Summary
Burkitt lymphoma (BL) is the most common histological subtype of non-Hodgkin lymphoma (NHL) in children and adolescents. Through the introduction of short intensive multi-agent chemoimmunotherapy, survival has improved significantly over the past 30 years. However, this successful approach is limited by significant chemotherapy-induced acute toxicity and risk of developing resistant disease, demonstrating the need to identify less toxic and targeted therapies. We analysed the comparative genomic signature and targetable signalling pathways in paediatric BL (PEBL) samples from the Children’s Oncology Group study (ANHL01P1) by genomic profiling and selected genes were confirmed by quantitative real time polymerase chain reaction. These results were compared to PEBL samples from public databases and utilised the Gene Expression Omnibus (GEO) Series (GSE) 10172 and 4475 (n=16), and 4732 (n=15). Three hundred and seventy-six genes (approximately 25%) were similarly expressed among three PEBL sample groups. Several target genes in Toll-like receptor signalling, JAK-STAT signalling and MAPK signalling were significantly overexpressed in PEBL. In addition, several tyrosine kinases, including Bruton tyrosine kinase (BTK), protein tyrosine phosphatase (PTP) and histone deacetylase inhibitor (HDACi) were highly expressed in PEBL. These pre-clinical results suggest that specific signal transduction pathways are overly expressed in PEBL and several pathways could serve as potential future therapeutic targets.
Keywords: Gene expression profiles, Paediatric Burkitt lymphoma, Signal transduction pathways
Introduction
Paediatric Burkitt lymphoma (PEBL) is an aggressive mature B-cell malignancy; it is the most common (about 40%) non-Hodgkin lymphoma (NHL) in children and adolescents in western countries. The prognosis of PEBL has steadily improved over the past 30 years through the introduction of short and intensive multi-agent chemotherapeutic regimens (Cairo, et al 2007, Cairo, et al 2012, Gerrard, et al 2008, Miles, et al 2012, Patte, et al 2007). The Children’s Oncology Group (COG), together with the United Kingdom Children’s Cancer Group (UKCCSG) and the French Paediatric Oncology Group (SFOP), conducted a prospective international study in PEBL (French-American-British [FAB] 96) that demonstrated 90% 5-year overall survival (OS) including patients with stage III/IV disease and those with bone marrow (BM) and central nervous system (CNS) disease (Cairo, et al 2007, Cairo, et al 2012, Gerrard, et al 2008, Patte, et al 2007). More recently, we have demonstrated the safety and efficacy of adding rituximab to the chemotherapy backbone (FAB 96) in children and adolescents with advanced mature Burkitt lymphoma (BL) (COG ANHL01P1) (Goldman, et al 2013, Goldman, et al 2014). However, this success has come at the cost of the significant chemotherapy-induced acute toxicity secondary to intensive chemotherapy, requiring the need to identify less toxic but targeted therapy. Furthermore, patients with PEBL who relapse or progress have chemotherapy-resistant disease and can rarely be salvaged (Cairo, et al 2007, Cairo, et al 2012, Miles, et al 2012). These results suggest the urgent need to identify alternative treatment approaches focusing on targeted therapy to circumvent chemotherapy-resistant disease by providing alternative therapeutic targets.
The rearrangement of proto-oncogene MYC on 8q24 is the hallmark cytogenetic feature of BL (Dave, et al 2006, Nelson, et al 2010, Poirel, et al 2009). The activation of MYC has been implicated in the promotion of aberrant cell proliferation (Shaffer, et al 2002). A set of MYC target genes were identified in both paediatric and adult BL samples by comparing all subtypes of diffuse large B-cell lymphoma (DLBCL) with samples of BL (Dave, et al 2006, Deffenbacher, et al 2012, Giulino-Roth, et al 2012, Hummel, et al 2006, Klapper, et al 2008). These results suggested that BL, in part, uses different oncogenic signalling pathway(s) compared to other B-cell lymphomas, including DLBCL. Unlike many other haematological malignancies, the nuclear factor (NF)-kB pathway was not constitutively activated in BL (Dave, et al 2006) and recently, the phosphatidylinositol 3-kinase (PI3K) pathway has been suggested as a key signalling pathway in BL lymphomagenesis (Sander, et al 2012). However, it is still unclear whether diverse signalling pathway(s) contribute to paediatric Burkitt lymphomagenesis. While it is important to know the degree of variability in the pathogenetic networks, a fuller understanding of these, as well as the key points in the pathways, could lead to more rational target selection than simply identifying the pathway of genes in the molecular signature. In this study we analysed the comparative genomic signatures of PEBL among COG PEBL samples from the most recent COG PEBL Study (COG ANHL01P1) and compared our results with the databases of Berlin-Frankfurt-Muenster (BFM) NHL samples (Klapper, et al 2008) and National Cancer Institute (NCI) samples (Dave, et al 2006). This approach demonstrated several potential targetable signalling pathways in PEBL.
Materials and methods
Patient samples
Primary tumour specimens were obtained from children and adolescents with newly diagnosed stage III/IV or BM ± CNS BL registered on the COG ANHL01P1 study (Clinicaltrials.gov NCT00057811) (Goldman, et al 2013, Goldman, et al 2014) that were submitted to the reference laboratory at Columbia University. The protocol was approved by the institutional review board of each COG centre participating in this trial. Samples (1–2 ml) of PEBL patient-involved BM, pleural fluid, peritoneal fluid and/or lymph node with tumour involvement were collected at the time of diagnosis. The specimens for molecular studies were immediately submerged in RNAlater reagent (Qiagen, Valencia, CA), and shipped overnight at room temperature to the central research laboratory at Columbia University. Upon receipt, the specimens in RNAlater were immediately processed to extract total RNA and frozen at −80°C.
Haematopathology
All cases underwent central pathology review for confirmation of diagnosis, consisting of review of morphology, immunophenotype and genetic data from the original diagnostic biopsy. If required, additional immunophenotyping was performed centrally to confirm the diagnosis according to the 2008 World Health Organization Classification (Swerdlow, et al 2008). In addition, CD20 expression of the tumour was confirmed by immunohistochemistry (L-26 clone, DAKO Cytomation, Carpinteria, CA) using standard methods and heat-induced epitope retrieval in citrate buffer (pH6.0) on an automated stainer (ES, Ventana Medical Systems, Tucson, AZ). A tumour was scored as CD20 positive if >80% of the tumour cells stained with the antibody.
