Abstract
Activation of T lymphocytes requires a tight regulation of microRNA (miRNA) expression. Terminal uridyltransferases (TUTases) catalyze 3′ nontemplated nucleotide addition (3′NTA) to miRNAs, which may influence miRNA stability and function. Here, we investigated 3′NTA to mature miRNA in CD4 T lymphocytes by deep sequencing. Upon T-cell activation, miRNA sequences bearing terminal uridines are specifically decreased, concomitantly with down-regulation of TUT4 and TUT7 enzymes. Analyzing TUT4-deficient T lymphocytes, we proved that this terminal uridyltransferase is essential for the maintenance of miRNA uridylation in the steady state of T lymphocytes. Analysis of synthetic uridylated miRNAs shows that 3′ addition of uridine promotes degradation of these uridylated miRNAs after T-cell activation. Our data underline post-transcriptional uridylation as a mechanism to fine-tune miRNA levels during T-cell activation.
Keywords: 3′ nontemplated nucleotides addition (3′NTA), CD4 T lymphocytes, microRNAs, uridylation, Zcchc11 (TUT4), Zcchc6 (TUT7), isomiRs
INTRODUCTION
MicroRNAs (miRNAs) are short (∼22 nt) noncoding RNAs involved in the control of gene expression in many biological processes (Filipowicz et al. 2008). To fulfill their function, their levels need to be tightly regulated, and dysregulated levels of miRNAs are associated with many diseases (Chang and Mendell 2007). MiRNA turnover is controlled through the precise regulation of both biogenesis and degradation (Filipowicz et al. 2008). The control of miRNA levels through transcriptional and processing mechanisms is well established, but regulation of miRNA degradation is still poorly understood, particularly in mammals. MiRNA stability has been shown to be affected by the cellular physiological status (Hwang et al. 2007; Krol et al. 2010) and the presence of mRNA targets (Ameres et al. 2010; Chatterjee et al. 2011). Moreover, the fate of mature miRNAs is determined by post-transcriptional modifications such as methylation (Li et al. 2005) and nucleotide additions (Landgraf et al. 2007).
In recent years, next-generation sequencing analysis has revealed variability in the individual mature miRNA sequences derived from the same precursor (pre-miRNA), giving rise to the term “isomiRs” (Ameres and Zamore 2013). This sequence diversification in mature miRNAs can be produced through a variety of mechanisms, including imprecise or alternative cropping of the precursor, post-transcriptional A-to-I editing mediated by the enzyme ADAR (adenosine deaminase acting on RNA), terminal trimming of nucleotides, or nontemplated nucleotide additions (NTA). These mechanisms increase the repertoire of cellular regulatory miRNAs and therefore provide additional flexibility to miRNA-mediated regulation of gene expression (Ameres and Zamore 2013).
The most common modification of mature miRNAs is the NTA of uridines and adenosines at the 3′ end (3′NTA) (Landgraf et al. 2007). Several noncanonical poly(A) polymerases (PAP) have been shown to add ribonucleotides to the 3′ end of pre-miRNAs and mature miRNAs in a template-independent manner. These enzymes are able to utilize not only adenine triphosphate (ATP) but also uridine triphosphate (UTP); hence, they are called terminal uridyltransferases (TUTases) (Heo et al. 2009, 2012). The identified nucleotidyl transferases in mammals are MTPAP, GLD2, PAPD5, PAPD7 (POLS), U6TUT, ZCCHC11 (TUT4), and ZCCHC6 (TUT7) (Wyman et al. 2011; Norbury 2013). Among these enzymes, TUT4 and TUT7 have been shown to preferentially uridylate mature miRNAs (Minoda et al. 2006; Jones et al. 2009, 2012; Thornton et al. 2015).
MiRNAs are tightly regulated during the activation of T lymphocytes (Monticelli et al. 2005; Cobb et al. 2006; Baumjohann and Ansel 2013). T cells are activated upon encounter with a specific antigen on the surface of an antigen presenting cell. Signaling through the T-cell receptor (TCR) and costimulatory molecules initiates an array of signaling programs that prepare the cell for differentiation, proliferation, and effector function, leading to profound changes in their metabolism and protein and RNA content. Recent studies link the T-cell activation process to a global down-regulation of miRNAs (Bronevetsky et al. 2013), although the specific regulatory mechanisms remain unknown.
