Abstract
Opioids are the gold standard for pharmacological treatment of neuropathic pain, but their analgesic effects are unsatisfactory in part due to nerve injury-induced downregulation of opioid receptors in dorsal root ganglia (DRG) neurons. How nerve injury drives such downregulation remains elusive. DNA methyltransferase-(DNMT-) triggered DNA methylation represses gene expression. We show here that blocking the nerve injury-induced increase in DRG DNMT3a (a de novo DNMT) rescued the expression of Oprm1 and Oprk1 mRNAs and their respective encoding mu opioid receptor (MOR) and kappa opioid receptor (KOR) proteins in the injured DRG. Blocking this increase also prevented the nerve injury-induced increase in DNA methylation in the promoter and 5′-untranslated region of the Oprm1 gene in the injured DRG, restored morphine or loperamide (a peripheral acting MOR preferring agonist) analgesic effects, and attenuated the development of their analgesic tolerance under neuropathic pain conditions. Mimicking this increase reduced the expression of Oprm1 and Oprk1 mRNAs and their coding MOR and KOR in DRG and augmented MOR-gated neurotransmitter release from the primary afferents. Mechanistically, DNMT3a regulation of Oprm1 gene expression required the methyl-CpG-binding protein 1, MBD1, as MBD1 knockout resulted in the decreased binding of DNMT3a to the Oprm1 gene promoter and blocked the DNMT3a-triggered repression of Oprm1 gene expression in DRG neurons. These data suggest that DNMT3a is required for nerve injury-induced and MBD1-mediated epigenetic silencing of the MOR and KOR in the injured DRG. DNMT3a inhibition may serve as a promising adjuvant therapy for opioid use in neuropathic pain management.
Keywords: DNA methyltransferase 3a, mu opioid receptor, kappa opioid receptor, MBD1, Dorsal root ganglion
Introduction
Nerve injury-induced neuropathic pain is a distressing and debilitating condition affecting approximately 7% of the world population [41]. Healthcare expenses and lost productivities account for over 100 billion dollars in spending each year. Although recent advances have been made in the therapeutic management of neuropathic pain, opioids, like morphine, remain the mainstay of prescribed medications in the treatment of this disorder. However, about two thirds of patients show unsatisfactory pain control [27]. Moreover, a majority of neuropathic pain patients require higher doses of opioids as well as repeated and prolonged administration of opioids to achieve pain relief. The consequence of such opioid regimens is the rapid onset of the adverse side effects including nausea, constipation, respiratory depression, opioid analgesic tolerance and hyperalgesia, and addiction liability [2], which greatly limits opioid use in neuropathic pain patients. Preclinical and clinical observations suggest that the reduced analgesic effects of opioids in neuropathic pain patients may result in part from nerve injury-induced downregulation of opioid receptor mRNA and protein expression in the first-order sensory neurons of the injured dorsal root ganglion (DRG) [11;14;29;32;40;51]. Therefore, understanding how nerve injury drives opioid receptor downregulation in DRG is essential for improving patient care.
Recent studies showed that the mechanism for gene regulation involves epigenetic modification, such as DNA methylation [16;31]. DNA methylation represses gene transcription through several mechanisms, such as (1) physically impeding the binding of transcription factors and/or (2) serving as docking sites for transcription repressors such as the family of methyl-CpG-binding domain (MBD) proteins, including MBD1-4 and MeCP2 [20;22]. DNA methylation is catalyzed primarily by the members of the DNA methyltransferase (DNMT) family, including DNMT1, DNMT3a and DNMT3b. DNMTs also contain DNMT2, an inactivated isoform and DNMT3L, which lacks the conserved catalytic domain [3;35]. Conventionally, DNMT3a and DNMT3b are classified as de novo DNMTs, whereas DNMT1 is considered to be the primary maintenance DNMT [22]. Whether and how DNMTs participate in nerve injury-induced opioid receptor downregulation in the DRG is elusive.
Here, we showed that the nerve injury-induced increase in DRG DNMT3a is responsible for the MBD1-mediated epigenetic silencing of mu opioid receptor (MOR) and kappa opioid receptor (KOR) in the injured DRG under neuropathic pain conditions. DNMT3a appears to be a potential target for improving opioid analgesia in neuropathic pain management.
Materials and Methods
Animal preparation
Sprague-Dawley rats were purchased from Charles River Laboratories (Willington, MA). Mbd1 KO mice [50] and Dnmt3afl/fl mice [12] had been bred onto C57B/L6 background for at least 6 generations and were bred by crossing the heterozygotes in our facility. All animals were kept in a controlled 12-h light/dark cycle, with free access to water and food pellet. Male rats weighing 250–300 g and male mice weighing 25–30 g were used for behavior testing. All procedures used were approved by the Animal Care and Use Committee at Rutgers New Jersey Medical School and were consistent with the ethical guidelines of the US National Institutes of Health and the International Association for the Study of Pain.
DRG microinjection
DRG microinjection was carried out as described [15;49]. Briefly, a midline skin incision was made in the lower lumbar back region, and the L5 DRG in rats or L4 DRG in mice was exposed. Viral solution (1 μl, 4 × 1012) was injected into the exposed DRG over a period of 10 minutes with a glass micropipette connected to a Hamilton syringe. After each injection, a 10-minute pipette retention was used before the glass pipette was removed. After injection, the surgical field was irrigated with sterile saline and the skin incision was closed with wound clips.
Neuropathic pain models
Spinal nerve ligation (SNL) in rats and mice was performed as previously reported [15;33;45]. Briefly, a midline skin incision was made on the lower back. The sixth lumbar transverse process on rats or fifth lumbar transverse process on mice was identified, and then was separated from its muscle attachments. The underlying fifth lumbar spinal nerve in rats or fourth lumbar spinal nerve in mice was isolated and ligated with a 4-0 silk suture in rats or 7-0 silk suture in mice. The ligated nerve was then transected at the distal end. Sham-operated groups received identical surgical procedures only exposing the nerves without ligation and transection. The surgical field was then irrigated with sterile saline and the skin incision was closed with wound clips.
Morphine- or loperamide-induced analgesia and tolerance
Analgesia was measured in rats receiving subcutaneous (s.c.) injection of 0.5 mg/kg morphine (WEST-WARD, Eatontown, NJ) or 3 mg/kg loperamide (Sigma, St. Louis, MO). Naltrexone (5 mg/kg. Sigma) or saline was given intraperitoneally 5 minutes before loperamide injection to confirm that loperamide’s effect is mediated by MOR. The thermal test as described below was performed before and 20 minutes after s.c. morphine or loperamide injection. Analgesic tolerance was induced in rats as previously described [18;46]. Briefly, morphine (10 mg/kg) was injected s.c. twice daily for 6 consecutive days in naïve rats or twice daily for 4 consecutive days starting on day 7 post-SNL. Loperamide (10 mg/kg, s.c. in naïve rats, and 0.75 mg/kg in SNL rats) was injected twice daily for 5 consecutive days in naïve rats or twice daily for 3 consecutive days starting on day 7 post-SNL. For morphine tolerance, the thermal test was performed before morphine injection and 20 minutes after s.c. injection of an inducing dose of morphine (5 mg/kg in naïve rats and 1.5 mg/kg in SNL rats) on the mornings of days 1, 3, 5, and 7 in naïve rats or on the mornings of days 7, 9, and 11 post-SNL. For loperamide tolerance, the thermal test was performed before and 20 minutes after the s.c. injection of loperamide on the mornings of days 1, 3 and 5 in naïve rats and days 7, 8, and 9 post-SNL. Cumulative morphine dose-response test was carried out as described previously [19;46] on the afternoon of day 7 in naïve rats or on the afternoon of day 11 post-SNL. Rats were injected s.c. with a low dose of morphine (1 mg/kg), and analgesia was assessed 20 minutes later by the thermal test. Rats that were not analgesic at the first dose then received a second dose (cumulative dosing increase by 0.3 log units) and were tested 20 minutes afterward. This procedure was repeated until the rats failed to move their tail within the cutoff time or no additional increase in paw withdrawal latency was noted from one dose to the next.
Thermal behavioral tests
Thermal test was carried out as previously described [15;46;49]. Briefly, each rat was placed in a plexiglas chamber on an elevated grid and acclimated for 30 minutes before behavioral test. Paw withdrawal latencies to noxious heat were measured with a Model 336 Analgesic Meter (IITC Inc./Life Science Instruments, Woodland Hills, CA, USA). A beam of light giving radiant heat was aimed to the middle of the plantar surface of each hind paw through the glass plate. When the animal withdrew its paw, the light beam was turned off. The length of time between the onset of the light beam and paw withdrawal was defined as the paw withdrawal latency. Each trial was repeated three times at 5-min intervals for each side. A cut-off time of 20 s was used to avoid tissue damage. Paw withdrawal latencies were converted to the percentage of the maximal possible antinociceptive effect (% MPAE): % MPAE = [(response latency − baseline latency)/(cut-off latency − baseline latency)] ×100%.
Reverse transcription (RT)-PCR
Two rat DRGs or four mouse DRGs were pooled together to achieve enough RNA. Total RNA was prepared using miRNeasy Mini Kit (Qiagen, Valencia, CA), following the manufacturer’s instruction. Briefly, the pooled DRGs were homogenized with 700 μl lysis reagent, extracted by 140 μl chloroform and centrifuged for 15 min at 12,000 × g at 4°C. RNA was then precipitated with 100% ethanol, treated with DNase I, and eluted with 35 μl RNase free water. After concentration measurement, the RNA was first reverse-transcribed to cDNA using the ThermoScript reverse transcriptase (Invitrogen/ThermoFisher Scientific, Grand Island, NY) and oligo (dT) primers (Invitrogen/ThermoFisher Scientific). Template (1 μl) was amplified by real-time PCR by using the primers listed in Table 1. Gapdh was used as an internal control for normalization. Each sample was run in triplicate in a 20 μL reaction with 250 nM forward and reverse primers, 10 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and 20 ng of cDNA. Reactions were performed in a BIO-RAD CFX96 real-time PCR system. Ratios of ipsilateral-side mRNA levels to contralateral-side mRNA levels were calculated using the ΔCt method (2−ΔΔCt). All data were normalized to Gapdh, which has been demonstrated to be stable even after peripheral nerve injury insult [15;49].
