Abstract
Recent studies have identified several new genes in Listeria monocytogenes which are positively or negatively affected by PrfA and grouped into three classes (E. Milohanic et al., Mol. Microbiol. 47:1613-1625, 2003). In vitro transcription performed with promoters of some class III genes showed strict SigB-dependent but PrfA-independent transcription initiation. Transcription starting at the prfA promoter PprfA2 was also optimal with SigB-loaded RNA polymerase, suggesting a direct link between SigB- and PrfA-dependent gene expression.
Virulence of Listeria monocytogenes is determined by a number of well-characterized genes (for a recent review, see reference 26), most of which are regulated by the transcriptional activator PrfA (14, 17), which recognizes a 14-bp sequence of dyad symmetry located 40 to 41 bp in front of the transcriptional start sites of the PrfA-regulated virulence genes (9, 25). PrfA belongs to the Crp/Fnr family of transcriptional activators (13, 16). There is evidence suggesting that PrfA activity is modulated in vivo by an as yet essentially unknown mechanism(s) (2-5, 7, 14).
Recently, we established an in vitro transcription system allowing PrfA-mediated transcription initiation at all known PrfA-dependent promoters in a dose-dependent fashion (2, 15, 18, 22). These studies revealed some features essential for PrfA-dependent promoters, including the requirement for a SigA-recognized −10 box, a fixed distance between the PrfA box and the −10 box of 22 or 23 bp, a purine start nucleotide 5 to 8 bp apart from the 3′ end of the −10 box, and high GTP concentration for transcription initiation (18).
A recent comparative transcriptome analysis (21) revealed listerial genes (divided into three groups) that were also affected in their expression by PrfA. Some of these genes contained putative PrfA binding sites (with one to two mismatches) and putative SigB-dependent promoters (group III genes) in their regulatory 5′ upstream regions (UTRs). Kazmierczak et al. also showed with microarrays using RNA from a sigB mutant and the wild-type strain that transcription of most of these genes, including lmo2067, is SigB dependent (12).
Here, we used the in vitro transcription system for studying the possible involvement of PrfA and SigB in transcription initiation at some of these newly identified PrfA-affected genes, concentrating in particular on the group III genes rsbV, lmo0596, and lmo2067 (bsh).
Sigma A and B proteins of L. monocytogenes were purified, and SigB-dependent in vitro transcription was established
For the isolation of sigma A and B proteins, His6 tags were placed at the N termini of both proteins. To generate the SigA and SigB constructs, the sigA and sigB gene sequences were amplified by PCR (Table 1) and cloned into the vector pQE-30 (QIAGEN). Purification of His6-tagged PrfA proteins (PrfA and PrfA* [24, 27]) and RNA polymerase (RNAP) of L. monocytogenes A42 (ΔprfA) grown in brain heart infusion at 37°C was performed as described previously (2, 3). To construct template plasmids for the in vitro transcription assays, the promoter region and adjacent sequences were cloned into pUC18 by using the oligonucleotides listed in Table 1. In vitro transcription and primer extension were carried out with various amounts of RNAP, PrfA protein, and/or additional SigA or SigB protein (2, 18).
TABLE 1.
PCR primers useda
| Sequence (5′→3′) | Application |
|---|---|
| GAGGAATACAGGGTACCATGAGTGATAAAACAAAAAACACAAAAC; GGTTTGTCC TCCTGCAGGGTTGTGCATTATTCCAGGAAG | Construct for His6-tagged SigA |
| AAAAGCAGGTGGGGTACCATGCCAAAAGTATCTCAACC; TTTGATTCAACTGCAGT GTTCATTTACTCC | Construct for His6-tagged SigB |
| AAGTGGGGAATTCTGTATTTATTATGTC; TTTGGCGCTGCAGAAGCATCGATCTC | pUC18*PrsbV, template for in vitro transcription |
| GTATTGTGGATCCAACTAAAGTAACACG; CCATCACTGCAGCAATACCAATAAGTG | pUC18*Pbsh, template for in vitro transcription |
| AAGGATTGGTACCTTTTCCGACTTATC; ATAATCACTGCAGTCCCCGCTAATAA | pUC18*Plmo0596, template for in vitro transcription |
| AATCCGTTTCTAGATATGTATGC; ATACAATAAGCTTGTGGATCCCA | pUC18*PprfA, template for in vitro transcription |
| CAACAAATATGTCTATGTGGTC | Primer extension of PrsbV |
| CGGCGTAACAACCACAACTTC | Primer extension of Pbsh |
| CCGCTAATAAAACAAGAATTCGTG | Primer extension of Plmo0596 |
| GGTTTTATCCCGTTAG | Primer extension of PprfA |
Primers were used for His6-tagged SigA and SigB constructs and cloning of in vitro transcription templates. Oligonucleotides were used for primer extension. Restriction sites that were used are underlined.
