Abstract
Caulobacter crescentus cells treated with amdinocillin, an antibiotic which specifically inhibits the cell elongation transpeptidase penicillin binding protein 2 in Escherichia coli, exhibit defects in stalk elongation and morphology, indicating that stalk synthesis may be a specialized form of cell elongation. In order to investigate this possibility further, we examined the roles of two other proteins important for cell elongation, RodA and MreB. We show that, in C. crescentus, the rodA gene is essential and that RodA depletion leads to a loss of control over stalk and cell body diameter and a stalk elongation defect. In addition, we demonstrate that MreB depletion leads to a stalk elongation defect and conclude that stalk elongation is a more constrained form of cell elongation. Our results strongly suggest that MreB by itself does not determine the diameter of the cell body or stalk. Finally, we show that cells recovering from MreB depletion exhibit a strong budding and branching cell body phenotype and possess ectopic poles, as evidenced by the presence of multiple, misplaced, and sometimes highly branched stalks at the ends of these buds and branches. This phenotype is also seen to a lesser extent in cells recovering from RodA depletion and amdinocillin treatment. We conclude that MreB, RodA, and the target(s) of amdinocillin all contribute to the maintenance of cellular polarity in C. crescentus.
Peptidoglycan is a large, shape-maintaining macromolecule that surrounds the cytoplasmic membrane of most eubacteria (32). It is known that peptidoglycan plays a critical role in maintaining the morphology of bacterial cells, not only because purified peptidoglycan retains the basic shape of the cell it surrounded (46) but also because cell shape mutants are often genetically linked to proteins involved in peptidoglycan synthesis (22). In addition to the importance of peptidoglycan in maintaining cell shape, research on bacterial actin homologs, namely MreB, strongly suggests that noncoccoid bacteria possess a protein scaffold that functions to maintain cell diameter during longitudinal growth (28). Unlike the proteins involved in peptidoglycan synthesis, which have enzymatic functions, MreB appears to have a recruitment and/or structural role, somehow constraining cellular diameter during growth along the long axis of the cell (16, 28, 31). It is not clear at present if MreB itself defines the diameter or if it associates with preexisting structures in a bacterium (6). It is of considerable interest to determine how proteins like MreB act in concert with enzymes involved in envelope synthesis to generate the final topology of a cell.
One specialized type of cell elongation occurs in the prosthecate bacteria, which produce cellular appendages by extending their cell envelopes. This diverse group of organisms includes, for example, Caulobacter crescentus, Asticcacaulis biprosthecum, and Hyphomonas neptunium. The appendage of C. crescentus is referred to as a stalk, and it is found at only one pole of a cell. Although the inner and outer membranes and peptidoglycan of the stalk and cell body are contiguous, the stalk appears to be devoid of ribosomes, DNA (41), and cytoplasmic proteins (25), an indication that the core of the stalk contains either no or very little cytoplasm. This is in contrast to H. neptunium, which generates a bud at the distal end of an elongated stalk. The bud receives a copy of the chromosome and becomes a new daughter cell after division (60). In this case, not only does the stalk contain cytoplasm, it also plays a role in reproduction.
C. crescentus produces two cell types. The daughter cell, or swarmer, is flagellated at one pole. The swarmer undergoes a differentiation process in which the polar flagellum is shed, and from the same pole, the cell elongates a stalk cylinder, which is approximately five times smaller in diameter than the cell body (40). Synthesis of new stalk material, including surface array, membranes, and peptidoglycan, has been shown to occur specifically at the junction where the stalk meets the cell body (47, 48, 50). Elongation occurs as cell body-proximal material, incorporated at the stalk-body junction, is pushed away from the cell body by new synthesis. Since stalk synthesis is a type of elongation, it is possible that proteins involved in general cell elongation are also involved in stalk synthesis. Alternatively, the prosthecate bacteria may have evolved entirely specialized sets of proteins in order to generate stalks.
Penicillin-binding protein 2 (PBP2), the cell elongation transpeptidase, is the killing target of the β-lactam antibiotic amdinocillin in Escherichia coli and other gram-negative bacteria (52). Cells treated with amdinocillin undergo a rod-to-sphere transition that eventually leads to cell lysis. In C. crescentus, amdinocillin treatment leads to a general increase in cell diameter, filamentation, bulging at the stalked pole, and the formation of abnormally short and widened stalks (30, 48). These results suggest that if the target of amdinocillin in Caulobacter is PBP2, as it is in E. coli and other gram-negative organisms, then PBP2 is involved in stalk elongation and morphogenesis.
In E. coli and C. crescentus, mrdA (the structural gene for PBP2) is cotranscribed with rodA (also called mrdB). RodA is a protein of unknown function, although overexpression of RodA has been shown to stimulate a peptidoglycan synthetic and cross-linking activity in membranes where PBP2 is also overexpressed (26). E. coli cells with inactivated RodA also become spherical (53). As with other rod-shaped organisms, C. crescentus RodA is predicted to have nine transmembrane-spanning domains, a small cytoplasmic loop, and a large periplasmic loop and is highly similar to the cell division protein FtsW (24, 33, 37). RodA may act to recruit proteins important for elongation. It has also been hypothesized to be the flippase responsible for passage of peptidoglycan precursors across the inner membrane to the periplasm, for incorporation into the cell wall by a nascent transglycosylase (13).