Table 1.
All primers used
| Names | Sequences (5′-3′) |
|---|---|
| PCR/Real-time PCR | |
| Rat-Oprm1-F | TTCCTGGTCATGTATGTGATTGT |
| Rat-Oprm1-R | GGGCAGTGTACTGGTCGCTAA |
| Rat-Oprd1-F | GGGTCTTGGCTTCAGGTGTT |
| Rat-Oprd1-R | ACGGTGATGATGAGAATGGG |
| Rat-Oprk1-F | TTTGTGGTGGGCTTAGTGGG |
| Rat-Oprk1-R | CTCTGGAAGGGCATAGTGGT |
| Rat-DNMT3A-F | CAGCGTCACACAGAAGCATATC |
| Rat-DNMT3A-R | GGTCCTCACTTTGCTGAACTTGG |
| Rat-DNMT3B-F | GAATTTGAGCAGCCCAGGTTG |
| Rat-DNMT3B-R | TGAGAAGAGCCTTCCTGTGCC |
| Rat-DNMT1-F | CGGCGGAGGTGTCCTAACTTGGC |
| Rat-DNMT1-R | GGGTGACGGCAACTCTGGTA |
| Rat-GAPDH-F | TCGGTGTGAACGGATTTGGC |
| Rat-GAPDH-R | CCTTCAGGTGAGCCCCAGC |
| Mouse-Oprm1-F | TCTTCACCCTCTGCACCATG |
| Mouse-Oprm1-R | TCTATGGACCCCTGCCTGTA |
| Mouse-Oprd1-F | TTTGGCATCGTCCGGTACAC |
| Mouse-Oprd1-R | AGAGCACAGCCTTGCACAGC |
| Mouse-Oprk1-F | AGTGTGGACCGCTACATTGCT |
| Mouse-Oprk1-R | CAATGACATCCACATCTTCCCTG |
| Mouse-DNMT3A-F | CGAATTGTGTCTTGGTGGATGAC |
| Mouse-DNMT3A-R | GGTGGAATGCACTGCAGAAGGA |
| Mouse-DNMT3B-F | CAGCCTTCTGAATTACACGCA |
| Mouse-DNMT3B-R | TCCCATTGCTATGTCGGGTT |
| Mouse-DNMT1-F | AGTCGGACAGTGACACCCTTT |
| Mouse-DNMT1-R | TGTGTCTACAACTCTGCGTTTCT |
| Mouse-MBD1-F | CTGCATCTGCGTCTTCACAT |
| Mouse-MBD1-R | CACACCCCACAGTCCTCTTT |
| Mouse-GAPDH-F | TCGGTGTGAACGGATTTGGC |
| Mouse-GAPDH-R | TCCCATTCTCGGCCTTGACT |
| ChIP-PCR | |
| rOprm1-ChIP-F1 | TTCTTGTTTCAGATACGCGGA |
| rOprm1-ChIP-R1 | GGTCGGTGTTTTCATCAGTTAGG |
| rOprm1-ChIP-F2 | CTAACTGATGAAAACACCGACCT |
| rOprm1-ChIP-R2 | GAGAGTCAGCCTCCTCGGTC |
| rOprm1-ChIP-F3 | CGACCGAGGAGGCTGACTCT |
| rOprm1-ChIP-R3 | CTTAGAAGTACACAGAGGCGCAT |
| rOprm1-ChIP-F4 | AGGTGGGAGGGGGCTACAAG |
| rOprm1-ChIP-R4 | GAGCACTCAGACTTTTCGGGT |
| rOprm1-ChIP-F5 | GAACCCGAAAAGTCTGAGTGC |
| rOprm1-ChIP-R5 | AACGTGGGACAAGTTGAGCC |
| rOprm1-ChIP-F6 | GCGACTGCTCAGACCCCTTA |
| rOprm1-ChIP-R6 | CGATAGAGTAGAGGGCCATGATG |
| rOprm1-ChIP-F7 | AGCCCTTCCATGGTCACAGC |
| rOprm1-ChIP-R7 | TCCTCTGTCACTCTCGGGCT |
| Clone Sequencing & Pyro-sequencing | |
| Oprm1-Methy-F1 | TATTGTTGTTTTGAATAGGTTTGTG |
| Oprm1-Methy-R1 | AACAATCAAAAAATCAACCTCC |
| Oprm1-Methy-F2 | AGGAGGTTGATTTTTTGATTGT |
| Oprm1-Methy-R2 | CAACCTCTTCCTCTCACAACTA |
| Oprm1-Methy-F3 | TAGGGTTGGTTTATGTAAGAATTT |
| Oprm1-Methy-R3 | AACATTCAAAACATTCCTTACCTTA |
| R1 Pyro1 | CTAATATTATCCTTTACTCTAATC |
| R1 Pyro2 | CTTATACTTAAACACTCTTC |
| R1 Pyro3 | CTTTATAACAACCATACATTAAAT |
| R2 Pyro1 | TTTTTTTTATTTAGAGAGTGG |
| R2 Pyro2 | GTTTTGGGAAGTTAGAGATG |
| R2 Pyro3 | GTTATAAGTAGAGGAGAATATTAG |
| R3 Pyro1 | TAGTTGTGAGAGGAAGAGGTTG |
| R3 Pyro2 | GTAGTAGGTTTTTAGTATTATGGAT |
| R3 Pyro3 | GTTGATGGTAATTAGTT |
| R3 Pyro4 | GGGAAYGATAGTTTGTGTTTTTAGAT |
| R3 Pyro5 | ATGGTTATAGTTATTATTATTATGG |
| Cloning | |
| mMBD1-F | CACTGGTGGAGGAGGAAGAG |
| mMBD1-R | TCCTGAATTCCAGGTTCAGC |
| mMBD1-N-F | ATATCCGGAGCCACCATGGCTGAGTCCTGGCA |
| mMBD1-N-R | CGCCAGCGGCCGCCTACAAAACTTCTTCTTTC |
| rDNMT3A-shRNA-F | TTACCGGTGGGCAGGAAGAGGGCCTATTTCCCATGA |
| rDNMT3A-shRNA-R | AATCTAGACATCCACTGTGAATGATAACGAATTCACTTATCATTCACAGTGGATGGGATCCTCGTCCTTTCCACAAGATAT |
PCR: polymerase chain reaction. F: Forward. R: Reverse. N-PCR: Nested PCR. Underlined letters: the restriction enzyme recognition sites.
Plasmid constructs and virus production
A Dnmt3a shRNA duplex corresponding to bases 2576-2594 from the open reading frame of rat Dnmt3a mRNA (GenBank accession number NM_001003958) was designed. A mismatch shRNA with a scrambled sequence and no known homology to a rat gene (scrambled shRNA) was used as a control. shRNAs were synthesized and were amplified by using AAV-shRNA-Ctrl-EYFP and the primers listed in Table 1. Fragments harboring Dnmt3a shRNA and Dnmt3a scrambled shRNA were ligated into pro-viral plasmids using AgeI and XbaI restriction sites. The resulting vectors expressed the genes under the control of the cytomegalovirus promoter. Rat full-length Dnmt3a cDNA was purchased from OriGene (Rockville, MD). Mouse full-length Mbd1 cDNA was amplified from total RNA of mouse DRG using the same Kit as described above and the primers listed in Table 1. Adeno-associated virus type 5 (AAV5) particles carrying the cDNA were produced at the UNC Vector Core (Chapel Hill, NC). AAV5-GFP and AAV5-Cre were purchased from the UNC Vector Core. Herpes simplex virus (HSV)-GFP and HSV-Dnmt3a were provided by Dr. Eric J Nestler.
DRG cell culture and viral transfection
Adult wildtype (WT) mice, Dnmt3a fl/fl mice, or Mbd1 KO mice were euthanized with isoflurane and all DRGs were collected in cold Neurobasal Medium (Gibco/ThermoFisher Scientific) with 10% fetal bovine serum (JR Scientific, Woodland, CA), 100 units/ml Penicillin, and 100 μg/ml Streptomycin (Quality Biological, Gaithersburg, MD). Collected DRGs were cut into smaller pieces and then treated with enzyme solution (5 mg/ml dispase, 1 mg/ml collagenase type I in Hanks’ balanced salt solution without Ca2+ and Mg2+ (Gibco/ThermoFisher Scientific)). After trituration and centrifugation, dissociated cells were resuspended in mixed Neurobasal Medium and plated in a six-well plate pre-coated with 50μg/ml poly-D-lysine (Sigma). The cultured neurons were incubated at 95% O2, 5% CO2, and 37 °C. After 24 h, 1–10 μl of virus (titer: 1 × 1012/ml) was added to each 2 ml-well. The cells were harvested 3 days later for RT-PCR.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were conducted using the EZ ChIP Kit (Upstate/EMD Millipore, Darmstadt, Germany), following the manufacturer’s instruction with minor modifications. Two DRGs from rats or five DRGs from mice were harvested and homogenized. The homogenization solution was then crosslinked with 1% formaldehyde for 10 min at room temperature. The reaction was quenched by adding 0.125 M glycine solution for 5 min at room temperature. After centrifugation, the collected pellet was lysed in sodium dodecyl sulfate lysis buffer containing protease inhibitor cocktail (Upstate Biotechnology) and sonicated to yield fragments with a length of 200~1,000 bp. After the samples were precleared with protein G agarose beads for 1 h at 4 °C with agitation, they were subjected to immunoprecipitation overnight with 2 μg of rabbit antibodies against DNMT3a (Abcam, Cambridge, MA) or MBD1 (Abcam) or with 2 μg of normal rabbit serum overnight at 4°C. Input (10–20% of the sample for immunoprecipitation) was used as a positive control. The DNA fragments were purified and identified using PCR/Real-time PCR with the primers listed in the Table 1.
