Abstract
Members of the family Chlamydiaceae possess at least 13 genes, distributed throughout the chromosome, that are homologous with genes of known type III secretion systems (TTS). The aim of this study was to use putative TTS proteins of Chlamydophila pneumoniae, whose equivalents in other bacterial TTS function as chaperones, to identify interactions between chlamydial proteins. Using the BacterioMatch Two-Hybrid Vector system (Stratagene, La Jolla, Calif.), lcrH-2 and sycE, positions 1021 and 0325, respectively, from C. pneumoniae CM-1 were used as “bait” to identify target genes (positions 0324, 0705, 0708, 0808 to 0810, 1016 to 1020, and 1022) in close proximity on the chromosome. Interaction between the products of the lcrH-2 (1021) and lcrE (copN) (0324) genes was detected and confirmed by pull-down experiments and enzyme immunoassays using recombinant LcrH-2 and LcrE. As further confirmation of this interaction, the homologous genes from Chlamydia trachomatis, serovar E, and Chlamydophila psittaci, Texas turkey, were also cloned in the two-hybrid system to determine if LcrH-2 and LcrE would interact with their orthologs in other species. Consistent with their genetic relatedness, LcrH-2 from C. pneumoniae interacted with LcrE produced from the three species of Chlamydiaceae; LcrH-2 from C. psittaci reacted with LcrE from C. pneumoniae but not from C. trachomatis; and C. trachomatis LcrH-2 did not react with LcrE from the other two species. Deletions from the N and C termini of LcrE from C. pneumoniae identified the 50 C-terminal amino acids as essential for the interaction with LcrH-2. Thus, it appears that in the Chlamydiaceae TTS, LcrH-2 interacts with LcrE, and therefore it may serve as a chaperone for this protein.
Type III secretion systems (TTS) are powerful virulence components shared by several pathogenic gram-negative bacteria (9). Described by some to be one of the most sophisticated devices evolved in nature, TTS allow for the efficient transfer of effector proteins from a bacterial cell into the host cytosol (6, 9, 24). Injected bacterial TTS proteins exert a variety of effects on the host to give selective advantage to the bacterial pathogen (11). TTS share common features, among them the clustering of the TTS genes in a pathogenicity island, low-molecular-weight chaperones dedicated to secreted effector proteins which oftentimes are located adjacent to one another on the chromosome, and a requirement for host cell contact for activation (9). In addition, there is similarity among structural proteins that compose the “syringe,” which spans the bacterial inner and outer membranes, and the translocon needle, which assembles upon contact with the host to allow the transfer of effector proteins. However, the effector proteins, which modulate host function, appear to be unique to the different bacterial pathogens, thus making their identification a challenge.
Evidence for a TTS in the obligate intracellular bacterial family Chlamydiaceae has been reported (8, 10, 16, 18, 19, 27). Gene sequences similar to those for TTS structural components and chaperones have been identified; however, unlike other TTS genes, these genes are scattered throughout the chromosome and are organized differently than those so far described for other bacterial pathogens (27). Studies on temporal expression of the Chlamydophila pneumoniae TTS genes, with few exceptions, show the majority to be expressed in the middle to late stage of the developmental cycle (26). Fields and Hackstadt (7), studying a Chlamydia TTS protein, CopN (LcrE), and more recently, Clifton et al. (4), studying CT456, a hypothetical protein with unknown function, have demonstrated that these chlamydial proteins can be secreted by using the Yersinia TTS. In addition, electron microscopy and proteomic analysis suggest that a TTS system is present in the Chlamydia membrane (15, 16, 18, 19).
The challenge is to find and fit the pieces of the Chlamydiaceae TTS puzzle together to determine how this system functions and the role it plays in entry into the host cell, growth, and survival. As a step to achieve this goal, we have employed genes that have similarity to bacterial TTS chaperone genes to identify interactions between Chlamydiaceae TTS components. Here we identified and characterized the interaction between the putative chaperone LcrH-2 and LcrE (CopN), which in Yersinia (YopN) plays a regulatory or “gating” function for the release of TTS effector proteins (24).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. pneumoniae CM-1, Chlamydophila psittaci Texas turkey, and Chlamydia trachomatis serovar E (Bour), obtained from the American Type Culture Collection (Manassas, Va.), were propagated in HEp-2 cells (American Type Culture Collection). HEp-2 cells were grown in Eagle's minimal essential medium (Irvine Scientific, Santa Ana, Calif.) supplemented with 10% fetal bovine serum (Atlanta Biological, Norcross, Ga.), 2 mM l-glutamine, and 50 μg of gentamicin (Irvine Scientific)/ml. Chlamydiaceae were prepared by inoculating HEp-2 cells in 1-dram glass vials which had been pretreated for 10 min at room temperature with DEAE-dextran (30 μg/ml). After inoculation, the monolayers were centrifuged at room temperature for 1 h at 800 × g followed by the addition of medium containing cycloheximide (1 μg/ml). Cultures were incubated for 36 to 72 h at 37°C, at which time the medium was removed and the cultures were sonicated in sucrose-phosphate-glutamate (pH 7.4) and centrifuged (500 × g, 10 min); the supernatant was then stored at −80°C.
