Abstract
This report describes a genetic locus associated with siderophore biosynthesis and transport in Corynebacterium diphtheriae. A BLAST search of the C. diphtheriae genome identified a seven-gene cluster that included four genes, designated ciuA, ciuB, ciuC, and ciuD, whose predicted products are related to ABC-type iron transporters. Downstream from ciuD is the ciuE gene, whose predicted product is similar to the aerobactin biosynthetic enzymes IucA and IucC. The CiuE protein, which has a predicted mass of 121,582 Da and is approximately twice the size of either IucC or IucA, is homologous to each of these proteins in both its N- and C-terminal regions. C. diphtheriae ciuE deletion mutants exhibited a defect in siderophore production, iron uptake, and growth in low-iron medium. Mutations in the ciuA gene, whose predicted product is a lipoprotein component of an iron transport system, resulted in a severe defect in iron uptake and reduced ability to use the C. diphtheriae siderophore as an iron source. Site-directed mutations in irp6A, a gene previously reported to be associated with siderophore transport, had no effect on iron uptake or the utilization of the C. diphtheriae siderophore as an iron source. Transcriptional analysis demonstrated that expression of ciuA and ciuE is DtxR and iron regulated, and DNase I protection experiments confirmed the presence of DtxR binding sites upstream from each of these genes. Thus, this iron- and DtxR-regulated gene cluster is involved in the synthesis and transport of the C. diphtheriae siderophore.
Iron is an essential element for almost all bacteria, including human pathogens. Under physiological conditions, iron is largely insoluble; however, microorganisms have developed a variety of mechanisms to obtain this limited iron. In the mammalian host, almost all of the iron is localized intracellularly, where it is primarily associated with heme and heme proteins. In the extracellular environment, inorganic iron and heme-associated iron is present at low levels on mucosal surfaces and in plasma, where iron is bound to transferrin and to a lesser extent to lactoferrin (22, 40, 44). Bacterial systems involved in acquiring mammalian iron sources often include surface proteins that bind various host iron sources, such as heme, heme proteins, transferrin, and lactoferrin (22, 32, 40). The subsequent steps in acquiring iron from host compounds frequently involve ATP-binding cassette (ABC) transport systems that facilitate the uptake of iron or heme into the cytosol (13, 22, 40). Many bacteria synthesize and secrete siderophores, low-molecular-weight iron chelators which have a very high and specific affinity for ferric iron (8, 45). Ferric-siderophore complexes are transported into bacteria through specific binding proteins present in the bacterial membrane. In gram-negative bacteria, the siderophore receptor is an outer membrane protein that binds the ferric-siderophore and transports it into the periplasm by an energy-requiring mechanism that is dependent on the TonB protein and the accessory proteins ExbB and ExbD (45). Transport of the ferric-siderophore into the cytosol involves a periplasmic protein-dependent ABC transport system. In gram-positive bacteria, the primary receptor for the siderophore is proposed to be a lipoprotein that is anchored in the cytoplasmic membrane through an N-terminal lipid moiety (37, 45). The gram-positive lipoprotein receptors share homology with gram-negative bacterial periplasmic binding proteins. Transport of the siderophore through the cytoplasmic membrane in gram-positive organisms requires an ABC-type transporter (37).
Iron acquisition systems, including high-affinity siderophore uptake systems, are virulence factors in many bacterial pathogens, including some recently described siderophore systems in gram-positive bacteria (6, 11). The initial evidence for a high-affinity iron uptake system in Corynebacterium diphtheriae, the causative agent of diphtheria (16), came from studies that identified chemically derived mutants, designated HC1, HC3, HC4, and HC5, that expressed high levels of diphtheria toxin (DT) in iron-replete medium, conditions under which DT is repressed in the wild-type strain (9). These mutants were also found to be severely defective in iron uptake and in growth in low-iron medium, which suggested they were defective in factors required for iron transport. Russell et al. (27) showed that C. diphtheriae strains secreted a siderophore under low-iron conditions, and a siderophore-deficient mutant, designated HC6, was isolated and shown to be defective in growth and iron transport. Growth of the HC6 mutant was stimulated by culture supernatants containing C. diphtheriae siderophore activity and also by purified aerobactin, a hydroxamate-type siderophore. A subsequent study showed that the PW8 strain, a C. diphtheriae isolate used for commercial vaccine production, was deficient in siderophore synthesis (28). The mutations responsible for the defect in HC6 and PW8 have not been identified, and the genes required for the synthesis of the siderophore activity in C. diphtheriae have not been reported. The iron uptake mutants HC1, HC3, HC4, and HC5 were all shown to express both DT and siderophore activity under elevated iron concentrations (41). Siderophore production in the C. diphtheriae C7 strain is coordinately regulated with DT, in which maximum levels are observed in low-iron medium and significant repression occurs under high-iron conditions (41). The DT repressor protein, DtxR, is required for the iron-dependent regulation of both DT and siderophore, since in the dtxR mutant C7(β)hm723, iron-dependent repression of siderophore and DT was markedly alleviated (33, 41).
In a report by Russell and Holmes (28), the siderophore activity present in low-iron culture supernatants in C. diphtheriae strains was designated corynebactin. The structure of the compound(s) associated with the C. diphtheriae siderophore activity has not been determined, and it is not known if C. diphtheriae produces more than one siderophore. C. diphtheriae siderophore activity is routinely detected using the chrome azurol S (CAS) assay, a method that is widely used to detect iron-chelating activity (38). Biochemical tests for the detection of hydroxamate and catechol (Arnow) siderophores are negative with culture supernatants containing C. diphtheriae siderophore activity (2, 9, 10), which indicates that the structure of the siderophore is distinct from these two common structural classifications. A recently described siderophore from Corynebacterium glutamicum was also named corynebactin (5), and structural analysis showed that this compound was a catechol-type siderophore similar to enterobactin which gives a positive reaction in the Arnow assay (4, 5). This finding indicates that the siderophore produced by C. diphtheriae is structurally different from the C. glutamicum siderophore. In an attempt to avoid confusion associated with the siderophore nomenclature, in this report the name corynebactin will be used for the structurally characterized catechol-type siderophore produced by C. glutamicum, while the siderophore activity produced by C. diphtheriae strains will be designated the C. diphtheriae siderophore.
A recent study by Qian et al. (26) reported the isolation of a novel iron- and DtxR-regulated promoter from C. diphtheriae termed IRP6. Downstream from IRP6 are three genes, irp6A, irp6B, and irp6C, which constitute an ABC-type iron transporter. Clones containing the irp6 genes were shown to correct the defect in growth and in siderophore regulation in the HC1, HC4, and HC5 iron uptake mutants. Sequence analysis of the irp6 allele in the HC1 strain revealed the presence of a single point mutation in the irp6A gene. It was concluded from this study that the specific mutant alleles in the irp6 operon present in the HC mutants were associated with a defect in the siderophore-dependent uptake system.
A recent study (18) which used a DtxR repressor titration assay, DRTA, to search for novel DtxR-binding sites in the C. diphtheriae genome identified 10 previously unknown DtxR binding regions. Downstream from one of these binding sites was a putative operon that encoded two genes, sidA and sidB, whose predicted products were similar to nonribosomal peptide synthetases that are associated with siderophore synthesis. Surprisingly, these genes were deleted in the well-studied C. diphtheriae C7 strain, but the complete open reading frames for the sidAB genes were present in the strain used for the genome sequence, NCTC 13129, and in strain 1737, a clinical isolate that is a member of the dominant clonal group associated with the recent Russian diphtheria epidemic (25, 43). Mutations in either the sidA or sidB gene in 1737 had no effect on siderophore activity (18), which indicated that the sid genes were either not involved in the production of a siderophore or were required for the synthesis of a second siderophore whose production was masked by the primary siderophore, which is presumably the siderophore produced by the C7 strain. Since the C7 strain produces high levels of siderophore activity and lacks functional sidA and sidB genes, it is clear that genes other than the sid genes are needed for siderophore production in the C7 strain.
