Abstract
We provide in this paper a detailed characterization of the human peripheral CD4+ CD127lowCD25+ regulatory T cell (Treg) compartment, with a particular emphasis in defining the population expressing higher levels of the IL-6 receptor (IL-6R). We provide a description of the phenotype of this population by assessing both the surface expression by flow cytometry as well as their transcriptional profile and functional features. In addition, we also present functional data describing the responsiveness of these subsets to IL-6 signalling in vitro and to IL-2 in vivo. The data presented in this paper support the research article “Human IL-6RhiTIGIT− CD4+CD127lowCD25+ T cells display potent in vitro suppressive capacity and a distinct Th17 profile” (Ferreira RC et al., 2017; doi: 10.1016/j.clim.2017.03.002) [1].
Keywords: IL-6 receptor, Regulatory T cells, Immunophenotyping, Human immunology
Specifications Table
| Subject area | Biology |
| More specific subject area | Human Immunology |
| Type of data | Tables (x3) and Figures (x12) |
| How data was acquired | Flow cytometry (Fortessa; BD biosciences); Gene expression profiling (NanoString Technologies) |
| Data format | Analyzed flow cytometry files and normalized gene expression counts (from Nanostring) |
| Experimental factors | Flow cytometry and gene expression profiling was performed in freshly isolated PBMCs or CD4+ T cells. Cytokine production was assessed following in vitro stimulation with PMA + ionomycin. Cell proliferation was assessed by flow cytometry by culturing cells in vitro with anti-CD3/anti-CD28 stimulation. |
| Experimental features | Delineation of the Treg compartment was performed in human peripheral blood cells using polychromatic flow cytometry. The global transcriptional profile of the assessed T cell subsets was assessed in sorted cells isolated ex vivo. |
| Data source location | Samples from human volunteers and T1D patients were collected in Cambridge, UK. |
| Data accessibility | All primary non-clinical data are available in this article. The DILT1D data from individuals prior to normalization as a group are available, however they cannot be anonymized sufficiently to be able to put into the public domain without risk of participant identification. Data are available on request, through the Cambridge University institutional repository (DOI link:https://doi.org/10.17863/CAM.832). |
| Related research article | The data presented in this paper support the research article “Human IL-6RhiTIGIT−CD4+CD127lowCD25+T cells display potent in vitro suppressive capacity and a distinct Th17 profile” (Ferreira RC et al., 2017)[1]. |
Value of the data
-
•
Data provide a detailed description of the human peripheral Treg compartment at both the protein and transcriptional level.
-
•
The flow cytometry data provides a valuable resource for other researchers to compare the expression levels of a number of classical Treg surface markers within the defined CD4+ T cell populations defined here.
-
•
The detailed information on the flow cytometry antibodies and polychromatic panel combinations will be a valuable tool for researchers in the field to help design their specific immunostaining panels.
-
•
Cell subset heterogeneity is one of the main challenges in the description of human Treg populations, and is clearly highlighted in the data presented in this paper.
-
•
Data provide precise quantitative information regarding the expression levels of a broad range of immune genes at the mRNA level in highly purified human Treg subsets both ex vivo and after in vitro stimulation.
1. Data
The dataset contained in this article provides a detailed characterization of the expression of the IL-6 receptor (IL-6R) on circulating human CD4+ CD127lowCD25high T cells ex vivo. These data also provide a functional characterization of the assessed T cell subsets, with a particular emphasis on a subset of IL-6Rhi regulatory T cells (Tregs) lacking the expression of the co-inhibitory receptor TIGIT. The Fig. 1 and Fig. 2 depict the delineation of the assessed immune subsets and their responsiveness to IL-6 signalling. Fig. 3 depicts data from a clinical study investigating the responsiveness of the assessed T cells subsets to IL-2 signalling in vivo. Fig. 4 depicts the proliferative capacity of the Treg subsets in vitro, in the absence of exogenous IL-2. Fig. 5 and Fig. 6 depict the expression at the protein level of Th17 surface markers. Fig. 7 depicts the differential mRNA expression of 579 immune genes between IL-6RhiTIGIT− and IL-6RhiTIGIT+ Tregs. Fig. 8, Fig. 9 and Fig. 10 depict the immunophenotyping of the Th17 transcription factor RORγt, and different tissue-homing receptors at the protein level. Fig. 11 depicts the expression of two cytokines, IL-17 and IL-10, in TIGIT+ Tregs and Fig. 12 depicts the variation of HELIOS-TIGIT- and HELIOS+TIGIT+ Tregs measured by intracellular flow cytometry on cryopreserved peripheral blood mononuclear cells (PBMCs) on 8 selected patients from the DILT1D clinical study. Table 1 provides the complete information on the fluorochrome-conjugated antibody panels used in tis study, and Table 2 and Table 3 contain the complete gene expression data of 579 immune genes on the assessed T cell subsets, obtained in ex vivo-isolated cells or following in vitro stimulation, respectively.
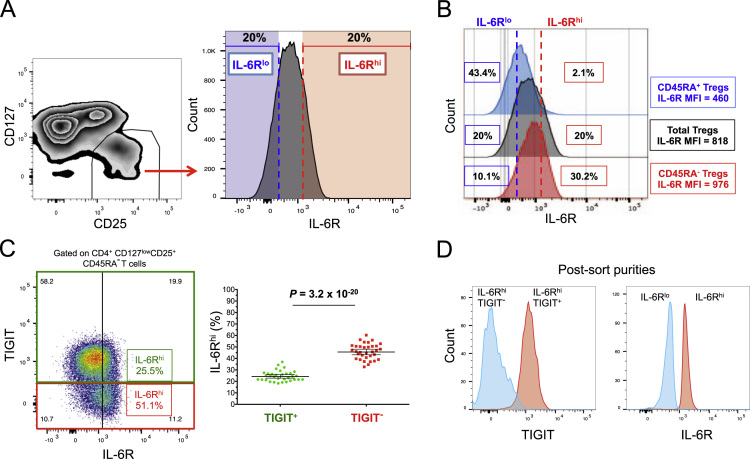
Fig. 1.