HEp-2 cells and the Chlamydiaceae stocks were determined to be free of Mycoplasma contamination by PCR using the Mycoplasma 16S rRNA primers 5′-GGG AGC AAA CAG GAT TAG ATA CCC T and 5′-TGC ACC ATC TGT CAC TCT GTT ACC CTC (20).
Escherichia coli strains employed were XL1Blue MRF′Kan with and without a reporter cassette (Stratagene, La Jolla, Calif.) and E. coli K-12 BL21(DE3) (Invitrogen Life Technologies, Carlsbad, Calif.). All plasmids used in this study are listed in Table 1. E. coli was cultured in either Luria-Bertani broth (1% NaCl, 0.5% yeast extract, 1% tryptone) with aeration or on Luria-Bertani agar (2% agar) at 30 to 37°C. Where appropriate, carbenicillin (250 μg/ml) (Research Products International Corp., Mt. Prospect, Ill.), ampicillin (100 μg/ml) (Sigma Chemical Co., St. Louis, Mo.), chloramphenicol (34 μg/ml) (Sigma), kanamycin (50 μg/ml) (Sigma), and tetracycline (12.5 μg/ml) (Sigma) were added.
TABLE 1.
Plasmids and primers
| Plasmid
|
Purposea | Forward primerb 5′-3′ | Reverse primerb 5′-3′ | |
|---|---|---|---|---|
| Vector | Insert | |||
| pBT | Cpn0325 | Bait, sycE | AAAGCGGATCCATGCAAAACCAATACGAGC | GTATTCCGCTCGAGTTACGCGACGTAGTAGATTCC |
| pBT | Cpn1021 | Bait, lcrH-2 | CTACGAATTCAATGTCACATTTAAATTATT | GCGGATCCTTATTTATGTTTTCGAATATC |
| pBT | Cps0740 | Bait, lcrH-2 | AACGCGGATCCATGTCATACTTAGCTTATTTAT | AAAGTTCTCGAGTTATGCGTGGTTTTTTATATC |
| pBT | CT862 | Bait, lcrH-2 | AGAATTCAATGCCACCAAGCAAGATCCAATGT | ATAGGATCCTTAGGCGTACGATTTGATCGC |
| pTRG | Cpn0324 | Target, lcrE | ATAGGGGATCCATGGCAGCATCAGGAGGC | TCGGCCTCGAGTTATGACCAAGGATAGGGT |
| pTRG | Cps0458 | Target, lcrE | AGGATCCATGGCTGCATCTGGAGGAGCT | ATAGAATTCTTAAGACCAGGGATAAGGTTT |
| pTRG | CT089 | Target, lcrE | AGGATCCATGACTGCATCAGGAGGAGCTGGAG | ATAGAATTCTTAGGGTGATGGAGGCTGTGT |
| pTRG | Cpn0705 | Target, HP | ATCAGGGCGAGCGGCCGCAATGGAATTAAAGAAAA | CGGCTCGAGTTATAAACGTGCTTCGAT |
| pTRG | Cpn0708 | Target, HP | TTTGGATCCATGATAGACCCTGTAGAATGT | GCCCTCGAGTTAACCTAATTGCGTTGTCAT |
| pTRG | Cpn0808 | Target, HP | AGGAGAATTCAGATGCCGCAATCAGCGGAG | GCCCTCGAGTTAATTAAAGATAGCAGATGT |
| pTRG | Cpn0809 | Target, HP | GATGGATCCATGTCTATTTCATCTTCTT | GCGAATTCTTAAAATTATTGGTTTTATGTGC |
| pTRG | Cpn0810 | Target, HP | GGAGGATCCATGAATAAAAAGCCCAAGA | GCGAATTCTTACTCAGCGCCTTTAACCAT |
| pTRG | Cpn1016 | Target, HP | GAGAGGGATCCATGAAAAAAGGGAAATTAGCA | GCGAATTCTTACGAAGCAGAAGTCGTTGT |
| pTRG | Cpn1017 | Target, HP | CCGTAGGATCCATGAGAAAACTTATTTTAT | ATGCCTCGAGTTAAGAACAACGGAGTTCT |
| pTRG | Cpn1018 | Target, HP | TGCTGGGATCCATGTGGTATTTCAATTACC | GCCCTCGAGTTAATTAAAGATAGCAGATGT |
| pTRG | Cpn1019 | Target, HP | CTTCTGGATCCATGACTGTATCTTACCAAT | GCCTCGAGTTACTGATACAGACTGCGAGC |
| pTRG | Cpn1020 | Target, HP | CTTTGGGATCCATGTCCTCTTGCTTATCTC | GCGAATTCTTATCCTCTTAAGGAGTGGAG |
| pTRG | Cpn1022 | Target, HP | TTATAGGATCCATGAATATGCCTGTTCCTTC | GCGAATTCTTAGGGGCGTAGGTTGTAAAT |
| pTRG | Cpn0324A | Target, −50N lcrE | AGGATCCCTGACAAATCCCGCAGCAGCAA | TCGGCCTCGAGTTATGACCAAGGATAGGGT |
| pTRG | Cpn0324B | Target, −100N lcrE | AGGATCCGCTTCTGGGAATTCTGAAATCT | TCGGCCTCGAGTTATGACCAAGGATAGGGT |
| pTRG | Cpn0324C | Target, −150N lcrE | AGGATCCCAAACGACTCCACCCTCCCAAG | TCGGCCTCGAGTTATGACCAAGGATAGGGT |
| pTRG | Cpn0324Z | Target, −50C lcrE | ATAGGGGATCCATGGCAGCATCAGGAGGC | ACTCGAGTTAAGAAAAGAATAAGTTCAAGACA |
| pTRG | Cpn0324Y | Target, −100C lcrE | ATAGGGGATCCATGGCAGCATCAGGAGGC | ACTCGAGTTAAGGAGTTTGGATTCCCTCAGCT |
| pTRG | Cpn0324X | Target, −150C lcrE | ATAGGGGATCCATGGCAGCATCAGGAGGC | ACTCGAGTTATTTTAATTCTGTTGCCATTCCT |
| pET-45b(+) | Cpn0324 | His6-rLcrE | GCGGATCCGGCAGCATCAGGAGGCACA | TCGGCCTCGAGTTATGACCAAGGATAGGGT |
| pET-45b(+) | Cpn0324T | His6-rLcrE, −50C | TTGGATCCGGCAGCATCAGGAGGCACA | ACCCTCGAGTTAAGAAAAGAATAAGTTCAAGAC |