In order to determine if proteins involved in general cell elongation are required for stalk synthesis, we tested the roles of RodA and MreB in C. crescentus. We show that rodA is essential in C. crescentus. To test whether RodA is involved in general cell elongation, shape determination, and/or stalk elongation, we constructed a C. crescentus strain in which rodA is under the control of an inducible promoter. When RodA is depleted, C. crescentus cells exhibit a pleomorphic phenotype, including rounding up, division defects, and lysis. In addition, we show that RodA is required for stalk elongation and morphogenesis. We demonstrate that depletion of MreB leads to a stalk elongation defect and propose a model in which proteins involved in general cell elongation also act at the C. crescentus pole but lead to the elongation of a structure of significantly smaller diameter than the cell proper. Finally, we show that cells depleted of MreB are unable to recover proper polarity when MreB is resynthesized; they exhibit branching and stalk synthesis at various places along the cell cylinder. Similar results were obtained during recovery from amdinocillin treatment or RodA depletion, indicating that MreB, RodA, and the target(s) of amdinocillin each possess an important role in maintaining cellular polarity during growth in C. crescentus, perhaps through their role in peptidoglycan synthesis.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or construction | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1 | E. coli 294::RP4-2(Tc::Mu)(Km::Tn7); strain used for conjugation of plasmids into C. crescentus | 49 |
| C. crescentus | ||
| NA1000 | syn-1000, previously called CB15N, synchronizable derivative of CB15 | 15 |
| CS606 | NA1000Δbla | 58 |
| JG5008 | JG5000 (16) ′recA::Tn5 | J. Gober (UCLA) |
| YB363 | Conjugation of pRodAxyl2 into NA1000 | This study |
| YB2089 | Conjugation of pMRGFP into YB363 | This study |
| YB3900 | Conjugation of pLacCC1547 into YB363 | This study |
| Plasmids | ||
| pBGST18 | pBGS18 derivative with RK2 oriT, Kanr | M. R. K. Alley |
| pBGxyl | 380-bp PstI-EcoRI fragment of xylose-dependent promoter from pNPT228XNE cloned into pBGST18/PstI-EcoRI | Y. Wang and Y. V. Brun |
| pLacCC1547 | Contains mrdA-rodA promoter, in-frame fusion of first 15 bases of mrdA and last 24 bases of mrdA, followed by entire rodA gene | This study |
| pJK75 | Contains mrdA-rodA genes, including promoter region, cloned into pGL10/HindIII-XbaI | P. J. Kang and L. Shapiro (Stanford) |
| pJK78 | Contains entire rodA gene except last 373 bp (BamHI-PvuII fragment) cloned into pBGST18/BamHI-blunt; also contains sacB cassette (PstI fragment) | P. J. Kang and L. Shapiro (Stanford) |
| pMRGFP | Contains entire E-GFPN2 cloned into HindIII-SacI site of pMR20 | This study |
| pRodAintBG | Internal fragment of rodA (bp 100 to 749) cloned into SalI-SmaI of pBGST18 in opposite orientation to lac promoter | This study |
| pRodAinttet | Contains Tetr cassette from pHP45-Tc (HindIII fragment) cloned into pRodAintBG (HindIII) | This study |
| pRodAxyl2 | First 500 bp of rodA cloned into NdeI/EcoRI site of pBGxyl to generate fusion of PxylX and first 500 bp of rodA gene | This study |
Generation of rodA lethal disruption.
The disruption plasmid pRodAintBG was introduced into C. crescentus NA1000 by conjugation. Cells were plated onto PYE (41) plates containing 20-μg/ml nalidixic acid and 20-μg/ml kanamycin. Plates were incubated at 30°C. NA1000 was used as a negative control for spontaneous kanamycin resistance, and S17-1/pJK78 was used as a positive control.
Rescuing the rodA lethal disruption phenotype.
The rescue plasmid pJK75 contains the mrdA-rodA genes, including the promoter region, in pGL10. pJK75 was introduced into C. crescentus NA1000 by conjugation. pJK75 confers kanamycin resistance and replicates in C. crescentus. The tetracycline resistance cassette from pHP45Ω-Tc (HindIII fragment) was cloned into pRodAintBG to add a tetracycline resistance marker to that plasmid, creating plasmid pRodAinttet. pRodAinttet was introduced into C. crescentus NA1000/pJK75 by conjugation. Cells were plated onto PYE containing 20-μg/ml nalidixic acid and 2-μg/ml tetracycline. NA1000, NA1000/pJK75, and S17-1/pBGST18 with no insert were used as negative controls for spontaneous tetracycline resistance, and S17-1/pRKlac290 was used as a positive control.
Construction of the RodA depletion strain, YB363.
To create pRodAxyl2, a truncated version of rodA was placed under the control of the xylose-dependent promoter PxylX in pBGST18. Oligonucleotides for amplification of rodA were designed so that the natural ATG start site of rodA was left intact with an NdeI site, and an EcoRI site was created about 500 bp downstream from the start site of the gene. The truncated version of rodA was placed immediately downstream of a 200-bp fragment of the xylX promoter. pRodAxyl2 was introduced into C. crescentus NA1000 by conjugation. Cells were plated onto PYE plates containing 20-μg/ml nalidixic acid, 20-μg/ml kanamycin, and either 0.3% xylose or 0.1% glucose. NA1000 was used as negative control for spontaneous kanamycin resistance, and S17-1/pBGxyl with no insert was used as a positive control for integration at the xylose locus on the chromosome. To test for xylose-dependent transconjugants, colonies were streaked onto PYE plates containing 20-μg/ml nalidixic acid, 20-μg/ml kanamycin, and either 0.3% xylose or 0.1% glucose. One xylose-dependent transconjugant was named YB363.
To determine if the YB363 depletion phenotype was due to polar effects on CC1548, the plasmid pLacCC1547, was conjugated into YB363, creating YB3900. YB3900 was grown overnight in PYE containing 1-μg/ml tetracycline, 5-μg/ml kanamycin, and either 0.3% xylose (PYE-X) or 0.3% glucose (PYE-G).
Microscopy.
With the exception of Fig. 5A, which was captured with Nikon Optiphot-2 microscope, phase-contrast microscopy was performed on a Nikon Eclipse E800 light microscope equipped with a ×100 Plan Apo oil objective and a Princeton Instruments cooled charge-coupled device camera model 3017. Images were captured and analyzed with MetaMorph imaging software package, version 4.6 (Universal Imaging Corporation).
FIG. 5.
MreB depletion in low phosphate. (A) JG5008 grown in HIGG-G containing 120 μM phosphate for 7.5 h. (B) JG5008 inoculated from culture in panel A after overnight growth in HIGG-X containing 30 μM phosphate. (C) JG5008 inoculated from the culture in panel A after overnight growth in HIGG-G containing 30 μM phosphate.
Whole mounts were prepared on Formvar-coated copper grids and stained with 7.5% uranyl magnesium acetate (Ted Pella). Cells were analyzed by transmission electron microscopy (TEM) in the Indiana Molecular Biology Institute's microscopy facility with a JEOL model JEM-1010 electron microscope at 60 kV.
RodA depletion and stalk length quantitation in mixed culture.
To test the role of RodA in stalk elongation, YB363 was grown in HIGG minimal medium (42) containing 5.0-μg/ml kanamycin, 0.3% xylose, and 0.03 mM phosphate to an optical density at 600 nm (OD600) of 0.554. When indicated, the glucose in each experiment is in addition to the 0.15% glucose used as carbon source in HIGG media. Cells were pelleted at 11,950 × g in a Sorvall SS-34 rotor, washed twice with fresh HIGG, and resuspended in HIGG containing 5.0-μg/ml kanamycin, 0.03 mM phosphate, and either 0.3% xylose (HIGG-X) or 0.3% glucose (HIGG-G). The cells were continually subcultured for 18 optical doublings, never reaching an OD600 higher than 0.6. Equal volumes of equal-density HIGG-X- and HIGG-G-grown cells were then used to inoculate respective fresh HIGG-X and HIGG-G, each containing 5-μg/ml kanamycin, and the cells were grown without subculturing for 13 h, reaching OD600s of 0.52 and 0.50, respectively. Images of cells were captured by phase-contrast microscopy at a magnification of ×1,000. Stalk lengths were measured with the “measure distance” function of MetaMorph.