Bisulfite sequencing
L5 DRGs were harvested 7 days after SNL or sham surgery. Two DRGs were pooled together for DNA extraction. Bisulfite treatment of genomic DNA was carried out using the EZ DNA Methylation-Gold kit (ZYMO Research, Irvine, CA) according to the manufacturer’s instructions. The region of the Oprm1 gene promoter from −670 to +450 bp consisted of 38 CpG sites was amplified. For the pyrosequencing assay, 3 pairs of primers of the Oprm1 gene with 5′-biotin listed in the Table 1 were used to amplifying the bisulfite DNA. The master mix of the binding buffer, streptavidin-sepharose beads, and PCR products were prepared for the binding reaction in a 96-well plate. After sealing the plate with sticker film, the plate was placed on the shaker for at least 15 minutes, but no longer than 1 hour. The pyrosequencing was then performed. For the clone-sequencing assay, the same promoter fragment was amplified. The PCR products were purified and sub-cloned into pMD T-19 vector (Takara/Clontech Laboratories, Mountain View, CA) and transformed into competent cells. After overnight bacterial culture, 20 sub-clones from each assay were subjected to PCR amplification and direct sequencing. The primers we used are listed in the Table 1.
Immunohistochemistry
Rats were anesthetized with isoflurane and perfused with 300 ml of 4% paraformaldehyde in 0.1 M PBS (pH 7.4). After perfusion, L4/5 DRGs were dissected, post-fixed at 4°C for 4 hours and cryoprotected in 30% sucrose overnight. The transverse sections were cut on a cryostat at a thickness of 25 μm. Three sets of the sections were collected. The sections were blocked for 1 h at 25 °C in 0.01M PBS containing 10% goat serum and 0.3% Triton X-100. One set of sections was incubated overnight at 4°C with rabbit anti-DNMT3a (1:500, Santa Cruz Biotechology, Inc., Dallas, TX) plus guinea pig anti-MOR (1:1,000, EMD Millipore, Darmstadt, Germany) and the second with rabbit anti-DNMT3a plus mouse anti-kappa opioid receptor (KOR; 1: 200, Neuromics, Edina, MN). The sections were then incubated with a mixture of goat anti-rabbit antibody conjugated to Cy3 (1:200, Jackson ImmunoResearch, West Grove, PA) and donkey anti-guinea pig antibody conjugated to Cy2 (1:200, Jackson ImmunoResearch) or with a mixture of goat anti-rabbit conjugated to Cy3 and donkey anti-mouse antibody conjugated to Cy2 (1:200, Jackson ImmunoResearch) for 2 hours at room temperature. The third set of sections was used for control experiments, which included preabsorption of the primary antibodies with excess of the corresponding antigens, substitution of the normal serum, and omission of the primary antibodies. Immunofluorescence-labeled images were generated using a Leica DMI4000 fluorescence microscope with a DFC365FX camera (Leica, Germany).
Western blotting
Two unilateral rat DRGs or four unilateral mouse DRGs were pooled together to obtain enough proteins. Tissues were prepared as previously described [15;49]. Briefly, DRGs were homogenized with ice-cold lysis buffer (10 mM Tris, 5 mM EGTA, 0.5% Triton X-100, 2 mM benzamidine, 0.1 mM phenylmethylsulfonyl fluoride, 40 μM leupeptin, 150 mM NaCl). The crude homogenate was centrifuged at 4°C for 15 minutes at 1,000g. The supernatants were collected for cytoplasmic protein detection. The pellets were further sonicated and dissolved in nucleus-soluble ice-cold buffer (1M Tris-HCl, 1% SDS, and 0.1% Triton X-100). After protein concentration was measured, the samples were heated at 99°C for 5 minutes and loaded onto a 4–15% stacking/7.5% separating SDS-polyacrylamide gel (Bio-Rad, Hercules, CA). The proteins were then electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with 3% nonfat milk in Tris-buffered saline (pH 7.40) containing 0.1% Tween-20 for 1 h and then incubated with primary antibodies overnight under gentle agitation. The primary antibodies used include rabbit anti-DNMT3a (1:500, Cell Signaling Technology, Danvers, MA), rabbit anti-DNMT3b (1:500, Santa Cruz Biotechology), rabbit anti-DNMT1 (1:500, Cell Signaling Technology), rabbit anti-mTOR (1:1000, Cell Signaling Technology), rabbit anti-MOR (1:500, Neuromics), rabbit anti-KOR (1:500, NOVUS Biologicals, Littleton, CO), mouse anti-MBD1 (1:200, provided by Dr. Xinyu Zhao), rabbit anti-mTOR (1:2,000, Cell Signaling Technology), rabbit anti-GAPDH (1:1000, Santa Cruz Biotechology), mouse anti-tubulin (1:1,000, Santa Cruz Biotechology), or rabbit anti–histone H3 (1:1,000, Cell Signaling Technology). The proteins were detected by horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (1:3,000, Jackson ImmunoResearch) and visualized by chemiluminescence regents (ECL, Bio-Rad). Images were generated using the ChemiDoc XRS System with Image Lab software (Bio-Rad). Intensities of protein bands were quantified using the Image Lab software. All cytosol protein bands were normalized to either GAPDH or tubulin, whereas nucleus protein bands were normalized to total histone H3.
Whole-cell patch clamp recording
Adult rat spinal cord slices were prepared as described [38]. Briefly, rats (age > 21 days) were deeply anesthetized with isoflurane. Laminectomy was applied from mid thoracic to lower lumbar levels. The spinal cord was quickly removed and placed in cold modified artificial cerebrospinal fluid (ACSF) containing: (in mM) 80 NaCl, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, 3.5 MgCl2, 25 NaHCO3, 75 sucrose, 1.3 Ascorbate, 3.0 Sodium Pyruvate, oxygenated with 95% O2 and 5% CO2 (pH 7.4, 310–320 mOsm). Transverse 350 μm slices for miniature EPSC (mEPSC) recording or 500 μm slices with attached L4 or L5 dorsal root for evoked EPSC (eEPSC) recording were cut by a vibratome VT-1200 (Leica). Slices were incubated at least 1 h at room temperature in the recording solution containing: (in mM) 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 26 NaHCO3, 25 D-glucose, 1.3 Ascorbate, 3.0 Sodium Pyruvate, oxygenated with 95% O2 and 5% CO2 (pH 7.4, 310–320 mOsm).
Whole-cell patch recording was carried out as described [38]. The slice was transferred to a recording chamber (Warner Instruments, Hamden, CT) and perfused with oxygenated recording solution at a rate of 3 ml/min at room temperature. Neurons were identified by infrared differential interference contrast (IR-DIC) with an upright microscope DM6000 (Leica) equipped 40 × 0.80 NA water-immersion objective and a CCD camera (Leica). Whole-cell patch clamp recording was carried out in the voltage-clamp mode. The electrode resistances of micropipettes ranged from 3 to 8 MΩ. Lamina II neurons were clamped with an Axopatch-700B amplifier (Molecular Devices, Downingtown, PA). The intracellular pipette solution contained: (in mM) 110 Cs2SO4, 5 TEA-Cl, 0.5 CaCl2, 2 MgCl2, 5 EGTA, 5 HEPES, 5 MgATP, 0.5 NaGTP, and 1 GDP-β-S. Cs and TEA was used as a K+-channel blocker and GDP-β-S used as a GTP binding protein blocker to prevent the postsynaptic μ-opioid mediated effect as previously described [1;8;9]. To record mEPSC, 500 nM TTX was presented at the recording solution. To record eEPSC, a suction electrode (A-M systems) was used for electrical stimulation of the attached dorsal root. Stimulation was delivered by the suction electrode with a constant current stimulator S88 (Grass, Rockland, MA). To verify different primary afferent inputs, Aβ fiber-evoked EPSCs were classified as monosynaptic by constant latency and absence of failures upon 25 μA/20 Hz stimulation; Aδ fiber-evoked EPSCs were classified as monosynaptic by constant latency and absence of failures upon 100 μA/2 Hz stimulation; C fiber-evoked EPSCs were classified as monosynaptic by absence of failures upon 500 μA/1 Hz stimulation. The conduction velocity was further measured to distinguish the primary afferent input (C < 0.8 m/s) [23;39]. Only C fiber input monosynaptic neurons were measured for further experiments. Measurements were made from only one neuron per slice. The pair pulse protocol was delivered by S88 with 20 Hz frequency and 50 ms pulse interval. The signals were filtered at 2kHz. Data were stored on a computer using the DigiData 1550 interface and were analyzed by the pCLAMP 10.4 software package (Molecular Devices). All experiments were performed at room temperature.
Statistical analysis
All of the results are given as means ± S.E.M. The data were statistically analyzed with two-tailed, paired Student’s t-test and a one-way or two-way ANOVA. When ANOVA showed a significant difference, pairwise comparisons between means were tested by the post hoc Turkey method (SigmaPlot 12.5, San Jose, CA). Significance was set at P < 0.05.
