Abstract
Levels of RpoS increase upon glucose starvation in Escherichia coli, which leads to the transcription of genes whose products combat a variety of stresses. RpoS stability is a key level of control in this process, as SprE (RssB)-mediated degradation is inhibited under glucose starvation. Starvation for ammonia or phosphate also results in increased stress resistance and induction of RpoS-dependent genes. However, we demonstrate that RpoS levels following ammonia starvation are only slightly increased compared to growing cells and are 10-fold below the levels observed under glucose or phosphate limitation. This difference is largely due to regulated proteolysis of RpoS, as its stability in ammonia-starved cells is intermediate between that in logarithmic-phase cells and glucose-starved cells. Use of an rpoS construct that is devoid of the gene's native transcriptional and translational control regions reveals that stability differences are sufficient to explain the different levels of RpoS observed in logarithmic phase, ammonia starvation, and glucose starvation. Under phosphate starvation, however, rpoS translation is increased. The cellular response to nutrient limitation is much more complex than previously appreciated, as there is not simply one response that is activated by starvation for any essential nutrient. Our data support the hypothesis that SprE activity is the key level at which ammonia and glucose starvation signals are transmitted to RpoS, and they suggest that carbon source and/or energy limitation are necessary for full inactivation of the SprE pathway.
Bacteria have evolved sophisticated stress response mechanisms to combat nutrient limitation and starvation. Escherichia coli cells starved for glucose mount a rapid response that ensures survival through the starvation period. This response, recently termed the general stress response, also protects cells from insults they may face during starvation, as these cells are much more resistant to heat, osmotic, and oxidative stresses than exponentially growing cells (12, 21).
Glucose starvation represents the best-characterized model for nutrient starvation, and the key regulator in this response is the sigma factor RpoS (σS, σ38). During glucose starvation RpoS protein levels increase dramatically, and RNA polymerase containing RpoS (EσS) transcribes over 70 genes involved in stress resistance and protection (12). In exponentially growing cells, RpoS levels are maintained at a low level due to degradation by the ClpXP protease (28, 32). This degradation is dependent on SprE (RssB), whose N-terminal domain is highly homologous to N-terminal domains of two-component response regulators, including a conserved aspartate (D58) that is traditionally phosphorylated to regulate this class of proteins (26, 28). Its C-terminal domain is unique and is believed to target RpoS to the ClpXP protease for degradation (25). The mechanism by which the cell senses starvation and transduces the starvation signal to affect SprE-dependent proteolysis is not currently understood. Recent evidence demonstrated that RpoS is still stabilized upon glucose starvation in cells with a mutation of the putative phosphorylation site (D58A), excluding regulation of SprE as a traditional two-component response regulator (27). SprE plays an important role in regulation of RpoS degradation, but how this activity is modulated in logarithmic phase and glucose starvation is not clear.
The role that RpoS synthesis plays under glucose starvation is also not well understood. There is roughly a twofold increase in activity of an RpoS-LacZ translational fusion as cells become limited for glucose (20, 22). The fusion used in these experiments lacked the region of RpoS that is necessary for SprE recognition, and so these results have been interpreted as evidence that synthesis of RpoS increases in the transition from growing exponentially to starving for glucose. Zgurskaya et al. (39) measured RpoS levels by quantitative Western blotting and degradation rates by [35S]methionine pulse-chase followed by immunoprecipitation. They calculated the synthesis rate of RpoS from these two values across a range of dilution rates in glucose-limited chemostat cultures and in starved and growing batch cultures. From these calculations it was found that the RpoS synthesis rate decreases two- to fourfold as cells starve for glucose (39).
Previous work suggested similarities between glucose and ammonia starvations. Common proteins are induced following starvation for ammonia or glucose as assayed by two-dimensional gel electrophoresis (9, 21). Additionally, ammonia-starved cells survive similarly to glucose-starved cells following treatment with heat or peroxide (15), and cells carrying an rpoS null allele fail to survive ammonia starvation as well as wild-type cells (16, 23). Further work has shown that transcription from the RpoS-dependent uspB promoter is induced following ammonia starvation and glucose starvation (5), suggesting further parallels between the two conditions.
It has been demonstrated that ppGpp levels increase upon ammonia starvation (14), and Gentry et al. (8) provided evidence that RpoS levels correlate with ppGpp levels. As such, they suggested that RpoS levels increase upon ammonia starvation due to the increase in ppGpp observed under those conditions. Although ppGpp has been reported to affect RpoS synthesis (3, 8, 13, 19), it has not been implicated in stability control, and the role that ppGpp plays under specific nutrient limitation remains unclear.
We focused our studies on ammonia starvation to understand how cells respond when challenged with limitation for a nutrient other than carbon. Previous work from our laboratory identified translational induction of RpoS following constitutive activation of the phosphate-scavenging regulon (31), and so it was of interest to compare and contrast another nutrient limitation with published data on glucose and phosphate limitation. Comparisons of starved cells versus growing cells are complicated by the difference in total protein synthesis rates under the two conditions, and so we reasoned that a more relevant approach to understand RpoS levels in the nongrowing state would be to compare RpoS levels following starvation for different nutrients.