In this study, we have investigated NTA to the 3′ of mature miRNAs during T-lymphocyte activation. Mature miRNA 3′ uridylation is predominantly observed in steady state (“naïve”) T cells, while T-cell activation promotes both the down-regulation of TUT4 and TUT7 enzymes and the degradation of uridylated microRNAs. Moreover, we have identified TUT4 to be essential for the maintenance of miRNA uridylation.
RESULTS
Uridylated miRNAs are decreased upon T-cell activation
To assess the dynamics of NTA during T-cell activation, we performed a deep-sequencing analysis on small RNAs from mouse CD4+ T cells in naïve conditions (CD25−, CD62L+) and upon stimulation with anti-CD3 and anti-CD28 antibodies. The homogeneity of the replicates for each condition was confirmed by their proximity on the principal components analysis (PCA) (Supplemental Fig. S1A). T-cell activation was confirmed by staining of the activation markers CD25 and CD69 (Supplemental Fig. S1B). Moreover, a global down-regulation of miRNA was detected between naïve and activated samples when miRNA count distributions were analyzed with the Kolmogorov–Smirnov test (P ≤ 4.66 × 10−12).
MiRNA modifications were classified according to the number of nucleotides added, i.e., mono addition (one nucleotide) and oligo addition (two or more nucleotides). The relative modification levels from miRNA to miRNA independent of their total expression levels was first examined (Fig. 1A). Uridylation and adenylation were the two most common modifications of CD4 T-cell miRNAs (Fig. 1A). A significant reduction of miRNA uridylation, both mono and oligo additions, was observed in activated T cells upon global examination of the data (Fig. 1B). Individual examination of each miRNA confirmed this observation (Fig. 1C; Supplemental Table S1). In contrast, adenylation seemed to be increased after activation when miRNAs were analyzed globally (Fig. 1A,B), but this was not confirmed in the individual analysis (Fig. 1D). This apparent contradiction is due to a highly expressed adenylated miRNA that must be dominating the global analysis but that does not reflect the general behavior of adenylated miRNAs that is better defined in the individual analysis (Fig. 1D).
FIGURE 1.
Uridylated miRNAs are decreased upon T-cell activation. Deep-sequencing libraries were generated from naïve CD4 T cells or cells activated for 48 h with anti-CD3 and anti-CD28 (n = 4). (A) 3′ NTAs to all mature miRNAs in naive and activation conditions. The fraction of modified reads that fall into one of eight categories was computed for each miRNA providing the normalized relative levels of modification. These were averaged across all miRNAs observed for both conditions. (B) Total normalized (raw) counts for uridylation and adenylation modifications observed across each sample (N: naïve; A: activated CD3/CD28) (upper panel) and averaged across replicates (lower panel). Error bars indicate the standard error between samples and P-values are computed using a Welch's two-sample t-test. (C,D) Comparison of the relative abundance of 3′ mono-uridine and oligo-uridines (C) and of 3′ mono-adenine or oligo-adenines (D) in individual miRNAs from naïve and activated CD4 T cells. Each dot represents an individual miRNA with the corresponding type of modification. (E) Pie charts for selected miRNAs depict the fraction of counts for each type of 3′NTA-modified and canonical miRNA detected. (A) Adenine, (C) cytosine, (G) guanine, and (U) uracil.
Abundance profiles corresponding to each 3′ NTA and the canonical (unmodified) sequence of example miRNAs are shown in Figure 1E. The fraction of counts that correspond to the uridylated forms decreases in activated samples as illustrated also in Supplemental Table S1. Moreover, the five example miRNAs with lower uridylation in activated samples (Fig. 1E) illustrate also that total counts of the miRNA can be either higher, lower, or not changing between naïve and activated conditions (Supplemental Fig. S2). Thus, these data indicate that in CD4 T cells uridylated miRNAs are decreased upon activation.
