Abstract
The AbsA1 sensor kinase and its cognate response regulator AbsA2 are important regulators of antibiotic synthesis in Streptomyces coelicolor. While certain point mutations in absA1 reduce or eliminate the synthesis of several antibiotics, null mutations in these genes bring about enhanced antibiotic synthesis. We show here that AbsA1, which is unusual in sequence and structure, is both an AbsA2 kinase and an AbsA2∼P phosphatase. The half-life of AbsA2∼P in solution is 68.6 min, consistent with a role in maintaining a relatively stable state of transcriptional repression or activation. We find that mutations in the absA locus that enhance antibiotic synthesis impair AbsA2 kinase activity and that mutations that repress antibiotic synthesis impair AbsA2∼P phosphatase activity. These results support a model in which the phosphorylation state of AbsA2 is determined by the balance of the kinase and phosphatase activities of AbsA1 and where AbsA2∼P represses antibiotic biosynthetic genes either directly or indirectly.
The streptomycetes are filamentous, spore-forming bacteria that produce a remarkable diversity of secondary metabolites. These compounds are not essential for viability, but as many of them possess antimicrobial activity they are presumed to confer a significant survival advantage in nature. These compounds have furthermore found numerous medical applications as antibiotics, chemotherapeutic agents, immune suppressants, and other drugs (11). Most streptomycetes produce multiple secondary metabolites. The model organism Streptomyces coelicolor, for example, produces the well-characterized antibacterial compounds actinorhodin, undecylprodigiosin, a calcium-dependent antibiotic (CDA) that is related to daptomycin (18), and methylenomycin (6). Actinorhodin and undecylprodigiosin are blue and red pigments, respectively, and can therefore serve as useful indicators for the onset of secondary metabolism. Antibiotic biosynthesis is carried out by groups of enzymes that are encoded by large gene clusters (5). The expression of the genes expressing these compounds is regulated in a complex manner that includes transcription factors specific to individual biosynthetic gene clusters (14, 29, 38) and others that appear to be master regulators controlling multiple gene clusters (6, 10, 12, 15, 19, 24).
The absA operon consists of the open reading frames (ORFs) absA1 (encoding a sensor kinase) and absA2 (encoding a response regulator). This locus was discovered through mutations in absA1 that severely compromised the production of the pigmented antibiotics (10). In contrast to the effect of these original alleles, strains bearing deletion mutations in either absA1 or absA2 produced enhanced levels of the antibiotics, suggesting that absA exerted negative control of the antibiotic biosynthetic genes (2, 10). Genetic experiments suggested that the phosphorylated form of AbsA2 carried out this negative regulation. For example, a mutation that changed the presumed site of AbsA1 autophosphorylation enhanced antibiotic synthesis, as did a mutation that changed the presumed site of phosphorylation in AbsA2 (2). In contrast, other mutations in absA1, including a leucine-to-arginine mutation at position 253, were characterized that had a strong inhibitory effect on antibiotic synthesis and these were interpreted as favoring the accumulation of AbsA2∼P (3). If correct, a mechanism where phosphorylation of AbsA2 serves to reduce or shut off antibiotic synthesis would be unusual: in most two-component systems, response regulator phosphorylation leads to activation of target genes (8, 16, 17, 30, 39). Alternatively, AbsA2∼P could activate transcription of a negative regulator of antibiotic synthesis.
The promoters bound by AbsA2 have not been identified. Transcript analysis demonstrated that expression of the act and red gene cluster-specific regulators actII-ORF4 and redD were enhanced in the absA deletion mutants and diminished in some of the absA1 inhibitory mutants—the structural genes in the act and red clusters were similarly affected by the absA mutants (1, 33). The expression of a putative CDA-specific regulatory gene called cdaR, which is also embedded in the CDA cluster, was not affected by absA mutations (33). During the sequencing of the S. coelicolor chromosome, it became apparent that the absA locus was embedded within the CDA gene cluster, suggesting that its primary role is to control production of this metabolite (2); indeed, strong effects of various absA mutations were observed at many CDA biosynthetic gene promoters (33). For example, the activity of the promoter that controls the production of the three peptide synthetases in the cluster was more active in absA deletion mutants and reduced in inhibitory absA mutants (33).
AbsA1 is an unusual sensor kinase. Typical sensor kinases have two transmembrane domains, one close to the amino terminus and one further into the protein, resulting in a topology that positions one major loop of the protein outside the cell where it can serve as a sensory domain. This topology positions the kinase domain and carboxy terminus inside the cell. In contrast, AbsA1 is predicted to have as many as five transmembrane domains, with four clustered close together between residues 29 and 162 and a fifth ending 82 amino acid residues from the C terminus (Fig. 1A). If AbsA1 has an extracellular sensory domain, it may therefore be at the carboxy terminus as the extracellular loops connecting the first and second pairs of transmembrane domains are predicted to be 33 amino acid residues and only 4 residues or smaller (34). Furthermore, AbsA1 appears to lack two of the conserved sequence motifs typically found in the catalytic domains of sensor kinases: while it has clear H, X, N, and G2 boxes, it lacks the so-called G1 and F boxes (37). These sequence peculiarities, the predicted topology, and the prediction that AbsA2 phosphorylation has a primarily negative influence on the transcription of antibiotic biosynthetic gene clusters suggest that AbsA1 may differ from other sensor kinases at the mechanistic level.
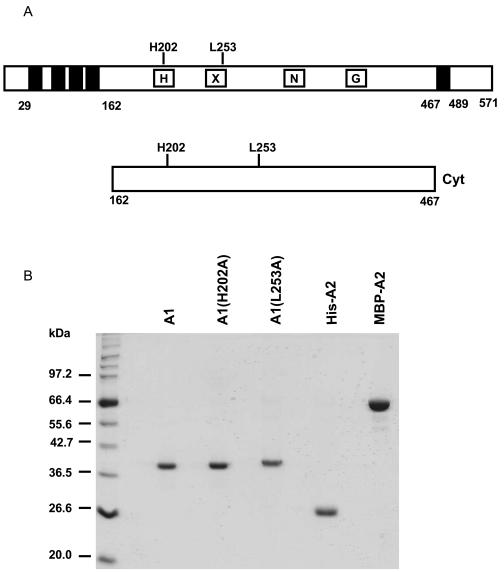
FIG. 1.
