Abstract
Chordomas are rare tumours of the bone arising along the spine from clivus to sacrum. We compared three chordoma cell lines of the clivus region including the newly established clivus chordoma cell line, U-CH14, with nine chordoma cell lines originating from sacral primaries by morphology, on genomic and expression levels and with patient samples from our chordoma tissue bank. Clinically, chordomas of the clivus were generally smaller in size at presentation and patients with sacral chordomas had more metastases and more often recurrent disease. All chordoma cell lines had a typical physaliphorous morphology and expressed brachyury, S100-protein and cytokeratin. By expression analyses we detected differentially expressed genes in the clivus derived cell lines as compared to the sacral cell lines. Among these were HOXA7, HOXA9, and HOXA10 known to be important for the development of the anterior-posterior body axis. These results were confirmed by qPCR. Immunohistologically, clivus chordomas had no or very low levels of HOXA10 protein while sacral chordomas showed a strong nuclear positivity in all samples analysed. This differential expression of HOX genes in chordomas of the clivus and sacrum suggests an oncofetal mechanism in gene regulation linked to the anatomic site.
Introduction
Chordomas are rare, malignant bone tumours thought to arise from notochordal remnants along the spine. This tumour entity is generally considered as a low-grade malignant tumour, but metastases and recurrences occur1, 2. Chordoma therapy usually consists of surgical resection in combination with radiotherapy. There is a lack of efficient standard chemotherapy, which may be due to the generally slow tumour growth3, 4. Therefore, there is a focus on novel therapeutic strategies for the treatment of chordoma, such as targeting the PDGF receptor5–7, VEGF receptor8 or mTOR-kinase9, 10. Overexpression of epidermal growth factor receptors (EGFR) has been reported to be a frequent event in chordoma11–14 and inhibition of the EGF receptor 1 has a significant effect on cellular growth15, 16. As the majority of chordomas show a deletion of the tumour suppressor gene CDKN2A (encoding for the protein p16)17, 18, we provided evidence that the CDK4/6 inhibitor palbociclib may be a promising therapeutic option for chordoma treatment19. Up to now, none of these strategies have made it to clinical usage.
A characteristic feature of chordoma is the overexpression of the transcription factor T encoding for the protein brachyury. Brachyury is essential for the development of the notochord and the formation of posterior mesodermal elements20–22.
Regarding developmental processes, an important role in the formation of the anterior-posterior body axis and other morphogenetic processes is assumed for members of the HOX gene family, a highly conserved gene family which encodes for several transcription factors23, 24.
In vertebrates, 39 HOX genes have been identified which are organized in four clusters, called HOXA, HOXB, HOXC, and HOXD. Each cluster is located on a different chromosome, namely 7p14, 17q21, 12q13, and 2q31, respectively. With regard to the formation of the anterior-posterior body axis, HOX gene expression follows the rules of the so-called temporal and spatial collinearity. This means that genes located at the 3′ end of the cluster are expressed earlier in the embryogenesis and have a more anterior boundary of expression than those located at the 5′ end25–27. As the two major predilection sites of chordoma are the cranial (clivus) and the caudal (sacral) ends of the spine, a comparison of these different entities might give new insights into the formation of chordomas. Due to the rareness of this tumour, there are a small number of chordoma cell lines and especially just two from clivus chordoma. In the following, we addressed the question whether differences between clival and sacral chordomas exist. We established a novel chordoma cell line, U-CH14, derived from a clivus chordoma, analysed this cell line together with two other clivus derived cell lines, MUG-CC1 and UM-Chor1, and compared them with nine sacral chordoma cell lines on genomic, gene expression and protein grounds to identify possible differences with focus on HOX genes. Finally, we validated these findings in our chordoma tissue bank in situ.
Results
Chordomas of the clivus differ in clinical features from sacral chordomas
Analyses of our chordoma tissue cohort (n = 43) with regard to eventual differences between chordomas of the clivus (n = 5) and the sacrum region (n = 24) showed that clivus chordomas were significantly smaller than sacral chordomas (p = 0.04). The median tumour size was 1.4 cm (inter quartile range (IQR): 0.65 cm; 2 cm) versus 7.0 cm (IQR: 5.125 cm; 14.65 cm). Clivus chordoma patients tended to be younger than sacral chordoma patients (median age at onset of disease: 49 years (IQR: 40; 80) versus 70 years (IQR: 51; 75.75)). Regardless of the different localisation along the spine, chordomas were of the ‘not otherwise specified’ (NOS)-subtype. The proliferation rate was lower in clivus chordoma samples, as all cases had a Ki-67 index lower than 10%. In sacral chordomas, 5/24 samples had Ki-67 indices above 10%. In p53 immunohistochemistry, 0/5 clivus and 7/23 sacrum chordoma cases showed a nuclear positivity in more than 1% of total cells (Supplementary Table 1, Supplementary Figure 1A+B). In deep targeted sequencing, all analysed patient probes (n = 22) harboured the common TP53 variant p.P72R (Supplementary Table 1). In addition, one clival and two sacral chordoma cases carried mutations, namely p.P72S (3/22) and p.A76V (1/22). In our cohort, metastases occurred in sacral (8/24; lung, liver and lymph node), but not in clivus chordoma patients (0/5). Recurrent disease was found in 13/24 sacral and in 0/5 clivus chordoma patients. Due to the anatomical complexity of the site of clival chordomas only an incomplete resection status (R2) was achieved in 4/5 cases; in one case the resection status was uncertain (Rx) (Table 1).
Table 1.
Summarized clinical data of the clival and sacral chordoma patients.
