “holds” in HIV-infected individuals. J Clin Exp Neuropsychol. 2014;36(9):992–1001. doi: 10.1080/13803395.2014.960370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford DB, Ances BM. HIV-associated neurocognitive disorder. The Lancet Infectious diseases. 2013;13(11):976–986. doi: 10.1016/S1473-3099(13)70269-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley SA, Paul RH, Fennema-Notestine C, Morgan EE, Vaida F, Deng Q, Chen JA, Letendre S, Ellis R, Clifford DB, Marra CM, Collier AC, Gelman BB, McArthur JC, McCutchan JA, Simpson DM, Morgello S, Grant I, Ances BM Group CHA-RTER. Apolipoprotein E epsilon4 genotype status is not associated with neuroimaging outcomes in a large cohort of HIV+ individuals. Journal of neurovirology. 2016 doi: 10.1007/s13365-016-0434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Lamers SL, Levine AJ, Valdes-Sueiras M, McGrath MS, Shapshak P, Singer EJ. Factors related to HIV-associated neurocognitive impairment differ with age. Journal of neurovirology. 2015;21(1):56–65. doi: 10.1007/s13365-014-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant I. Neurocognitive disturbances in HIV. Int Rev Psychiatry. 2008;20(1):33–47. doi: 10.1080/09540260701877894. [DOI] [PubMed] [Google Scholar]
- Grant I, Franklin DR, Jr, Deutsch R, Woods SP, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Collier AC, Marra CM, Clifford DB, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Smith DM, Heaton RK. Asymptomatic HIV-associated neurocognitive impairment increases risk for symptomatic decline. Neurology. 2014;82(23):2055–2062. doi: 10.1212/WNL.0000000000000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75(23):2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, Leblanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, Collier AC, Marra CM, Morgello S, Mindt MR, Taylor MJ, Marcotte TD, Atkinson JH, Wolfson T, Gelman BB, McArthur JC, Simpson DM, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. Journal of neurovirology. 2011;17(1):3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Franklin DR, Jr, Deutsch R, Letendre S, Ellis RJ, Casaletto K, Marquine MJ, Woods SP, Vaida F, Atkinson JH, Marcotte TD, McCutchan JA, Collier AC, Marra CM, Clifford DB, Gelman BB, Sacktor N, Morgello S, Simpson DM, Abramson I, Gamst AC, Fennema-Notestine C, Smith DM, Grant I. Neurocognitive change in the era of HIV combination antiretroviral therapy: the longitudinal CHARTER study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2015;60(3):473–480. doi: 10.1093/cid/ciu862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekens CH, Buring JF. In: Epidemiology in Medicine. Mayrent, editor. Lippincott Williams and Wilkins; 1987. [Google Scholar]
- Hill CH, Ashwell CM, Nolin SJ, Keeley F, Billingham C, Hinek A, Starcher B. Dietary iron deficiency compromises normal development of elastic fibers in the aorta and lungs of chicks. J Nutr. 2007;137(8):1895–1900. doi: 10.1093/jn/137.8.1895. [DOI] [PubMed] [Google Scholar]
- Hoenigl M, Chaillon A, Little SJ. CD4/CD8 Cell Ratio in Acute HIV Infection and the Impact of Early Antiretroviral Therapy. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2016 doi: 10.1093/cid/ciw293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. The Journal of infectious diseases. 2015;212(10):1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liao J, Sathanoori M, Kochmar S, Sebastian J, Yatsenko SA, Surti U. CNTN6 copy number variations in 14 patients: a possible candidate gene for neurodevelopmental and neuropsychiatric disorders. J Neurodev Disord. 2015;7(1):26. doi: 10.1186/s11689-015-9122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GJ, Smith AL, Gray DH, Cosgrove C, Singer BH, Edwards A, Sims S, Parent JM, Johnsen A, Mott R, Mathis D, Klenerman P, Benoist C, Flint J. A genetic and functional relationship between T cells and cellular proliferation in the adult hippocampus. PLoS Biol. 2010;8(12):e1000561. doi: 10.1371/journal.pbio.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulgan T, Samuels DC, Bush W, Ellis RJ, Letendre SL, Heaton RK, Franklin DR, Straub P, Murdock DG, Clifford DB, Collier AC, Gelman BB, Marra CM, McArthur JC, McCutchan JA, Morgello S, Simpson DM, Grant I, Kallianpur AR, Group C. Mitochondrial DNA Haplogroups and Neurocognitive Impairment During HIV Infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2015;61(9):1476–1484. doi: 10.1093/cid/civ527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur AR, Jia P, Ellis RJ, Zhao Z, Bloss C, Wen W, Marra CM, Hulgan T, Simpson DM, Morgello S, McArthur JC, Clifford DB, Collier AC, Gelman BB, McCutchan JA, Franklin D, Samuels DC, Rosario D, Holzinger E, Murdock DG, Letendre S, Grant I. Genetic variation in iron metabolism is associated with neuropathic pain and pain severity in HIV-infected patients on antiretroviral therapy. PloS one. 2014;9(8):e103123. doi: 10.1371/journal.pone.0103123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur AR, Levine AJ. Host genetic factors predisposing to HIV-associated neurocognitive disorder. Current HIV/AIDS reports. 2014;11(3):336–352. doi: 10.1007/s11904-014-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur AR, Wang Q, Jia P, Hulgan T, Zhao Z, Letendre SL, Ellis RJ, Heaton RK, Franklin DR, Barnholtz-Sloan J, Collier AC, Marra CM, Clifford DB, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Grant I, Group CS. Anemia and Red Blood Cell Indices Predict HIV-Associated Neurocognitive Impairment in the Highly Active Antiretroviral Therapy Era. The Journal of infectious diseases. 2016;213(7):1065–1073. doi: 10.1093/infdis/jiv754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashevarova AA, Nazarenko LP, Skryabin NA, Salyukova OA, Chechetkina NN, Tolmacheva EN, Sazhenova EA, Magini P, Graziano C, Romeo G, Kucinskas V, Lebedev IN. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene. 2014;536(1):145–150. doi: 10.1016/j.gene.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Kelder W, McArthur JC, Nance-Sproson T, McClernon D, Griffin DE. Beta-chemokines MCP-1 and RANTES are selectively increased in cerebrospinal fluid of patients with human immunodeficiency virus-associated dementia. Ann Neurol. 1998;44(5):831–835. doi: 10.1002/ana.410440521. [DOI] [PubMed] [Google Scholar]
