Abstract
The bldC locus, required for formation of aerial hyphae in Streptomyces coelicolor, was localized by map-based cloning to the overlap between cosmids D17 and D25 of a minimal ordered library. Subcloning and sequencing showed that bldC encodes a member of a previously unrecognized family of small (58- to 78-residue) DNA-binding proteins, related to the DNA-binding domains of the MerR family of transcriptional activators. BldC family members are found in a wide range of gram-positive and gram-negative bacteria. Constructed ΔbldC mutants were defective in differentiation and antibiotic production. They failed to form an aerial mycelium on minimal medium and showed severe delays in aerial mycelium formation on rich medium. In addition, they failed to produce the polyketide antibiotic actinorhodin, and bldC was shown to be required for normal and sustained transcription of the pathway-specific activator gene actII-orf4. Although ΔbldC mutants produced the tripyrrole antibiotic undecylprodigiosin, transcripts of the pathway-specific activator gene (redD) were reduced to almost undetectable levels after 48 h in the bldC mutant, in contrast to the bldC+ parent strain in which redD transcription continued during aerial mycelium formation and sporulation. This suggests that bldC may be required for maintenance of redD transcription during differentiation. bldC is expressed from a single promoter. S1 nuclease protection assays and immunoblotting showed that bldC is constitutively expressed and that transcription of bldC does not depend on any of the other known bld genes. The bldC18 mutation that originally defined the locus causes a Y49C substitution that results in instability of the protein.
A special feature of the developmental cycle of the filamentous bacteria Streptomyces is the formation at the start of differentiation of an aerial mycelium. This structure consists of hyphae that grow out of the aqueous environment of the substrate mycelium into the air, giving the developing colonies their characteristic fuzzy appearance. Subsequently, each multigenomic aerial hypha undergoes a synchronous septation event, giving rise to ∼50 to 100 unigenomic prespore compartments that ultimately develop into mature exospores (10, 16, 26, 52).
Genetic analysis of differentiation in Streptomyces coelicolor has revealed at least three classes of genes required for the formation of an aerial mycelium: the ram, chp, and bld genes. The ram and chp genes specify hydrophobic structural components required for aerial hyphae to escape surface tension and grow into the air, while the majority of bld genes encode regulatory proteins (16).
The ram genes were discovered by virtue of their ability to induce rapid aerial mycelium formation when overexpressed in the wild type (32), and they have recently been shown to specify production of the morphogenetic peptide SapB (29). SapB is a 21-amino-acid peptide that plays a vital role in aerial mycelium formation during growth on rich medium (16, 26, 51, 52). It functions as a surfactant, releasing surface tension at the air-water interface to allow nascent aerial hyphae to escape into the air (47). The structure of SapB has recently been determined, revealing it to be a lantibiotic-like peptide containing two eight-residue loops formed by lanthionine bridges between Cys and dehydroalanine residues (29). Aerial mycelium formation on minimal medium is SapB independent, and SapB is not produced (51). The ram cluster consists of five genes, the SapB biosynthetic operon itself (ramCSAB) and the divergently encoded response regulator, RamR, which activates transcription of the ramCSAB operon on rich medium (25, 38, 40). SapB is derived from the 42-amino-acid primary translation product of the ramS gene through extensive posttranslational modification (29). RamC is likely to be the SapB synthetase involved in this posttranslational processing, and the ramAB genes encode components of an ABC transporter that may function in SapB export (29). A ramR mutant is blocked in SapB production and cannot erect aerial hyphae on rich medium (38, 40). Conversely, overexpression of ramR results in SapB overproduction and the biosynthesis of SapB by wild-type strains under conditions when its production is normally repressed (38). The ram genes are not transcribed during growth on minimal medium (25).
The chaplins are a family of eight hydrophobic cell wall-associated proteins that confer hydrophobicity on aerial hyphae and spores, and constructed strains lacking most or all of the chaplin (chp) genes fail to form aerial hyphae on all media tested (12, 13, 19). The chaplins can self assemble to form amyloid-like fibers and are believed to heteropolymerise into filaments on the surface of the aerial hyphae (12, 13, 19). Like SapB, the chaplins are very surface active and can reduce surface tension from 72 to 26 mJ/m2 (12), an ability likely to facilitate the escape of aerial hyphae into the air and to prevent them from collapsing back through surface tension.
bld mutants are a class of developmental mutants that cannot erect aerial hyphae and therefore appear bald, lacking the characteristic fuzzy morphology of the wild type (10, 16, 26, 52). Many of the characterized bld genes encode proteins with regulatory functions; bldB, bldD, and bldM encode known or putative DNA-binding proteins (15, 17, 18, 27, 35, 42), bldN encodes a sigma factor (1, 2), and bldG specifies an anti-anti-sigma factor (4). Expression of the chaplin genes is developmentally regulated, and their transcription is blocked in all the bld mutants tested, including bldC (19). Expression of the ram genes is also developmentally regulated, and ramR and ramCSAB transcription is blocked in bldA, bldB, bldD, and bldH mutants (bldC has not been tested) (25). Further, almost all bld mutants regain the ability to form aerial structures when purified SapB is applied to the colony surface (51). However, these aerial filaments fail to metamorphose into chains of spores, implying that SapB plays a purely structural role and that the bld genes control other functions vital for differentiation in addition to SapB production (47). Similarly, the engineered overexpression of ramR in bld mutant strains induces SapB biosynthesis and restores aerial hyphae formation (38).
In addition to their morphological complexity, streptomycetes are also renowned for their ability to produce a diverse range of secondary metabolites, many of medical and veterinary importance. As well as causing loss of aerial mycelium formation, mutations in some bld loci have pleiotropic effects on antibiotic production. Certain S. coelicolor bld mutants are deficient in the production of actinorhodin, undecylprodigiosin, methylenomycin, and calcium-dependent antibiotic (7, 9, 34).
bldC was one of four loci identified in the first genetic screen for S. coelicolor mutants defective in aerial mycelium formation (34) and it is the only remaining uncharacterized locus from that study. Only one mutant allele of bldC (bldC18, carried by strain J660) was identified. Phenotypic characterization of bldC strains showed that, as with many of the bld mutants, aerial mycelium formation depended on the medium; although bald on many media, aerial hyphae formation and sporulation were restored on minimal medium containing mannitol, galactose, or maltose (34). Here, we use a map-based cloning strategy to isolate bldC and find that it encodes a member of a family of small DNA-binding proteins related to the DNA-binding domain found in members of the MerR family of transcriptional activators. We show that the bldC18 allele carries a point mutation that results in loss of the bldC product, causing the same phenotype as constructed null mutants. We also show that bldC is required for expression of the pathway-specific activators of the actinorhodin and undecylprodigiosin biosynthetic genes during differentiation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. coelicolor strains used are listed in Table 1 and were cultured on R2YE, SMMS, MS, or DNA plates and in yeast extract-malt extract (YEME) or tryptic soy broth liquid media (28). Escherichia coli BL21λDE3(pLysS) (46) was used to overproduce BldC, and BW25113 (14) was used for PCR-targeted disruptions (21). Plasmids used were pSET152 (3), pIJ2925 (24), and pET15b (Novagen).
TABLE 1.
