Abstract
The design and synthesis of metal complexes that can specifically target DNA secondary structure has attracted considerable attention. Chiral metallosupramolecular complexes (e.g. helicates) in particular display unique DNA-binding behavior, however until recently few examples which are both water-compatible and enantiomerically pure have been reported. Herein we report that one metallohelix enantiomer Δ1a, available from a diastereoselective synthesis with no need for resolution, can enantioselectively stabilize human telomeric hybrid G-quadruplex and strongly inhibit telomerase activity with IC50 of 600 nM. In contrast, no such a preference is observed for the mirror image complex Λ1a. More intriguingly, neither of the two enantiomers binds specifically to human telomeric antiparallel G-quadruplex. To the best of our knowledge, this is the first example of one pair of enantiomers with contrasting selectivity for human telomeric hybrid G-quadruplex. Further studies show that Δ1a can discriminate human telomeric G-quadruplex from other telomeric G-quadruplexes.
INTRODUCTION
G-quadruplexes are highly dynamic four-stranded non-canonical DNA structures formed in guanine-rich sequences via stacking of G·G·G·G quartets (1). They are involved in a range of biological processes such as telomere maintenance (2–6), replication, transcription, epigenetic regulation and recombination (7–13). As a consequence, the study of specific and selective targeting of the G-quadruplex structure is an exciting avenue for exploration of the biological function of this motif, and of course the possibility of regulation of the corresponding processes. Human telomeric DNA sequences can form antiparallel G-quadruplexes in Na+ buffer and hybrid G-quadruplexes in K+ buffer (14–18). However, up to now only few ligands have been demonstrated to selectively target these highly challenging target structures: an acyclic oligoheteroaryle (TOxaPy) shows selectivity to antiparallel G-qaudruplex, while N-methyl mesoporphyrin IX shows selectivity to hybrid G-quadruplex with a low binding constant (19–21).
Helical bimetallic complexes (helicates and related structures) resemble α-helices in terms of their diameter and charge. Recently, great attention has been paid to their diagnostic and therapeutic applications (22–24). This has been accelerated by recent advances in their enantioselective self-assembly from simple components, thus avoiding chromatographic resolution or lengthy syntheses and the new ability to make low symmetry systems. They have been shown to interact with different DNA structures and showed admirable chiral selectivity (25–29). Our recent works showed that one pair of di-nickel helicates exhibited chiral selective binding with human telomeric G-quadruplex the right-handed helix selectively stabilized antiparallel G-quadruplex, whilst the left-handed compound did not, and both enantiomers stabilized hybrid G-quadruplex (27). Since high K+ concentration is existed in cells, hybrid G-quadruplex has been considered more predominant than antiparallel G-quadruplex under physiological conditions (30). Thus, the development of chiral compounds to enantioselectively target human telomeric hybrid G-quadruplex remains an important objective. To the best of our knowledge, this has not been achieved.
In this work, we investigated the interactions of human telomeric G-quadruplex with a pair of enantiomers of metallohelices (abbreviated to Δ1a and Λ1a) (Figure 1A), which are water-compatible and enantiomerically pure (22,31). Our results showed that enantiomer Δ1a could enantioselectively stabilize human telomeric hybrid G-quadruplex and strongly inhibit telomerase activity. In contrast, Λ1a showed no such a preference. More intriguingly, neither enantiomer displayed notable binding with human telomeric antiparallel G-quadruplex. This is the first example of one pair of enantiomers with contrasting selectivity for human telomeric hybrid G-quadruplex.
Figure 1.
