Abstract
Products from the degradation of the branched-chain amino acids valine, leucine, and isoleucine contribute to the production of a number of important cellular metabolites, including branched-chain fatty acids, ATP and other energy production, cell-cell signaling for morphological development, and the synthesis of precursors for polyketide antibiotics. The first nonreversible reactions in the degradation of all three amino acids are catalyzed by the same branched-chain α-keto acid dehydrogenase (BCDH) complex. Actinomycetes are apparently unique among bacteria in that they contain two separate gene clusters, each of which encodes a BCDH enzyme complex. Here, we show that one of these clusters in Streptomyces coelicolor is regulated, at least in part, at the level of transcription by the product of the bkdR gene. The predicted product of this gene is a protein with similarity to a family of proteins that respond to leucine and serve to activate transcription of amino acid utilization operons. Unlike most other members of this class, however, the S. coelicolor bkdR gene product serves to repress transcription, suggesting that the branched-chain amino acids act as inducers rather than coactivators of transcription. BkdR likely responds to the presence of branched-chain amino acids. Its role in transcriptional regulation may be rationalized by the fact that transition from vegetative growth to aerial mycelium production, the first stage of morphological development in these complex bacteria, is coincident with extensive cellular lysis generating abundant amounts of protein that likely serve as the predominant source of carbon and nitrogen for metabolism. We suggest that bkdR plays a key role in the ability of Streptomyces species to sense nutrient availability and redirect metabolism for the utilization of branched-chain amino acids for energy, carbon, and perhaps even morphogen synthesis. A null mutant of bkdR is itself defective in morphogenesis and antibiotic production, suggesting that the role of the bkdR gene product may be more global than specific nutrient utilization.
Soil bacteria display amazing versatility in their ability to use relatively poor sources of carbon and nitrogen. They thrive on a mixture of complex carbohydrates and proteins that result from the degradation of organic material. The streptomycetes are particularly proficient in the use of complex nutrient sources and contain numerous enzymes for the specific degradation of complex carbohydrates such as cellulose, chitin, and xylan. These common soil bacteria grow vegetatively as a multicellular, multinucleoid mycelium; as the substrate mycelia run out of food in their local environment, they begin to lyse, providing nutrients for developmental structures called aerial hyphae that coil and septate into spores. The spores provide a method of dispersal to a new location and new sources of food. At this stage in its life cycle, the organism undergoes dramatic changes in both its morphology and its metabolism. As development begins, the likely substrates for growth are glycogen stored by the substrate mycelia (3), and protein is released from the lysing substrate mycelia. Coincident with this change in cell type and metabolism is the production of a number of secondary metabolites, including most of the natural product antibiotics used in human and animal health care. These compounds serve to protect the nutrients provided by the lysing mycelia from other soil organisms and are likely critical to the production and survival of the spores in nature. The regulation of carbon utilization has, in fact, been shown to play a key role in the initiation of morphological development in Streptomyces (24).
So where does the carbon and nitrogen used for morphogenesis and antibiotic production come from? What regulates carbon flow to these processes and how is flux through these various pathways controlled? The products generated from the degradation of amino acids are known to be important, as many soil bacteria make the transition from vegetative growth to stationary phase and morphogenesis. Degradation of branched-chain amino acids are used for ATP and energy production in Pseudomonas putida (7) and Enterococcus faecalis (32), branched-chain fatty acids for membrane biosynthesis during sporulation in Bacillus subtilis (12), and cell-cell signaling in Myxococcus development (8, 13). In Streptomyces, branched-chain amino acid degradation supplies precursors for branched-chain fatty acids and are the exclusive source of some of the precursors for polyketide antibiotic production (6). The gene clusters that encode enzymes that degrade protein and amino acids for carbon and nitrogen may, in fact, provide a regulatory link between primary and secondary metabolism.
Degradation of the branched-chain amino acids valine, leucine, and isoleucine begins with three reactions common to each pathway. The first reaction is transamination to the corresponding α-keto acid (α-ketoisovalerate, α-keto-β-methyvalerate, and α-ketoisocaproate, respectively). The second is oxidative decarboxylation to the corresponding acyl-coenzyme A (CoA) derivative. The third is dehydrogenation by NAD to form a double bond. The second and third reactions are carried out by a common branched-chain α-keto acid dehydrogenase (BCDH) complex. The remainder of the isoleucine degradation pathway is identical to that of fatty acid oxidation yielding acetyl-CoA and propionyl-CoA, which is subsequently converted into succinyl-CoA. Valine degradation is a variation on this theme, yielding propionyl-CoA. Leucine degradation yields both acetyl-CoA and acetoacetate.
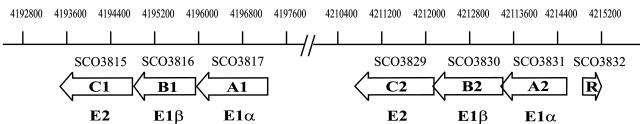
The BCDH complex consists of subunits E1-α and E1-β (containing dehydrogenase and decarboxylase functions), E2 (containing an acyltransferase function), and E3 (containing a dehydrogenase function). In streptomycetes, the E3 dihydrolipoamide dehydrogenase enzyme function is not linked to this cluster and likely resides elsewhere in the genome. The actinomycetes are apparently unique among bacteria in that they contain two sets of clustered genes, bkdA1B1C1 and bkdA2B2C2, each of which encodes E1-α, E1-β, and E2 subunits. A diagram of the organization of these gene clusters in Streptomyces coelicolor is shown in Fig. 1. Homologues of these gene clusters in Streptomyces avermitilis, where the gene functions were first described (6, 29), are designated bkdABC, corresponding to bkdA2B2C2, and bkdFGH, corresponding to bkdA1B1C1.