RodA depletions in synchronous populations.
YB363 was grown overnight at 30°C with shaking in HIGG-G containing 1.0 mM phosphate and 5-μg/ml kanamycin. The overnight culture was used to inoculate HIGG-G containing 1.0 mM phosphate and 5-μg/ml kanamycin, and grown to an OD600 of 0.6. Cells were then pelleted at 4°C and resuspended to an OD600 of 0.3 in PYE containing 5-μg/ml kanamycin and either 0.3% xylose (PYE-X) or 0.3% glucose (PYE-G). Cells were then placed at 30°C with shaking to an OD600 of 0.6 (about 2 h). The cultures were then synchronized (15), resuspended in PYE-X or PYE-G, each containing 5-μg/ml kanamycin, and placed at 30°C with shaking. Samples were taken every 15 min and fixed in 1.7% formaldehyde until cell division was observed (about 120 min). The number of cells with stalks was quantitated by scoring predivisional cells from the 120-min time point of each culture for the presence or absence of a “stalk-like projection.” This was defined as any projection from the pole of the cell on the phase-contrast image, even if it could not be definitively identified as a stalk.
For the temperature shift experiments, YB363 was grown overnight at 30°C with shaking in HIGG-G containing 1.0 mM phosphate and 5-μg/ml kanamycin. The overnight culture was used to inoculate HIGG-G containing 1.0 mM phosphate and 5-μg/ml kanamycin and grown at 30°C with shaking to an OD600 of 0.6. Cells were then pelleted at 11,950 × g in a Sorvall SS-34 rotor at 4°C and resuspended to an OD600 of 0.3 in PYE-G or PYE-X, each containing 5-μg/ml kanamycin. Cells were then allowed double once at 30°C with shaking to an OD600 of 0.6 (about 2 h). The cultures were then synchronized, resuspended in PYE-X or PYE-G, each containing 5-μg/ml kanamycin, and placed at 16°C with shaking. Aliquots of the culture were taken every 30 min and fixed in 1.7% formaldehyde (Ted Pella) prior to imaging. To determine if the bulbs contained cytoplasm, a plasmid was generated which placed green fluorescent protein (GFP) under control the β-galactosidase promoter of E. coli. This promoter is constitutively active in C. crescentus. The plasmid pMRGFP was introduced into YB363, generating YB2089, and the temperature shift experiment was repeated. The bulbs were assayed for the presence of absence of the cytoplasmically expressed GFP by epifluorescent microscopy.
MreB depletion.
JG5008 was grown in HIGG-X containing 0.12 mM phosphate and 1-μg/ml tetracycline at 30°C with shaking to an OD600 of 0.53. Cells were pelleted at 4°C in a Sorvall SS-34 rotor at 11,950 × g, and resuspended in HIGG-G containing 0.12 mM phosphate and 1-μg/ml tetracycline. They were then placed at 30°C with shaking for 7.5 h. Cells were then pelleted (as before) and resuspended in a small volume of HIGG without phosphate. This suspension was used to inoculate HIGG-X or HIGG-G, each containing 0.030 mM phosphate and 1-μg/ml tetracycline. The cultures were then grown overnight at 30°C with shaking.
Recovery experiments. (i) MreB recovery.
JG5008 was grown in HIGG-G containing 0.120 mM phosphate and 1-μg/ml tetracycline for 11.5 h to deplete the MreB protein. Cells were then subcultured into HIGG-X or HIGG-G containing 0.005 mM phosphate and 1-μg/ml tetracyline. Aliquots of cells were taken from each culture over the course of 28.5 h for microscopy.
(ii) Amdinocillin recovery.
CS606, a β-lactam-sensitive derivative of NA1000, was grown overnight in PYE liquid medium at 30°C with shaking. A small volume (100 μl) of the overnight culture was used to inoculate 5 ml of PYE containing 12.5-μg/ml amdinocillin (a gift from P. Sorter, Hoffmann-La Roche), and the cells were grown at 30°C with shaking for 10 h. The cells were washed three times with HIGG containing no phosphate, resuspended in HIGG containing 0.005 mM phosphate, and placed at 30°C with shaking. Aliquots of cells were taken for microscopy over the course of 39 h.
(iii) RodA recovery.
YB363 was grown in PYE-G containing 5-μg/ml kanamycin at 23°C (room temperature) with shaking for a total of 24 h. Cells were washed and resuspended in HIGG-X containing 0.005 mM phosphate and placed at 30°C with shaking for a total of 36 h.
RESULTS
rodA is essential and required for the maintenance of cell shape.
Treatment of C. crescentus cells with amdinocillin led to an inability to elongate wild-type stalks (30, 48), suggesting that PBP2 (the product of mrdA) plays a role in stalk synthesis. Unfortunately, attempts to knock out or generate a chromosomal PBP2 depletion strain in order to study its role in stalk synthesis directly were unsuccessful, possibly due to insufficient expression of PBP2 from the xylose promoter (data not shown). Since RodA is required for the activity of PBP2 (26), we hypothesized that inactivation of RodA might also lead to a stalk elongation or morphogenesis defect. mrdA and rodA are transcriptionally coupled by a promoter found just upstream of the structural gene for mrdA (data not shown). Directly downstream of rodA is a gene encoding a putative, conserved hypothetical protein, annotated CC1548, which may also be part of the mrdA operon.
In order to create a disruption of the rodA gene, an internal fragment of the rodA gene was cloned into a suicide vector pRodAintBG carrying a kanamycin resistance marker. The frequency of kanamycin-resistant colonies obtained from mating the rodA knockout plasmid into NA1000 was statistically indistinguishable from mock matings using NA1000 with no plasmid, indicating that the disruption of rodA is lethal. Transconjugants were successfully recovered when pRodAinttet (a rodA knockout plasmid with a different resistance marker from pRodAintBG) was mated into NA1000 harboring a trans copy of rodA on a plasmid, YB3900. It was therefore concluded that the inability to recover transconjugants in the disruption matings without a trans copy of rodA indicates that disruption of rodA is lethal to the cell.
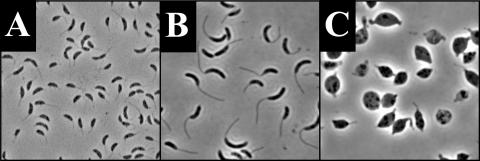
Since rodA was essential in the NA1000 background, a conditional mutant was constructed which placed rodA under the control of the xylose-inducible promoter PxylX (36). The resulting strain, YB363, was capable of growth in the presence of xylose (the PxylX inducer) but was not viable when grown with glucose. To examine the effect of RodA depletion on cell morphology, YB363 was grown in PYE (a complex medium) containing either xylose (PYE-X) or glucose (PYE-G). After overnight growth in PYE-G, approximately 75% of the YB363 population consisted of lysed, misshapen, or spherical cells (Fig. 1B). YB363 cells grown overnight in PYE-X appeared normal (Fig. 1A). The depletion phenotype of YB363 was rescued by the rodA plasmid pLacCC1547, indicating that the depletion phenotype was due to the depletion of RodA.