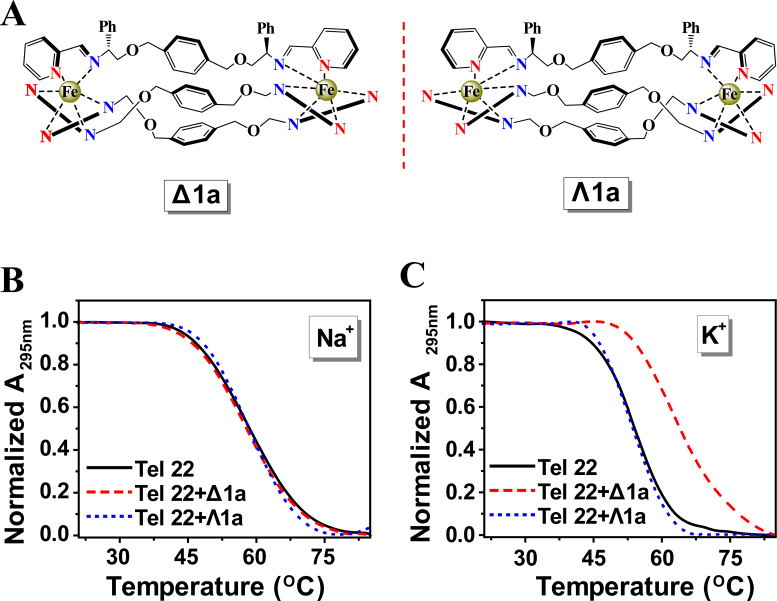
(A) Structure of Δ1a and Λ1a cations. UV melting profiles of the Tel22 in the absence and presence of Δ1a or Λ1a in Na+ buffer (B) and K+ buffer (C). Melting assays were measured in 10 mM Tris buffer containing 10 mM KCl or 100 mM NaCl, pH = 7.2. The concentration of Δ1a/Λ1a was equivalent with the concentration of Tel22 (1 μM in strand).
MATERIALS AND METHODS
DNA sequences
Tel22: 5΄-AGGGTTAGGGTTAGGGTTAGGG-3΄
HT: 5΄-TTAGGGTTAGGGTTAGGGTTAGGGA-3΄
Ap7: 5΄-AGGGTTApGGGTTAGGGTTAGGG-3΄
Ap13: 5΄-AGGGTTAGGGTTApGGGTTAGGG-3΄
Ap19: 5΄-AGGGTTAGGGTTAGGGTTApGGG-3΄ (Ap was 2-aminopurine)
Oxytricha: 5΄-TTTTGGGGTTTTGGGGTTTTGGGGTTTTGGGG-3΄
Tetrahymena: 5΄-GGGGTTGGGGTTGGGGTTGGGG-3΄
i-motif: 5΄- CCCTAACCCTAACCCTAACCCT-3΄
Tel22-TTT: 5΄-AGGGTTTGGGTTTGGGTTTGGG-3΄
DNAs were synthesized by Shanghai Sangon Biological Engineering Technology & Services (Shanghai, China). Concentrations of the oligomers were determined by measuring the absorbance at 260 nm after melting. Extinction coefficients were estimated by the nearest-neighbor method by using mononucleotide and dinucleotide values. Chemicals were purchased from Sigma-Aldrich and used without further purification. All water used to prepare buffer solutions was obtained by using a Milli-Q water system.
Synthesis of metallo-supramolecular complexes
Complex 1 and complex 2 were synthesized according to previous report (22).
The stability of the metal complexes in cells
Hela cells were plated in Dulbecco's modified Eagle's medium, supplemented with 10% (v/v) heat inactivated fetal bovine serum in a 6-well plate grown for 24 h at 37°C and 5% CO2. Then, different amounts of metal complexes were added for further incubation of 72 h. Then, the cell culture media was separated from the well for further analysis and cell lysis solution was added in the well. The lysates were incubated for 30 min on ice and centrifuged 20 min at 12 000 rpm (4°C) to obtain cleared lysate for further analysis.
Thermodynamic analysis
Absorbance measurements and melting experiments were performed on a Cary 300 UV/Vis spectrophotometer equipped with a Peltier temperature control accessory. All UV/Vis spectra were measured in 1.0-cm path-length cell. For metal complex-containing melting assays, the same concentration of corresponding metal complex aqueous solution was used as the reference solution. Absorbance changes at 295 nm versus temperature were collected at a heating rate of 0.5°C min−1. Before measurement, the DNA samples were heated at 95°C for 5 min, and gently cooled from 95°C to room temperature, following by incubation at 4°C overnight. To prepare the final DNA-complex samples, the corresponding metal complexes were added to the annealed DNA samples and incubated at 4°C for 3 h.