Results
DNMT3a regulation of MOR and KOR expression in the DRGs of naïve or SNL rats
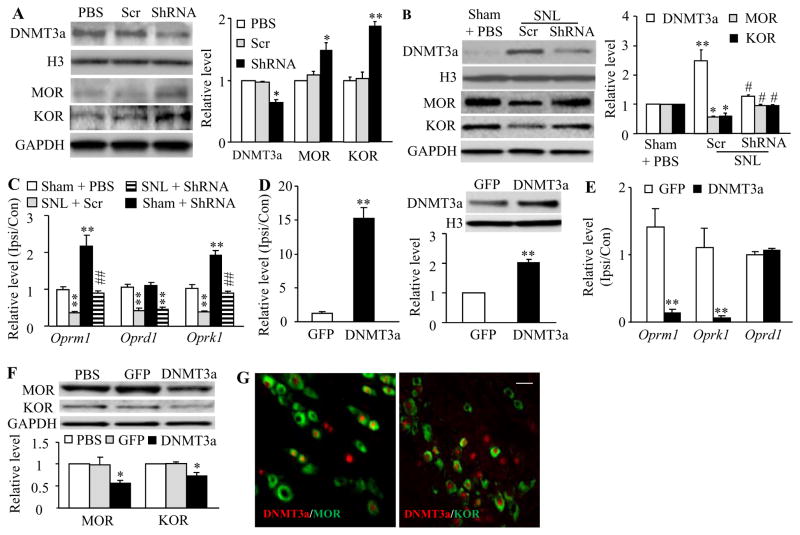
Our recent study demonstrated a significant increase in the level of DNMT3a protein in the injured DRG neurons following peripheral nerve injury [48]. Given that DNMT3a is a repressor of gene expression [16;48], we first asked whether this increased DNMT3a participated in nerve injury-induced downregulation of DRG opioid receptors. shRNA strategy was carried out to knock down DRG DNMT3a expression through unilateral DRG microinjection of AAV5 that expresses Dnmt3a shRNA [48]. AAV5 that expresses scrambled shRNA was used as a control. Since AAV5 transduces the neurons, but not the satellite cells, after viral microinjection into the DRG [49], the delivered Dnmt3a shRNA expresses predominantly in the DRG neurons, in which DNMT3a is exclusively distributed [48]. Specific and selective effects of Dnmt3a shRNA were verified by its ability to knock down DNMT3a, but not DNMT1, DNMT3b, or an intracellular protein mTOR in the injected DRGs of naïve rats 5 weeks after viral microinjection (Fig. 1A; Supplementary Fig. 1). The basal levels of MOR and KOR proteins in the injected DRGs from the AAV5-Dnmt3a shRNA-injected group were higher than those from either the PBS- or AAV5-scrambled shRNA-treated group (Fig. 1A). Consistent with the previous studies [14;29], the amounts of MOR and KOR proteins (Fig. 1B) and Oprm1, Oprd1, and Oprk1 mRNAs (Fig. 1C) were significantly reduced in the ipsilateral L5 (injured) DRG on day 7 post-unilateral L5 SNL in the AAV5-scrambled shRNA plus SNL group. The reductions in the levels of Oprm1 and Oprk1 mRNAs and their coding proteins (but not Oprd1 mRNA) in the injured DRG were abolished in the AAV5-Dnmt3a shRNA plus SNL group (Figs. 1B and 1C). As expected, basal levels of Oprm1 and Oprk1 mRNAs in the ipsilateral L5 DRG from the AAV5-Dnmt3a shRNA plus sham group increased (Fig. 1C).
Figure 1.
DNMT3a represses MOR and KOR expression in DRG. ShRNA: AAV5-Dnmt3a shRNA. Scr: AAV5-scambled shRNA. DNMT3a: AAV5-Dnmt3a. GFP: AAV5-Gfp. A. Expression of DNMT3a, MOR and KOR proteins in the ipsilateral L4/5 DRGs of naïve rats 5 weeks after DRG microinjection of PBS, ShRNA or Scr. Left: representative Western blots. Right: statistical summary of the densitometric analysis. n = 6 rats/group. *P < 0.05, **P < 0.01 vs the corresponding PBS-treated group. B and C. Expression of MOR and KOR proteins in the ipsilateral L5 DRG (B) and expression of Oprm1, Oprd1, and Oprk1 mRNAs in the ipsilateral (Ipsi) and contralateral (Con) L5 DRGs (C) from the treatment groups indicated 7 days after sham or SNL surgery. n = 6 rats/group. *P < 0.05, **P < 0.01 vs the corresponding sham plus PBS group. #P < 0.05, ##P < 0.01 vs the SNL plus Scr group. D and E. Expression of Dnmt3a (left in D), Oprm1 (E), Oprd1 (E) and Oprk1 (E) mRNAs in the ipsilateral (Ipsi) and contralateral (Con) L4/5 DRGs and expression of DNMT3a protein in the ipsilateral L4/5 DRGs (right in D) 5 weeks after DRG microinjection of GFP or DNMT3a. n = 5 rats/group. **P < 0.01 vs the corresponding GFP group. F. Expression of MOR and KOR proteins in the ipsilateral L4/5 DRGs 5 weeks after DRG microinjection of PBS, GFP or DNMT3a. n = 5 rats/group. *P < 0.05, **P < 0.01 vs the corresponding PBS-treated group. G. Co-localization of DNMT3a (red) with MOR (green) or KOR (green) in the DRG neurons of naïve rats. Scale bar: 40 μm.
Given that shRNA may have off-target effects, we further confirmed DNMT3a regulation of opioid receptors in neuropathic pain using microinjection of AAV5-Cre into the ipsilateral L4 DRG of Dnmt3afl/fl mice 5 weeks before unilateral L4 SNL or sham surgery [33]. AAV5-GFP was used as a control. Injection of AAV5-Cre, but not AAV5-GFP, prevented SNL-induced upregulation of Dnmt3a mRNA and its coding protein and reversed downregulation of Oprm1 and Oprk1 mRNAs and their coding proteins (but not Oprd1 mRNA) in the ipsilateral L4 DRG of Dnmt3afl/fl mice on day 7 post-SNL (Supplementary Fig. 2A and 2B). As expected, in the ipsilateral L4 DRG of sham Dnmt3afl/fl mice, AAV5-Cre injection markedly decreased the basal level of Dnmt3a mRNA and increased basal amounts of Oprm1 and Oprk1 mRNAs on day 7 post-sham surgery (Supplementary Fig. 2A and 2B).
We further tested whether mimicking the nerve injury-induced increase in DRG DNMT3a via microinjection of AAV5-Dnmt3a (that expresses full-length DNMT3a) into the unilateral L4/5 DRGs affected DRG opioid receptor expression. As shown in Figures 1D and 1E, the levels of Dnmt3a mRNA and its coding protein in the AAV5-Dnmt3a-treated group markedly increased, compared to the control AAV5-GFP group, in the injected DRG 5 weeks post-injection. The levels of Oprm1 and Oprk1 mRNAs and their coding proteins, but not Oprd1 mRNA, in the injected DRG from the AAV5-Dnmt3a-injected group significantly decreased compared to those from the PBS- or AAV5-GFP-injected group (Fig. 1F). The evidence suggests that DNMT3a directly regulates Oprm1 and Oprk1 gene expression. Indeed, immunohistochemical double-labelling staining showed that approximately 83.7% of MOR-labeled DRG neurons and 75.5% of KOR-labeled DRG neurons were positive for DNMT3a (Fig. 1G). Knockdown of DNMT3a through transduction of AAV5-Cre in in vitro cultured DRG neurons of Dnmt3afl/fl mice substantially increased Oprm1 and Oprk1 mRNA expression (Supplementary Fig. 3). These increases could be prevented by co-transduction of AAV5-Cre and HSV-Dnmt3a (Supplementary Fig. 3). As expected, HSV-Dnmt3a transduction alone reduced the levels of Oprm1 and Oprk1 mRNAs (Supplementary Fig. 3). Oprd1 mRNA expression was not altered in the cultured DRG neurons transduced by single virus or their combinations (Supplementary Fig. 3). The evidence described above strongly suggests the contribution of DNMT3a to nerve injury-induced downregulation of MOR and KOR, but not DOR, in the injured DRG.
Effect of DRG DNMT3a knockdown on morphine/loperamide analgesia and their tolerance in naïve or SNL rats
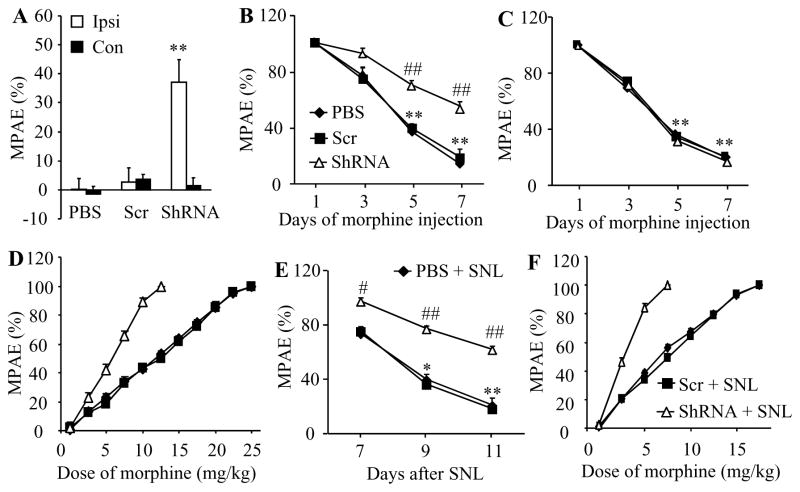
Next, we examined the role of DRG DNMT3a in opioid analgesia. Given that morphine binding predominantly to MOR is often used in pain management, we observed whether DNMT3a knockdown in DRG altered morphine analgesia and morphine-induced tolerance in naïve animals. As indicated in Fig. 2A, the AAV5-Dnmt3a shRNA-treated rats, but not the PBS- or AAV5-scrambled shRNA-treated rats, displayed a significant increase in morphine’s maximal possible analgesic effect (MPAE) on the ipsilateral (not contralateral) side after subcutaneous (s.c.) administration of a single low dose of morphine (0.5 mg/kg). Consistent with previous studies [46], repeated s.c. injection of morphine (10 mg/kg, twice daily) in naïve rats produced morphine analgesic tolerance as indicated by time-dependent decreases in morphine MPAEs in the PBS-injected group (Fig. 2B and 2C). These decreases were markedly attenuated on days 5 and 7 after morphine injection on the ipsilateral (but not contralateral) side of the AAV5-Dnmt3a shRNA-treated, but not AAV5-scrambled shRNA-treated, groups (Fig. 2B and 2C). This effect of AAV5-Dnmt3a shRNA was further verified by a significant leftward shift in the cumulative dose-response curve of morphine in the AAV5-Dnmt3a shRNA-treated group compared to the PBS- or AAV5-scrambled shRNA-treated group on the last day after repeated s.c. administration of morphine (Fig. 2D). The EC50 value of morphine in the AAV5-Dnmt3a shRNA-treated group (5.65 mg/kg) was substantially smaller than those in the PBS-treated (10.36 mg/kg) and AAV5-scrambled shRNA-treated (10.59 mg/kg) groups (Fig. 2D).
Figure 2.