TUT4 and TUT7 are down-regulated during T-cell activation
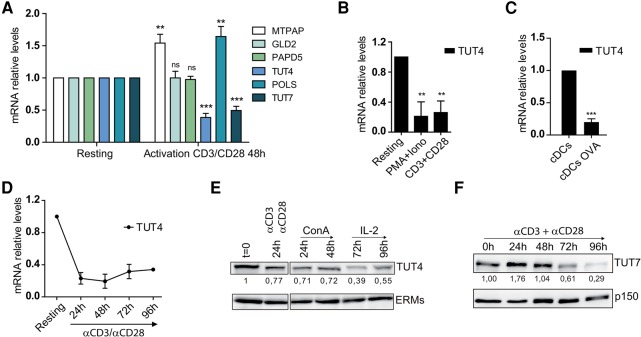
To investigate the molecular mechanism underlying the post-transcriptional modification of miRNAs during T-cell activation, we studied the regulation of terminal uridyl transferases (TUTases) during this process. The mRNA transcript levels of MTPAP, GLD2 (TUT2), PAPD5, TUT4 (ZCCHC11), POLS, and TUT7 (ZCCHC6) in CD4 T cells were determined before and after polyclonal stimulation with anti-CD3 and anti-CD28 antibodies for 48 h. TUT4 and TUT7, the two enzymes mainly responsible for uridylation, were decreased upon T-cell activation, whereas MTPAP and POLS were up-regulated, and GLD2 and PAPD5 remained unaltered (Fig. 2A). The reduction of TUT4 mRNA levels was confirmed in human T lymphoblasts stimulated in a polyclonal manner (Fig. 2B) and in antigen-specific experiments in which CD4 T cells from transgenic OT-II mice were co-cultured with bone marrow-derived dendritic cells loaded with OVA peptide (Fig. 2C). TUT4 mRNA was decreased after 24 h of stimulation and remained low at 96 h (Fig. 2D). Western blot analysis of TUT4 protein showed a decrease at 24 h after activation with anti-CD3 and anti-CD28 antibodies and upon treatment with concanavalin A, followed by expansion with recombinant interleukin 2 (an equivalent activating stimulus), where a maximal decrease was observed after 72 h of stimulation (Fig. 2E). Immunofluorescence and subcellular fractionation experiments showed a predominant cytoplasmic confinement of TUT4, and T-cell activation did not elicit any significant subcellular redistribution (Supplemental Fig. S3A,B). The levels of TUT7 protein were also decreased after stimulation with anti-CD3 and anti-CD28 antibodies, albeit with a slower kinetic profile compared to TUT4 (Fig. 2F). These data indicate that T-cell activation triggers a decrease in the cellular levels of TUT4 and TUT7, the main TUTases accounting for miRNA uridylation. The conjoined down-regulation of these enzymes, concomitant with the decrease in miRNA uridylation, suggests that the decrease of uridylated miRNA plays an important role during T-cell activation.
FIGURE 2.
TUT4 and TUT7 expression is down-regulated after T-cell activation: (A) RNA levels of MTPAP, GLD2, PAPD5, TUT4, POLS, and TUT7 were assessed by RT-qPCR in freshly isolated mouse naïve CD4 T cells or cells activated with anti-CD3 and anti-CD28 for 48 h. mRNA levels were normalized to β-actin and Yhwaz housekeeping genes and are presented in arbitrary units (n = 7). (B) TUT4 mRNA levels in human T lymphoblasts measured by RT-qPCR after two different activation stimuli: phorbol myristate acetate (PMA) with ionomycin and anti-CD3 plus anti-CD28 (n = 3). (C) Mouse naïve CD4 T cells from OT-II mice specific to ovalbumin peptide were co-cultured with dendritic cells in the absence or presence of the peptide. TUT4 mRNA levels were determined by RT-qPCR as in A (n = 5). (D) Long-term time course of TUT4 mRNA levels in activated mouse CD4 T cells (n = 3). (E) Western blot analysis of TUT4 protein content in CD4 T cells after activation with anti-CD3 plus anti-CD28 or with concanavalin A (ConA) followed by expansion with interleukin 2 (IL-2). Representative immunoblots (n = 3). ERMs were included as a loading control. (F) Western blot analysis of TUT7 protein at different time points after antibody activation of CD4 T cells. Representative immunoblots (n = 3). p150 was included as a loading control. Numbers below blots show normalized densitometry values relative to naïve T cells. Error bars in A–C represent standard deviation; (***) P < 0.001; (**) P < 0.05; ns, nonsignificant.