Gating strategy for the delineation of IL-6Rhi CD127lowCD25+ T cells (Tregs). (A) Gating strategy for the delineation of IL-6Rlo and IL-6Rhi CD127lowCD25+ Tregs, defined as the lower and upper 20th percentile, respectively, of the IL-6R mean fluorescence intensity (MFI) distribution in total CD127lowCD25+ Tregs. (B) Histograms depict the distribution of IL-6R expression and the respective frequency of IL-6Rlo and IL-6Rhi cells among: (i) CD45RA+ naïve (depicted in blue); (ii) total (depicted in black); and (iii) CD45RA− memory (depicted in red) CD127lowCD25+ Tregs. Frequencies shown in the figure represent the average frequency of the IL-6Rlo and IL-6Rhi cells in the three assessed Treg subsets from 33 healthy donors. (C) Data shown depict the expression of TIGIT versus IL-6R in one illustrative donor and the frequency (GeoMean +/− 95% CI) of IL-6Rhi cells in the TIGIT+ (depicted in green) and TIGIT− (depicted in red) subsets of CD127lowCD25+CD45RA− mTregs in 33 healthy donors. P value was calculated using a two-tailed paired non-parametric Wilcoxon signed rank test, comparing the frequency of IL-6Rhi cells in the TIGIT+ and TIGIT− subsets. (D) Histograms illustrate the distribution of the two sorting markers TIGIT and IL-6R following flow-cytometric sorting.
Fig. 2.
Memory Tregs are sensitive to IL-6 signalling in vitro. (A) Gating strategy for the delineation of the CD45RA− memory T effector (Teff) and Treg subsets. (B) Frequency of pSTAT3+ cells following stimulation of freshly isolated PBMCs with 0, 1 or 10 ng/ml of IL-6. Intracellular levels of pSTAT3 were measured by flow cytometry in CD4+ memory Teffs (depicted in black) and in CD4+ memory Tregs (depicted in red) from three healthy volunteers. (C) Histograms depict the distribution of the pSTAT3 mean fluorescence intensity (MFI) in memory Teffs (top panel) and memory Tregs (bottom panel) in response to stimulation with 0, 1 or 10 ng/ml of IL-6 in one illustrative donor.
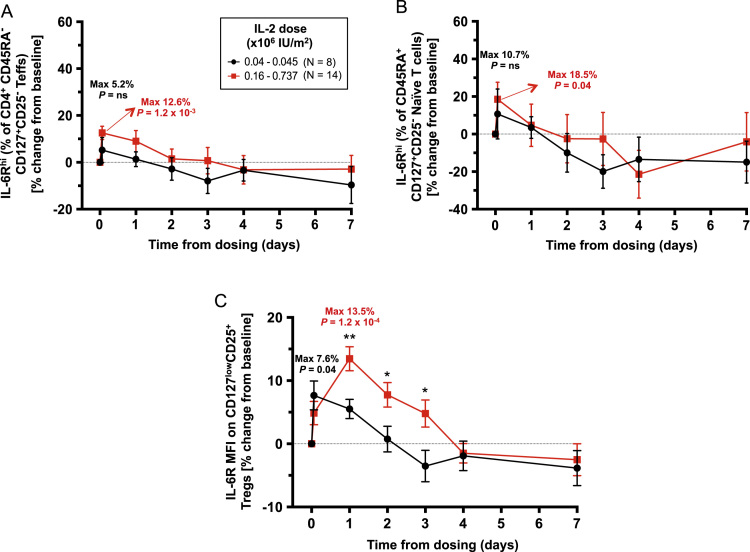
Fig. 3.
Effect of single-dose IL-2 treatment on the expression of IL-6R on the CD4+ T cell compartment in vivo. (A,B) Data shown depict the variation (Mean +/− SEM) of the frequency of IL-6Rhi cells on: (i) CD127+CD25− CD45RA− Teffs (A); and (ii) IL-6Rhi CD127+CD25− CD45RA+ naïve T cells (B) compared to the pre-treatment baseline, following IL-2 treatment in 22 T1D patients enrolled in the “Adaptive study of IL-2 dose on regulatory T cells in type 1 diabetes” (DILT1D) [2], [3]. Median pre-treatment baseline frequencies of IL-6Rhi cells were 39.3% (range: 21.5–59.9%) and 5.39% (range: 1.49–30.4%) within CD4+ CD45RA− Teffs and CD4+ CD45RA+ naïve T cells, respectively. (C) Data shown depict the variation (Mean +/− SEM) of the IL-6R mean fluorescence intensity (MFI) levels on the surface of total CD127lowCD25+ Tregs compared to the pre-treatment baseline MFI levels (median = 600; range: 392–836). Patients were stratified based on whether they received: (i) the lower IL-2 doses of 0.04–0.045 x 106 U/ml (N = 8; depicted in black); or (ii) the higher IL-2 doses of 0.16–0.737×106 U/ml (N = 14; depicted in red). The MFI cuttoff to define IL-6Rhi cells was the same as the one used for the Treg subset (detailed in Fig. 1). The maximum increases over the baseline pre-treatment frequencies achieved during the course of the study are indicated for each IL-2 dosing group. P values for the maximum increase in the frequency of the assessed parameter in response to a single dose of IL-2 was calculated using a two-tailed paired non-parametric Wilcoxon signed rank test comparing the frequencies observed at the timepoint where the maximal increase was achieved with the respective baseline pre-treatment frequencies. P values for the IL-2 dose-dependent effects were calculated using a two-tailed non-parametric a two-tailed non-parametric Mann-Whitney test comparing the frequency of IL-6Rhi cells between the low and high dose groups at each timepoint. The DILT1D data from individuals prior to normalization as a group are available, however they cannot be anonymized sufficiently to be able to put into the public domain without risk of participant identification. Data are available on request, through the Cambridge University institutional repository (DOI link: https://doi.org/10.17863/CAM.832). *P < 0.05; **P < 0.01; ns = not significant.