Cytogenetics and fluorescence in situ hybridization (FISH) analysis
Cytogenetic analysis was performed, as previously described, with nomenclature according to the International System for Human Chromosome Nomenclature (Poirel, et al 2009, Shaffer and Tommerup 2005). Interphase FISH analysis for MYC rearrangement was performed on a portion of the specimen submitted for cytogenetic analysis. A dual-colour MYC/IGH translocation probe designed to detect t(8;14)(q24.1;q32) or a dual-colour MYC break-apart probe, designed to detect rearrangements of the MYC gene region at 8q24.1 with various partner chromosomes, was utilised according to the COG reference laboratories standard protocols. Slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI) in Antifade solution and the nuclei were visualized and captured using a fluorescence microscope equipped with appropriate filters and imaging software.
Clinical characteristics
Specimens were collected from 11 PEBL patients registered on the COG ANHL01P1 study and the clinical and demographic data on these patients are described in Table I. All patients were treated with chemoimmunotherapy with modified FAB B4 and C1 chemotherapy with the addition of rituximab in induction and consolidation as previously described (Barth, et al 2013, Galardy, et al 2013, Goldman, et al 2013, Goldman, et al 2014, Shiramizu, et al 2011).
Table I.
Clinical Characteristics of COG Patients
| Patient | Age (Years) |
Gender | Histology | Stage | FAB Group |
Bone Marrow (≥25% blasts) |
CNS Status |
|---|---|---|---|---|---|---|---|
| 001 | 7.5 | Male | Burkitt | Stage IV | Group C | Positive | Positive |
| 002 | 1.9 | Female | Burkitt | Stage III | Group B | Negative | Negative |
| 003 | 14.5 | Female | Burkitt | Stage IV | Group C | Positive | Negative |
| 004 | 9.5 | Male | Burkitt | Stage III | Group B | Negative | Negative |
| 005 | 15.0 | Male | Burkitt | Stage III | Group B | Negative | Negative |
| 006 | 11.9 | Male | Burkitt | Stage IV | Group C | Positive | Negative |
| 007 | 8.1 | Male | Burkitt | Stage IV | Group C | Positive | Negative |
| 008 | 10.5 | Male | Burkitt | Stage III | Group B | Negative | Negative |
| 009 | 18.8 | Male | Burkitt | Stage IV | Group C | Positive | Negative |
| 010 | 11.8 | Male | Burkitt | Stage IV | Group C | Positive | Negative |
| 011 | 15.0 | Male | Burkitt | Stage IV | Group C | Positive | Negative |
COG, Children’s Oncology Group; FAB, French-American-British classification; CNS, central nervous system.
Oligonucleotide cDNA microarray analysis
RNA purification and hybridization for the microarray analyses were performed as previously described (Jiang, et al 2009). In brief, total RNA from patient tumour specimens was isolated by Trizol reagents (Invitrogen, Carlsbad, CA) and followed by affinity column purification (Qiagen, Valencia, CA). cDNA was generated from 100 ng total RNA using a poly dT oligonucleotide (Affymetrix, Santa Clara, CA). Biotinylated labelling of cRNA was performed by in vitro transcription according to manufacturer’s protocol. Fragmented cRNA (15 µg) was hybridized to HG_U133 A2 Gene Chip® arrays (Affymetrix) using Fluidics Station 450 (Affymetrix).
Gene expression profiling
Signal intensities of gene expression were calculated by Expression Console (Affymetrix). Data were imported into GeneSpring GX 10 (Agilent Technologies, Foster City, CA) or Partek Genomics Suite (St. Louis, MI) and normalised and presented as log2 values. Fold changes of target expression were compared to the average of 530 median expressed genes. The identified gene information and accession numbers were first obtained through NetAffx, provided by Affymetrix, and then each entity was further confirmed by a search of GenBank. Data has been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; accession number GSE64905).
Quantitative real time polymerase chain reaction (qRT-PCR)
Selected genes (PIM1, STAT1, MAP2K1, RAF1, PTPN11, DLEU1, and MYC), based on being a BL classifier gene or representing different signalling transduction pathways, were examined for their expression levels by qRT-PCR using qScript™ cDNA Synthesis Kit (Quantas, Gaithersburg, MD) and SsoFast™ EvaGreen® Supermix (Bio-rad, Hercules, CA). The primer sequences are listed in Supplementary Table I. Real time detection of the RT-PCR product was monitored using the CFX96 Real-time system (Bio-rad). Relative quantification (ddCt) of mRNA expression of each gene was determined by normalizing to the housekeeping gene GAPDH. cDNA amplification efficiencies of target and reference genes were confirmed to be equivalent and the absolute values of the slope of log input amount vs. ΔCT were < 0.1. Fold changes were obtained by being normalised to the qRT-PCR results of GAPDH expression.
Comparative genomic signature
For the genomic comparison in this study, paediatric genome expression results from three different research groups were utilised (Supplementary Table II); COG ANHL01P1 in the current study (n=11), GEO GSE10172 and GSE4475 (Klapper, et al 2008) (n=16), and GSE4732 (Dave, et al 2006) (n=15) (≤13 years of age).
Pathway and function analysis
Functional interpretation of the classified genes with similar expression from the comparative genomic data of COG, NCI and BFM, and relevance to molecular and cellular functions and pathways was generated by Ingenuity Pathways Analysis (IPA®), version 6.5, (Qiagen, Redwood City, CA). P-values less than 0.05 indicate a statistically significant and non-random association between a set of computational selected genes from the 376 PBL genes and a set of all genes related to a given function in Ingenuity's knowledge base.
Gene functional classification
The genes using Affymetrix ID as the identifier were uploaded into the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) at the National Institute of Health, and then analysed by functional annotation clustering. Gene lists obtained from each functional group were then re-uploaded into DAVID, and then searched for specific pathways.
Statistical analysis
Statistical differences between the three genomic expression profiles were analysed by Student t test; differences between multiple groups were analysed by one-way ANOVA followed by the Tukey multiple comparison test. Results are expressed as mean±standard error of the mean (SEM) with probability values less than or equal to 0.05 considered significant.
Results
Demographics
The mean age of the COG BL patients was 11.3 years, with a male/female ratio of 9:2. There were 3 stage III and 8 stage IV patients. Seven patients had BM involvement (≥25% L3 blasts) and one patient had CNS disease. Four patients were treated on the group B arm and seven patients were treated on the group C arm (Table I) (Goldman, et al 2013, Goldman, et al 2014).
Haematopathology
All of the cases used in this study showed typical morphological and immunophenotypic features of BL, including CD20, CD10 and BCL6 positivity and lack of expression of T-cell markers and BCL2 protein. In all cases, the tumours showed a high proliferative rate (>95%) by Ki-67 immunohistochemistry.
