Abstract
The F-plasmid-encoded TraI protein, also known as DNA helicase I, is a bifunctional protein required for conjugative DNA transfer. The enzyme catalyzes two distinct but functionally related reactions required for the DNA processing events associated with conjugation: the site- and strand-specific transesterification (relaxase) reaction that provides the nick required to initiate strand transfer and a processive 5′-to-3′ helicase reaction that provides the motive force for strand transfer. Previous studies have identified the relaxase domain, which encompasses the first ∼310 amino acids of the protein. The helicase-associated motifs lie between amino acids 990 and 1450. The function of the region between amino acids 310 and 990 and the region from amino acid 1450 to the C-terminal end is unknown. A protein lacking the C-terminal 252 amino acids (TraIΔ252) was constructed and shown to have essentially wild-type levels of transesterase and helicase activity. In addition, the protein was capable of a functional interaction with other components of the minimal relaxosome. However, TraIΔ252 was not able to support conjugative DNA transfer in genetic complementation experiments. We conclude that TraIΔ252 lacks an essential C-terminal domain that is required for DNA transfer. We speculate this domain may be involved in essential protein-protein interactions with other components of the DNA transfer machinery.
Conjugative DNA strand transfer is a highly conserved mechanism for unidirectional transfer of genetic information among bacteria of the same species, from one species to another and, in some instances, between kingdoms (for reviews, see references 15, 22, 25, 30, 37, and 44). Importantly, this is the mechanism used by most plasmids and conjugative transposons to facilitate their spread throughout a bacterial population, and the same underlying mechanism is used to transfer T-DNA from Agrobacterium tumefaciens to plant cells (23, 45).
The process of conjugative DNA transfer (CDT) is initiated with the formation of a stable mating pair between donor and recipient to establish the close cell-cell contact required for physical transfer of single-stranded DNA (ssDNA) from one cell to another. This is followed by the production of a site- and strand-specific nick at a locus called nic within the origin of transfer (oriT) or at the ends of T-DNA and the unwinding/replication of the duplex DNA molecule to provide the ssDNA that enters the recipient. Within the recipient cell, host enzymes (primarily) convert the transferred ssDNA into double-stranded DNA that either circularizes to form a plasmid or is recombined into the recipient chromosome (for reviews, see references 15 and 44).
Although reasonably well understood at this “macroscopic” level, the molecular details of the process by which DNA is transferred from donor to recipient are still being resolved. Key players are the proteins that initiate the physical transfer of ssDNA, the conjugative initiator proteins (7, 22, 30). These proteins introduce a site- and strand-specific break in the phosphodiester backbone at nic via a transesterification reaction that leaves the initiator protein covalently bound on the 5′ end of the cleaved DNA strand via a phosphotyrosyl linkage. The DNA is subsequently unwound, through the action of a helicase or as a function of DNA replication extending from the newly formed 3′-OH, to produce the ssDNA molecule that can be transferred to the recipient cell. In the case of plasmid DNA transfer, the 3′-OH subsequently acts as a nucleophile to reverse the original transesterification. This restores the integrity of the ssDNA transferred to the recipient and releases the bound initiator protein. The action of the initiator protein at oriT is governed by several auxiliary proteins that act to alter the architecture of the DNA molecule in the vicinity of nic to allow binding of the initiator protein. These auxiliary proteins are both host encoded and plasmid encoded in origin (7, 22, 29).
A small subset of the conjugative initiator proteins contain a second domain harboring a functional DNA helicase (16, 19, 39). These proteins include, among others, the TrwC protein from plasmid R388 (16, 24) and the TraI protein from the F plasmid (6, 10, 39). Both of these proteins have been shown to be bifunctional proteins with a transesterase (relaxase) activity encoded in the first ∼300 amino acids of the protein and a helicase residing in the C-terminal portion of the protein (6, 24). Moreover, in both cases, both biochemical activities have been shown to be required for DNA transfer. In the case of F-plasmid-encoded TraI, in addition to these two defined functional domains, there are additional regions of the protein with no known function (Fig. 1). For example, there is a region between the C-terminal end of the active transesterase domain (amino acid 306) and the beginning of the helicase domain (amino acid 990 as defined by conserved helicase-associated motifs) that has no known function. However, previous studies have suggested that the active helicase begins before amino acid 348 (6), so this region may play a role in the processive helicase reaction catalyzed by TraI. In addition, there are approximately 300 amino acids at the C-terminal end of the protein with no known function.
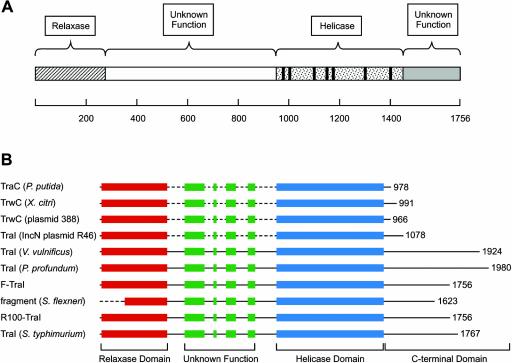
FIG. 1.
Diagram of TraI and ClustalW alignments. (A) Schematic diagram of TraI. The relaxase domain is wholly contained within the region spanned by residues 1 to 310 (6, 11). The helicase-associated motifs are located within the region spanned by residues 990 to 1450. The functional role of the central region (residues 310 to 990) and the C-terminal region (residues 1450 to 1756) is unknown. (B) ClustalW alignment of selected sequences obtained from a BLAST search using TraI (NP_862951.1) as the query sequence. The relaxase domain is shown in red, the central region with unknown function is shown in green, and the region containing the helicase-associated motifs is in blue. The sequences used in the alignment shown are TraI (F plasmid), TraI (plasmid R100) (NP_052981.1), TraI (Salmonella enterica serovar Typhimurium) (AAM90727.1), oriT nicking/unwinding protein fragment (Shigella flexneri) (NP_085415.1), putative conjugative transfer protein TraI (Vibrio vulnificus YJ016) (NP_932226.1), putative DNA helicase (TraI) (Photobacterium profundum SS9) (YP_015476.1), putative TraC protein (Pseudomonas putida) (AAP57243.1), TrwC (Xanthomonas citri) (NP_942625.1), TrwC protein (IncW plasmid R388) (S43878), and TraI [IncN plasmid R46] (NP_511201.1).