Strains and oligonucleotides used in this study
| Strain or primer | Genotype or sequence | Reference or source |
|---|---|---|
| Strains | ||
| S. coelicolor | ||
| M600 | SCP1− SCP2− | 28 |
| M145 | SCP1− SCP2− | 28 |
| J660 | bldC18 mthB2 cysD18 agaA7 SCP1NF SCP2* | 34 |
| J2161 | ΔbldC::apr derivative of M600 | This work |
| J2162 | ΔbldC::apr derivative of J660 | This work |
| J2166 | ΔbldC derivative of M600 | This work |
| J2167 | ΔbldC::apr derivative of M145 | This work |
| J2168 | ΔbldC derivative of M145 | This work |
| J2169 | bldC+ derivative of J660 | This work |
| J3210 | bldC18 derivative of M600 | This work |
| J3211 | bldC18 derivative of M145 | This work |
| J1700 | bldA39 hisA1 uraA1 strA1 SCP1− SCP2− | 30 |
| J669 | bldB43 mthB2 cysD18 agaA7 SCP1NF SCP2* | 34 |
| J774 | bldD53 cysA15 pheA1 mthB2 strA1 SCP1NF SCP2* | 34 |
| 166 | bldF hisD3 pheA1 strA1 SCP1+ SCP2+ | 43 |
| WC103 | bldG103 hisA1 uraA1 strA1 Pgl−SCP1− SCP2− | 7 |
| WC109 | bldH109 hisA1 uraA1 strA1 Pgl−SCP1− SCP2− | 7 |
| HU261 | bldJ261 hisA1 uraA1 strA1 Pgl− SCP1NF SCP2* | 50 |
| NS17 | bldK::aadA derivative of M145 | 39 |
| J2151 | ΔglkA119 bldM::hyg SCP1− SCP2− | 35 |
| J2177 | ΔglkA119 bldN::hyg SCP1− SCP2− | 2 |
| Primers | ||
| Cloning and mutagenesis primers | ||
| BLDC1 | CGTCGCTTGGAGCTGTGGTTCC | |
| BLDC2 | TGCCGACAAGGGACGTAGGTCC | |
| BLDC5 | CCGCATATGACCGCTCGTACCCCGGACGCTGAGCCGCTGCTGACC | |
| BLDC6 | CCCGGTTATTCAGTTGTTCAGG | |
| Y49F FOR | TTCCGCGAAGCCGAGGTCCG | |
| Y49F REV | GCGGCGATGCCCGCCGAGCG | |
| Gene disruption primers | ||
| BLDC K-OUT FOR | ATGAACCGAGAAGGTTCGGTTCTCCCGAGGAGGCCGCTCATTCCGGGGATCGTCGACC | |
| BLDC-K-OUT REV | GGTTGGGGGACCCGCCGTTCTGCCCGGTTATTCAGTTGTTGTAGGCTGGAGCTGCTTC | |
| S1 nuclease mapping and runoff primers | ||
| BLDC2 | See cloning and mutagenesis primers | |
| BLDC3a | GAACATGGTGGCGACCTCAGCC | |
| BLDC4 | TATATACTGTGACTGATCGTCACAGAGG | |
| BLDC17 | GAGCGGCCTCCTCGGGAG | |
| BLDC25a | GCACCTTGACCCGCGTTGCC | |
| ACTIIORF4.1a | GTACGTCTGCAGCGTCGTCATGGCGCT | |
| ACTIIORF4.2 | ATCGGAGATCGCTTGTGACGGCA | |
| HRDB FOR | CGCAAGGTACGAGTTGATGACC | |
| HRDB REVa | CCATGACAGAGACGGACTCGG | |
| REDD.1a | GATCGATACGGGTCCCAATA | |
| REDD.JW2 | CATGGATCCTGCTTCGTTTGCGTCGTTCAGTTC | |
| REDZ.1a | CTCCAGCAATGCGCGAAT | |
| REDZ.2 | CGTGCACGTGTCCTTCTG |
Labeled primers.
Protoplast transformation and conjugation from E. coli into S. coelicolor.
To bypass the methyl-specific restriction system of S. coelicolor, cosmids and plasmids were passed through the dam dcm hsdS E. coli strain ET12567 (33) prior to protoplast transformation or conjugation. E. coli ET12567 carrying the nontransmissible, oriT-mobilizing “driver” plasmid pUZ8002 (41) was used for conjugation. Streptomyces protoplast transformation and conjugation from E. coli to Streptomyces were carried out as described by Kieser et al. (28).
PCR-based site-directed mutagenesis.
The oligonucleotides BLDC1 and BLDC2 were used to amplify bldC and its promoter region from M600. The PCR product was cloned into the SmaI site of pIJ2925 to create pIJ6839. A single base-pair substitution was engineered by PCR-based site-directed mutagenesis. Two abutting oligonucleotides, Y49F FOR and Y49F REV, were used to amplify the whole of the plasmid, introducing a change from TAC to TTC into the Y49 codon. The PCR program was 95°C for 5 min; then 10 cycles of 95°C for 1 min, 60°C for 30 s, and 72°C for 8 min 30 s; 10 cycles of 95°C for 1 min, 60°C for 30 s, and 72°C for 12 min 30 s; followed by a final elongation step of 72°C for 15 min. The resulting PCR product was circularized by self ligation to create pIJ6840 and sequenced to ensure that only the intended mutation had been introduced. The bldC alleles were subcloned as BamHI-EcoRI fragments into the conjugative vector pSET152 to create pIJ6842 (encoding wild-type BldC) and pIJ6844 (encoding BldCY49F). The bldC18 allele was amplified from the chromosome of J660 with the oligonucleotides BLDC1 and BLDC2 and cloned into the EcoRV site of pSET152 to create pIJ6843 (encoding BldCY49C). The constructs were introduced into J660 by conjugation.
Overexpression of His-tagged BldC and production of a BldC polyclonal antibody.
A BldC overexpression construct was produced by amplifying the bldC-coding region with M600 chromosomal DNA as a template and the oligonucleotides BLDC5 and BLDC6. These oligonucleotides introduced an NdeI site overlapping the ATG start codon and changed the fourth, sixth, seventh, and eighth codons to synonymous codons commonly associated with highly expressed genes in E. coli. The resulting PCR fragment was cloned into the SmaI site of pIJ2925 to create pIJ6837 and sequenced. The bldC coding region was removed from pIJ6837 as a NdeI-BamHI fragment and ligated to pET15b (Novagen) that had been digested with NdeI and BamHI to produce pIJ6838. pIJ6838 was introduced into E. coli BL21λDE3(pLysS) (46), and overexpression of BldC was induced in exponentially growing cells (optical density at 600 nm, 0.5) by addition of 0.1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) for 4 h at 30°C. The cell pellet was resuspended in 10 ml of binding buffer (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 5 mM imidazole) before lysis was completed by four 15-s cycles of sonication at 1-min intervals on ice. The cell lysate was centrifuged at 18,000 rpm for 30 min at 4°C in a Sorvall SS34 rotor. His-tagged BldC was purified on a HiTrap Chelating HP column (Amersham Pharmacia Biotech) with a Biocad Sprint perfusion chromatography system. A total of 2 mg of His-tagged BldC was used to raise a polyclonal antiserum in rabbit (Genosys).