Cytogenetics
Six of 11 patients had evaluable cytogenetics. One patient was FISH positive for IGH/MYC but had a failed karyotype. Three patients had a complex karyotype, defined as the presence of 3 or more chromosomal aberrations, and three patients had a t(8;14)(q24;q32) translocation (Supplementary Table III).
Comparative genomic signature
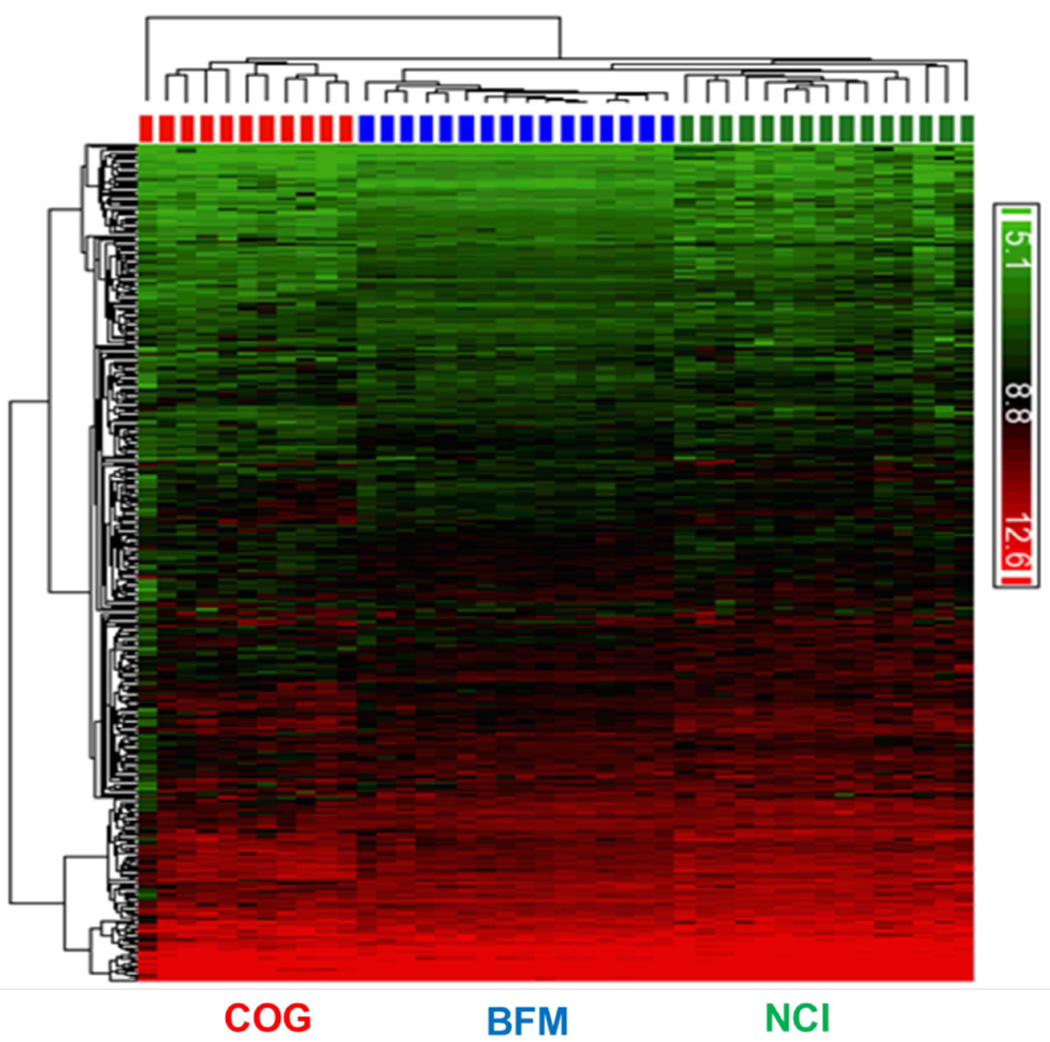
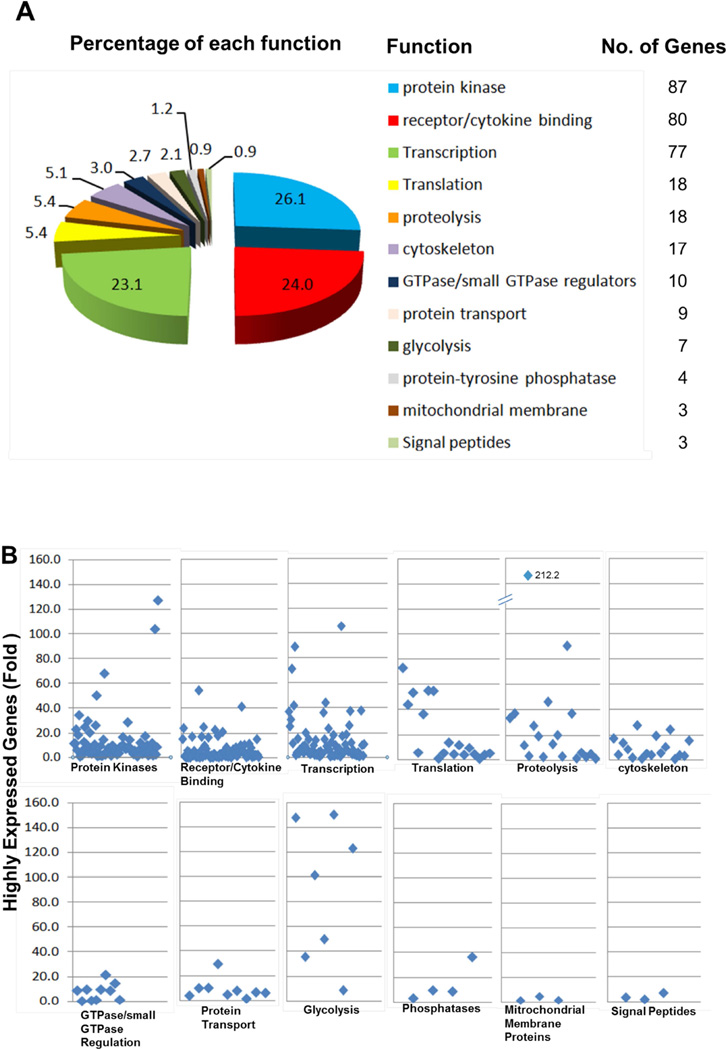
A total of 1565 genes were identified (p<0.05), among which 376 genes showed a similar genomic expression pattern within the PEBL signature among the three clinical subgroups (Fig. 1). MYC, a molecular identifier for BL, showed 27-fold greater expression in these subgroups, consistent with the previous BL signature described by Dave et al. (2006). We further classified these 376 genes and identified 12 major functions (333 genes with known functions and 43 genes with unknown functions) that were involved in PEBL (Figs. 2A and 2B). Detailed classifications and descriptions of genes in each 12 functional group are listed in Supplementary Table IV. The lead functional categories were serine/threonine kinases and tyrosine kinases (87 genes), cytokine binding receptors (80 genes) and transcription (77 genes) (Figs. 2A and 2B). PEBL tumour samples also demonstrated significantly higher levels of expression of genes, including the protein tyrosine phosphatases (PTP), PTPN11 (also called SHP2; 36.6-fold), and PTPN6 (8.6-fold) SHP1). Small GTPases and regulators were also significantly expressed in PEBL (10 genes), including RALBP1 (8.9-fold), RHOG (9.7-fold), ARHGAP19 (9.7-fold), ARHGEF18 (21.5-fold) and ARHGAP17 (8.9-fold). Seven glycolysis genes were also significantly overexpressed (8.1- to 109.5-fold).