To understand how the cell senses starvation and transduces a signal to RpoS based on specific nutrient limitation, we examined the stability and synthesis of RpoS under ammonia starvation. We report that in contrast to glucose starvation and phosphate limitation, cells starved for ammonia fail to demonstrate a dramatic increase in RpoS levels relative to logarithmic-phase cells. Signaling through SprE/ClpXP was intermediate between that observed in growing cells and that under glucose starvation, indicating that without exhaustion of the carbon and/or energy source significant proteolysis of RpoS continued. We discuss the relevance of these results to ammonia starvation specifically and in the larger context of E. coli's response to nutrient deprivation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains used in this study are listed in Table 1. Standard microbial techniques (33) were used to construct strains. The rpoS::kan, sprE::tet, and rssA2::cam alleles are published (30). The ΔclpX::kan and ΔclpP::cam alleles were transduced from a strain kindly provided by S. Gottesman. Strain YMC12 (1) was kindly provided by A. Ninfa.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 4 |
| MG1655 | 34 | |
| MJM211 | MC4100 sprE::tet | |
| MJM221 | MC4100 ΔclpX::kan ΔclpP::cam | |
| MJM282 | MC4100 rssA2::cam | |
| MJM294 | MC4100 rpoS::kan malE::Tn10 | |
| MJM302 | MJM294/pBAD18 | |
| MJM310 | MJM294/pRpoS | |
| MJM312 | MJM294/pMalE | |
| YMC12 | thi endA hsr ΔlacU169 hutCKlebsiella glnG::Tn5 | 1 |
Plasmids pRpoS and pMalE were constructed for this study. For pRpoS, colony PCR was performed on MC4100 cells with the primers MJM287F (5′-GAGGATGTCGCTAGCCAAGGACCATAGATTATGAGTCAGAATACGCTGAAAGTTC ) and MJM307R (5′-GTGACGTCAGGTACCTTCTGACAGATGCTTACTTACTC ). These primers amplified the entire rpoS open reading frame, adding the NheI and KpnI restriction sites (underlined), respectively. MJM287F also added the last 15 bp of the malE 5′ untranslated region (UTR) (bold) directly upstream of the rpoS open reading frame. For pMalE, colony PCR was performed on MC4100 cells with the primers MJM288F (5′-GAGGATGTCGCTAGCCAAGGACCATAGATTATGAAAATAA ) and MJM308R (5′-GTGACGTCAGGTACCTTACTTGGTGATACGAGTCTG ). These primers amplified the entire malE open reading frame and 15 bp of the 5′ UTR (bold), adding the NheI and KpnI restriction sites (underlined), respectively. After digestion with NheI and KpnI, each PCR product was introduced into the NheI and KpnI sites of pBAD18 (10). In the resulting plasmids, both rpoS and malE were under the transcriptional control of the pBAD promoter with the Shine-Dalgarno sequence provided by the malE 5′ UTR. The constructs are identical except for the open reading frame they contain. Oligonucleotide synthesis and plasmid sequencing were conducted by the Princeton University Department of Molecular Biology Synthesis and Sequencing Facility.
Media and growth conditions.
Cells were grown in Luria-Bertani (LB) broth and M63 minimal 0.4% glucose as described previously (33) with modifications as discussed below. Unless indicated, all bacterial strains were grown under aeration at 37°C and growth was monitored by measuring the optical density at 600 nm (OD600). Mid-logarithmic phase refers to cultures with an OD600 of 0.3 to 0.5.
For starvation experiments, full M63 minimal glucose (N+C+) contained 0.2% (NH4)2SO4 and 0.4% glucose. N− medium lacked (NH4)2SO4; C− medium lacked glucose; and N−C− medium lacked both nutrients.
Cells for experiments in minimal medium were grown first by selecting single colonies from LB plates and growing them in liquid LB for 3 to 5 h. Cells were then subcultured 1:100 into minimal glucose and grown overnight. Cells were subcultured 1:100 into minimal glucose medium and grown to mid-logarithmic phase for the appropriate treatment. There have been reports of variation in RpoS levels when cells were not permitted to undergo at least 10 generations between exit from stationary phase and mid-logarithmic phase (8). Control experiments in which cells were diluted 1:105 from LB into minimal glucose yielded identical results as the 1:100 dilutions (data not shown). For standardization, the 1:100 dilutions were used in all cases.
Antibiotics used in strain construction included chloramphenicol (20 μg/ml), kanamycin (50 μg/ml), and tetracycline (25 μg/ml). Ampicillin (125 μg/ml) was used for growth of strains containing the pBAD18-based plasmids.
Starvation treatments.
Cells were grown to mid-logarithmic phase in minimal glucose and then pelleted for 7 min in 15-ml conical tubes at 1,800 × g (3,500 rpm) in a Durafuge 200 centrifuge (Precision Scientific). After resuspension in prewarmed starvation medium, cells were pelleted again and resuspended in prewarmed starvation medium. Time after starvation refers to the time since the first resuspension. Starvation was confirmed by observing a stable OD600, and addition of the missing nutrient in various forms [e.g., glutamine, glutamate, NH4Cl, or (NH4)2SO4 for nitrogen] was sufficient for resumption of growth.
SDS-PAGE and Western blot analysis.
Cells were assayed for the OD600, and then 1-ml samples were removed at given time points and added to 50 μl of cold trichloroacetic acid on ice to precipitate proteins. After an incubation of at least 30 min, proteins were pelleted, washed with 500 μl of cold acetone, and then resuspended in a volume of sodium dodecyl sulfate (SDS) sample buffer (18) equal to the OD600/6 (in milliliters). Samples were boiled for 10 min, and equal volumes were subjected to SDS-12% polyacrylamide gel electrophoresis (PAGE) as described by Laemmli (18). The proteins were transferred to nitrocellulose membranes (Schleicher & Schuell), and Western blot analyses were performed. Polyclonal sera (rabbit) against ClpX or against ClpP, both kindly provided by S. Gottesman, were used as primary antibodies at dilutions of 1:5,000. Polyclonal sera (rabbit) against RpoS or against SprE, both from our lab stock, were used as primary antibodies at dilutions of 1:6,000 and 1:4,000, respectively. The anti-SprE serum was preincubated with membrane bound by whole-cell lysate from MJM211 cells (which lack SprE) to reduce background. Polyclonal serum (rabbit) against MalE was kindly provided by D. Isaac and was used as a primary antibody at a dilution of 1:3,000. Donkey anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Amersham Biosciences) was used as a secondary antibody at a dilution of 1:6,000. For visualization of bands, the ECL antibody detection kit (Amersham Biosciences) and X-Omat film (Kodak) were used. Analysis of bands was performed using the public domain ImageJ program (developed at the National Institutes of Health and available at http://rsb.info.nih.gov/ij/).