| pGEX-4T-1 | Cpn1021 | GST-rLcrH-2 | GCGGATCCATGTCACATTTAAATTAT | AGAATTCTCATTTATGTTTTCGAATAT |
| pBT | Empty bait | TCCGTTGTGGGGAAAGTTATC | GGGTAGCCAGCAGCATCC | |
| pTRG | Empty target | CAGCCTGAAGTGAAAGAA | ATTCGTCGCCCGCCATAA | |
| pGEX-4T-1 | GST vector | GGGCTGGCAAGCCACGTTTGGTG | CCGGGAGCTGCATGTGTCAGAGG | |
| pET-45b(+) | His6 vector | TAATACGACTCACTATAGGG | GCAGCAGCCTAGGTTAATT | |
HP, hypothetical protein. −50N and −50C (and analogous designations), 50 aa deleted from the N and C termini, respectively, of the protein. r, recombinant.
Bases underlined are the restriction endonuclease site used for cloning into the multiple cloning site of the plasmid vector.
DNA techniques.
Chlamydiaceae elementary bodies harvested as described previously (2) were used to isolate genomic DNA, using the Wizard Genomic DNA Purification kit (Promega Corporation, Madison, Wis.). Routine isolation of plasmid DNA from E. coli strains was performed by using the QIAprep Miniprep kit (QIAGEN, Germantown, Md.) and HiSpeed Plasmid Midi kit (QIAGEN), as described by the manufacturer. Standard DNA techniques, including the use of the restriction enzymes, in vitro DNA cloning, and agarose gel electrophoresis, were performed as described by Sambrook and Russell (25). Chemical transformation of E. coli was performed by the method of Inoue et al. (12). Recovery of DNA fragments from agarose gels was achieved using either the Wizard SV Gel and the PCR Clean-Up system (Promega) or the QIAquick Gel Extraction kit (QIAGEN) as detailed by the manufacturers.
Plasmid construction.
DNA fragments used for cloning were obtained by PCR. DNA primers were synthesized by Integrated DNA Technologies Inc. (Coralville, Ia). Reaction mixtures consisted of a 0.25 mM concentration of each deoxynucleoside triphosphate (Promega), a 0.2 μM concentration of each primer, 150 ng of Chlamydiaceae genomic DNA template, 5 U of Native Pfu DNA polymerase (Stratagene), and Native Plus Pfu buffer (Stratagene) supplemented with 1% dimethyl sulfoxide in a final volume of 0.1 ml. Cycling conditions were as follows: denaturation at 96°C for 1 min, annealing at the primer melting temperature minus 5°C for 45 s, and extension at 72°C for 2 to 3 min (25 to 35 cycles) in a DNA thermal cycler-Mastercycler gradient (Eppendorf Scientific Inc., Westbury, N.Y.). The amplification products were purified, digested with appropriate endonucleases, and, using T4 DNA ligase (New England Biolabs, Beverly, Mass.), ligated into the corresponding vector. Recombinant plasmids were used to transform competent E. coli. Resulting clones were then selected and checked by PCR using Taq DNA polymerase for the appropriate insert. To confirm the proper sequence of the insert, selected clones were sequenced by using the ABI Prism BigDye Terminator v 3.1 Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase, FS (Applied Biosystems, Foster City, Calif.), as instructed by the manufacturer.
Bacterial two-hybrid system.
The BacterioMatch Two-Hybrid Vector system (Stratagene) was used according to manufacturer's instructions. Briefly, the two plasmids, pBT and pTRG, containing the “bait” and target genes, respectively, were used to simultaneously transform E. coli. Protein-protein interactions were screened for based on activation of the bla ampicillin resistance gene in the E. coli reporter gene cassette and the resulting resistance to carbenicillin (250 μg/ml). A second reporter gene, for β-galactosidase, expressed in the same reporter cassette, was used to validate the bait and target interaction. A modification of the method described by Miller (17) was employed to quantitate and compare β-galactosidase activities among different clones.