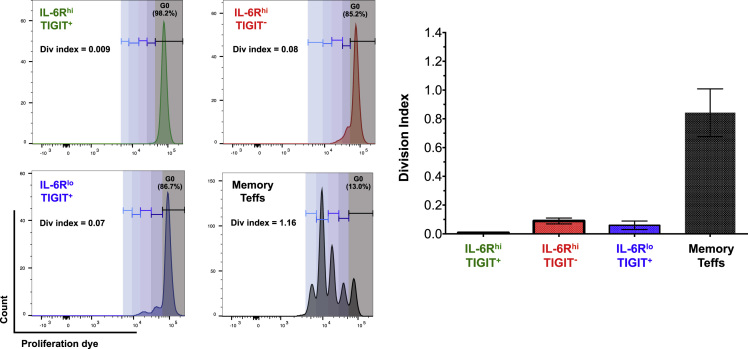
Fig. 4.
Proliferative capacity of IL-6RhiTIGIT− mTregs is dependent on exogenous IL-2. Proliferative capacity of sorted (i) IL-6RhiTIGIT+ (depicted in green), (ii) IL-6RhiTIGIT− (depicted in red), (iii) IL-6RloTIGIT+ (depicted in blue) mTregs, and (iv) CD127+CD25−CD45RA− Teff cells (depicted in black) was assessed in response to in vitro stimulation with anti-CD3/CD28 beads, in the absence of exogenous IL-2. Data (mean +/− SEM) were obtained from cells sorted from three independent donors. Proliferation and suppressive capacity were calculated using the Division Index in FlowJo, setting 0% suppression as the condition with the respective Teffs cultured in the absence of Tregs.
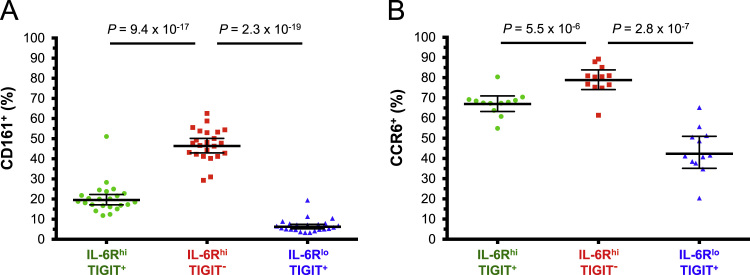
Fig. 5.
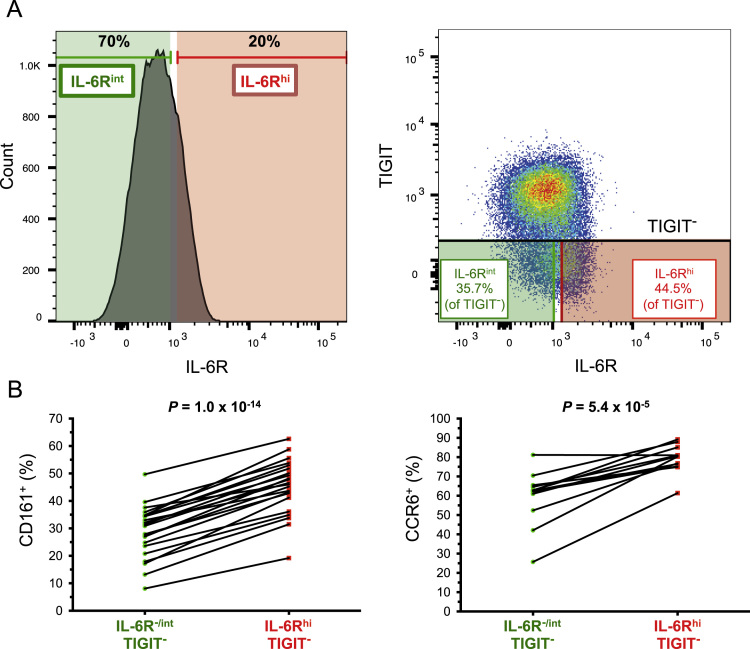
CD161 and CCR6 expression is increased on IL-6RhiTIGIT− mTregs. (A,B) Frequency (GeoMean +/− 95% CI) of CD161 (N = 23) (A) and CCR6 (N = 12) (B) was assessed by flow cytometry in freshly isolated PBMCs from healthy donors. Data were stratified according to the three assessed mTreg subsets: IL-6Rlo (depicted in blue), IL-6RhiTIGIT+ (depicted in green) and IL-6RhiTIGIT− (depicted in red). P values were calculated using a two-tailed paired non-parametric Wilcoxon signed rank test.
Fig. 6.
Elevated IL-6R expression delineates a subset of TIGIT− mTregs with increased expression of the canonical Th17 markers CD161 and CCR6. (A) Gating strategy to delineate IL-6R−/int (depicted in green) and IL-6Rhi (depicted in red) TIGIT− mTregs. (B) Data shown depict the frequencies of the Th17 markers CD161 (N = 23) and CCR6 (N = 12) in IL-6R−/int and IL-6Rhi mTregs, obtained by flow cytometry in freshly isolated PBMCs from healthy donors. P values were calculated using a two-tailed paired non-parametric Wilcoxon signed rank test.
Fig. 7.
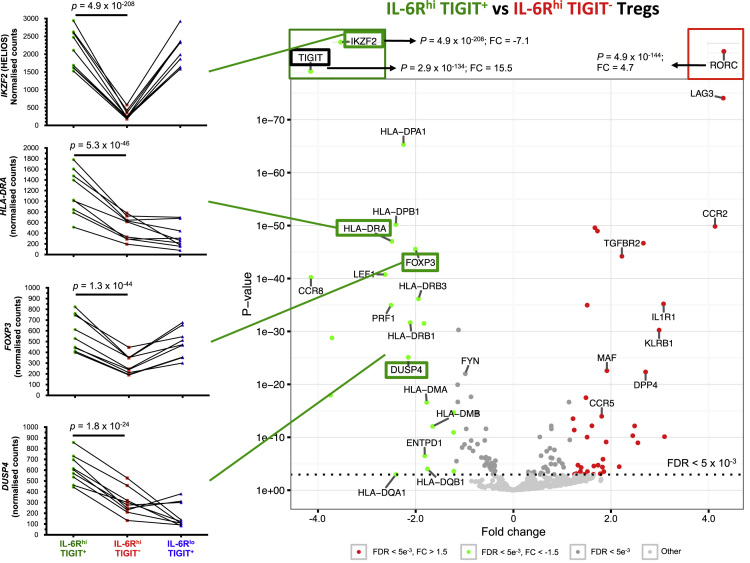
ex vivo isolated IL-6RhiTIGIT+ mTregs show an activated Treg transcriptional profile. Volcano plot depicts the differential expression of 579 immune genes in IL-6RhiTIGIT+ and IL-6RhiTIGIT− mTregs sorted ex vivo from nine independent healthy donors using NanoString. Plots depicting the normalized read counts from four differentially expressed activated Treg signature genes (marked in green), HELIOS, HLA-DRA, DUSP4 and FOXP3 are also shown. The flow sorting marker TIGIT, used for the isolation of the assessed Treg subsets, is marked in black. P values were calculated using two-tailed paired non-parametric Wilcoxon signed rank tests, comparing the normalized NanoString read counts between IL-6RhiTIGIT+ and IL-6RhiTIGIT− mTregs.