DRG DNMT3a knockdown enhanced morphine analgesia and attenuated morphine tolerance in naïve and SNL rats. All of the following experiments were carried out 5 weeks after microinjection of AAV5-Dnmt3a shRNA (ShRNA), AAV5-scambled shRNA (Scr), or PBS into unilateral L4/5 DRGs (A–D) or L5 DRG (E and F). A. Morphine maximal possible analgesic effects (MPAE) measured 20 min after subcutaneous injection of 0.5 mg/kg morphine on the ipsilateral (Ipsi) and contralateral (Con) sides of naïve rats. n = 5 rats/group. **P < 0.01 vs the corresponding PBS-treated group. B and C. Morphine analgesic tolerance developed by subcutaneous injection of 10 mg/kg morphine twice daily for 6 days on the ipsilateral (B) and contralateral (C) sides of naïve rats. MPAEs were measured 20 min after subcutaneous injection of 5 mg/kg morphine on the mornings of days 1, 3, 5, and 7. n = 5 rats/group. **P < 0.01 vs the corresponding MPAE on day 1. ## P < 0.01 vs the PBS-treated group at the corresponding time points. D. Dose-response curve on the ipsilateral side of naïve rats from the PBS-, ShRNA- and Scr-treated groups on day 7 after subcutaneous injection of 10 mg/kg morphine twice daily for 6 days. E. Morphine analgesic tolerance developed by subcutaneous injection of 5 mg/kg morphine twice daily for 4 days starting at day 7 post-SNL on the ipsilateral side. MPAEs were measured 20 min after subcutaneous injection of 1.5 mg/kg morphine on the mornings of days 7, 9, and 11 post-SNL. n = 5 rats/group. *P < 0.05, **P < 0.01 vs the corresponding MPAE on day 7 post-SNL. #P < 0.05, ##P < 0.01 vs the PBS plus SNL group at the corresponding time points. F. Dose-response curve on the ipsilateral side on day 11 post-SNL after subcutaneous injection of 5 mg/kg morphine twice daily for 4 days starting at day 7 post-SNL from the PBS-, ShRNA- and Scr-injected rats. n = 5 rats/group.
Furthermore, we examined whether DNMT3a knockdown in the injured DRG improved morphine analgesia and alleviated morphine-induced tolerance under neuropathic pain conditions. As expected, when the first dose of morphine was administered s.c. on day 7 after SNL, morphine MPAE was much greater in the AAV5-Dnmt3a shRNA-treated group than that in the PBS- or AAV5-scrambled shRNA-treated group on the ipsilateral side (Fig. 2E). Repeated s.c. morphine administration twice daily for 5 days starting from day 7 after SNL also led to a time-dependent reduction in morphine MPAE in the PBS- or AAV5-scrambled shRNA-treated groups (Fig. 2E). However, this reduction was significantly prohibited in the AAV5-Dnmt3a shRNA-treated group (Fig. 2E). This effect of AAV5-Dnmt3a shRNA was also confirmed by a significant leftward shift in the cumulative dose-response curve of morphine in the AAV5-Dnmt3a shRNA-treated group compared to the PBS- or AAV5-scrambled shRNA-treated group on the last day after repeated s.c. administration of morphine (Fig. 2F). The EC50 values of morphine were 3.50 mg/kg in the AAV5-Dnmt3a shRNA-treated group, 7.02 mg/kg in the PBS-treated group, and 7.32 mg/kg in the AAV5-scrambled shRNA-treated group (Fig. 2F). It is evident that blocking increased DNMT3a improves morphine analgesia and prevents morphine analgesic tolerance development under neuropathic pain conditions.
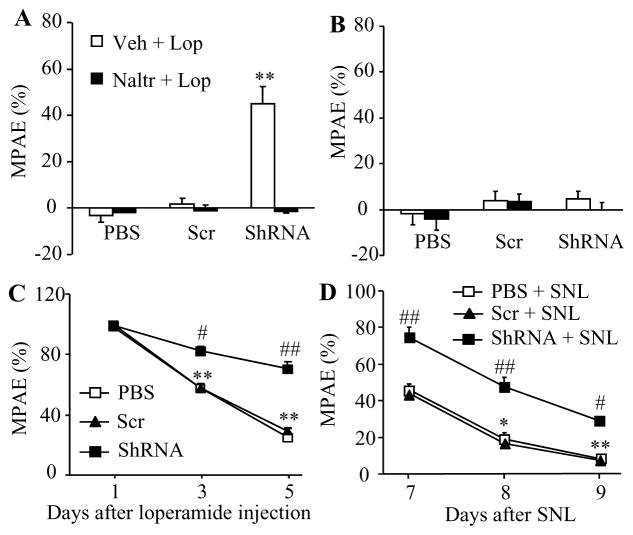
Analgesic effects caused by subcutaneous injection of morphine are mediated by MOR located in both peripheral and central nervous systems. To further demonstrate the role of DRG DNMT3a in peripheral MOR-mediated analgesia in naïve and SNL animals, we subcutaneously injected loperamide, which is a peripheral acting MOR preferring agonist that does not penetrate the blood-brain barrier [7]. Like morphine, loperamide produced similar behavioral responses seen in naïve and SNL rats from the PBS- and virus-treated groups (Fig. 3A–3D).
Figure 3.
DRG DNMT3a knockdown enhanced loperamide analgesia and alleviated loperamide tolerance in naïve and SNL rats. All of the following experiments were carried out 5 weeks after microinjection of AAV5-Dnmt3a shRNA (ShRNA), AAV5-scambled shRNA (Scr), or PBS into unilateral L5 DRG. A and B. Loperamide maximal possible analgesic effects (MPAE) measured 20 min after subcutaneous co-injection of 3 mg/kg loperamide (Lop) plus vehicle (Veh) or 5 mg/kg naltrexone (Naltr) on the ipsilateral (A) and contralateral (B) sides. n = 5 rats/group. **P < 0.01 vs the corresponding PBS group. C. Loperamide analgesic tolerance developed by subcutaneous injection of 10 mg/kg loperamide twice daily for 5 days on the ipsilateral side of naïve rats. MPAEs were measured 20 min after loperamide injection on the mornings of days 1, 3, and 5 post-injection. n = 5 rats/group. **P < 0.01 vs the corresponding MPAE on day 1 post-injection. #P < 0.05, ##P < 0.01 vs the PBS-treated group at the corresponding time points. D. Loperamide analgesic tolerance developed by subcutaneous injection of 0.75 mg/kg loperamide twice daily for 3 days starting on day 7 post-SNL on the ipsilateral side. n = 5 rats/group. *P < 0.05, **P < 0.01 vs the corresponding MPAE on day 7 post-SNL. #P < 0.05, ##P < 0.01 vs the PBS plus SNL group at the corresponding time points.
Effect of increased DNMT3a in the DRG on MOR-gated primary afferent neurotransmitter release
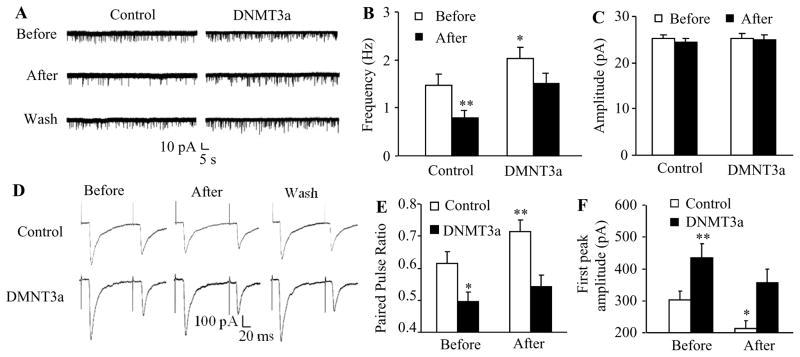
MOR expressed in small primary afferents controls neurotransmitter release from their central terminals [8;37]. MOR downregulation in the injured DRG augments the release of primary afferent neurotransmitters in neuropathic pain [34;36]. We further tested whether mimicking the nerve injury-induced increase in DRG DNMT3a via microinjection of AAV5-Dnmt3a into the unilateral L4/5 DRGs affected primary afferent neurotransmitter release. To this end, we first measured the changes in miniature excitatory postsynaptic current (mEPSC) frequencies, which reflected the alteration in presynaptic neurotransmitter release [44;47], recorded in lamina II neurons from L4/5 spinal cord slices 5 weeks post-viral injection. mEPSC frequency in the AAV5-Dnmt3a-treated group was significantly higher than that in the control AAV5-GFP-treated group (Fig. 4A and 4B). To verify whether this alteration was attributed to the reduction of MOR function in primary afferents, we applied the selective MOR agonist DAMGO. DAMGO (1 μM) led to a greater decrease in mEPSC frequency in the control group (46.93 ± 4.38%) than that in the AAV5- Dnmt3a-treated group (26.23 ± 3.79%) (Fig. 3D). This effect was abolished after DAMGO wash-out (Fig. 4B) or by the addition, prior to DAMGO treatment, of the selective MOR antagonist CTOP (data not shown). mEPSC amplitude was not altered in the two groups with or without DAMGO application (Fig. 4C). The evidence suggests that increased DNMT3a expression in DRG results in an increase in MOR-gated presynaptic neurotransmitter release.
Figure 4.
Overexpression of DNMT3a in DRG promotes neurotransmitter release from primary afferents. All of the following experiments were carried out 5 weeks after microinjection of AAV5-Dnmt3a (DNMT3a), or AAV5-GFP (Control) into unilateral L4/5 DRGs. A. Example traces of miniature EPSC (mEPSC) recorded in lamina II dorsal horn neurons before and after DAMGO application and its wash out. B and C. Effects of DAMGO (1 μM) on the frequency (B) and amplitude (C) of mEPSC in lamina II neurons from the AAV5-Dnmt3a-treated group (n = 19 neurons, 9 rats) and control group (n = 26 neurons, 12 rats). *P < 0.05, **P < 0.01 vs the control group before DAMGO application. D. Example traces of C-fiber input-evoked EPSC in lamina II neurons before and after DAMGO application and its wash out. E and F. Effects of DAMGO (1 μM) on the pair pulse ratio (E) and first peak amplitude of EPSC (F) in lamina II neurons from the AAV5-Dnmt3a-treated group (n = 16 neurons, 19 rats) and control group (n = 23 neurons, 20 rats). *P < 0.05, **P < 0.01 vs the control group before DAMGO application.