CD spectroscopy
Circular dichroism (CD) spectra were recorded with a JASCO J-810 spectropolarimeter. CD spectra were recorded in 1 nm increments with an average time of 2 s and three scans were accumulated and automatically averaged. The various concentration of complexes was scanned as a control and subtracted from the spectra of complex/DNA mixture to eliminate its influence on CD signal of DNA.
Fluorescence spectroscopy
Fluorescence measurements were carried out on a JASCO FP-6500 spectrofluorometer at 25°C. For 2-Ap-labeled oligonucleotides, fluorescence spectra were measured by using an excitation wavelength of 305 nm. DNA concentration was fixed at 0.5 μM in strand.
Binding constants obtained by fluorescence titration
Binding constants were measured by fluorescence titration methods, in which fixed concentrations of 2-Ap-labeled DNA was titrated with increasing Δ1a or Λ1a concentrations. To obtain the proper fluorescence intensity values, fluorescence data has been corrected for the inner filter effect caused by attenuation of the excitation beam and emission signal because of the absorption by quencher and fluorophore which led to artifical decreases in the fluorescence intensities. This effect was corrected with knowledge of the absorbance values from the corresponding spectra (31–33). The fluorescence of the system can be corrected using the following equation:
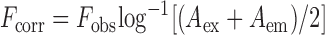 |
where Fcorr and Fobs were the corrected and observed fluorescence intensity. Aex was the absorbance value at the excitation wavelength and Aem was the absorbance value at the emission wavelength.
Absorption spectra titrations
Absorption spectra titrations were carried out at 25°C to determine the binding affinity between DNA and enantiomers. Initially, 500 μl solutions of blank buffer and the enantiomers (5 μM) were placed in the reference and sample cuvettes (1 cm path length), respectively. Then, we measured the absorption spectra in the range of 220–800 nm. During the titration, aliquot of buffered DNA solutions was added to each cuvette to eliminate the absorbance of DNA itself, and the solutions were mixed thoroughly. Afterward, the absorption spectra were recorded. The titration processes were repeated until there was no change in the spectra for four titrations at least, which indicating binding saturation was achieved. The changes in the enantiomers concentration due to dilution at the end of each titration were negligible.
The binding constant obtained from UV titration experiment was calculated from the following equation:
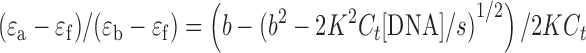 |
(1a) |
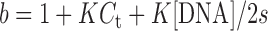 |
(1b) |
[DNA] is the strand concentration of Tel22, εa, εf and εb are the apparent extinction coefficient, the extinction coefficient for the free metal complex and the extinction coefficient in the fully bound form, respectively. K is the equilibrium binding constant in M−1, Ct is the total metal complex concentration and s is the binding site size (34,35).
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) assays were performed on a NANO ITC System (TA Instruments Inc., New Castle, DE, USA). Titrations were performed in buffer (10 mM Tris–HCl buffer, 10 mM KCl, pH = 7.2). Injections of 10 μl of 0.25 mM Δ1a/Λ1a was added from a microsyringe at an interval of 600 s into Tel22 DNA (20 μM) solution with stirring at 400 rpm at 25°C. The experimental data were analyzed with NanoAnalyze software (TA Instruments Inc.).
NMR spectroscopy
Samples for nuclear magnetic resonance (NMR) were incubated in 10mM Tris–KCl buffer (pH 7.2) at 25°C with 10% D2O added. The final concentration of Tel22 was 140 μM. The enantiomer was incubated with Tel22 at 25°C before measurement. NMR experiment was carried out on a Bruker 600 MHz AVANCE NMR spectrometer equipped with a triple-channel cryoprobe at 5°C.