Pulse-labeling and immunoprecipitation following pulse-chase.
For pulse-labeling alone, samples were pulsed with 50 μCi of [35S]methionine (MP Biomedicals) per ml of culture for 15 or 60 s. One-milliliter samples were removed at given time points and treated as above for SDS-PAGE, and the results were visualized by autoradiography.
For immunoprecipitation following pulse-chase, cells were pulse-labeled for 2 min as described above and then chased for 0, 15, or 30 min after addition of 150 μl of 5% unlabeled methionine. One-milliliter samples were removed at given time points and added to 50 μl of cold trichloroacetic acid on ice to precipitate proteins. After an incubation of at least 30 min, proteins were pelleted, washed with 500 μl of cold acetone, pelleted again, and resuspended in 150 μl of 50 mM Tris-1 mM EDTA-1% SDS. The samples were boiled for 5 min and then pelleted. A 140-μl aliquot of the supernatant was incubated with 1 μl of anti-RpoS polyclonal antiserum in 1 ml of 50 mM Tris (pH 8.0)-0.1 mM EDTA-0.15 M NaCl-2% Triton X-100. These reaction mixtures were gently rotated for at least 1 h at 4°C. A 50-μl aliquot of a 50% protein A-Sepharose bead slurry (prepared in the same buffer) was added to the reaction mixture, followed by gentle rotation for at least 1 h. After pelleting the reaction mixture, the pelleted immunoprecipitated samples were washed with 50 mM Tris (pH 7.5)-0.5 M NaCl-0.25% Triton X-100 for 30 min. The pelleted samples were then washed with 50 mM Tris (pH 7.5)-0.15 M NaCl-0.25% Triton X-100 for 30 min. The samples were again pelleted and then resuspended in a volume of SDS sample buffer (18) equal to the OD600 at the start of the experiment divided by 6 (in milliliters). Samples were run on SDS-PAGE, and the results were visualized by autoradiography.
Protein stability analysis following inhibition of protein synthesis.
Similar to the method used by Zhou and Gottesman (40), cells were treated with 200 μg of chloramphenicol per ml of culture and then 1-ml samples were removed at given time points and treated as above for Western blot analysis.
Calculation of relative rates of RpoS synthesis and degradation.
Since the degradation of RpoS approximates first-order exponential decay in logarithmic phase, ammonia starvation, and glucose starvation, the rates of RpoS synthesis and degradation were calculated based on the following differential equation: dN/dt = kS − kDN, where N represents the RpoS level, kS is the synthesis rate, and kD is the degradation rate. This degradation rate can be calculated from the measured half-life (t1/2) as follows: kD = ln2/t1/2. Lastly, under steady-state conditions dN/dt = 0, and so the synthesis rate can be calculated as follows: kS = kDN0.
RESULTS
RpoS levels fail to increase dramatically following ammonia starvation.
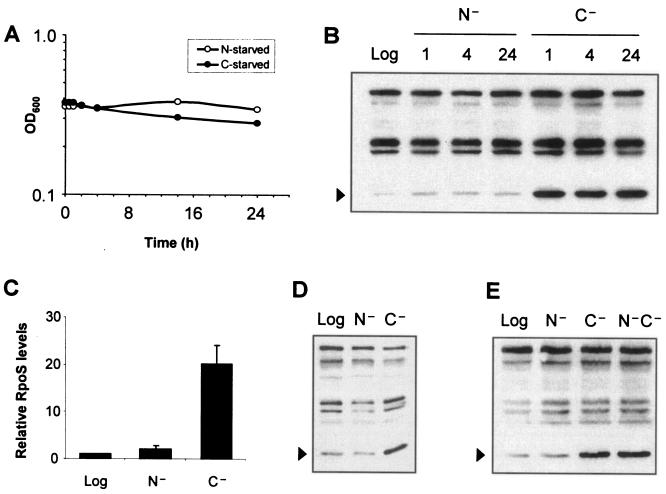
RpoS levels increase upon a number of cellular stresses, most notably under glucose depletion (12). As this response is believed to be necessary to achieve the multiple stress resistance found in starved cells, and as ammonia-depleted cells are reported to be resistant to challenge with heat or peroxide (15), we expected to see an increase in RpoS levels in cells upon ammonia starvation. To better understand the regulation of RpoS in ammonia-starved cells, we shifted logarithmic-phase cells into medium lacking ammonia and monitored RpoS levels. Since RpoS regulation is best characterized in response to glucose starvation, we used that condition as a control for our studies. As Fig. 1A shows, cells shifted to ammonia-free medium arrested growth as did cells similarly starved for glucose.
FIG. 1.
RpoS fails to accumulate significantly under ammonia starvation as it does under glucose starvation. Starvation was performed in M63 minimal glucose as described in Materials and Methods. Time points refer to hours after cells were first exposed to medium lacking the indicated nutrient, and RpoS levels were analyzed by Western blotting. Cross-reacting bands recognized by the polyclonal anti-RpoS serum served as loading controls and were not observed to vary based on growth phase or medium. (A) Optical densities of ammonia-starved and glucose-starved MC4100 cultures following starvation. (B) RpoS levels (arrowheads) in MC4100 in logarithmic phase (Log), under ammonia starvation (N−), and under glucose starvation (C−). Time points indicating hours poststarvation are shown above the lanes. (C) Quantification of the relative levels of RpoS from Western blotting, using NIH ImageJ and normalizing to the levels measured in logarithmic phase (set to 1.0). (D) RpoS levels (arrowheads) in MG1655 cells in log phase or starved for 1 h. (E) Double starvation for both ammonia and glucose mimics glucose starvation alone. RpoS levels were analyzed by Western blotting. MC4100 cells starved for both nutrients (N−C−) displayed the high levels of RpoS characteristic of glucose starvation, but not of ammonia starvation. Samples were obtained from cells that had been starved for 1 h.