Crude cell extract preparation and immunoblot analysis.
Surface-grown Streptomyces cultures were harvested from cellophane-covered R2YE plates into 1 ml of complete protease inhibitor buffer (Roche) in 0.1 M HEPES (pH 7.2). Samples were sonicated at half power for three cycles of 10 s at 1-min intervals on ice. Cell debris was removed by centrifugation at 14,000 rpm for 10 min at 4°C, and the protein concentration of the supernatant was determined with Bradford reagent (Bio-Rad). Tricine sodium dodecyl sulfate-polyacrylamide gels (45) were used to enhance resolution of proteins in the range of 5 to 20 kDa. Samples (each, 20 μg) were separated by electrophoresis, transferred to a Hybond-C Extra nylon membrane (Amersham Pharmacia Biotech), and probed with a 1:1,000 dilution of rabbit anti-BldC antibody. Horseradish peroxidase-coupled secondary antibody (Amersham Pharmacia Biotech) was used at a 1:5,000 dilution and detected by chemiluminescence with ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
Construction of bldC null mutants and genetic manipulations with cosmid SCD25.
Null mutants were constructed by replacing the entire bldC coding sequence with a cassette carrying the apramycin resistance gene (apr) and oriT of RK2 by PCR targeting (21). Cosmid SCD25 was introduced into E. coli BW25113 (14) carrying plasmid pIJ790 (21), and bldC was disrupted by electroporation of the cells with the PCR-amplified apr-oriT cassette, generated with the primers BLDC K-OUT FOR and BLDC K-OUT REV (Table 1). The resulting cosmid (SCD25ΔbldC::apr) was introduced into E. coli ET12567 carrying pUZ8002 (41) and transferred into S. coelicolor M600, M145, or J660 by conjugation; apramycin-resistant (Aprr), kanamycin-sensitive (Kans) exconjugants were identified and purified (J2161, J2167, and J2162, respectively) (Table 1). The disruptions were confirmed by PCR and Southern analysis. An in-frame deletion allele of bldC was also constructed; cosmid SCD25ΔbldC::apr was introduced into E. coli BT340 (11), and deletion of the apr-oriT cassette (which is flanked by FRT sites) was obtained after induction of FLP recombinase (21). The mutant cosmid thus obtained, SCD25ΔbldC, was introduced into J2161 and J2167 by protoplast transformation, and Kanr transformants were selected. After growth in the absence of antibiotic selection, colonies that had lost both apramycin and kanamycin resistance were selected and purified (J2166 and J2168, respectively) (Table 1) and analyzed by PCR to confirm replacement of apr-oriT cassette by the nonpolar “scar” sequence (21).
A bldC+ derivative of J660 was constructed by introducing the SCD25 cosmid into J2162 (the ΔbldC::apr derivative of J660) (Table 1) by protoplast transformation and selecting for Kanr transformants. After nonselective growth, a strain sensitive to both apramycin and kanamycin was obtained and purified (J2169) (Table 1).
bldC18 derivatives of M600 and M145 were obtained by first rescuing the bldC18 allele from the chromosome of J660 into the SCD25 cosmid. This was done by introducing the SCD25ΔbldC::apr cosmid into J660 by conjugation and selection for Aprr Kanr exconjugants; these were pooled and grown in 100 ml of liquid YEME medium without antibiotics until late exponential phase and used to purify covalently closed circular DNA (28). The DNA was concentrated in a small volume and used to transform E. coli DH5α to kanamycin resistance; about 20% of all Kanr colonies were Aprs and carried cosmids whose restriction pattern was indistinguishable from that of the wild-type SCD25 cosmid. One of these cosmids (SCD25bldC18) was purified; DNA sequencing confirmed that it carried the bldC18 allele. SCD25bldC18 was introduced into J2161 (the ΔbldC::apr derivative of M600) (Table 1) and J2167 (the ΔbldC::apr derivative of M145) (Table 1) by protoplast transformation, and Kanr colonies were selected. After growth in the absence of antibiotics, Kans Aprs colonies were selected and purified, resulting in strains J3210 and J3211, respectively (Table 1). Amplification and sequencing of bldC from the chromosome of these strains confirmed that they carried the bldC18 allele.
RNA isolation, S1 nuclease protection analysis, and in vitro transcription.
Mycelium was harvested from S. coelicolor strains grown on cellophane-covered R2YE plates, and RNA was extracted as described by Kieser et al. (28). Quantitation of RNA was carried out by UV spectroscopy and agarose gel electrophoresis. For each S1 nuclease reaction mixture, 30 μg of RNA was hybridized to a 5′ end-labeled probe at 45°C for 4 to 15 h following denaturation at 65°C for 10 min. S1 nuclease (Amersham Pharmacia Biotech) digestions were performed as described by Kieser et al. (28), and the reaction products were separated on 6% polyacrylamide denaturing sequencing gels. Uniquely end-labeled probes were generated by PCRs with a 5′ end-labeled primer internal to the coding sequence and an upstream, unlabeled primer. Primers were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Transgenomic or Invitrogen).
In vitro transcription was carried out as described by Buttner et al. (6). Runoff templates were generated by PCR with oligonucleotide BLDC4 as the upstream primer and oligonucleotide BLDC3 (template 1; 327 bp) or BLDC17 (template 2; 264 bp) as the downstream primer. RNA polymerase was purified as described by Kieser et al. (28) from strain M600 and grown to late exponential phase in YEME medium.
Microscopy.
Scanning electron microscopy of Streptomyces colonies was performed as described previously (36).
DNA sequence analysis.
BLAST and PSI-BLAST searches were done online at the National Center for Biotechnology Information (NCBI) site (http://www.ncbi.nlm.nih.gov). Sequence comparison and alignment were also done online with ClustalW and EMBOSS at the European Bioinformatics Institute sites (http://www.ebi.ac.uk). BLAST searches on draft actinomycete genome sequences were done at the respective web sites (http://genome.ornl.gov/microbial/tfus for Thermobifida fusca, http://www.bcgsc.ca/gc/rhodococcus for Rhodococcus sp. RHA1, and http://nocardia.nih.go.jp/for Nocardia farcinica).
RESULTS
Identification of the bldC gene by complementation.