SigA- and SigB-dependent in vitro transcription of PrsbV
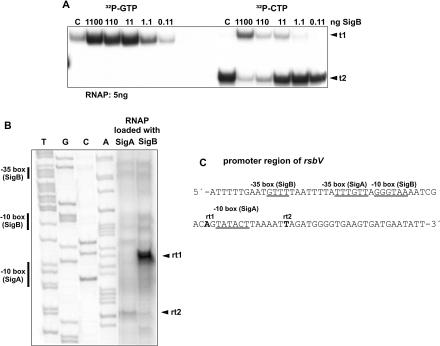
In vitro transcription was first performed with the promoter of rsbV, which is the first gene of an operon encoding the anti-anti-sigma (RsbV), the anti-sigma (RsbW), and the sigma B factor (RsbB) itself and was shown to be transcribed by SigB-loaded RNA polymerase in Bacillus subtilis (10, 11). The UTR of rsbV in L. monocytogenes contains the sequence 5′-ATGTTT-N15-GGGTAA-3′, which is closely related to the B. subtilis SigB recognized consensus sequence AGGTTT-N16-GGGTAT (1). Using RNAP loaded with SigA (15) and [32P]CTP as the labeled nucleoside triphosphate (NTP) (0.08 μM; the other three NTPs, 200 μM), we obtained a major transcript (t2), while labeling with [32P]GTP led to the synthesis of a weaker transcript (t1) (Fig. 1A). The absence of t2 in the presence of labeled [32P]GTP suggested that this transcript contains a G within the first 4 nucleotides (15, 18). Addition of increasing amounts of purified sigma B protein to the assay resulted in a dose-dependent increase of t1 and a corresponding decrease of t2, confirming that t1 starts at the anticipated SigB-dependent promoter, whereas t2 probably starts at a SigA-dependent promoter. The start sites of t1 and t2 were determined by primer extension with RNA transcripts as templates. These transcripts were obtained with SigA- and SigB-loaded RNAP in the presence of the 4 nucleotides in equally high concentrations (200 μM each) (Fig. 1B). With SigA-loaded RNAP, we obtained a major cDNA (reverse transcript 2 [rt2]) (Fig. 1B), suggesting nucleotide T as the start site, 7 bp downstream from the anticipated SigA-dependent −10 box (TATACT). With SigB-loaded RNAP, the reverse transcript rt1 becomes the major cDNA, while rt2 decreased. The size of rt1 suggests A as the start site, 8 bp downstream of the anticipated SigB-dependent −10 box (GGGTAA).
FIG. 1.
(A) In vitro transcription with the promoter region of rsbV. RNAP was loaded with different amounts of purified SigB protein, as indicated. The obtained transcripts are indicated as t1 and t2. C indicates control without addition of SigB protein. (B) Analysis of in vitro derived transcripts by primer extension. (C) The promoter sequence of rsbV. RNAP loaded either with SigA or with SigB was used for in vitro transcription. The main cDNA fragments obtained after reverse transcription are labeled as rt1 and rt2. Putative −35 and −10 boxes as well as transcription start sites are indicated. A sequence ladder was used for determining the transcription start site.
These data clearly indicate that the two purified sigma proteins function as expected.
SigB-dependent but PrfA-independent in vitro transcription of bsh and lmo0596
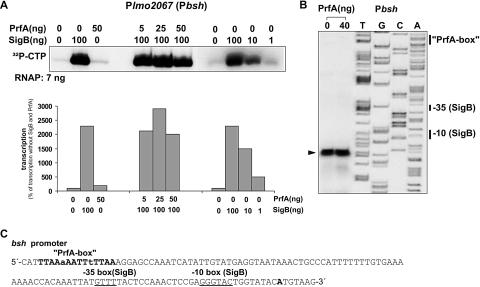
To test whether SigB-loaded RNAP also drives transcription of lmo2067 (bsh) and lmo0596 (possibly in a PrfA-dependent fashion), we carried out in vitro transcription with UTR-DNA fragments of these genes (containing the putative SigB-dependent promoters and PrfA boxes) and SigB-loaded RNAP in the presence or absence of PrfA. As shown in Fig. 2A, efficient in vitro transcription was obtained with the SigB-loaded RNAP from the bsh promoter, but this transcription was not further enhanced by increasing amounts of PrfA. Primer extension with SigB-loaded RNAP (Fig. 2B) confirmed that transcription of bsh is indeed initiated at the anticipated SigB-dependent promoter (Fig. 2C), which is apart from the previously proposed PrfA box (6).