FIG. 1.
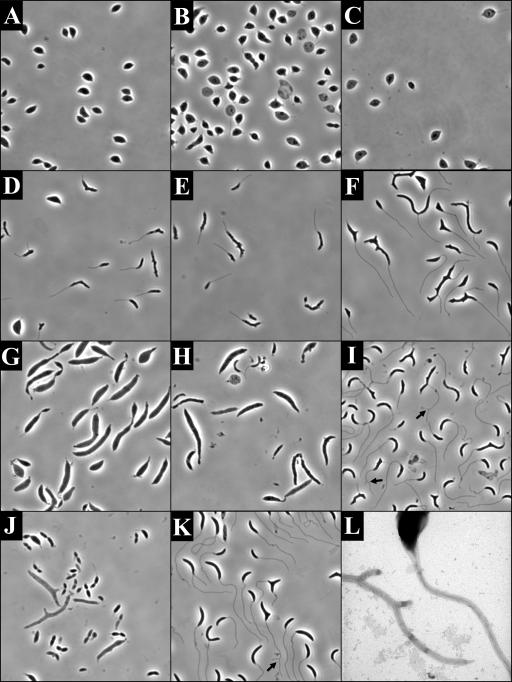
Effect of RodA depletion on cell morphology. All images are phase micrographs taken at an original magnification of ×1,000. (A) YB363 grown overnight in PYE-X. (B) YB363 grown overnight in PYE-G. (C) YB363 grown in HIGG-X with 30 μM phosphate. (D) YB363 grown in HIGG-G with 30 μM phosphate.
RodA is involved in stalk elongation.
When cells were depleted of RodA in a mixed culture in PYE-G, a stalk defect was not obvious, possibly due to an incomplete depletion of RodA before cell lysis. Since there are slow-growth mutants of E. coli that allow cells to survive inactivation of PBP2 (3), we reasoned that slower growth conditions could allow a more complete depletion of RodA without cell lysis in C. crescentus. In order to test this hypothesis, YB363 was grown in the absence of xylose in minimal medium that both decreases growth rate and dramatically increase the rate of stalk synthesis (20). As expected, YB363 cells grown in HIGG-G were able to survive and exhibited a pleomorphic phenotype. YB363 cells grown in HIGG-G were smaller, were less crescentoid in shape, and/or had more variability in stalk length (Fig. 1D) than the control cells grown in HIGG-X (Fig. 1C).
The pleomorphic phenotype of YB363 appeared between 9 and 13 doublings after the beginning of the depletion. In contrast, the cells grown only in HIGG-X appeared wild type. Cells grown in the presence of xylose (with RodA) possessed stalks which were on average 55% longer (n = 65) than cells grown in glucose (n = 113) (RodA depleted). These results indicate that depletion of RodA leads to a stalk elongation defect in a mixed population of cells.
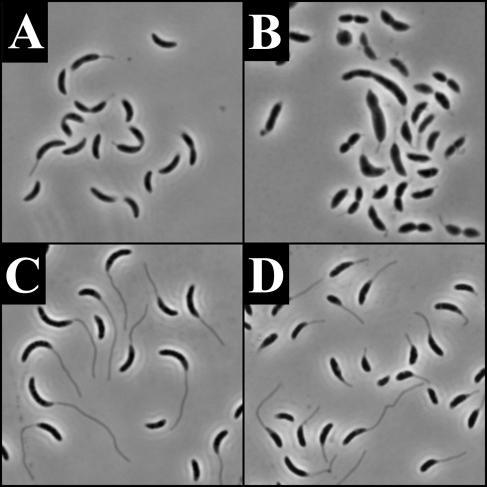
In order to determine if RodA is required for stalk elongation or if it simply modulates final stalk length, we examined stalk synthesis in a synchronized population of swarmer cells. Swarmer cells were synchronized and grown at 30°C in PYE-X or PYE-G. Phase-contrast images for 0 (swarmers only), 60, and 120 min (1 cell cycle) for cells grown in the presence of xylose and glucose are shown in Fig. 2. Cells grown in PYE-X appeared morphologically wild type and were clearly able to elongate stalks (Fig. 2E). The RodA-depleted cells became lemon shaped, and most failed to elongate stalks (Fig. 2F). At 120 min, 55% (n = 67) of nondepleted predivisional cells possessed stalks. In contrast, only 6% (n = 87) of RodA-depleted cells had stalk-like projections. TEM analysis indicated that in the rare instances where stalks were observed in the RodA-depleted cells, they were shorter and nearly two times wider at the base than cells grown under the same conditions in xylose (data not shown). These results indicate that RodA plays a role in stalk elongation, but they do not establish whether RodA's role is direct or indirect.
FIG. 2.
RodA depletion in a synchronous population of cells grown at 30°C. All images are phase micrographs taken at an original magnification of ×1,000. (A, C, and E) YB363 grown in PYE-X. (B, D, and F) YB363 grown in PYE-G. (A and B) 0 min; (C and D) 60 min; (E and F) 120 min. Arrows in panel E indicate cells with representative stalk-like projections.
RodA is required for proper stalk morphogensis.
Since a small proportion of RodA-depleted cells can elongate wide stalks in PYE-G but are able to make wild-type diameter stalks when grown in minimal medium under slower-growing conditions, we speculated that shifting cells from faster to slower growth conditions might produce an intermediate stalk phenotype. In order to test this hypothesis, cells were treated exactly as described in the previous experiment, but following synchronization, the cells were shifted to 16°C (which slows the doubling time of C. crescentus by about threefold). Aliquots of cells were fixed for microscopy over the course of one optical doubling (360 min).
The synchronized cells grown in PYE-G at 16°C showed a different phenotype from cells which were always grown at 30°C in PYE-G (compare Fig. 2F and Fig. 3F). Twenty-one percent of RodA-depleted predivisional cells possessed stalk-like projections; however, the projections appeared wider than the stalks of cells grown in the presence of xylose. In addition, 12% of the cells at the 360-min time point possessed bulbous structures that appeared first by phase-contrast microscopy at 180 min (Fig. 3F and 4B and C).
FIG. 3.