FIG. 1.
Organization of the S. coelicolor branched-chain amino acid dehydrogenase gene clusters. Gene designations are based on the nomenclature used by the Sanger Centre Genome Sequencing Projects (www.sanger.ac.uk/Projects/S_coelicolor). The numbers above the diagram indicate the base pair positions of the genes in the cosmid DNA sequence. The function of each open reading frame is shown below the arrow, indicating its position.
While little is known about the regulation of these clusters in Streptomyces, regulation of the bkd clusters of P. putida and B. subtilis have been partially characterized. The single cluster in these bacteria is transcribed apparently from a single start site upstream of the first open reading frame (4, 18). In each case, a regulatory gene, bkdR, located upstream of the bkd cluster acts as a transcriptional activator (4, 18). Homologues of bkdR in P. putida and B. subtilis have been most extensively characterized. In both Bacillus and Pseudomonas, mutations in bkdR eliminate transcription of the bkd cluster but have no affect on amino acid transport (21). In B. subtilis, regulation by BkdR is mediated by an alternative sigma factor, σL, a σ54 homologue, which responds to nitrogen regulation. B. subtilis BkdR activates transcription in response to the presence of branched-chain amino acids, but it is not an Lrp (leucine-responsive regulatory protein)-type transcriptional regulator and mediates activation by a completely different mechanism from that of other BkdR proteins (4, 5). In P. putida, the BkdR protein has been shown to bind a promoter directly upstream of the first open reading frame of the bkd cluster, and transcription of the cluster is activated by the binding of branched-chain amino acids to the BkdR protein (4, 5, 18). The P. putida BkdR is a member of the Lrp class of transcriptional regulators. The Lrps are small DNA-binding proteins with molecular masses of about 15 kDa and are widely distributed among prokaryotes (recently reviewed by Brinkman et al.) (2). The archetype Lrp is a global regulator of transcription in Escherichia coli and is apparently involved in the regulation of at least 10% of all E. coli genes (31).
Here, we report a preliminary analysis of the role of the bkdR gene of S. coelicolor. A null mutation in the bkdR gene resulted in a defect in sporulation (a whi phenotype) and loss of the pigment associated with actinorhodin production, a type II polyketide. Loss of the bkdR gene product also resulted in constitutive expression of one of the bkd clusters compared to regulated expression in the wild type, suggesting that it acts as a repressor of transcription.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and microbiological techniques.
General techniques for bacterial growth and isolation and manipulation of nucleic acids were as in Sambrook et al. (28) and Kieser et al. (15) for E. coli and S. coelicolor, respectively. S. coelicolor strains were grown in tryptic soy broth medium for genomic DNA isolation and in YEME (1% glucose, 0.5% Bacto peptone, 0.3% malt extract, 0.3% yeast extract, 10% sucrose, and 5 mM MgCl2) for protoplast preparation. R2YE plates were used for protoplast regeneration and soy agar medium with the addition of 10 mmol per liter of MgCl2 for mating experiments. Antibiotic selections were applied by overlay with soft nutrient agar. Streptomyces RNA was isolated from cells grown on cellophane disks placed on top of maltose yeast extract medium (MYM) agar. Minimal medium, R2YE, and MYM were used in the analysis of antibiotic production and morphogenesis. NMMP medium (15) was used for analysis of growth in liquid medium with either 0.5% glucose or 0.5% leucine as the sole carbon source and 0.5% ammonium sulfate as the sole nitrogen source. Cells were grown in 1 liter of medium, and 30-ml samples were taken as indicated. Samples were washed with 0.9% NaCl and desiccated to determine dry cell weight. PCRs were carried out according to the manufacturer's instructions (Finnzymes) with the addition of 5% dimethyl sulfoxide.
Construction of a bkdR deletion mutant.
The wild-type allele of the bkdR gene from S. coelicolor A(3)2 was cloned as a PCR-generated (3,174 bp) fragment with primers bkdR-UP1Sc and bkdR-DS1Sc with DyNAzyme EXT high-fidelity DNA polymerase (Finnzymes) and genomic DNA of S. coelicolor as a template. This fragment was treated with the Klenow fragment of DNA polymerase to generate blunt ends, digested with BspEI, and cloned into pBluescript IISK (Stratagene) that had been digested with EcoRV and XmaI, resulting in plasmid pbkdR. A deletion was constructed in a series of steps replacing the entire bkdR open reading frame with a spectinomycin resistance gene, aadA. A fragment containing the addA gene was cloned from pHP45Ω (25) by digestion with HindIII, treated with Klenow, and ligated pbkdR that had been digested with SphI and BamHI and treated with Klenow. To restore the genomic DNA sequence downstream of bkdR, the 912-bp SmaI fragment removed in the previous step was restored. After the orientation of the insert DNA was confirmed, the deletion cassette was cloned from pΔbkdR by digestion with NotI, treated with Klenow, then digested with HindIII, and ligated to the SphI-HindIII fragment of pSET152 (1) that had been treated with Klenow before digestion with HindIII. The resulting plasmid contained the apramycin resistance gene, aac(3)IV, and oriT from pSET152 but not the μC31 integrase or attachment site. PCR-generated fragments were sequenced to ensure fidelity. Plasmid pSETΔbkdR::aadA was transformed into E. coli ET12567 for plasmid DNA used to transform S. coelicolor M145 protoplasts and into E. coli ET12567 containing pUB307 and then conjugated into S. coelicolor M145. Transformations and conjugations were carried out as previously described (15) with selection for spectinomycin resistance. Double-crossover events were identified by screening for apramycin sensitivity. The absence of the bkdR open reading frame was confirmed by Southern hybridization and restriction analysis.