Fig. 8.
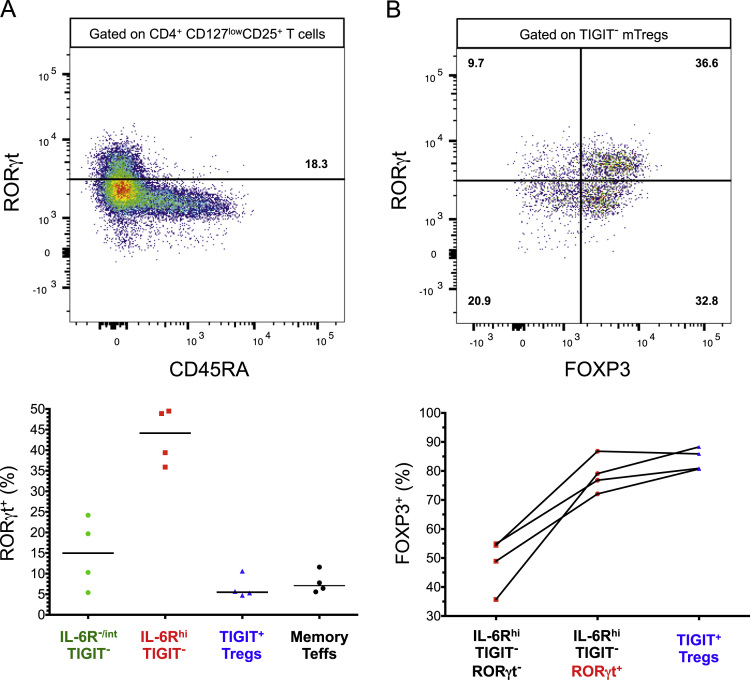
Elevated expression of IL-6R on TIGIT− mTregs marks a subset of FOXP3+ RORγt+ TIGIT− mTregs. (A) Expression of the canonical Th17 transcription factor RORγt was assessed on four healthy donors using intracellular flow cytometry. Data shown depicts the distribution of RORγt+ cells in CD45RA− IL-6R−/intTIGIT−; IL-6RhiTIGIT− and TIGIT+ CD127lowCD25+ T cells; and in memory T effector cells (see gating strategy on Fig. 6). The histogram shown depicts an illustrative example of the expression of RORγt (stratified by CD45RA) on total CD4+ CD127lowCD25+ T cells. Gating of RORγt+ cells as detected by mAb clone AFKJS-9 followed that of Ayyoub M et al. [4]. (B) Frequency of FOXP3+ cells was assessed within the RORγt− and RORγt+ fractions of IL-6RhiTIGIT− mTregs, and in TIGIT+ mTregs. Illustrative histogram depicts the co-expression of FOXP3 and RORgt with TIGIT- CD45RA− CD127lowCD25+ T cells. Horizontal bars represent the median distribution of the assessed immune phenotypes in each group.
Fig. 9.
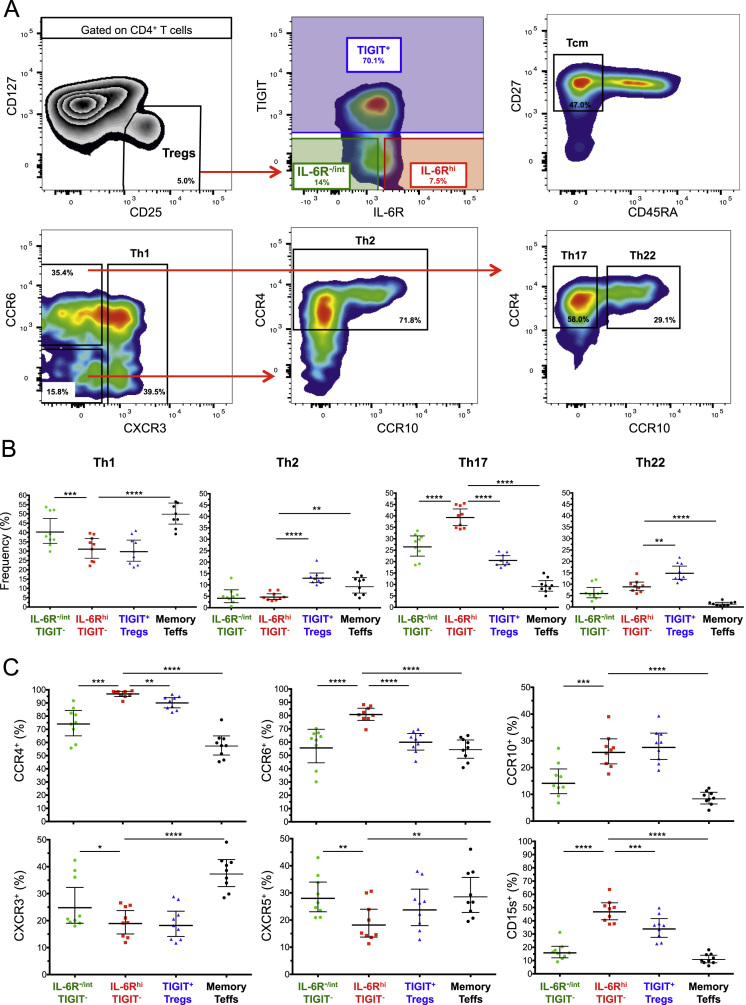
IL-6RhiTIGIT+ mTregs chemokine receptor profile is consistent with a tissue-homing Treg subset. (A) Gating strategy for the delineation of the Th1, Th2, Th17 and Th22 Treg subsets. The delineation of the Th Treg subsets was based on the definition previously described by Duhen et al. [5]. (B) Data shown depicts the frequencies (GeoMean +/− 95% CI) of the Th subsets in CD45RA− IL-6R−/intTIGIT−; IL-6RhiTIGIT− and TIGIT+ CD127lowCD25+ T cells; and in memory T effector cells. (C) Data shown depicts the frequency of the individual chemokine receptors and cell adhesion markers in the same T cell subsets. P values were calculated using a two-tailed paired non-parametric Wilcoxon signed rank test, comparing the frequency of the assessed phenotypes between IL-6RhiTIGIT− mTregs and the other T cells subsets. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 10−4.