Proteins used in this work. (A) Schematic representation of AbsA1 (top) and the cytoplasmic portion of AbsA1 (bottom). The black boxes represent putative transmembrane domains. The site of autophosphorylation (H202), and a residue involved in phosphatase activity (L253) are indicated. (B) A total of 2 μg of each purified protein was resolved by SDS-10% PAGE and stained with Coomassie brilliant blue. The following abbreviations are used in the figures and figure legends: A1, AbsA1-cyt162-467; A1(H202A), AbsA-cyt162-467H202A; A1(L253A), AbsA1-cyt162-467L253A; His-A2, His6-AbsA2; MBP-A2, MBP-AbsA2.
We have tested several predictions concerning absA by reconstituting the function of AbsA1 and AbsA2 in vitro and correlating all the effects we observed in vivo. Our results are generally consistent with the view that AbsA2∼P is a negative regulator of the synthesis of the four S. coelicolor antibiotics, including CDA, although we found no evidence for direct interaction of this protein with any of three promoters in the CDA biosynthetic gene cluster.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. Escherichia coli strain XL-1 Blue was used to propagate all plasmids. Plasmids were demethylated in E. coli ET12567 and introduced into Streptomyces by protoplast transformation. E. coli strains BL21(DE3) and ER2058 were used for protein overexpression. E. coli strains were grown at 37°C in Luria-Bertani medium. Antibiotic concentrations used for plasmid selection were 100 μg of ampicillin/ml, 50 μg of kanamycin/ml, and 50 μg of apramycin/ml. S. coelicolor strains were grown in liquid YEME and on solid R2YE (26).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal met λ(DE3) | Novagen |
| ER2058 | F−ara-14 leuB6 fhuA2 Δ(argF-lac)U169 lacY1 lon::miniTn10(Tetr) glnV44 galK2 rpsL20(Strr) | New England Biolabs |
| ET12567 | xyl-5 mtl-5 Δ(malB. zjc::Tn5(Kanr) Δ(mcrC-mrr)HB101 dam dcm hsdS | 28 |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacqZΔM15 Tn10(Tetr)] | Stratagene |
| S. coelicolor | ||
| C530 | J1501 hisA1 uraA1 strA1 pgl absA1 Δ530 | 2 |
Plasmids, primers, and sequencing.
The plasmids used in this study are listed in Table 2. Primers were purchased from either Sigma Aldrich or the Mobix Laboratory at McMaster University. PCR was performed using VentR, DEEP VentR, and Pfu DNA polymerases (New England Biolabs and Stratagene). DNA sequencing was performed by the Mobix Laboratory.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Phenotypeb | Reference or source |
|---|---|---|---|
| S. coelicolor | |||
| pSET152 | lacZα MCS reppUCoriT fC31 int attP aac(IV)3 | Aprr | 7 |
| pAbsA1 | absA1 in pSET152 | Aprr | This work |
| pAbsA1(H202A) | absA1H202A in pSET152 | Aprr | This work |
| pAbsA1(L253A) | absA1L253A in pSET152 | Aprr | This work |
| E. coli | |||
| pBluescript II SK(+) | Phagemid cloning vector | Ampr | Stratagene |
| pPCR-Script Amp | Phagemid cloning vector; variant of pBluescript II SK(+) containing a Srfl site for insertion of blunt-end amplicons | Ampr | Stratagene |
| pET21-a(+) | Protein overexpression vector resulting in a C-terminal His6-tagged protein; T7 promoter | Ampr | Novagen |
| pET21A1 | absA1 cytoplasmic fragment in pET21-a(+) | Ampr | This work |
| pET21HA | absA1-cyt162-467H202A cytoplasmic fragment in pET21-a(+) | Ampr | This work |
| pET21LA | absA1-cyt162-467 L253A fragment in pET21-a(+) | Ampr | This work |
| pET28-a(+) | Protein overexpression vector resulting in a N-terminal His6-tagged protein; T7 promoter | Kanr | Novagen |
| pET28A2 | AbsA2 in pET28-a(+) | Kanr | This work |
| pMAL-c2X | Protein overexpression vector resulting in an N-terminal maltose binding protein fusion; Tac promoter | Ampr | New England Biolabs |
| pMALA2 | AbsA2 in pMAL-c2X | Ampr | This work |
MCS, multiple cloning site.
Antibiotic resistance markers are apramycin (Aprr), ampicillin (Ampr), and kanamycin (Kanr).
Construction of absA1 complementation plasmids pAbsA1, pAbsA1(H202A), and pAbsA1(L253A).
The 1.846-kbp absA1 gene, including the promoter region, was amplified by PCR with primers absA1-1 (5′ GAATTCGTGGCTCCTTCCAAAGTC 3′, with an EcoRI site at positions 1 to 6) and absA1-2 (5′ TCTAGAGCAGTACGCGAATCATCC 3′, with an XbaI site at positions 1 to 6). The absA1 fragment was ligated to pSET152 to generate pAbsA1. Site-directed mutagenesis was used to change histidine 202 and leucine 253 to alanine residues. The H202A (5′ CAC 3′ → 5′ GCC 3′) site-directed mutation was created with primers absA1H202A-1 (5′ CAGGATCGCGCAGGATATCGCCGACTCCCTG 3′) and absA1H202A-2 (5′ GAGGGAGTCGGCGATATCCTGCGCGATCCTG 3′). The L253A (5′ CTG 3′ → 5′ GCC 3′) site-directed mutation was created using primers absA1L253A-1 (5′CTGCACGAGGTGATCGGGGCCCTGCGGGAGGAC 3′) and absA1L253A-2 (5′ GTCCTCCCGCAGGGCCCCGATCACCTCGTGCAG 3′). Bold letters indicate altered base pairs. The template for the PCRs was pBluescriptAbsA1. Following the PCRs, DpnI was added to digest the parental DNA template. The sample was transformed into E. coli XL-1 Blue, and candidate DNA was isolated and sequenced. pBluescriptAbsA1H202A and pBluescriptAbsA1L253A were digested with EcoRI and XbaI and ligated to pSET152 to generate pAbsA1(H202A) and pAbsA1(L253A), respectively. The constructs were introduced into S. coelicolor strain C530 by protoplast transformation.