| Patient | Localisation | Age at diagnosis | Gender | Tumour Size (cm) | Histological subtype | Ki-67 | Metastases | Recurrences | R-Status |
|---|---|---|---|---|---|---|---|---|---|
| C1 | Clivus | 35 | male | 0.4 | NOS | ≤10% | — | — | R2 |
| C2 | Clivus | 45 | female | 2.3 | NOS | ≤10% | — | — | Rx |
| C3 | Clivus | 49 | female | 0.9 | NOS | ≤10% | — | — | R2 |
| C4 | Clivus | 81 | male | 1.4 | NOS | ≤10% | — | — | R2 |
| C5 | Clivus | 79 | male | 1.7 | NOS | ≤10% | — | — | R2 |
| S1 | Sacrum | 60 | female | 12 | NOS | ≤10% | — | — | Rx |
| S2 | Sacrum | 78 | female | 18 | NOS | ≤10% | — | — | R1 |
| S3 | Sacrum | 50 | female | 10 | NOS | ≤10% | — | — | R0 |
| S4 | Sacrum | 74 | male | 7 | NOS | ≤10% | — | — | Rx |
| S5 | Sacrum | 45 | female | 7 | NOS | ≤10% | — | 1 | R1 |
| S6 | Sacrum | 68 | male | 6 | NOS | ≤10% | — | 1 | R1 |
| S7 | Sacrum | 69 | female | 17 | NOS | ≤10% | 1 | — | R1 |
| S8 | Sacrum | 45 | male | 20 | NOS | ≥10% | 1 | 1 | R1 |
| S9 | Sacrum | 70 | female | 13.6 | NOS | ≤10% | — | 1 | R0 |
| S10 | Sacrum | 76 | female | 6 | NOS | ≤10% | 1 | 1 | R0 |
| S11 | Sacrum | 74 | female | 13 | NOS | ≤10% | — | 1 | R1 |
| S12 | Sacrum | 71 | female | 18 | NOS | ≤10% | — | 1 | R1 |
| S13 | Sacrum | 70 | male | 3 | NOS | ≤10% | 1 | — | R2 |
| S14 | Sacrum | 67 | male | 6 | NOS | ≥10% | 1 | >1 | Rx |
| S15 | Sacrum | 75 | male | 6.5 | NOS | ≤10% | — | 1 | R1 |
| S16 | Sacrum | 49 | male | 5.2 | NOS | ≥10% | >1 | >1 | NA |
| S17 | Sacrum | 77 | male | 2.5 | NOS | ≤10% | 1 | 1 | R2 |
| S18 | Sacrum | 78 | female | 5.1 | NOS | ≤10% | — | 1 | R1 |
| S19 | Sacrum | 54 | male | 4.5 | NOS | ≥10% | — | 1 | R2 |
| S20 | Sacrum | 47 | male | 2 | NOS | ≤10% | — | — | R2 |
| S21 | Sacrum | 29 | male | 2.9 | NOS | ≤10% | 1 | — | R1 |
| S22 | Sacrum | 71 | male | 8 | NOS | ≤10% | — | — | R0 |
| S23 | Sacrum | 83 | male | 50 | NOS | ≥10% | — | — | R2 |
| S24 | Sacrum | 79 | female | 15 | NOS | ≤10% | — | — | R0 |
NOS not otherwise specified, R-Status residual tumour status, R0 no residual tumour, R1 tumour cells detected microscopically at the resection margin, R2 macroscopic detectable residual tumour, Rx R-status uncertain.
Characterization of the clivus chordoma cell line U-CH14
After a cultivation period of about 2 years, U-CH14 was established as a stable chordoma cell line. Comparable to the primary tumour, the cells showed the typical physaliphorous appearance in vitro (Fig. 1c) and a generation time of about 10 days. The Ki-67 index representing the proliferation rate was below 5% both in situ and in vitro (Fig. 1b,d). Therefore, the long population doubling time of the cell line fitted to the slow tumour growth.
Figure 1.
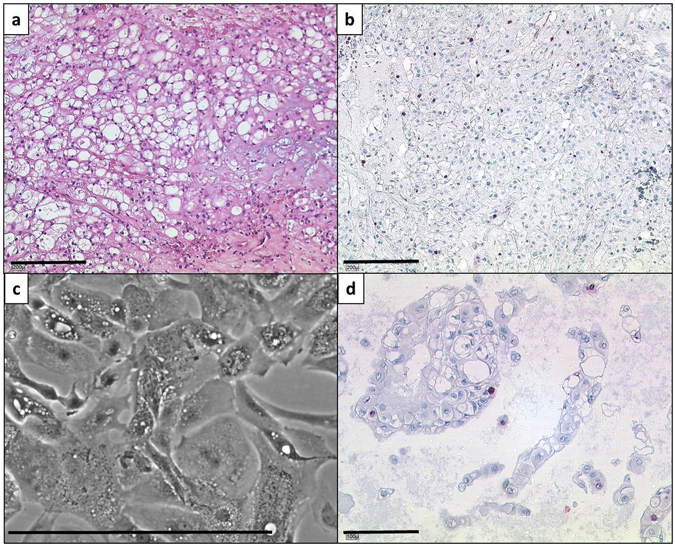
Morphology and proliferation rate detection of the parental tumour and the U-CH14 cell line. (a) Section of the parental tumour. The cells show the typical physaliphorous appearance. The tumour is classified as not otherwise specified (NOS) subtype. (b) Ki-67 staining of a parental tumour section. The proliferation index is below 5%. (c) Phase contrast image of the U-CH14 cell line. The typical morphology is conserved in vitro. (d) Proliferation index of U-CH14 (<5%; Ki-67 staining). Lines indicate 200 µm (a+b) or 100 µm (c+d).
Immunocytochemically, the tumour cells showed a strong nuclear expression of brachyury and a cytoplasmatic positivity of vimentin, cytokeratins, and S100-protein (Fig. 2a–d). Hence, all typical chordoma markers were conserved in the cell line. The expression of brachyury was verified by western blot analyses (Fig. 2e). The origin of the cell line was confirmed by STR comparison with its parenteral tumour. The same alleles could be confirmed in 10/12 markers. In U-CH14, the markers D18S51 and TH01 showed only one of the two alleles detected in the primary tumour indicating a loss of heterozygosity (Supplementary Table 2). Since CDKN2A gene loss is frequent in chordomas, the CDKN2A status was analysed by fluorescence in situ hybridization (FISH). A loss of CDKN2A in the chordoma cells was found as compared to the co-hybridized centromere 12 probe (Supplementary Table 3). Additional chromosomal gains and losses were determined by array comparative genomic hybridization (aCGH) analyses. Ten chromosomes showed alterations. We detected gains on chromosomes 2, 7, 9, 11, 14, 18, and 22 and losses on chromosomes 8, 9, 11, 17, 18, 20, and 22 (Supplementary Figure 2). The complete list of genes in the affected chromosomal regions is given in Supplementary Table 4. A total loss of chromosomal material was solely detected for chromosome 9p21.3-ter which confirms the total CDKN2A loss as identified by FISH analyses.
Figure 2.