In this report, we have investigated the role of the C-terminal end of F-TraI, using a truncation mutant lacking the last 252 amino acid residues. This protein (TraIΔ252) is an active transesterase/relaxase that binds and catalyzes a transesterification reaction at the F plasmid oriT as well as an active DNA helicase. In addition, the protein interacts with the other components of the minimal relaxosome, integration host factor (IHF), and the F-plasmid-encoded TraY protein. Thus, the protein is fully competent for the two known biochemical activities of TraI. However, genetic complementation studies reveal the inability of this protein to support DNA transfer. We conclude that TraI contains a third functional domain at the C-terminal end of the protein that is required for DNA strand transfer. Further, we hypothesize that the C-terminal end of TraI may play a critical role in the next step in the process of DNA transfer—interaction with the export machinery at the interior surface of the donor cell.
MATERIALS AND METHODS
Protein purification.
Wild-type (full-length) TraI protein and TraIΔ252 were expressed and purified as described previously (28). Briefly, each protein was expressed in Escherichia coli HMS174(DE3) [F− recA1 hsdR (rK12− mK12+) (DE3) (Rifr)] with a pET-derived expression plasmid. Cells were grown at 37°C in Luria-Bertani (LB) medium to an optical density at 600 nm of ∼1.0, and protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. Growth was continued for 3 h at 37°C, and cells were harvested by centrifugation. After cell lysis, a cleared lysate was prepared, nucleic acids were precipitated with Polymin P, and, after a series of ammonium sulfate backwashes, the proteins were purified with phosphocellulose, MonoQ, and Superose 6 chromatography columns. The TraI concentration was determined by measuring A280 with an extinction coefficient for TraI of 129,795 M−1 cm−1. The TraIΔ252 concentration was determined with a Bio-Rad protein assay with purified TraI protein as the standard. Protein concentrations are expressed as equivalents of protein monomers. IHF and TraY were purified as described previously (18, 29).
DNA substrates.
Plasmid pBSoriT has been previously described (26) and contains the F-plasmid oriT region from the BglII site to the SalI site cloned in compatible sites in pBluescript. Plasmid DNA was purified by double banding on CsCl gradients in the presence of ethidium bromide (17). This was necessary to reduce the background of nicked DNA molecules to an acceptable level. Partial duplex unwinding substrates were prepared essentially as described previously (21). Briefly, a 91-nucleotide oligonucleotide or the complementary strand of an 849-bp M13mp18 replicative form DNA HaeIII restriction fragment was annealed to its complementary sequence on purified M13mp18 ssDNA at a molar ratio of 1:1. The 3′ end of the annealed DNA strand was extended with E. coli DNA polymerase I (large fragment) and [α-32P]dCTP. The final length of the oligonucleotide was 93 or 851 nucleotides. The preparation was phenol-chloroform extracted and further purified with a Biogel A5 M column equilibrated with 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM NaCl. The void volume fractions were pooled and used directly in DNA unwinding reaction mixtures. The DNA concentration of the pooled fractions is estimated to be 20 μM (expressed as DNA-Pi).
The gene expressing TraIΔ252 was constructed by using PCR to amplify the traI gene on pET11d-traI (28). The upstream primer 5′-TTTTTCATATGATGAGTATTGCGC-3′ contained an NdeI site (underlined), and the downstream primer 5′-TTTGGATCCTCACTACGCCTGACGGAGAACAGC-3′ contained a BamHI site (underlined). In addition, the downstream PCR primer contained two stop codons immediately following codon 1504 in the traI sequence to produce the C-terminal truncation of the traI gene. The amplified fragment of traI was digested with NdeI and BamHI and inserted into the expression vector pET3c, digested with the same two enzymes. The gene was sequenced in its entirety to ensure the absence of unintended mutations. The protein was expressed and purified as indicated above. Oligonucleotides were synthesized by IDT Inc. Each oligonucleotide was purified on a 20% denaturing polyacrylamide gel. Plasmids containing point mutations in traI (pET11d-traIK998M and pET11d-traIY16V) were constructed by the QuikChange mutagenesis procedure (Stratagene) with the appropriate oligonucleotide primers. The construction of pET11d-traIK998M has been described previously (28), and the construction of pET11d-traIY16V will be described elsewhere.
DNA helicase assays.
DNA unwinding reaction mixtures (typically 20 μl) contained 25 mM Tris-HCl (pH 7.5), 20 mM NaCl, 3 mM MgCl2, 5 mM β-mercaptoethanol, approximately 2 μM (DNA-Pi) partial duplex DNA substrate, and 2 mM ATP. Reaction mixtures were assembled at room temperature, and the reaction was initiated by the addition of enzyme. Incubation was at 37°C for 5 min. The reactions were quenched with 10 μl of stop solution (37.5% glycerol, 50 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue, 0.3% sodium dodecyl sulfate [SDS]), and the reaction products were resolved on a nondenaturing polyacrylamide gel (10% for the 93-bp substrate and 8% for the 851-bp substrate; 20:1 cross-linking ratio) run at 200 V for 2 h (93-bp substrate) or overnight (851-bp substrate). The results were visualized with a PhosphorImager and analyzed with ImageQuant software (Molecular Dynamics).
Duplex DNA relaxation assays.
Assays were done in a manner slightly modified from those previously described (26). A typical reaction mixture (16 μl) contained 7 nM supercoiled pBSoriT (or pBS) DNA, 40 mM Tris-HCl (pH, 7.5), 4 mM MgCl2, 15% glycerol, and the indicated concentration of either TraI or TraIΔ252. Reaction mixtures were assembled at room temperature, the reaction was initiated by the addition of enzyme, and the mixtures were incubated at 37°C for 20 min. Reactions were stopped by the addition of proteinase K (Boehringer-Mannheim Biochemicals) and SDS to final concentrations of 0.2 mg/ml and 0.1%, respectively, and the mixtures were allowed to incubate at 37°C for an additional 20 min. The products were resolved on 0.8% agarose gels and visualized by ethidium bromide staining (0.5 μg/ml). For the duplex DNA relaxation assays involving the addition of IHF and TraY, the reaction mixture was altered to include NaCl at a final concentration of 75 mM as previously described (29).
Oligonucleotide cleavage assays.