Construction of absA1-cyt486-1401 and absA2 overexpression vectors.
The 915-bp absA1 cytoplasmic gene fragment was amplified by PCR with primers absA1cyt-1 (5′ CCCGCATATGCCCCACGTTGCCC 3′, with a NdeI site at positions 5 to 10) and absA1cyt-2 (5′ CTCGAGCCGCCGGGCGTCCCGCCTGA 3′, with a XhoI site at positions 1 to 6) and ligated to pET21-a(+) to generate pET21A1. The 669-bp absA2 gene was amplified with primers absA2-1 (5′ CATATGATTCGCGTACTGCTCGC 3′, with an NdeI site at positions 1 to 6) and absA2-2 (5′AAGCTTCTAACGGTTGAGGTCGGCG 3′, with a HindIII site at positions 1 to 6) to generate fragment absA2-1. absA2 was also amplified with primers absA2-3 (5′ GAATTCATGATTCGCGTACTGCTCGC 3′, with an EcoRI site at positions 1 to 6) and absA2-2 to generate fragment absA2-2. The absA2-1 and absA2-2 fragments were ligated to pET28-a(+) and pMAL-c2X to generate pET28A2 and pMALA2, respectively.
Site-directed mutagenesis of absA1-cyt486-1401.
pBluescriptAbsA1H202A was used as the template in the PCR to change histidine 41 to alanine (5′ CAC 3′ → 5′ GCC 3′). Primers absA1cyt-1 and absA1cyt-2 were used to generate the absA1-cytH202A DNA fragment. This fragment was ligated to pET21-a(+) to generate pET21HA. Site-directed mutagenesis was used to change leucine 253 to an alanine residue (5′ CTG 3′ → 5′ GCC 3′). This mutation was created with primers absA1L253A-1 and absA1L253A-2, with pPCRA1 (AbsA1 in pPCR-Script AMP) as the template. pPCRA1L253A was digested with NdeI and XhoI and ligated to pET21-a(+) to generate pET21LA.
Overexpression of AbsA1-cyt162-467, AbsA1-cyt162-467H202A, AbsA1-cyt162-467L253A, and His6-AbsA2.
Plasmids pET21A1, pET21HA, and pET21LA were transformed into E. coli BL21(DE3). The purification procedure for all three of the proteins was similar. Following induction with 1 mM isopropyl-β-d-thiogalactopyranoside and growth for 3.5 h at 30°C, the two 1-liter cultures were harvested, washed with 0.85% saline, resuspended in lysis buffer A (50 mM phosphate buffer [pH 7.4], 0.5 M NaCl, and 10% glycerol plus 1× protease inhibitor cocktail [Roche]) and passed twice through a French pressure cell. The lysed cells were centrifuged, and the clarified lysate was applied to a 1-ml nickel sulfate column (Amersham Pharmacia) and washed with wash buffer A (50 mM phosphate buffer [pH 7.4], 0.5 M NaCl, 50 mM imidazole, 10% glycerol). The bound protein was eluted with elution buffer A (50 mM phosphate buffer [pH 7.4], 0.5 M NaCl, 0.8 M imidazole, 10% glycerol) with a gradient up to 100%. The eluted protein was dialyzed overnight into SP Sepharose wash buffer B (50 mM phosphate buffer [pH 7.2], 50 mM NaCl, 0.1 mM dithiothreitol [DTT], 10% glycerol) and further purified with either SP Sepharose resin (Amersham Biosciences) or a 1-ml prepacked SP Sepharose column (Amersham Pharmacia). A gradient of elution buffer B (50 mM phosphate buffer [pH 7.0], 1.0 M NaCl, 0.1 mM DTT, 10% glycerol) was used to elute the bound protein. The eluted protein was dialyzed into storage buffer A (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 0.1 mM DTT, 0.1 M NaCl, 30% glycerol) and concentrated. The above conditions were also used to purify His6-AbsA2 but with the following exceptions. The protein that eluted from the nickel column was dialyzed into Q Sepharose wash buffer C, which consists of 20 mM Tris-HCl (pH 7.8), 50 mM NaCl, 0.1 mM DTT, 0.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 10% glycerol, and further purified with Q Sepharose resin (Amersham Biosciences). A gradient of elution buffer C (20 mM Tris-HCl [pH 7.8], 2 M NaCl, 0.1 mM DTT, 0.5% CHAPS, 10% glycerol) was used to elute the bound protein. The eluted protein was dialyzed into storage buffer B (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 0.1 mM DTT, 0.3 M NaCl, 0.1% CHAPS, 30% glycerol) and concentrated. Protein concentrations were determined with the Bio-Rad protein assay dye reagent with bovine serum albumin as the standard. The protein was flash frozen in liquid nitrogen and stored at −80°C.
Overexpression of maltose binding protein (MBP)-AbsA2 fusion.
Plasmid pMALA2 was transformed into E. coil ER2058. Two 1-liter cultures were induced with 0.6 mM isopropyl-β-d-thiogalactopyranoside and grown for 22.5 h at 16°C. The cells were harvested by centrifugation, washed with 0.85% saline, and resuspended in column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) containing 1× protease inhibitor cocktail. The sample was passed twice through a French pressure cell, and the clarified lysate was applied to amylose resin (New England Biolabs). The bound protein was eluted with column buffer plus 10 mM maltose. The protein was diluted to one-quarter strength with 20 mM Tris-HCl (pH 7.8) and 10% glycerol to reduce the NaCl concentration, applied to Q Sepharose resin, washed with wash buffer D (20 mM Tris-HCl [pH 7.8], 50 mM NaCl, 0.1 mM DTT, 10% glycerol), and eluted with a gradient of elution buffer D (20 mM Tris-HCl [pH 7.8], 2 M NaCl, 0.1 mM DTT, 10% glycerol). Glycerol was added to 15%, and the protein was flash frozen in liquid nitrogen and stored at −80°C.
Alpha-chymotrypsin limited proteolysis.
A total of 8 μg of AbsA1-cyt162-467 was incubated in 85 μl of storage buffer with 0.4 μg of alpha-chymotrypsin (Miles Laboratories Pye, Ltd.) at room temperature for 2, 15, or 60 min. Proteolysis was stopped by the addition of 3× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Samples were resolved by 13% SDS- PAGE and visualized by Coomassie brilliant blue staining.