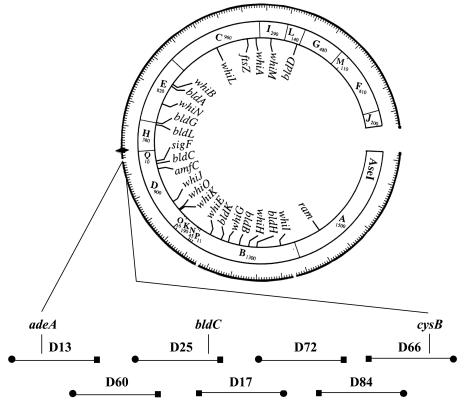
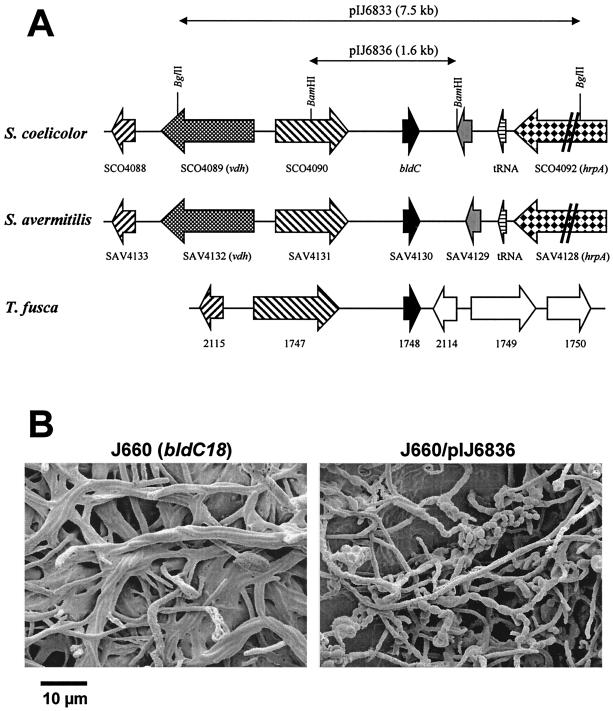
The bldC18 mutation was previously mapped genetically to a position between adeA and cysB in the 8 o'clock region of the S. coelicolor chromosome (34). This region is spanned by seven cosmids of the ordered library of Redenbach et al. (44), as illustrated in Fig. 1. Although these cosmids cannot replicate autonomously in S. coelicolor, they can integrate into the chromosome via insert-directed homologous recombination. Each of the seven cosmids shown in Fig. 1 was individually introduced into bldC18 strain J660 by protoplast transformation, selecting for kanamycin resistance. Transformants were patched onto R5 medium and analyzed for restoration of aerial hyphae formation, sporulation, and pigmented antibiotic production. Two overlapping cosmids, D25 and D17, complemented the bldC18 mutation, indicating that bldC resides in the 27.8-kb overlap between these two cosmids that extends from SCO4075 to SCO4097 and encompasses 23 protein-encoding genes and five tRNA genes. Subcloning identified a 7.5-kb BglII fragment that complemented the bldC18 mutation when cloned into the integrating vector pSET152 and introduced into J660 (pIJ6833) (Fig. 2A). Further rounds of subcloning localized the complementing region to a 1.6-kb BamHI fragment, which contained only one complete open reading frame (ORF), SCO4091 (pIJ6836) (Fig. 2A). Scanning electron micrographs of J660 with and without pIJ6836 confirmed that this plasmid complemented the bldC18 mutation, restoring the ability to produce aerial hyphae, spores (Fig. 2B), and pigmented antibiotics. SCO4091 was therefore designated bldC.
FIG. 1.
A simplified version of the combined physical and genetic map of the S. coelicolor chromosome, showing the locations of developmental genes and the seven ordered cosmids that span the adeA to cysB interval in the 8 o'clock region. The location of bldC is shown on the overlap between cosmids D17 and D25, as determined by complementation. The locations of other developmental genes on the cosmid contig are shown on the inside of the circle. The cosmids and their overlaps are arbitrarily shown to be of equal length. The sizes of the AseI fragments are given in kilobases. •, T3 end; ▪, T7 end; ♦, oriC.
FIG. 2.
(A) Conservation of genetic organization of the bldC region between S. coelicolor, S. avermitilis, and T. fusca. The positions of protein- encoding genes and tRNA genes are indicated by arrows, and restriction sites referred to in the text are marked. Equivalent shading of arrows indicates homology. The extent of the pIJ6833 and pIJ6836 subclones described in the text is shown above the S. coelicolor map. (B) Scanning electron micrographs showing the developmental phenotype of J660 (bldC18) and its complementation by bldC carried on pIJ6836. Strains were grown on R2YE plates.
BldC is a member of a previously unrecognized family of small DNA-binding proteins related to the DNA-binding domains of the MerR family.
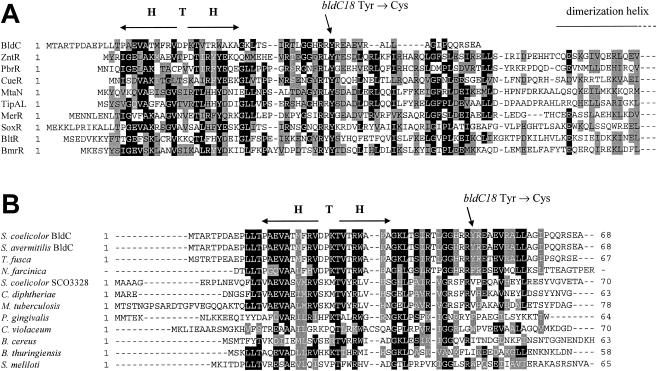
bldC encodes a protein only 68 amino acids long. BLAST searches of the NCBI databases showed that BldC is most similar to the DNA-binding domains of members of the MerR family of transcriptional activators (Fig. 3A). MerR family members function as homodimers. Each subunit consists of a conserved N-terminal DNA-binding domain and a nonconserved C-terminal effector recognition domain, and transcriptional activation is triggered by effector binding (5, 8, 20, 37, 53). Between the N-terminal and C-terminal domains lies a long α-helix that interacts with the same helix in the other subunit, forming a coiled coil responsible for dimerization (5, 8, 20, 37, 53). However, BldC apparently lacks this dimerization helix and a C-terminal effector recognition domain, with sequence similarity limited to the N-terminal DNA-binding domain of the MerR family (Fig. 3A).
FIG. 3.
Amino acid sequence alignments. (A) Alignment of BldC with the N-terminal DNA-binding domains of MerR family proteins. Note that the complete BldC sequence is shown but that the MerR family proteins are arbitrarily truncated halfway through the dimerization helix. Proteins and their corresponding NCBI accession numbers are as follows: S. coelicolor BldC (CAB56371), E. coli ZntR (BAB37580), Ralstonia metallidurans PbrR (CAC28872), E. coli CueR (AAC73589), Bacillus subtilis MtaN (CAB15677), S. lividans TipAL (CAB42766), Pseudomonas aeruginosa MerR (CAA83897), E. coli SoxR (CAA42162), B. subtilis BltR (CAB14599), and B. subtilis BmrR (CAB14333). (B) Alignment of BldC with other members of the BldC family of small DNA-binding proteins. The complete sequence of each protein is shown. Proteins and their corresponding NCBI accession numbers are as follows: S. coelicolor BldC (CAB56371), S. avermitilis BldC (= SAV4130) (BAC71842), T. fusca (ZP_00293028), N. farcinica (partial sequence), S. coelicolor SCO3328 (CAB45362), C. diphtheriae (CAE48899), M. tuberculosis (CAE55290), P. gingivalis (AAQ66571), C. violaceum (AAQ61522), B. cereus (AAP12345), B. thuringiensis bacteriophage Bam35c (CAD59942), and S. meliloti (CAC41930). H T H, helix-turn-helix motif. The position of the BldC Y49C amino acid substitution caused by the bldC18 point mutation is shown.