The mEPSCs recorded above may reflect overall excitatory inputs from both primary afferents and interneurons to the recorded neurons [44;47]. To further examine the role of DNMT3a in the release of primary afferent neurotransmitters, we recorded the EPSCs evoked by stimulation of the dorsal root (C-fiber: 500 μA intensity and 1 Hz) with a paired-pulse protocol. The pair-pulse ratio (PPR), which reflects the probability of primary afferent neurotransmitter release [44;47], was determined by dividing the second peak amplitude by the first one. PPR significantly declined in the AAV5-Dnmt3a-treated group compared to the control group (Figs. 4D and 4E). Additionally, the first peak amplitude of the EPSC in the AAV5-Dnmt3a-treated group was higher than that in the control group (Figs. 4D and 4F). DAMGO application markedly increased the PPR by 15.85% and decreased the first peak amplitude by 29.56% in the control group, but only 8.85% and 17.78%, respectively, in the AAV5-Dnmt3a-treated group (Figs. 4D and 4F). DAMGO effects could be abolished after its wash out (Fig. 4F) or by prior treatment with CTOP (data not shown). These results suggest that increased DNMT3a in DRG promotes neurotransmitter release from primary afferent terminals possibly through presynaptic MOR downregulation in DRG.
DNMT3a binds to the Oprm1 gene and is required for SNL-induced DNA methylation within this gene
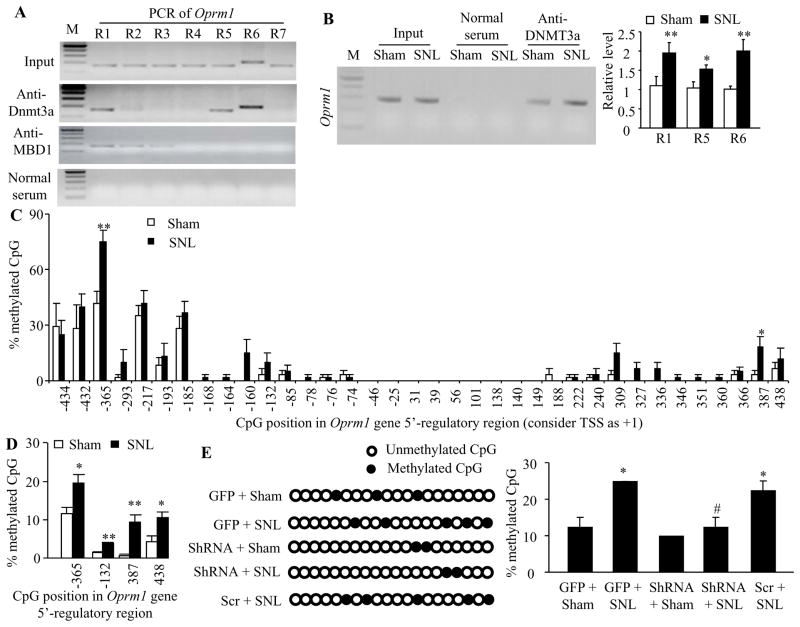
We further examined whether SNL altered the DNA methylation level of the Oprm1 gene and whether DNMT3a was involved in this alternation in the injured DRG. We first used chromatin immunoprecipitation (ChIP) assays and identified that DNMT3a bond to three regions (−450/−288 bp, +142/+312 bp, and +238/+439 bp) of the Oprm1 gene as demonstrated by the amplification of only these three regions (out of 7 regions from −450 to +558 bp) from the complexes immunoprecipitated with DNMT3a antibody in nuclear fractions from sham DRG (Fig. 5A). The binding activities in these three regions in the injured DRG on day 7 after SNL increased by 1.8-fold, 1.5-fold, and 2-fold, respectively, compared to those after sham surgery (Fig. 5B). Furthermore, these increased binding activities affected the DNA methylation pattern and level in the promoter and 5′end untranslated regions of the Oprm1 gene after SNL. The bisulfite clone-sequencing assay showed increases in DNA methylation levels at −365 and +387 CpG sites (Fig. 5C), which are located within the DNMT3a binding regions described above. The bisulfite pyro-sequencing assay further confirmed the increases in DNA methylation in these two CpG sites, although the increases in DNA methylation were detected at additional −132 and +438 CpG sites (Fig. 5D). More importantly, this increase in DNA methylation may be dependent on DNMT3a expression, as DRG DNMT3a knockdown abolished the SNL-induced increase in the methylation at the −365 CpG site (Fig. 5E).
Figure 5.
DNMT3a is required for the nerve injury-induced increase in Oprm1 DNA methylation in the injured DRG. A. Rabbit anti-DNMT3a immunoprecipitated region 1 (R1, −450/−288), region 5 (R5, +142/+312) and region 6 (R6, +238/+439), but not the remaining 4 regions (R2, −310/−143; R3, −164/−6; R4, −7/+166; R7: +391/+558), within the Oprm1 gene and the rabbit anti-MBD1 immunoprecipitated regions 1–3, but not regions 4–7, within the Oprm1 gene in rat DRGs. Input, total purified fragments. M, ladder marker. n = 3 repeats. B. The binding activity of DNMT3a to regions 1 (R1), 5 (R5) and 6 (R6) within the Oprm1 gene in the injured DRGs of rats on day 7 post-SNL or sham surgery. Left: representative binding between DNMT3a and R1. Right: statistical summary of the quantitative RT-PCR analysis in the binding activity. n = 9 rats/group. *P < 0.05, **P < 0.01 vs the corresponding sham group. C and D. DNA methylation levels at CpG sites of the Oprm1 gene in the injured DRG on day 7 post-SNL or sham surgery detected by bisulfite clone-sequencing assay (C) and bisulfite pyro-sequencing assay (D). n = 6 rats/group. *P < 0.05, **P < 0.01 vs the corresponding sham group. E. The percentage of methylation at the −365 CpG site of the Oprm1 gene in the injured DRG on day 7 post-SNL or sham surgery from the AAV5-GFP (GFP) plus sham group, the AAV5-GFP plus SNL group, AAV5-scrambled shRNA (Scr) plus SNL group, AAV5-Dnmt3a shRNA (ShRNA) plus SNL group, and AAV5-Dnmt3a ShRNA plus sham group. n = 6 rats/group. *P < 0.05 vs the AAV5-GFP plus sham group. #P < 0.05 vs the GFP plus SNL group.
MBD1 participates in DNMT3a regulation of MOR expression in DRG neurons
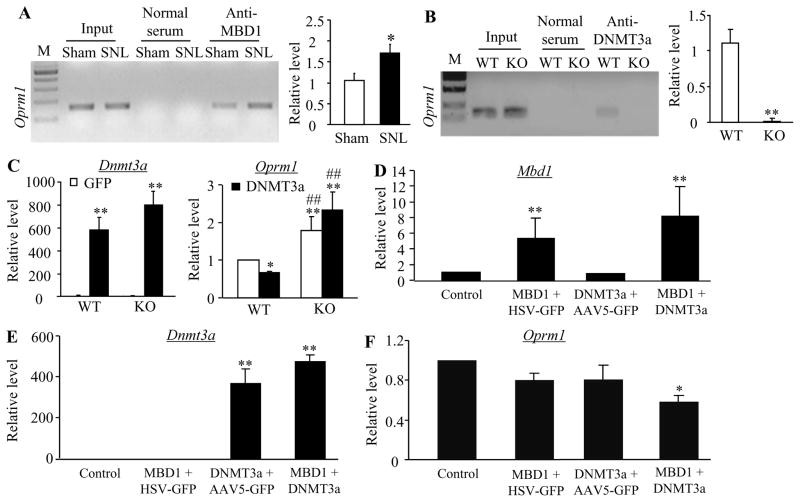
DNA methylation-gated gene expression also requires the participation of the family of Methyl-CpG-binding domain (MBD) proteins including MBD1-3 and MeCP2 [20;22]. Except for MBD3, the remaining proteins function as methylation-dependent transcriptional repressors [24–26]. Here, we demonstrated that MBD1 bond to three regions (−450/−288 bp, −310/−143 bp, and −164/−6 bp) (out of 7 regions from −450 to +558 bp) of the Oprm1 gene in sham DRG (Fig. 5A). MBD1 and DNMT3a have an overlapping binding region (−450/−288) within the Oprm1 gene. Moreover, MBD1 binding activity at this overlapping region in the injured DRG on day 7 after SNL increased by 1.64-fold compared to that after sham surgery (Fig. 6A). This suggests that MBD1 is involved in DNMT3a regulation of Oprm1 gene expression in DRG neurons. To address this conclusion, we used MBD1 knockout (KO) mice (Supplementary Fig. 4) and revealed the requirement of MBD1 for DNMT3a binding to the Oprm1 gene in DRG as demonstrated by almost failure to the amplification of the Oprm1 gene from the complexes immunoprecipitated with DNMT3a antibody in nuclear fractions from MBD1 KO DRG, compared to that from the WT DRG (Fig. 6B). We then over-expressed DNMT3a in the DRG neurons by transducing HSV-Dnmt3a into the cultured DRG neurons from the WT and KO mice (Fig. 6C). HSV-GFP was used as a control. As expected, the level of Oprm1 mRNA in the HSV-Dnmt3a-treated group significantly decreased compared to the AAV5-GFP-treated group from the WT mice (Fig. 6C). In contrast, the amount of Oprm1 mRNA was highly elevated in both viral treatment groups from the KO mice (Fig. 6C). Finally, we co-expressed DNMT3a and MBD1 in the DRG neurons by co-transduction of HSV-Dnmt3a and AAV5-Mbd1 (that expresses full-length MBD1) into the cultured DRG neurons from the WT mice (Fig. 6D and 6E). This co-transduction markedly reduced the Oprm1 mRNA expression although single transduction of each gene did not alter the level of the Oprm1 mRNA (Fig. 6F). These in vitro findings strongly support the involvement of MBD1 in DNMT3a regulation of the Oprm1 gene.
Figure 6.