Assay of telomerase activity
Telomerase activity was assayed using a conventional telomere repeat amplification protocol (TRAP) assay. A total of 5 μl of telomerization products with corresponding complex were added into 45 μl of solution which contains 1 × PCR buffer, 200 μM dNTPs, 3U of Taq DNA polymerase, 0.1 μg of TS primer and 0.1 μg of ACX primer. PCR was carried out in an Eppendorf AG thermal cycler with the following program: 94°C for 4 min, 30 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, 72°C for 5 min, 4°C cold. PCR products were analyzed on a Bio-Rad (Bio-Rad Laboratories, USA) slab electrophoresis system. The 10 μl samples were loaded onto a 12% native polyacrylamide gel (29:1 acryl/bisacryl) in 0.5 × Tris borate ethylenediaminetetraacetic acid. Gels were run at room temperature for 1 h at 120 V. The gel was confirmed by silver staining.
RESULTS AND DISCUSSION
Enantioselectivity to hybrid G-quadruplex of Tel22
The stability of the metallohelix enantiomers (Δ1a and Λ1a) was firstly studied. UV and CD spectra showed that cation concentration and type had negligible effects on their CD and UV spectra, implying that Δ1a and Λ1a were stable in Na+/K+ containing buffer (experimental conditions of this work) (Supplementary Figure S1A–D). In addition, for testifying whether the complexes can be applied in vivo or in vitro studies, the stability of metal complexes in cells was also investigated. As shown in Supplementary Figure S1E and F, Δ1a and Λ1a were stable both in cell culture media and cell lysate. These results indicated the high stability of such complexes (22).
UV-melting experiments were employed to study the effects of the enantiomers on the melting temperature (Tm) of human telomeric DNA (Tel22). As shown in Figure 1B and C, the chiral selectivity of the enantiomers on stabilization of Tel22 was remarkable. Δ1a stabilized Tel22 in K+ containing buffer and Tm increased by ∼10.8°C at 1:1 ratio of [Δ1a]/[Tel22] whilst there was no measureable stabilization in Na+ containing buffer (Figure 1B and Supplementary Figure S2). In contrast, Λ1a did not stabilize Tel22 either in K+ or Na+ buffer. According to previous studies, Tel22 adopted antiparallel G-quadruplex in Na+ buffer but hybrid G-quadruplex in K+ buffer (15,36). Thus, we concluded that Δ1a strongly stabilized the former structure but not the latter, while Λ1a stabilized neither. As far as we know, this is the first example of one pair of enantiomers with contrasting chiral selectivity for hybrid G-quadruplex.
Further studies were next carried out to confirm and extend these unusual observations. Melting studies showed that neither of the constituent ligands (LΔ1a and LΛ1a, used for synthesizing Δ1a and Λ1a) could stabilize Tel22 G-quadruplex (Table 1). These data confirmed that the selectivity of Δ1a for Tel22 G-quadruplex originated from the three-dimensional structure of ligand/metal assembly. Further, Tm of Tel22 in low Na+ concentration (10 mM) was shown to be unaffected in the presence or absence of Δ1a/Λ1a, thus excluding the possible effect of cation concentration (Supplementary Figure S3B). The selectivity of the enantiomers to i-motif and double-strand DNA (formed by Tel22 and i-motif) was also found to be negligible (Table 1 and Supplementary Figure S3), demonstrating that Δ1a not only recognizes G-quadruplex but also discriminates against other DNA motifs. Similarly, the stabilization effect of the enantiomers to calf thymus DNA (ct-DNA) and Tel22 was compared. After addition of Δ1a, Tm of Tel22 G-quadruplex increased by ∼10.4°C, however just ∼1.4°C increase for ct-DNA (Supplementary Figure S4). Therefore, from the aspect of stabilization effect, Δ1a had a 7.4-fold selectivity for Tel22 DNA against ct-DNA.