In vitro phosphorylation analysis. Twenty-microliter autokinase reactions were carried out in kinase buffer (50 mM Tris-HCl [pH 8.0], 2 mM DTT, 50 mM KCl, 10% glycerol, and 0.1 mM MgCl2) and included 0.9 μM AbsA1-cyt162-467, 60 μM ATP, and 0.022 μM (2 μCi) [γ-32P]ATP (4,500 Ci/mM, 10 mCi/ml; ICN). The reaction mixtures were incubated at 30°C for 30 min, and the reaction was stopped by the addition of 3× SDS-PAGE loading buffer plus DTT. Samples were resolved by SDS-10% PAGE, and the wet gels were exposed to X-Omat Blue Kodak film. The phosphotransfer reaction mixture was identical to the autokinase reaction mixture, except that 0.9 μM AbsA2 was present in the former.
In vitro dephosphorylation analysis. MBP-AbsA2 (1.8 μM) was phosphorylated by 1.8 μM AbsA1-cyt162-467L253A at 30°C for 30 min in a 1-ml phosphotransfer reaction mixture that included 11 nM ATP and 29 nM (132.5 μCi) [γ-32P]ATP (4,500 Ci/mM, 10 mCi/ml; ICN). A total of 450 μl of amylose resin resuspended in kinase buffer was added to the completed kinase reaction mixture and incubated on ice for 10 min. The amylose resin was centrifuged for 1 min at 14,000 rpm, and the supernatant was decanted. The resin was washed three times with 1.2 ml of kinase buffer and then resuspended in 400 μl of kinase buffer containing 10 mM maltose. Following a 10-min incubation on ice, the sample was pelleted and the supernatant was decanted. A total of 15 μl of the supernatant containing phosphorylated MBP-AbsA2 was added to 1.15 μM AbsA1-cyt162-467. The 24.3-μl reaction mixture was incubated at 30°C for 1, 4, 20, 60, or 300 min, and the reaction was stopped by the addition of 3× SDS-PAGE loading buffer plus DTT. The reactions were resolved by SDS-10% PAGE. Products were visualized by autoradiography and quantified by a PhosphorImager with ImageQuant software.
Cross-linking of AbsA1-cyt162-467 and AbsA2.
Prior to cross-linking, the proteins were exchanged into the cross-linking buffer (50 mM phosphate [pH 7.4], 0.3 M NaCl, 1 mM DTT, 1 mM EDTA, 20% glycerol). In a 20-μl reaction mixture, 5 μM of AbsA1-cyt162-467, His6-AbsA2, or AbsA1-cyt162-467 and His6-AbsA2 were incubated for 30 min at 30°C in the presence of 1,000 μM of dimethyl suberimidate. The reactions were stopped by the addition of 1 μl of 1 M Tris-HCl (pH 8) and resolved by SDS-10% PAGE. Products were visualized by silver staining.
RESULTS AND DISCUSSION
Effects of absA1 mutations in vivo.
To test and confirm previous genetic work on absA1, we introduced the absA1 ORF and promoter region into the single-copy integrating vector pSET152. Using site-directed mutagenesis, we constructed mutant absA1 alleles in which the presumed site of autophosphorylation, H202, and the residue L253, shown previously to be important for AbsA1 function, were changed to alanines.
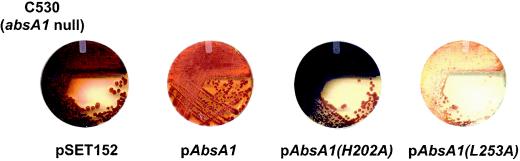
We introduced pSET152, pAbsA1, pAbsA1(H202A), and pAbsA1(L253A) into an in-frame absA1 deletion mutant constructed in the Champness laboratory to determine the effects of the mutations on function in vivo. The absA1 null mutant produced a greater overall amount of the pigmented antibiotics than congenic wild-type strain J1501 (2). As expected, introducing a wild-type allele reduced the level of the pigmented antibiotics considerably, relative to level in the strain containing the pSET152 control vector (Fig. 2). To our surprise, the strain containing the H202A allele of absA1 produced even more of the pigmented antibiotics than the strain that lacked a functional absA1 gene (Fig. 2). In contrast and consistent with previous work with an L253R mutant of absA1 (3), the L253A allele virtually eliminated production of the pigmented antibiotics. These data are generally consistent with the previously reported genetics of the absA genes and provided us with a basis for further biochemical experiments.
FIG. 2.
Effect of AbsA1 (pAbsA1), AbsA1H202A [pAbsA1(H202A)], and AbsA1L253A [pAbsA1(L253A)] on production of actinorhodin and undecylprodigiosin in the absA1 null C530.
Phosphorylation of AbsA2 by AbsA1-cyt162-467.
Genetic analysis of the absA genes strongly supports a model in which AbsA2∼P inhibits antibiotic synthesis (2); however, the signals and mechanisms that establish the level of phosphorylation of the protein in the cell are not well understood. To begin to address these questions, we reconstituted AbsA1 function in vitro and tested a number of the predictions concerning the most well-characterized absA1 mutants. We amplified an absA1 DNA fragment encoding the predicted intracellular domain of the protein, which we referred to as the cyt fragment (residues 162 to 467), and introduced it into pET21-a(+) to produce the expression plasmid pET21A1. We generated a mutant allele of absA1-cyt486-1401, changing the predicted site of AbsA1 autophosphorylation (H202A) in pET21A1 to produce the plasmid pET21HA. To follow up on our in vivo experiment with the absA1-L253A mutant, we also introduced an L-to-A mutation at the corresponding position in absA1-cyt486-1401 L253A to produce plasmid pET21LA. We also amplified the absA2 ORF and introduced it into vectors pET28-a(+) and pMAL-c2X to produce expression plasmids pET28A2 and pMALA2, respectively. The first of these fused absA2 to a His6 tag and the second fused it to a malE gene encoding the E. coli maltose binding protein. We introduced these expression vectors into E. coli expression strains and found that all wild-type and variant proteins were expressed in a soluble form and could be readily purified (Fig. 1B). We are not able to explain the slight reduction in mobility of AbsA1-cyt162-467L253A; however, all gene fragments used in this work have been sequenced end to end, so we are certain that the amino acid sequence is correct.