Synchronous population of YB363 shifted from 30°C to 16°C at the time of synchronization. All images are phase micrographs taken at an original magnification of ×1,000. YB363 was grown at 30°C in PYE-G, synchronized, and then shifted to 16°C in PYE-X (A, C, and E) or PYE-G (B, D, and F). (A. and B) 0 min; (C and D) 180 min; (D and F) 360 min. Arrows in panels D and F and the inset in panel F indicate cells with representative stalk-like projections. The inset shows a 3× enlargement of the cell with the arrow.
FIG. 4.
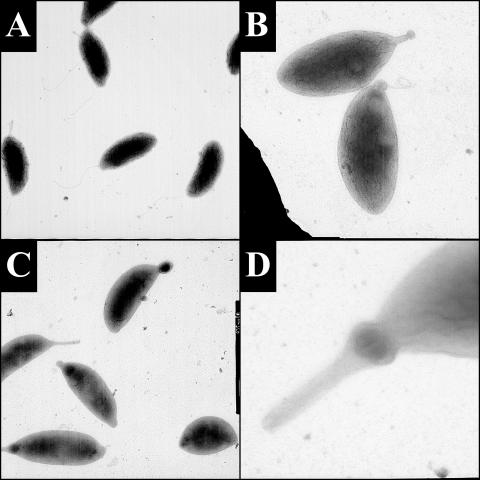
Transmission electron micrographs of YB363 grown in PYE-G at 30°C, synchronized, and shifted to 16°C in PYE-G. Micrographs were taken at the original magnifications indicated: panels A and B, swarmer cells at 0 min at ×20,000 (A) and at 180 min at ×25,000 (B); panels C and D, representative images of stalk-like projections at 360 min at ×12,000 (C) and ×75,000 (D).
TEM analysis showed that swarmer cells at time zero possessed flagella and sometimes a slight extension of the membrane at the pole opposite the flagellum (Fig. 4A). After 180 min, projections became visible on a subset of the cells. These projections were sometimes consistent with the appearance of wild-type stalk cylinders, but more often they were round and variable in diameter. For example, in Fig. 4B, a structure which appears to be a stalk is found at one pole of the cell and it is tipped by a small bulb. After 360 min, various types of projections could be seen (Fig. 4C and D). The majority of cells examined possessed either no stalk or a small bulb at one pole. Some cells, presumably corresponding to the 12% with large bulbous structures visible under phase-contrast microscopy, had bulbs of larger diameter. There were also cells that possessed stalk-like projections but with a bulb at the base (Fig. 4D). The larger bulbs contained cytoplasm, as evidenced by the presence of cytoplasmically expressed GFP, indicating that the bulging is not due simply to plasmolysis (data not shown). Rarely, cells were observed with a structure that appeared to be a wild-type stalk. We conclude that RodA is required for proper stalk morphogenesis.
MreB is involved in stalk elongation.
mreB mutants in E. coli and C. crescentus show a similar rounded-up phenotype to mrdA and rodA mutants (16, 55, 56). It has been proposed that the peptidoglycan-synthesizing machinery responsible for cell elongation tracks along a scaffold of MreB polymers present inside the cells (16, 28). If MreB is required for either PBP2 or RodA activity or localization, and PBP2 and/or RodA are involved in stalk elongation and morphogenesis, depletion of MreB should lead to a stalk synthesis defect.
To test this hypothesis, we used an MreB depletion strain, JG5008, which contains the mreB gene under the control of the xylose-inducible promoter PxylX. JG5008 was depleted of MreB for 7.5 h in PYE-G (Fig. 5A). These cells were then shifted to HIGG-X or HIGG-G, each containing 30 μM phosphate. Under these conditions, the rate of stalk synthesis per cell cycle is dramatically increased; thus, any stalk synthesis defect will be more easily detected (20, 48). The phenotype of the cells was examined after 16 h (approximately three doublings). JG5008 grown in the presence of xylose appeared essentially wild type and possessed long stalks of normal diameter (Fig. 5B). However, the majority of the cells depleted of MreB in low phosphate were lysed, possessed short or no stalks, and polar extensions and bulbs very similar to those seen with amdinocillin treatment (48) and RodA depletion (Fig. 4C). The stalk phenotype of the MreB-depleted cells was not an effect of rapid lysis since these cells were able to increase their size considerably (compare Fig. 5A and C). These results demonstrate that MreB plays a role (direct or indirect) in stalk biogenesis.
MreB is required for maintenance of cellular polarity, including stalk placement.
Recent findings suggest that the MreB-like proteins play a critical role, not only in the maintenance of cell shape during growth but also in the proper segregation of chromosomes (17, 18, 31, 51) and the localization of developmental regulators (18). MreB may act as a track for the locomotion of chromosomes and polarly localized proteins (18), analogous to the establishment of polarity in eukaryotic organisms via the cytoskeleton (44, 45, 57). An alternative interpretation is that MreB is involved in the maintenance of cellular polarity through a role in peptidoglycan synthesis (16, 18) and shaping and that the absence of MreB allows polar markers (such as the inert cell wall of polar caps) to be created at aberrant sites.
We hypothesized that if the absence of MreB leads to the formation of aberrant polar material and/or ectopic poles, then cells allowed to recover from the depletion of MreB would manifest gross defects in morphology, such as creation of ectopic poles (12, 39, 59) and that stalk synthesis could be used as a convenient marker of ectopic poles. In order to test this hypothesis, we used the MreB depletion strain, JG5008. JG5008 was first depleted of MreB for 11.5 h and then shifted to HIGG-X containing 5 μM phosphate in order to restore MreB synthesis. Cells were shifted to phosphate-limiting medium to induce stalk elongation and to inhibit cell division (20) because we reasoned that polarity defects might be rapidly lost in viable cells after the shift to inducer (as the cell envelope of the daughter cells would be synthesized in the presence of MreB). Since phosphate-starved cells have a limited capacity for division, a majority of the cells in the recovery population constitute the original population of viable cells.