A more thorough PSI-BLAST search was carried out to look for BldC-like proteins consisting exclusively of a MerR-like DNA-binding domain. This search revealed several such proteins from both gram-positive and gram-negative bacteria (Fig. 3B). The most similar proteins of this kind were found in other actinomycetes. The product of Streptomyces avermitilis gene SAV4130 is 100% identical to BldC and is the predicted orthologue in this species (see below). The Thermobifida fusca genes TFU627 and TFU1748 encode proteins with 60 and 91% identity to BldC, respectively, the Rhodococcus sp. RHA1 ORF 19459 encodes a protein with 50% identity, and BLAST searches on the draft genome sequence of N. farcinica revealed a partial protein sequence with 50% identity to BldC. A second BldC-like protein, SCO3328, is present in S. coelicolor and shows 42% identity to BldC; this also has an orthologue in S. avermitilis. Other proteins consisting exclusively of a MerR-like DNA-binding domain, but with lower levels of identity to BldC (usually around 30%), were identified in a wide range of bacteria, including actinomycetes (e.g., Mycobacterium tuberculosis, Mycobacterium leprae, Corynebacterium diphtheriae, Corynebacterium glutamicum, and Corynebacterium efficiens), other gram-positive bacteria (e.g., Bacillus cereus and Bacillus thuringiensis), and gram-negative bacteria (e.g., Sinorhizobium meliloti, Porphyromonas gingivalis, Chromobacterium violaceum, and Leptospirillum ferrooxidans) (Fig. 3B and data not shown). One member of the BldC family was also identified in the genome of the B. thuringiensis bacteriophage, Bam35c (Fig. 3B).
bldC gene organization is conserved in some other actinomycetes.
Analysis of the SAV4130 locus showed that the genetic organization in the region surrounding bldC is conserved between S. coelicolor and S. avermitilis (Fig. 2A). In S. coelicolor, there appears to be a large (901-bp) noncoding region downstream of bldC, whereas in S. avermitilis there is an ORF for an additional hypothetical protein (SAV4129) annotated in this region. This additional ORF is also conserved in the equivalent locus of “Streptomyces diversa” (Mervyn Bibb, Diversa Corporation, personal communication), and closer examination of the S. coelicolor sequence suggests that there is indeed an ORF downstream of bldC, not annotated in the genome sequence, that would encode a protein of unknown function similar to SAV4129 and the “S. diversa” protein (Fig. 2A and data not shown). A comparison with the T. fusca genome sequence shows some degree of conservation in gene organization between the bldC locus from S. coelicolor (and S. avermitilis) and the T. fusca region surrounding TFU1748, which encodes the protein showing 91% identity to BldC (Fig. 2A). Gene 1748 in T. fusca is preceded by homologues of two genes found upstream of bldC, although the intervening vdh gene for valine dehydrogenase is absent. There is, however, no conservation of gene organization downstream of gene 1748 in T. fusca. The regions surrounding gene TFU627 in T. fusca and ORF 19459 in Rhodococcus sp. RHA1 do not show any conservation of gene organization with the bldC locus.
The bldC18 mutation causes a Y49C change that results in BldC instability.
To determine the nature of the bldC18 mutation, a 750-bp region encompassing bldC was amplified by PCR from J660 and sequenced. A single-nucleotide difference from the wild-type sequence was identified: bldC18 has an AT-to-GC transition that results in a tyrosine-to-cysteine substitution at position 49 in the primary amino acid sequence of BldC. This tyrosine is very highly conserved among the N-terminal DNA-binding domains of MerR family members (Fig. 3A). The loss of BldC activity resulting from substitution of a tyrosine raised the possibility that BldC might be regulated by phosphorylation of this residue. To examine this possibility, we constructed a bldC allele encoding Y49F, a conservative substitution that should eliminate the potential for phosphorylation of the residue without grossly affecting protein structure. The integrative vector pSET152 was used to introduce the wild-type bldC allele (pIJ6842), the Y49C bldC18 allele (pIJ6843), and the Y49F allele (pIJ6844) into J660 (bldC18) by conjugation, and the phenotype of exconjugants was analyzed on SMMS. As expected, the wild-type bldC allele restored sporulation to J660, whereas the bldC18 allele did not. J660 carrying pIJ6844 (Y49F) sporulated, although at a slightly lower level compared to J660 carrying pIJ6842 (wild-type bldC allele) (data not shown). Thus, Y49 is not essential for BldC function, making regulation of BldC activity through phosphorylation of this residue extremely unlikely.
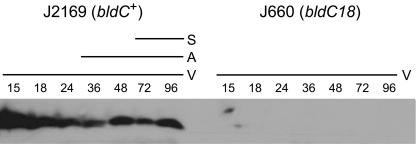
To determine whether the bldC18 mutation affects the stability of BldC, immunoblots were performed. His-tagged BldC was overexpressed in E. coli, purified, and used to generate a polyclonal anti-BldC antiserum. For valid comparison, congenic strains were required. Unfortunately, J650, the bldC+ parent strain from which J660 was derived by N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis (34), has been lost. Therefore, we used the SCD25 cosmid to create J2169, a new congenic bldC+ parent of J660, as described in Materials and Methods. Crude cell extracts prepared from R2YE surface-grown cultures of the J2169 and J660 (bldC18) strains were subjected to immunoblot analysis with the anti-BldC antibodies (Fig. 4). The BldC protein was readily detected throughout development in the wild-type strain, but no BldC protein could be detected at any time point in the bldC18 mutant. Thus, it appears that the bldC18 Y49C mutation results in instability of the BldC protein.
FIG. 4.
Immunoblot analysis of BldC expression during developmental time courses of S. coelicolor J2169 (bldC+) and J660 (bldC18), grown on R2YE solid medium. The time points in hours at which mycelium was harvested and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S) are shown, as judged by microscopic examination.
Construction and phenotypic characterization of bldC null mutants.
A series of bldC deletion mutants was constructed in morphologically wild-type strains of S. coelicolor using the PCR targeting method of Gust et al. (21), in order to compare their phenotypes to that of J660 (bldC18). The widely used strains M600 and M145 (plasmid-free derivatives of wild-type S. coelicolor) were chosen. These two strains, which until recently had been considered genetically similar, are in fact different, as the chromosome of M600 carries a 1.06-Mbp-long terminal inverted repeat absent from M145 (48). Analysis of strain J660 (bldC18) showed that it lacks the long terminal inverted repeats present in M600 (data not shown). The null mutations constructed in the M600 and M145 backgrounds replaced bldC either with an apramycin-resistance cassette (containing the apr gene and oriT) or with an in-frame deletion. No difference in phenotype could be detected between null mutants carrying the resistance cassette and those carrying the in-frame deletion, and so the in-frame deletion mutants were chosen for analysis. After 4 days of growth on minimal medium, SMMS, colonies of both M600 and M145 had produced a dense covering of aerial mycelium and grey-pigmented spores. In contrast, the ΔbldC null mutants derived from them (J2166 and J2168, respectively) had not differentiated to form aerial hyphae but had a bald phenotype after 4 days, and they remained bald even on prolonged incubation (data not shown). When the phenotypes of the same strains were analyzed on the rich medium, R2YE, M600, and M145 readily sporulated within 4 days, whereas the bldC deletion mutants did not (Fig. 5). However, upon prolonged incubation on R2YE (>1 week), both J2166 and J2168 did go on to produce aerial mycelium and spores (data not shown). The phenotype of these strains contrasted with that of J660 (bldC18), which, even on prolonged incubation, did not make an aerial mycelium on R2YE. pIJ6836 fully complemented the sporulation defects of the bldC null mutants J2166 and J2168 on both SMMS and R2YE; no complementation was observed when only the vector pSET152 was introduced (data not shown).