This deletion of the bkdR open reading frame was also introduced into M145 by recombination using the cosmid-based PCT targeting system for S. coelicolor (9).
Genetic complementation.
Two versions of the wild-type allele of bkdR were generated. An NcoI-BamHI fragment containing the entire bkdR open reading frame and 800 bp of DNA upstream of the annotated translation start site was cloned from plasmid pbkdR, incubated with the Klenow fragment of DNA polymerase, and ligated to pSET152 that had been digested with BamHI and EcoRV. The resulting plasmid was introduced into S. coelicolor ΔbkdR::aadA by conjugation selecting apramycin resistance. A PCR fragment containing only the bkdR open reading frame was generated with primers bkdR-SC2Fwd and bkdR-SCRev (Table 1) and cloned downstream of the tipA promoter. For this construction, the forward primer contained base changes that introduced an NdeI site overlapping the bkdR ATG translation start codon. The fragment was then digested with BclI to generate a fragment with a 5′ blunt end and a 3′ BclI cloning site and ligated to pBluescript II SK that had been digested with BamHI and EcoRIV. The bkdR gene was subcloned from this plasmid as an NdeI-XbaI fragment, cloned into pIJ8600 (30) digested with the same enzymes placing the bkdR open reading frame downstream of the tipA promoter, and introduced into ΔbkdR::aadA by conjugation, selecting for apramycin resistance.
TABLE 1.
Oligonucleotides used in the study
| Primer> | Sequence |
|---|---|
| bkdR-related constructions | |
| bkdR-UP1Sc | 5′-TGAGGGCCTGCGCCATGGTG-3′ |
| bkdR-DS1Sc | 5′-CCGCTCACGTCGTCGTCGAG-3′ |
| bkdR-SC2Fwd | 5′-CATATGCTCCAGGCGGACGGCCG-3′ |
| bkdR-SCRev | 5′-CTCGTCTTCGGCATGATCAACTC-3′ |
| Probes for S1 nuclease mapping | |
| BkdR-S1-1 | 5′-TTGATGCGGGCGTAGGCGTTC-3′ |
| BkdA-2S1-1 | 5′-TTGGTGCCGAGGACGCGGTAG-3′ |
| ScGD319S1-1 | 5′-TCGCCGGAGAGCAGATCGTGC-3′ |
| bkdA1-S1-1 | 5′-GTACGGGTGGTGCTGGTGGTC-3′ |
S1 nuclease mapping.
S1 nuclease mapping was carried out as previously described (16). RNA was isolated from cells after 16, 24, 36, 48, or 60 h of growth on MYM agar plates overlaid with cellophane disks, and 40 μg was hybridized to approximately 0.05 pmol of a 32P-labeled 5′end-labeled DNA probe and incubated with 100 U of S1 nuclease (Roche). Probes were generated by PCR with Taq DNA polymerase (QIAGEN) using S. coelicolor genomic DNA as a template. Oligonucleotides bkdR-S1-1 and bkdA2-S1-1 (labeled) were used to generate a probe for the bkdA2 transcript. ScGD319S1-1 and bkdA1-S1-1 (labeled) oligonucleotides were used for the bkdA1-specific probe. The hrdB promoter probe was described in Ryding et al. (26). The same labeled oligonucleotides were used in the fmol DNA Cycle Sequencing System (Promega) to generate the G, A, T, and C sequencing ladders. DNA fragments were separated on a 6% denaturating polyacrylamide gel, and bands were visualized by autoradiography.