Recombinant proteins.
PCR-amplified DNA harboring wild-type lcrE or truncated lcrE (minus 50 amino acids [aa] from the C terminus) from C. pneumoniae was cloned into the BamHI-XhoI sites of the pET-45b(+) vector (Novagen, Madison, Wis.) under the control of the T7 promoter, using lcrE-specific primers (Table 1). C. pneumoniae lcrH-2 was amplified by PCR and cloned in fusion with a glutathione S-transferase (GST) tag into the prokaryotic expression vector pGEX-4T-1 (Amersham Bioscience, Piscataway, N.J,) as instructed by the manufacturer. The ligated vectors were then used to transform E. coli XL1Blue chemically competent cells.
E. coli BL21 harboring either pET-45b(+)-lcrE, pET-45b(+)-lcrE(−50 aa), or pGEX-4T-1-lcrH-2 was cultivated at 37°C with aeration and induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 32°C for 3 to 4 h. Sonication was used to disrupt bacteria suspended in phosphate-buffered saline (PBS; 135 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]) containing 5% glycerol followed by centrifugation at 20,000 × g for 30 min at 4°C. His-tagged LcrE and truncated LcrE were purified over an Ni-nitrilotriacetic acid (NTA) resin (QIAGEN) column and dialyzed in PBS with 5% glycerol. GST-LcrH-2 was purified in one step by passing the cell-free lysate over a glutathione-agarose resin (Sigma) column, eluting with glutathione, and dialyzing against PBS containing 5% glycerol. The same procedure was applied to prepare free GST protein from E. coli BL21 cells carrying the pGEX-4T-1 plasmid without an insert. The purity and stability of the fusion proteins were assessed by polyacrylamide gel electrophoresis (PAGE) and Western blotting using polycolonal antisera raised to LcrH-2 and LcrE.
Enzyme immunoassay.
For enzyme immunoassay (EIA), high-protein-binding-capacity 96-well microtiter plates (Corning, Fountain Valley, Calif.) were coated with His-LcrE and truncated His-LcrE (5 μg/well in 50 μl of PBS, pH 8.0) at 4°C overnight. The plates were then washed three times with PBS-0.05% Tween 20, blocked with 0.2 ml of BLOTTO (Bovine Lacto Transfer Technique Optimizer) (13) overnight at 4°C, and washed three times with PBS-Tween. GST-LcrH-2 or GST alone, which served as the negative control, was added (5 μg/well in 0.05 ml of PBS, pH 8.0) and incubated overnight at 4°C. After incubation, wells were washed five times with PBS-Tween 20. An anti-GST mouse monoclonal antibody (MAb) (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) diluted 1:1,000 in BLOTTO was added, and plates were incubated for 1 h at 37°C. Subsequently, horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (IgG, IgA, and IgM) (ICN Biomedical, Inc., Aurora, Ohio), diluted 1:1,000 in BLOTTO, was added and plates were incubated for 1 h at 37°C and washed as described above. The reaction was developed for 30 min using a solution of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) diammonium salt (Sigma) and hydrogen peroxide. Plates were read on a Multiskan RC reader (Thermo Electron, Milford, Mass.) at 405 nm. Differences between the measurements after the background was subtracted were analyzed by a Student unpaired t test.
Pull-down experiments.
Pull-down experiments were performed with purified recombinant proteins. Briefly, 2 mg of each GST- or His-tagged recombinant protein was bound to GST-agarose or Ni-NTA columns (1.5 ml of resin), respectively. Columns were then washed with PBS containing 5% glycerol, pH 7.4, until the optical density of the eluate at 280 nm (OD280) was <0.01. The opposite fusion protein, GST or His tagged, was applied to each column (2 mg of purified recombinant protein in PBS containing 5% glycerol, pH 7.4). Both columns were washed again with the same buffer as that described above. Protein complexes were eluted with 2 ml each of 250 mM imidazole (Sigma) for the Ni-NTA columns or 15 mM oxidized glutathione (Sigma) for the GST-agarose columns. Eluted protein samples were used for subsequent sodium dodecyl sulfate (SDS)-PAGE and Western blot analysis. Purified GST (2 mg) applied to an Ni-NTA column with bound His-LcrE protein and treated as described above served as a control for nonspecific binding.
SDS-PAGE and Western blotting.
Proteins to be analyzed by immunoblotting were resolved by SDS-PAGE (10% [vol/vol] polyacrylamide} and subsequently transferred to NitroBind nitrocellulose membranes (0.45 μm) (Osmonics Inc., Minnetonka, Minn.) by using a semidry transfer procedure (10 V for 45 min) with a Trans-Blot SD kit (Bio-Rad Laboratories, Hercules, Calif.) (14). Following blocking of the membrane with BLOTTO at 4°C overnight, a 1;1,000 dilution of primary antibody in PBS was added to the membranes. His-tagged mouse MAb (Novagen) or GST(B-14):sc138 anti-GST mouse MAb (Santa Cruz Biotechnology) was used for His-LcrE or GST-LcrH-2 detection, respectively. Blots were incubated with primary antibody for 45 min at 37°C, followed by washing three to five times with PBS-Tween 20. Subsequently, HRP-conjugated goat anti-mouse Pan Ig (ICN) diluted 1:1,000 in BLOTTO was added and blots were incubated and washed as described above. Blots were developed with 2-chloronaphthol and hydrogen peroxide in PBS for 10 to 20 min.