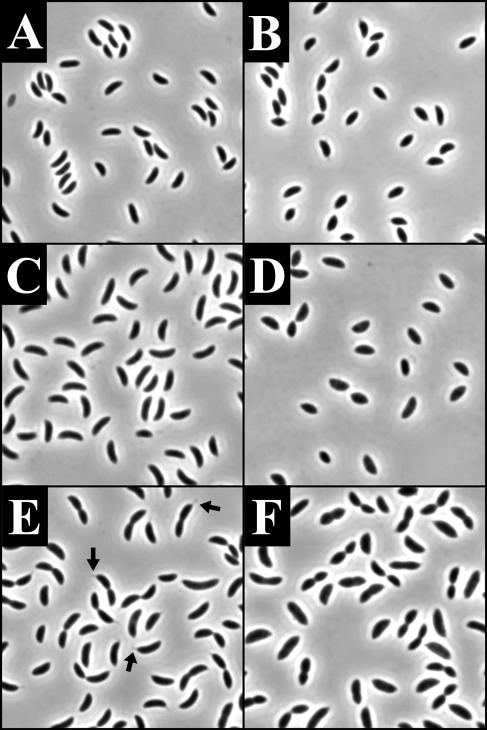
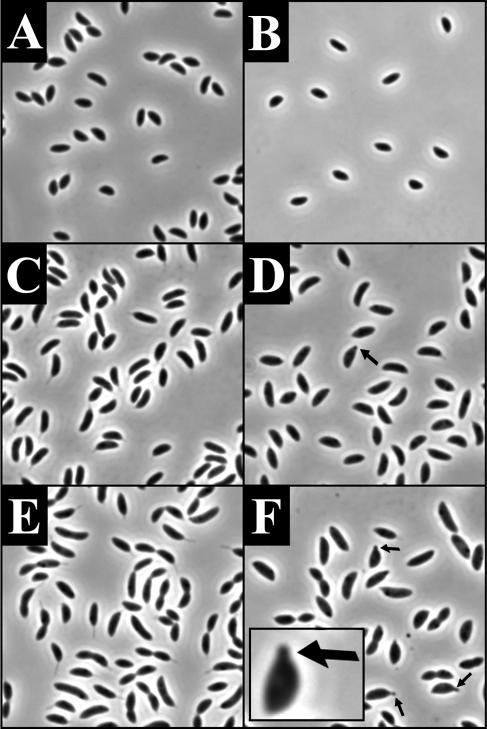
Images of the cells were captured during the course of the recovery (Fig. 6D to F). Initially, the cells were rounded up and lemonoid in shape (similar to Fig. 6A). After 2 h, the cells were smaller in diameter, elongated, and often displayed a bumpy appearance (Fig. 6D). After 11.5 h, the bumps on the surface of the cell became more apparent (Fig. 6E), and after 28.5 h, the bumps began to elongate into branches (Fig. 6F). The branches were often tipped by stalks, with some cells possessing five or more stalks. Some of the stalks themselves also exhibited a branching phenotype (Fig. 6L). When MreB-depleted cells were shifted to HIGG-G containing 5 μM phosphate, they continued to round up, eventually lysing (Fig. 6A to C). JG5008 always grown in HIGG-X with 5 μM phosphate did not display a branching morphology (data not shown). The severity of the recovery phenotype was dependent on the length of the depletion and the recovery time and was also observed in PYE medium (Fig. 7). This indicates that the branching phenotype is not an indirect consequence of the combination of MreB depletion and low-phosphate growth conditions. The distorted morphology and misplaced stalks seen in the recovered cells indicate that the presence of MreB is critical to the maintenance of cellular polarity and stalk localization in C. crescentus.
FIG. 6.
Morphology of cells recovering from MreB depletion (D to F and L), amdinocillin treatment (G to I), and RodA depletion (J and K). Images in panels A to K are phase micrographs taken at an original magnification of ×1,000. Panel L is a transmission electron micrograph taken at an original magnification of ×30,000. All cells underwent recovery in HIGG containing 5 μM phosphate. (A to C) Phenotypes of JG5008 after 9.5 (A), 19 (B), and 36 (C) h of depletion in HIGG-G. (D to F) Phenotypes of JG5008 grown in HIGG-G for 11.5 h of depletion and then recovered in HIGG-X for 2 (D), 11.5 (E), and 28.5 (F) h. (G to I) Recovery after amdinocillin treatment. Phenotype of cells after 10 h of growth in the presence of 12.5-μg/ml amdinocillin (G) with 16 (H) and 39 (I) h of recovery. (J and K) Morphology of YB363 grown for 24 h at room temperature in PYE-G (J) and shifted to HIGG-G for 36 h (K). (L) Transmission electron micrograph of branching stalk phenotype observed in JG5008 recovery (compare to panel F). Arrows in panels I and K indicate cells with obvious stalk forks.
FIG. 7.
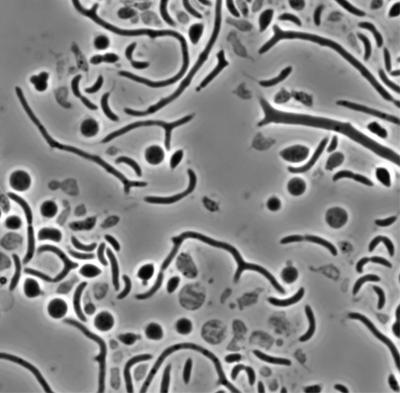
Morphology of cells recovering from MreB depletion. The image is phase micrograph taken at an original magnification of ×1,000. JG5008 was depleted of MreB for 11.5 h in PYE-G and then shifted to PYE-X for 9 h for recovery.
The targets of amdinocillin and RodA are required for maintenance of cellular polarity, including stalk placement.
It has been previously suggested that branching in rod-shaped bacteria is the result of “asymmetries” in the cell wall generated during the elongation phase of growth (21). If this is the case, conditions other than MreB depletion that perturb cell elongation, such as RodA depletion and amdinocillin treatment, should result in a similar branching or budding phenotype when cells are allowed to recover. To test this hypothesis, a β-lactam-sensitive strain of C. crescentus, CS606, was treated with amdinocillin for a period of 10 h. The concentration of amdinocillin used in this experiment affects stalk synthesis and causes C. crescentus to grow as wide filaments (48) (Fig. 6G). The cells were then washed to remove excess amdinocillin and allowed to recover in phosphate-limiting medium, as in the MreB recovery. After 16 h of recovery, cells were smaller in diameter, possessed stalks, and also exhibited some branching (Fig. 6H). After 39 h of recovery, the phenotypes observed were strikingly similar to the MreB recovery experiment in that cells were elongated and possessed forked and misplaced stalks (Fig. 6I). The cells also displayed budding and branching but to a lesser extent than cells recovering from the MreB depletion. The budding and branching phenotype is not seen when CS606 is grown under the same conditions but with no amdinocillin treatment (data not shown).
Similarly, to examine the role of RodA in maintenance of polarity, we grew YB363 in PYE-G for 24 h to deplete the RodA protein (Fig. 6J) and then shifted cells to the same phosphate-limiting medium for recovery. The phenotype of YB363 after 36 h of recovery was less dramatic than those in either the MreB or amdinocillin experiments; however, approximately 10% of cells still exhibited cell branching and/or forked and misplaced stalks (Fig. 6K). These results suggest that RodA and the target(s) of amdinocillin both have a function in the preservation of cellular polarity and stalk placement in C. crescentus.