TUT4-dependent uridylation of mature microRNA
To assess the role of TUT4 in the uridylation of mature miRNAs in T lymphocytes, we examined CD4 T cells of TUT4-deficient mice in steady state. The lymphoid organs of these mice presented no significant alteration in the percentage of CD4 and CD8 T lymphocytes in thymus (Supplemental Fig. S4A), and CD4 and CD8 T lymphocytes as well as B lymphocytes in spleen or peripheral lymph nodes (Supplemental Fig. S4B,C). Levels of miRNA mono- and oligo-uridylation were lower in naive TUT4-deficient CD4 T cells compared with wild-type cells (Fig. 3A,B). Interestingly, miRNA mono- and oligo-adenylation were higher in TUT4-deficient T cells (Fig. 3C,D). Putative miRNA targets of TUT4 were identified in T cells. We considered as targets both mono-uridylated and oligo-uridylated species that were significantly less uridylated in TUT4-deficient CD4 T cells compared with wild-type cells (Supplemental Table S2A,B). Moreover, miR-seq data showed no significant differences in the levels of canonical miRNAs corresponding to TUT4 targets between TUT4-deficient and wild-type CD4 T cells (Fig. 3E) in accordance with previous reports (Jones et al. 2012; Thornton et al. 2015). Interestingly, analysis of these identified putative targets of TUT4 during T-cell activation of wild-type T cells revealed that the majority of these uridylated miRNAs were down-regulated (Fig. 3F; Supplemental Tables S3, S4). Thus, our data reveal that putative TUT4 targets account for a substantial proportion of the uridylated miRNAs down-regulated upon T-cell activation. These results indicate that TUT4 contributes to the turnover control of a specific set of modified miRNAs during T-cell activation.
FIGURE 3.
TUT4-dependent uridylation of mature microRNA. Small RNAs from naïve wild-type or TUT4-deficient CD4 T cells were analyzed by deep sequencing. (A–D) Fold changes between wild-type and TUT4−/− CD4 T cells calculated from oligo- (A) and mono-uridylated miRNAs (B) as well as mono- (C) and oligo-adenylated miRNA counts (D). Only modified miRNAs whose expression level is above log2 > 2 are presented. A negative fold change indicates lower levels in TUT4-deficient compared to wild-type. (E) Comparison of the levels of the individual canonical miRNAs corresponding to the identified TUT4 uridylation targets between wild-type and TUT4-deficient CD4 T cells. (F) Comparison of the levels of individual uridylated miRNAs identified as TUT4 targets between naïve and activated wild-type CD4 T cells.
Although uridylated miRNAs are clearly less abundant in TUT4-deficient CD4 T cells, we could still detect some miRNAs with 3′ nontemplated uridines. This may reflect the activity of TUT7, which can uridylate miRNAs (Thornton et al. 2015). TUT4-deficient T cells showed no compensatory increase in the levels of TUT7 or any other TUTase (Supplemental Fig. S5A,B), but given the presence of TUT7 protein, it is likely that the remaining miRNA uridylation in TUT4-deficient mice is attributable to this enzyme.
Uridylation directs mature miRNA to activation-dependent degradation
Uridylation of mRNA is associated with both stabilization [when 1 or 2 nt are added to histone mRNAs and poly(A) RNAs] (Lackey et al. 2016; Scheer et al. 2016; Zuber et al. 2016) and decay (Shen and Goodman 2004; Mullen and Marzluff 2008; Schmidt et al. 2011; Lim et al. 2014). To investigate whether uridylation targets mature miRNAs for degradation during T-cell activation, we transfected naïve CD4 T cells with synthetic RNAs (miR-151-3p and miR-25-3p) corresponding to canonical or di-uridylated miRNA sequences. The cells were subsequently either activated with anti-CD3 and anti-CD28 or maintained in resting conditions with IL-7 to prevent naïve T-cell apoptosis. Levels of the synthetic miRNAs were measured with a variation of miQPCR (Fig. 4A, and see Materials and Methods; Benes et al. 2015), which distinguishes single-nucleotide changes in the miRNA sequence. The levels of synthetic di-uridylated forms of both miR-151-3p and miR-25-3p dropped sharply in activated T cells, whereas the levels of their canonical (unmodified) counterparts remained much more stable (Fig. 4B), indicating that uridylated miRNAs are declined faster than the unmodified miRNAs, specifically upon T-cell activation. Moreover, these results suggest that the decay of uridylated miRNAs comprises total degradation of the molecule rather than the trimming of merely the U tail since there is no increase of canonical synthetic miRNA after activation. That would only occur if the U tail had been removed from the synthetic di-uridylated miRNA while preserving the rest of the molecule. Finally, it is worth mentioning that the synthetic ssRNAs that we are using might not be behaving exactly as endogenous miRNAs in terms of their incorporation to the RISC complex, a process that could affect their decay, Nevertheless, these data indicate that miRNA uridine tailing is an important mechanism to target a specific set of miRNAs for degradation, and that this process is tightly regulated upon T-cell activation.