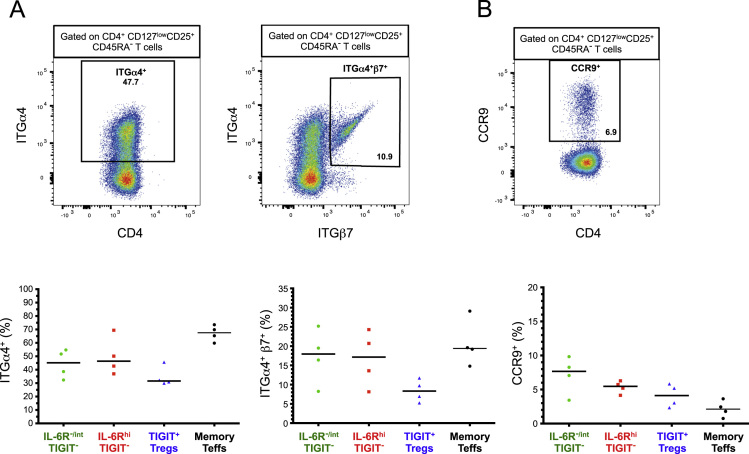
Fig. 10.
IL-6RhiTIGIT+ mTregs display the capacity to migrate to the gut. (A,B) Gating strategy for the delineation of the colon-homing receptors ITGα4+ and ITGα4+β7+ cells (A) and the small intestine-homing receptor CCR9+ cells (B). Illustrative plots depict the expression of the assessed gut-homing receptors on CD45RA− CD4+ CD127lowCD25+ T cells (C) Data shown depicts the distribution (median) of ITGα4+, ITGα4+β7+ and CCR9+ cells in CD45RA− IL-6R−/intTIGIT−; IL-6RhiTIGIT− and TIGIT+ CD127lowCD25+ T cells; and in memory T effector cells.
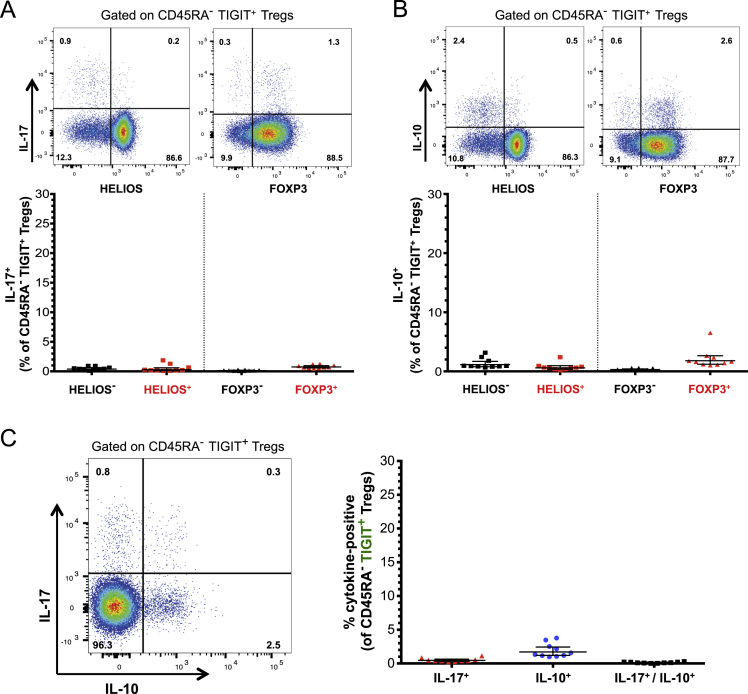
Fig. 11.
CD45RA− TIGIT+ memory Tregs produce limited amounts of IL-17 and IL-10 upon in vitro activation. (A,B) Data shown depict the frequency (GeoMean +/- 95% CI) of IL-17+ (A) and IL-10+ (B) cells among CD45RA−TIGIT+ mTregs, stratified by the expression of HELIOS and FOXP3. IL-17 and IL-10 production was assessed by intracellular flow cytometry in freshly isolated PBMCs from 10 healthy donors, following in vitro activation with PMA + ionomycin. (C) Data depict the frequency (GeoMean +/− 95% CI) of IL-17 and IL-10 single-producers as well as IL-17/IL-10 double producers among the CD45RA−TIGIT+ Treg subset.