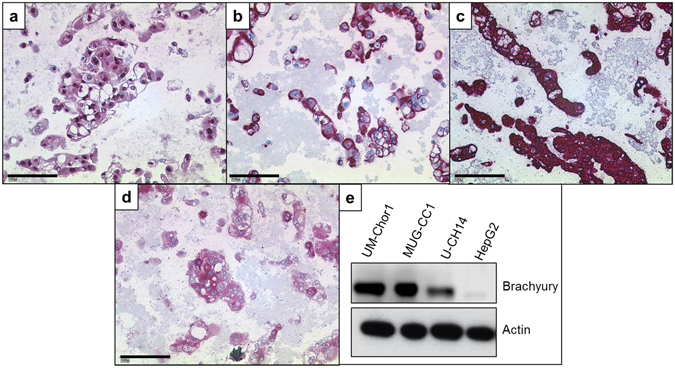
Immunocytochemistry and western blot analysis of U-CH14. The cells are strongly positive for brachyury (a), vimentin (b), cytokeratin (AE1/AE3) (c), and S-100-protein (d). (e) Western blot confirmation of the brachyury expression in the three clivus chordoma cell lines (UM-Chor1, MUG-CC1, and U-CH14). HepG2 served as a negative control. Lines indicate 100 µm (a–d).
Clivus chordoma cell lines differ in gene expression from sacral chordoma cell lines
Measurement of the expression of ~20000 transcripts showed differences between the clival and the sacral chordoma cell lines. Hierarchical cluster analysis of the three clivus (GEO accession number GSE95084) and the nine sacral chordoma cell lines (GEO accession number: GSE68497) revealed that the clivus chordoma cell lines clustered together in one group against the background of the sacral chordoma cell lines (Fig. 3). Filtering for most significant differentially expressed genes (p < 0.005), we identified 4572 genes being differently regulated in clivus chordoma cell lines. Pathway analysis (IPA) showed significant differences in the expression of more than 1000 genes related to physiological system development and function such as organismal development (282), tissue development (548), reproductive system development (85) and embryonic development (155). Gene ontology (GO) analyses revealed these differently expressed genes belonging to 25 distinct GO terms. Interestingly, in 12 of the 25 GO terms we found several members of the HOX gene family being down regulated in the clivus chordoma cell lines as compared to the sacral chordoma cell lines. Furthermore, in seven of the remaining GO terms at least one member of the HOX gene family was dysregulated. The common feature of all these GO terms was their impact in developmental processes, involving the anterior-posterior pattern formation and the skeletal development (Table 2).
Figure 3.
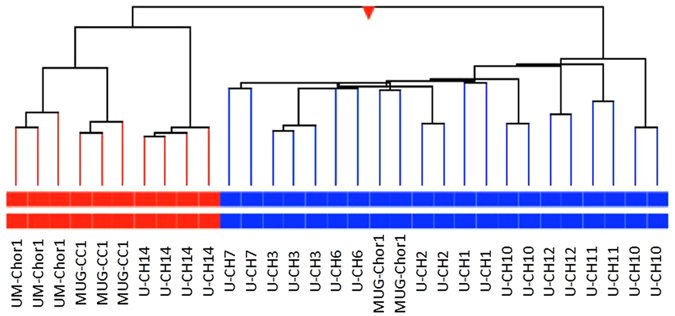
Hierarchical cluster analysis of nine sacral (blue) and three clivus (red) chordoma cell lines. The gene expression was tested using the 4 × 44 K whole genome microarray (Agilent Technologies). Each cell line was tested at least in duplicates.
Table 2.
HOX genes down regulated in clivus chordoma cell lines compared to sacrum chordoma cell lines and the related gene ontology (GO) terms.
| GO Term | Downregulated HOX genes in clivus cell lines |
|---|---|
| Anterior-posterior pattern formation | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Skeletal development | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Regionalization | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Multicellular organismal development | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Embryonic skeletal system morphogenesis | HOXA2, HOXA4, HOXA5, HOXA7 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Developmental processes | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Anatomical structure development | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| System development | HOXA2, HOXA4, HOXA5, HOXA7, HOXA9, HOXA10 |
| HOXB3, HOXB7 | |
| HOXC9 | |
| Tissue development | HOXA5 |
| HOXB3 |
Sacral chordomas have higher expression levels of HOXA7, HOXA9, and HOXA10 mRNA than clivus chordomas
We analysed the expression of the different HOX genes in our two subgroups (Fig. 4a). As indicated by the corrected p-values, the most promising differentially expressed genes were HOXA7, HOXA9 and HOXA10 (pcorr 8.91 × 10−10; 3.31 × 10−4; 7.14 × 10−12). Compared to the sacrum lines HOXA7 and HOXA10 were downregulated in all clivus lines. For HOXA9 expression we found a broad range of expression levels within the sacral subgroup. All three clivus cell lines, however, had very low expression levels. Therefore, these 3 HOX family members were further analysed in detail.
Figure 4.
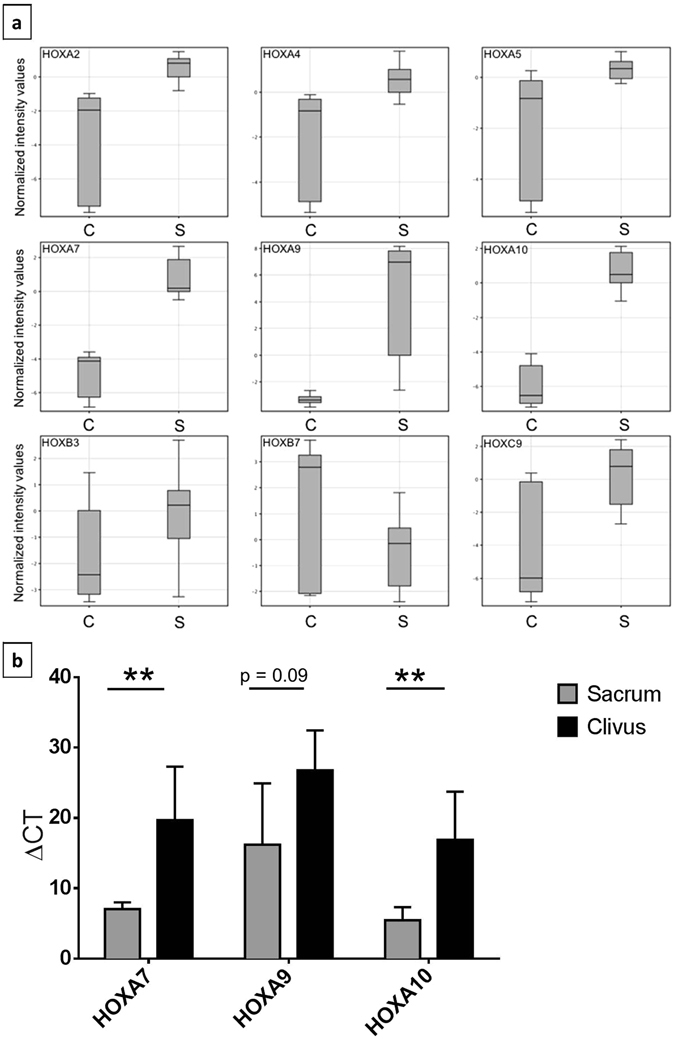
Expression of several HOX gene family members. (a) Box plot illustration of the differences in the expression of HOXA2, -A4, -A5, -A7, -A9, -A10, -B3, -B7, and -C9 between clivus (c) and sacrum (S) chordoma cell lines. Measurements represent the results of microarray expression profiling of three clivus and nine sacrum chordoma cell lines (each cell line was tested at least in duplicates). (b) Validation of the expression of HOXA7, -A9 and -A10 in the chordoma cell lines via qPCR shown as measured ΔCT values. High ΔCT values represent lower expression of the corresponding gene. Measurements were performed in triplicates. Differences in expression were tested by t-tests. p < 0.05 was regarded as significant difference (*). **p < 0.01.