In this study, we report the identification of a cluster of genes in C. diphtheriae that are involved in iron transport and siderophore biosynthesis. The first four genes in this seven-gene cluster, ciuA, ciuB, ciuC, and ciuD (named for corynebacterium iron uptake), share sequence similarity with ABC-type iron transport systems. The predicted product of the fifth gene, ciuE, is similar to the aerobactin biosynthetic enzymes IucA and IucC, and mutations in ciuE greatly reduced siderophore production. C. diphtheriae ciuA mutants were strongly defective in iron transport and in their ability to use the C. diphtheriae siderophore as an iron source. We show that the ciu gene cluster is regulated in an iron-dependent manner from two separate DtxR-regulated promoters. These findings indicate that the products of the iron-regulated ciu gene cluster are involved in the synthesis and transport of the major C. diphtheriae siderophore.
MATERIALS AND METHODS
Bacterial strains and media.
Luria-Bertani medium was used for culturing of Escherichia coli, and heart infusion broth (Difco, Detroit, Mich.) containing 0.2% Tween 80 (HIBTW) was used for routine growth of C. diphtheriae strains. Bacterial stocks were maintained in 20% glycerol at −70°C. E. coli and C. diphtheriae strains used in this study are listed in Table 1. Antibiotics were added to Luria-Bertani medium at 34 μg/ml for chloramphenicol and at 50 μg/ml for kanamycin; to HIBTW for C. diphtheriae cultures, chloramphenicol was at 2 μg/ml, nalidixic acid was at 35 μg/ml, and kanamycin was at 50 μg/ml. HIBTW was depleted of iron by the addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) at the indicated concentrations.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli and Shigella strains | ||
| E. coli DH5α | Routine cloning and lacZ expression | Invitrogen |
| E. coli TOP10 | Cloning PCR products | Invitrogen |
| E. coli S17-1 | RP4 mobilization functions | 39 |
| S. flexneri 2457T | Aerobactin production | D. Kopecko |
| C. diphtheriae strains | ||
| C7(−) | Wild type, tox | 3 |
| C7(β)hm723 | dtxR mutant, tox+ | 17 |
| PW8 | tox+, siderophore mutant | 23 |
| 1737 | Russian clinical strain, tox+ | 25 |
| HC1 | Chemically derived mutant C7(β), irp6A/G240D | 9 |
| C7A | ciuA integration mutant of C7(−) | This study |
| C7E | ciuE deletion mutant of C7(−) | This study |
| C7p6A | irp6A deletion mutant of C7(−) | This study |
| C7A-1 | Site-specific mutant of C7(−), irp6A/G240D | This study |
| 1737A | ciuA deletion mutant of 1737 | This study |
| 1737E | ciuE deletion mutant of 1737 | This study |
| 1737p6A | irp6A deletion mutant of 1737 | This study |
| 1737sidA/ciuE | sidA ciuE double mutant of 1737 | This study |
| Plasmids | ||
| pCR-BluntII-TOPO | Routine E. coli cloning plasmid, Knr | Invitrogen |
| pCm2.6sacB | C. diphtheriae shuttle vector with sacB, Cmr | This study |
| pCmSB-ORI6 | Allelic exchange vector, Cmr | This study |
| pKN2.6 | C. diphtheriae shuttle vector, Knr | 33 |
| pCM502 | Promoter-probe vector, Cmr | 31 |
| pKE | 1737 ciuE gene on pKN2.6 | This study |
| pciuEP-lacZ | ciuE promoter lac fusion in pCM502 | This study |
| pciuAP-lacZ | ciuA promoter lac fusion in pCM502 | This study |
PGTH medium is a low-iron medium that was prepared as described previously (18) but with the following changes: the Chelex treatment of the HIBTW component was extended for an additional hour at room temperature to further reduce the iron levels in the medium, and the trace metals MnCl2, CuSO4, and ZnSO4 were omitted, since additional supplementation of the PGTH medium with these metals was not required for growth.
Plasmid construction and DNA manipulation.
Plasmids used in this study are listed in Table 1. All PCR-generated DNA fragments were initially cloned into the pCR-Blunt II-TOPO vector (Invitrogen). The promoter probe vector pCM502 (31) contains a promoterless lacZ gene and was used for the construction of the lacZ promoter fusions. The pCM502 plasmid is derived from a C. diphtheriae-E. coli shuttle vector and replicates at low copy in C. diphtheriae strains. Plasmid pKE was constructed using PCR-generated DNA fragments encoding the ciuE gene from 1737. These fragments were ligated into the C. diphtheriae-E. coli shuttle vector pKN2.6 at the SpeI and BamHI sites (introduced into the PCR primers) to generate plasmid pKE. The sequence of the 1737 ciuE gene on plasmid pKE was shown to be identical to that reported for the genome strain, NCTC 13129 (7).
Construction of the allelic exchange plasmid pCmSB-ORI6 was as follows: a 2.4-kb PstI fragment containing the sacB gene was excised from plasmid pCVD442 (12) and ligated into the PstI site in plasmid pCM2.6 (33) to generate plasmid pCM2.6sacB. PCR was used to amplify a 3.4-kb DNA fragment from pCm2.6sacB that contained the Cmr gene and the sacB gene. PCR was also used to amplify a 1.8-kb DNA fragment which contained the origin of replication (RepA) from the C. diphtheriae-E. coli shuttle vector pKN2.6 (33). Relevant restriction sites were incorporated into the various PCR primers, and the 3.4- and 1.8-kb PCR products were cloned into plasmid pCR-BluntII-TOPO. The 3.4- and 1.8-kb fragments were excised from pCR-BluntII-TOPO with NotI and then ligated together to generate plasmid pCmSB-ORI6.
Mutant construction.
The C7 ciuA gene was disrupted by the integration of a suicide vector into the 5′ region of the ciuA gene. A 699-bp DNA fragment containing an internal segment of the 5′ region of ciuA was amplified by PCR from the C. diphtheriae C7 genome and then ligated into the pKN2.6 plasmid. Construction of vector integration mutations in the C. diphtheriae chromosome was done as previously described (32).
Deletions in the C7 irp6A and ciuE genes and in the 1737 ciuA, ciuE, and irp6A genes were constructed by using an allelic replacement technique. Vector pCmSB-ORI6 was used in the construction of the various C. diphtheriae deletion mutants and contained the origin of DNA replication from plasmid pKN2.6 and the sacB gene, which was used for counterselection, since strains carrying sacB are sucrose sensitive. PCR was used to amplify DNA fragments ranging in size from 893 to 1,050 bp that contained regions upstream (5′) or downstream (3′) of the genes that were targeted for deletion. The PCR-amplified fragments also contained a segment from either the 5′ or 3′ coding region of the targeted gene, and restriction sites were introduced into the primers such that ligation of the upstream fragment to the downstream fragment created an in-frame deletion of the targeted gene with the introduction of a 6-bp restriction site at the fusion junction. The percentage of the coding region that was deleted in the various in-frame gene deletions was as follows: C7 and 1737 irp6A, 70%; C7 and 1737 ciuE, 94%; 1737 ciuA, 75%. The pCmSB-ORI6 plasmid that contained the fused upstream and downstream fragments was moved into C. diphtheriae, and the plasmid DNA was isolated. The plasmid origin of replication was excised with NotI, and the resulting linear fragment was self-ligated under conditions that would favor the formation of circular molecules (32). The ligated DNA (suicide plasmid) was moved into the relevant C. diphtheriae strains by electroporation, where the circular DNA recombined into the chromosome by homologous recombination. Single-crossover recombinants were confirmed by PCR, and these isolates were plated onto HIBTW plates containing 10% sucrose, which resulted in a second recombination event that generated either the wild-type chromosomal sequence or an in-frame deletion of the targeted gene. A 1737 sidA ciuE double mutant was constructed in 1737 sidA (18) by deletion of the ciuE gene as described above. PCR was used to identify isolates that had deleted the gene of interest.