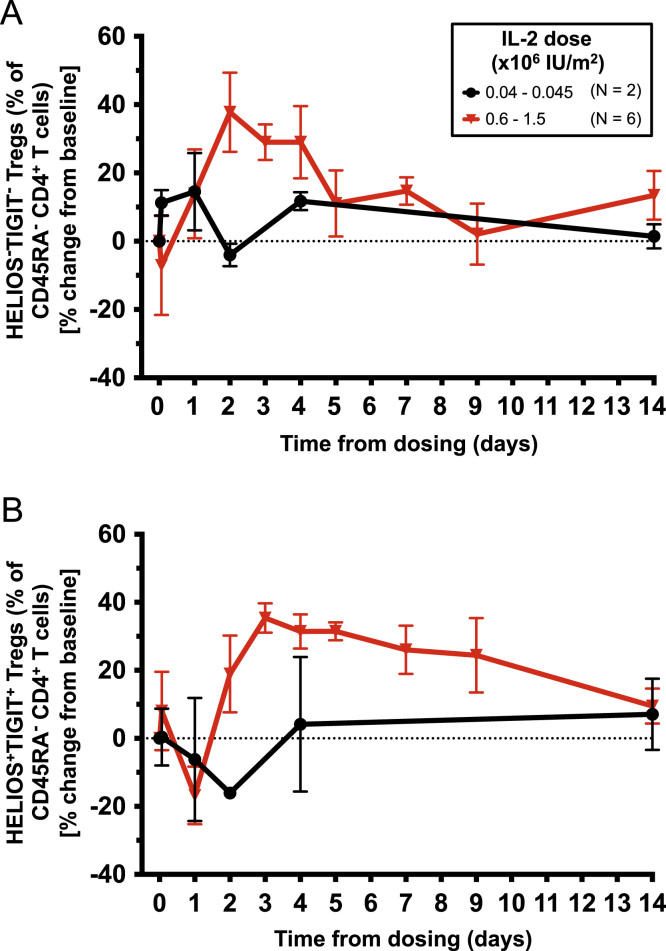
Fig. 12.
Single low dose of IL-2 does not preferentially expand CD45RA− HELIOS−TIGIT− mTregs in vivo. (A,B) Data depict the variation (Mean +/− SEM) of the frequency of CD45RA− HELIOS−TIGIT− (A) or conventional CD45RA− HELIOS+TIGIT+ mTregs (B), following IL-2 treatment in T1D patients receiving: (i) the lower IL-2 doses of 0.04–0.045×106 U/ml (N = 2; depicted in black); or (ii) the higher IL-2 doses of 0.6–1.5 x 106 U/ml (N = 6; depicted in red). Data were obtained from intracellular staining of cryopreserved PBMCs from eight selected T1D patients enrolled in the “Adaptive study of IL-2 dose on regulatory T cells in type 1 diabetes” (DILT1D). The DILT1D data from individuals prior to normalization as a group are available, however they cannot be anonymized sufficiently to be able to put into the public domain without risk of participant identification. Data are available on request, through the Cambridge University institutional repository (DOI link: https://doi.org/10.17863/CAM.832).
Table 1.
Antibodies and immunostaining panels used for flow cytometry. Detailed description of the fluorochrome-conjugated antibodies and immunostaining panels used to immunophenotype and flow sort the assessed CD4+ T cell populations.
| Immunostaining Panel | Antibody | Fluorochrome | Clone | Manufacturer |
|---|---|---|---|---|
| DILT1D Treg immunophenotyping (Whole blood) | CD4 | AF700 | RPA-T4 | BioLegend |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| CD14 | PB | M5E2 | BioLegend | |
| CCR6 | AF488 | G034E3 | BioLegend | |
| IL-6R | PE | UV4 | BioLegend | |
| CD8 | APC-Cy7 | RPA-T8 | BioLegend | |
| CXCR3 | PerCP Cy5.5 | G025H7 | BioLegend | |
| CD62L | BV605 | DREG-56 | BioLegend | |
| IL-6RhiTreg immunophenotyping (Freshly isolated PBMCs) | CD3 | BV510 | UCHT1 | BioLegend |
| CD4 | BUV398 | SK3 | BD Biosciences | |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| CTLA-4 | PE-CF594 | BNI3 | BD Biosciences | |
| CD8 | APC-Cy7 | RPA-T8 | BioLegend | |
| CD62L | BV605 | DREG-56 | BioLegend | |
| PD-1 | BV711 | EH12.2 | BD Biosciences | |
| IL-6R | BV421 | M5 | BD Biosciences | |
| HELIOS | FITC | 22F6 | BioLegend | |
| Ki-67 | PerCP Cy5.5 | B59 | BD Biosciences | |
| FOXP3 | PE | 259D | BioLegend | |
| IL-6RhiTIGIT- Treg immunophenotyping (Freshly isolated PBMCs) | CD3 | BV510 | UCHT1 | BioLegend |
| CD4 | BUV737 | SK3 | BD Biosciences | |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| HLA-DR | AF700 | L243 | BioLegend | |
| CD8 | APC-Cy7 | RPA-T8 | BioLegend | |
| CD62L | BV605 | DREG-56 | BioLegend | |
| CCR6 | BV711 | 11A9 | BD Biosciences | |
| TIGIT | PerCP eFluor710 | MBSA43 | eBioscience | |
| CD161 | PE | HP-3G10 | BioLegend | |
| IL-6R | BV421 | M5 | BD Biosciences | |
| HELIOS | FITC | 22F6 | BioLegend | |
| Ki-67 | BUV398 | B59 | BD Biosciences | |
| FOXP3 | PE-CF594 | 259D | BioLegend | |
| FACS Sorting (Freshly isolated PBMCs) | CD3 | FITC | UCHT1 | BioLegend |
| CD4 | AF700 | RPA-T4 | BioLegend | |
| CD25 | APC | M-A251 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| IL-6R | PE | M5 | BD Biosciences | |
| TIGIT | PerCP eFluor710 | MBSA43 | eBioscience | |
| Proliferation dye | eFluor450 | – | eBioscience | |
| IL-17/IL-10 production (Freshly isolated CD4+T cells) | CD3 | BV510 | UCHT1 | BioLegend |
| CD4 | BUV737 | SK3 | BD Biosciences | |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| TIGIT | PerCP eFluor710 | MBSA43 | eBioscience | |
| HELIOS | FITC | 22F6 | BioLegend | |
| FOXP3 | PE-CF594 | 259D | BioLegend | |
| IL-17 | AF700 | BL168 | BioLegend | |
| IC | AF700 | MOPC-21 | BioLegend | |
| IL-10 | PE | JES-9D7 | BioLegend | |
| IC | PE | RTK2071 | BioLegend | |
| Viability Dye | eFluor780 | – | eBioscience | |
| Chemokine receptor profiling (Freshly isolated PBMCs) | CD4 | BUV737 | SK3 | BD Biosciences |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| TIGIT | PerCP eFluor710 | MBSA43 | eBioscience | |
| CD27 | BUV398 | L128 | BD Biosciences | |
| CD8 | APC-Cy7 | RPA-T8 | BioLegend | |
| IL-6R | BV421 | M5 | BD Biosciences | |
| CCR4 | BV510 | L291H4 | BioLegend | |
| CCR6 | AF488 | G034E3 | BioLegend | |
| CCR10 | PE | 6588-5 | BioLegend | |
| CXCR3 | AF700 | G025H7 | BioLegend | |
| CXCR5 | PE-CF594 | J252D4 | BioLegend | |
| CD15s | BV711 | CSLEX1 | BD Biosciences | |
| RORγt / gut-homing immunophenotyping (Freshly isolated CD4+T cells) | CD4 | BUV737 | SK3 | BD Biosciences |
| CD25a | APC | M-A251+2A3 | BD Biosciences | |
| CD127 | PE-Cy7 | eBioRDR5 | eBioscience | |
| CD45RA | BV785 | HI100 | BioLegend | |
| TIGIT | PerCP eFluor710 | MBSA43 | eBioscience | |
| CD3 | BV510 | UCHT1 | BioLegend | |
| CCR6 | BV711 | 11A9 | BD Biosciences | |
| CD8 | APC-Cy7 | RPA-T8 | BioLegend | |