We verified the expression differences using qPCR (Fig. 4b). Whereas the low expression of HOXA7 and HOXA10 was an invariable feature of the clivus cell lines, a low expression of HOXA9 was detected in all clivus lines and in 4/9 sacrum cell lines. The sacrum lines U-CH1, U-CH6, U-CH12, and MUG-Chor1 expressed HOXA9 at low levels comparable to the clivus lines U-CH14, MUG-CC1, and UM-Chor1 (Supplementary Figure 3). We further analysed the in situ protein expression of HOXA10 in the 24 sacral and five clivus chordoma patients from our chordoma tissue bank by immunohistochemistry (as shown in Fig. 5). A strong nuclear positivity (+++) was detected in the majority of tumour cells in 20/24 sacral chordoma cases whereas the clivus cases had low levels of HOXA10 or were completely negative (Table 3).
Figure 5.
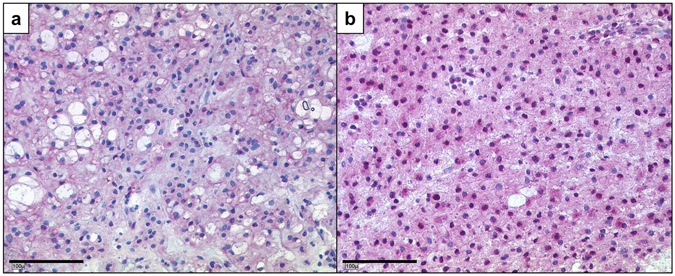
Differences of HOXA10 protein levels in clival and sacral chordomas. Immunohistochemistry of HOXA10 shows a very weak cytoplasmic staining of HOXA10 protein in clivus (a) and a nuclear positivity in the sacrum chordomas (b).
Table 3.
Expression of HOXA10 in correlation to the localisation of the tumour.
| Patient Number | Localisation | Expression of HOXA10 |
|---|---|---|
| C1 | Clivus | − |
| C2 | Clivus | − |
| C3 | Clivus | + |
| C4 | Clivus | + |
| C5 | Clivus | + |
| S1 | Sacrum | ++ |
| S2 | Sacrum | ++ |
| S3 | Sacrum | +++ |
| S4 | Sacrum | +++ |
| S5 | Sacrum | +++ |
| S6 | Sacrum | +++ |
| S7 | Sacrum | +++ |
| S8 | Sacrum | +++ |
| S9 | Sacrum | ++ |
| S10 | Sacrum | ++ |
| S11 | Sacrum | +++ |
| S12 | Sacrum | +++ |
| S13 | Sacrum | +++ |
| S14 | Sacrum | +++ |
| S15 | Sacrum | +++ |
| S16 | Sacrum | +++ |
| S17 | Sacrum | +++ |
| S18 | Sacrum | +++ |
| S19 | Sacrum | +++ |
| S20 | Sacrum | +++ |
| S21 | Sacrum | +++ |
| S22 | Sacrum | +++ |
| S23 | Sacrum | +++ |
| S24 | Sacrum | +++ |
Clivus chordomas (n = 5) have a weaker expression of HOXA10 than sacrum chordomas (n = 24) shown by immunohistochemistry.
Discussion
Chordoma is a primary bone tumour with characteristic genomics, mRNA profiles, and expression of proteins such as brachyury, S100-protein, and the epithelial markers epithelial membrane antigen (EMA), and cytokeratins. Chordomas may arise all along the spine, ranging from the cranial to the caudal ends. Here, we present a comparison of chordomas of the clivus and the sacrum region. Analysing the clinical aspects of our chordoma tissue cohort, no significant differences could be detected with regard to morphology of the tumours and age of onset of disease. In contrast, the tumour entities differed clearly in size, proliferation rate, and the propensity to metastasize and recur. Obviously, clival chordomas are smaller than the sacral tumours explained by the localisation itself and earlier symptoms they provoke28. In contrast, sacral chordomas remain longer asymptomatic, serious limitations in the daily life, e.g. incontinency or vast ulcerations usually occur, but in late stages of the disease29. Otherwise, the small size of the clival tumours may be in parts explained by the lower proliferation rate detected in our cohort, suggesting a less aggressive behaviour. This hypothesis fits to our data concerning the absence of metastases and recurrent disease although in all cases the resection was rated as incomplete. This is in line with published data stating that sacrum chordoma metastasize two to fourfold more often as clivus chordoma30–32. The p53 staining of our cohort revealed that only sacral cases showed higher expression indices matching the finding that a strong nuclear labeling of p53 in IHC negatively correlates with progression free survival33. All examined cases showed the same TP53 variant (p.P72R), as previously described by Fischer et al.34. Furthermore, we identified 2 additional mutations in 3 chordoma cases (resulting in p.P72S in C4, S23 and S16 and additionally p.A76V in the latter). These variants were graded neutral in 8/8 protein prediction tools (Supplementary Table 4). A correlation between the variants and enhanced nuclear p53 accumulation could not be found. Therefore, the differences in p53 protein expression in clival and sacral cases in our cohort are not due to the TP53 mutation status.
Our results argue for biological differences between clival and sacral chordomas as two different tumour entities. To further elucidate this, we established a novel cell line (U-CH14) derived from a clival chordoma in addition to the only two further clival chordoma cell lines available, MUG-CC1 and UM-Chor1. Morphology and the expression of typical chordoma markers are conserved in U-CH14 indicating that this cell line is a suitable in vitro model. Hence, we were able to compare three clivus with nine sacral chordoma cell lines on the expression profile level. In an unsupervised approach the clivus chordoma cell lines clustered together in one group and clearly separated from the sacral cell lines. This confirms that, indeed, there are differences between these two groups. Taking a closer look on the differently expressed genes it is remarkable that many of the downregulated genes in clival cell lines can be assigned to developmental processes. As chordoma cell lines have shown to express typical stem cell markers35, a connection to developmental processes seems to make sense. Amongst the downregulated genes in clivus chordomas, there were genes of the HOX family. HOX genes play a fundamental role in the development of the anterior posterior body axis24. The downregulation of HOX genes was validated in qPCR and confirmed in our tissue cohort.