FIG. 5.
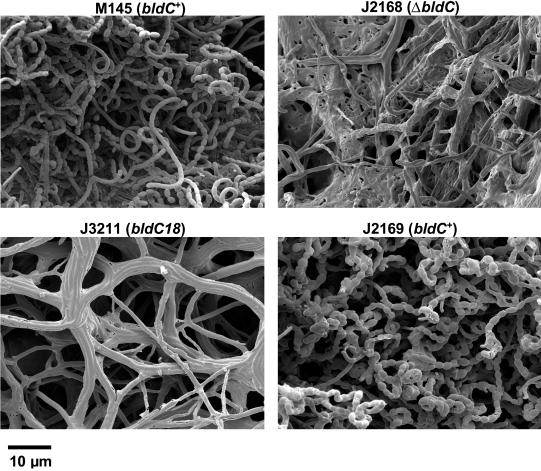
Scanning electron micrographs showing the developmental phenotypes of M145 (a plasmid-free derivative of wild-type S. coelicolor), J2168 (M145ΔbldC), J3211 (M145bldC18), and J2169 (bldC+ derivative of J660). Strains were grown on R2YE plates.
These experiments showed that deletion of bldC in M600 or M145 resulted in a less severe phenotype than that of J660 (bldC18) when grown on R2YE, raising the possibility that the severe bald phenotype exhibited by J660 was allele specific. To assess this possibility, we deleted bldC from the chromosome of J660 by PCR targeting, resulting in strain J2162. The phenotype of J2162 was indistinguishable from that of J660, suggesting that the severe bald phenotype of J660 is not bldC18 specific but is due to the genetic background of this strain. Confirmation of this conclusion was obtained by constructing bldC18 derivatives of M600 (J3210) and M145 (J3211), as described in Materials and Methods; these bldC18 derivatives had phenotypes indistinguishable from that of the bldC deletion mutants constructed in the same backgrounds, i.e., they were bald on SMMS and differentiation was delayed to the same extent on R2YE (Fig. 5). These results suggest that bldC18 is a null allele, consistent with the lack of detectable BldC protein in a bldC18 background (Fig. 4).
A consistent feature of the bldC18 and ΔbldC null mutants when viewed under the scanning electron microscope was the aggregation of the hyphae into cable-like bundles (Fig. 2B and 5). It seems possible that this phenotype might be connected to the leather-like quality of bldC mycelium grown on plates: for example, when confluent lawns of bldC mutants are grown on cellophane disks for RNA isolation, the mycelium can be peeled off in one piece, a phenomenon not seen with wild-type strains or other bld mutants we have worked with extensively.
Extracellular complementation.
Previous studies have provided evidence for an extracellular signaling cascade between different bld mutants, based on the ability of some bld strains to induce aerial mycelium formation in certain other bld strains when they are grown in close proximity but without actual contact (35, 39, 50). These studies placed bldC between bldG and bldD in the extracellular complementation cascade, based on the behavior of J660 (bldC18), the only bldC mutant previously available (50). We therefore carried out experiments in an attempt to determine if the ΔbldC null mutant derivatives of M600 (J2166) and M145 (J2168) constructed in this work showed the same extracellular complementation properties as the original bldC strain, J660 (bldC18). Unfortunately, no clear-cut results could be obtained in these experiments, which were complicated in part by the fact that the constructed bldC null mutants eventually sporulate in R2YE, the rich medium on which extracellular complementation is tested.
Antibiotic production in the bldC mutants.
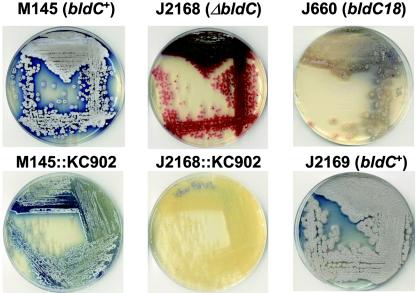
In addition to the differences in sporulation between J660 (bldC18) and the bldC null mutants constructed in the M600 and M145 backgrounds, there appeared to be significant differences in production of the pigmented antibiotics actinorhodin and undecylprodigiosin. J660 remained largely unpigmented when grown on R2YE (Fig. 6), whereas the wild-type strains M600 and M145 produced significant levels of both antibiotics, as did J2169, the bldC+ derivative of J660 (Fig. 6). Strains J2166 and J2168, in which the bldC gene had been deleted, produced significant amounts of red pigment when grown on R2YE, but produced less blue pigment (Fig. 6); the same was true of the constructed bldC18 derivatives of M600 (J3210) and M145 (J3211). Confirmation that the red pigment was indeed undecylprodigiosin was obtained by lysogenizing J2166 and J2168 with the aHP φC31 derivative KC902 (22), which disrupts the redX gene and therefore eliminates undecylprodigiosin production. These lysogens were unpigmented (Fig. 6), confirming that the red pigment observed in the bldC null mutant derivatives of M600 (J2166) and M145 (J2168) was indeed undecylprodigiosin and that these strains were strongly affected in actinorhodin production (KC902 lysogens of M600 and M145, by contrast, produced significant amounts of blue pigment) (Fig. 6). The defect in actinorhodin production shown by J2166 and J2168 was fully complemented by introduction of plasmid pIJ6836 (data not shown).
FIG. 6.
Phenotypes of M145, J2168 (M145ΔbldC), J660 (bldC18), J2169 (bldC+ derivative of J660), and KC902 lysogens of M145 and J2168 on R2YE. Note that J2168 and J660 cannot raise an aerial mycelium and that they fail to synthesize the blue-pigmented antibiotic actinorhodin. Lysogenization by KC902 disrupts the redX gene and therefore eliminates production of the red-pigmented antibiotic undecylprodigiosin (22).
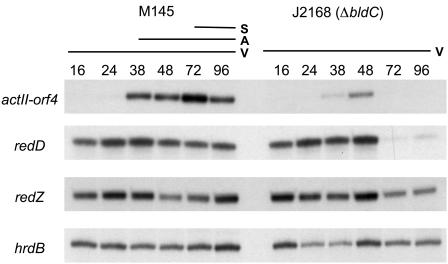
To further understand the effect of bldC deletion on antibiotic production, M145 and J2168 (ΔbldC) were grown on R2YE plates, and RNA was isolated at different time points. These RNA preparations were used to determine the level of transcription of actII-orf4, redD, and redZ, which encode pathway-specific activators of the actinorhodin and undecylprodigiosin biosynthetic gene clusters. The results shown in Fig. 7 indicate that deletion of bldC does indeed affect production of both antibiotics. In the case of actinorhodin, there is clearly a delay and a reduction in the levels of actII-orf4 transcripts, disappearing after 48 h, when the highest levels are observed in the wild-type strain. Therefore, bldC is required for normal and sustained transcription of actII-orf4 throughout development. In contrast, early transcription of redD was not affected by lack of bldC. However, redD transcripts were reduced to almost undetectable levels after 48 h in the bldC mutant, in contrast to M145 in which redD continued to be transcribed during aerial mycelium formation and sporulation (Fig. 7). Since transcription of redD depends in turn on RedZ (49), redZ transcription levels were also analyzed; redZ transcripts were present at all time points in both M145 and in J2168 (Fig. 7).