| IL-6R | BV421 | M5 | BD Biosciences | |
| ITGB7 | FITC | FIB504 | BioLegend | |
| HELIOS | FITC | 22F6 | BioLegend | |
| ITGA4 | BV605 | 9F10 | BioLegend | |
| CD62L | BV605 | DREG-56 | BioLegend | |
| CCR9 | PE | L053E8 | BioLegend | |
| RORγ/γt | PE | AFKJS-9 | eBioscience | |
| FOXP3 | PE-CF594 | 259D | BioLegend | |
| Ki-67 | BUV398 | B59 | BD Biosciences |
Two clones of anti-CD25 that bind to different epitopes were used simultaneously to enhance CD25 staining. IC, Isotype control.
2. Experimental design, materials and methods
2.1. Patient selection
Patient selection and the protocol for the “Adaptive study of IL-2dose on regulatory T cells in type 1 diabetes” (DILT1D) has been published previously [2], [3]. A subset of 22 T1D patients (median age = 26, range 18–48) were selected for this study, and assessed for the expression of IL-6R on Tregs. A blood sample was taken before treatment to establish baseline Treg frequencies and phenotypes, followed by subcutaneous administration of a single dose of recombinant human IL-2 (Proleukin/aldesleukin; dose range 45,000–737,000 IU/m2) on day 0. The patients were bled 90 min after treatment, and then daily to day 4 and at days 7, 9, 14, 21 and 60.
Study participants for all further immunophenotyping and functional assays included in this study were adult healthy volunteers recruited from the Cambridge BioResource (http://www.cambridgebioresource.org.uk/). All samples were collected after approval from the relevant research ethics committees, and written informed consent was obtained from the participants.
2.2. Flow cytometry
Treg immunophenotyping in healthy donors was performed in fresh peripheral blood mononuclear cells (PBMCs) isolated by Ficoll gradient centrifugation (Lymphoprep; STEMCELL Technologies) from whole blood within 2 h of phlebotomy. Cells were stained with fluorochrome-conjugated antibodies against surface receptors (see Table 1 for details) for 45 min at 4 °C. Fixation and permeabilisation was performed using FOXP3 Fix/Perm Buffer Set (BioLegend) and cells were then stained with the respective intracellular antibodies for 45 min at room temperature (Table 1).
For the DILT1D clinical trial participants, 30 ml whole blood were collected into lithium heparin tubes and processed within 4 h of phlebotomy. Immunostaining was performed in whole blood with specific fluorochrome-conjugated antibodies (listed in Table 1) at room temperature for 45 min. IL-6R expression was assessed using a phycoerythrin (PE)-conjugated antibody, which provided the better resolution in our flow cytometric setting. This was critical to increase the sensitivity of the assay, and assess quantitative differences in IL-6R expression in different T cell subsets.
2.3. Cell sorting
Cell sorting was performed using a BD FACSAria Fusion flow cytometer (BD Biosciences) after pre-enrichment of CD4+ T cells from whole blood by negative selection. Fluorescence-conjugated antibodies used for sorting are described in Table 1. Sorting efficiencies were determined in four donors, based on IL-6R and TIGIT expression and ranged between 90–99%.
2.4. Intracellular pSTAT3 immunostainings
PBMCs were isolated from three healthy donors by Ficoll gradient centrifugation from whole blood within 2 h of phlebotomy. IL-6 sensitivity of the memory Treg and Teff subsets was determined by intracellular pSTAT3 immunostaining in freshly isolated PBMCs in response to IL-6 stimulation in vitro, as previously described [6].
2.5. in vitro proliferation assays
To assess the proliferative capacity of IL-6RhiTIGIT−, IL-6RhiTIGIT+ and IL-6RloTIGIT+ Tregs and memory Teffs, 104 sorted cells from each subset were labelled with eFluor450 Cell Proliferation Dye (eBioscience), and cultured in the presence of exogenous IL-2 (100 U/ml; Proleukin) and anti-CD3/CD28 activation beads, at a 1:1 bead:Teff ratio in X-VIVO 15 + 5% human AB serum. Cells were cultured in X-VIVO 15 + 5% human AB serum for 84 h at 37 °C in V-bottom 96-well cell culture plates (CELLSTAR, Greiner) in the presence of exogenous IL-2 (100 U/ml; Proleukin) and anti-CD3/CD28 activation beads (Life Technologies), at a 1:1 bead:Teff ratio. Proliferation of the responder cells was assessed by the dilution of the proliferation dye by flow cytometry.
Proliferation capacity was calculated using the Division Index (DI) in FlowJo (Tree Star). The DI represents the average number of cell divisions that each seeding Teff cell has undergone and was obtained using the following equation: DI = Total number of Cell Divisions / Initial number of Teff cells in culture = , where n represents the number of divisions and G(n) represents the number of cells that have undergone n divisions.