Whether the differential HOX gene expression between clival and sacral chordoma is solely oncofetal in origin or has a pathologic role in the phenotype of the more aggressive sacral chordomas is an open question. Considering the redundancy within paralogue groups and the temporal and spatial collinearity, HOX expression effects solely caused by the localisation of the tumour would result in most of the 3′- HOX genes being up and 5′-HOX genes (including further members of the paralogue groups) being downregulated in clival chordoma and vice versa. The downregulated HOX genes in clivus cell lines (HOXA2, -A4, -A5, -A7, -A9, -A10, -B3, -B7, and -C9, Table 2) are distributed along the whole cluster and do not belong to corresponding paralogue groups. Therefore, it can be assumed that these HOX gene dysregulations cannot be explained by typical embryogenesis expression patterns and therefore not by the localisation of the originating tumour. Nonetheless, a final proof of this assumption can only be provided by systematically changing the expression of the identified HOX genes by overexpression or knockdown in vitro. Notably, most of the dysregulated HOX genes in clivus chordomas belong to the HOXA cluster located on chromosome 7p, a chromosomal region which frequently shows gains in chordoma cell lines36, 37. Hence, we analysed whether the clivus chordoma cell lines lack the typical gain of 7p, as explanation of a lower expression of HOXA cluster genes compared to sacral chordoma cell lines. Two of three clivus cell lines showed a gain of 7p fitting to published data on five of six sacral chordoma cell lines19. Therefore, the downregulation of the described HOX genes is not caused by a loss of chromosomal material.
An altered expression of HOX genes has been observed in various cancer types. As postulated in the so called “oncology recapitulates ontology” hypothesis, the development of malignancies is directly linked to differentiation and development of cells38, 39. Fitting to this, the reexpression of HOX genes in tumours is a sign for dedifferentiation and reprogramming towards stem cell features in cancer cells. For example, expression profiling of 39 HOX genes in head and neck squamous cell carcinoma (HNSCC) compared to normal tissue revealed that 18 of the 39 genes showed an increased expression40. High expressions of HOXA7, HOXA9, and HOXA10 are frequently seen in acute myeloid leukaemia (AML) and the overexpression of HOXA9 is linked to a poor prognosis41–43. An increased proliferation in human hepatocellular carcinoma has been described for the overexpression of HOXA7 44. Here, we present the first study of HOX genes in chordomas of different localisations along the spine. As a consequence of the rareness of this tumour disease, the low number of cases (n = 29) and cell lines (n = 12) may be a limiting factor of this study. Nevertheless, the expression of various HOX genes in sacral chordoma fits well with the results observed in other tumour entities. Furthermore, it was reported that the expression pattern of HOX genes can differ between cancer subtypes of one tumour entity. Abe et al. showed that in both squamous cell carcinoma (SCC) and adenocarcinoma of the lung, HOXA5 and HOXA10 are expressed at high levels, while HOXA1 and HOXC6 were only overexpressed in SCC45. The difference in HOX gene expression between clival and sacral chordomas buttresses our hypothesis of two separated subentities of chordomas. Sacral chordomas have a higher expression of HOX genes indicating a lower differentiation status which may be linked to tumour aggressiveness presenting itself by higher proliferation, the occurrence of metastases and recurrent disease, as seen in our chordoma cohort. As HOX overexpression is a feature in various cancer types, there is a high effort in trying to take advantage of HOX transcription factors as a potential therapeutic target. It is known that PBX forms dimers with many HOX proteins and potentiates the DNA binding capacity and specificity46. The interaction between HOX and PBX proteins was reduced by using a cell-permeable 18-amino acid peptide called HXR9 which induced apoptosis in cancer cell lines47–49 and tumour growth reduction in melanoma xenograft models50. Hence, HXR9 may also be a potential agent for treating sacral chordomas.
In summary, we here describe the newly established clivus chordoma cell line U-CH14, to increase the number of the existing clivus chordoma cell lines to three. In addition, we compared clival and sacral chordomas with regard to clinical features of our chordoma cohort and gene expression profiles of the existing cell lines. We found the expression of various HOX genes as a discriminator between these two subtypes.
Material and Methods
Cell culture and establishment of a novel chordoma cell line
A new clival chordoma cell line, U-CH14, was established from a 79-year-old, male chordoma patient. The tumour measured 1.7 × 1.4 × 1.2 cm and was localised in the clivus region and was resected via a transnasal endoscopic approach at the Department of Neurosurgery, Günzburg, Germany. Only a partial resection was achieved and the patient received proton beam therapy hereafter. Histologically, the tumour presented as a clivus chordoma of the not otherwise specified (NOS) subtype according to the WHO. The diagnosis was confirmed as the tumour showed immunopositivity for brachyury, S100-protein, and epithelial membrane antigen (EMA). The proliferation rate determined by the anti-Ki-67 antibody was below 5%.
The tumour tissue was immediately processed for cell culture at the Institute of Pathology at the University Hospital Ulm. The material was mechanically minced into small pieces and partially digested with collagenase. Isolated cells were cultured in Iscove’s Modified Dulbecco’s Medium/RPMI 1640 (4:1; Lonza, Basel, Switzerland) with 10% fetal bovine serum (Biochrom AG, Berlin, Germany), 2 mM glutamine, and penicillin/streptomycin. Before reaching a state of complete confluence, the cells were detached using trypsin/EDTA (0.25%/0.02%, Lonza). Another continuous chordoma cell lines, MUG-CC1 (Clivus) was contributed by Beate Rinner37. UM-Chor1 (Clivus) and the sacral chordoma cell line MUG-Chor151 were provided by the Chordoma Foundation (www.chordomafoundation.org), the other sacral cell lines U-CH1, U-CH2, U-CH3, U-CH6, U-CH7, U-CH10, U-CH11, and U-CH12 were established at the Institute of Pathology, University Hospital Ulm, as previously described19, 36, 52. All cell lines were cultured applying the same protocol as mentioned above. All the performed methods and experiments were in line with the guidelines of the ethics committee of the Federal General medical Council (Dtsch Arztebl 2003;100: A1632 [Heft 23]; Dtsch Arztebl 2003; 100: A 2251 [Heft 34–35]). The research was carried out in compliance with the Helsinki Declaration. All experimental protocols were conducted and approved in accordance with the local ethics committee of the University of Ulm (votum for usage of archived human material 03/2014). The patient gave his written informed consent for scientific usage of his tumour cells.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded tissue via the avidin-biotin-complex-method using the K005 AP/RED Detection System (Dako, Glostrup, Denmark). The following antibodies were used: monoclonal antibodies against brachyury (H-210, Santa Cruz, Dallas, USA, 1:100), epithelial membrane antigen (EMA; E29, Dako, 1:500), vimentin (VIM3B4, Dako, 1:300), cytokeratin (AE1 + AE3, Dako, 1:100), Ki-67 (MIB-1, Dako, 1:200), p53 (DO-7, Dako, 1:500) and the polyclonal antibodies against S100-protein (Dako, 1:1000) and HOXA10 (OriGene Technologies, Rockville, USA, 1:100). Appropriate positive and negative controls were included.