RESULTS
S. coelicolor contains a bkdR homologue adjacent to the bkdA2B2C2 gene cluster.
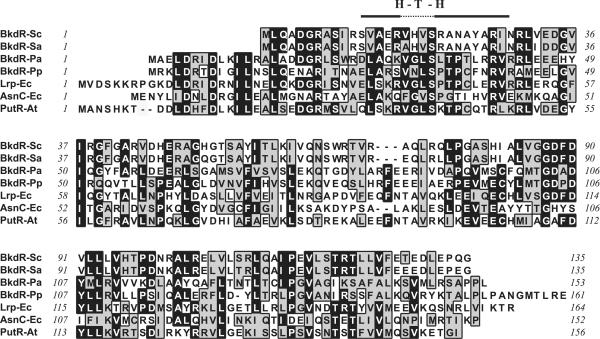
Inspection of the DNA sequence surrounding the BCDH clusters of S. coelicolor (Fig. 1) revealed an open reading frame whose predicted gene product had striking similarity to members of the Lrp class of transcription factors. A comparison of BkdR with members of this class is shown in Fig. 2. These proteins contain a DNA-binding domain with a typical helix-turn-helix fold, and they interact with DNA as multimers from two to as many as eight subunits. Often multiple-binding sites are present in target promoters and binding to multiple sites within the same promoter is apparently cooperative. The binding sites themselves consist of imperfect inverted repeat sequences with no obvious sequence homology (2). The ligand-binding domain contains a typical αβ-sandwich fold and resides in the C-terminal end of the protein. The ligand-binding site of these proteins is somewhat novel and has been designated RAM. Database searches have also identified RAM domains as stand-alone modules of unknown function. The DNA-binding and RAM domains are separated by a flexible hinge region, suggesting a general allosteric switch mechanism for activation (2). The predicted protein sequence of the S. coelicolor bkdR contains homology to these conserved domains as well as general similarity throughout the sequence of the protein.
FIG. 2.
End-to-end alignment of conserved (black background) and functionally similar (grey background) amino acids in the sequences of inferred and known proteins with similarity to the predicted protein encoded by S. coelicolor BkdR. Abbreviations: Sc, S. coelicolor; Sa, S. avermitilis; Pa, Pseudomonas aeruginosa; Pp, P. putida; Ec, E. coli; At, Agrobacterium tumeofaciens. Protein alignment was performed with the ClustalW program, and the graphic was generated with MacVector 7.0.
Null mutants of bkdR have a whi phenotype and fail to produce actinorhodin.
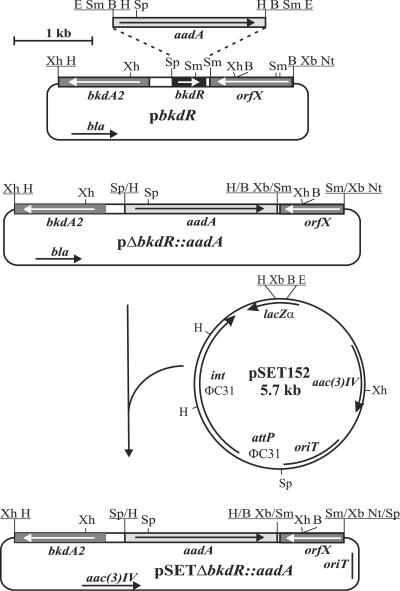
A complete deletion of the bkdR open reading frame was constructed in the S. coelicolor chromosome by marker replacement. Details of that construction are outlined in Fig. 3. The entire bkdR open reading frame was replaced with a spectinomycin resistance gene, aadA, and placed in a plasmid containing an apramycin resistance gene, oriT from pSET152, but not the μC31 integrase or attachment site. The presence of the deletion construction was selected by spectinomycin resistance, and double-crossover events were identified by screening apramycin sensitivity. All constructions involving PCR-generated fragments were sequenced to ensure fidelity. The replacement of the bkdR open reading frame with the spectinomycin cassette in the chromosome was confirmed by Southern hybridization and restriction analysis (data not shown).
FIG. 3.
Construction of plasmid pSETΔbkdR::aadA used to generate a deletion of the bkdR open reading frame in the S. coelicolor genome. Abbreviations: E, EcoRI; Sm, SmaI; B, BamHI; H, HindIII; Sp, SphI; Xh, XhoI; Nt, NotI; Xb, XbaI.
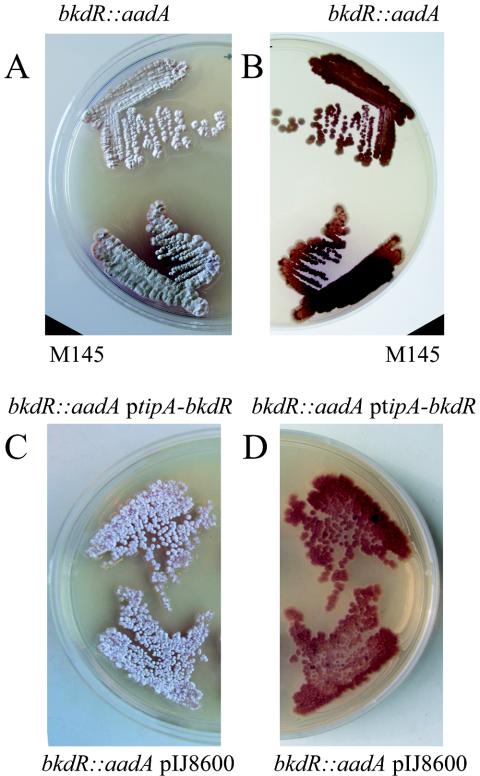
To investigate the effect of a deletion of bkdR on development, the mutant was compared to wild-type cells on a number of solid media that support sporulation. As shown in Fig. 4, the null mutant of bkdR is able to erect aerial hyphae on MYM medium but fails to produce the grey pigment associated with mature spores (a whi phenotype) and fails to make the blue pigment associated with actinorhodin production. Some spores were apparently produced by the bkdR mutant, but the plating efficiency of spore stocks from the mutant was reduced compared to that of M145, suggesting that either fewer spores were produced or the spores had reduced viability.
FIG. 4.