As expected, RpoS levels increased dramatically upon glucose starvation (Fig. 1B). Surprisingly, we found that wild-type cells (strain MC4100) failed to increase RpoS levels significantly when shifted from logarithmic phase into ammonia starvation (Fig. 1B). This was not due to a delayed response, as glucose-starved cells and ammonia-starved cells displayed maximal levels of RpoS after starvation for 1 h. Quantification of RpoS levels identified an approximately twofold increase in ammonia starvation versus logarithmic cells (Fig. 1C). Under glucose starvation, RpoS levels were consistently 10-fold above those observed under ammonia starvation (Fig. 1C).
We were concerned that the observed phenotype could be peculiar to the MC4100 strain background, since this strain contains the relA1 allele, an almost complete loss-of-function mutation in relA (4, 24). Therefore, we assayed the starvation phenotypes in the stringent strain MG1655. As shown in Fig. 1D, MG1655 cells similarly failed to accumulate RpoS upon ammonia starvation as they did under glucose starvation. We also replaced the relA1 allele of MC4100 with a relA+ allele. Although the baseline levels of RpoS were slightly higher in logarithmic phase, the increases observed under ammonia starvation or glucose starvation were comparable to those in MC4100 (data not shown). Therefore, the difference between ammonia starvation and glucose starvation was present even when cells were able to mount a stringent response.
glnG encodes NtrC, the transcriptional activator of nitrogen assimilation and scavenging genes under conditions of nitrogen-limited growth (41). Control experiments with a glnG::Tn5 allele demonstrated that the Ntr system did not affect RpoS levels or ammonia-induced stress resistance under these conditions (data not shown). Results presented by Zimmer et al. indicated that RpoS-dependent promoters were not expressed upon genetic activation of GlnG (supplementary data in reference 41), supporting the idea that the RpoS-mediated ammonia starvation response is distinct from the Ntr-mediated nitrogen-scavenging response.
It is important to note that the low levels of RpoS observed under ammonia starvation are not a trivial consequence of ammonia starvation physiology. For example, were ammonia-starved cells limited in their ability to synthesize proteins, then the limited RpoS levels could have been due to biochemical constraints rather than signal transduction differences. When cells were starved for both ammonia and glucose together, RpoS levels increased commensurate with the increase observed under glucose starvation alone (Fig. 1E). Therefore, glucose starvation transduces a signal that is absent under ammonia starvation. Since there are factors known to affect RpoS transcription, translation, and protein stability, we inquired as to what step(s) is regulated differently between ammonia starvation and glucose starvation.
RpoS stability following ammonia starvation is higher than in logarithmic phase but lower than under glucose starvation.
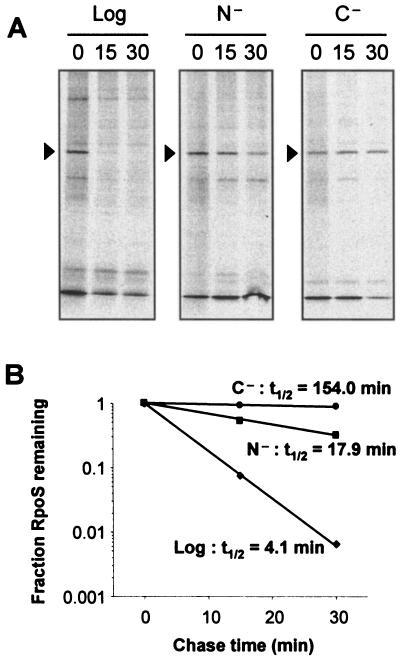
RpoS levels increase markedly following glucose starvation, and there is consensus that RpoS stability is an important level at which the glucose starvation signal is transmitted (12, 28, 39). Therefore, we assayed RpoS stability following ammonia starvation by pulse-chase analysis with [35S]methionine, followed by immunoprecipitation for RpoS. The data presented in Fig. 2 are consistent with published results demonstrating that the half-life of RpoS under glucose starvation was significantly higher than in logarithmic phase (154 ≫ 4.1 min). The half-life of RpoS under ammonia starvation was found to be intermediate between these values at 17.9 min (Fig. 2). Therefore, ammonia-starved cells demonstrated elevated RpoS stability compared to logarithmic-phase cells, but the protein was still degraded at a much higher rate than in glucose-starved cells.
FIG. 2.
RpoS stability following ammonia starvation is intermediate between the labile RpoS in logarithmic phase and the stable RpoS observed under glucose starvation. (A) Logarithmic-phase (Log) or 1-h-starved MC4100 cells, starved for ammonia (N−) or glucose (C−), were pulse-labeled with [35S]methionine for 2 min and then chased with cold methionine for the times indicated (in minutes). RpoS was then immunoprecipitated and analyzed by SDS-12% PAGE and autoradiography. (B) Quantification of the levels of RpoS in panel A by using NIH ImageJ, with each sample's zero-minute time point set to 1.0. Half-lives (t1/2) were calculated by regression analysis of the exponential decay using Microsoft Excel.
It is important to note that assaying RpoS stability by pulse-chase analysis was possible despite E. coli being able to utilize methionine as a nitrogen source in the presence of glucose (35). During the relatively short time course of this experiment (2 min for the pulse, 30 min for the chase), the optical density of the culture remained constant (data not shown). The doubling time of these cells growing on methionine as a nitrogen source was over 12 h (data not shown), and ammonia-starved cells failed to degrade RpoS as logarithmic cells do (Fig. 2), further suggesting that the presence of methionine did not confound these results. Additionally, measuring stability by an independent method—protein synthesis arrest by antibiotic addition, followed by monitoring of RpoS levels by Western blot analysis—similarly showed ammonia starvation stability to be intermediate between that of logarithmic cells and glucose-starved cells (data not shown).