Oligonucleotide cleavage assays were similar to those used previously to analyze the cleavage reaction catalyzed by TraI from the IncPα plasmid RP4 (31). The complete reaction mixture (10 μl) contained 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 6 mM MgCl2, 20% glycerol, 1 pmol of 5′-end-labeled 22-mer (5′-TTTGCGTGGGGTGTGGTGCTTT-3′), and 1 pmol of TraI or TraIΔ252 (unless otherwise stated). Reaction mixtures were assembled at room temperature and incubated at 37°C for 20 min. Reactions were stopped by the addition of SDS to 0.2%, and incubation was continued at 37°C for 10 min. Ten microliters of 85% formamide-50 mM EDTA-0.1% dyes was added to each reaction mixture, and the products were denatured at 100°C for 3 min and analyzed on a 16% polyacrylamide-8 M urea denaturing gel. The gels were electrophoresed at 25 W in 1 × TBE (90 mM Tris-borate, 2 mM EDTA) and visualized with a PhosphorImager (Molecular Dynamics). Markers were prepared as described previously (36).
Strand transfer reactions.
Strand transfer reactions (31) were performed in a manner similar to the oligonucleotide cleavage assay, except after the 20-min incubation, 1 pmol of a second unlabeled oligonucleotide (5′-CTTGTTTTTGCGTGGGGTGTGGTGCTTTTG-3′) of different length containing the F-plasmid nic site was added to the reaction mixture and incubation was continued at 37°C for 1 h. The reaction was stopped and analyzed by the procedure described above.
Genetic assays.
The liquid mating assay protocol was followed as previously described (28). Briefly, the donor strain (JS10) was HMS174 [F− recA1 hsdR (rK12− mK12+) (Rifr)] containing pOX38T, a mini-F plasmid fully competent for conjugative DNA transfer, or HMS174 containing pOX38TΔtraI (JS11), a derivative of pOX38T lacking the traI gene (28), and the appropriate complementing plasmid. The recipient strain was HB101 or JS4 (Strr). Donor and recipient strains were diluted 1:50 into LB medium from saturated overnight cultures grown under antibiotic selection and allowed to grow to mid-log phase in the absence of selection at 37°C. Donors and recipients were mixed at a volume ratio of 1 donor to 9 recipients and incubated at 37°C for 5 min. The mating mixtures were then diluted 1:10 in prewarmed medium and incubated for 60 min at 37°C. The mating mixtures were then vigorously vortexed to disrupt mating pairs, and 10-fold serial dilutions were prepared in phosphate-buffered saline for analysis. Appropriate dilutions were plated onto LB plates containing streptomycin and tetracycline to counterselect donors and unmated recipients while selecting for transconjugants. Aliquots of the unmated donor and recipient cultures were subjected to 10-fold serial dilution and plated onto LB plates containing the appropriate antibiotics to determine viable donor cell count and viable recipient cell count. Mating frequency was calculated as the number of transconjugants per 100 viable donor cells.
RESULTS
The F-plasmid-encoded TraI protein, also known as DNA helicase I (3), is essential for CDT (41). Recently, we and others have shown this protein to be capable of catalyzing two distinct but functionally related biochemical reactions (6, 10, 39). The protein is a site- and strand-specific transesterase that makes the site- and strand-specific strand break required to initiate CDT (26, 33, 38). This reaction is also referred to as a relaxation reaction since the product of the reaction (after protein denaturation) is a relaxed plasmid (7). As a result of the transesterification, TraI is covalently bound to the 5′ end of the nicked DNA strand that will ultimately be transferred into the recipient cell (27). TraI is also a processive 5′-to-3′ DNA helicase (1, 2, 21). Both of these catalytic activities are essential for CDT (6, 28). It is known that TraI binds the F-plasmid oriT sequence, with the aid of the F-plasmid-encoded TraY protein and the host-encoded IHF, to form the relaxosome that produces the site- and strand-specific nick at oriT (18, 29). It is presumed that the helicase activity of the protein then unwinds the plasmid for transfer into the recipient. This assumption is based on two facts: (i) a single strand of the F plasmid is transferred to the recipient, and (ii) the helicase activity of TraI is essential for CDT (28).
Interestingly, TraI contains significant regions that do not appear to correlate with the known biochemical activities of the protein (Fig. 1). Biochemical studies have demonstrated the transesterase domain to be wholly contained within the first 310 amino acids of the protein (6, 38). Recent structural studies are consistent with this conclusion (11). The conserved motifs associated with superfamily I DNA helicases are found in the region of the protein from amino acid 990 to amino acid 1450. The large region between amino acids 310 and 990 and the region beyond amino acid 1450 are not defined in terms of function within this large protein. However, it should be noted that previous studies suggested that the residues between amino acids 310 and 990, in addition to the region encompassing the helicase-associated motifs, were important for helicase activity (6). More recent work (E. G. Gibbons and S. W. Matson, unpublished data) indicates that the region of the protein from amino acid 903 to amino acid 1756 does exhibit a very weak DNA helicase activity and a poor ATPase activity that went undetected in the earlier studies. It is apparent that the region from amino acid 310 to amino acid 990 is critical for wild-type levels of helicase and ATPase activity. However, the region harboring the helicase-associated motifs can support a basal helicase activity. Additional functional roles for the region of the protein from residue 310 to residue 903 remain undefined.
The aim of these studies was to examine the functional role of the C-terminal ∼250 amino acids of TraI. This region of the protein is conserved in several, but not all, conjugative initiator proteins that harbor both a relaxase domain and a helicase domain (Fig. 1B). It is interesting to note that this region is not present in the IncW plasmid initiator protein TrwC, which, in addition, to F-TraI, has both relaxase and helicase activity and has been extensively characterized. To examine the functional role of the last 252 amino acids, a truncated protein lacking the C-terminal 252 amino acids (TraIΔ252) was expressed, purified, and characterized in both genetic and biochemical assays.
TraIΔ252 expression and purification.
The traIΔ252 gene was constructed as described in Materials and Methods and lacks the C-terminal 252 amino acid residues associated with wild-type TraI. The protein contains the relaxase domain (amino acids 1 to 310) as well as all seven of the helicase motifs associated with superfamily I DNA helicases. The protein also contains approximately 60 residues beyond helicase motif VI. The truncation site was chosen based on the distance between other superfamily I DNA helicases (i.e., UvrD, Rep, and helicase IV) and the C terminus of the protein and a prediction of secondary structure that indicated the possibility of a loop in this region.
TraI and TraIΔ252 were expressed in both HMS174 and HMS174(DE3) (Fig. 2). Expression in HMS174(DE3) was the result of IPTG-dependent induction, and these cells were used for purification of the protein as described in Materials and Methods. The TraIΔ252 protein was purified by the same procedure used for purification of the full-length protein and is shown in Fig. 2A (lane 3). The purified protein was truncated relative to the wild-type protein, as expected, and was approximately 90% homogeneous and was fully soluble. The relatively minor contaminant that migrates faster than TraIΔ252 is a degradation product, as evidenced by its reaction with antisera directed against TraI protein on Western blots (data not shown). This protein was used for the biochemical studies described below.