Comparison of the bkdR deletion strain (ΔbkdR::aadA) with the wild type (M145), the deletion mutant containing a wild-type copy of bkdR under the control of the tipA promoter (ΔbkdR::aadA ptipA-bkdR), and the deletion mutant containing the pIJ8600 vector without the bkdR wild-type allele (ΔbkdR::aadA pIJ8600) grown on MYM agar plates (A and B) or on MYM agar with the addition of 2 μg of thiostrepton/ml (C and D). Photographed from the top of the plate (A) to visualize morphological development or from the bottom of the plate (B) to visualize the pigments associated with antibiotic production.
The bkdR null mutant is apparently not complemented in trans by the wild-type allele.
To test the ability of the wild-type bkdR allele to complement the bkdR null mutant, the bkdR gene and upstream region were amplified by PCR from S. coelicolor M145 chromosomal DNA and cloned into an integrational vector, pSET152. This placed the wild-type allele in trans at the φC31 chromosomal attachment site. The bkdR mutant strain containing this vector had the same phenotype as the mutant itself or with the vector without the wild-type allele inserted. In an attempt to eliminate possible problems that might have arisen from insufficient expression to complement the mutant in trans, the bkdR wild-type open reading frame was cloned downstream of a strong inducible promoter, tipA (10). As shown in Fig. 4, the mutant strain containing this construction was indistinguishable from the mutant itself or the mutant containing the vector without the wild-type allele, even under conditions that allowed full induction of tipA.
While it is possible that the strain containing the null mutant allele contains a second site mutation that is responsible for part or all of the mutant phenotype, it is unlikely given that several independent constructions of the null mutant were made by two different methods. In addition, the null mutant had no detectable growth defects or nutritional requirements when compared to the wild type; while the lack of other detected mutations does not argue against the possibility of second site mutations in the constructed deletion, there is no evidence that the procedures used were highly mutagenic.
In most cases, including S. coelicolor, E. coli, and P. putida, genes that encode Lrps like bkdR are adjacent to and transcribed in the opposite orientation from the gene clusters they regulate. In E. coli, the open reading frame that encodes the Lrp itself contains binding sites for the Lrp, and these sites are required in cis for regulation of ilvIH transcription (27). One explanation for the failure of the wild-type allele to complement the bkdR deletion mutant is that deletion of the bkdR open reading frame in S. coelicolor eliminates sites for BkdR interaction so that complementation in trans is not possible. A prediction of this hypothesis is that point mutations within the bkdR open reading frame that did not affect putative binding sites for BkdR protein would allow complementation; attempts to isolate such mutants are in progress.
A null mutant of bkdR is able to utilize leucine as sole carbon source and has no detectable growth defect.
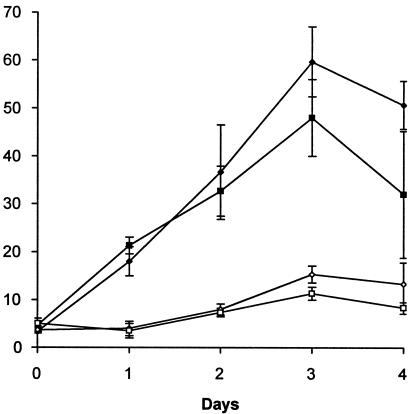
Denoya et al. (6) showed that a deletion in the 5′ region of one of the bkd clusters in S. avermitilis (corresponding to bkdA1B1C1) resulted in a failure to utilize valine, leucine, or isoleucine as the sole carbon source. To investigate whether a deletion of bkdR affected growth on branched-chain amino acids or had a general defect in growth, we compared growth of the mutant and wild type in NMMP (15) liquid medium with glucose or leucine as the sole carbon source. As shown in Fig. 5, growth of the bkdR mutant was essentially the same as that of the wild type with glucose as the sole carbon source. While growth on leucine as the sole carbon source is very poor, growth was detectable in both the mutant and the wild type to approximately the same degree.
FIG. 5.
A comparison of the growth of the bkdR mutant (squares) with wild-type cells (circles and diamonds) grown in minimal (NMMP) medium with either glucose (closed symbols) or leucine (open symbols) as the sole carbon source. One-liter cultures were sampled (each sample, 30 ml) at 24-h time intervals, and changes in cell dry weight were measured.
BkdR is a repressor of bkdA2B2C2 transcription but has no apparent effect on transcription of bkdA1B1C1.