The point mutation in the irp6A gene in strain HC1 was moved into the wild-type C7 strain by a previously described technique for allelic replacement (42). PCR was used to amplify a 1.4-kb DNA fragment carrying the HC1 irp6A allele, and the fragment was subsequently ligated into the conjugation-proficient C. diphtheriae suicide plasmid pK19mobsacB (Kanr) (30). The pK19mobsacB plasmid carrying the HC1 irp6A allele was transformed into E. coli S17-1 (39), and the recombinant plasmid was moved into C. diphtheriae C7 by conjugation as described elsewhere (42). Nalidixic acid (35 μg/ml) was used to select against the donor E. coli S17-1 strain. Since the pK19mobsacB plasmid cannot replicate in C. diphtheriae strains, all Kanr transconjugates were generated by recombination between homologous regions on the plasmid and the chromosome. To isolate C7 strains carrying the point mutation, corynebacterial cointegrates were plated onto sucrose medium as described above. The C7 strain carrying the irp6A point mutation was designated C7A-1, and the presence of the point mutation was confirmed. Primers and primer sequences will be made available on request.
Growth analysis of C. diphtheriae strains.
Overnight cultures of C. diphtheriae strains grown in HIBTW medium were inoculated 1:2 into fresh HIBTW medium containing 8 μM EDDA and grown for 6 h. The EDDA-containing cultures were used to inoculate PGTH medium (1:100), which were grown for 18 h at 37°C with shaking. The 18-h culture was diluted with fresh PGTH medium (3 ml) to an optical density at 650 nm (OD650) of 0.2. (In some experiments, as indicated, the 18-h cultures were diluted to an OD650 of 0.02, which extended the growth period for the bacteria.) The PGTH medium contained either 5 μM FeSO4, no added supplement, or 1 μM EDDA. Aliquots were removed at various times for measurement of the OD650.
Siderophore assays.
Siderophore determinations were done using the culture supernatants from the 24-h cultures described above in the growth analysis section. Siderophore measurements were done on 0.5 ml or appropriate dilutions of the culture supernatants by using the CAS assay as described previously (33). Reactions were allowed to develop for 2 h at room temperature, and the OD630 was measured. Siderophore units were determined as described previously (33) and were normalized for variations in growth.
Plate bioassays.
C. diphtheriae strains that were tested for growth stimulation were grown as described above for the growth analysis. HIBTW agar plates were depleted for iron by the addition of 140 μM EDDA, and test strains (106 bacteria/ml) were added to the EDDA-agar (45°C). Iron sources in 25-μl volumes were added to wells in the plates, and zones of bacterial growth were scored after 18 h at 37°C. Culture supernatants containing C. diphtheriae siderophore activity were obtained from C. diphtheriae C7 or 1737 after 24 h of growth in PGTH medium. Aerobactin was obtained from culture supernatants of Shigella flexneri 2457T after growth in low-iron medium as previously described (24). The presence of aerobactin in culture supernatants was confirmed by the perchloric acid assay, a test that is specific for hydroxamate-type siderophores (2). Supernatants were filter sterilized prior to use in the bioassay.
55Fe uptake assays.
C. diphtheriae cultures were grown as described above for the growth analysis section. The 18-h cultures were diluted into 1 ml of fresh PGTH medium (with or without EDDA) to an OD650 of 0.2 and then grown to an OD650 between 0.5 and 0.7. Assays were initiated by the addition of 1 μl of 0.1-μCi/μl 55FeCl3 (Perkin-Elmer, Boston, Mass.; stock solution in 0.5 M HCl is 19.4 mCi/ml and has a specific activity of 94.6 mCi/mg), and 50-μl aliquots were removed at intervals and applied to nitrocellulose filters (Millipore, Bedford Mass.; 0.45-μm pore size). Filters were washed twice with 1 ml of 100 mM sodium citrate and dried, and 55Fe levels were determined using a liquid scintillation counter.
In experiments that assessed the ability of siderophore to stimulate iron transport, cultures were centrifuged and the supernatant was discarded and replaced with an equivalent volume of C7 culture supernatant obtained from strains grown in low-iron PGTH medium. Supernatants were filter sterilized prior to use in the 55Fe uptake assays.
LacZ assays.
Eighteen-hour cultures of C. diphtheriae strains grown in HIBTW medium were inoculated 1:2 into fresh HIBTW medium, grown for 6 h, and then inoculated 1:100 into PGTH medium that contained either 5 μM FeSO4 or no added supplement and grown for 16 to 18 h at 37°C with shaking. LacZ activity was determined for C. diphtheriae as previously described (34).
DNase I protection experiments.
Fragments used for footprinting experiments were generated by PCR. DNase I protection experiments were done as described elsewhere (35). Briefly, DNA fragments at 0.5 nM were end labeled at their 3′ termini with [α-32P]dCTP using the Klenow fragment (29). Purified DtxR was prepared as described previously (35) and used at the indicated concentrations, and CoSO4 was included at 100 μM in all reaction mixtures to activate DtxR binding (Co2+ was used in place of Fe2+ to activate DtxR, since it is not rapidly oxidized under aerobic conditions).
Computer analysis.
Nucleic acid and amino acid sequences were analyzed using DNA analysis software provided by the Genetics Computer Group, Wisconsin Package version 10.3. Amino acid sequence similarity searches were done using the BLAST program (1) at the National Center for Biotechnology Information and also by using the BLAST server provided at the online site for the Sanger Institute (http://www.sanger.ac.uk/Projects/C_diphtheriae). The genome sequence for C. diphtheriae strain NCTC 13129 was recently published (7), and the annotated sequence is accessible in the EMBL/GenBank database at accession number BX248353" LOCATOR-TYPE="GEN">BX248353.
RESULTS
Identification of a C. diphtheriae iron uptake and siderophore biosynthesis gene cluster.
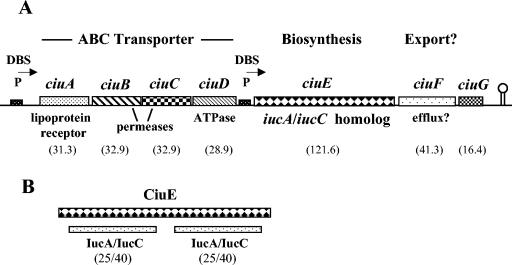
It was shown 20 years ago that C. diphtheriae can use the siderophore aerobactin as an iron source (27). Studies also revealed that aerobactin and the C. diphtheriae siderophore might be transported in C. diphtheriae by a common mechanism (26, 27), which suggests that these siderophores might share structural similarities and utilize similar enzymes in their biosynthetic pathways. In light of these earlier observations, we conducted a BLAST search of the C. diphtheriae genome sequence and identified a gene, which we termed ciuE (DIP0586), whose predicted product shares sequence similarity to the aerobactin biosynthetic enzymes IucA and IucC (Fig. 1). The C. diphtheriae CiuE protein has a predicted size of 121,582 Da (1,107 amino acids), which is almost twice the mass of IucA and IucC (predicted sizes of 65 to 66 kDa [21]). Both IucA and IucC showed almost 25% identity and 40% similarity to CiuE over approximately 400-amino-acid segments in both the N-terminal and C-terminal regions of CiuE (Fig. 1B). This 400-amino-acid region of similarity in both IucA and IucC is located in the central portion of the protein. The IucA and IucC proteins have related enzymatic activities in the synthesis of aerobactin and have 24% identity and 38% similarity to each other in this 400-amino-acid internal region. No obvious homologs for the two other proteins involved in aerobactin synthesis were found in the C. diphtheriae genome (IucB and IucD).
FIG. 1.