2.6. Cytokine secretion assays
To assess cytokine production, CD4+ T cells were isolated from whole blood by negative selection using RosetteSep (STEMCELL Technologies) within 2 h of phlebotomy. Cells were resuspended in X-VIVO 15 (Lonza) + 5% heat-inactivated, filtered human AB serum (Sigma), and cultured (1–2×106 CD4s/well) in a 24-well flat-bottom cell culture plate (CELLSTAR, Greiner) at 37 °C in the presence or absence of the 1X Cell Stimulation Cocktail (eBiosiences), containing phorbol myristate acetate (PMA), ionomycin, and protein transport inhibitors (brefeldin A and monensin).
After 6 h culture, cells were harvested and immunostained with surface and intracellular antibodies (listed in Table 1). The unstimulated cells were used to determine background levels of cytokine production. Dead-cell exclusion was performed using the eFluor780 Fixable Viability Dye (eBiosciences).
2.7. Transcriptional profiling of the Treg subsets
Gene expression profiling was performed by NanoString, using the pre-designed nCounter Human Immunology v2 Panel (NanoString Technologies). The four assessed immune cell subsets were flow sorted as described above, and 25,000 cells were collected into RLT lysis buffer (Qiagen) either: (i) directly ex vivo; or (ii) following in vitro stimulation for 165 min in the presence or absence of 50 ng/ml PMA (Sigma-Aldrich) and 500 ng/ml ionomycin (Sigma-Aldrich), without addition of protein transport inhibitors. RNA from the flow-sorted T cell subsets was extracted using the RNAeasy Micro Plus kit (Qiagen), with gDNA cleanup, following manufacturer׳s instructions. Total RNA samples were then hybridized to the NanoString CodeSets, following manufacturer׳s instructions. Expression levels were assessed using an nCounter Flex instrument (NanoString Technologies). Data were processed using the nSolver Analysis Software following normalization of the raw read counts to the geometric mean of positive control spike-ins, and the gene expression of 15 selected housekeeping genes (ATG10, C14orf166, CD3E, CD46, G6PD, GPI, POLR1B, POLR2A, PSMB5, PSMB10, PTPRC, SDHA, SKI, TOLLIP and TUBB) that were found to have low variability on both the samples collected ex vivo and following in vitro stimulation.
2.8. Statistical analyses
Statistical analyses were performed using Prism software (GraphPad) and R (www.r-project.org.com). Statistical significance was assessed using a two-tailed non-parametric Mann-Whitney test. Comparison of immune phenotypes between the assessed Treg subsets from the same individual was performed using a two-tailed paired non-parametric Wilcoxon signed rank test.
Differential expression of normalized NanoString transcriptional data was calculated using a paired analysis with DESeq. 2 v1.12.3 [7], with pre-set size factors equal to one for all samples. Adjusted P values correspond to the false discovery rates (FDR) for differential expression, computed after correcting P values for multiple testing. A missing FDR is reported for genes that were found to contain an expression outlier by DESeq. 2 Cook׳s distance-based flagging of P values, and thus excluded from multiple testing.
Acknowledgements
We wish to thank the staff of the Cambridge NIHR BRC Cell Phenotyping Hub, in particular Anna Petrunkina Harrison, Simon McCullum, Christopher Bowman, Natalia Savinykh and Esther Perez for their advice and support in cell sorting. We thank Neil Walker and Helen Schuilenburg from the JDRF/Wellcome Trust Diabetes and Inflammation Laboratory, University of Cambridge for data management. We also thank Helen Stevens, Pamela Clarke, Gillian Coleman, Sarah Dawson, Simon Duley, Meeta Maisuria-Armer and Sumiyya Mahmood from the JDRF/Wellcome Trust Diabetes and Inflammation Laboratory, University of Cambridge for their help in processing blood samples.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.04.043.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.04.043.
Transparency document. Supplementary material
Supplementary material
.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Ferreira R.C., Rainbow D.B., García A. Rubio, Pekalski M.L., Porter L., Oliveira J.J., Waldron-Lynch F., Wicker L.S., Todd J.A. Human IL-6RhiTIGIT− CD4+CD127lowCD25+ T cells display potent in vitro suppressive capacity and display a distinct Th17 profile. Clin. Immunol. 2017;179:25–39. doi: 10.1016/j.clim.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldron-Lynch F., Kareclas P., Irons K., Walker N.M., Mander A., Wicker L.S., Todd J.A., Bond S. Rationale and study design of the Adaptive study of IL-2 dose on regulatory T cells in type 1 diabetes (DILT1D): a non-randomised, open label, adaptive dose finding trial. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2014-005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd J., Evangelou M., Cutler A., Pekalski M., Walker N., Stevens H., Porter L., Smyth D., Rainbow D., Ferreira R., Esposito L., Hunter K., Loudon K., Irons K., Yang J., Bell C., Schuilenburg H. Regulatory T cell responses in patients with Type 1 diabetes after a single-dose of interleukin-2: a non-randomized, open label, adaptive dose-finding trial. PLoS Med. 2016;13:e1002139. doi: 10.1371/journal.pmed.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayyoub M., Deknuydt F., Raimbaud I., Dousset C., Leveque L., Bioley G., Valmori D. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the TH17 lineage-specific transcription factor RORγt. Proc. Natl. Acad. Sci. 2009;106:8635–8640. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duhen T., Duhen R., Lanzavecchia A., Sallusto F., Campbell D.J. Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells. Blood. 2012;119:4430–4440. doi: 10.1182/blood-2011-11-392324. 〈http://www.bloodjournal.org/content/119/19/4430.abstract〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira R.C., Freitag D.F., Cutler A.J., Howson J.M.M., Rainbow D.B., Smyth D.J., Kaptoge S., Clarke P., Boreham C., Coulson R.M., Pekalski M.L., Chen W.-M., Onengut-Gumuscu S., Rich S.S., Butterworth A.S., Malarstig A., Danesh J., Todd J.A. Vol. 9. 2013. Functional IL6R 358Ala Allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases; p. e1003444. (PLoS Genet.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material