FIG. 2.
(A) In vitro transcription assay with the promoter region of lmo2067 (bsh gene). RNAP of L. monocytogenes was loaded with different amounts of purified SigB protein, as indicated. (B) The corresponding in vitro transcripts were taken for primer extension to determine the transcriptional start sites. The putative −35 and −10 boxes of the SigB-recognized promoters are indicated. (C) The transcription start site is shown in bold letters.
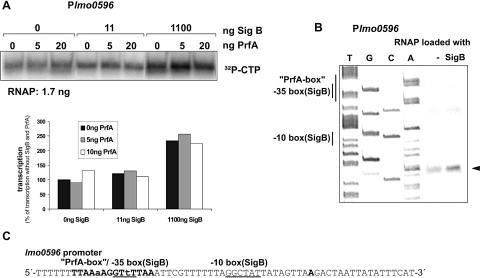
In vitro transcription with the UTR-DNA fragment of lmo0596 also resulted in PrfA-independent transcription, which was again highly activated with SigB-loaded RNAP. The SigB-dependent transcription starts at A, 9 bp downstream of the −10 box of the anticipated SigB-dependent promoter (Fig. 3). There is a faint but reproducible band 24 bp upstream from the transcription start site of the SigB-dependent promoter, the intensity of which also seems to increase in the presence of SigB. Whether this band represents a transcriptional start site of another weak SigB-dependent promoter or simply a primer extension artifact cannot be determined (Fig. 3B).
FIG. 3.
(A) In vitro transcription assay with the promoter region of lmo0596. RNAP of L. monocytogenes was loaded with different amounts of purified SigB protein, as indicated. (B) The corresponding in vitro transcripts were taken for primer extension to determine the transcriptional start sites. The putative −35 and −10 boxes of the SigB-recognized promoters are indicated. (C) The transcription start site is shown in bold letters.
SigB-dependent in vitro transcription starting at PprfA2
One possible link between PrfA- and SigB-dependent gene expression is the recently suggested SigB-dependent transcription of the prfA gene itself (23). The prfA gene is transcribed together with plcA in a bicistronic mRNA from an autoregulated promoter located in front of plcA (9, 20) and in addition from two promoters (PprfA1 and PprfA2) located in the UTR of prfA; the latter ones seem to be essential for basal-level transcription of prfA in the absence of PrfA.
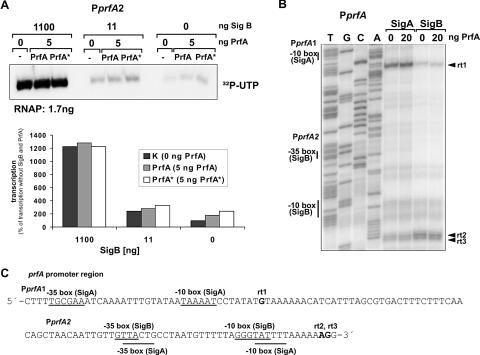
SigB-dependence of PprfA2 was directly demonstrated by in vitro transcription with RNAP in the presence and absence of SigB by using as template a DNA fragment containing PprfA1 and PprfA2. With [32P]UTP, we obtained, in accord with previously reported data (15), mainly a prfA transcript starting at PprfA2, the amount of which was significantly enhanced by increasing the SigB concentration (Fig. 4). No difference in the size of this transcript was observed, however, in the presence of SigA (and the absence of SigB), suggesting that PprfA2 may actually consist of two overlapping promoters, one being SigA dependent and the other being SigB dependent (Fig. 4C).
FIG. 4.
In vitro transcription with the 5′ upstream region of prfA containing the two previously mapped promoters PprfA1 (P1) and PprfA2 (P2) (8). (A) Wild-type PrfA and hyperactive PrfA* (24, 27) proteins and RNAP loaded with different amounts of purified SigB protein were used as indicated. (B) The primer extensions were carried out to determine the transcriptional start sites. (C) The putative −35 and −10 boxes of SigB- and SigA-recognized promoters and the transcription start sites (rt1, rt2, and rt3) are indicated.