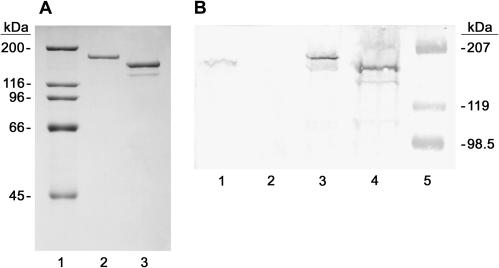
FIG. 2.
SDS-polyacrylamide gel analysis of the purification and expression of TraI and TraIΔ252. (A) One microgram each of purified TraI (lane 2) and purified TraIΔ252 (lane 3) was resolved on an 8% polyacrylamide gel run in the presence of SDS and stained with Coomassie brilliant blue. High-range molecular mass standards (Bio-Rad) with sizes in kilodaltons indicated to the left are shown in lane 1. (B) Whole-cell lysates of HMS174/pOX38T (lane 1), HMS174/pOX38TΔtraI (lane 2), HMS174/pET11d-traI (lane 3), and HMS174/pET3c-traIΔ252 (lane 4) were resolved on an 8% polyacrylamide gel run in the presence of SDS, transferred to nitrocellulose, and probed with anti-TraI antisera. Prestained molecular mass standards (Bio-Rad) with sizes in kilodaltons are shown on the right. The slight difference in migration between TraI in lane 1 and TraI in lane 3 is due to a gel artifact.
The level of expression of TraI and TraIΔ252 determined with expression plasmids in HMS174 was evaluated and compared with the level of TraI expression from its native promoter on plasmid pOX38T (mini-F plasmid that supports CDT). Expression of both proteins in HMS174 results from the adventitious use of an E. coli promoter on the pET expression plasmid since this strain lacks the lambda lysogen (DE3) required for expression of T7 RNA polymerase and IPTG-dependent induction of TraIΔ252. The expression of TraI and TraIΔ252 in HMS174 (Fig. 2B) was evaluated by Western blotting (i) to ensure the protein was stably expressed in strains to be used for genetic complementation studies (see below) and (ii) to ensure the protein was not significantly overexpressed under these conditions. It is clear that both TraI and TraIΔ252 are stably expressed in HMS174 (Fig. 2B, lanes 3 and 4) and that expression is elevated only slightly relative to expression of TraI from pOX38T (Fig. 2B, lane 1). It is also clear that TraI is not expressed in the pOX38TΔtraI derivative that lacks the traI gene (Fig. 2B, lane 2).
The C-terminal end of TraI is required for conjugative DNA transfer.
Using genetic complementation assays, we have shown that both the transesterase activity and the helicase activity of TraI are essential for DNA transfer (6, 28). These experiments utilized a mini-F plasmid (pOX38T) that expresses all the genes required for CDT and its derivative (pOX38TΔtraI) that contains a traI deletion/insertion. Since TraI is required for CDT, pOX38TΔtraI cannot be transferred to a recipient cell unless a complementing plasmid expressing TraI (pET11d-TraI) is also provided (Table 1, compare lines 2 and 3). The complemented transfer frequency is somewhat lower than the transfer frequency obtained with cells that contain pOX38T (Table 1, line 1) because the recipient in a pOX38T transfer event becomes a donor and can continue to transfer DNA to other recipients, while a recipient in a complemented transfer event does not become a donor since the plasmid it receives (pOX38TΔtraI) cannot support further transfer.
TABLE 1.
Conjugative DNA transfer complementationa
| Strain | Relevant genotype | Transfer frequency (no. of transconjugants/ 100 donors) |
|---|---|---|
| JS10 | HMS174/pOX38T | 0.2 |
| JS11 | HMS174/pOX38TΔtraI | <2 × 10−6b |
| HMS174/pOX38TΔtraI/pET11c-traI | 0.03 | |
| HMS174/pOX38TΔtraI/pET11d-traIY16V | <1 × 10−5b | |
| HMS174/pOX38TΔtraI/pET11d-traIK998M | <7 × 10−4b | |
| HMS174/pOX38TΔtraI/pET3c-traIΔ252 | <8 × 10−6b |
Complementation assays were performed as described in Materials and Methods, using the indicated donor cell strain and JS4 as the recipient cell strain. All donor strains are HMS174 [F−recA1 hsdR (rK12− mK12+) (Rifr)] containing the indicated plasmid(s).
No transconjugants were detected in any experiment. This value represents the sensitivity of the complementation assay and was determined by assuming there was a single transconjugant in the experiment. The actual transfer frequency may be much lower.
As expected, the wild-type TraI protein, expressed from the pET11d plasmid (Table 1, line 3) complements DNA transfer. As indicated above, expression in this case is due to an adventitious promoter on the plasmid and is not due to induction of T7 RNA polymerase. Therefore, expression is relatively low and we estimate less than threefold greater than expression from the mini-F plasmid (Fig. 2B). Thus, high-level expression of TraI is not required for genetic complementation. A point mutation in the active site tyrosine of the transesterase domain (TraIY16V) or a point mutation in helicase motif I of the helicase domain (TraIK998M) abolishes genetic complementation as reported previously (6, 28; unpublished results). Interestingly, deletion of the C-terminal 252 amino acids also abolishes genetic complementation, suggesting that this region of the protein is essential for some aspect of the DNA transfer event. It is important to note that this truncation leaves all helicase-associated motifs intact.
TraIΔ252 maintains essentially wild-type helicase and transesterase activity.