(A) Genetic map of the ciu gene cluster (DIP0582 to DIP0588). The predicted sizes of the gene products are indicated in parentheses (in kilodaltons) below the map. P, location of putative promoter; DBS, DtxR binding site; arrow, direction of transcription. Stem and loop indicates a putative transcriptional terminator. (B) Alignment of CiuE with an approximately 400-amino-acid internal region from IucA/IucC. IucC and IucA share similarity to CiuE in both the N-terminal region and C-terminal region. Homologies are indicated below the alignment (percent identity/percent similarity).
Upstream of ciuE are four genes, which we designated ciuA, ciuB, ciuC, and ciuD (DIP0582 to DIP0585), whose predicted products have significant similarity to ABC-type iron transport systems (Fig. 1A). The genes appeared to be organized into an operon, since the gaps between the start and stop codons were relatively small, with the exception of a 34-bp gap between the stop of ciuA and the start codon for ciuB (there is no gap between ciuB and ciuC, and there is a 3-bp gap between ciuC and ciuD). The predicted product of the ciuA gene, the first gene in this putative operon, is related to a diverse group of periplasmic binding proteins associated with ABC-type iron transporters from gram-negative bacteria and is also similar to iron uptake proteins from gram-positive bacteria. The amino acid similarities range between 25 and 35% identity and 45 and 55% similarity. The N terminus of CiuA has a putative 26-amino-acid leader sequence that contains a possible signal peptidase II recognition sequence, which suggests that CiuA is a lipoprotein. The ciuB and ciuC genes are predicted to encode the permease components of an ABC transport system, while the predicted product of ciuD has similarities to the ATPase component. Downstream from ciuE are two open reading frames, which we have designated ciuF (DIP0587) and ciuG (DIP0588), and a putative rho-independent termination sequence. The predicted product of ciuF has weak similarity to integral membrane efflux proteins found in various gram-positive bacteria. The predicted product of ciuG has no significant similarities to any known proteins.
CiuE is required for production of the C. diphtheriae siderophore.
Since CiuE has sequence similarity to the aerobactin biosynthetic proteins IucA and IucC, we wanted to determine if CiuE is involved in the synthesis of the C. diphtheriae siderophore. In-frame deletion mutations that removed 94% of the ciuE gene were constructed in C. diphtheriae strain C7(-), designated C7E, and in the Russian clinical strain 1737, 1737E. Siderophore activity was determined by the CAS method after growth of C. diphtheriae strains in PGTH medium that contained either 5 μM FeSO4 (+Fe) or no added iron (-Fe). Siderophore production was iron regulated in the wild-type strains, with optimal activity detected under low-iron conditions (Table 2). In the ciuE mutant strains, C7E and 1737E, a significant decrease in siderophore activity was observed relative to the parental strains under low-iron conditions, which indicates that these mutants have a defect in siderophore production. Plasmid pKE, which carries a copy of the wild-type ciuE gene, restored siderophore synthesis to 1737E and C7E (Table 2). The higher level of siderophore activity observed in the complemented mutants (1737E/pKE and C7E/pKE) relative to that in the wild-type parent strains may be due to the multiple copies and/or high expression of the cloned ciuE gene. The ciuE gene did not complement the siderophore defect in PW8, which suggests that the diminished siderophore production in this strain is not due to a defective ciuE gene (data not shown).
TABLE 2.
Siderophore production by C. diphtheriae strains
| Supplement in PGTH mediumb | Siderophore unitsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1737 (wt) | 1737A | 1737E | 1737E/pKE | C7 (wt) | C7A | C7E | C7E/pKE | PW8 | |
| +Fe | 2.6 | 3.7 | 2.5 | 7.7 | 10.1 | 10.7 | 10.8 | 11.1 | 9.4 |
| −Fe | 23.6 | 22.9 | 4.8 | 45.2 | 56.2 | 45.7 | 15.7 | 71.2 | 19.5 |
Siderophore units were determined as described previously (33). Values are the means of three independent experiments, and the results of each experiment varied by less than 15% from the mean.
+Fe contained 5 μM FeSO4; −Fe contained no added supplement.
In a previous report, it was shown that mutations in the putative siderophore biosynthetic genes sidA and sidB in 1737 had no effect on siderophore production (18). However, the possibility that a second siderophore was produced by the sidA-B system could not be eliminated. To further examine the role of the sid genes in siderophore synthesis, we constructed a sidA ciuE double mutant in 1737 and observed that it produced similar results in the siderophore assay to that seen with the ciuE single mutant (data not shown). These observations suggest that it is unlikely that the sid genes have a role in siderophore synthesis under the conditions examined in this study. C7 and 1737 ciuA mutants exhibited wild-type levels of siderophore synthesis and regulation (Table 2).
C. diphtheriae ciuE and ciuA mutants have reduced growth in low-iron medium.
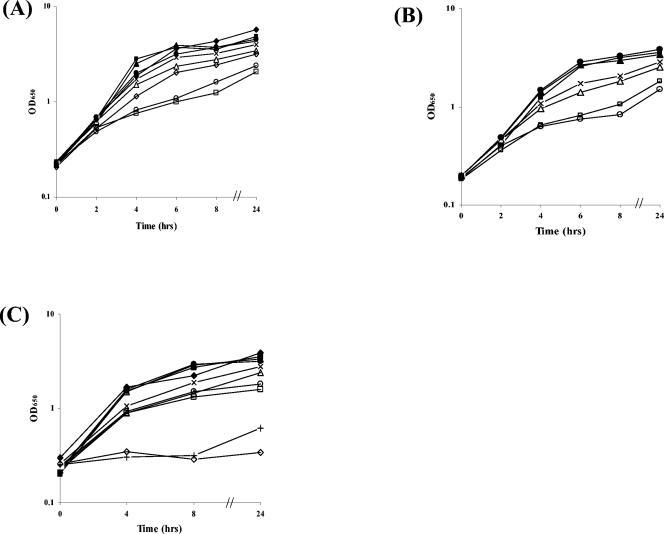
Bacterial strains that are defective in siderophore synthesis and transport frequently exhibit diminished growth under iron-limiting conditions. To determine if the ciuE and ciuA genes are required for growth under low-iron conditions, we compared the growth of wild-type C. diphtheriae strains C7(-) and 1737 with their isogenic mutants C7E (ciuE), C7A (ciuA), 1737E (ciuE), and 1737A (ciuA). Growth was assessed in three media: (i) PGTH medium with 5 μM FeSO4 (high iron), (ii) no added supplements (low iron), and (iii) a 1 μM concentration of the iron chelator EDDA (very low iron). All of the mutant strains exhibited similar growth rates and final cell densities as the wild-type parental strains in PGTH medium that contained 5 μM FeSO4 (Fig. 2A and B) or no added supplement (data not shown). However, in the presence of 1 μM EDDA, which further reduced the iron content of the PGTH medium, C7E, C7A, 1737E, and 1737A grew at slower rates and reached lower cell densities than their isogenic parent strains (Fig. 2A and B). In the presence of the cloned ciuE gene (pKE), 1737E grew at wild-type levels in PGTH medium that contained 1 μM EDDA (Fig. 2A). These findings indicate that the ciuE and ciuA genes are required for wild-type levels of growth in PGTH medium that contains 1 μM EDDA, and they suggest that the products of these genes are needed for sufficient acquisition of iron. Although the ciuE and ciuA mutants showed diminished growth relative to the parent strains in low-iron medium, these mutants were able to achieve cell densities exceeding an OD650 of 1.5 by 24 h, which suggests that additional iron uptake systems may be active in these mutants (7, 18, 26, 36).
FIG. 2.