RESULTS
Identification of interaction.
Two putative TTS chaperone genes, lcrH-2 and sycE, from C. pneumoniae were used as bait in a bacterial two-hybrid system. Twelve genes in proximity to the TTS gene clusters were used as targets (Table 1). No interaction between SycE and any of the target proteins was detected. In contrast, interaction between LcrH-2 and LcrE was identified by the ability of the cotransformants containing both the bait and target plasmids to grow on selective medium containing carbenicillin.
To determine if this protein-protein interaction occurred in C. psittaci and C. trachomatis, thus corroborating our results with C. pneumoniae, the lcrH-2 and lcrE genes from both C. trachomatis, serovar E, and C. psittaci, Texas turkey, were used in the bacterial two-hybrid system. As with C. pneumoniae, both homologous pairs of bait and target proteins were shown to interact by the ability of the cotransformed E. coli to grow on selective medium.
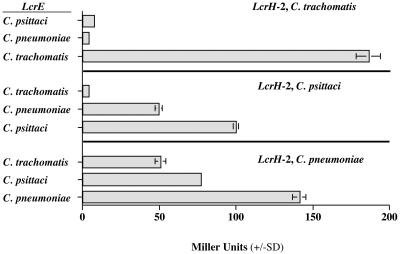
The three lcrH-2 genes from C. pneumoniae, C. psittaci, and C. trachomatis were then used as bait with the two heterologous lcrE genes in order to determine if the interaction between the two proteins was interchangeable and thus conserved among the species or whether it evolved within the species. LcrH-2 from C. pneumoniae was able to interact with LcrE from both C. psittaci and C. trachomatis. LcrH-2 from C. psittaci interacted with LcrE from C. pneumoniae but failed to interact with the C. trachomatis LcrE protein. In contrast, LcrH-2 from C. trachomatis did not interact with LcrE from either C. pneumoniae or C. psittaci, as judged by failure of the cotransformants to grow on carbenicillin selective medium. Therefore, these findings are consistent with the phylogenetic relatedness of these organisms and suggest that, once acquired, these two proteins evolved independently within the species of the family Chlamydiaceae.
To verify the interaction of these proteins, a secondary marker, β-galactosidase, expressed in the same reporter cassette was employed. In Fig. 1 the level of β-galactosidase activities for all combinations of Chlamydiaceae lcrH-2 and lcrE gene transformants are shown. The levels of activity corroborated the results obtained using carbenicillin resistance as the marker of a protein-protein interaction.
FIG. 1.
Measurement of β-galactosidase activity in the bacterial two-hybrid system using all combinations of the lcrH-2 (bait) and lcrE (target) genes from C. pneumoniae, C. psittaci, and C. trachomatis. A standard assay using IPTG induction of lacZ was used (17). Enzyme activity is expressed in Miller units.
Mapping the site of interaction.
To determine the region of LcrE that is critical for the binding of the LcrH-2 chaperone, deletions of C. pneumoniae LcrE were made. Deletions of 50, 100, and 150 aa were made from both the N and C termini (Table 1). Deleted lcrE genes were then used to cotransform the two-hybrid E. coli host (Table 2). Deletions from the N terminus of the LcrE protein resulted in an equivalent or even a stronger signal than that seen with the intact LcrE protein. However, deletions from the C terminus of the LcrE protein resulted in greatly reduced β-galactosidase activity.
TABLE 2.
β-Galactosidase activity of the C. pneumoniae lcrE deletion mutants in the two-hybrid bacterial system, using LcrH-2 as a bait protein
| LcrE> | β-Galactosidase activityb |
|---|---|
| Complete protein | 141 (4.3) |
| −50 aa, N terminus | 133 (1.2) |
| −100 aa, N terminus | 230 (7.8) |
| −150 aa, N terminus | 195 (4.8) |
| −50 aa, C terminus | 33 (0.4) |
| −100 aa, C terminus | 22 (0.3) |
| −150 aa, C terminus | 15 (0.9) |
| No proteina | 18 (3.3) |
lcrE was omitted from the target plasmid; therefore, cotransformants contained the bait plasmid with the lcrH-2 insert and the target plasmid minus lcrE.
β-Galactosidase activity is given in Miller units, with the standard deviation shown in parentheses.
Confirmation of the interaction.