Table 1. Stabilization effects of different ligands on Tel22 G-quadruplex DNA, i-motif DNA and dsDNAa.
| Δ T m (°C) | |||
|---|---|---|---|
| Ligand | G-quadruplex | i-motif | dsDNA |
| LΔ1a | 0.2 ± 0.3 | −0.1 ± 0.2 | 0.3 ± 0.1 |
| LΛ1a | 0.1 ± 0.1 | 0.1 ± 0.4 | 0.4 ± 0.2 |
| Δ1a | 10.8 ± 0.3 | 0.2 ± 0.3 | 0.3 ± 0.2 |
| Λ1a | 0.2 ± 0.2 | 0.1 ± 0.1 | 0.3 ± 0.4 |
aThe dsDNA is duplex DNA formed by complementary sequence of Tel22 G-quadruplex DNA and i-motif DNA. LΔ1a and LΛ1a are constituent ligands that used for synthesizing Δ1a and Λ1a. Melting assays were carried out in 10 mM Tris buffer containing 10 mM KCl at pH 7.2 or pH 5.5 (only for i-motif DNA). DNA is 3 μM in strand. The concentration of the Ligands is equivalent with corresponding DNA. The values were the average of three independent measurements.
For further studying the structural effect of 1a on the chiral selectivity for hybrid G-quadruplex of Tel22, the interactions of other enantiomeric metallohelices (1b and 2a) with Tel22 were compared (Supplementary Figures S5 and 6). Although all enantiomers of 1b and 2a had a comparable size and identical charge with 1a, none of them exhibited obvious stabilization effect on either hybrid or antiparallel G-quadruplex of Tel22. It is particularly interesting when compared with the enantiomers of 1b because the difference between 1b and 1a is just additional hydroxyl groups at the termini. This suggests that the structure of end groups of 1a may play an important role in chiral recognization of human telomeric hybrid G-quadruplex. Considering the large size of 1a (length ∼2.2 nm), we propose that the end of 1a could be the binding site for Tel22.
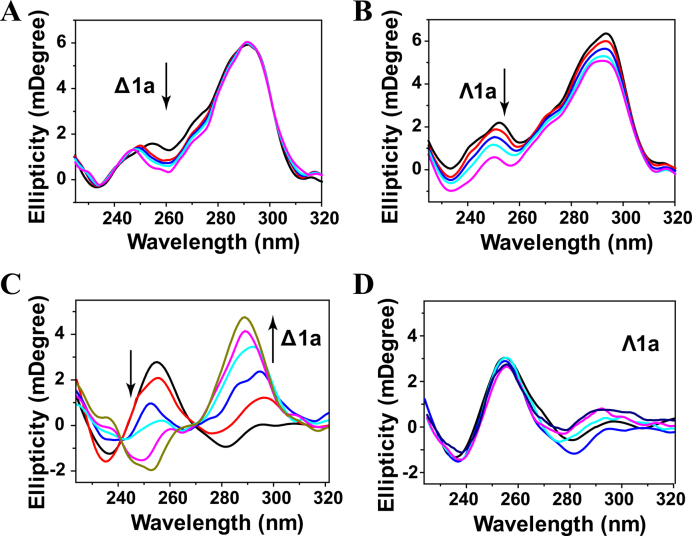
The conformation changes of Tel22 induced by Δ1a and Λ1a were determined by CD studies. In K+ buffer, Tel22 alone formed hybrid G-quadruplex characterized by two positive bands at 294 and 255 nm and a negative band around 235 nm. After addition of Δ1a, no obvious signal change was observed at band of 294 and 235 nm, while a notable shift was observed at trough of 255 nm (Figure 2A and Supplementary Figure S7). In contrast, Λ1a induced a hypochromism at band of 294 and 255 nm (Figure 2B). These results implied that, both Δ1a and Λ1a disturbed slightly the conformation of Tel22, but could not alter the hybrid conformation. Therefore, CD signal of Tel22 displayed different change after treated with different enantiomer. This implied that Δ1a and Λ1a might employ different binding mode when interacting with Tel22. In addition, the CD spectra of Tel22 in Na+ buffer were also determined in the absence and presence of the enantiomers (Supplementary Figure S8). No new bands or band shifts were observed, indicating that both enantiomers showed no impact on the antiparallel conformation of Tel22.
Figure 2.