Growth analysis of C. diphtheriae strains. C. diphtheriae strains were grown in PGTH medium containing various supplements as described in Materials and Methods, and the OD650 of cultures was determined at the indicated times. Strains were grown in PGTH medium that contained 5 μM FeSO4 (+Fe), no supplement (PGTH), or 1 μM EDDA (PGTH-E). (A) 1737 strains. Closed triangle, 1737 wild type (wt) (+Fe); X, 1737 wt (PGTH); open triangle, 1737 wt (PGTH-E); closed diamond, 1737E/pKE (+Fe); open diamond, 1737E/pKE (PGTH-E); closed square, 1737E (ciuE) (+Fe); open square, 1737E (ciuE) (PGTH-E); closed circle, 1737A (ciuA) (+Fe); open circle, 1737A (ciuA) (PGTH-E). (B) C7(-) strains. Closed triangle, C7(-) wt (+Fe); X, C7(-) wt (PGTH); open triangle, C7(-) wt (PGTH-E); closed square, C7E (ciuE) (+Fe); open square, C7E (ciuE) (PGTH-E); closed circle, C7A (ciuA) (+Fe); open circle, C7A (ciuA) (PGTH-E). (C) Closed triangle, C7(-) wt (+Fe); X, C7(-) wt (PGTH); open triangle, C7(-) wt (PGTH-E); closed square, C7A-1 (irp6A pt.) (+Fe); open square, C7A-1 (PGTH-E); closed circle, C7p6A (irp6A Δ) (+Fe); open circle, C7p6A (PGTH-E); closed diamond, HC1 (+Fe); +, HC1 (PGTH); open diamond, HC1 (PGTH-E). Results represent the means of three independent experiments, and values varied by less than 15% from the mean.
Growth analysis of irp6A mutants under high- and low-iron conditions.
A previous study indicated that genes in the C7 irp6ABC operon were involved in the utilization of the C. diphtheriae siderophore as an iron source (26). The HC1 strain, a chemically derived mutant of C7 which contains a missense mutation in irp6A (G240D), was previously shown to have a severe defect in iron uptake and in growth in low-iron medium (9, 26). HC1 also expressed iron-repressible promoters in high-iron medium, which was attributed to the severe iron uptake defect in this strain (26). To better define the role of the irp6 operon in iron uptake and in growth in low-iron medium, we constructed deletion mutants in the irp6A gene in the C7 and 1737 strains and measured the growth of these strains in high- and low-iron PGTH medium. The irp6A deletion mutants C7p6A and 1737p6A and the irp6A point mutant, HC1, grew to similarly high cell densities in the presence of 5 μM FeSO4 (Fig. 2C; data not shown for 1737p6A). However, the HC1 strain grew poorly in PGTH medium that had no added supplement (low iron) or contained 1 μM EDDA (very low iron) (Fig. 2C), which is consistent with previous reports showing that HC1 has a severe growth defect in low-iron medium (9, 26). The C7p6A deletion mutant had similar growth characteristics to those of the C7 parent strain in PGTH medium without added supplements (data not shown) but showed a slight reduction in growth at 24 h in the presence of EDDA compared to the isogenic C7 parent strain (very low iron) (Fig. 2C). The growth difference between C7p6A and C7 wild type in EDDA medium after 24 h was more apparent if the growth period was extended by diluting the overnight (18-h) cultures to 0.02 instead of 0.2 OD650. Under these modified growth conditions, the OD650 after 24 h of growth in 1 μM EDDA was as follows: C7 wild type, 2.5; C7p6A, 1.33; and C7A-1, 1.42 (the values are the means of three experiments). When grown under the modified conditions, the C7 irp6A mutants showed similar growth kinetics to that of the wild type during log phase, and growth differences between the mutant and wild type were not apparent until the late stages of growth (data not shown). Surprisingly, the 1737p6A mutant showed similar growth rates to that of the parent strain under the standard conditions (data not shown) and using the modified conditions described above. (The OD650 after 24 h of growth in 1 μM EDDA was as follows: 1737 wild type, 3.25; 1737p6A, 3.15.) These observations suggest that while the 1737 irp6A gene is not needed for wild-type levels of growth under the low-iron conditions examined, the C7 irp6A gene is required for wild-type levels of growth.
In light of the striking growth differences between the HC1 strain (a strain that was derived from C7 after chemical mutagenesis) and C7p6A (irp6AΔ), we constructed strain C7A-1, which is isogenic with the C7 wild-type strain, with the exception that C7A-1 contains the identical point mutation that is present in the HC1 irp6A gene. The C7A-1 strain has growth properties that are indistinguishable from those of C7p6A (irp6AΔ) (Fig. 2C and data above in the text). This observation suggests that the point mutation in irp6A is not the sole cause of the severe growth phenotype in HC1, and it is likely that additional mutations in the HC1 chromosome contribute to the severe growth defect. The presence of multiple mutations in HC1 would be expected, since this strain was subjected to chemical mutagenesis (9).
An in-frame deletion of the irp6B gene was also constructed in C7(-) and 1737 and showed growth characteristics that were very similar to those observed with the irp6A deletion mutants (M. P. Schmitt, unpublished observation).
Siderophore bioassay.
A siderophore bioassay was developed to determine if the various C. diphtheriae strains were able to use the C. diphtheriae siderophore as an iron source. The addition of 2.5 μM FeCl3 to the siderophore-containing supernatants enhanced the growth of all the strains except the ciuA mutants and HC1 (Table 3). The addition of 25 μM iron to the supernatant resulted in only a trace amount of growth in the ciuA mutants, but it caused a significant increase in growth with HC1 and heavy growth with the remaining strains, including the irp6A mutants (Table 3). Low-iron culture supernatants containing only the C. diphtheriae siderophore or to which only 25 μM FeCl3 was added were not sufficient to stimulate growth of any of the strains except the irp6A mutants, which were weakly stimulated by supernatant alone (Table 3). The reason for this weak stimulation is not known. The inability of 25 μM FeCl3 to stimulate growth was due to the high levels of the iron chelator EDDA present in the medium (140 μM). The growth inhibition caused by EDDA could be partially overcome by higher concentrations of FeCl3 (50 μM) (Table 3). The addition of iron to supernatants containing the C. diphtheriae siderophore greatly facilitated the use of the siderophore as an iron source. The findings suggest that the iron-free form of the C. diphtheriae siderophore, the predominant form present in low-iron culture supernatants, is unable to compete effectively with high levels of EDDA for iron. As seen in previous studies (27, 28), C. diphtheriae culture supernatants required added iron to stimulate growth when a chelator was present.
TABLE 3.
Growth stimulation of C. diphtheriae strains by iron and siderophore
| Strain | Growth with various iron sources
|
|||||
|---|---|---|---|---|---|---|
| Supa + 2.5 μM Fe3+ | Sup + 25 μM Fe3+ | Sup | 25 μM Fe3+ | 50 μM Fe3+ | Aerobactinb | |
| C7 (−) wt | +c | ++ | − | − | + | + |
| C7E (ciuE) | + | ++ | − | − | +/− | + |
| C7A (ciuA) | − | +/− | − | − | +/− | + |
| C7p6A (irp6AΔ) | + | ++ | +/− | − | + | − |
| C7A-1 (irp6A pt.) | + | ++ | +/− | − | + | − |
| HC1 | − | + | − | − | + | − |
| 1737 wt | + | ++ | − | − | + | − |
| 1737A (ciuA) | − | +/− | − | − | +/− | − |
| 1737E (ciuE) | + | ++ | − | − | +/− | − |
Sup, supernatant from C7 cultures grown for 24 h in low-iron PGTH medium.
Aerobactin was from S. flexneri 2457T supernatant.
++, heavy growth; +, light growth; +/−, trace growth; −, no growth.
Low-iron culture supernatants derived from the ciuE mutant strains showed no enhancement of growth of strains C7(-) or C7E in the presence of 25 μM iron, which suggests that the ciuE mutants do not produce biologically active siderophores (data not shown). Siderophore-containing culture supernatants obtained from either C7(-) or 1737 gave identical results in the bioassay, suggesting there is no obvious difference in the growth-stimulating properties between the siderophores secreted by these two strains. The results from this bioassay provide strong support for a role of the CiuA protein in transport of the C. diphtheriae siderophore, and they also suggest that Irp6A is not required for transport or for utilization of the C. diphtheriae siderophore, since the irp6A deletion mutant was able to utilize the siderophore as an iron source.