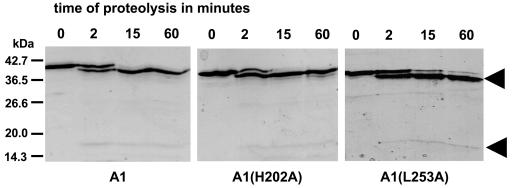
To compare the activities of AbsA1-cyt162-467, AbsA1-cyt162-467H202A and AbsA1-cyt162-467L253A in vitro, we needed to demonstrate that the mutations had not caused substantial protein misfolding. We therefore carried out the limited proteolysis experiment in which we incubated 0.4 μg of alpha-chymotrypsin with 8 μg of each protein for 2, 15, and 60 min and assessed the extent of proteolysis by SDS-PAGE. As shown in Fig. 3, all three proteins were extremely resistant to proteolysis and exhibited virtually identical cleavage patterns consistent with one principal region of cleavage. We concluded from this that the three proteins had folded in a similar and probably correct manner.
FIG. 3.
Limited proteolysis of A1, A1(H202A), and A1(L253A). Proteins were incubated with alpha-chymotrypsin for the indicated time in minutes, resolved by SDS-13% PAGE, and stained with Coomassie brilliant blue.
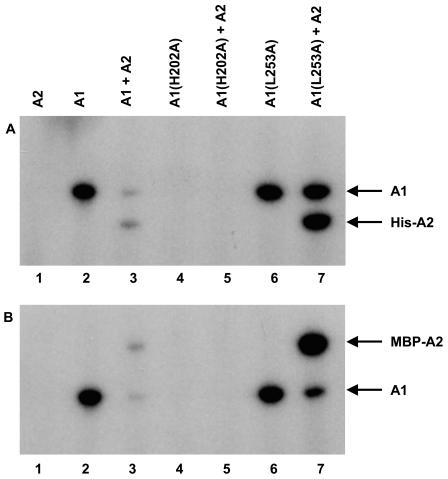
We then tested purified AbsA1-cyt162-467 for autophosphorylation activity and for phosphotransfer to AbsA2. We incubated 0.9 μM AbsA1-cyt162-467 with [γ-32P]ATP, ran the reaction products on SDS-PAGE, and located radiolabeled bands by autoradiography. As shown in Fig. 4, our purified AbsA1-cyt162-467 was able to autophosphorylate under these conditions (lane 2). To determine whether AbsA1-cyt162-467∼P could transfer phosphate to AbsA2, we incubated AbsA2 with AbsA1-cyt162-467 and [γ32P]ATP. AbsA2 was not labeled when incubated with ATP on its own but was labeled when AbsA1-cyt162-467 was present (lanes 1 and 3). The intensity of the AbsA1-cyt162-467 band was greatly reduced in the presence of AbsA2, consistent with direct phosphate transfer from AbsA1-cyt162-467 to AbsA2. We confirmed the specificity of this phosphorylation by incubating AbsA1-cyt162-467 with the unrelated response regulator RamR, showing that AbsA1-cyt162-467 did not phosphorylate this protein to any measurable extent (data not shown). As another test for the specificity of these reactions, we determined the acid-base sensitivity of AbsA1-cyt162-467∼P and AbsA2∼P. As expected, the phosphate in AbsA2∼P was hydrolyzed during incubation at low or high pH, consistent with phosphorylation of an aspartate residue while the phosphate in AbsA1-cyt162-467∼P was only hydrolyzed at low pH, consistent with phosphorylation of a histidine residue (36).
FIG. 4.
Autophosphorylation and phosphotransfer activity of A1, A1(H202A), and A1(L253A). A total of 0.9 μg of each protein was incubated with [γ-32P]ATP for 30 min at 30°C. The reaction products were resolved by SDS-10% PAGE and visualized by autoradiography.
Sequence alignments predicted that the histidine residue at position 202 in the full-length protein was the likely site of AbsA1 autophosphorylation, and genetic experiments have suggested that mutations altering this residue compromised the function of the absA genes in vivo. We therefore carried out an autophosphorylation reaction with purified AbsA1-cyt162-467 bearing an alanine residue at position 202 and found that this protein did not show any autophosphorylation activity under conditions in which the wild-type protein was active (Fig. 4, lane 4). To determine whether this histidine residue was required for phosphorylation of AbsA2 by AbsA1-cyt162-467, we carried out phosphotransfer reactions with the H202A variant with purified AbsA2. We found that the H202A mutation abolished phosphorylation of AbsA2 by AbsA1-cyt162-467 (lane 5). In sum, our results are consistent with H202 in AbsA1 serving as the site of autophosphorylation and confirm that this residue is necessary for normal phosphotransfer to AbsA2.
An absA1 allele that changed residue 253 from a leucine to an arginine caused greatly reduced antibiotic synthesis in vivo; this was interpreted as evidence for enhanced phosphorylation of AbsA2 in the presence of this allele (3). We found that changing the residue from a leucine to an alanine caused a similar change in antibiotic synthesis in vivo (Fig. 2) and pursued the L253A protein biochemically. If the reduction in antibiotic synthesis was due to enhanced AbsA2∼P levels in the cell, this could have been brought about either by more active AbsA1-cyt162-467 kinase activity or by compromised AbsA2∼P dephosphorylation activity. To determine whether either of these was the case, we purified the L253A variant of the AbsA1-cyt162-467 fragment and characterized its activity in vitro. When incubated with [γ-32P]ATP, the L253A variant (Fig. 4, lane 6, and Table 3) exhibited autophosphorylation that was similar to that of the wild-type protein (3.9% phosphorylated protein after electrophoresis versus 3.3% for the wild-type variant). This suggests that the L253 residue has little or no role to play in the autophosphorylation reaction of AbsA1. The L253A variant was also capable of carrying out phosphotransfer to AbsA2 and, consistent with the allele causing enhanced levels of AbsA2∼P in vivo, the extent of phosphorylation of the AbsA2 protein was approximately sixfold greater than that of the wild type (Fig. 4, lane 7, and Table 3).
TABLE 3.