- Koiliari E, Roussos P, Pasparakis E, Lencz T, Malhotra A, Siever LJ, Giakoumaki SG, Bitsios P. The CSMD1 genome-wide associated schizophrenia risk variant rs10503253 affects general cognitive ability and executive function in healthy males. Schizophrenia research. 2014;154(1–3):42–47. doi: 10.1016/j.schres.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Vardarajan B, Lantigua R, Reyes-Dumeyer D, Ortmann W, Graham RR, Bhangale T, Behrens TW, Medrano M, Jimenez-Velazquez IZ, Mayeux R. Genetic Modifiers of Age at Onset in Carriers of the G206A Mutation in PSEN1 With Familial Alzheimer Disease Among Caribbean Hispanics. JAMA Neurol. 2015;72(9):1043–1051. doi: 10.1001/jamaneurol.2015.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Service S, Miller EN, Reynolds SM, Singer EJ, Shapshak P, Martin EM, Sacktor N, Becker JT, Jacobson LP, Thompson P, Freimer N. Genome-wide association study of neurocognitive impairment and dementia in HIV-infected adults. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2012;159B(6):669–683. doi: 10.1002/ajmg.b.32071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte TD, Deutsch R, Michael BD, Franklin D, Cookson DR, Bharti AR, Grant I, Letendre SL, Group C. A concise panel of biomarkers identifies neurocognitive functioning changes in HIV-infected individuals. J Neuroimmune Pharmacol. 2013;8(5):1123–1135. doi: 10.1007/s11481-013-9504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavigner M, Cazabat M, Dubois M, L’Faqihi FE, Requena M, Pasquier C, Klopp P, Amar J, Alric L, Barange K, Vinel JP, Marchou B, Massip P, Izopet J, Delobel P. Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. J Clin Invest. 2012;122(1):62–69. doi: 10.1172/JCI59011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EE, Woods SP, Letendre SL, Franklin DR, Bloss C, Goate A, Heaton RK, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Ellis RJ, Abramson I, Gamst A, Fennema-Notestine C, Smith DM, Grant I, Vaida F, Clifford DB Group CHATER. Apolipoprotein E4 genotype does not increase risk of HIV-associated neurocognitive disorders. Journal of neurovirology. 2013;19(2):150–156. doi: 10.1007/s13365-013-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JP, Fellows RP, Rivera-Mindt M, Morgello S, Byrd DA Manhattan HIVBB. Reading Ability as an Estimator of Premorbid Intelligence: Does It Remain Stable Among Ethnically Diverse HIV+ Adults? Clin Neuropsychol. 2015;29(7):1034–1052. doi: 10.1080/13854046.2015.1122085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcerisas A, Rubio SE, Muhaisen A, Gomez-Ramos A, Pujadas L, Puiggros M, Rossi D, Urena J, Burgaya F, Pascual M, Torrents D, Rabano A, Avila J, Soriano E. Somatic signature of brain-specific single nucleotide variations in sporadic Alzheimer’s disease. J Alzheimers Dis. 2014;42(4):1357–1382. doi: 10.3233/JAD-140891. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS genetics. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LA, Stone E, Price P, van Bockxmeer F, Brew BJ. The relationship between ApoE, TNFA, IL1a, IL1b and IL12b genes and HIV-1-associated dementia. HIV medicine. 2008;9(8):677–680. doi: 10.1111/j.1468-1293.2008.00614.x. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Sui Y, Murphey-Corb MA, Reinhart TA. Association between decreased CXCL12 and CCL25 expression and increased apoptosis in lymphoid tissues of cynomolgus macaques during SIV infection. J Med Primatol. 2008;37(Suppl 2):46–54. doi: 10.1111/j.1600-0684.2008.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers L, Kangwantas K, Rodriguez-Grande B, Denes A, Penny J, Kielty C, Pinteaux E. Activation of brain endothelial cells by interleukin-1 is regulated by the extracellular matrix after acute brain injury. Mol Cell Neurosci. 2013;57:93–103. doi: 10.1016/j.mcn.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Thornton-Wells TA, Fennema-Notestine C, Hulgan T, Letendre S, Ellis R, Kallianpur AR. Iron-Regulatory Genes are Associated with Neuroimaging Traits in HIV-Infected Persons: A CHARTER Study. Poster presentation at the 2015 Conference on Retroviruses and Opportunistic Infections; Seattle, WA. February 2015; 2015. Abstract 461. [Google Scholar]
- Zeyer KA, Reinhardt DP. Engineered mutations in fibrillin-1 leading to Marfan syndrome act at the protein, cellular and organismal levels. Mutat Res Rev Mutat Res. 2015;765:7–18. doi: 10.1016/j.mrrev.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Zuko A, Oguro-Ando A, van Dijk R, Gregorio-Jordan S, van der Zwaag B, Burbach JP. Developmental role of the cell adhesion molecule Contactin-6 in the cerebral cortex and hippocampus. Cell Adh Migr. 2016:1–15. doi: 10.1080/19336918.2016.1155018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.