Since earlier studies indicated that aerobactin could be used as an iron source by C. diphtheriae, we used the bioassay to determine whether the growth of various C. diphtheriae strains could be enhanced by aerobactin. Growth of the wild-type C7 strain, as well as the C7E and C7A mutant strains, was moderately stimulated by aerobactin (Table 3). This observation is consistent with earlier reports that indicated the C7 strain is able to use aerobactin iron (27), but the findings further suggest that the transport system encoded by ciuABCD is not needed for aerobactin transport in the C7 strain, since the ciuA mutant showed no diminished ability to use aerobactin as an iron source. Surprisingly, the 1737 strain was unable to use aerobactin as an iron source and, furthermore, growth of the C7 irp6A mutant strains, C7p6A, C7A-1, and HC1, was also not stimulated by aerobactin (Table 3), which suggests that the iron uptake system encoded by the irp6 operon may be involved in the utilization of aerobactin iron.
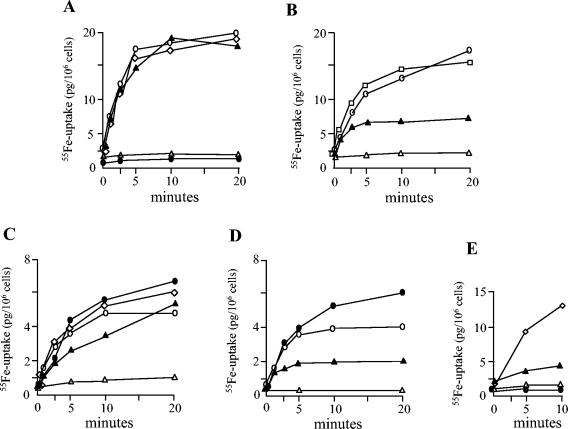
55Fe uptake assays.
55Fe uptake assays were done to determine the ability of the various C. diphtheriae mutant strains to transport iron and to use the C. diphtheriae siderophore for iron uptake. The C. diphtheriae ciuA mutants and HC1 were severely deficient in iron transport under all conditions tested (Fig. 3A to D). The ciuE mutants, when grown in PGTH medium without any supplements, showed similar iron uptake kinetics to that of the parent strain (Fig. 3A and C); however, in the presence of 1 μM EDDA, the ciuE mutants exhibited significantly reduced ability to transport iron relative to the wild-type strains (Fig. 3B and D). The presence of the cloned ciuE gene on plasmid pKE in the ciuE mutants restored wild-type levels of iron uptake in the presence of 1 μM EDDA, which indicates that the reduced iron uptake in the ciuE mutants is due to the deletion of the ciuE gene (Fig. 3B and D).
FIG. 3.
55Fe uptake in C. diphtheriae. 55Fe uptake assays were done as described in Materials and Methods. Strains were passaged into 1 ml of PGTH medium in the presence or absence of EDDA, and the assays were initiated by the addition of 1 μl of 0.1-μCi/μl 55FeCl3 to cultures that had attained an OD650 of 0.5 to 0.8. (A) C7(-) strains assayed in unsupplemented PGTH medium. Open circle, C7(-) wild type (wt); closed triangle, C7E (ciuE); closed circle, HC1; open triangle, C7A (ciuA); open diamond, C7p6A (irp6AΔ). (B) C7(-) strains assayed in PGTH medium that contained 1 μM EDDA. Strain designations are the same as those indicated for panel A. Open square, C7E/pKE. (C) 1737 strains assayed in unsupplemented PGTH medium. Open circle, 1737 wt; closed triangle, 1737E; closed circle, 1737E/pKE; open triangle, 1737A; open diamond, 1737p6A (irp6AΔ). (D) 1737 strains assayed in PGTH medium that contained 1 μM EDDA. Strain designations are the same as those indicated for panel C. (E) Stimulation of 55Fe uptake by culture supernatants. Experiments were done as described in Materials and Methods. C7E was grown in 3 μM EDDA, and culture supernatants from C7(-) wt (open diamond) or C7E (closed triangle) were added to washed cells prior to initiation of the assay. C7A (open triangle) and HC1 (closed circle) were assayed after growth in unsupplemented PGTH medium to which culture supernatants from C7(-) wt were added prior to initiation of the assay. Results are the means of at least three independent experiments, and values for each time point varied by less than 10% from the mean.
The addition of low-iron culture supernatants containing the C. diphtheriae siderophore did not enhance iron transport in the ciuA mutants C7A or 1737A or in HC1 (Fig. 3E and data not shown), which suggests these strains are unable to utilize the C. diphtheriae siderophore for iron uptake. The ciuE mutant showed only low levels of iron uptake activity when grown in the presence of 3 μM EDDA (Fig. 3E); however, when supernatants containing the C. diphtheriae siderophore were added, the ciuE mutant showed a marked stimulation in iron uptake (Fig. 3E), which suggests that the defect in iron uptake in the ciuE mutants is due to the inability to produce siderophore. The irp6A deletion mutants C7p6A and 1737p6A showed wild-type levels of iron uptake when grown in PGTH medium that contained no added supplement or 1 or 3 μM EDDA (Fig. 3A and C and data not shown).
Transcriptional analysis of the ciu gene cluster.
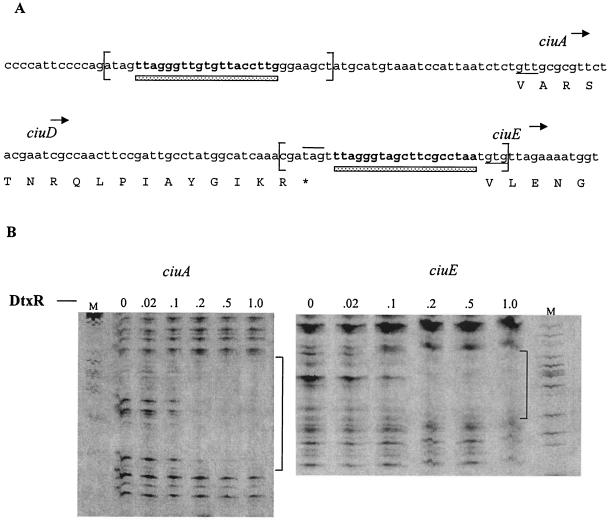
Most genes involved in siderophore biosynthesis and transport are repressed by increased iron concentration in the growth medium (14). To analyze the transcriptional regulation of the ciu gene cluster, we constructed promoter-lacZ fusions using the promoter probe vector pCM502. DNA sequences upstream of the ciuA and ciuE genes share homology to the 19-bp consensus DtxR binding site, including a 12-of-19-bp match at ciuA (with eight of nine of the most highly conserved residues present [18]) and a 15-of-19-bp match at ciuE (Fig. 4A). Several putative −10 and −35 promoter elements that showed limited homology to the consensus promoter sequences could be identified (data not shown). PCR was used to amplify DNA fragments from the 1737 chromosome that contained these putative DtxR binding regions. These fragments were ligated into pCM502 and assessed for promoter activity in C. diphtheriae strains. Transcription from both the ciuA and ciuE promoters in C7(-) and in 1737 was regulated by the iron content in the growth medium, with maximum expression observed only under low-iron conditions (Table 4). The ciuA promoter was more strongly expressed and showed less iron repression than the ciuE promoter in the C7(-) strain (Table 4). Furthermore, transcription of the ciuA promoter in 1737 was more strongly expressed and iron repressible than the same ciuA promoter fragment in the C7 strain. The DNA sequence of the DtxR binding sites upstream of ciuA and ciuE is identical between C7 and 1737 (data not shown), and transcription from the ciuA promoter region obtained from C7 showed similar regulation and expression to those seen with the ciuA promoter from 1737 (Table 4 and data not shown). This finding indicates that the differences in expression and iron-dependent regulation at the ciuA promoter between C7 and 1737 are not due to differences in DNA sequence at these promoters but rather to other strain differences.