Therefore, by two different methods we conclude that RpoS degradation under ammonia starvation is intermediate between that in logarithmic phase and that in glucose starvation.
Stabilization of RpoS under glucose starvation is the key difference between ammonia starvation and glucose starvation.
The above results highlight differing RpoS degradation rates between ammonia and glucose starvation. Since RpoS synthesis is regulated at the levels of transcription and translation, we sought to study RpoS dynamics in the absence of this control to see if the observed stability difference alone can account for the divergent RpoS levels between ammonia starvation and glucose starvation.
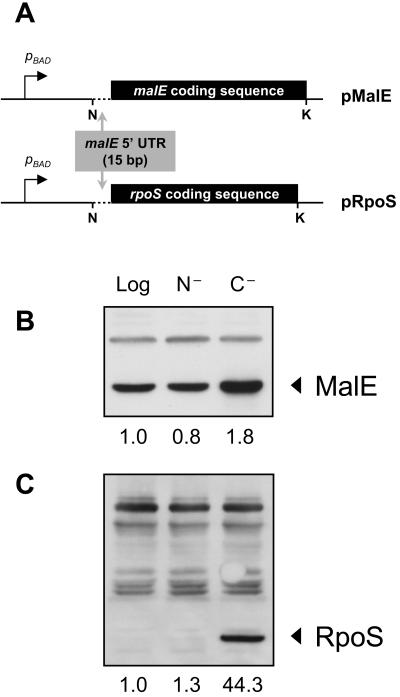
To accomplish this, the rpoS open reading frame, in the absence of its native cis control elements, was placed downstream from the pBAD promoter and the malE Shine-Dalgarno sequence on the pBAD18 plasmid (Fig. 3A). Since there are no known cis-acting elements within the rpoS open reading frame that are both necessary and sufficient for transcriptional or translational control, we expected that this construct would allow us to evaluate the role of protein stability only. As a control for background expression under each experimental condition tested, the malE coding sequence was introduced into the same vector. Therefore, the experimental rpoS construct and the control malE construct differ only in the open reading frame that they contain. The relative constancy of MalE levels in logarithmic, ammonia-starved, and glucose-starved cells, its similarity in size to RpoS, and its cellular localization in the periplasm (making it inaccessible to SprE and ClpXP) make it an appropriate expression control.
FIG. 3.
Stability differences explain the low levels of RpoS observed in ammonia-starved cells and the high levels in glucose-starved cells. (A) pRpoS and pMalE (control) were constructed to study RpoS stability in the absence of the native cis elements that control rpoS synthesis. Transcription is controlled by the pBAD promoter, and translation is regulated by pBAD plasmid sequences and the last 15 bp of the malE 5′ UTR. N, NheI; K, KpnI. The regions outside of these restriction sites belong to the pBAD18 backbone. Therefore, stability differences between the two open reading frames are responsible for differences in levels. (B) MalE levels from MJM312 (MJM294/pMalE) in logarithmic (Log) and cells starved for 1 h for ammonia (N−) or glucose (C−). The slight increase in glucose-starved cells is likely due to increased transcription from the pBAD promoter. (C) RpoS levels from MJM310 (MJM294/pRpoS) in logarithmic-phase and 1-h-starved cells. Quantification of bands was performed using NIH ImageJ, and shown below each panel are levels relative to those observed in logarithmic phase of the respective strain.
Figure 3B shows expression of the control MalE under each condition. Ammonia-starved cells had 20% less MalE than logarithmic-phase cells, whereas glucose-starved cells contained 80% more than logarithmic-phase cells, likely as a result of increased synthesis from the pBAD promoter owing to relief of catabolite repression. These numbers served as the basal expression levels of the constructs under each condition, and the levels of RpoS were normalized to these levels to infer the role that stability played under each condition. From pRpoS, RpoS was barely detectable in logarithmic or ammonia-starved cells but was remarkably high following glucose starvation (Fig. 3C). When normalized to the MalE levels, RpoS levels were 1.0 in logarithmic phase, 1.6 under ammonia starvation, and 24.6 under glucose starvation (values in Fig. 3C divided by those in Fig. 3B). These values are strikingly similar to those observed in wild-type cells under each condition (1.0, 2.0, and 20.1, respectively) (Fig. 1C), confirming that the elevated RpoS stability under glucose starvation is sufficient to explain its high levels. Since the only difference between the malE and rpoS constructs is the sequence of the open reading frame, these data demonstrate that the difference in RpoS levels between ammonia-starved cells and glucose-starved cells is at the level of protein stability and not synthesis.
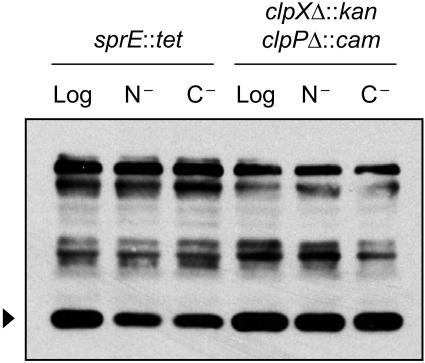
SprE activity controls the stability of RpoS.
It is of interest to know whether the stability phenotype observed in ammonia starvation is the result of either partial inactivation of the SprE pathway or of full inactivation of the SprE pathway and degradation via a distinct pathway. Using Western blot analysis on starved cultures lacking SprE or ClpXP, we compared levels of RpoS in ammonia-starved versus glucose-starved cells (Fig. 4). It is clear that in the absence of key SprE pathway members, RpoS levels are identical under either starvation condition, strongly suggesting that the lability of RpoS observed under ammonia starvation is due to partial inactivation of the SprE pathway and not due to degradation via an alternate pathway. We also note that RpoS levels in the starved cultures were slightly lower than in the logarithmic sample in the sprE::tet strain, but not in the ΔclpX::kan ΔclpP::cam background (Fig. 4). Therefore, there appears to be slight degradation of RpoS via ClpXP under starvation that is SprE independent.