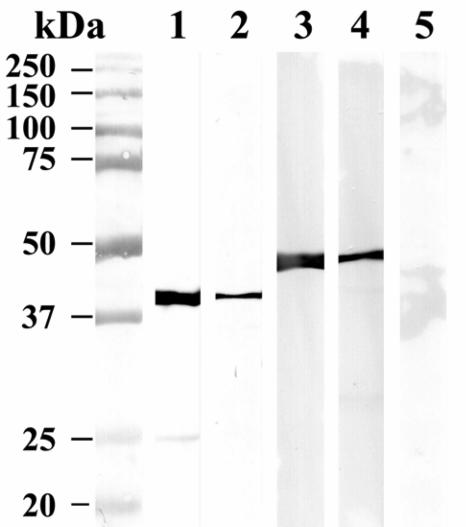
As further evidence of the protein-protein interaction between C. pneumoniae LcrH-2 and LcrE, His-tagged LcrH-2 and LcrE proteins were expressed and purified. In addition, a GST-LcrH-2 recombinant was made. A GST pull-down assay using GST-LcrH-2 bound to GSH-agarose showed the binding of His-LcrE when this recombinant protein was incubated and subsequently eluted from the GST column. After a washing, bound protein was eluted with glutathione. As seen by Western blotting using antibody to the His tag, His-LcrE was coeluted with GST-LcrH-2 (Fig. 2). Additionally, His-LcrE was bound to a Ni-NTA column and subsequently GST-LcrH-2 was passed through the nickel column. Upon elution of His-LcrE with imidazole, GST-LcrH-2 was coeluted (Fig. 2). To control for nonspecific binding, GST alone was used and, in contrast to GST-LcrH-2, GST alone was not specifically bound to the His-LcrE column (Fig. 2).
FIG. 2.
Western blot of pull-down assays using recombinant GST-LcrH-2 and His-LcrE. Molecular mass markers are shown on the left. Purified GST-LcrH-2 is shown in lane 1 and was visualized using anti-GST MAb. Lane 2 contains the eluate from a His-LcrE Ni-NTA affinity column to which GST-LcrH-2 was applied and the column was washed and eluted with imidazole and reacted with anti-GST MAb. Purified His-LcrE is shown in lane 3, visualized using anti-His MAb. Lane 4 contains the eluate from a GST-LcrH2 affinity column to which His-LcrE was applied and the column was washed and eluted with glutathione and reacted with anti-His MAb. Lane 5 shows the negative control for lane 2, where GST alone was passed through a His-LcrE affinity column and the column was washed and eluted with imidazole and incubated with anti-GST MAb.
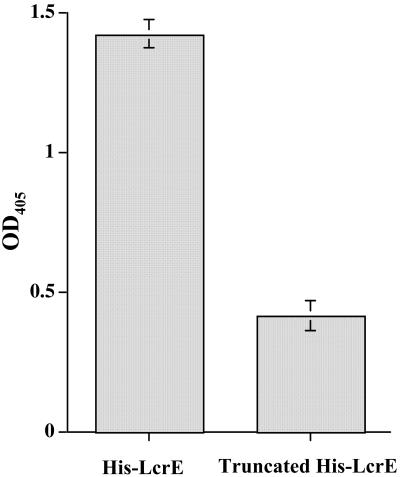
An EIA was also performed to capture GST-LcrH-2, using microtiter plates coated with His-LcrE or truncated His-tagged LcrE that lacked the 50 C-terminal aa. As a control for nonspecific binding (background), GST alone was added to microtiter plates coated with both His-tagged full-length and truncated LcrE. After incubation and washing of the plates, a mouse MAb to GST was added and binding was assessed by color development using substrate and an HRP-labeled second antibody. The resulting average OD405 from five EIA determinations showed a significant difference between the reactions when GST-LcrH-2 (OD, 1.43 ± 0.05 [standard deviation]) was added to the His-LcrE-coated wells compared to the truncated His-LcrE (OD, 0.42 ± 0.05) (P < 0.001) (Fig. 3).
FIG. 3.
Results of EIA using His-tagged full-length LcrE and His-tagged LcrE minus the C-terminal 50 amino acids (truncated LcrE). Both His-tagged proteins were used to coat a microtiter plate to which GST-LcrH-2 and GST alone were added, incubated, washed, and reacted with anti-GST MAb. Shown are the averages (bars represent ± standard deviations) of five reactions each between GST-LcrH-2 and His-LcrE and the truncated His-LcrE. The background binding for both reactions using His-LcrE (0.3 ± 0.01) and truncated His-LcrE (0.64 ± 0.12) was subtracted by using the average of five determinations using GST alone. Differences between the two reactions were statistically different (P < 0.001).
Homology to other chaperones.