The extent of phosphorylation of purified AbsA1-cyt162-467 and AbsA2a
| Lane | % AbsA1-cyt162-467 | % AbsA2 |
|---|---|---|
| 1 | NA | 0 |
| 2 | 3.3 | NA |
| 3 | 0.85 | 0.95 |
| 4 | 0 | NA |
| 5 | 0 | 0 |
| 6 | 3.9 | NA |
| 7 | 3.3 | 5.9 |
The data shown in Fig. 4 were the basis for this quantification. The duration of each reaction was 30 min.
We quantified the degree of phosphorylation of each protein in the reactions shown in Fig. 4 (Table 3 and Materials and Methods). By the time SDS-PAGE and autoradiography were completed, 3.3% of AbsA1-cyt162-467 and 3.9% of AbsA1-cyt162-467L253A were phosphorylated, suggesting that the kinase activities of the two proteins were quite similar. When phosphotransfer to AbsA2 was possible, reactions containing wild-type AbsA1-cyt162-467 yielded a total of 0.85% phosphorylated AbsA1-cyt162-467 and 0.95% phosphorylated AbsA2. The reaction mixture containing the AbsA1-cyt162-467L253A protein yielded a greater degree of phosphorylation of both proteins: 3.3% phosphorylated AbsA1-cyt162-467-L253A and 5.9% phosphorylated AbsA2. We carried out a number of experiments of this type in an effort to enhance the total yield of AbsA2∼P; however, we were never able to exceed this level. Note that these measurements were made several hours after the reaction had occurred. The initial phosphorylation state prior to electrophoresis must have been substantially higher.
Dephosphorylation of AbsA2∼P by AbsA1-cyt162-467.
To understand how absA controls antibiotic synthesis, it was necessary to determine whether AbsA1-cyt162-467 has an AbsA2∼P phosphatase activity and, if so, whether H202A or L253A altered this activity. In addition, we also needed to determine whether AbsA2 possessed autophosphatase activity. In some two-component systems, the sensor kinase acts as a phosphatase and dephosphorylates the response regulator whereas in others, removal of the phosphoryl group is either solely or partially dependent upon the autophosphatase activity of the response regulator (37). To address these questions, we needed to first establish an AbsA2∼P dephosphorylation assay; for this, we therefore took advantage of our purified MBP-AbsA2 fusion protein. We first confirmed that this protein could be phosphorylated by AbsA1-cyt162-467 and AbsA1-cyt162-467L253A. As shown in Fig. 4B, the proteins exhibited virtually identical capacities to phosphorylate the MBP fusion to AbsA2 as they did to the protein bearing the much smaller His6 fusion, demonstrating that the MBP moiety did not significantly alter the structure of AbsA2 in the fusion. Furthermore, none of the AbsA1-cyt162-467 proteins were able to phosphorylate an unrelated MBP fusion protein (data not shown), so we were confident that this was a specific reaction.
To establish the dephosphorylation assay, we used AbsA1-cyt162-467L253A to phosphorylate MBP-AbsA2 because it yielded the most highly phosphorylated product. The phosphorylated AbsA2 fusion was then separated from AbsA1-cyt162-467L253A by incubating the completed kinase reaction mixture with amylose resin. The MBP-AbsA2 fusion protein bound the resin efficiently as in the initial purification, whereas AbsA-cyt162-467L253A did not. The resin was then washed, and AbsA2∼P was eluted with kinase buffer containing 10 mM maltose. We confirmed that there was no detectable AbsA1-cyt162-467L253A in the eluate by running the protein from a nonradioactive trial experiment by SDS-PAGE and staining it with Coomassie brilliant blue.
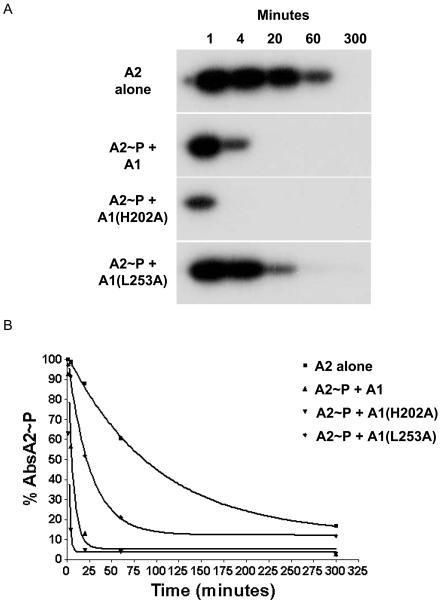
Once it was established that AbsA2∼P could be efficiently separated from AbsA1-cyt162-467L253A, we labeled MBP-AbsA2 with [γ-32P]ATP by AbsA1-cyt162-467L253A, purified the AbsA2 fusion as above, and incubated it on its own or in the presence of 1.15 μM AbsA1-cyt162-467, AbsA1-cyt162-467H202A, and AbsA1-cyt162-467L253A. As shown in Fig. 5A, the AbsA2 protein dephosphorylated spontaneously in solution with a half-life (t1/2) of 68.6 min. Acyl phosphates typically have half-lives of ∼5.0 h at pH 7 and 37°C (31), and many response regulators have an autophosphatase activity that greatly reduces the stability of the phosphorylated form of the protein. CheY∼P, for example, which regulates the direction of rotation of the E. coli flagellum, has a t1/2 of 20 s, consistent with the need for rapid changes in the direction of swimming during bacterial chemotaxis (27). In contrast, OmpR, which controls porin gene expression and must maintain stable patterns of gene expression unless there are changes in environmental osmolarity, has a t1/2 of 1 to 2 h (23). The half-life of AbsA2∼P was less than that of phosphoaspartate, suggesting that the protein has a modest autophosphatase activity, but the relative stability of the phosphorylated form of the protein is also consistent with the presumed role of the protein: maintaining an expression pattern for antibiotic biosynthetic genes until environmental or developmental conditions necessitate a change.
FIG. 5.
Phosphatase activity of A1, A1(H202A), and A1(L253A). (A) Phosphorylated MBP-A2 was incubated alone or in the presence of A1, A1(H202A), or A1(L253A) for the times indicated (in minutes). Reaction products were resolved by SDS-10% PAGE and visualized by autoradiography. (B) The dephosphorylation of MBP-A2 in the absence and presence of A1 is represented graphically.