The ratio of positive chordoma cells was characterized as follows: “no immunoreactivity detected” (−), “immunoreactivity in up to 30% (+), “immunoreactivity in more than 30% and up to 70%” (++) and “immunoreactivity in more than 70%” (+++) of the total number of chordoma cells in the section53.
Short tandem repeats (STR) analysis
For genotyping of the cell lines the GenomeLab STR Primer Set Kit (Beckman Coulter, Krefeld, Germany) and the Amplitaq GOLD DNA Polymerase (Life Technologies, Carlsbad, USA) were used, respectively. The analysis was performed according to the manufacturer’s handbook. PCR products were separated on the CEQ GeXP capillary electrophoresis system (Beckman Coulter). 12 alleles were determined using the Gene marker software (Softgenetics, State College, USA).
Chordoma tissue bank
We analysed five clival and 24 sacral chordoma samples of our chordoma tissue bank (n = 43)54 with regard to eventual differences between clival and sacral chordomas concerning the age of the patients, tumour size, presence or absence of metastases and/or recurrences, R-status, proliferation rate, TP53 status, and histological subtype.
Gene Expression Analysis
Total RNA was extracted from the three clival chordoma cell lines (U-CH14, UM-Chor1, and MUG-CC1) and from the 9 sacral chordoma cell lines (U-CH1, U-CH2, U-CH3, U-CH6, U-CH7, U-CH10, U-CH11, U-CH12, and MUG-Chor1) using the RNeasy Mini-Kit (Qiagen, Hilden, Germany). Quality and quantity control was performed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). Gene expression analysis was performed using the Human Gene Expression 4 × 44 K Microarray Kit (Design ID 014850, Agilent Technologies). 200 ng of each sample was used and processed according to the standard protocol (Agilent Technologies). Each sample was analysed in duplicates at least. The signals were captured by a Microarray Scanner (G2565, Agilent Technologies, Santa Clara, USA). The generated data was analysed using the software Genespring 12.1 (Agilent Technologies) and Ingenuity Pathway Analysis (IPA; Qiagen). Hierarchical clustering was performed with an euclidean similarity measure and centroid linkage rule. Differentially expressed genes with statistical significance (p-value cut-off ≤ 0,005) between the two groups (clivus versus sacrum) were identified using scatter plot filtering. Classification of the differently expressed genes was performed via gene ontology (GO) analysis.
Real-time polymerase chain reaction (qPCR)
Verification of the microarray results was performed using qPCR. Single-strand cDNA was synthesized from total RNA using the QuantiNova Reverse-Transcription Kit (Qiagen). qPCR was performed using the QuantiNova SYBR Green PCR Kit (Qiagen) and the Light-cycler Rotor Gene Q (Qiagen). The expression of the following genes was analysed: HOXA7, HOXA9, and HOXA10 using the suitable QuantiTect Primer Assays (Qiagen). β-Actin and GAPDH served as an internal control. The PCR reaction was performed as follows: the initial denaturation step was 95 °C for 5 min, followed by 60 cycles at 95 °C for 10 sec and at 60 °C for 30 sec. All experiments were performed as triplicates.
Array comparative genomic hybridization (aCGH)
Array comparative genomic hybridization was carried out using the SurePrint G3 Human CGH Microarray 8 × 60 K (Design ID 021924, Agilent Technologies). DNA isolation of the cell lines was performed using the QiAmp DNA Mini Kit (Qiagen). Quantity and quality of the DNA were assessed on the NanoDrop platform (Thermo Fisher Scientific, Rockford, USA). DNA hybridization was performed according to the standard procedures after labelling of 500 ng of the sample DNA and the control DNA (Human Reference DNA Female or Human Reference DNA Male, Agilent Technologies). Microarray data was analysed using Agilent Genomic Workbench (Agilent Technologies).
Protein isolation and Western blot
Protein isolation was performed using TNT-buffer (20 mM Tris (pH 8,0), 200 mM sodium chloride, 1%-Triton-X100, 1 mM DTT) containing both phosphatase and protease inhibitor cocktails (Sigma-Aldrich, St. Louis, USA). Lysates were incubated in liquid nitrogen for 3 min and then centrifuged at 14 000 rpm at 4 °C to remove debris. The isolated proteins were quantified using the Biorad protein assay (Biorad, Munich, Germany) applying the Bradford method. SDS-PAGE was performed using NuPAGE 4–12% Bis-Tris protein gels (ThermoFisher Scientific). 10 µg of the protein lysate was used; after gel electrophoresis the proteins were transferred to a nitrocellulose membrane applying a standard semi-dry electro-transfer method. The membrane was blocked with TBS, 0.1% Tween 20, 10% skim milk powder (Sigma-Aldrich) for 60 min. Following, the membrane was washed thrice (each 10 min) with TBS containing 0,1% Tween 20 and then incubated with the primary antibody (Brachyury, H-210, Santa Cruz, 1:1000) overnight at 4 °C. After three washing steps the membrane was incubated for 1 hr with the appropriate secondary horseradish-peroxidase-conjugated antibody. After further washing, ECL detection was performed using the SuperSignal West Dura Extended Duration Substrate (ThermoFisher Scientific). Anti-β-actin antibody (AC-74, Sigma-Aldrich, St. Lewis, USA, 1:10000) was used to detect β-actin as a house keeping protein.
FISH-Analysis
FISH was performed using the commercially available ZytoLight SPEC CDKN2A/CEN9 Dual Colour Probe (Zytovision, Bremerhaven, Germany) according to the manufacturer’s protocol.