FIG. 4.
(A) DNA sequences of ciuA and ciuE upstream regions, including the DtxR binding sites. DtxR binding sites are in bold and underlined with a bar. Brackets indicate regions protected by DtxR from DNase I digestion. Underlined nucleotides indicate putative start codons, and the line above the sequence indicates the ciuD stop codon. (B) DNase I protection at the ciuA and ciuE upstream sequences. DNase I footprinting was done as described in Materials and Methods. Brackets indicate sequences protected from DNase I digestion by DtxR. DtxR was included at the concentrations indicated above the autoradiogram (micromolar concentrations). M, DNA sequencing ladder used to identify the protected sequences; G, ladder for ciuE; C, ladder for ciuA. (Several independent footprint experiments, including the gels shown, were used to define the protected region.)
TABLE 4.
Transcriptional analysis of promoter-lacZ fusions in C. diphtheriae strains under high- and low-iron conditions
| Promoter fusionb | β-Galactosidase activity unitsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| C7(−) (dtxR+)
|
1737 (dtxR+)
|
C7(β)hm723 (dtxR)
|
|||||||
| +Fec | −Fe | Ratio (−/+) | +Fe | −Fe | Ratio (−/+) | +Fe | −Fe | Ratio (−/+) | |
| pciuEP-lacZ | 1.7 | 21.6 | 12.7 | 0.7 | 16.6 | 23.7 | 10.2 | 30.5 | 3.0 |
| pciuAP-lacZ | 27.4 | 100.0 | 3.6 | 5.9 | 179.3 | 30.4 | 106.0 | 165.2 | 1.6 |
| ptox-lacZ | 0.7 | 139.4 | 199.1 | 0.5 | 80.1 | 160.2 | 38.1 | 153.5 | 4.0 |
β-Galactosidase units were determined as described previously (34). Values are the means of three experiments, and results from each assay varied by less than 25% from the mean.
DNA fragments containing the 1737 ciuE and ciuA upstream regions were assessed for promoter activity in pCM502.
+Fe, PGTH medium with 5 μM FeSO4; −Fe, unsupplemented PGTH medium.
The iron repression at both promoters was largely alleviated in the dtxR mutant C7(β)hm723, which suggests these promoters are DtxR regulated. No obvious transcriptional termination signal was seen in the 21-bp intergenic region between ciuE and ciuD and, in fact, 19 of the 21 bases within this small intergenic region were part of the putative DtxR binding site (Fig. 4A). The lack of any clearly defined termination signals in this region suggests that ciuE may be transcribed from both the ciuA and ciuE promoters. The tox promoter was included as a control for iron and DtxR regulation (18).
DtxR binds to the ciuA and ciuE upstream regions.
DNase I protection experiments were done with DNA fragments carrying the regions upstream of the ciuA and ciuE genes to determine if DtxR was able to bind to these putative DtxR binding sites. Figure 4B shows the results of the DNase I protection experiments in which the DtxR protein was able to protect an approximately 30-bp region from DNase I digestion at both the ciuA and ciuE upstream regions. The protected area is indicated in the sequence shown in Fig. 4A. The region that showed homology to the consensus DtxR binding site at both the ciuA and ciuE upstream sequences is located within the DNase I-protected region. These studies confirm that DtxR is able to bind to the regions upstream of both ciuA and ciuE, and they provide strong evidence that DtxR mediates the iron-dependent regulation of the ciu gene cluster.
DISCUSSION
Until this report, genes associated with the synthesis of the C. diphtheriae siderophore had not been described. We report here the identification of the ciu gene cluster, which encodes products involved in the transport (CiuABCD) and synthesis (CiuE) of the major C. diphtheriae siderophore. The predicted product of ciuE is similar to the aerobactin biosynthesis enzymes IucC and IucA, and mutations in ciuE greatly reduce siderophore production, which suggests that CiuE has a role in siderophore synthesis. Based on what is known regarding the synthesis of bacterial siderophores and, more specifically, those siderophores that utilize enzymes that share similarity with the IucC and IucA-like proteins (6, 11, 20, 21), it seems likely that additional enzymes are involved in the production of the C. diphtheriae siderophore. Genes that are predicted to encode siderophore biosynthetic enzymes were not identified adjacent to the ciu gene cluster, nor was any clustering of siderophore biosynthetic genes identified in the DRTA study (18), with the exception of the sid genes. The sid genes, however, do not appear to be associated with the synthesis of a siderophore based on the findings from the previous report (18) and from this study, in which we showed that the levels of siderophore production in a ciuE sidA double mutant were similar to those of the ciuE single mutant. It is possible, nevertheless, that the sid genes may be involved in siderophore production under other conditions not examined in these studies. Since the DRTA system did not identify all DtxR binding sites on the C. diphtheriae chromosome (for example, the DtxR-regulated ciu or irp6 promoters were not identified using DRTA), it is possible that additional DtxR- and iron-regulated genes involved in siderophore synthesis may be present on the chromosome. These additional genes are likely to be associated with the synthesis of a precursor siderophore molecule, since IucC/IucA-like proteins are involved in the final steps of siderophore synthesis (20, 21).
One of the more unusual features of ciuE is that it appears to be a fusion of two iucC/iucA-like genes. The predicted product of ciuE shows similar relatedness to both IucC and IucA in both halves of the protein, and CiuE sequences do not exhibit significantly higher amino acid similarity to either IucC or IucA in either the N-terminal region or C-terminal section. While CiuE appears to be a fusion protein, it remains to be determined if the protein has two distinct enzymatic activities within a single peptide. For example, does the N-terminal region possess an IucC-like function while the C-terminal portion has an IucA-like activity? Another unusual feature of this siderophore system is that while CiuE has homology to enzymes involved in the synthesis of aerobactin, a hydroxamate siderophore, the C. diphtheriae siderophore gives a negative result in tests for the hydroxamate-type siderophores (27). Similar observations have been made in studies with siderophores produced by Bacillus anthracis (6) and Legionella pneumophila (15, 19). Synthesis of the siderophore anthrachelin in B. anthracis appears to require two genes that share homology to iucC and iucA, yet anthrachelin gives a negative result in the Csaky test for hydroxamates. The predicted product of the frgA gene in L. pneumophila is an IucC/IucA homolog, and mutations in frgA resulted in impaired intracellular growth (15). Although a siderophore has not been specifically associated with frgA, L. pneumophila does not appear to produce a hydroxamate-type siderophore (15, 19). Interestingly, the frgA gene is not clustered with any other siderophore biosynthetic genes, a situation that is similar to what we have observed with ciuE in C. diphtheriae. The structures for the siderophores in B. anthracis and L. pneumophila have not been determined.
Although the C. diphtheriae ciuE mutants exhibited reduced siderophore production, a low level of iron chelating activity was detected in the C7 and 1737 ciuE mutants grown under low-iron conditions, and this activity was higher than that seen in iron-replete cultures. The source of this activity in low-iron cultures is not known but may be caused by a siderophore precursor or by some other weak iron-chelating compound secreted under low-iron conditions. It is also possible that certain metabolites that are secreted under low-iron conditions, such as the previously described coproporphyrin III (27), may interfere with the CAS assay or function as weak chelators.