When we incubated AbsA2∼P with 1.15 μM AbsA1-cyt162-467, we observed rapid dephosphorylation of the response regulator: under these conditions, the t1/2 of AbsA2∼P was reduced to 4.28 min. The H202A variant of AbsA1-cyt162-467 dephosphorylated AbsA2∼P even more rapidly, conferring a t1/2 of 1.25 min. As with all of the experiments in this work, we repeated this observation at least three times and found the effects of the H202 mutation on phosphatase activity to be highly reproducible. This difference cannot simply be due to a change in the balance between opposing phosphorylation and phosphatase reactions, because ATP was not added to these reactions. This could therefore suggest that a subtle structural shift brought about by the H202A mutation not only disallowed autophosphorylation and phosphotransfer to AbsA2 but also enhanced the phosphatase activity of AbsA1-cyt162-467. In contrast, altering the site of autophosphorylation usually reduces the phosphatase activity of the kinase, as is the case of EnvZ (21), or has no affect on this activity, as is the case for NtrB (25).
The L253A mutant caused a clear and reproducible defect in the AbsA1-cyt162-467 phosphatase activity compared to wild-type and AbsA1-cyt162-467-H202A phosphatase activity. The t1/2 of AbsA2∼P in the presence of this variant was 17.46 min or about four times slower than that of the wild-type protein. It was clear, however, that the protein retained residual activity; this may explain our inability to generate more than 6% phosphorylated AbsA2 in the reactions shown above (Table 3). In vivo, this change must shift the level of AbsA2∼P to favor the inhibition of antibiotic synthesis, since the kinase reaction of the protein is normal. We do not imagine that L253 plays a catalytic role in dephosphorylation of AbsA2∼P; it is difficult to imagine how a leucine side chain could do such a thing. Instead, we imagine that AbsA1 may shift its conformation when switching between the two activities and that the L253A mutation may inhibit the shift to the dephosphorylation conformation. This would have to be a rather subtle rearrangement, as there was no obvious difference between the sensitivities of AbsA1-cyt162-467 and the two AbsA1-cyt162-467 variants to proteases (Fig. 3). Interestingly, L253 is located in the putative X region of AbsA1, a sequence motif that has been the target of mutations that reduced phosphatase activity in other sensor kinases such as EnvZ (20) and NtrB (4).
absA1-cyt486-1401 mutations have no effect on the stability of AbsA1-cyt162-467-AbsA2 complexes.
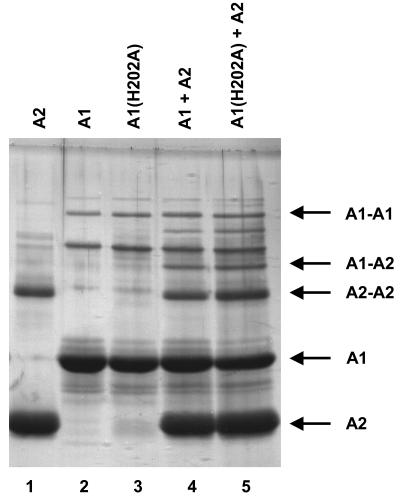
The fact that the H202A variant of AbsA1-cyt162-467 possesses a functional AbsA2∼P phosphatase activity could explain the effect of the H202A allele on pigment production in an absA1 null strain provided that there is an AbsA1-independent mechanism for phosphorylating AbsA2 in vivo. Alternatively, perhaps AbsA2 possesses a reduced affinity for its target promoters such that it is still able to exert some effect on transcription even in the unphosphorylated form. In this scenario, the effect of the H202A allele might be to bind up AbsA2 in stable complexes that are normally separated as a result of phosphorylation. To test this, we carried out a chemical cross-linking experiment to explore the interactions of AbsA1-cyt162-467 and AbsA2 (Fig. 6).
FIG. 6.
Cross-linking of A1, His-A2, and A1 plus His-A2. AbsA2 (5 μM) alone (lane 1), AbsA1-cyt162-467 alone (lane 2), AbsA1-cyt162-467H202A alone (lane 3), AbsA1-cyt162-467 plus AbsA2 (lane 4), or AbsA1-cyt162-467H202A plus AbsA2 (lane 5) were incubated in the presence of dimethyl suberimidate for 30 min at room temperature, resolved by SDS-10% PAGE, and visualized by silver staining.
We first found that both AbsA1cyt162-467 and AbsA2 form cross-linked species in the absence of other proteins (Fig. 6, lanes 1 and 2), suggesting that both form dimers or other higher-order complexes. This is the norm for most sensor kinases and response regulators (13, 32). The H202A mutation had no effect on the capacity of AbsA1-cyt162-467 to form a complex with itself (lane 3). When cross-linking was carried out in the presence of both proteins, we observed AbsA1-cyt162-467-AbsA2 species in addition to the two homodimeric species (lanes 4 and 5). The formation of these three complexes was not affected by the inclusion of ATP in the reactions (data not shown), suggesting that phosphorylation does not have a major effect on the oligomeric state of either protein. These data suggest that the effect of the H202A allele on pigment production in the absA1 null mutant (Fig. 2) cannot be explained simply by AbsA2 sequestration by AbsA1. We suggest therefore that the effect may be due to the presence of an alternative AbsA2 phosphorylating activity in S. coelicolor and that the H202A variant of AbsA1 accordingly lowers the level of AbsA2∼P in cells, thereby increasing pigment production over and above that of the absA1 deletion. Many response regulators can be phosphorylated by acetyl phosphate or other low-molecular-weight phosphodonors in vitro; for some this may occur in vivo as well (9, 22, 35). The enhanced pigment production by the absA1 null mutant containing the absA1-H202A allele may therefore be due to the dephosphorylation of AbsA2∼P that had been modified by acetyl phosphate or some other molecule.
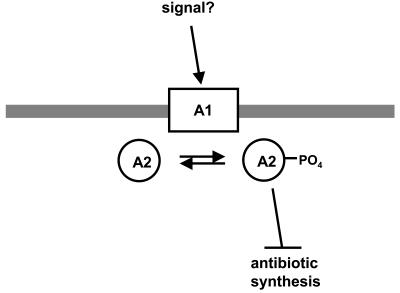
In sum therefore, these experiments demonstrated that AbsA1 possesses both kinase and phosphatase activities that are specific for AbsA2. The concordance between the effects of the H202 and L253 mutations in vivo are supportive of the hypothesis for absA function in which phosphorylation of AbsA2 represses transcription of antibiotic biosynthetic genes (Fig. 7). We assume that some kind of extracellular or perhaps intracellular signaling molecule controls the kinase and phosphatase activities of AbsA1 in vivo; however, the nature of this mechanism is unknown at present.