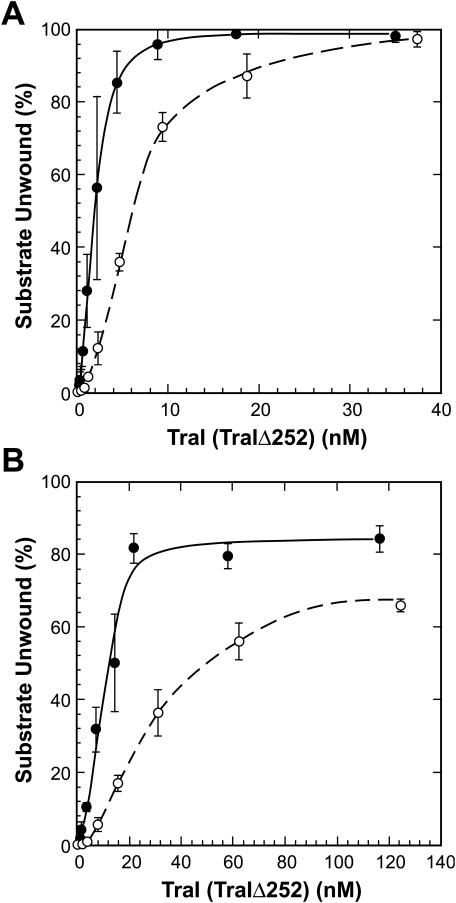
To eliminate the possibility that TraIΔ252 was unable to support CDT due to an impact on either helicase activity or relaxase activity, the purified protein was evaluated in biochemical assays for both activities. The helicase activity of the protein was tested with a 93-bp partial duplex DNA substrate and an 851-bp partial duplex DNA substrate (Fig. 3). Helicase activity measured with a 93-bp partial duplex substrate was reduced slightly compared to the activity of the full-length protein at low protein concentrations. However, at higher protein concentrations, the activity of TraIΔ252 was virtually indistinguishable from that of full-length protein. Similarly, TraIΔ252-catalzyed helicase activity on the longer partial duplex substrate (851 bp) was reduced relative to that of full-length protein at low protein concentrations but was only slightly lower than that of full-length protein at higher protein concentrations. Importantly, the protein exhibits robust helicase activity on both substrates, suggesting the failure to complement in genetic assays is not due to a loss of DNA helicase activity.
FIG. 3.
DNA helicase activity of TraI and TraIΔ252. DNA unwinding assays using a 93-bp partial duplex substrate (A) or an 851-bp partial duplex substrate (B) were performed as described in Materials and Methods, using the indicated amount of TraI (•) or TraIΔ252 (○). Reaction mixtures were incubated for 10 min at 37°C, and the products were resolved on a native polyacrylamide gel. The fraction of the substrate unwound was calculated by the formula [(U − Bu)/(S − Bs + U − Bu)] × 100, where U represents the DNA in the product band, S represents DNA in the substrate band and Bu and Bs represent background levels of DNA at the position of the product (Bu) or the substrate (Bs) determined from reaction mixtures containing no protein that were either incubated as described above or incubated at 95°C for 5 min to denature the substrate.
The transesterase activity of the protein was also evaluated in a series of biochemical activity assays. Previous studies have shown that TraI catalyzes the “nicking” of a plasmid containing its cognate oriT sequence (7, 26, 33). This reaction is not a classical endonuclease cleavage reaction but rather a transesterification in which TraI becomes covalently attached to the 5′ end of the cleaved DNA strand via a phosphotyrosyl linkage. This reaction does not require the participation of the other components of the relaxosome (F-plasmid-encoded TraY protein and IHF), provided the reaction is done at low ionic strength. Under these conditions, the plasmid must be negatively supercoiled (26), with negative supercoiling introducing sufficient ssDNA character to the DNA to allow binding and transesterification by TraI.
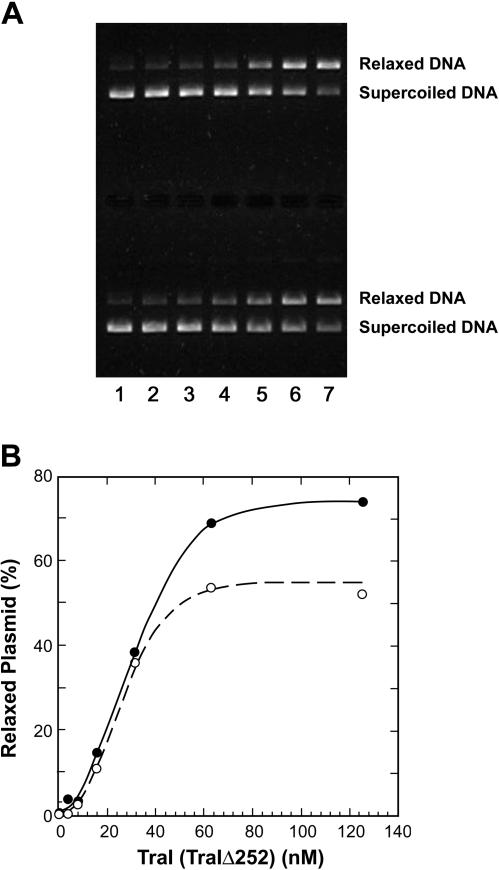
TraIΔ252 was compared with full-length TraI protein in a plasmid nicking assay. A titration of TraI (Fig. 4A upper) and TraIΔ252 (Fig. 4A, lower) clearly demonstrated that both proteins were capable of nicking a plasmid containing oriT at similar protein concentrations. The reaction was specific for DNA containing the F-plasmid oriT, since a plasmid lacking this region was not nicked by either protein (data not shown). Quantitation of the nicking activity (Fig. 4B) indicated that TraIΔ252 was as active as the full-length protein at low concentrations and only slightly less active at higher concentrations. The reduction in activity at higher protein concentrations may reflect a shift in the equilibrium of the transesterase reaction with the truncated protein at a high protein concentration. The transesterase reaction catalyzed by TraI is isoenergetic and reversible (7). One reason the nicking reaction fails to go to completion is the fact that the reverse reaction (ligation of the nicked strand) is in equilibrium with the forward (nicking) reaction. It is possible that this equilibrium is altered somewhat in reaction mixtures containing the TraIΔ252 protein, resulting in a lower fraction of the plasmid in the nicked form at higher protein concentrations. Nonetheless, it is clear the truncated protein retained essentially wild-type levels of relaxase activity and this activity is specific for the F-plasmid oriT.
FIG. 4.
Relaxase activity of TraI and TraIΔ252. (A) The relaxase activity assays using pBSoriT DNA were performed as described in Materials and Methods. A representative experiment is shown. The upper half of the gel represents a titration of TraI, and the lower half represents a titration of TraIΔ252. Lane 1, no-enzyme control; lane 2, 3.9 nM TraI (TraIΔ252); lane 3, 7.8 nM TraI (TraIΔ252); lane 4, 15.6 nM TraI (TraIΔ252); lane 5, 31.3 nM TraI (TraIΔ252); lane 6, 62.5 nM TraI (TraIΔ252); and lane 7, 125 nM TraI (TraIΔ252). (B) The gel shown in panel A was quantified as described previously (29) for TraI (•) and TraIΔ252 (○), and the data were plotted as a fraction of total plasmid in the relaxed species.