FIGURE 4.
Uridylation of mature miRNAs determines their turnover during T-cell activation. (A) Diagram representing the qPCR method used to distinguish between di-uridylated synthetic miRNAs and unmodified ones. After a linker ligation and reverse transcription, custom probes specific for the 3′ end were used in combination with reverse and forward primers for the miRNA. (B) Synthetic miRNAs were nucleofected into naïve CD4 T cells, which were then activated with anti-CD3 and anti-CD28 or left resting in the presence of IL-7. Levels of the synthetic miRNAs were analyzed by a variation of MiQPCR. Synthetic RNA levels were normalized to endogenous controls snoRNA 420 and snoRNA 412 and are presented in arbitrary units (n = 3). (C) Scheme of the proposed mechanism of miRNA post-transcriptional regulation during T-cell activation. Naïve cells under steady-state contain basal levels of uridylated miRNAs that are down-regulated after T-cell activation. This decrease is accompanied by the down-regulation of uridyltransferases TUT4 and TUT7. Additionally, we hypothesize that a putative exonuclease might be induced in activated T cells responsible for uridylated miRNAs degradation.
DISCUSSION
The present study demonstrates that miRNA 3′ NTA are modifications tightly regulated that can modulate the stability of these small RNA species. Our data indicate that the enzymes responsible for miRNA uridylation as well as uridylated miRNAs themselves are regulated during T-cell activation. We further report TUT4 as an enzyme responsible for uridylation of mature miRNA in T cells, and show that the stability of uridylated miRNAs differs in T cells between naïve and activated conditions. The proposed mechanism for the post-transcriptional regulation of miRNAs during T-cell activation is illustrated in the scheme (Fig. 4C).
The addition of poly(U) tails to the 3′ ends of pre-miRNAs has been reported to trigger their degradation in stem cells (Heo et al. 2009), while in somatic cells lacking Lin28, TUT-mediated mono-uridylation of pre-miRNAs facilitates their processing (Heo et al. 2012). The impact of 3′ NTA on mature miRNAs varies depending on the organism and the miRNA examined (Katoh et al. 2009; Lu et al. 2009; Burroughs et al. 2010). Uridylation of miRNAs leads to degradation in Chlamydomonas (Ibrahim et al. 2010) and plants, where it prevents their methylation (Zhao et al. 2012). In mammals, uridylation of mature miRNA has been shown to specifically reduce the functionality of miR-26a, miR-126-5p, and miR-379 (Jones et al. 2009, 2012). Our data demonstrate that the stability of mature miRNAs is decreased in the context of T-cell activation when they bear uracils added to their 3′ end. This is supported by the fact that synthetic di-uridylated miRNAs are decreased more rapidly than the corresponding synthetic canonical miRNAs, but only when T cells are activated. Nevertheless, since these conclusions arise from exogenously added miRNAs, we cannot undoubtedly predict that the exact same process is happening for endogenous uridylated miRNAs.
Our data are consistent with a decrease in the uridylation of miRNA due to a reduction in the levels of the uridyl transferases TUT4 and TUT7 during T-cell activation. Additionally, a specific subset of uridylated miRNAs has been identified as targets of TUT4. In this regard, it is important to mention that TUT4-deficient T cells still contain low levels of uridylated miRNAs that are likely to be substrates of other TUTases, e.g., TUT7.
According to data from previous work (Jones et al. 2009, 2012) and the present study, it is conceivable that uridylation changes the targeting specificity of the miRNA, its functionality, and/or labels miRNAs for degradation. Thus, the degradation of a defined set of uridylated miRNAs specifically during T-cell activation may represent an additional mechanism for the cell to eliminate certain miRNAs at this specific stage. However, our data do not allow us to rule out additional mechanisms of miRNA biogenesis, like their transcription and processing, accounting for miRNA levels’ regulation and the imbalance of the 3′ uridylated miRNA pool during T-cell activation.
A key outstanding question about miRNA decay is the identity of the enzymes that catalyze the degradation of these small RNAs during T-cell activation. Several mammalian exoribonucleases related to miRNAs have been described that show a certain substrate specificity (Ruegger and Grosshans 2012). Further research will be needed to identify the specific exoribonucleases that mediate the specific degradation of uridylated miRNAs observed after T-cell activation. Defining the mechanisms and purpose of miRNA 3′ nucleotide additions will be important for understanding the post-transcriptional regulation of miRNA function and turnover, and could also inform strategies for therapeutic modulation of miRNA activity.