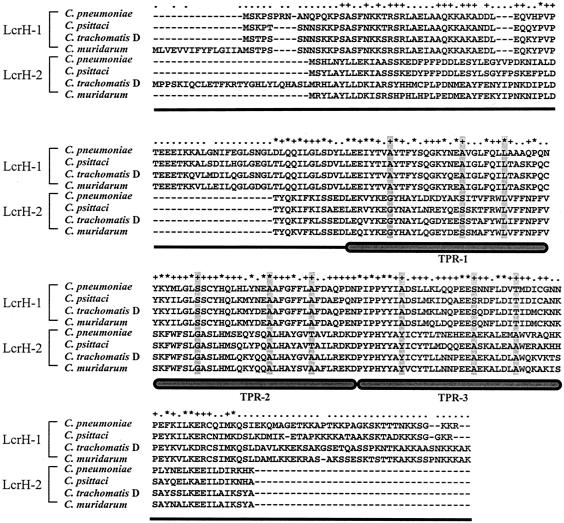
In aligning the amino acid sequences of LcrH-2 and LcrH-1 from C. pneumoniae, C. psittaci, C. trachomatis, and Chlamydia muridarum, it is clear that both proteins possess a 34-aa tetratricopeptide repeat (TPR) that has been reported to be present in some TTS and eukaryotic chaperones (Fig. 4). The three canonical positions, 8, 20, and 27, within the TPR were identical among all the LcrH-1 proteins for these four organisms. Only two of these positions were identical among all the LcrH-1 and LcrH-2 proteins. Within the TPR of LcrH-2 the amino acids for these three positions (8, 20, 27) were GSL, GAT, and YAA, respectively. All four organisms shared the same sequence at these positions, with the exception of a substitution of an A for a T for amino acid 20 in the second repeat of C. trachomatis and C. muridarum.
FIG. 4.
Alignment of LcrH-1 and LcrH-2 of four strains of Chlamydiaceae. TPRs are indicated as well as positions 8, 20, and 27 within each repeat, which appear shaded in grey.
DISCUSSION
The TTS of Chlamydiaceae may play a pivotal role in the ability of these organisms to gain entry into the host cell, multiply, and in some cases remain latent within the host. As a step in unraveling this potentially important system, we have shown an interaction between two of the Chlamydiaceae TTS proteins. Based on the results shown here it is possible that LcrH-2 may function as a chaperone for LcrE (CopN). In Yersinia, LcrE (YopN), the homologue of LcrE of the Chlamydiaceae, has a gating or regulatory role in plugging the TTS pore (24). Upon contact with the host, or a decrease in the calcium concentration, the pore is opened upon the release of LcrE, giving way to the translocation of effector proteins into the host cell. While we do not know the function of LcrE in Chlamydiaceae, Fields and Hackstadt (7) have established that C. trachomatis LcrE can be secreted by the Yersinia TTS apparatus.
If we assume that LcrH-2 is functioning as a chaperone for LcrE, this interaction is surprising since the genes encoding these two proteins in C. pneumoniae are located distant from one another, at positions 1021 and 0324, respectively. In other bacterial TTS it is characteristic for chaperone genes to be located in close proximity to the gene encoding their substrate (6). In C. pneumoniae, lcrE is adjacent to sycE, a putative chaperone gene; however, using the bacterial two-hybrid system we were unable to detect the interaction of the proteins encoded by these genes.
To further confirm our results showing an interaction between C. pneumoniae LcrH-2 and LcrE, we performed the same experiment using the corresponding genes from C. psittaci and the more distantly related C. trachomatis. The positive results with homologous gene pairs corroborated the findings with C. pneumoniae. Furthermore, the nonreciprocity of the interaction between C. trachomatis and the other two species of Chlamydiaceae was in accordance with the previously reported genetic distance between these organisms (1).
In addition to their gene proximity, TTS chaperones share other characteristics, including their relatively low molecular mass (∼15 kDa), acidic pI, and predicted amphipathic helix near the C terminus. LcrH-2 fits this description in being 172 amino acids in length (20.3 kDa), having a pI of 5.83, and a predicted amphipathic helix at its C terminus (6, 23). At one time all TTS chaperones were thought to interact with only one TTS protein. However, more recently this dogma has been challenged as it appears that in addition to chaperones that are devoted to only one protein substrate, referred to as class 1 chaperones, others interact with more than one protein, referred to as class 2 chaperones (21). Pallen et al. (22) described the presence of 34-amino-acid TPRs in TTS class 2 chaperones but not in class 1 chaperones, which reinforces the concept of differences among TTS chaperones. These investigators have proposed that there are basically two types of chaperones that are distinct as determined by sequence homology and structure, those that fit the description of a SycE-like chaperone with an alpha-beta peptide binding groove (class 1) and the SycD/LcrH-like chaperones with a TPR that forms an all-alpha-helical-array peptide binding groove (class 2). All LcrH-2 proteins of the Chlamydiaceae that we examined have a TPR-like repeat, highly suggestive that they belong to the class 2 TTS chaperones. Class 2 chaperones have been described as the chaperones of translocators, as opposed to the chaperone of one effector (22, 23). This description might give insight into the functions of both LcrH-2 and LcrE in Chlamydiaceae.
In general, TTS chaperones bind to the N terminus of their substrate (6). Here we report that the C terminus of LcrE is important for binding to LcrH-2. For the Yersinia TTS it has been shown that TyeA binds with the C terminus (residues 242 to 293) of LcrE (YopN) (3, 5). Therefore, it appears that not all TTS chaperones bind to the N terminus of their substrate, perhaps a reflection of remnants of the flagellar system (23). In Yersinia, TyeA is thought to function to limit LcrE secretion and translocation. It has been suggested that TyeA may act in concert with other Yersinia effector protein chaperones to ensure a hierarchy of delivery of effectors to the host cell. Sundberg and Forsberg (28) have postulated that the main role of TyeA is to mediate rapid delivery of those effectors which are essential at initial stages of infection to promote extracellular infection of pathogenic Yersinia. While there are distinct difference between the Chlamydiaceae LcrH-2 and Yersinia TyeA, it will nevertheless be interesting to see if LcrH-2 also plays a role in regulating the “gating” of the Chlamydiaceae TTS pore.