To prove this assumption, primer extension was performed with transcripts produced in the presence of the four unlabeled NTPs and RNAP loaded with either SigA or SigB. With SigA-RNAP, we obtained as major reverse transcript mainly rt1 (Fig. 4B), suggesting a transcriptional start at G, 8 bp downstream of the −10 box (TAAAAT) of PprfA1, and at a lower concentration rt2 and rt3, two reverse transcripts which suggested transcription starts from PprfA2, presumably 6 and 7 bp downstream of a possible SigA-recognized −10 box (TATTTT) (Fig. 4C). Transcription with RNAP loaded with additional SigB resulted mainly in two reverse transcripts apparently identical to rt2 and rt3, suggesting that SigB-dependent transcription is initiated 9 and 10 bp downstream of the SigB-specific −10 box of PprfA2 (GGGTAT) (Fig. 4B). These data suggested that PprfA1 is a typical SigA-dependent promoter, whereas PprfA2 seems to consist of two overlapping promoters, one being SigB dependent and the other being SigA dependent. Transcription from neither of these prfA promoters was affected by PrfA (Fig. 4B).
In conclusion, our in vitro transcription data indicate that the tested group III genes (21) are controlled by SigB-dependent promoters but that PrfA has no direct influence on their transcription. There appears to be, however, a link between PrfA and SigB through the partial transcription of prfA by SigB-RNAP. In vitro transcription (data not shown) with several other newly identified genes (lmo0788, lmo2219, lmo0178, and lmo0278), all belonging to class I or II, which are positively or negatively affected by PrfA in vivo (21), indicated transcription by SigA-loaded RNAP which was also not directly activated or repressed by PrfA, suggesting that transcription of (at least most of) these newly identified PrfA-affected genes is not directly regulated by PrfA, as in the case of the PrfA-dependent virulence genes, but possibly by activators or repressors whose expression or activity might be influenced by PrfA (19).
Acknowledgments
We thank D. Beier, R. Gross, and B. Joseph for critical reading of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 479-B1) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böckmann, R., C. Dickneite, W. Goebel, and J. Bohne. 2000. PrfA mediates specific binding of RNA polymerase of Listeria monocytogenes to PrfA-dependent virulence gene promoters resulting in a transcriptionally active complex. Mol. Microbiol. 36:487-497. [DOI] [PubMed] [Google Scholar]
- 3.Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 4.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 5.Dickneite, C., R. Böckmann, A. Spory, W. Goebel, and Z. Sokolovic. 1998. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol. Microbiol. 27:915-928. [DOI] [PubMed] [Google Scholar]
- 6.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 7.Ermolaeva, S., S. Novella, Y. Vega, M. T. Ripio, M. Scortti, and J. A. Vázquez-Boland. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol. Microbiol. 52:601-611. [DOI] [PubMed] [Google Scholar]
- 8.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 9.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreft, J., J. Bohne, R. Gross, H. Kestler, Z. Sokolovic, and W. Goebel. 1995. Control of Listeria monocytogenes virulence by the transcriptional regulator PrfA, p. 129-142. In R. Rappuoli, V. Scarlato, and B. Aricó (ed.), Signal transduction and bacterial virulence. R. G. Landes Company, Austin, Tex.
- 14.Kreft, J., and J. A. Vázquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 15.Lalic-Mülthaler, M., J. Bohne, and W. Goebel. 2001. In vitro transcription of PrfA-dependent and -independent genes of Listeria monocytogenes. Mol. Microbiol. 42:111-120. [DOI] [PubMed] [Google Scholar]
- 16.Lampidis, R., R. Gross, Z. Sokolovic, W. Goebel, and J. Kreft. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol. Microbiol. 13:141-151. [DOI] [PubMed] [Google Scholar]
- 17.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, Q., M. Rauch, A. K. Marr, S. Müller-Altrock, and W. Goebel. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39-52. [DOI] [PubMed] [Google Scholar]
- 19.Marr, A. K., B. Joseph, and W. Goebel. Submitted for publication.
- 20.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vázquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 21.Milohanic, E., P. Glaser, J. Y. Coppée, L. Frangeul, Y. Vega, J. A. Vázquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 22.Müller-Altrock, S., and N. Mauder. Personal communication.
- 23.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ripio, M. T., G. Domínguez-Bernal, M. Lara, M. Suárez, and J. A. Vázquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vega, Y., C. Dickneite, M. T. Ripio, R. Böckmann, B. González-Zorn, S. Novella, G. Domínguez-Bernal, W. Goebel, and J. A. Vázquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]