FIG. 7.
S1 nuclease protection analysis of actII-ORF4, redD, redZ, and hrdB (positive control) transcription during development of S. coelicolor M145 and J2168 (M145ΔbldC) on R2YE solid medium. The time points in hours at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S) are shown, as judged by microscopic examination.
Transcriptional analysis of bldC.
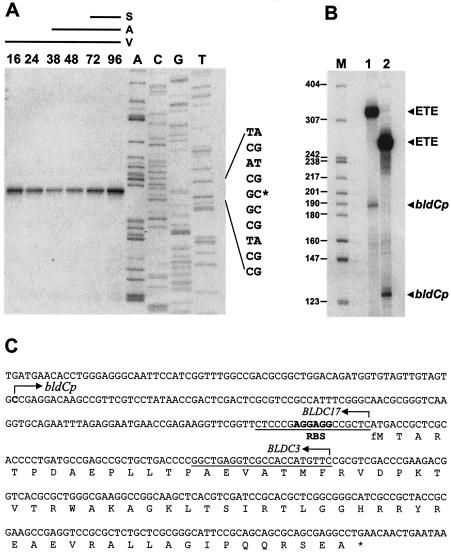
High-resolution S1 nuclease mapping of the bldC promoter region was performed with a PCR-generated probe and RNA isolated from either M600 or M145. A single bldC promoter (bldCp) was identified, initiating transcription 125 bp upstream from the ATG start codon (Fig. 8A). In vitro transcription experiments carried out with highly purified S. coelicolor RNA polymerase confirmed the location of the single bldC promoter (Fig. 8B).
FIG. 8.
Transcriptional analysis of bldC. (A) High-resolution mapping of the 5′ end of the bldC message and analysis of bldC transcription levels during development of S. coelicolor M145 on R2YE solid medium. The time points in hours at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S) are shown, as judged by microscopic examination. Lanes labeled A, C, G, and T represent a dideoxy sequencing ladder generated with the same radiolabeled oligonucleotide that was used to make the S1 mapping probe. (B) In vitro transcription of the bldCp promoter by S. coelicolor RNA polymerase holoenzyme. Transcripts were generated from template 1 (327 bp) or template 2 (264 bp) (see Materials and Methods). The expected sizes of the runoff transcripts from the bldCp promoter were 188 nucleotides (template 1) and 125 nucleotides (template 2). The size markers (M) were a 32P-end-labeled HpaII digest of pBR322. ETE, end-to-end transcription of the linear template. (C) Nucleotide sequence of the bldC gene showing the bldCp transcription start point, the putative ribosome-binding site (RBS), the complete bldC encoding sequence, and the sequences of the BLDC3 and BLDC17 downstream oligonucleotide PCR primers used to generate the templates.
Transcription of bldC was monitored by S1 nuclease mapping during development of S. coelicolor M145 on R2YE. bldC transcripts were detectable at all of the time points analyzed, representing growth from the early vegetative stage through to sporulation (Fig. 8A). The levels of bldC transcript did not change markedly during this time, showing that bldC is transcribed constitutively from a single promoter throughout development in wild-type S. coelicolor. Although the bldC transcript was approximately equally abundant at all time points, the possibility that bldC transcription may be spatially restricted, for example to the vegetative mycelium, is not excluded because no attempt was made to fractionate the harvested cell material used for RNA preparation. Transcription of bldC was analyzed in the bldC18 point mutant, J660, and in the bldC null mutant, J2161. bldC transcripts were readily detectable throughout development and were present at the same level as in the corresponding congenic bldC+ parent strains (data not shown), showing that bldC is not autoregulated.
To determine whether transcription of bldC depends on any of the other known bld genes, transcription from bldCp was examined in RNA samples isolated from bldA, bldB, bldD, bldF, bldG, bldH, bldJ, bldK, bldM, and bldN mutants grown for 36 h on R2YE. As these bld mutations exist in a complicated variety of genetic backgrounds (Table 1), only striking effects would be considered potentially significant. However, bldC transcripts were readily detected in all of the bld mutants analyzed, showing that transcription of bldC does not depend on any of the other known bld genes (data not shown).
DISCUSSION
The BldC family of small DNA-binding proteins is related to the N-terminal DNA-binding domain of transcriptional activators belonging to the MerR family. The basic structure of MerR-like proteins is a dimer consisting of two identical subunits, each composed of an N-terminal DNA-binding domain containing a winged helix-turn-helix motif, a C-terminal effector recognition domain, and an interconnecting linker region which consists of a long α-helix that interacts with the same helix in the other subunit, forming a coiled coil responsible for dimerization (5, 8, 20, 37, 53). MerR family proteins share similarity only within their DNA-binding domains; as different family members bind different effectors, their C-terminal domains are variable and show little, if any, similarity to one another. BldC consists exclusively of a MerR-like DNA-binding domain and lacks the dimerization and C-terminal effector recognition domains. These observations raise interesting questions about the evolution of BldC and the MerR family. Brown et al. (5) have suggested that MerR family members might have arisen from gene fusion events that brought together N-terminal DNA-binding domains with separate effector recognition domains. This suggestion was prompted in part by analysis of two members of the MerR family, TipA and NolA. Two separate proteins are synthesized from the tipA gene of Streptomyces lividans: TipAL, the typical full-length MerR-like protein with an N-terminal DNA-binding domain and a C-terminal effector recognition domain, and TipAS, a protein consisting only of the C-terminal effector recognition domain, translated from an in-frame initiation site 110 amino acids downstream from the TipAL start codon (23). Similarly, three proteins are synthesized from alternative start codons of the Bradyrhizobium japonicum nolA gene, only one of which (NolA1) contains the helix-turn-helix DNA-binding motif (31). The characterization of BldC and the discovery of other members of the BldC family is the first evidence that proteins equivalent to the N-terminal DNA-binding domains of MerR family regulators can function independently of a C-terminal effector recognition domain. The presence of bldC family genes in many different bacterial genera might contribute significantly to the evolution of new members of the MerR family by allowing reshuffling of the DNA-binding region with different effector-binding domains, as proposed by Brown et al. (5).