MATERIALS AND METHODS
Mice
C57BL/6 and OT-II mice and C57BL/6 Zcchc11-/+ mice (TUT4-deficient mice) were bred under specific pathogen-free conditions according to European Commission recommendations. The TUT4-deficient strain was maintained in heterozygosis as described previously (Jones et al. 2012).
T-cell isolation, culture, and activation
Naïve CD4+ T cells were obtained from cell suspensions of lymph nodes or spleen. Cell suspensions were incubated with biotinylated antibodies against CD8, CD19, CD25, CD11b, CD45R, major histocompatibility complex (MHC) class II (I-Ab), DX5, IgM, Gr-1, and F4/80 and subsequently with streptavidin microbeads (MACS; Miltenyi Biotec). CD4+ T cells were negatively selected in auto-MACS Pro Separator (Miltenyi Biotec) according to the manufacturer's instructions. The cells were then labeled with antibodies to CD4, CD62L, and CD25 (BD Biosciences) and analyzed by flow cytometry to confirm their purity and naïve status (data not shown). Naïve CD4+ T cells (106 cells mL−1) were either directly lysed or cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U mL−1 of penicillin and streptomycin, 10 mM Hepes, 50 µM 2-mercaptoethanol, and 1 mM sodium pyruvate and 2 µg mL−1 of anti-CD28 on plates previously coated with 10 µg mL−1 of anti-CD3 (BD Biosciences). Alternatively, cells were maintained in the naïve state in the presence of 4 ng mL−1 of interleukin 7 (Peprotech). The activation status of CD4 T cells was assessed at the indicated time points by flow cytometry after staining with antibodies against CD4, CD25, and CD69 (BD Biosciences). Data were acquired and analyzed with a FACSFortessa flow cytometer and FACSDiva (BD Biosciences) or FlowJo software.
Antigen-specific T-cell stimulation
When indicated, dendritic cells (DCs) were used to stimulate OT-II CD4 T cells. DCs were derived from wild-type bone marrow cell suspensions after culture on nontreated 150-mm Petri dishes in complete RPMI 1640 medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U mL−1 penicillin and streptomycin, 50 µM 2-mercaptoethanol and 20 ng mL−1 recombinant GM-CSF (Peprotech). Cells were collected on day 9 and subsequently stimulated with 100 ng mL−1 LPS from Escherichia coli (Sigma). At this point, DC preparations were characterized as CD11c+ MHCII+ Ly6G−. OT-II CD4 T cells were co-cultured with DCs (8:1 T-cell/DC ratio) in the presence of ovalbumin (OVA) peptide for 18 h. Cells were stained for CD4 and MHCII for subsequent sorting of the CD4 T-cell fraction on a FACSAria flow cytometer (BD Biosciences).
Human T lymphoblasts
Human T lymphoblasts were obtained as previously described (Mittelbrunn et al. 2011). Briefly, peripheral blood mononuclear cells were isolated from buffy coats from healthy donors. Isolated nonadherent cells were cultured for 2 d in the presence of phytohemagglutinin (5 µg mL−1) and subsequently interleukin 2 (50 U mL−1) was added to the medium every 2 d for a period of 8 d. T lymphoblasts were later stimulated either with anti-CD3 and anti-CD28 coated beads (Gibco) or with phorbol myristate acetate (PMA) and ionomycin.
RNA isolation
Total RNA was extracted with QIAzol lysis reagent (Qiagen) and the miRNeasy mini kit (Qiagen). Purity and concentration were measured in a Nanodrop-1000 spectrophotometer (Thermo Scientific), and RNA integrity was assessed using the Agilent 2100 Bioanalyzer.
Small RNA cloning and sequencing (miR-seq library preparation, sequencing, and generation of FastQ files)
Total RNA (1 µg) was used to generate barcoded miR-seq libraries using the TruSeq Small RNA Sample Preparation Kit (Illumina). Briefly, 3′ and 5′ adapters were first ligated to the RNA sample. Next, reverse transcription followed by PCR amplification was used to enrich cDNA fragments with adapters at both ends. Adapter-ligated cDNA fragments from different samples were pooled and run on a 6% polyacrylamide gel. The band between 147 and 180 base pairs, corresponding to the pooled miRNA libraries including slightly longer fragments (potential IsomiRs with 3′NTAs), was purified from the gel. Finally, the quantity and quality of the pooled miR-seq libraries were determined using the Agilent 2100 Bioanalyzer High Sensitivity DNA chip. Libraries were sequenced on a Genome Analyzer IIx (Illumina). FastQ files for each sample were obtained using CASAVA v1.8 (Illumina). Library preparation and sequencing was performed by the CNIC genomic unit.