FIG. 4.
Ammonia-starved cells with a defective SprE pathway are identical to glucose-starved cells. Results shown are for logarithmic-phase (Log) or 1-h-starved cells, starved for ammonia (N−) or glucose (C−), of strain MJM211 (MC4100 sprE::tet) or MJM221 (MC4100 ΔclpX::kan ΔclpP::cam). RpoS levels (arrowheads) were assayed by Western blotting.
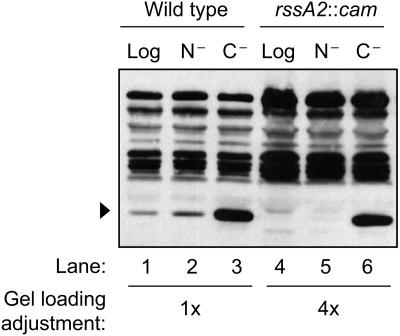
The rssA2::cam allele (30) leads to overexpression of sprE from the chromosome. RpoS levels in logarithmic phase were lower in this strain than in the wild type—below our level of detection by Western blot analysis (Fig. 5, lane 4)—presumably owing to the high levels of active SprE that were present. Upon glucose starvation, SprE was inactivated and RpoS levels accumulated as expected, even in the rssA2::cam background (Fig. 5, lane 6). Levels of glucose-starved RpoS were fourfold lower in this strain than in the wild-type background, presumably due to the lower levels of RpoS in logarithmic phase that were present at the onset of starvation to be stabilized. RpoS was stable during glucose starvation in this strain, as assayed by antibiotic protein synthesis arrest followed by Western blot analysis (data not shown), demonstrating that SprE activity is inhibited upon glucose starvation, even with elevated SprE levels.
FIG. 5.
Overexpression of SprE exacerbates differences between ammonia starvation and glucose starvation. Levels of RpoS (arrowheads) were assayed in logarithmic-phase (Log) or 1-h-starved MC4100 or MJM282 (MC4100 rssA2::cam) cells, starved for ammonia (N−) or glucose (C−). To facilitate comparisons, we loaded fourfold-higher amounts of protein from rssA2::cam cells than from wild-type cells.
Next, we compared the ratio of RpoS under ammonia starvation to that under glucose starvation in both strains. To facilitate analysis of relative RpoS levels, four times as much of the rssA2::cam samples were loaded on the gel compared to wild-type samples (Fig. 5). This adjustment allowed us to directly compare the ratio of RpoS present under ammonia starvation and glucose starvation in the two strains. In wild-type cells, there were 10-fold-higher levels of RpoS under glucose starvation compared to levels under ammonia starvation (Fig. 5, lane 3 versus 2). This ratio is significantly higher when making the same comparison under SprE overexpression (lane 6 versus lane 5). SprE acts catalytically, so overexpression of SprE should increase the reaction rate only when the catalyst is active. Figure 5 shows this is indeed the case under ammonia starvation, strongly suggesting that SprE retains some activity under ammonia starvation.
From this we conclude that SprE retains some activity under ammonia starvation and is fully inactivated only upon glucose starvation, and that regulation of SprE activity is the basis for the different levels of RpoS.
The differences in SprE activity are not due to differences in SprE levels.
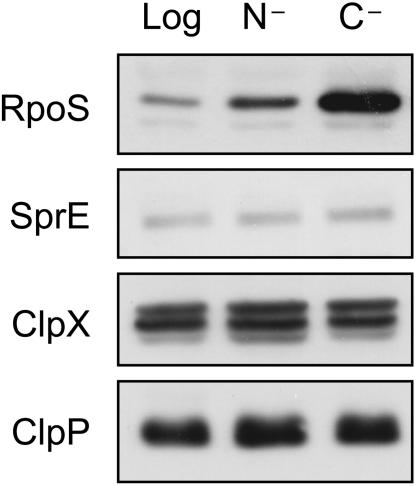
Given that SprE activity is the key difference between ammonia starvation and glucose starvation, we assayed levels of SprE pathway proteins to determine if levels of SprE, ClpX, or ClpP were significantly different between the two starvation conditions. The data in Fig. 6 exclude levels of SprE pathway members as the regulated step in distinguishing glucose starvation from ammonia starvation. Levels of each of the proteins were the same in ammonia-starved versus glucose-starved cells, so the higher degradation rate is not due to increased levels of any of the known pathway members.
FIG. 6.
Levels of SprE pathway proteins are identical in ammonia-starved (N−) and glucose-starved (C−) cells. Logarithmic-phase cells (Log) are also shown for comparison. For ClpX, the lower of the two dark bands corresponds to ClpX, by comparison with a ΔclpX::kan strain (data not shown).
It is also of note that SprE levels were subtly higher under both ammonia starvation and glucose starvation relative to levels in logarithmic cells. This is consistent with data presented by Ruiz et al. (30), in which sprE is transcriptionally activated by RpoS, and with data discussed above in which RpoS activity is higher under ammonia starvation than in logarithmic cells.
As the levels of known components in the SprE pathway are constant in ammonia starvation and glucose starvation, direct modulation of SprE activity is the most likely mechanism by which the cell regulates RpoS stability in response to cues for limited ammonia or glucose.
DISCUSSION
The developmental program that E. coli cells undergo upon starvation is critical to their survival, and this has largely been modeled by studying glucose starvation. Our results illustrate the shortcomings of using a single nutrient limitation as a model for all starvation conditions. These results also identify ammonia starvation as a valuable tool for identifying common themes and divergent pathways in starvation signal transduction. Ammonia starvation represents the only nutrient limitation, to our knowledge, in which RpoS levels do not increase dramatically. Furthermore, the difference in stability observed between glucose starvation and phosphate limitation (27) exposes different bases for the high levels of RpoS in those cases, arguing for even more breadth in the E. coli starvation repertoire. Therefore, there is not solely one response that is engaged upon starvation for any nutrient, as has been believed for some time (8, 11, 12, 37). This is not unprecedented in nature, as Saccharomyces cerevisiae, for example, monitors both the nitrogen and carbon status of the growth medium in determining whether to sporulate, enter stationary phase, or initiate pseudohyphal growth (36).