Figure 1. Hierarchical clustering displaying gene expression patterns in paediatric Burkitt lymphoma.
Similarly expressed genes were compared between three paediatric Burkitt lymphoma clinical subgroups (COG, Children’s Oncology Group (ANHL01P1 study); BFM (Klapper, et al 2008), NCI (Dave, et al 2006) demonstrating green (decreased expression) and red colour (increased expression) according to the scale shown. Detailed procedures are described in the Methods.
COG, Children’s Oncology Group; BFM, Berlin-Franfürt-Münster; NCI, National Cancer Institute
Figure 2. Expression of genes in paediatric Burkitt lymphoma samples stratified by functional group.
A total of 376 similarly expressed genes were grouped into 12 functional categories. The percentage (A) and fold changes and numbers (B) of genes in each functional category are demonstrated.
Comparison of genomic profiling and qRT-PCR
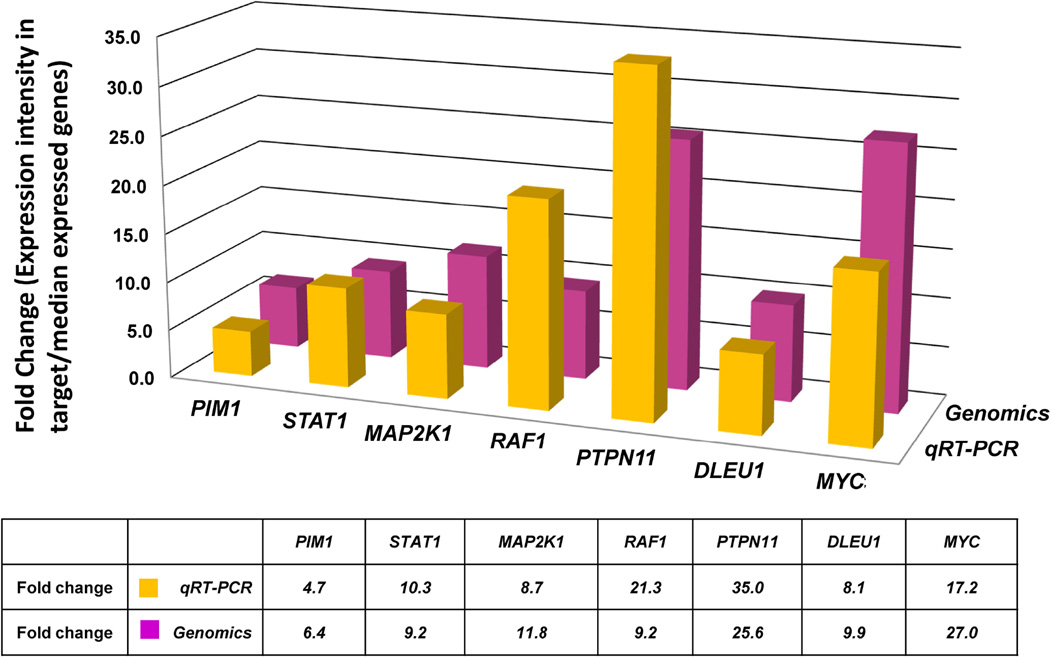
To validate the level of gene expression generated from oligonucleotide microarray studies, we analysed a selected number of target genes with significant clinical implication and/or biological function by qRT-PCR. The qRT-PCR results were then compared to the results of microarray analysis. These genes were PIM1, STAT1, MAP2K1, RAF1, PTPN11, DLEU1 and MYC, and the fold changes by qRT-PCR were 4.7, 10.3, 8.7, 21.3, 35.0, 8.1 and 17.2, respectively (Fig. 3). These qRT-PCR results were consistent with gene expression studies generated from the microarray studies.
Figure 3. Comparison of fold changes in gene expression in paediatric Burkitt lymphoma samples between Affymetrix Genechip profiling and real time qRT-PCR analyses of PIM1, STAT1, MAP2K1, RAF1, PTPN11, DLEU1 and MYC genes.
Quantitative real time polymerase chain reaction (qRT-PCR) expression levels of each selected genes are consistent with the results of genechip expression profiling analysis on Paediatric Burkitt Lymphoma samples. The fold changes were calculated from 27 and 9 PEBL samples for genomics and qRT-PCR, respectively, and histogram show the data of qRT-PCR with the means of three independent experiments performed in duplicate. GAPDH was used as endogenous control for qRT-PCR normalization.