Next-generation sequencing analysis and statistics
Raw FASTQ files were trimmed for adapter contamination using REAPER from the Kraken package (Davis et al. 2013). All reads were mapped against miRNA precursors obtained from miRBase release 21 (Kozomara and Griffiths-Jones 2014) using BLASTn. A custom pipeline interrogated each read to determine which miRNAs originated from which precursor and which arm (either 5′ or 3′). Extra nucleotides that could not be explained by the associated miRNA precursor sequence were flagged as noncanonical 5′ or 3′ extensions. Count data for canonical matches and noncanonical extensions were stored for each sample in the tabular form. Count data were normalized and analyzed using R/BioConductor with the DeSeq2 package. DESeq2 automatically excludes outliers using a Cook′s distance for all genes.
Changes to either global miRNA levels or individual miRNA modifications were viewed as significant if they had an absolute log fold-change >0.5 and an adjusted P-value <0.05. miRNAs were considered into DE analysis when having a mean expression level of log2 > 2.
All data and scripts used are available. NGS analysis data have been deposited in European Nucleotide Archive and are accessible through ENA accession number PRJEB15528.
mRNA reverse transcription and quantitative real-time PCR
cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Expression analysis by quantitative PCR was performed with SYBRgreen PCR master mix (Applied Biosystems), and the corresponding primers are listed in Supplemental Table S5. Data were acquired and analyzed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems) and Biogazelle QBasePlus software (Biogazelle). B-actin and Yhwaz (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ) genes were used as endogenous controls for presentation of relative mRNA levels.
Reverse transcription and RT-qPCR of mature canonical miRNA
cDNA was synthesized and mature miRNAs were quantified by miRCURY LNA Universal RT microRNA PCR (Exiqon), using miRNA LNA primers (Exiqon) and SYBRgreen PCR master mix (Applied Biosystems). Quantitative miRNA expression data were acquired and analyzed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Data were further analyzed using BiogazelleQBasePlus software (Biogazelle). RNU1A1 and RNU5G RNAs were used as endogenous controls and results are expressed in arbitrary units.
Nucleofection of synthetic RNAs
Synthetic RNAs that correspond to canonical or di-uridylated miRNAs (Integrated DNA Technologies) were nucleofected into freshly isolated naïve CD4 T cells using the Amaxa Mouse T-cell Nucleofector Kit and Amaxa Nucleofector II (Lonza) according to the manufacturer's instructions.
Reverse transcription and qPCR of synthetic miRS and isomirs
Levels of the synthetic miRNAs previously nucleofected into T cells were analyzed by a variation of the miQPCR method (Benes et al. 2015). In brief, 60 ng of total RNA were ligated to an oligonucleotide adapter (miLINKER) followed by retrotranscription to cDNA. Quantitative PCR was then performed with Taqman PCR master mix (Applied Biosystems) using both specific primers and LNA Taqman probes. Quantitative data of synthetic RNA levels were acquired and analyzed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Data were further analyzed using BiogazelleQBasePlus software (Biogazelle). snoRNA 420 and snoRNA 412 were used as endogenous controls for normalization and results are expressed in arbitrary units.
Immunofluorescence
Cells were plated onto slides coated with poly-L-lysine (50 µg mL−1), incubated for 30 min, fixed, blocked, and stained with antitubulin-FITC (Sigma-Aldrich) and rabbit polyclonal anti-Zcchc11 (TUT4) (Proteintech) (5 µg mL−1) followed by Rhodamine Red X-labeled secondary antibody (5 µg mL−1) (Life Technologies). Samples were examined with a Leica SP5 confocal microscope (Leica) fitted with a ×63 objective, and images were processed and assembled using Leica software.