The accumulation of RpoS following glucose starvation is mainly due to a tremendous increase in stability, as has been reported previously (39). However, RpoS stability under ammonia starvation or phosphate limitation is approximately an order of magnitude lower than under glucose starvation, in which SprE is fully inactivated. The mechanism by which SprE becomes inactivated is not clear, and Peterson et al. (27) presented strong evidence that it is not via canonical two-component phosphorylation as previously thought. The hypothesis that growth rate controls SprE activity is also excluded, since growth is inhibited in response to ammonia starvation, glucose starvation, or chloramphenicol addition, yet each state exhibits different levels of RpoS. Instead, we propose that the signal to inactivate SprE is more directly tied to carbon source and/or energy source limitation. Ballesteros et al. (2) presented evidence that glucose metabolism continues after the optical density plateaus from limited ammonia or phosphate, supporting the hypothesis that full inactivation of SprE is linked to glucose metabolism.
In addition, there are conflicting data regarding how RpoS synthesis responds upon glucose starvation, in that RpoS-LacZ translational fusions (lacking the stability control region) indicate a slight increase in RpoS synthesis upon glucose starvation (20, 22), whereas chemostat experiments have suggested that RpoS synthesis decreases as cultures become more glucose limited (39). We attempted to use LacZ reporter fusions to dissect synthesis control of RpoS under glucose and ammonia starvation, but these attempts proved unsuccessful as the LacZ was degraded under ammonia starvation in as short a period as 1 h (data not shown). Matin's group previously reported conflicting findings between transcriptional fusion data and direct measurements of RNA synthesis under glucose starvation (39). Given the limitations of using reporter fusions to study protein dynamics under conditions of starvation, we turned to more direct methods.
We attempted to measure RpoS synthesis rates directly by pulse-labeling with [35S]methionine followed by immunoprecipitation. Quantitative estimates were complicated by the rapid turnover of RpoS in logarithmic phase (Fig. 2). Since reporter fusion and direct labeling attempts were confounded by technical constraints, we inferred synthesis rates as was done by Zgurskaya et al. (39) and as described in Materials and Methods. The results of these calculations are listed in Table 2.
TABLE 2.
Relative rates of RpoS synthesis and degradation under different nutrient limitationsa
| Nutrient limitation | Measured
|
Calculated
|
||
|---|---|---|---|---|
| Level (relative units), N0 | t1/2 (min) | kD (min−1) | ks (relative units) | |
| Log phase | 1.0 | 4.1 | 0.1691 | 0.1691 |
| N− | 2.0 | 17.9 | 0.0387 | 0.0774 |
| C− | 20.1 | 154.0 | 0.0045 | 0.0900 |
| P− | 20.1 | 21.7b | 0.0319b | 0.6420b |
Details of the calculations are discussed in Materials and Methods.
The phosphate starvation data do not clearly approximate first-order exponential decay, and so this “half-life” represents the time in which half of the initial population decays, as measured previously (27). As a result, the true degradation rate is more complex than the one calculated here. Therefore, the values for phosphate limitation should be interpreted only as rough estimates.
First, these calculations agreed with published data that RpoS synthesis is reduced in glucose starvation compared to logarithmic phase. Zgurskaya et al. (39) demonstrated a two- to fourfold decrease in RpoS synthesis in glucose-limited cultures compared to logarithmic cultures, which is consistent with the twofold decrease that we calculated (Table 2).
Second, we note that RpoS synthesis rates under ammonia starvation and glucose starvation are remarkably similar. Under glucose starvation, the RpoS level is an order of magnitude higher and the degradation rate is an order of magnitude lower, thereby resulting in comparable rates of synthesis as under ammonia starvation (based on the equation described above, kD = ln 2/t1/2). We point out, though, that protein synthesis in ammonia-starved and glucose-starved cells is reduced compared to that in logarithmic cells. To approach this semiquantitatively, we pulse-labeled cultures with [35S]methionine (without immunoprecipitation) to examine total protein synthesis. The amount of protein labeled after 15 s in logarithmic cultures was comparable to that labeled after 60 s in ammonia-starved or glucose-starved cultures (data not shown). Therefore, the decrease in RpoS synthesis under starvation is likely to reflect the lower level of basal protein synthesis rather than regulation of RpoS synthesis specifically. Unless more precise measurements identify a discrepancy between RpoS synthesis and total protein synthesis, it is unnecessary to invoke regulated synthesis to explain differences in RpoS levels among logarithmic phase, ammonia starvation, and glucose starvation.
Third, the hypothesis that increases in RpoS synthesis overwhelm SprE, thereby increasing RpoS levels indirectly (29), does not seem to be supported under ammonia starvation or glucose starvation. Both have decreased rates of synthesis compared to logarithmic phase, whereas levels of SprE are actually higher under either starvation condition than in logarithmic phase. Therefore, RpoS levels would be expected to be lower during starvation than in logarithmic phase. Furthermore, rates of synthesis under ammonia starvation and glucose starvation are comparable, thus reinforcing that dramatically different RpoS stability can be achieved under conditions with similar rates of RpoS synthesis. This is an example where the comparison of two starved states provides information that cannot be gleaned by comparison of growing cells to starved cells: ammonia starvation provides intrinsic controls for the low protein synthesis rate and growth arrest that are present under glucose starvation but absent in logarithmic cells.