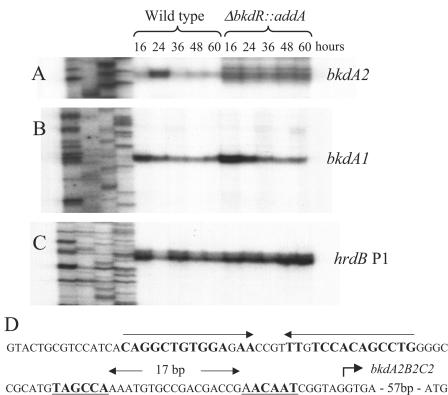
To test whether the bkdR gene product was involved in transcriptional regulation of either of the bkd gene clusters, we performed S1 nuclease mapping experiments with probes that would detect transcription originating upstream of either bkdA1 or bkdA2 (Fig. 6). RNA was isolated from wild-type and bkdR mutant cells after 16, 24, 36, 48, and 60 h of growth on MYM agar plates. This solid medium supports morphological development of the wild type, and the stages of development are clearly detectable. Cells harvested after 16 h grew vegetatively (no aerial mycelia were present). Aerial mycelia began to appear between 24 and 36 h and were abundant between 36 and 48 h. The grey pigment associated with mature spores was evident at 60 h. As shown in Fig. 6, in wild-type cells a transcript originating upstream of the bkdA1B1C1 cluster was abundant in vegetatively growing cells (16 h) and apparently decreased in abundance as development proceeded. A null mutation in bkdR had no obvious affect on this transcript. A transcript originating upstream of the bkdA2B2C2 cluster was detected in wild-type cells as the cells began aerial mycelium production and was apparently tightly regulated. Virtually no transcript was detected during vegetative growth (16 h) but was readily detected at 24 h just as aerial mycelia were being produced. The transcript virtually disappeared after aerial mycelium production was complete. A null mutation in bkdR resulted in constitutive expression of this transcript, suggesting that BkdR acts as a repressor of transcription. The pattern of expression of this transcript further suggests that repression by BkdR is induced transiently at the initiation of morphogenesis.
FIG. 6.
S1 nuclease mapping of transcripts originating upstream of bkdA2 (A) and bkdA1 (B) in wild-type and bkdR mutant cells, harvested at 16, 24, 36, 48, and 60 h of growth on MYM agar plates. The hrdB transcript (C) was used as a control for RNA. The sequence of the predicted bkdA2 promoter region (D) with the putative RNA polymerase-binding site (underlined and in boldface type) and 14-bp inverted repeat sequence (boldface type with arrows above the sequence) are shown.
While it is not known whether one or both clusters in S. coelicolor are required for utilization of branched-chain amino acids, the fact that a null mutation in bkdR had no effect on the ability to utilize leucine as sole carbons is consistent with the notion that the bkdR gene product acts as a repressor, rather than an activator, of transcription.
The putative bkdA2B2C2 promoter contains a prototypical RNA polymerase-binding site and an almost perfect 14-bp inverted repeat sequence.
S1 nuclease mapping experiments identified an apparent transcription start site for the bkdA2B2C2 gene cluster approximately 60-bp upstream of the translation start site (Fig. 6D). Upstream of this apparent start site are sequences centered at −10 and −35 that are prototypical for prokaryotic promoters. TAGCCA is a four out of six match to the TTGACA consensus sequence at −35, and AACAAT is a five out of six match for the TTCAAT consensus sequence at −10 (Streptomyces hrdB promoters often contain a C in the third position rather than a T, which is typical in E. coli.) In addition, there is an almost perfect (one-base mismatch) 14-bp inverted repeat sequence just upstream of the −35 RNA polymerase recognition sequence. While BkdR proteins are known to bind inverted repeats within target promoters, the binding sites are typically not perfect and exist in more than one copy to allow cooperative protein binding. The presence of an almost perfect inverted repeat sequence is more typical of regulatory proteins such as the cAMP receptor protein and the regulator of fumarate and nitrate reduction; and if this site is involved in bkdR binding, the interaction between the S. coelicolor BkdR protein and its target promoters would be very different from previously characterized members of the Lpr class.
DISCUSSION
Actinomycetes are apparently unique among bacteria in that they contain two separate gene clusters, each of which encodes an apparently equivalent BCDH enzyme complex. Here, we show that one of these clusters is regulated, at least in part, at the level of transcription by the product of the bkdR gene. Transcription of bkdA2B2C2 is regulated in the wild-type strain and constitutive in a bkdR mutant, suggesting that the bkdR gene product directly or indirectly serves to repress transcription of this cluster. The simplest hypothesis is that BkdR protein makes direct contact with the bkdA2B2C2 promoter. By analogy with similar Lrp-type transcriptional regulators from other bacteria, we speculate that the BkdR protein likely responds to the presence of one or more of the branched-chain amino acids, perhaps causing a conformational change in the protein to relieve repression. The fact that bkdR mutants themselves are defective in morphogenesis and antibiotic production suggests that the role of bkdR may be more global, perhaps through the action of the BCDH cluster it controls or other gene clusters that respond to the presence of branched-chain amino acids or in combination with other transcriptional regulators.
Perhaps the most relevant homologue of the S. coelicolor bkdR gene is the bkdR gene of P. putida. Transcription of the single bkd cluster in P. putida is completely eliminated in a bkdR mutant, regardless of the presence of branched-chain amino acids. This suggests that it plays a role in activation (21). In vitro DNA-binding studies showed that the BkdR protein made direct contact with the bkdA1 promoter to form an inactive transcription complex. The presence of l-valine resulted in a conformational change in the BkdR protein, and this active form of the protein bends the DNA to facilitate RNA polymerase interaction and transcriptional activation (21). With in vitro transcription experiments, BkdR responded to any of the l-branched-chain amino acids as well as d-leucine, but not d-valine or d-isoleucine (17). The binding sites for BkdR proteins and Lrps in general are difficult to identify in target promoters because while these proteins bind inverted repeat sequences, the repeats are not perfect or even near-perfect inverted repeat sequences (2). It is interesting that the target promoter for the putative BkdR protein of S. coelicolor contains an almost-perfect 14-bp inverted repeat very atypical of BkdR-binding sites. The same inverted repeat (with the same single-base-pair mismatch and the same four nucleotides in the spacer) is present in the S. avermitilis genome upstream of the bkdABC cluster (which corresponds to the bkdA2B2C2 cluster of S. coelicolor). A search for this inverted repeat in other bacteria (BLASTN 2.2.9 at http://www.ncbi.nlm.nih.gov/BLAST) found no matches in E. coli, but several were identified in Bacillus species. There are 12 such inverted repeats (with perfect symmetry) in Bacillus thuringiensis, one in Bacillus cereus, and one in Bacillus anthracis 68-bp upstream of a putative transposase (GI:4894336) located on the pXO1 virulence plasmid. This type of perfect repeat sequence is more typical of activator-binding sites, but our data clearly indicate that the S. coelicolor bkdR gene serves to repress transcription of bkdA2B2C2. If this inverted repeat sequence is a binding site for BkdR, its interaction with BkdR protein may be unusual; the presence of this potential binding site in other bacteria may suggest a role for BkdR homologues in gene regulation for those organisms.