Immunoblotting
Cells were lysed in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1%NP-40, 5 mM EDTA) containing a protease inhibitor cocktail (Complete, Roche). Proteins were separated on 8% acrylamide/bisacrylamide gels and transferred to a nitrocellulose membrane. Membranes were incubated with specific primary antibodies (5 µg mL−1) and peroxidase-conjugated secondary antibodies (5 µg mL−1), and chemoluminescence was measured with LAS-3000 (Fujifilm). The following antibodies were used: rabbit polyclonal anti-Zcchc11 (TUT4), rabbit polyclonal anti-Zcchc6 (TUT7) (Proteintech), goat polyclonal anit-Zcchc11 (ProSci), anti-p150glued (BD Transduction Laboratories), monoclonal anti-EZH2 (Cell Signaling), polyclonal anti ezrin/moesin (ERMs) (90/3) (provided by Heinz Furthmayr, Stanford University, Stanford, CA). Secondary antibodies were goat anti-mouse peroxidase (31446, Thermo Scientific) (1:5,000) and goat anti-rabbit peroxidase (37460, Thermo Scientific) (1: 10,000). Band intensities were quantified with Image Gauge (Fujifilm). The Proteo Extract subcellular proteome extraction kit (Calbiochem) was used for subcellular fractionation of proteins followed by Western blot.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate help from Carolina Villarroya-Beltri and Marta Ramirez-Huesca. We also thank the CNIC Genomics and Cellomics Units for technical support, Simon Bartlett for help with English editing, and Salvador Iborra, David Sancho, and Miguel Vicente Manzanares for helpful discussions. This work was supported by grants SAF2014-55579R from the Ministerio de Economía y Competitividad-Spain, ERC-2011-AdG 294340-GENTRIS, COST-Action BN1202, CIBERCV (Instituto de Salud Carlos III) PIE-13-00041, and INDISNET S2011-BMD-2332 (F.S.-M.). This research has been cofinanced by FEDER. The Centro Nacional de Investigaciones Cardiovasculares (CNIC, Spain) is supported by the Ministerio de Economía y Competitividad-Spain and the Pro-CNIC Foundation. A.R.R. was funded by research grants SAF2013-42767-R and SAF2016-75511-R (Plan Estatal de Investigación Científica y Técnica y de Innovación 2013-2016 Programa Estatal de I+D+i Orientada a los Retos de la Sociedad Retos Investigación: Proyectos I+D+i 2016, Ministerio de Economía, Industria y Competitividad) and cofunded by Fondo Europeo de Desarrollo Regional (FEDER). M.M. is supported by grant MS14/00219 from the Instituto de Salud Carlos III. A.R.-G. is supported by the FPU program (Spanish Ministry of Education).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.060095.116.
REFERENCES
- Ameres SL, Zamore PD. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14: 475–488. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Ansel KM. 2013. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol 13: 666–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes V, Collier P, Kordes C, Stolte J, Rausch T, Muckentaler MU, Haussinger D, Castoldi M. 2015. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci Rep 5: 11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, et al. 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 210: 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. 2007. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215–239. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Fasler M, Bussing I, Grosshans H. 2011. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell 20: 388–396. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, et al. 2006. A role for Dicer in immune regulation. J Exp Med 203: 2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151: 521–532. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. 2007. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci 107: 3906–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 11: 1157–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, Powers JT, Daley GQ, Quinton LJ, Mizgerd JP. 2012. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS Genet 8: e1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. 2010. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141: 618–631. [DOI] [PubMed] [Google Scholar]
- Lackey PE, Welch JD, Marzluff WF. 2016. TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA 22: 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. 2009. Adenylation of plant miRNAs. Nucleic Acids Res 37: 1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoda Y, Saeki K, Aki D, Takaki H, Sanada T, Koga K, Kobayashi T, Takaesu G, Yoshimura A. 2006. A novel Zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem Biophys Res Commun 344: 1023–1030. [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. 2011. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol 6: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ. 2013. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol 14: 643–653. [DOI] [PubMed] [Google Scholar]
- Ruegger S, Grosshans H. 2012. MicroRNA turnover: when, how, and why. Trends Biochem Sci 37: 436–446. [DOI] [PubMed] [Google Scholar]
- Scheer H, Zuber H, De Almeida C, Gagliardi D. 2016. Uridylation earmarks mRNAs for degradation…and more. Trends Genet 32: 607–619. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, West S, Norbury CJ. 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. 2004. Uridine addition after microRNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, Sliz P, Zon LI, Gregory RI. 2015. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res 42: 11777–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. 2012. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22: 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber H, Scheer H, Ferrier E, Sement FM, Mercier P, Stupfler B, Gagliardi D. 2016. Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep 14: 2707–2717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.