Next generation sequencing
For isolation of genomic DNA from the FFPE tissue samples, 5 µm tissue slices were transferred to glass slides. To spot the area containing the tumour, HE stained FFPE tissue slices (2 µm) were marked by an expert pathologist. The tumour-harboring areas of the FFPE tissue were subjected to a DNA extraction procedure using the GeneRead DNA FFPE kit (Qiagen) according to manufacturer’s instruction. DNA isolation of the cell lines was performed using the QiAmp DNA Mini Kit (Qiagen). DNA purity and concentration was determined fluorometrically (Qubit 2.0; Invitrogen). For molecular characterization of both tumour tissue and cell line DNA, we employed a targeted re-sequencing methodology using the GeneRead V2 chemistry (Qiagen) and a custom-made re-sequencing panel including primers for all exons of TP53. Target enrichment, amplicon processing, and library generation were performed according to the manufacturer’s handbook. For target enrichment, we included 1 ng to 40 ng DNA. Successful target enrichment and library generation was checked using the High Sensitivity DNA kit on a bioanalyzer device (Agilent Technologies). Libraries were diluted to 10 pM solutions and the sequencing was performed on a MiSeq platform (Illumina, San Diego, CA, USA) using the V2 chemistry. Mean read depth on target region was 200–4000 fold and 99% of bases were covered at 96–100% on average. The resulting fastq files were subjected to further analysis using the GeneRead web based analysis tool (http://ngsdataanalysis.sabiosciences.com/NGS2/), the Biomedical Workbench software package (Qiagen), and the Variant Studio software (Illumina). All identified mutations were manually re-analyzed using the Integrated Genome Viewer Software (Broad Institute, MA, USA). The impact of confirmed mutations on protein functions were classified using 8 online protein prediction tools (Polyphen-2, FATHMM, SIFT, PANTHER, Provean, PHD-SNP, SNAP, and Meta-SNP) according to the providers’ instructions.
Statistics
For statistical analysis, student’s t-tests were performed. A p-value ≤ 0.05 was considered as significant.
Electronic supplementary material
Acknowledgements
The technical help of Michaela Buck, Lena Kelsch, Elena Moser, Ulrike Kostezka, and Juliane Nell is greatly acknowledged. We thank the Chordoma Foundation for financial support.
Author Contributions
D.J., T.F.E., and K.M. conceived and carried out the experiments and analysed the data. A.v.W., A.S., B.R., R.M.S., M.S. and C.R.W. collected material and primary data. A.L. and Pa.M. analysed data. S.B., Pe.M., T.F.E. and K.M. contributed in the study design. D.J. and K.M. wrote the manuscript. All authors had final approval of the submitted and published versions.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Daniela Jäger and Thomas F. E. Barth contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02174-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Walcott BP, et al. Chordoma: current concepts, management, and future directions. The Lancet. Oncology. 2012;13:e69–76. doi: 10.1016/S1470-2045(11)70337-0. [DOI] [PubMed] [Google Scholar]
- 2.Stacchiotti S, et al. Chordoma of the mobile spine and sacrum: a retrospective analysis of a series of patients surgically treated at two referral centers. Annals of surgical oncology. 2010;17:211–219. doi: 10.1245/s10434-009-0740-x. [DOI] [PubMed] [Google Scholar]
- 3.Angelini A, Pala E, Calabro T, Maraldi M, Ruggieri P. Prognostic factors in surgical resection of sacral chordoma. Journal of surgical oncology. 2015;112:344–351. doi: 10.1002/jso.23987. [DOI] [PubMed] [Google Scholar]
- 4.Park L, et al. Sacral chordomas: Impact of high-dose proton/photon-beam radiation therapy combined with or without surgery for primary versus recurrent tumor. International journal of radiation oncology, biology, physics. 2006;65:1514–1521. doi: 10.1016/j.ijrobp.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 5.Stacchiotti S, et al. Phase II study of imatinib in advanced chordoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:914–920. doi: 10.1200/JCO.2011.35.3656. [DOI] [PubMed] [Google Scholar]
- 6.Casali PG, et al. Imatinib mesylate in chordoma. Cancer. 2004;101:2086–2097. doi: 10.1002/cncr.20618. [DOI] [PubMed] [Google Scholar]
- 7.Hindi N, et al. Imatinib in advanced chordoma: A retrospective case series analysis. European journal of cancer. 2015;51:2609–2614. doi: 10.1016/j.ejca.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 8.Akhavan-Sigari R, et al. Expression of vascular endothelial growth factor receptor 2 (VEGFR-2), inducible nitric oxide synthase (iNOS), and Ki-M1P in skull base chordoma: a series of 145 tumors. Neurosurgical review. 2014;37:79–88. doi: 10.1007/s10143-013-0495-5. [DOI] [PubMed] [Google Scholar]
- 9.Han S, et al. Aberrant hyperactivation of akt and Mammalian target of rapamycin complex 1 signaling in sporadic chordomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:1940–1946. doi: 10.1158/1078-0432.CCR-08-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Presneau N, et al. Potential therapeutic targets for chordoma: PI3K/AKT/TSC1/TSC2/mTOR pathway. British journal of cancer. 2009;100:1406–1414. doi: 10.1038/sj.bjc.6605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhavan-Sigari R, et al. Immunohistochemical expression of receptor tyrosine kinase PDGFR-alpha, c-Met, and EGFR in skull base chordoma. Neurosurgical review. 2015;38:89–98. doi: 10.1007/s10143-014-0579-x. [DOI] [PubMed] [Google Scholar]
- 12.Akhavan-Sigari R, Gaab MR, Rohde V, Abili M, Ostertag H. Expression of PDGFR-alpha, EGFR and c-MET in spinal chordoma: a series of 52 patients. Anticancer research. 2014;34:623–630. [PubMed] [Google Scholar]
- 13.Dewaele B, et al. Frequent activation of EGFR in advanced chordomas. Clinical sarcoma research. 2011;1:4. doi: 10.1186/2045-3329-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalaby A, et al. The role of epidermal growth factor receptor in chordoma pathogenesis: a potential therapeutic target. The Journal of pathology. 2011;223:336–346. doi: 10.1002/path.2818. [DOI] [PubMed] [Google Scholar]
- 15.Scheipl S, et al. EGFR inhibitors identified as a potential treatment for chordoma in a focused compound screen. The Journal of pathology. 2016;239:320–334. doi: 10.1002/path.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaly M, Seelemann C, Jahani-Asl A. A focused compound screen highlights the significance of EGFR signalling in chordoma pathogenesis. The Journal of pathology. 2016;240:381–383. doi: 10.1002/path.4780. [DOI] [PubMed] [Google Scholar]