Fourth, phosphate limitation identifies a nutrient deprivation that behaves differently from either ammonia starvation or glucose starvation. The high levels of RpoS under phosphate limitation are comparable to those in glucose starvation (8, 27, 31), whereas the degradation rate is similar to that observed in ammonia starvation (27) (Fig. 2). The resulting synthesis rate is approximately sevenfold higher than under the other starvation conditions and fourfold higher than in logarithmic phase. This makes sense, based on two known aspects of phosphate limitation. First, the cells continue to grow at a slow rate following removal of external phosphate, suggesting that the basal level of protein synthesis is higher than under the other starvation conditions (31). Second, there is a PhoBR-regulated process that increases translation of RpoS (31). The high synthesis rate of RpoS under phosphate limitation likely results from these two processes. As shown in Table 2, though, RpoS decay under phosphate limitation is not a good fit to first-order exponential decay, suggesting that its degradation is more complex. SprE is still active in the presence of high levels of RpoS in phosphate limitation, and so it is possible that SprE availability is what is limiting RpoS degradation under this condition, not regulation of SprE activity per se.
What emerges from the data in Table 2 is that starvations for different nutrients do not elicit identical responses as has been assumed for a number of years. Reviews of RpoS regulation have highlighted the diversity of ways in which RpoS is regulated, at the levels of transcription, translation, stability, and activity. However, starvation for any essential nutrient had been thought to elicit a conserved response to fully inactivate regulated proteolysis through SprE and ClpXP. This is clearly not the case, and different starvation responses may facilitate our understanding of what aspects of physiology the SprE pathway is using to sense glucose limitation.
Finally, ammonia starvation is certainly unique in its response, in that levels of RpoS remain close to logarithmic levels instead of rising to those observed under glucose starvation or phosphate limitation. It has been known that cultures under long-term starvation in LB and in minimal glucose acquire partial loss-of-function mutations in rpoS (6, 7, 38). These so-called GASP (growth advantage in stationary phase) mutations result in moderate levels of RpoS activity that balance individual cells' needs for protection with their ability to continue growth at a slow rate. Furthermore, a recent report demonstrated that rpoS mutant strains were able to utilize a wider range of carbon sources than their isogenic rpoS+ parents (17). Therefore, it is possible that modulation of RpoS activity may provide additional benefits under ammonia starvation that counteract the negative consequences of not fully activating the RpoS regulon.
Acknowledgments
Special thanks to Thomas Elliott for his valuable comments that focused our approach. We are grateful to members of the Silhavy lab for critical review of the manuscript, to N. Ruiz for preliminary work, to N. Ruiz, K. C. Huang, J. Paulsson, C. Peterson, and N. Wingreen for helpful discussions, to S. DiRenzo for manuscript assistance, to D. Siegele for sharing unpublished results, and to S. Gottesman, D. Isaac, and A. Ninfa for gifts of strains and reagents.
This work was supported by a grant from the National Institute of General Medical Sciences (GM65216) to T.J.S. and by a National Science Foundation Graduate Research Fellowship to M.J.M.
REFERENCES
- 1.Backman, K., Y. M. Chen, and B. Magasanik. 1981. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 78:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballesteros, M., A. Fredriksson, J. Henriksson, and T. Nystrom. 2001. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20:5280-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 5.Farewell, A., K. Kvint, and T. Nystrom. 1998. uspB, a new σS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 180:6140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel, S. E., E. R. Zinser, and R. Kolter. 2000. Long-term survival and evolution in the stationary phase, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 8.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groat, R. G., J. E. Schultz, E. Zychlinsky, A. Bockman, and A. Matin. 1986. Starvation proteins in Escherichia coli: kinetics of synthesis and role in starvation survival. J. Bacteriol. 168:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 12.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch, M., and T. Elliott. 2002. Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli. J. Bacteriol. 184:5077-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irr, J. D. 1972. Control of nucleotide metabolism and ribosomal ribonucleic acid synthesis during nitrogen starvation of Escherichia coli. J. Bacteriol. 110:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabir, M. S., T. Sagara, T. Oshima, Y. Kawagoe, H. Mori, R. Tsunedomi, and M. Yamada. 2004. Effects of mutations in the rpoS gene on cell viability and global gene expression under nitrogen starvation in Escherichia coli. Microbiology 150:2543-2553. [DOI] [PubMed] [Google Scholar]
- 17.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 21.Matin, A. 1991. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 5:3-10. [DOI] [PubMed] [Google Scholar]
- 22.McCann, M. P., C. D. Fraley, and A. Matin. 1993. The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J. Bacteriol. 175:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173:4188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzger, S., G. Schreiber, E. Aizenman, M. Cashel, and G. Glaser. 1989. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J. Biol. Chem. 264:21146-21152. [PubMed] [Google Scholar]
- 25.Moreno, M., J. P. Audia, S. M. Bearson, C. Webb, and J. W. Foster. 2000. Regulation of sigma S degradation in Salmonella enterica var. Typhimurium: in vivo interactions between sigma S, the response regulator MviA(RssB) and ClpX. J. Mol. Microbiol. Biotechnol. 2:245-254. [PubMed] [Google Scholar]
- 26.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, C. N., N. Ruiz, and T. J. Silhavy. 2004. RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J. Bacteriol. 186:7403-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruteanu, M., and R. Hengge-Aronis. 2002. The cellular level of the recognition factor RssB is rate-limiting for σS proteolysis: implications for RssB regulation and signal transduction in σS turnover in Escherichia coli. Mol. Microbiol. 45:1701-1713. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, N., C. N. Peterson, and T. J. Silhavy. 2001. RpoS-dependent transcriptional control of sprE: regulatory feedback loop. J. Bacteriol. 183:5974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz, N., and T. J. Silhavy. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J. Bacteriol. 185:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 34.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyler, B. 1978. Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 47:1127-1162. [DOI] [PubMed] [Google Scholar]
- 36.Werner-Washburne, M., E. L. Braun, M. E. Crawford, and V. M. Peck. 1996. Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 37.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]
- 38.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 39.Zgurskaya, H. I., M. Keyhan, and A. Matin. 1997. The σS level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]