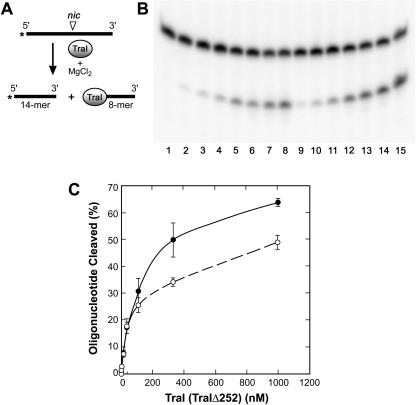
As indicated above, the nicking reaction is, in fact, a transesterification reaction that utilizes the -OH group on a specific tyrosine in the relaxase active site as a nucleophile to disrupt a specific scissile phosphodiester bond in oriT (7). To ensure that the nicking reaction observed with TraIΔ252 was the result of transesterification, two experiments were done. First, a cleavage assay (31) using a specific 22-nucleotide oligonucleotide that contains the 11-nucleotide sequence recognized by the transesterase active site of TraI was performed (Fig. 5A and B). The oligonucleotide was labeled on the 5′ end, and cleavage at the specific scissile phosphodiester bond releases a 14-nucleotide fragment. It is evident that both full-length TraI protein and TraIΔ252 cleave the oligonucleotide at the same location and with nearly equal efficiency. The 8-mer is covalently bound to TraI (TraIΔ252) as a result of the transesterification and is not observed since it is not radioactively labeled. Protein titrations suggest that the full-length protein is somewhat more active than TraIΔ252 at high protein concentrations (Fig. 5C). Again, this may be a manifestation of a shift in the equilibrium of the transesterase reaction.
FIG. 5.
Cleavage activity of TraI and TraIΔ252. (A) Schematic illustration of the oriT cleavage reaction catalyzed by TraI. The 22-nucleotide substrate was 5′-end labeled and contained the binding site recognized by TraI and the scissile phosphodiester bond (nic). The reaction requires the addition of MgCl2 and results in the release of a 5′-end-labeled 14-mer and an 8-mer that is covalently bound to TraI (TraIΔ252). (B) Cleavage assays were performed as described in Materials and Methods and were incubated for 20 min at 37°C prior to denaturation and resolution of products on a 16% denaturing polyacrylamide gel. Lane 1, no-protein control; lane 2, 1.7 nM TraI; lane 3, 4.1 nM TraI; lane 4, 12.3 nM TraI; lane 5, 37 nM TraI; lane 6, 111 nM TraI; lane 7, 333.3 nM TraI; lane 8, 1,000 nM TraI; lane 9, 1.7 nM TraIΔ252; lane 10, 4.1 nM TraIΔ252; lane 11, 12.3 nM TraIΔ252; lane 12, 37 nM TraIΔ252; lane 13, 111 nM TraIΔ252; lane 14, 333.3 nM TraIΔ252; and lane 15, 1,000 nM TraIΔ252. This is a representative experiment. (C) Quantitation of multiple (three to five) oligonucleotide cleavage assays using TraI (•) and TraIΔ252 (○) by PhosphorImager analysis using ImageQuant software.
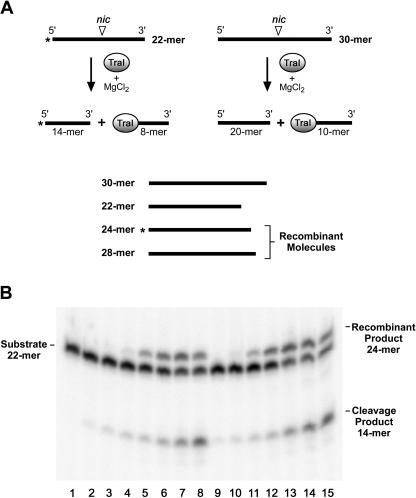
As indicated above, the transesterase reaction is reversible and a strand transfer reaction has been used to demonstrate this reversibility (31, 36). In this reaction, TraI protein is incubated with a 5′-end-labeled oligonucleotide containing the nic site (Fig. 6A; 22-mer) to allow cleavage to occur. Subsequently, an unlabeled oligonucleotide of different length that also contains the nic site (Fig. 6A; 30-mer) is added. The reversibility of the transesterase reaction is demonstrated by the formation of “recombinant” products when the labeled DNA fragment resulting from cleavage of the 22-mer (14-mer) is ligated to the 10-mer that is part of the covalent protein-DNA complex resulting from cleavage of the 30-mer. In the experiment shown in Fig. 6, the labeled oligonucleotide was 22 nucleotides in length, the unlabeled oligonucleotide was 30 nucleotides in length, and the expected recombinant product is 24 nucleotides in length. It is clear that both TraI protein and TraIΔ252 catalyze an efficient exchange reaction, as evidenced by the appearance of the expected recombinant product. We conclude that TraIΔ252 is a fully functional transesterase capable of both cleavage and ligation at the nic sequence.
FIG. 6.
Cleavage/ligation activity of TraI and TraIΔ252. (A) Schematic representation of the strand exchange assay. See text for details. (B) Strand exchange assays were performed as described in Materials and Methods. The substrate (22-mer) is indicated on the left, and the cleavage product (14-mer) and expected recombinant product (24-mer) are indicated on the right. Lane 1, no-protein control; lane 2, 1.7 nM TraI; lane 3, 4.1 nM TraI; lane 4, 12.3 nM TraI; lane 5, 37 nM TraI; lane 6, 111 nM TraI; lane 7, 333.3 nM TraI; lane 8, 1,000 nM TraI; lane 9, 1.7 nM TraIΔ252; lane 10, 4.1 nM TraIΔ252; lane 11, 12.3 nM TraIΔ252; lane 12, 37 nM TraIΔ252; lane 13, 111 nM TraIΔ252; lane 14, 333.3 nM TraIΔ252; and lane 15, 1,000 nM TraIΔ252. This is a representative experiment.