- 17.Choy E, et al. Genotyping cancer-associated genes in chordoma identifies mutations in oncogenes and areas of chromosomal loss involving CDKN2A, PTEN, and SMARCB1. PloS one. 2014;9:e101283. doi: 10.1371/journal.pone.0101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallor KH, et al. Frequent deletion of the CDKN2A locus in chordoma: analysis of chromosomal imbalances using array comparative genomic hybridisation. British journal of cancer. 2008;98:434–442. doi: 10.1038/sj.bjc.6604130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Witzleben A, et al. Preclinical Characterization of Novel Chordoma Cell Systems and Their Targeting by Pharmocological Inhibitors of the CDK4/6 Cell-Cycle Pathway. Cancer research. 2015;75:3823–3831. doi: 10.1158/0008-5472.CAN-14-3270. [DOI] [PubMed] [Google Scholar]
- 20.Nibu Y, Jose-Edwards DS, Di Gregorio A. From notochord formation to hereditary chordoma: the many roles of Brachyury. BioMed research international. 2013;2013:826435–14. doi: 10.1155/2013/826435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Presneau N, et al. Role of the transcription factor T (brachyury) in the pathogenesis of sporadic chordoma: a genetic and functional-based study. The Journal of pathology. 2011;223:327–335. doi: 10.1002/path.2816. [DOI] [PubMed] [Google Scholar]
- 22.Hsu W, et al. Generation of chordoma cell line JHC7 and the identification of Brachyury as a novel molecular target. Journal of neurosurgery. 2011;115:760–769. doi: 10.3171/2011.5.JNS11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis EB. The 1991 Albert Lasker Medical Awards. Clusters of master control genes regulate the development of higher organisms. Jama. 1992;267:1524–1531. doi: 10.1001/jama.1992.03480110100042. [DOI] [PubMed] [Google Scholar]
- 24.Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 25.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- 26.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. The EMBO journal. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends in genetics: TIG. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 28.Harbour JW, et al. Clivus chordoma: a report of 12 recent cases and review of the literature. Skull base surgery. 1991;1:200–206. doi: 10.1055/s-2008-1057099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeys L, Gibbins R, Evans G, Grimer R. Sacral chordoma: a diagnosis not to be sat on? International orthopaedics. 2008;32:269–272. doi: 10.1007/s00264-006-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergh P, et al. Prognostic factors in chordoma of the sacrum and mobile spine: a study of 39 patients. Cancer. 2000;88:2122–2134. doi: 10.1002/(SICI)1097-0142(20000501)88:9<2122::AID-CNCR19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Volpe R, Mazabraud A. A clinicopathologic review of 25 cases of chordoma (a pleomorphic and metastasizing neoplasm) The American journal of surgical pathology. 1983;7:161–170. doi: 10.1097/00000478-198303000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Gay E, et al. Chordomas and chondrosarcomas of the cranial base: results and follow-up of 60 patients. Neurosurgery. 1995;36:887–896. doi: 10.1227/00006123-199505000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Tauziede-Espariat A, et al. Prognostic and Therapeutic Markers in Chordomas: A Study of 287 Tumors. Journal of neuropathology and experimental neurology. 2016;75:111–120. doi: 10.1093/jnen/nlv010. [DOI] [PubMed] [Google Scholar]
- 34.Fischer C, et al. Mutation Analysis of Nine Chordoma Specimens by Targeted Next-Generation Cancer Panel Sequencing. Journal of Cancer. 2015;6:984–989. doi: 10.7150/jca.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aydemir E, et al. Characterization of cancer stem-like cells in chordoma. Journal of neurosurgery. 2012;116:810–820. doi: 10.3171/2011.12.JNS11430. [DOI] [PubMed] [Google Scholar]
- 36.Scheil S, et al. Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes, chromosomes & cancer. 2001;32:203–211. doi: 10.1002/gcc.1184. [DOI] [PubMed] [Google Scholar]
- 37.Gellner V, et al. Establishment of clival chordoma cell line MUG-CC1 and lymphoblastoid cells as a model for potential new treatment strategies. Scientific reports. 2016;6:24195. doi: 10.1038/srep24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner JF, et al. Human lung cancer cells and tissues partially recapitulate the homeobox gene expression profile of embryonic lung. Lung cancer. 2002;37:41–47. doi: 10.1016/S0169-5002(02)00036-3. [DOI] [PubMed] [Google Scholar]
- 39.Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. The Ulster medical journal. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 40.Darda L, et al. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PloS one. 2015;10:e0122285. doi: 10.1371/journal.pone.0122285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreeff M, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia. 2008;22:2041–2047. doi: 10.1038/leu.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drabkin HA, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 43.Gao L, Sun J, Liu F, Zhang H, Ma Y. Higher expression levels of the HOXA9 gene, closely associated with MLL-PTD and EZH2 mutations, predict inferior outcome in acute myeloid leukemia. OncoTargets and therapy. 2016;9:711–722. doi: 10.2147/OTT.S95279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. HOXA7 stimulates human hepatocellular carcinoma proliferation through cyclin E1/CDK2. Oncology reports. 2015;33:990–996. doi: 10.3892/or.2014.3668. [DOI] [PubMed] [Google Scholar]
- 45.Abe M, et al. Disordered expression of HOX genes in human non-small cell lung cancer. Oncology reports. 2006;15:797–802. [PubMed] [Google Scholar]
- 46.Chang CP, et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes & development. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 47.Morgan R, et al. Targeting HOX transcription factors in prostate cancer. BMC urology. 2014;14:17. doi: 10.1186/1471-2490-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS. Targeting HOX and PBX transcription factors in ovarian cancer. BMC cancer. 2010;10:89. doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) British journal of cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan R, et al. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer research. 2007;67:5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- 51.Rinner B, et al. Establishment and detailed functional and molecular genetic characterisation of a novel sacral chordoma cell line, MUG-Chor1. International journal of oncology. 2012;40:443–451. doi: 10.3892/ijo.2011.1235. [DOI] [PubMed] [Google Scholar]
- 52.Bruderlein S, et al. Molecular characterization of putative chordoma cell lines. Sarcoma. 2010;2010:630129–14. doi: 10.1155/2010/630129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barth TF, et al. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood. 2003;101:3681–3686. doi: 10.1182/blood-2002-08-2577. [DOI] [PubMed] [Google Scholar]
- 54.von Witzleben A, et al. In chordoma, metastasis, recurrences, Ki-67 index, and a matrix-poor phenotype are associated with patients’ shorter overall survival. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2015;25:4016–4024. doi: 10.1007/s00586-015-4242-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