The proximity of the ciuABCD transport genes to ciuE initially suggested to us that this transport system might be involved in aerobactin uptake, since the predicted product of ciuE is similar to aerobactin biosynthetic enzymes. Surprisingly, however, we showed that a ciuA mutant utilized aerobactin as an iron source, while a mutation in the C7 irp6A gene abolished aerobactin utilization. Theses findings suggest that the C7 Irp6 transporter is needed for the utilization of aerobactin iron, while the CiuABCD transport system is involved in the transport of the major C. diphtheriae siderophore and is not required for aerobactin uptake. The 1737 clinical isolate was unable to use aerobactin as an iron source, and while it is not known why the 1737 and C7 strains differ in their abilities to use aerobactin iron, we have noted several amino acid differences between the predicted irp6 gene products for 1737 and C7. It is possible that differences in the amino acid sequence in the irp6 gene products between C7 and 1737 may be responsible for their differences in the ability to use aerobactin iron.
A previous study suggested that specific mutant alleles in the irp6 operon were responsible for the iron transport defect in the HC1 (irp6A), HC4 (irp6C), and HC5 (irp6B. C. diphtheriae mutants (26). While these mutants are chemically derived and likely carry mutations outside the irp6 operon, it was shown, nevertheless, that clones carrying the relevant irp6 gene could complement the mutant phenotype of the HC strains (26). (Complementation of the defect in HC1 by clones carrying the irp6A gene has also been done in our laboratory [Schmitt, unpublished]). In this study, we constructed a defined deletion in the irp6A gene and also an irp6A point mutant, C7A-1, that carried the identical mutation as that present in the HC1 irp6A gene. Analysis of the site-directed irp6A mutant strains revealed that they were unaffected in iron transport and in the utilization of the C. diphtheriae siderophore as an iron source. The irp6A mutants also regulated siderophore synthesis in a manner similar to that of the wild-type strain (Schmitt, unpublished). The irp6A gene was also not required for growth in low-iron medium in the clinical strain, 1737, but mutations in the C7 irp6A gene resulted in reduced growth relative to the parent strain, indicating that the irp6A gene is required for normal growth in low-iron medium in the C7 strain. However, mutations in the C7 irp6A gene do not result in the severe growth defect or iron uptake deficiency seen with the HC1 strain. These findings indicate that the severe growth and iron uptake phenotype in HC1 cannot be attributed solely to the point mutation in irp6A, and they suggest that additional mutations in the HC1 chromosome contribute to the severe growth defect in this strain.
The ciuA mutation in C7 and 1737 resulted in a severe defect in iron uptake and in the ability to use the C. diphtheriae siderophore as an iron source, which strongly suggests that the CiuA protein has an important role in the transport of the siderophore. We also noted a marked difference in the iron uptake activity between the ciuE and ciuA mutant strains, and we suspect this is largely a result of differences in siderophore production. Although the ciuA mutant produces siderophore at wild-type levels, it is unable to utilize the siderophore as an iron source, and therefore the siderophore activity present in the culture supernatant of the ciuA mutant is sequestering much of the available iron and is potentially inhibiting iron uptake by iron transport systems that are distinct from the ciu system. Recent studies have revealed that C. diphtheriae has several putative ABC iron transporters, including irp6 (26), frg (18), irp1 (36), and others identified from the genome sequence (7), and it is almost certain that alternative iron uptake systems are functioning to transport iron in the ciu mutants. This is supported by the observation that the ciu mutants, while exhibiting diminished growth in low-iron medium relative to wild type, are still able to grow to relatively high densities, which suggests that additional systems involved in iron uptake are active in the absence of the ciu system. It is likely that mutations in multiple iron transporters are needed to fully abolish growth of C. diphtheriae under low-iron conditions.
Iron uptake through these alternate iron transporters would be unaffected in the ciuE mutants, since these mutants do not produce siderophore. The ciuE mutant shows transport activity comparable to wild type only in the unsupplemented PGTH medium (Fig. 3A). In the presence of EDDA, the ciuE mutant shows a significant decrease in iron uptake relative to that of the wild-type strain (Fig. 3B). Since ciuE mutants do not produce siderophore, the iron transport that is observed presumably occurs by a siderophore-independent mechanism. It is not known which iron uptake systems (or how many) have a role in iron transport in the absence of siderophore. Future studies are needed to determine the role of the various iron transport systems in C. diphtheriae.
Acknowledgments
We thank Olaf Schneewind for strains and plasmids, Tanja Popovic for providing the C. diphtheriae clinical strain 1737, and Dennis Kopecko for S. flexneri 2457T. We are also grateful to Karen Meysick and Scott Stibitz for critical reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., G. Warren, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atkin, C. L., J. B. Neilands, and H. J. Pfaff. 1970. Rhodotorulic acid from species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a new alanine-containing ferrichrome from Cryptococcus melibiosum. J. Bacteriol. 103:722-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barksdale, W. L., and A. M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluhm, M. E., B. P. Hay, S. S. Kim, E. A. Dertz, and K. N. Raymond. 2002. Corynebactin and a serine trilactone based analogue: chirality and molecular modeling of ferric complexes. Inorg. Chem. 41:5475-5478. [DOI] [PubMed] [Google Scholar]
- 5.Budzikiewicz, H., A. Bossenkamp, K. Taraz, A. Pandey, and J. M. Meyer. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterum sp. DSM 20411). Z. Naturforsch. Sect. C 52:551-554. [Google Scholar]
- 6.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 7.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC 13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryz, S. J., L. M. Russell, and R. K. Holmes. 1983. Regulation of toxinogenesis in Corynebacterium diphtheriae: mutations in the bacterial genome that alter the effects of iron on toxin production. J. Bacteriol. 154:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csaky, T. Z. 1948. On the estimation of bound hydroxylamine in biological materials. Acta Chem. Scand. 2:450-454. [Google Scholar]
- 11.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from hemin and hemoglobin are homologous to ABC hemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 14.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 15.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and Fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181:56-67. [DOI] [PubMed] [Google Scholar]
- 17.Kanei, C., T. Uchida, and M. Yoneda. 1977. Isolation from Corynebacterium diphtheriae C7(β) of bacterial mutants that produce toxin in medium with excess iron. Infect. Immun. 18:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liles, M. R., T. A. Scheel, and N. P. Cianciotto. 2000. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 182:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez, J. L., M. Herrero, and V. de Lorenzo. 1994. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J. Mol. Biol. 238:288-293. [DOI] [PubMed] [Google Scholar]
- 22.Otto, B. R., V.-V. Vught, A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18:217-233. [DOI] [PubMed] [Google Scholar]
- 23.Park, W. H., and A. W. Williams. 1896. The production of diphtheria toxin. J. Exp. Med. 1:164-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne, S. M. 1980. Synthesis and utilization of siderophores by Shigella flexneri. J. Bacteriol. 143:1420-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, I. K. Wachsmuth, and J. D. Wenger. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 174:1064-1072. [DOI] [PubMed] [Google Scholar]
- 26.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell, L. M., S. J. Cryz, and R. K. Holmes. 1984. Genetic and biochemical evidence for a siderophore-dependent iron transport system in Corynebacterium diphtheriae. Infect. Immun. 45:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, L. M., and R. K. Holmes. 1985. Highly toxinogenic but avirulent Park-Williams 8 strain of Corynebacterium diphtheriae does not produce siderophore. Infect. Immun. 47:575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schafer, A., A. Tauch, W. Jager, J. Kalinowski G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 183:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt, M. P., and R. K. Holmes. 1993. Analysis of diphtheria toxin repressor-operator interactions and characterization of a mutant repressor with decreased binding activity for divalent metals. Mol. Microbiol. 9:173-181. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor and iron. J. Bacteriol. 176:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider, R., and K. Hantke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111-121. [DOI] [PubMed] [Google Scholar]
- 38.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biol. Chem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 39.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. BioTechnology 1:784-794. [Google Scholar]
- 40.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 41.Tai, S.-P. S., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 42.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 43.Vitek, C. R., and M. Wharton. 1998. Diphtheria in the former Soviet Union: reemergence of a pandemic disease. Emerg. Infect. Dis. 4:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkelmann, G. 2002. Microbial siderophore-mediated transport. Biochem. Soc. Trans. 30:691-696. [DOI] [PubMed] [Google Scholar]