Loss of bldC function results in a complex phenotype. When bldC was deleted in the M600 or M145 backgrounds, the resulting strains exhibited a bald phenotype on SMMS minimal medium. However, on the widely used rich medium R2YE, bldC null mutants still managed to sporulate, although differentiation was severely delayed with respect to the corresponding wild-type strains. In contrast, the original bldC18 point mutant, J660, failed to differentiate on R2YE, even with prolonged incubation. We showed that this phenotypic difference between the constructed null mutants and J660 (bldC18) was not due to allele specificity but was due to the genetic background. In fact, our results suggest that bldC18 is a null allele. J660 (bldC18) failed to accumulate detectable levels of BldC, presumably because Y49 is essential for proper folding and stability. This suggestion is supported by studies of three MerR-like proteins whose structures have recently been determined, BmrR, MtaN, and CueR (8, 20, 37, 53). BmrR, MtaN, and CueR contain three regions that contact their target promoters: the helix-turn-helix motif and two wing domains (20, 53). BmrR Y45, MtaN Y41, and CueR Y39, the tyrosine residues equivalent to Y49 in BldC, fall within wing 1, which consists of two β-sheets and the connecting loop between them. A hydrophobic core stabilizes the structure of the MtaN DNA-binding domain (20), and Y41 forms part of this core. The fact that a Y49C change in BldC is highly disruptive but a Y49F change is tolerated suggests that Y49 may serve the equivalent hydrophobic core function in BldC.
Apart from their effects on sporulation, bldC mutations also affect production of actinorhodin and undecylprodigiosin. Thus, bldC is required not only for the normal differentiation of aerial hyphae, but also for antibiotic production in the substrate mycelium. Our results show that transcription of actII-orf4 depends on bldC, explaining the lack of actinorhodin production in a bldC null mutant. In contrast, bldC does not appear to be necessary for normal transcription of the undecylprodigiosin pathway-specific activators redD and redZ during vegetative growth. However, levels of redD transcripts were high in the wild type during aerial mycelium formation and sporulation but were severely reduced at the equivalent time points in the bldC mutant, while redZ transcription continued unaffected. This suggests that bldC may be required for maintenance of redD transcription during differentiation. RedZ is required for transcription of redD, and RedD in turn activates the red biosynthetic genes (49), yet bldC mutations appear to affect transcription of redD and not redZ. These results show that regulatory inputs relating to differentiation can be integrated at the level of redD transcription, at least in the case of bldC.
Acknowledgments
We thank Nigel Brown, Marie Elliot, Mervyn Bibb, Mike Merrick, and David Hopwood for helpful discussion and for their comments on the manuscript.
This work was supported by a BBSRC studentship (to A.C.H.) and by a grant-in-aid to the John Innes Centre from the BBSRC. L. S.-G. received a sabbatical scholarship from the DGAPA of the National University of Mexico.
REFERENCES
- 1.Bibb, M. J., and M. J. Buttner. 2003. The Streptomyces coelicolor developmental transcription factor σBldN is synthesized as a pro-protein. J. Bacteriol. 185:2338-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Bignell, D. R., J. L. Warawa, J. L. Strap, K. F. Chater, and B. K. Leskiw. 2000. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146:2161-2173. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 6.Buttner, M. J., I. M. Fearnley, and M. J. Bibb. 1987. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol. Gen. Genet. 209:101-109. [DOI] [PubMed] [Google Scholar]
- 7.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Changela, A., K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran, and A. Mondragon. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383-1387. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F., and M. J. Bibb. 1997. Regulation of bacterial antibiotic production, p. 57-105. In H.-J. Rehm and G. Reed (ed.), Products of secondary metabolism. VCH, Weinheim, Germany.
- 10.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 12.Claessen, D., R. Rink, W. de Jong, J. Siebring, P. de Vreugd, F. G. H. Boersma, L. Dijkhuizen, and H. A. B. Wösten. 2003. A novel class of secreted hydrophobic protein is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claessen, D., I. Stokroos, H. J. Deelstra, N. A. Penninga. C. Bormann, J. A. Salas, L. Dijkhuizen, and H. A. B. Wösten. 2004. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53:433-443. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliot, M., F. Damji, R. Passantino, K. F. Chater, and B. Leskiw. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliot, M. A., and N. J. Talbot. 2004. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 7:594-601. [DOI] [PubMed]
- 17.Elliot, M. A., and B. K. Leskiw. 1999. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J. Bacteriol. 181:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliot, M. A., M. J. Bibb, M. J. Buttner, and B. K. Leskiw. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269. [DOI] [PubMed] [Google Scholar]
- 19.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of secreted, hydrophobic proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17:1727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178-47184. [DOI] [PubMed] [Google Scholar]
- 21.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. Gene replacement by PCR targeting in Streptomyces and its use to identify a protein domain involved in the biosynthesis of the sesquiterpene odour geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie, E. P., and K. F. Chater. 1990. The level of a transcript required for production of a Streptomyces coelicolor antibiotic is conditionally dependent on a tRNA gene. J. Bacteriol. 172:6189-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, D. J., J. L. Caso, and C. J. Thompson. 1993. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 12:3183-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a multiple cloning site and that retain ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 25.Keijser, B. J., G. P. van Wezel, G. W. Canters, and E. Vijgenboom. 2002. Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J. Bacteriol. 184:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 27.Kelemen, G. H., P. H. Viollier, J. Tenor, L. Marri, M. J. Buttner, and C. J. Thompson. 2001. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40:804-814. [DOI] [PubMed] [Google Scholar]
- 28.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 29.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 31.Loh, J., M. G. Stacey, M. J. Sadowsky, and G. Stacey. 1999. The Bradyrhizobium japonicum nolA gene encodes three functionally distinct proteins. J. Bacteriol. 181:1544-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, H., and K. Kendall. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacNeil, D. J., J. L. Occi, K. M. Gewain, T. MacNeil, P. H. Gibbons, C. L. Ruby, and S. L. Danis. 1992. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115:119-125. [DOI] [PubMed] [Google Scholar]
- 34.Merrick, M. J. 1976. A morphological and genetic mapping study of bald mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 35.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 36.Molle, V., W. J. Palframan, K. C. Findlay, and M. J. Buttner. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:1286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newberry, K. J., and R. G. Brennan. 2004. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 279:20356-20362. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, K. T., J. M. Willey, L. D. Nguyen, L. T. Nguyen, P. H. Viollier, and C. J. Thompson. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46:1223-1238. [DOI] [PubMed] [Google Scholar]
- 39.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 41.Paget, M. S. B., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor, σE, is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pope, M. K., B. D. Green, and J. Wespheling. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180:1556-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puglia, A.-M., and E. Capelletti. 1984. A bald, superfertile, UV-resistant stain in Streptomyces coelicolor A3(2). Microbiologica 7:263-266. [PubMed] [Google Scholar]
- 44.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 45.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 46.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 47.Tillotson, R. D., H. A. B. Wösten, M. Richter, and J. M. Willey. 1998. A surface active protein involved in aerial mycelium formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595-602. [DOI] [PubMed] [Google Scholar]
- 48.Weaver, D., N. Karoonuthaisiri, H. H. Tsai, C. H. Huang, M. L. Ho, S. Gai, K. G. Patel, J. Huang, S. N. Cohen, D. A. Hopwood, C. W. Chen, and C. M. Kao. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535-1550. [DOI] [PubMed] [Google Scholar]
- 49.White, J., and M. J. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 51.Willey, J., R. Santamaria, J. Guijarro, M. Geistlich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641-650. [DOI] [PubMed] [Google Scholar]
- 52.Wosten, H. A., and J. M. Willey. 2000. Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology 146:767-773. [DOI] [PubMed] [Google Scholar]
- 53.Zheleznova-Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]