FIG. 7.
AbsA1 responds to unknown cellular or environmental cues by phosphorylating AbsA2. AbsA2∼P then inhibits antibiotic synthesis by interacting with the promoters of currently unknown genes.
Finally, transcript analysis carried out in the Champness laboratory suggested a number of possible genes that might be targets of AbsA2. The promoters upstream of absA1 and the CDA peptide synthetase genes, for example, appeared to be up- and down-regulated, respectively, by AbsA2∼P (33). The gene encoding a putative cluster-specific regulator or SARP, cdaR, appeared to be expressed independently of absA. We amplified DNA fragments containing these three promoters, labeled them with [α-32P]dATP, and attempted mobility shifts with purified AbsA2. Surprisingly, we did not observe any direct interactions between any of these DNA fragments using either the His6-AbsA2 or the MBP-AbsA2 fusion proteins. Phosphorylating AbsA2 did not change this outcome. This result could be due to the fact that we could not phosphorylate all of the AbsA2 in vitro or due to spontaneous dephosphorylation of AbsA2 during electrophoresis. If so, the interactions would have to be entirely dependent on the phosphorylation state of AbsA2. It is also possible that absA, although located in the CDA biosynthetic gene cluster, acts on antibiotic synthesis indirectly or that AbsA2 binds DNA cooperatively with another protein—an obvious candidate being CdaR. We are currently testing these hypotheses.
Acknowledgments
We are especially grateful to Wendy Champness for providing bacterial strains and for critical appraisal of the manuscript. We also thank Eric Brown for helpful suggestions and comments. We thank Tamara O'Connor for providing purified RamR protein for control experiments and for useful suggestions during the work.
N.L.S. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship, and J.R.N. was supported by a New Investigator award and operating grant MOP-57684 from the Canadian Institutes for Health Research.
REFERENCES
- 1.Aceti, D. J., and W. Champness. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulation by the absA and absB loci. J. Bacteriol. 180:3100-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, T., P. Brian, and W. Champness. 2001. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol. Microbiol. 39:553-556. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, T., P. Brian, P. Riggle, R. Kong, and W. Champness. 1999. Genetic suppression analysis of non-antibiotic-producing mutants of the Streptomyces coelicolor absA locus. Microbiology 145:2343-2353. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson, M. R., and A. J. Ninfa. 1992. Characterization of Escherichia coli glnL mutations affecting nitrogen regulation. J. Bacteriol. 174:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. D. Bentley, K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bibb, M. 1996. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. [DOI] [PubMed] [Google Scholar]
- 7.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 8.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 9.Bouché, S., E. Klauck, D. Fischer, M. Lucassen, K. Jung, and R. Hengge-Aronis. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27:787-795. [DOI] [PubMed] [Google Scholar]
- 10.Brian, P., P. J. Riggle, R. A. Santos, and W. C. Champness. 1996. Global negative regulation of Streptomyces coelicolor antibiotic synthesis is mediated by an absA-encoded putative signal transduction system. J. Bacteriol. 178:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champness, W. 2000. Actinomycete development, antibiotic production, and phylogeny: questions and challenges, p. 11-31. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 12.Champness, W., P. Riggle, T. Adamidis, and P. Vandervere. 1992. Identification of Streptomyces coelicolor genes involved in regulation of antibiotic synthesis. Gene 115:55-60. [DOI] [PubMed] [Google Scholar]
- 13.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Moreno, M. A., A. J. Martin-Triana, E. Martinex, J. Niemi, H. M. Kieser, D. A. Hopwood, and F. Malpartida. 1992. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J. Bacteriol. 174:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forst, S., J. Delgado, A. Rampersaud, and M. Inouye. 1990. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J. Bacteriol. 172:3473-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadner, R. 1995. Expression of the Uhp sugar-phosphate transport system of Escherichia coli, p. 263-274. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 18.Hojati, Z., C. Milne, B. Harvey, L. Gordon, M. Borg, F. Flett, B. Wilkinson, P. J. Sidebottom, B. A. Rudd, M. A. Hayes, C. P. Smith, and J. Micklefield. 2002. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 9:1175-1187. [DOI] [PubMed] [Google Scholar]
- 19.Hong, S. K., M. Kito, T. Beppu, and S. Horinouchi. 1991. Phosphorylation of the AfsR product, a global regulatory protein for secondary-metabolite formation in Streptomyces coelicolor A3(2). J. Bacteriol. 173:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsing, W., F. D. Russo, K. K. Bernd, and T. J. Silhavy. 1998. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 180:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsing, W., and T. Silhavy. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchings, M., and M. Buttner. Personal communication.
- 23.Igo, M. M., A. J. Ninfa, J. B. Sock, and T. J. Silhavy. 1989. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 3:1725-1734. [DOI] [PubMed] [Google Scholar]
- 24.Ishizuka, H., S. Horinouchi, H. M. Kieser, D. A. Hopwood, and T. Beppu. 1992. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J. Bacteriol. 174:7585-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamberov, E. S., M. R. Atkinson, P. Chandran, and A. J. Ninfa. 1994. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. 269:28294-28299. [PubMed]
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 27.Lukat, G. S., B. H. Lee, J. M. Mottonen,A. M. Stock, and J. B. Stock. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 266:8348-8354. [PubMed] [Google Scholar]
- 28.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Narva, K. E., and J. Feitelson. 1990. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J. Bacteriol. 172:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninfa, A. J., and B. Magasanik. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Post, R. L., and S. Kume. 1973. Evidence for an aspartyl phosphate residue at the active site of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 248:6993-7000. [PubMed] [Google Scholar]
- 32.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626-633. [DOI] [PubMed] [Google Scholar]
- 33.Ryding, J., T. Anderson, and W. Champness. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184:794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheeler, N., and J. Nodwell. Unpublished data.
- 35.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sickmann, A., and H. E. Meyer. 2001. Phosphoamino acid analysis. Proteomics 1:200-206. [DOI] [PubMed] [Google Scholar]
- 37.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 38.White, J., and M. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, G. D., T. R. Holman, and C. T. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]