MBD1 is required for DNMT3a repression of MOR expression. A. The binding activity of MDB1 to region 1 (R1) within the Oprm1 gene in the injured DRGs of rats on day 7 post-SNL or sham surgery. Left: representative binding between MBD1 and R1. Right: statistical summary of the quantitative RT-PCR analysis in the binding activity. Input, total purified fragments. M, ladder marker. n = 9 rats/group. *P < 0.05 vs the corresponding sham group. B. The binding activity of DNMT3a to region 1 (R1) within the Oprm1 gene in the DRGs from wild type (WT) and Mbd1 knockout (KO) mice. Left: representative bindings between DNMT3a and R1. Right: statistical summary of the quantitative RT-PCR analysis in the binding activity. Input, total purified fragments. M, ladder marker. n = 12 mice/group. *P < 0.05 vs the corresponding WT mice. C. Expression of Dnmt3a (left) and Oprm1 (right) mRNAs in DRG cultured neurons from WT and Mbd1 KO mice transfected with HSV-GFP (GFP) or HSV-Dnmt3a (DNMT3a). *P < 0.05, **P < 0.01 vs the corresponding HSV-GFP-transfected neurons from WT mice. ##P < 0.01 vs the corresponding HSV-Dnmt3a-transfected neurons from WT mice. D–F. Expression of Mbd1 (D), Dnmt3a (E), and Oprm1 (F) mRNAs in WT DRG cultured neurons co-transfected with HSV-GFP plus AAV5-GFP (control), AAV5-Mbd1 (MBD1) plus AAV5-GFP, HSV-Dnmt3a (DNMT3a) plus AAV5-GFP, or AAV5-Mbd1 plus HSV-Dnmt3a. *P < 0.05, **P < 0.01 vs the control group.
Discussion
The present study demonstrated that nerve injury-induced increase in DRG DNMT3a is responsible for epigenetic silencing of DRG Oprm1 and Oprk1 genes and opioid analgesic reduction under neuropathic pain conditions. Mechanistically, the increased DNMT3a repressed MOR expression by promoting the MBD1-mediated DNA methylation in the promoter and 5′-untranslated region of the Oprm1 gene. These findings suggest that inhibition of DRG DNMT3a may improve opioid analgesic effects in treating neuropathic pain.
The Dnmt3a gene could be transcriptionally activated in the injured DRG neurons following peripheral nerve injury. The amounts of Dnmt3a mRNA and its coding DNMT3a protein significantly and time-dependently increased through the activation of the transcription factor octamer transcription factor 1 in the injured DRG after SNL or chronic construction injury (CCI) [48]. Given that DNMT3a is a gene repressor [16;31] and expressed exclusively in DRG neurons [48], the increased DNMT3a likely participates in nerve injury-induced downregulation in gene expression in the injured DRG neurons. It should be pointed out that DNMT3a has two isoforms, the long DNMT3a1 and the shorter DNMT3a2, which differ in that a 219-amino-acid N-terminal tail is present only in DNMT3a1[4;43]. The DNMT3a antibody used in the present study was generated against DNMT3a amino acids 15–126 and thus actually detected the expressional increase of DNMT3a1 isoform, although our quantitative RT-PCR assay displayed the expressional elevations of both DNMT3a1 and DNMT3a2 mRNAs in the injured DRG. Whether peripheral nerve injury alters DRG DNMT3a2 isoform expression remains to be determined after the primary antibody specifically and selectively against DNMT3a2 is available commercially.
We demonstrated that DNMT3a is essential for epigenetic silencing of Oprm1 and Oprk1 genes in the injured DRG under neuropathic pain conditions. A previous study showed that CCI-induced MOR downregulation might be attributed to increases in the levels of DNA methylation within the Oprm1 gene at −388, −344, and −255 CpG sites in the injured mouse DRG [51], because blocking these increases via intrathecal 5-aza-dC (a non-specific DNMT inhibitor) rescued the expression of Oprm1 mRNA and its coding protein in the injured DRG and restored morphine analgesic effects following CCI [51]. However, how nerve injury causes an increase in Oprm1 gene methylation is unknown. Moreover, which members of the DNMT family are responsible for this increase are unclear. The present study demonstrated that DNMT3a bound to three regions (−450/−288, +142/+312, and +238/+439) within the Oprm1 gene in rat DRG. The binding activities in these three regions were increased in the injured DRG following SNL. Furthermore, the DNMT3a-triggered increases in DNA methylation were identified at −365 and +387 CpG sites within the Oprm1 gene in the injured rat DRG. These two methylated sites were different from ones reported previously [51]. The reasons in the difference between the present and previous studies are unknown, but may be related to the use of distinct neuropathic pain models (SNL vs CCI) and animal species (rats vs mice). In additional to Oprm1 mRNA, DNMT3a is involved in nerve injury-induced downregulation of Oprk1 mRNA and its coding KOR, but not Oprd1 mRNA, in the injured DRG. How DNMT3a specifically and selectively targets Oprm1 and Oprk1 genes is still elusive, but DNMT3a participation in nerve injury-induced DRG Oprm1 silencing may require MBD1. MBD1 as a transcriptional repressor binds to more efficiently to methylated DNA with a specific sequence context [5]. Selective and specific targeting of DNMT3a on the Oprm1 and Oprk1 genes is likely associated with the characteristics of MBD1 sequence-specific binding to the genes. It is worth noted that DNMT3a may participate in nerve injury-induced silencing of other genes (such as Kcna2) in the injured DRG [48].
The mechanisms by which nerve injury causes a reduction in systemic, spinal, and peripheral opioid analgesia are complicated and may involve DRG opioid receptor downregulation, descending spinal facilitation, enhanced excitatory neurotransmitter release in primary afferents, glutamate receptor and glial cell activation, and cytokine release in spinal cord and brain regions under neuropathic pain conditions [6;10;13;28–30;32;40;42;51]. The data from the present study demonstrated that rescuing MOR downregulation by blocking increased DNMT3a in the injured DRG significantly restored analgesia to systemically administered morphine or loperamide and attenuated the development of their analgesic tolerance following SNL. This suggests that the nerve injury-induced reduction in opioid analgesia could be due at least in part to DNMT3a-triggered DRG MOR silencing. The previous studies revealed that other epigenetic modifications might also regulate MOR-gated analgesic actions under neuropathic pain conditions. Peripheral nerve injury increased the enrichment of the G9a-catalyzed histone 3 at lysine 9 dimethylation in the promoter of the Oprm1 gene and decreased neuron-restrictive silencer factor-mediated histone 3/4 acetylation within the Oprm1 gene, both of which result in DRG Oprm1 gene silencing and morphine analgesic reduction [17;40;51]. It is evident that nerve injury-induced downregulation of DRG MOR and the reduction of morphine analgesia may be controlled by multiple epigenetic mechanisms.
DNMT3a-triggered silencing of the Oprm1 gene in the injured DRG is implicated in the mechanism of neuropathic pain development. MOR downregulation in the injured DRG augments the release of primary afferent neurotransmitters following peripheral nerve injury [34;36]. MOR knockout mice displayed increased mechanical pain hypersensitivity under neuropathic pain conditions [21]. These findings imply that nerve injury-induced MOR downregulation in the injured DRG might participate in the mechanism of neuropathic pain through a loss of tonic MOR-mediated inhibition at central terminals of the primary afferents. Present study showed that the increased DRG DNMT3a augmented MOR-gated neurotransmitter release from the primary afferents. Our recent work demonstrated that blocking increased DNMT3a in the injured DRG attenuated the development of nerve injury-induced pain hypersensitivity [48]. The evidence suggests that DRG DNMT3a participates in neuropathic pain development at least in part through the mechanism of its triggered DRG MOR downregulation.
In summary, we demonstrated a DNMT3a-triggered epigenetic mechanism of how MOR is downregulated in the injured DRG under neuropathic pain conditions. We also reported that blocking increased DRG DNMT3a rescues morphine analgesia and prevents morphine-induced analgesic tolerance following peripheral nerve injury. DNMT3a is likely a novel target for adjunctive use with opioids in treating neuropathic pain.
Supplementary Material
Acknowledgments
We thank Han-Rong Weng (University of Georgia College of Pharmacy) for electrophysiological data analysis. This work was supported by NIH grants (NS094664, NS094224, DA033390, NS 072206, and HL117684).
Footnotes
AUTHOR CONTRIBUTIONS
Y.X.T. conceived the project and supervised all experiments. L.S., J.Y.Z., X.G., and Y.X.T. designed the project. L.S., J.Y.Z., L.L., S.W., K.M., and J.Z. performed molecular, biochemical, and behavioral experiments. X.G. performed patch clamp recordings. J.F. and E.J.N. prepared for HSV virus and generated DNMT3afl/fl mice. W.G. and X.Z. generated MBD knockout mice. L.S., J.Y.Z., X.G., S.W., A.B., and Y.X.T. analyzed the data. L.S. and Y.X.T. wrote the manuscript. All of the authors read and discussed the manuscript.
CONFLICT OF INTEREST
The authors declared no conflicts of interest.