Temporal expression work with C. pneumoniae previously reported from our laboratory found that lcrH-2 was one of the few TTS genes expressed early in the developmental cycle of C. pneumoniae (7, 26). This may suggest an early function for this chaperone in the entry stages of Chlamydiaceae. Clifton et al. (4) have recently presented evidence suggesting that Chlamydia use a TTS secreted effector protein, a tyrosine-phosphorylated protein (Tarp), in the initial stages of infection. They have shown, using complementation with Yersinia TTS, that Tarp is rapidly translocated by a TTS and phosphorylated on attachment to the host cell. Therefore, Tarp translocation may serve as the initial signal transduced from the pathogen to the host to facilitate entry of this obligate intracellular pathogen into the host. Building on this information and that established with the Yersinia system, one may speculate that LcrH-2 functions early in the TTS to regulate the gating function of LcrE, which when activated by cell contact would then lead to the rapid release of Tarp into the host cell.
The TTS of Chlamydiaceae differs from many other bacterial TTS in not being clustered on the chromosome and appears to have two genes that code for proteins that resemble the lcrH chaperone gene in other systems, referred to as lcrH-1 and lcrH-2. We have previously reported that lcrH-1 is expressed in the middle to late stage of the developmental cycle, in contrast to the early expression of lcrH-2 (26). Other differences between these two putative chaperones are their molecular masses of 25.9 kDa (231 aa) and 20.3 kDa (172 aa) and pIs of 9.35 and 5.83, respectively. As stated earlier both have a TPR with different amino acids in positions 8, 20, and 27 of the triple repeat. Therefore, while they have homology among themselves and with the lcrH of other systems, they clearly are different from one another. It is intriguing to speculate as to why Chlamydiaceae have two lcrH genes and, in light of the temporal expression data, whether they function at different times in the developmental cycle of these pathogens.
Acknowledgments
We thank Robin L. Bush for constructive comments concerning the manuscript.
REFERENCES
- 1.Bush, R. M., and K. D. E. Everett. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51:203-220. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, L. W., O. Kay, and O. Schneewind. 2001. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183:5293-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day, J. B., F. Ferracci, and G. V. Plano. 2003. Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol Microbiol. 47:807-823. [DOI] [PubMed] [Google Scholar]
- 6.Feldman, M. F., and G. R. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151-158. [DOI] [PubMed] [Google Scholar]
- 7.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048-1060. [DOI] [PubMed] [Google Scholar]
- 8.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671-683. [DOI] [PubMed] [Google Scholar]
- 9.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 10.Hsia, R. C., Y. Pannekoek, E. Ingerowski, and P. M. Bavoil. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25:351-359. [DOI] [PubMed] [Google Scholar]
- 11.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. A., J. W. Gautsch, J. R. Sportsman, and J. H. Elder. 1984. Improved technique utilizing nonfat dry milk and nucleic acids transferrred to nitrocellulose. Gene Anal. Tech. 1:3-8. [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, A. 1981. Electron microscopic observations of surface projections and related intracellular structures of Chlamydia organisms. J. Electron Microsc. 30:315-320. [PubMed] [Google Scholar]
- 16.Matsumoto, A., E. Fujiwara, and N. Higashi. 1976. Observations of the surface projections of infectious small cell of Chlamydia psittaci in thin sections. J. Electron Microsc. 25:169-170. [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Molestina, R. E., J. B. Klein, R. D. Miller, W. H. Pierce, J. A. Ramirez, and J. T. Summersgill. 2002. Proteomic analysis of differentially expressed Chlamydia pneumoniae genes during persistent infection of HEp-2 cells. Infect. Immun. 70:2976-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols, B. A., P. Y. Setzer, F. Pang, and C. R. Dawson. 1985. New view of the surface projections of Chlamydia trachomatis. J. Bacteriol. 164:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossewaarde, J. M., A. de Vries, T. Bestebroer, and A. F. Angulo. 1996. Application of a Mycoplasma group-specific PCR for monitoring decontamination of Mycoplasma-infected Chlamydia sp. strains. Appl. Environ. Microbiol. 62:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Pallen, M. J., M. S. Francis, and K. Futterer. 2003. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53-60. [DOI] [PubMed] [Google Scholar]
- 23.Parsot, C., C. Hamiaux, and A. L. Page. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7-14. [DOI] [PubMed] [Google Scholar]
- 24.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Slepenkin, A., V. Motin, L. M. de la Maza, and E. M. Peterson. 2003. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect. Immun. 71:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 28.Sundberg, L., and A. Forsberg. 2003. TyeA of Yersinia pseudotuberculosis is involved in regulation of Yop expression and is required for polarized translocation of Yop effectors. Cell. Microbiol. 5:187-202. [DOI] [PubMed] [Google Scholar]