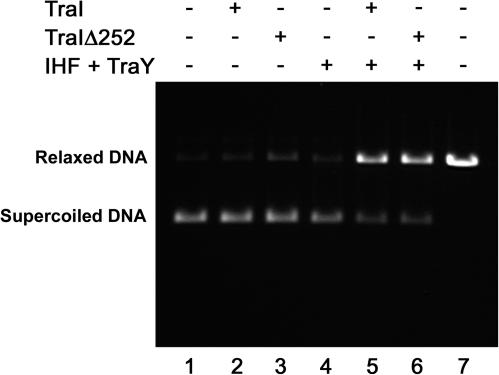
Finally, we tested the ability of TraIΔ252 to interact with the other components of the relaxosome in a relaxosome reconstitution experiment. Previous studies have shown that TraI assembles with the host-encoded IHF and the F-plasmid-encoded TraY to form a relaxosome at oriT (18, 29). Importantly, the addition of both IHF and TraY is required, and together these two proteins markedly stimulate the nicking reaction catalyzed by TraI. We considered the possibility that the defect in CDT observed with TraIΔ252 might be due to its inability to form a fully functional relaxosome with IHF and TraY. To test this directly, relaxosomes were assembled with TraI and TraIΔ252 and tested for nicking activity (Fig. 7). As expected, neither TraI nor TraIΔ252 exhibited significant nicking activity under the conditions of this assay (Fig. 7, lanes 2 and 3), which includes a significantly higher NaCl concentration than the relaxase assay shown in Fig. 4. The addition of both IHF and TraY markedly stimulated the TraI-catalyzed nicking reaction under these conditions (Fig. 7, lane 5). The addition of both IHF and TraY also stimulated the nicking reaction catalyzed by TraIΔ252 (Fig. 7, lane 6). We conclude that TraIΔ252 is fully capable of participating in the formation of an active relaxosome at the F-plasmid oriT.
FIG. 7.
TraIΔ252 is capable of assembling into a functional relaxosome. The relaxosome reconstitution experiments were performed as described in Materials and Methods with the indicated proteins at the following concentrations: TraI (TraIΔ252), 62.5 nM; IHF, 49.4 nM; and TraY, 170 nM. The positions of relaxed and supercoiled DNA are indicated on the left. Lane 7 contains a linear DNA marker that migrates just slightly faster than nicked DNA on an agarose gel.
DISCUSSION
Previous studies (6, 10, 33, 39) have shown that TraI is a bifunctional protein capable of catalyzing both a processive helicase reaction and a site- and strand-specific transesterase reaction. Both of these biochemical activities are critical for the process of DNA transfer, and the protein is fully functional only when both catalytic domains reside on the same polypeptide (6). The relaxase domain is wholly contained within the first ∼310 amino acids of the protein, and the recognized helicase-associated motifs are found in the region from amino acid 990 to amino acid 1450. Together, these two activities account for the major DNA processing events known to be required in CDT. However, while necessary for the DNA processing events associated with DNA transfer, these two activities are not sufficient and several other proteins, including IHF, TraY, TraM, and TraD, are known to have essential roles in the process (15, 44).
In addition to the two functional domains mentioned above, TraI has two regions that are not shared by all CDT initiator proteins and that may have a significant and specific functional role in DNA transfer. The central region of the protein, encompassing residues 310 to 990, has no well-characterized function but is known to be important for wild-type levels of DNA helicase activity (6). The functional role of the C-terminal 250 amino acids was explored in this study, and the results clearly indicate this region of TraI is essential for CDT. Deletion of the C-terminal domain eliminates DNA transfer while having little or no effect on either the relaxase or the helicase activity of TraI. We conclude that the protein lacking the C-terminal domain, TraIΔ252, is properly folded but lacks a region of the protein that is critical for some aspect of DNA transfer. Thus, this region of the protein represents a third functional domain that is essential for CDT but whose specific role in the events leading to DNA transfer remains to be fully elucidated.
The process of CDT is highly regulated, even in the derepressed F plasmid. Since the F plasmid is derepressed, due to an insertion in the finOP regulatory region (9, 43), all the components of the relaxosome are available in the cell at all times and it seems likely that the relaxosome is assembled at oriT in advance of the formation of a stable mating pair. It has been suggested that a “mating signal” triggers the relaxation of the conjugative plasmid in the cell upon formation of a stable mating pair (42). The nature of this mating signal and its propagation in the cell is unknown. The TraM protein, which is known from genetic studies to be required for DNA transfer yet has a poorly defined role in the process (5), may be a component of this mating signal. It is known that the tetrameric TraM binds DNA at multiple sites within oriT (4, 12, 14, 32, 35, 40). The two highest-affinity binding sites overlap the traM promoter and act to regulate the expression of TraM. The third, lower-affinity binding site is located upstream of the traM promoter, and TraM binding at this site is presumed to be important for the DNA transfer event. In addition, TraM has been demonstrated to interact with TraD, a protein involved in coupling the relaxosome to the secretion apparatus at the membrane (10). It is possible that TraM, bound at the low-affinity site within oriT, interacts specifically with TraI to trigger relaxation of the plasmid. Such an interaction would be essential for DNA transfer and could be mediated by the C-terminal domain of TraI. At this point, it is not known if F-TraI interacts with F-TraM. However, experiments using the F-plasmid-like plasmid R1 have demonstrated involvement of TraM in the formation of an active relaxosome (20).
An alternative, and not mutually exclusive, possibility envisions an interaction between TraI and TraD that involves the C-terminal domain of TraI. TraD is the F-plasmid-encoded representative of the TraG family of proteins that have been defined as conjugative coupling proteins (34). These proteins are thought to mediate an interaction between the relaxosome formed on oriT and the membrane-spanning protein complex responsible for translocation of DNA into the recipient cell. The F-plasmid-encoded TraD protein contains a C-terminal extension that is not present in several other members of the TraG family and that appears to be important for this interaction (34). The C-terminal extension on TraD renders the protein highly specific in its interaction with the F-plasmid relaxosome, and the full-length protein fails to support the transfer or mobilization of other conjugative plasmids. When the C-terminal extension is removed, the specificity is reduced and the truncated TraD is able to promote the transfer of other conjugative and mobilizable plasmids (34). It is tempting to speculate that there is an interaction between the C-terminal extension of TraI and the C-terminal extension on TraD that is, at least in part, responsible for the coupling of the F-plasmid relaxosome to the membrane-spanning complex in the cell envelop. If this were the case, then loss of either C-terminal extension would be expected to inhibit CDT. We have shown here that loss of the C-terminal extension on TraI does indeed abate DNA transfer. Previous studies (34) have shown that loss of the C-terminal extension on TraD also significantly reduces F-plasmid DNA transfer. Further studies will be required to determine if TraD and TraI do, in fact, interact and if the C-terminal extensions on each protein are responsible for that interaction.
In summary, the C-terminal region on F-plasmid-encoded TraI represents a third functional domain of the protein that is required for CDT. The relaxase domain and the helicase domain are necessary but not sufficient to support the DNA transfer event. We speculate that the C-terminal domain is a protein-protein interaction domain and that either TraM or TraD (or both) interacts with TraI to relax the plasmid in preparation for transfer of ssDNA and to mediate the interaction of the active relaxosome with the secretion apparatus at the cell envelop.