Reference List
- 1.Anesti AM, Coffin RS. Delivery of RNA interference triggers to sensory neurons in vivo using herpes simplex virus. Expert Opin Biol Ther. 2010;10:89–103. doi: 10.1517/14712590903379486. [DOI] [PubMed] [Google Scholar]
- 2.Ataka T, Kumamoto E, Shimoji K, Yoshimura M. Baclofen inhibits more effectively C-afferent than Adelta-afferent glutamatergic transmission in substantia gelatinosa neurons of adult rat spinal cord slices. Pain. 2000;86:273–282. doi: 10.1016/S0304-3959(00)00255-4. [DOI] [PubMed] [Google Scholar]
- 3.Bekhit MH. Opioid-induced hyperalgesia and tolerance. Am J Ther. 2010;17:498–510. doi: 10.1097/MJT.0b013e3181ed83a0. [DOI] [PubMed] [Google Scholar]
- 4.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 6.Clouaire T, de Las Heras JI, Merusi C, Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res. 2010;38:4620–4634. doi: 10.1093/nar/gkq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao YJ, Ji RR. Chemokines, neuronal-glial interactions, and central processing of neuropathic pain. Pharmacol Ther. 2010;126:56–68. doi: 10.1016/j.pharmthera.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Johanek LM, Hartke TV, Shim B, Tao YX, Ringkamp M, Meyer RA, Raja SN. Peripherally acting mu-opioid receptor agonist attenuates neuropathic pain in rats after L5 spinal nerve injury. Pain. 2008;138:318–329. doi: 10.1016/j.pain.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinke B, Gingl E, Sandkuhler J. Multiple targets of mu-opioid receptor-mediated presynaptic inhibition at primary afferent Adelta- and C-fibers. J Neurosci. 2011;31:1313–1322. doi: 10.1523/JNEUROSCI.4060-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikoma M, Kohno T, Baba H. Differential presynaptic effects of opioid agonists on Adelta- and C-afferent glutamatergic transmission to the spinal dorsal horn. Anesthesiology. 2007;107:807–812. doi: 10.1097/01.anes.0000286985.80301.5e. [DOI] [PubMed] [Google Scholar]
- 11.Kim YS, Chu Y, Han L, Li M, Li Z, Lavinka PC, Sun S, Tang Z, Park K, Caterina MJ, Ren K, Dubner R, Wei F, Dong X. Central terminal sensitization of TRPV1 by descending serotonergic facilitation modulates chronic pain. Neuron. 2014;81:873–887. doi: 10.1016/j.neuron.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohno T, Ji RR, Ito N, Allchorne AJ, Befort K, Karchewski LA, Woolf CJ. Peripheral axonal injury results in reduced mu opioid receptor pre- and post-synaptic action in the spinal cord. Pain. 2005;117:77–87. doi: 10.1016/j.pain.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 13.LaPlant Q, Vialou V, Covington HE, III, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain. 2009;10:895–926. doi: 10.1016/j.jpain.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CY, Perez FM, Wang W, Guan X, Zhao X, Fisher JL, Guan Y, Sweitzer SM, Raja SN, Tao YX. Dynamic temporal and spatial regulation of mu opioid receptor expression in primary afferent neurons following spinal nerve injury. Eur J Pain. 2011;15:669–675. doi: 10.1016/j.ejpain.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Gu X, Sun L, Wu S, Liang L, Cao J, Lutz BM, Bekker A, Zhang W, Tao YX. Dorsal root ganglion myeloid zinc finger protein 1 contributes to neuropathic pain after peripheral nerve trauma. Pain. 2015;156:711–721. doi: 10.1097/j.pain.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L, Lutz BM, Bekker A, Tao YX. Epigenetic regulation of chronic pain. Epigenomics. 2015;7:235–245. doi: 10.2217/epi.14.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang L, Zhao JY, Gu X, Wu S, Mo K, Xiong M, Bekker A, Tao YX. G9a inhibits CREB-triggered expression of mu opioid receptor in primary sensory neurons following peripheral nerve injury. Mol Pain. 2016 doi: 10.1177/1744806916682242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liaw WJ, Zhang B, Tao F, Yaster M, Johns RA, Tao YX. Knockdown of spinal cord postsynaptic density protein-95 prevents the development of morphine tolerance in rats. Neuroscience. 2004;123:11–15. doi: 10.1016/j.neuroscience.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Liaw WJ, Zhu XG, Yaster M, Johns RA, Gauda EB, Tao YX. Distinct expression of synaptic NR2A and NR2B in the central nervous system and impaired morphine tolerance and physical dependence in mice deficient in postsynaptic density-93 protein. Mol Pain. 2008;4:45. doi: 10.1186/1744-8069-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL. Epigenetic Mechanisms: Critical Contributors to Long-Term Memory Formation. Neuroscientist. 2011 doi: 10.1177/1073858411386967. [DOI] [PubMed] [Google Scholar]
- 22.Mansikka H, Zhao C, Sheth RN, Sora I, Uhl G, Raja SN. Nerve injury induces a tonic bilateral mu-opioid receptor-mediated inhibitory effect on mechanical allodynia in mice. Anesthesiology. 2004;100:912–921. doi: 10.1097/00000542-200404000-00022. [DOI] [PubMed] [Google Scholar]
- 23.Mifsud KR, Gutierrez-Mecinas M, Trollope AF, Collins A, Saunderson EA, Reul JM. Epigenetic mechanisms in stress and adaptation. Brain Behav Immun. 2011;25:1305–1315. doi: 10.1016/j.bbi.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Nakatsuka T, Park JS, Kumamoto E, Tamaki T, Yoshimura M. Plastic changes in sensory inputs to rat substantia gelatinosa neurons following peripheral inflammation. Pain. 1999;82:39–47. doi: 10.1016/S0304-3959(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 25.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 26.Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 28.O’Connor AB. Neuropathic pain: quality-of-life impact, costs and cost effectiveness of therapy. Pharmacoeconomics. 2009;27:95–112. doi: 10.2165/00019053-200927020-00002. [DOI] [PubMed] [Google Scholar]
- 29.Obara I, Makuch W, Spetea M, Schutz J, Schmidhammer H, Przewlocki R, Przewlocka B. Local peripheral antinociceptive effects of 14-O-methyloxymorphone derivatives in inflammatory and neuropathic pain in the rat. Eur J Pharmacol. 2007;558:60–67. doi: 10.1016/j.ejphar.2006.11.037. [DOI] [PubMed] [Google Scholar]
- 30.Obara I, Parkitna JR, Korostynski M, Makuch W, Kaminska D, Przewlocka B, Przewlocki R. Local peripheral opioid effects and expression of opioid genes in the spinal cord and dorsal root ganglia in neuropathic and inflammatory pain. Pain. 2009;141:283–291. doi: 10.1016/j.pain.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Obara I, Przewlocki R, Przewlocka B. Local peripheral effects of mu-opioid receptor agonists in neuropathic pain in rats. Neurosci Lett. 2004;360:85–89. doi: 10.1016/j.neulet.2004.01.056. [DOI] [PubMed] [Google Scholar]
- 32.Poetsch AR, Plass C. Transcriptional regulation by DNA methylation. Cancer Treat Rev. 2011;37(Suppl 1):S8–12. doi: 10.1016/j.ctrv.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Rashid MH, Inoue M, Toda K, Ueda H. Loss of peripheral morphine analgesia contributes to the reduced effectiveness of systemic morphine in neuropathic pain. J Pharmacol Exp Ther. 2004;309:380–387. doi: 10.1124/jpet.103.060582. [DOI] [PubMed] [Google Scholar]
- 34.Rigaud M, Gemes G, Barabas ME, Chernoff DI, Abram SE, Stucky CL, Hogan QH. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain. 2008;136:188–201. doi: 10.1016/j.pain.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehgal N, Smith HS, Manchikanti L. Peripherally acting opioids and clinical implications for pain control. Pain Physician. 2011;14:249–258. [PubMed] [Google Scholar]
- 36.Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006;53:245–256. [PubMed] [Google Scholar]
- 37.Stein C, Lang LJ. Peripheral mechanisms of opioid analgesia. Curr Opin Pharmacol. 2009;9:3–8. doi: 10.1016/j.coph.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Taddese A, Nah SY, McCleskey EW. Selective opioid inhibition of small nociceptive neurons. Science. 1995;270:1366–1369. doi: 10.1126/science.270.5240.1366. [DOI] [PubMed] [Google Scholar]
- 39.Tao YX, Rumbaugh G, Wang GD, Petralia RS, Zhao C, Kauer FW, Tao F, Zhuo M, Wenthold RJ, Raja SN, Huganir RL, Bredt DS, Johns RA. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J Neurosci. 2003;23:6703–6712. doi: 10.1523/JNEUROSCI.23-17-06703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torsney C, MacDermott AB. Disinhibition opens the gate to pathological pain signaling in superficial neurokinin 1 receptor-expressing neurons in rat spinal cord. J Neurosci. 2006;26:1833–1843. doi: 10.1523/JNEUROSCI.4584-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchida H, Ma L, Ueda H. Epigenetic gene silencing underlies C-fiber dysfunctions in neuropathic pain. J Neurosci. 2010;30:4806–4814. doi: 10.1523/JNEUROSCI.5541-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van HO, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155:654–662. doi: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, King T, De FM, Guo W, Ossipov MH, Porreca F. Descending facilitation maintains long-term spontaneous neuropathic pain. J Pain. 2013;14:845–853. doi: 10.1016/j.jpain.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene. 2002;298:91–99. doi: 10.1016/s0378-1119(02)00976-9. [DOI] [PubMed] [Google Scholar]
- 45.Weng HR, Chen JH, Pan ZZ, Nie H. Glial glutamate transporter 1 regulates the spatial and temporal coding of glutamatergic synaptic transmission in spinal lamina II neurons. Neuroscience. 2007;149:898–907. doi: 10.1016/j.neuroscience.2007.07.063. [DOI] [PubMed] [Google Scholar]
- 46.Xu JT, Sun L, Lutz BM, Bekker A, Tao YX. Intrathecal rapamycin attenuates morphine-induced analgesic tolerance and hyperalgesia in rats with neuropathic pain. Transl Perioper Pain Med. 2015;2:27–34. [PMC free article] [PubMed] [Google Scholar]
- 47.Xu JT, Zhou X, Zhao X, Ligons D, Tiwari V, Lee CY, Atianjoh FE, Liang L, Zang W, Njoku D, Raja SN, Yaster M, Tao YX. Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J Clin Invest. 2014;124:592–603. doi: 10.1172/JCI70236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan X, Weng HR. Endogenous interleukin-1beta in neuropathic rats enhances glutamate release from the primary afferents in the spinal dorsal horn through coupling with presynaptic N-methyl-D-aspartic acid receptors. J Biol Chem. 2013;288:30544–30557. doi: 10.1074/jbc.M113.495465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao JY, Liang L, Gu X, Li Z, Wu S, Sun L, Atianjoh FE, Feng J, Mo K, Jia S, Lutz BM, Bekker A, Nestler EJ, Tao YX. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun. 2016 doi: 10.1038/ncomms14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, Gao YJ, Hoffman PN, Cui H, Li M, Dong X, Tao YX. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou XL, Yu LN, Wang Y, Tang LH, Peng YN, Cao JL, Yan M. Increased methylation of the MOR gene proximal promoter in primary sensory neurons plays a crucial role in the decreased analgesic effect of opioids in neuropathic pain. Mol Pain. 2014;10:51. doi: 10.1186/1744-8069-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.