The BkdR members of the Lrp family that have been characterized experimentally from gram-negative bacteria appear to have a specific rather than a global role in gene expression. Without exception, these proteins are involved in the regulation of amino acid metabolism. The mode of action is primarily activation by complex with branched-chain amino acids; however, some members that function in autorepression of bkdR itself appear to be ligand independent (11, 14, 20, 21). As pointed out by Brinkman et al., however, it cannot be excluded that nonenteric bacteria use Lrp-like proteins as global regulators. Even in E. coli, Lrp has been shown to interact with global regulators such as the cAMP receptor protein (22, 33) and the integration host factor (23). Of particular interest is the Lrp of E. coli, which is adjacent to and transcribed in an orientation opposite to the genes it regulates, the ilvIH operon. Transcriptional regulation relies on binding of the Lrp to sites upstream of the ilvIH transcription start site and within the open reading frame of the Lrp itself (27). These sites are required in cis for transcriptional regulation; the deletion of the Lrp open reading frame was not complemented in trans by the wild-type allele (27). Interestingly, our attempts to complement the S. coelicolor bkdR null mutant were unsuccessful, and this may suggest that deletion of the bkdR open reading frame somehow interferes with bkdR interaction with the bkdA2 promoter region. In support of this hypothesis is the fact that a deletion of the bkdR open reading frame was constructed by two independent methods, making the existence of a second site mutation that affects the mutant phenotype unlikely.
Isobutyryl-CoA, 2-methylbutyryl-CoA, and isovaleryl-CoA, the products of BCDH enzyme complex activity, have been shown to play crucial a role in several important biological processes. These molecules serve as precursors for branched-chain fatty acid biosynthesis that contribute to membrane fluidity and solute transport (12). In Myxococcus xanthus, BCDH generates branched-chain fatty acids that act as signals for morphological development (13). Isobutyryl-CoA and 2-methylbutyryl-CoA also serve as starter units for the biosynthesis of the polyketide antibiotic avermectin in S. avermitilis. BCDH mutants have, in fact, been used to generate novel avermectins by alternative precursor feeding (6). Interestingly, a bkdR null mutant is itself defective in sporulation and antibiotic production. Such mutants are able to erect aerial hyphae but are defective in mature, wild-type spore formation, which is a whi mutant phenotype. Whether bkdR plays a specific or global role in the regulation of gene expression in Streptomyces will be partially elucidated by transcriptional and metabolic profiling of the bkdR mutant; these experiments are in progress.
Finally, it is interesting that transcription of bkdA2B2C2 is transiently induced at the onset of aerial mycelium production. l-Leucine is the most abundant amino acid found in proteins (http://www.cmbi.kun.nl/gvteach/aainfo/), and we speculate that as cells within the mycelium exhaust carbohydrate carbon sources and development begins, there is a shift to protein as the primary source of carbon. At this stage the concentration of amino acids, especially leucine, would be high. Studies with P. putida show that the conformational change in BkdR requires millimolar concentrations of branched-chain amino acids in vitro (19) and that activation of the bkd cluster in vivo requires high exogenous levels of branched-chain amino acids in the growth medium (17). We suggest that at this time in the life cycle of Streptomyces, the concentration of branched-chain amino acids is sufficient to cause transient induction of bkdA2B2C2 transcription. Recent microarray analysis revealed that the Lrp regulator in E. coli affects transcription of at least 10% of all E. coli genes, most of which are expressed as cells enter stationary phase (31). While E. coli exhibits no obvious morphological development, it is known to cannibalize protein from lysing cells as it enters stationary phase.
Many of the most important compounds produced by Streptomyces, including antibiotics, are made after the proliferation of the substrate mycelia, when the primary growth phase has ended. As nutrients become limiting at this stage of the life cycle, the substrates for carbon and nitrogen utilization change dramatically, and the gene clusters that encode enzymes that degrade protein and amino acids as a sources of carbon and nitrogen play an important role in metabolism. Some of the precursors for antibiotic biosynthesis come exclusively from the degradation of amino acids; it is likely that these substrates provide other important molecules for signaling and completion of morphogenesis. The regulation of pathways responsible for amino acid degradation may, in fact, provide a link between primary and secondary metabolism, and understanding of the regulation of these pathways will likely contribute to an understanding of the biological regulatory circuitry that allows for redirection of carbon and nitrogen for post growth phase metabolism.