DISCUSSION
In eukaryotes, cell shape is primarily a consequence of the arrangement of cytoskeletal fibers, namely microfilaments, microtubules, and especially intermediate filaments that form a dynamic network inside the cell. Research on cell shape in bacteria has focused primarily on peptidoglycan and other components of the cell envelope and the enzymes associated with its synthesis (19, 23, 43). However, recent exciting studies of bacterial actin homologs, namely MreB and Mbl, have produced strong evidence for the presence of a bacterial cytoskeleton and have changed the entire context in which cell shape is studied in prokaryotes (34, 35, 54).
The MreB protein forms a spiral along the inner membrane of bacterial cells and is critical for the proper control of cell diameter during elongation (5, 16, 28, 31). MreB may be interacting directly or indirectly with peptidoglycan-synthesizing proteins to localize, guide, and/or stabilize these proteins as new cell wall is laid down (16, 28). Is the role of the MreB-like proteins to physically constrain the diameter of the cell through force generation, or are they instead associating with unidentified structures of a defined diameter in the cell and then recruiting proteins required for new envelope synthesis? In order to begin dissecting this question, we chose to study the synthesis of a specific structure, a bacterial stalk. We showed previously that stalk elongation and morphogenesis are affected by treatment of cells with amdinocillin (48), a β-lactam antibiotic that specifically inhibits the cross-linking reaction of PBP2 in E. coli (27, 52). Aside from the obvious fact that stalks grow longitudinally, the amdinocillin phenotype was the first indication that stalk elongation might simply be an iteration of cell body elongation.
To investigate this possibility further, we examined the role of RodA and MreB, two other proteins involved in cell elongation. We show that RodA depletion in C. crescentus causes cells to become lemon shaped or round and eventually lyse. In addition, depletion of RodA leads to a stalk elongation defect and the formation of polar bulbous structures. These structures are often tipped by or are found at the tip of stalks. Unlike the stalks of cells following amdinocillin treatment which are wide at the cell body junction (48), the RodA depletion stalk morphology is more rounded and regular. Growth of the bulbous structures is still somewhat constrained at the junction with the body but with outgrowth exhibiting a loss of restraint over diameter. The difference in the two phenotypes may be due to some unknown effects of amdinocillin treatment in addition to the inhibition of PBP2. Another possibility is that differences in growth and depletion/inactivation conditions lead to differences in the manifestation of the phenotypes.
MreB depletion in C. crescentus also leads to a stalk elongation defect, even under conditions that normally stimulate stalk elongation. Although some cells possess wild-type-looking stalks, they are generally much shorter than the stalks of control cells and often are found at the end of misshapen cell extensions. These structures may be attempts at stalk synthesis, as is the case with the amdinocillin treatment (48). The role of MreB in stalk elongation raises some interesting questions. Since the MreB spiral encircles the body of the cell, how does it also act at such a defined place at the cell pole? The stalk cylinder is five times smaller than the cell cylinder. If MreB is involved in physically constraining the cell envelope during growth, parallel to the proposed role of FtsZ during cell division (1, 4, 14), it might be expected that the diameter of the cell would be inherent to the shape of the MreB spiral. Curvature intrinsic to the MreB polymer would not be anticipated to generate two structures of such different diameters (the stalk and the cell body). Of course, it is still possible that other proteins act to modulate MreB in order to make different shapes, as may be the function of the recently identified CreS protein that imparts to C. crescentus its distinctive cell curvature (2).
We show that cells recovering from MreB depletion exhibit a budding and branching phenotype. This phenotype is remarkably similar to that of E. coli cells in which multiple low-molecular-weight PBPs have been deleted (12, 39, 59). It was recently shown in E. coli that budding and branching sites have the biochemical properties of polar caps (39). At these sites, the cell wall is inert (inert peptidoglycan [iPG]) (12), covered by a cap of immobile outer membrane proteins (9), and is recognized by polarly localizing proteins (39). In C. crescentus, the branches and buds seen after MreB recovery are often tipped by stalks, a strong indication that these sites are also behaving as ectopic poles in C. crescentus. These results suggest that MreB may have a function in preventing insertion of iPG during elongation. This role would be indirect if the localization of the iPG-synthesizing protein(s) requires normal cell shape and polarity cues (59). Another possibility is that MreB is directly required for the localization and/or activity of the low-molecular-weight PBPs (8, 38, 59).
It has also been shown that in Bacillus subtilis overexpression of the MreB-like protein Mbl leads to a propensity to branch (5). Perhaps the increased copy number of Mbl in these cells leads to a titration of the low-molecular-weight PBPs from sites where they are normally required. The formation of branches in cells recovering from amdinocillin treatment and RodA depletion, albeit at a lower frequency than during recovery from MreB depletion, suggests that these proteins may also function (directly or indirectly) in preventing the insertion of iPG along the cylinder of the cell.
Another implication of these results concerns the mislocalization of chromosomes and developmental regulators in MreB-depleted cells (17, 18, 31, 51). As stated in the introduction, one proposed role for MreB is to traffic bacterial macromolecules within the cell. In light of the results presented in this work, an equally valid alternative interpretation is that cells depleted of MreB create ectopic poles which could then be recognized by proteins that localize to poles without direct interaction with MreB. More studies will be needed in order to investigate this possibility further. For example, if the above hypothesis is correct, it would be expected that chromosomes and polarly localizing proteins will also be mislocalized in cells treated with amdinocillin, which also show a strong propensity to branch upon recovery.
What determines bacterial shape? One attractive model is that a given shape is defined by cell growth away from iPG (59). Consider the example of E. coli, in which inert polar caps are generated at division. In the iPG model, the cell cylinder is created by insertion of new envelope along the long axis of the cell (7, 10, 11), away from the inert caps. The diameter defined by the edge of the iPG is hypothesized to be maintained along the cell cylinder by turgor pressure (29) and/or internal scaffolding (59). This model is supported by some experimental evidence which shows buds and branches are always tipped by iPG (12). A branch would be generated when iPG is incorporated into the elongating cylinder of the cell, followed by incorporation of new peptidoglycan at the edge of the iPG (59).
If bacterial shapes are generated by growth away from iPG, is it plausible that stalks are formed through the same mechanism? Assuming C. crescentus poles generated at division are inert, this presents a special problem for the synthesis of a polar stalk. How could synthesis of iPG in a cap that is already inert lead to the generation of a stalk cylinder? In the context of the iPG model for cell shape determination, either C. crescentus poles are not inert (this remains to be determined) or a distinction must be made between septal peptidoglycan (which is generated in an FtsZ-dependent manner and is itself inert) and the edge of the iPG, from which elongation would take place. Of course, C. crescentus stalks may be generated by another shape-determining mechanism that still utilizes cell elongation proteins such as MreB and RodA. Experimental examination of these questions will shed light on the still mysterious area of shape determination in bacteria and help us understand the underlying mechanisms that create the myriad of morphologies in the bacterial world.