Targetable signalling pathways
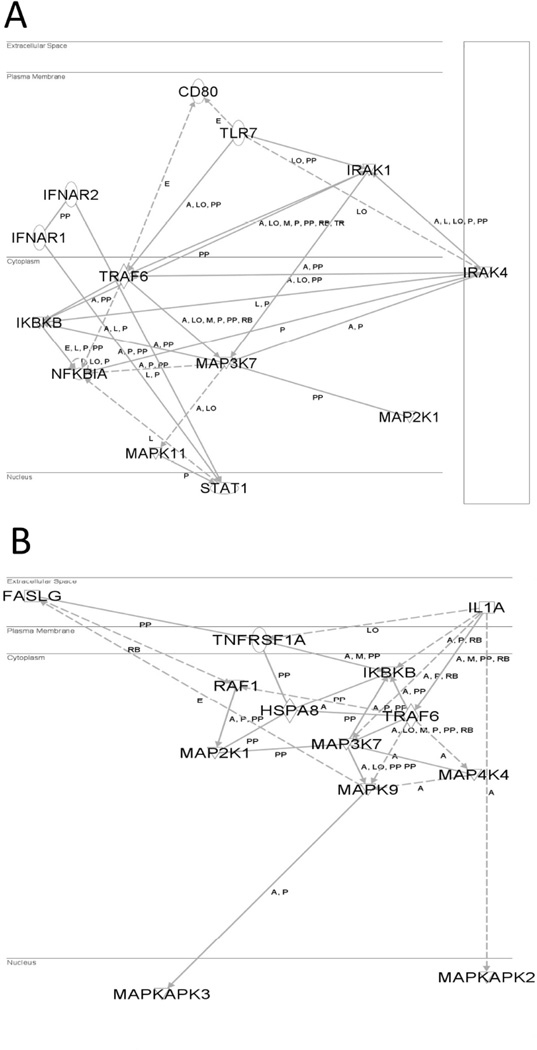
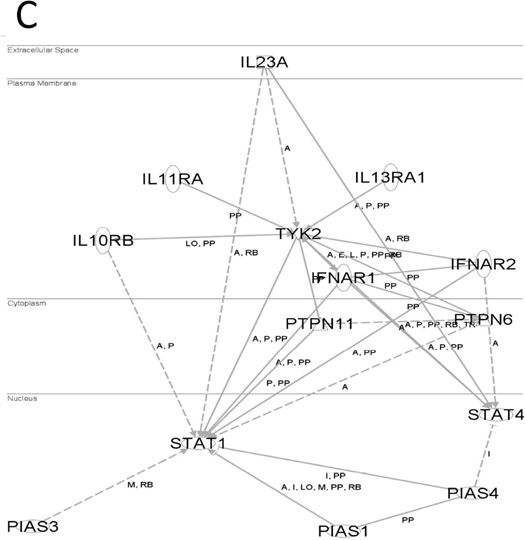
Within the 376 genes identified as common in all three data sets, IPA® demonstrated a significant association with three signalling pathways - Toll-like receptor (TOLL) signalling (p<0.01), Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signalling (p<0.01), and mitogen-activated protein kinase (MAPK) signalling (p<0.01) pathways were significantly overexpressed (Fig. 4 and Supplementary Table V). We identified gene overexpression of 7 kinases involved in the TOLL pathway and 13 kinases involved in the MAPK pathway from 87 serine/threonine kinases and tyrosine kinases (Supplementary Table VI), and 6 receptor genes involved in the TOLL pathway, JAK-STAT pathway and cytokine binding receptors pathway. Expression of genes involved in the TOLL signalling (Supplementary Table VII) included TLR7 (2.9-fold), IRAK1 (34.1-fold), NFKBIA (18.1-fold) and IKBKB (7.2-fold). In addition, the genes encoding interleukin (IL)-10 receptor (IL10RA; 3.1-fold) and IL-21 receptor (IL21R; 2.3-fold) were both expressed at a similar level to TLR7. Expression of genes involved in JAK-STAT signalling (Supplementary Table VII) included PIM1 (8.7-fold), STAT1 (11.1-fold), TYK2 (4.1-fold) and IFNAR2 (6.2-fold) and expression of genes involved in the MAPK signalling in PEBL included MAP2K1 (11.1-fold), MAP3K7 (7.6-fold), MAPK9 (10.5-fold), MAP4K4 (8.3-fold), RAF1 (11.3-fold), PTPN11 (36.6-fold) and PTPN6 (8.6-fold). Most interestingly, the B cell receptor signalling (BCR) pathway had a significant overexpression of BTK (9.1-fold) and a significant increase in HDAC1 (42-fold) and HDAC2 (15.5-fold) (Supplementary Table V).
Figure 4. Ingenuity pathway analysis of gene networks in TOLL-like receptor (A), MAPK (B), and JAK-STAT3 (C) pathways on paediatric Burkitt lymphoma samples.
Three significant signalling pathways were identified by the ingenuity pathway analysis tool (www.ingenuity.com) using up-regulated genes in paediatric Burkitt lymphoma samples in Toll-like receptors (TLRs), Mitogen-activated protein kinase (MAPK) and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathways.
A, activation; E, expression; I, inhibition; L, proteolysis; LO, localisation; M, biochemical modification; P, phosphorylation/dephosphorylation; PP, Protein-protein binding; RB, regulation of binding; TR, translocation.
Discussion
This is the first report using comparative genomic identification to compare the genomic expression signature in PEBL. We compared our COG PEBL samples with PEBL samples reported by the NCI (Dave, et al 2006) and the BFM (Klapper, et al 2008). These data suggested that PEBL gene expression signatures are consistent among three cooperative groups/laboratories. The fold changes in PEBL gene expression profiles in these three sets of samples were highly similar, if not identical. In addition, the comparative data was remarkable given that these three groups used different designated genechips for microarray. The samples investigated by Dave, et al (2006) were processed by custom-made cDNA Lymphochips, while Klapper et al (2008) and our group both utilised Affymetrix Oligonucleotide Genechips (Jiang, et al 2009).
Our results indicated that TOLL, MAPK, and JAK-STAT signalling pathways are overexpressed in PEBL samples, including threonine kinases, tyrosine kinases and cytokine binding receptors that regulate cellular proliferation (Supplementary Table VI). Interestingly, seven genes involving the regulation of glycolysis (Vander Heiden, et al 2009) were significantly overexpressed in PEBL (Fig. 3), and several target genes involved in the TLR signalling pathway (Supplementary Table VI), TLR7 (2.9-fold), IRAK1 (34.1-fold), NFKBIA (18.1-fold) and IKBKB (7.2-fold), were significantly overexpressed. In addition, IL10RA (3.1-fold) and IL21R (2.3-fold) were both expressed at a similar level to TLR7. The TLRs and the IL1 receptor (IL1R) are known to possess a highly conserved intracellular Toll-IL-1R (TIR) domain (Barton and Medzhitov 2003). Furthermore, TLR triggering has been shown to regulate cellular growth in BL (Noack, et al 2012) and TLR activation has been implicated in tonic BCR activation in DLBCL (Akhter, et al 2015). Ligand activated TLRs cause dimerization of the cytoplasmic TIR domains, which significantly activate the IL-1R associated kinase 1 (i.e. IRAK1) and then further activates the downstream NF-κB signalling. The consequence of this TLR pathway is to enhance the production of interferons (IFNα and IFNβ) in PEBL. MAPK has been demonstrated to be overexpressed in EBV-positive vs EBV-negative BL (Cerimele, et al 2005).
Target genes involved in JAK-STAT signaling (Supplementary Table VI) included PIM1 (8.7-fold), STAT1 (11.1-fold), TYK2 (4.1-fold) and IFNAR2 (6.2-fold), which were also significantly overexpressed. The oncogenic PIM1 has been implicated as a critically important gene with MYC in the pathogenesis of a number of haematological malignancies (Wang, et al 2010). Furthermore, these preclinical data suggest that the interferons may activate IFNAR2 in PEBL and, in turn, IFNAR2 activates TYK2. The activated TYK2 then phosphorylates STAT1 and STAT4 and may result in dimerization of these transcription factors. The STAT1 and STAT4 dimers may then translocate into the nucleus where they bind on the promoter region of PIM1. This pathway may be partly responsible for PEBL lymphomagenesis.