Acknowledgments
We thank Hamish McArthur for advice and support throughout the course of this work and Larry Shimkets, Mark Buttner, and Brandan Hillerich for critical review of the manuscript.
K.S. was supported in part by an NIH Genetics Training Grant to the University of Georgia Department of Genetics. The work was funded by a grant to J.W. from Pfizer, Inc.
REFERENCES
- 1.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Mendez, and J. D. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59:133-143. [DOI] [PubMed] [Google Scholar]
- 4.Debarbouille, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debarbouille, M., I. Martin-Verstraete, F. Kunst, and G. Rapoport. 1991. The Bacillus subtilis sigL gene encodes an equivalent of s54 from gram-negative bacteria. Proc. Natl. Acad. Sci. USA 88:9092-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denoya, C. D., R. W. Fedechko, E. W. Hafner, H. A. McArthur, M. R. Morgenstern, D. D. Skinner, K. Stutzman-Engwall, R. G. Wax, and W. C. Wernau. 1995. A second branched-chain α-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J. Bacteriol. 177:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolin, M. L. 1961. Cytochrome-independent electron transporst enzymes of bacteria, vol. 2. Academic Press, New York, N.Y.
- 8.Downard, J., and D. Toal. 1995. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol. Microbiol. 16:171-175. [DOI] [PubMed] [Google Scholar]
- 9.Gust, B., T. Kieser, and K. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes, D. J., J. L. Caso, and C. J. Thompson. 1993. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 12:3183-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jafri, S., S. Evoy, K. Cho, H. G. Craighead, and S. C. Winans. 1999. An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol. 288:811-824. [DOI] [PubMed] [Google Scholar]
- 12.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearns, D. B., A. Venot, P. J. Bonner, B. Stevens, G. J. Boons, and L. J. Shimkets. 2001. Identification of a developmental chemoattractant in Myxococcus xanthus through metabolic engineering. Proc. Natl. Acad. Sci. USA 98:13990-13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keuntje, B., B. Masepohl, and W. Klipp. 1995. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J. Bacteriol. 177:6432-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser, T., and the John Innes Foundation. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 16.Kormanec, J. 2001. Analyzing the developmental expression of sigma factors with S1-nuclease mapping. Methods Mol. Biol. 160:481-494. [DOI] [PubMed] [Google Scholar]
- 17.Madhusudhan, K. T., Luo, J., and, J. R. Sokatch. 1999. In vitro transcriptional studies of the bkd operon of Pseudomonas putida: l-branched-chain amino acids and d-leucine are the inducers. J. Bacteriol. 181:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madhusudhan, K. T., K. L. Hester, V. Friend, and J. R. Sokatch. 1997. Transcriptional activation of the bkd operon of Pseudomonas putida by BkdR. J. Bacteriol. 179:1992-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madhusudhan, K. T., N. Huang, E. H. Braswell, and J. R. Sokatch. 1997. Binding of l-branched-chain amino acids causes a conformational change in BkdR. J. Bacteriol. 179:276-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhusudhan, K. T., N. Huang, and J. R. Sokatch. 1995. Characterization of BkdR-DNA binding in the expression of the bkd operon of Pseudomonas putida. J. Bacteriol. 177:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madhusudhan, K. T., D. Lorenz, and J. R. Sokatch. 1993. The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli. J. Bacteriol. 175:3934-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathew, E., J. Zhi, and M. Freundlich. 1996. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 178:7234-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, L., R. M. Blumenthal, and R. G. Matthews. 2001. Activation from a distance: roles of Lrp and integration host factor in transcriptional activation of gltBDF. J. Bacteriol. 183:3910-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon unilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 25.Prentki, P., A. Binda, and A. Epstein. 1991. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene 103:17-23. [DOI] [PubMed] [Google Scholar]
- 26.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flardh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343-357. [DOI] [PubMed] [Google Scholar]
- 27.Sacco, M., E. Ricca, R. Marasco, R. Paradiso, and M. D. Felice. 1993. A stereospecific alignment between the promoter and cis-acting sequence is required for Lrp-dependent actibation of IlvIH transcription in Escherichia coli. FEMS Microbiol. Lett. 107:331-336. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Skinner, D. D., M. R. Morgenstern, R. W. Fedechko, and C. D. Denoya. 1995. Cloning and sequencing of a cluster of genes encoding branched-chain α-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [αβ] component in Escherichia coli. J. Bacteriol. 177:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takano, E., J. White, C. J. Thompson, and M. J. Bibb. 1995. Construction of thiostrepton-indicible, high-copy-number expression vectors in Streptomyces spp. Gene 166:133-137. [DOI] [PubMed] [Google Scholar]
- 31.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, D. E., C. C. van Der Weijden, M. J. van Der Merwe, H. V. Westerhoff, A. Claiborne, and J. L. Snoep. 2000. Branched-chain α-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. J. Bacteriol. 182:3239-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weyand, N. J., B. A. Braaten, M. van der Woude, J. Tucker, and D. A. Low. 2001. The essential role of the promoter-proximal subunit of CAP in pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol. Microbiol. 39:1504-1522. [DOI] [PubMed] [Google Scholar]