Acknowledgments
We thank members of the Brun laboratory for critical reading of the manuscript, J. Gober for sending JG5008 prior to publication, P. J. Kang and L. Shapiro for sending pJK75 and pJK78 prior to publication, and B. Stein for technical assistance with TEM.
This work was supported by grants from the National Institutes of Health (GM51986 to Y.V.B. and GM61336 to J. Reilly and Y.V.B.) and by a National Institutes of Health Predoctoral Fellowship (GM07757) to J.K.W.
REFERENCES
- 1.Addinall, S. G., and B. Holland. 2002. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 2.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705-713. [DOI] [PubMed] [Google Scholar]
- 3.Bouloc, P., A. Jaffe, and R. D'Ari. 1989. The Escherichia coli lov gene product connects peptidoglycan synthesis, ribosomes and growth rate. EMBO J. 8:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bramhill, D. 1997. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13:395-424. [DOI] [PubMed] [Google Scholar]
- 5.Carballido-Lopez, R., and J. Errington. 2003. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell. 4:19-28. [DOI] [PubMed] [Google Scholar]
- 6.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 7.Den Blaauwen, T., M. E. Aarsman, N. O. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 8.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Pedro, M. A., C. G. Grünfelder, and H. Schwarz. 2004. Restricted mobility of cell surface proteins in the polar regions of Escherichia coli. J. Bacteriol. 186:2594-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Pedro, M. A., J. C. Quintela, J. -V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pedro, M. A., H. Schwarz, and A. L. Koch. 2003. Patchiness of murein insertion into the sidewall of Escherichia coli. Microbiology 149:1753-1761. [DOI] [PubMed] [Google Scholar]
- 12.de Pedro, M. A., K. D. Young, J. V. Holtje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehlert, K., and J. -V. Höltje. 1996. Role of precursor translocation in coordination of murein and phospholipid synthesis in Escherichia coli. J. Bacteriol. 178:6766-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errington, J. , R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes, K., J. Moller-Jensen, G. Ebersbach, T. Kruse, and K. Nordstrom. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 18.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glauner, B., and J. -V. Holtje. 1990. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 265:18988-18996. [PubMed] [Google Scholar]
- 20.Gonin, M., E. M. Quardokus, D. O'Donnol, J. Maddock, and Y. V. Brun. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182:337-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gullbrand, B., T. Akerlund, and K. Nordstrom. 1999. On the origin of branches in Escherichia coli. J. Bacteriol. 181:6607-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heijenoort, J. V. 1994. Biosynthesis of the bacterial peptidoglycan unit. In J. M. Ghuysen and R. Hakenback (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands.
- 23.Holtje, J. -V., and U. Schwarz. 1985. Biosynthesis and growth of the murein sacculus, p. 77-119. In N. Nanninga (ed.), Molecular cytology of Escherichia coli. Academic Press, London, United Kingdom.
- 24.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ireland, M. M., J. A. Karty, E. M. Quardokus, J. P. Reilly, and Y. V. Brun. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 45:1029-1041. [DOI] [PubMed] [Google Scholar]
- 26.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. Spratt, and M. Matsuhashi. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 261:7024-7031. [PubMed] [Google Scholar]
- 27.Ishino, F., S. Tamaki, B. G. Spratt, and M. Matsuhashi. 1982. A mecillinam-sensitive peptidoglycan crosslinking reaction in Escherichia coli. Biochem. Biophys. Res. Commun. 109:689-696. [DOI] [PubMed] [Google Scholar]
- 28.Jones, L. J. , R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 29.Koch, A. L. 1983. The surface stress theory of microbial morphogenesis. Adv. Microb. Physiol. 24:301-366. [DOI] [PubMed] [Google Scholar]
- 30.Koyasu, S., A. Fukuda, Y. Okada, and J. Poindexter. 1983. Penicillin-binding proteins of the stalk of Caulobacter crescentus. J. Gen. Microbiol. 129:2789-2799. [Google Scholar]
- 31.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labischinski, H., and H. Maidhof. 1994. Bacterial peptidoglycan: overview and evolving concepts, p. 23-38. In J. M. Ghuysen and R. H. Hakenback (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands.
- 33.Lara, B., and J. A. Ayala. 2002. Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol. Lett. 216:23-32. [DOI] [PubMed] [Google Scholar]
- 34.Margolin, W. 2003. Bacterial shape: growing off this mortal coil. Curr. Biol. 13:R705-R707. [DOI] [PubMed] [Google Scholar]
- 35.Mayer, F. 2003. Cytoskeletons in prokaryotes. Cell Biol. Int. 27:429-438. [DOI] [PubMed] [Google Scholar]
- 36.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercer, K. L. N., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsen, T., A. S. Ghosh, M. B. Goldberg, and K. D. Young. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poindexter, J. L. S., and G. C. Bazire. 1964. The fine structure of stalked bacteria belonging to the family Caulobacteraceae. J. Cell Biol. 23:587-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poindexter, J. S. 1978. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J. Bacteriol. 135:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popham, D. L., and K. D. Young. 2003. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr. Opin. Microbiol. 6:594-599. [DOI] [PubMed] [Google Scholar]
- 44.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 45.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 46.Salton, M. R. J. 1994. The bacterial envelope—a historical perspective, p. 1-22. In J. M. Ghuysen and R. Hakenback (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands.
- 47.Schmidt, J. M., and R. Y. Stanier. 1966. The development of cellular stalks in bacteria. J. Cell Biol. 28:423-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seitz, L. C., and Y. V. Brun. 1998. Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J. Bacteriol. 180:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, R., U. Prieffer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 50.Smit, J. , and N. Agabian. 1982. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J. Cell Biol. 95:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soufo, H. J. , and P. L. Graumann. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916-1920. [DOI] [PubMed] [Google Scholar]
- 52.Spratt, B. G. 1977. The mechanism of action of mecillinam. J. Antimicrob. Chemother. 3(Suppl. B):13-19. [DOI] [PubMed] [Google Scholar]
- 53.Tamaki, S., H. Matsuzawa, and M. Matsuhashi. 1980. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J. Bacteriol. 141:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 55.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wachi, M., and M. Matsuhashi. 1989. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol. 171:3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wasteneys, G. O. 2000. The cytoskeleton and growth polarity. Curr. Opin. Plant Biol. 3:503-511. [DOI] [PubMed] [Google Scholar]
- 58.West, L., D. Yang, and C. Stephens. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]
- 60.Zerfas, P. M., M. Kessel, E. J. Quintero, and R. M. Weiner. 1997. Fine-structure evidence for cell membrane partitioning of the nucleoid and cytoplasm during bud formation in Hyphomonas species. J. Bacteriol. 179:148-156. [DOI] [PMC free article] [PubMed] [Google Scholar]