Many MAP kinase signalling pathway genes were significantly overexpressed; target genes involved in MAPK signalling in PEBL (Supplementary Table VII) included MAP2K1 (11.1-fold), MAP3K7 (7.6-fold), MAPK9 (10.5-fold), MAP4K4 (8.3-fold), RAF1 (11.3-fold), PTPN11 (36.6-fold) and PTPN6 (8.6-fold). MAPKs are serine-threonine kinases that regulate a wide variety of cellular functions (Johnson and Lapadat 2002). RAF1 encodes a MAP kinase kinase kinase (MAP3K). Mitogen-activated protein kinase (MAPK9), also known as a c-Jun N-terminal kinase (JNK2), blocks the ubiquitination of tumour suppressor TP53 (Johnson and Lapadat 2002). PTPN11 and PTPN6 are protein tyrosine phosphatases (Alonso, et al 2004, Ostman, et al 2006). From these studies we hypothesise that PEBL cells express IFNα or IFNβ following induction of TOLL signalling pathways and the overexpression of IRAK1 results in an increase of the TOLL signalling pathway. In addition, an increase of MAP3K7 may enhance the phosphorylation of IKK. These augment phosphorylation of IκBα and IκBβ and release of NFκB, then dimerized NFκB bind to the IFNα and IFNβ promoters in the nucleus and promote IFNα and IFNβ production. These interferons then activate the JAK-STAT and MAPK signalling pathways. Although the functions of these interferons are to enhance apoptosis and reduce proliferation, a highly activated RAS-MAPK signalling pathway enhances oncogene activation, including MYC (Roux and Blenis 2004). The interferon-activated IFNAR2-mediated MAPK signalling pathway in PEBL, via RAS proteins, phosphorylates MYC and enhances cellular proliferation and promote anti-apoptosis. Furthermore, serine/threonine kinases and tyrosine kinases were significantly overexpressed in this PEBL genomic study. From these comparative genomic signatures, we have demonstrated that IRAK1, RAF1, MAP4K4, BTK, MAP3K7, MAP2K1, MAPK9, NFKBIA, IKBKB, PIM1, STAT1, PTPN6, and PTPN11 may be candidate targets in the treatment of PEBL. Currently, there are several clinical trials using inhibitors/siRNA of targets in BL in phase I studies, including BTK by ibrutinib (NCT0119069), RAF1 inhibition by regorafenib (NCT02693535) and MYC inhibition by siRNA DCR-MYC (NCT02110563). Additionally, we recently demonstrated that proteins with cellular functions including NF-kB and MAPK signalling pathways are significantly overexpressed in different epidemiological subtypes of BL cell lines compared to normal B cells from healthy donors by proteomic expression profile studies (ElMallawany, et al 2014).
More interestingly, we have observed a significant increase of BCR signalling pathway genes, including BTK, and have recently demonstrated that the inhibition of PI3K/Akt is a key in a Burkitt lymphomagenesis (Frys, et al 2014) (Ippolito, et al 2015). BTK is a regulator of normal B-cell development and is activated upon BCR stimulation. Mutations of TCF3 and it’s negative regulator ID3, including MYC, are prevalent in BL and contribute to activation of the BCR pathway (Schmitz, et al 2012) (Love, et al 2012) and interestingly, MYC transcriptional expression suppressed small molecule inhibitors of the BET family of chromatin adaptors (Mertz, et al 2011). However, BL tumourigenesis may be secondary to dysregulation of un-mutated genes via transcription factors versus mutations in the targets of transcription factors or alteration of the function of other genes on the signalling pathway.
Chronic active BCR signalling through BTK activation can be inhibited by the selective, covalent BTK inhibitor, ibrutinib (Davis, et al 2010, Young and Staudt 2013). Ibrutinib is an active agent in activated B cell like DLBCL (Mathews Griner, et al 2014), and preclinical studies in chronic lymphocytic leukaemia (CLL) and mantle cell lymphoma (MCL) suggested an inhibitory effect on cell proliferation (Cinar, et al 2013, Dasmahapatra, et al 2013, Honigberg, et al 2010, Ponader, et al 2012). Furthermore, we have recently demonstrated that ibrutinib has a significant inhibitory effect of on cell proliferation and increase in caspase 3/7 and apoptosis in BL (Lee, et al 2015). Clinically, ibrutinib has been highly effective in the treatment of refractory patients with CLL and MCL and has been approved by the Food and Drug Administration for patients with CLL or MCL who have received at least one prior therapy (Byrd, et al 2013, Wang, et al 2010, Wang, et al 2013).
In summary, this is the first study comparing the genomic signature of PEBL from three completely independent databases. Among the 1565 genes identified, 376 genes (approximately 25%) were similarly expressed between the 3 groups. Furthermore, there was significant overexpression of genes involved in the Toll receptor, JAK-STAT and MAPK signalling pathways. Among these pathways, several genes, including IRAK1, PTPN6, BTK, RAF1, HDAC1 and HDAC2 may be attractive targets for alternate targeted therapeutic strategies in PEBL.
Supplementary Material
Acknowledgments
We dedicate this manuscript to Warren G. Sanger, PhD, Human Genetics Laboratories, Munroe Meyer Institute for Genetics and Rehabilitation, University of Nebraska Medical Center, Omaha, NE, for his contribution of the cytogenetic methods and results. Dr. Sanger, a dedicated physician-scientist, passed away during the final preparation of this manuscript. Additionally, we would like to acknowledge Erin Morris, RN and Virginia Moore, RN, for their assistance in the preparation of this manuscript.
Financial support was provided by The Division of Cancer Treatment, National Cancer Institute, and National Institutes of Health, Department of Health and Human Services (CA98543-09 and CA98413-09), Pediatric Cancer Research Foundation, St. Baldrick’s Foundation and Hyundai Hope on Wheels.
Footnotes
All authors declare no conflicts of interest.
S.L. and N.D. performed the research, analysed the data and wrote the paper, R.M., S.P., J.A., L.H. N.E. and S.G. performed the research, C.V. analysed the data, and M.S.C. designed the research study, analysed the data and wrote the paper.