Acknowledgments
We thank Susan Whitfield for help with artwork preparation.
This work was supported by U.S. National Institutes of Health grant GM 61020 (to S.W.M.).
REFERENCES
- 1.Abdel-Monem, M., and H. Hoffmann-Berling. 1976. Enzymic unwinding of DNA. 1. Purification and characterization of a DNA-dependent ATPase from Escherichia coli. Eur. J. Biochem. 65:431-440. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Monem, M., H. Dürwald, and H. Hoffmann-Berling. 1976. Enzymic unwinding of DNA. 2. Chain separation by an ATP-dependent DNA unwinding enzyme. Eur. J. Biochem. 65:441-449. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Monem, M., G. Taucher-Scholz, and M. Klinkert. 1983. Identification of Escherichia coli DNA helicase I as the traI gene product of the F sex factor. Proc. Natl. Acad. Sci. USA 80:4659-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abo, T., S. Inamoto, and E. Ohtsubo. 1991. Specific DNA binding of the TraM protein to the oriT region of plasmid R100. J. Bacteriol. 173:6347-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achtman, M., N. Willetts, and A. J. Clark. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd, D. R., J. K. Sampson, H. M. Ragonese, and S. W. Matson. 2002. Structure-function analysis of Escherichia coli DNA helicase I reveals nonoverlapping transesterase and helicase domains. J. Biol. Chem. 277:42645-42653. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalyzed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 8.Cabezón, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein-family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 9.Cheah, K.-C., and R. Skurray. 1986. The F plasmid contains an IS3 insertion within finO. J. Gen. Microbiol. 132:3269-3275. [DOI] [PubMed] [Google Scholar]
- 10.Dash, P. K., B. A. Traxler, M. M. Panicker, D. D. Hackney, and E. G. Minkley. 1992. Biochemical characterization of Escherichia coli DNA helicase I. Mol. Microbiol. 6:1163-1172. [DOI] [PubMed] [Google Scholar]
- 11.Datta, S., C. Larkin, and J. F. Schildbach. 2004. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11:1369-1379. [DOI] [PubMed] [Google Scholar]
- 12.Di Laurenzio, L., L. S. Frost, and W. Paranchych. 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6:2951-2959. [DOI] [PubMed] [Google Scholar]
- 13.Disqué-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fekete, R. A., and L. S. Frost. 2002. Characterizing the DNA contacts and cooperative binding of F plasmid TraM to its cognate sites at oriT. J. Biol. Chem. 277:16705-16711. [DOI] [PubMed] [Google Scholar]
- 15.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 16.Grandoso, G., M. Llosa, J. C. Zabala, and F. de la Cruz. 1994. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur. J. Biochem. 226:403-412. [DOI] [PubMed] [Google Scholar]
- 17.Heilig, J. S., K. L. Elbing, and R. Brent. 1998. E. coli, plasmids and bacteriophages, p. 1.7.1-1.7.16.In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 18.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 270:28381-28386. [PubMed] [Google Scholar]
- 19.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 20.Kupelwieser, G., M. Schwab, G. Högenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 21.Lahue, E. E., and S. W. Matson. 1988. Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem. 263:3208-3215. [PubMed] [Google Scholar]
- 22.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 23.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 24.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz.. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 25.Llosa, M., F. X. Gömis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Matson, S. W., and B. S. Morton. 1991. DNA helicase I catalyzes a site and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 266:16232-16237. [PubMed] [Google Scholar]
- 27.Matson, S. W., W. C. Nelson, and B. S. Morton. 1993. Characterization of the reaction product of the oriT nicking reaction catalyzed by Escherichia coli DNA helicase I. J. Bacteriol. 175:2599-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matson, S. W., J. K. Sampson, and D. R. Byrd. 2001. F plasmid conjugative DNA transfer: the traI helicase activity is essential for DNA strand transfer. J. Biol. Chem. 276:2372-2379. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 30.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 31.Pansegrau, W., W. Schroder, and E. Lanka. 1993. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. USA 90:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penfold, S. S., J. Simon, and L. S. Frost. 1996. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol. Microbiol. 20:549-558. [DOI] [PubMed] [Google Scholar]
- 33.Reygers, U., R. Wessel, H. Müller, and H. Hoffman-Berling. 1991. Endonuclease activity of Escherichia coli DNA helicase I directed against the transfer origin of the F factor. EMBO J. 10:2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sastre, J. I., E. Cabezón, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab, M., H. Gruber, and G. Högenauer. 1991. The TraM protein of plasmid R1 is a DNA-binding protein. Mol. Microbiol. 5:439-446. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, J. A., and S. W. Matson. 1994. Escherichia coli DNA helicase I catalyzes a sequence specific cleavage/ligation reaction at the F plasmid origin of transfer. J. Biol. Chem. 269:26220-26226. [PubMed] [Google Scholar]
- 37.Sprague, G. F., Jr. 1991. Genetic exchange between kingdoms. Curr. Opin. Genet. Dev. 1:530-533. [DOI] [PubMed] [Google Scholar]
- 38.Street, L. M., M. J. Harley, J. C. Stern, C. Larkin, S. K. Williams, D. L. Miller, J. A. Dohm, M. E. Rodgers, and J. F. Schildbach. 2003. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta 1646:86-99. [DOI] [PubMed] [Google Scholar]
- 39.Traxler, B. A., and E. G. Minkley, Jr. 1988. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J. Mol. Biol. 204:205-209. [DOI] [PubMed] [Google Scholar]
- 40.Verdino, P., W. Keller, H. Strohmaier, K. Bischof, H. Lindner, and G. Koraimann. 1999. The essential transfer protein TraM binds to DNA as a tetramer. J. Biol. Chem. 274:37421-37428. [DOI] [PubMed] [Google Scholar]
- 41.Willetts, N., and J. Maule. 1979. Investigations of the F conjugation gene traI:traI mutants and lambdatraI transducing phages. Mol. Gen. Genet. 169:325-336. [DOI] [PubMed] [Google Scholar]
- 42.Willetts, N. S., and B. Wilkins. 1984. Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 48:24-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshioka, Y., H. Ohtsubo, and E. Ohtsubo. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 in F finO. J. Bacteriol. 169:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, et al. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 45.Zupan, J. R., and P. Zambryski. 1995. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 107:1041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]