References
- Akhter A, Masir N, Elyamany G, Phang KC, Mahe E, Al-Zahrani AM, Shabani-Rad MT, Stewart DA, Mansoor A. Differential expression of Toll-like receptor (TLR) and B cell receptor (BCR) signaling molecules in primary diffuse large B-cell lymphoma of the central nervous system. J Neurooncol. 2015;121:289–296. doi: 10.1007/s11060-014-1655-3. [DOI] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Barth MJ, Goldman S, Smith L, Perkins S, Shiramizu B, Gross TG, Harrison L, Sanger W, Geyer MB, Giulino-Roth L, Cairo MS. Rituximab pharmacokinetics in children and adolescents with de novo intermediate and advanced mature B-cell lymphoma/leukaemia: a Children's Oncology Group report. Br J Haematol. 2013;162:678–683. doi: 10.1111/bjh.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, Johnson AJ, Sukbuntherng J, Chang BY, Clow F, Hedrick E, Buggy JJ, James DF, O'Brien S. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo MS, Gerrard M, Sposto R, Auperin A, Pinkerton CR, Michon J, Weston C, Perkins SL, Raphael M, McCarthy K, Patte C. Results of a randomized international study of high-risk central nervous system B non-Hodgkin lymphoma and B acute lymphoblastic leukemia in children and adolescents. Blood. 2007;109:2736–2743. doi: 10.1182/blood-2006-07-036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo MS, Sposto R, Gerrard M, Auperin A, Goldman SC, Harrison L, Pinkerton R, Raphael M, McCarthy K, Perkins SL, Patte C. Advanced stage, increased lactate dehydrogenase, and primary site, but not adolescent age (>/= 15 years), are associated with an increased risk of treatment failure in children and adolescents with mature B-cell non-Hodgkin's lymphoma: results of the FAB LMB 96 study. J Clin Oncol. 2012;30:387–393. doi: 10.1200/JCO.2010.33.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerimele F, Battle T, Lynch R, Frank DA, Murad E, Cohen C, Macaron N, Sixbey J, Smith K, Watnick RS, Eliopoulos A, Shehata B, Arbiser JL. Reactive oxygen signaling and MAPK activation distinguish Epstein-Barr Virus (EBV)-positive versus EBV-negative Burkitt's lymphoma. Proc Natl Acad Sci U S A. 2005;102:175–179. doi: 10.1073/pnas.0408381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar M, Hamedani F, Mo Z, Cinar B, Amin HM, Alkan S. Bruton tyrosine kinase is commonly overexpressed in mantle cell lymphoma and its attenuation by Ibrutinib induces apoptosis. Leuk Res. 2013;37:1271–1277. doi: 10.1016/j.leukres.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra G, Patel H, Dent P, Fisher RI, Friedberg J, Grant S. The Bruton tyrosine kinase (BTK) inhibitor PCI-32765 synergistically increases proteasome inhibitor activity in diffuse large-B cell lymphoma (DLBCL) and mantle cell lymphoma (MCL) cells sensitive or resistant to bortezomib. Br J Haematol. 2013;161:43–56. doi: 10.1111/bjh.12206. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, Greiner TC, Weisenburger DD, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Delabie J, Rimsza LM, Braziel RM, Grogan TM, Campo E, Jaffe ES, Dave BJ, Sanger W, Bast M, Vose JM, Armitage JO, Connors JM, Smeland EB, Kvaloy S, Holte H, Fisher RI, Miller TP, Montserrat E, Wilson WH, Bahl M, Zhao H, Yang L, Powell J, Simon R, Chan WC, Staudt LM. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, Xu W, Shaffer AL, Wright G, Xiao W, Powell J, Jiang JK, Thomas CJ, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Johnson NA, Rimsza LM, Campo E, Jaffe ES, Wilson WH, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pierce SK, Staudt LM. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbacher KE, Iqbal J, Sanger W, Shen Y, Lachel C, Liu Z, Liu Y, Lim MS, Perkins SL, Fu K, Smith L, Lynch J, Staudt LM, Rimsza LM, Jaffe E, Rosenwald A, Ott GK, Delabie J, Campo E, Gascoyne RD, Cairo MS, Weisenburger DD, Greiner TC, Gross TG, Chan WC. Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling. Blood. 2012;119:3757–3766. doi: 10.1182/blood-2011-05-349662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElMallawany NK, Day N, Ayello J, van de Ven C, Conlon K, Fermin D, Basrur V, Elenitona-Johnson K, Lim M, Cairo MS. Differential proteomic analysis of endemic and sporatic Epstein-Barr Virus positive and negative Burkitt Lymphoma. Eur J Cancer. 2014;51:92–100. doi: 10.1016/j.ejca.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Frys S, Czuczman N, Mavis C, Rolland D, Lim M, Tiwari A, Cairo MS, Miles R, Barth M. Deregulation of the PI3K/Akt Signal Transduction Pathway Is Associated with the Development of Chemotherapy Resistance and Can be Effectively Targeted to Improve Chemoresponsiveness in Burkitt Lymphoma Pre-Clinical Models. Blood. 2014;124:1769. [Google Scholar]
- Galardy PJ, Hochberg J, Perkins SL, Harrison L, Goldman S, Cairo MS. Rasburicase in the prevention of laboratory/clinical tumour lysis syndrome in children with advanced mature B-NHL: a Children's Oncology Group Report. Br J Haematol. 2013;163:365–372. doi: 10.1111/bjh.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard M, Cairo MS, Weston C, Auperin A, Pinkerton R, Lambilliote A, Sposto R, McCarthy K, Lacombe MJ, Perkins SL, Patte C. Excellent survival following two courses of COPAD chemotherapy in children and adolescents with resected localized B-cell non-Hodgkin's lymphoma: results of the FAB/LMB 96 international study. Br J Haematol. 2008;141:840–847. doi: 10.1111/j.1365-2141.2008.07144.x. [DOI] [PubMed] [Google Scholar]
- Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F, Pedrosa M, Teruya-Feldstein J, Bhagat G, Alobeid B, Leoncini L, Bellan C, Rogena E, Pinkney KA, Rubin MA, Ribeiro RC, Yelensky R, Tam W, Stephens PJ, Cesarman E. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–5184. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Smith L, Anderson JR, Perkins S, Harrison L, Geyer MB, Gross TG, Weinstein H, Bergeron S, Shiramizu B, Sanger W, Barth M, Zhi J, Cairo MS. Rituximab and FAB/LMB 96 chemotherapy in children with Stage III/IV B-cell non-Hodgkin lymphoma: a Children's Oncology Group report. Leukemia. 2013;27:1174–1177. doi: 10.1038/leu.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Smith L, Galardy P, Perkins SL, Frazer JK, Sanger W, Anderson JR, Gross TG, Weinstein H, Harrison L, Shiramizu B, Barth M, Cairo MS. Rituximab with chemotherapy in children and adolescents with central nervous system and/or bone marrow-positive Burkitt lymphoma/leukaemia: a Children's Oncology Group Report. Br J Haematol. 2014;167:394–401. doi: 10.1111/bjh.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, Buggy JJ. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, Hansmann ML, Haralambieva E, Harder L, Hasenclever D, Kuhn M, Lenze D, Lichter P, Martin-Subero JI, Moller P, Muller-Hermelink HK, Ott G, Parwaresch RM, Pott C, Rosenwald A, Rosolowski M, Schwaenen C, Sturzenhofecker B, Szczepanowski M, Trautmann H, Wacker HH, Spang R, Loeffler M, Trumper L, Stein H, Siebert R. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Ippolito T, Czuczman N, Mavis C, Rolland D, Lim M, Tiwari A, Cairo MS, Miles R, Barth M. Targeting the PI3K/Akt/mTOR pathway improves chemoresponsiveness in cell line models of chemotherapy sensitive and resistant Burkitt lymphoma. Pediatr Blood Cancer. 2015;62:S143–S418. [Google Scholar]
- Jiang H, van de Ven C, Baxi L, Satwani P, Cairo MS. Differential gene expression signatures of adult peripheral blood vs cord blood monocyte-derived immature and mature dendritic cells. Exp Hematol. 2009;37:1201–1215. doi: 10.1016/j.exphem.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Klapper W, Szczepanowski M, Burkhardt B, Berger H, Rosolowski M, Bentink S, Schwaenen C, Wessendorf S, Spang R, Moller P, Hansmann ML, Bernd HW, Ott G, Hummel M, Stein H, Loeffler M, Trumper L, Zimmermann M, Reiter A, Siebert R. Molecular profiling of pediatric mature B-cell lymphoma treated in population-based prospective clinical trials. Blood. 2008;112:1374–1381. doi: 10.1182/blood-2008-01-136465. [DOI] [PubMed] [Google Scholar]
- Lee S, Yin C, O'Connell T, Barth M, Ayello J, Harrison L, van de Ven C, Miles R, Galardy P, Goldman SC, Lim MS, Hermiston M, McAllister-Lucas L, Roth LG, Perkins SL, Cairo MS. Ibrutinib significantly improves survival in a human Burkitt lymphoma (BL) xenograft NSG mouse model: Ibrutinib may be a potential adjuvant agent in the treatment of BL. American Association for Cancer Research. 2015;56:655. (abstract) [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers CR, Naresh KN, Evens AM, Chadburn A, Gordon LI, Czader MB, Gill JI, Hsi ED, Greenough A, Moffitt AB, McKinney M, Banerjee A, Grubor V, Levy S, Dunson DB, Dave SS. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, Duveau DY, Jiang JK, Michael S, Mierzwa T, Huang W, Walsh MJ, Mott BT, Patel P, Leister W, Maloney DJ, Leclair CA, Rai G, Jadhav A, Peyser BD, Austin CP, Martin SE, Simeonov A, Ferrer M, Staudt LM, Thomas CJ. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111:2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles RR, Arnold S, Cairo MS. Risk factors and treatment of childhood and adolescent Burkitt lymphoma/leukaemia. Br J Haematol. 2012;156:730–743. doi: 10.1111/j.1365-2141.2011.09024.x. [DOI] [PubMed] [Google Scholar]
- Nelson M, Perkins SL, Dave BJ, Coccia PF, Bridge JA, Lyden ER, Heerema NA, Lones MA, Harrison L, Cairo MS, Sanger WG. An increased frequency of 13q deletions detected by fluorescence in situ hybridization and its impact on survival in children and adolescents with Burkitt lymphoma: results from the Children's Oncology Group study CCG-5961. Br J Haematol. 2010;148:600–610. doi: 10.1111/j.1365-2141.2009.07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack J, Jordi M, Zauner L, Alessi D, Burch A, Tinguely M, Hersberger M, Bernasconi M, Nadal D. TLR9 agonists induced cell death in Burkitt's lymphoma cells is variable and influenced by TLR9 polymorphism. Cell Death Dis. 2012;3:e323. doi: 10.1038/cddis.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- Patte C, Auperin A, Gerrard M, Michon J, Pinkerton R, Sposto R, Weston C, Raphael M, Perkins SL, McCarthy K, Cairo MS. Results of the randomized international FAB/LMB96 trial for intermediate risk B-cell non-Hodgkin lymphoma in children and adolescents: it is possible to reduce treatment for the early responding patients. Blood. 2007;109:2773–2780. doi: 10.1182/blood-2006-07-036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel HA, Cairo MS, Heerema NA, Swansbury J, Auperin A, Launay E, Sanger WG, Talley P, Perkins SL, Raphael M, McCarthy K, Sposto R, Gerrard M, Bernheim A, Patte C Committee, F.L.I.S. Specific cytogenetic abnormalities are associated with a significantly inferior outcome in children and adolescents with mature B-cell non-Hodgkin's lymphoma: results of the FAB/LMB 96 international study. Leukemia. 2009;23:323–331. doi: 10.1038/leu.2008.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, Keating MJ, O'Brien S, Chiorazzi N, Burger JA. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, Bullinger L, Rajewsky K. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell. 2012;22:167–179. doi: 10.1016/j.ccr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Ogwang MD, Reynolds SJ, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pittaluga S, Wilson W, Waldmann TA, Rowe M, Mbulaiteye SM, Rickinson AB, Staudt LM. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- Shaffer LC, Tommerup N. An International System for Human Cytogenetic Nomenclature. Switzerland: S. Karger, Basel; 2005. [Google Scholar]
- Shiramizu B, Goldman S, Kusao I, Agsalda M, Lynch J, Smith L, Harrison L, Morris E, Gross TG, Sanger W, Perkins S, Cairo MS. Minimal disease assessment in the treatment of children and adolescents with intermediate-risk (Stage III/IV) B-cell non-Hodgkin lymphoma: a children's oncology group report. Br J Haematol. 2011;153:758–763. doi: 10.1111/j.1365-2141.2011.08681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th. Lyon, France: International Agency for Research on Cancer Press; 2008. [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML, Abdulkadir SA. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477–2487. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, Jurczak W, Advani RH, Romaguera JE, Williams ME, Barrientos JC, Chmielowska E, Radford J, Stilgenbauer S, Dreyling M, Jedrzejczak WW, Johnson P, Spurgeon SE, Li L, Zhang L, Newberry K, Ou Z, Cheng N, Fang B, McGreivy J, Clow F, Buggy JJ, Chang BY, Beaupre DM, Kunkel LA, Blum KA. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369:507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12:229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.