CD titration of Tel22 with Δ1a (A) and Λ1a (B) in 10 mM KCl, 10 mM Tris buffer, pH = 7.2. Enantiomer was varied from 0 to 2 μM. CD titration of Tel22 with Δ1a (C) and Λ1a (D) in 10 mM Tris buffer, pH = 7.2. The concentration of the enantiomer was varied fom 0.5 to 4μM.
Inducing human telomeric G-quadruplex formation is important for telomeric DNA to exert the desired biological function. Thus, we next studied whether the enantiomer could induce G-quadruplex formation under Na+ or K+-deficient conditions. As shown in Figure 2C, Tel22 was in an unfolded single strand DNA in the absence of Na+ or K+. After titration with Δ1a, the intensity of the CD band at 256 nm decreased gradually from positive to negative, and at the same time, a distinct increase at 288 nm and an emergence of a shoulder peak at 270 nm were observed. This induced CD spectrum was similar to that of the hybrid G-quadruplex in K+ buffer, indicating that Δ1a can induce the formation of hybrid G-quadruplex under Na+ or K+-deficient conditions (37). In contrast, no inducing process was observed after adding of Λ1a (Figure 2D). These results further supported the chiral selectivity of Δ1a for human telomeric hybrid G-quadruplex.
Binding constant and binding mode
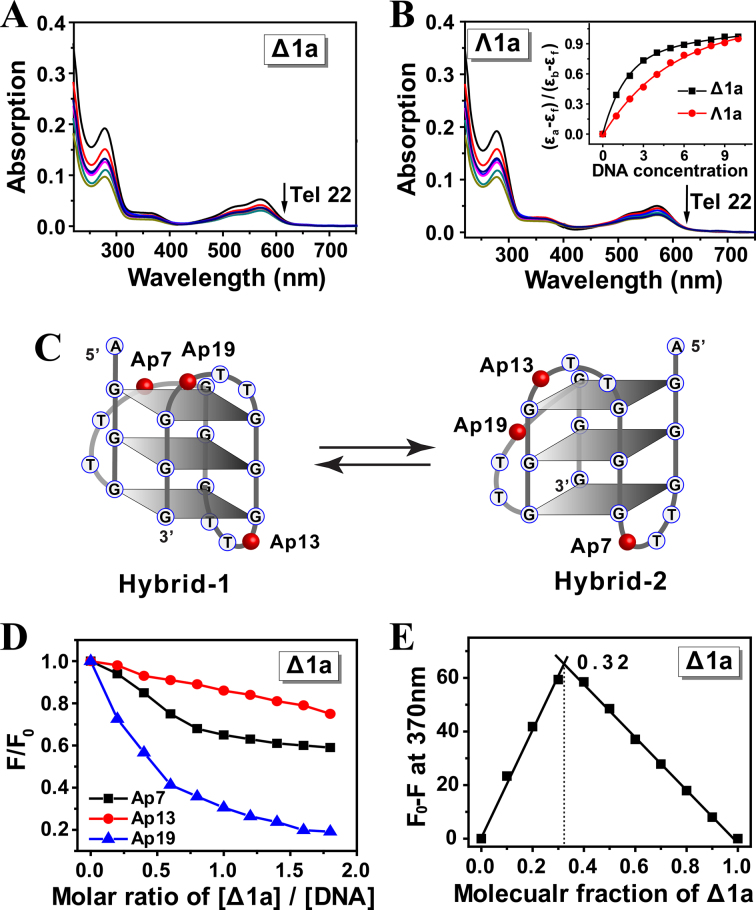
Absorption changes of Δ1a/Λ1a upon addition of Tel22 DNA were investigated by titration experiments. The UV spectra of the enantiomers were monitored on addition of aliquots of preformed Tel22 hybrid G-quadruplex DNA (Figure 3A and B). When treated with DNA, both Δ1a and Λ1a showed notable hypochromism at band of 280 and 570 nm. However, obviously, a stronger hypochromism was observed for Δ1a rather than Λ1a in the presence of low concentration of DNA (Figure 3B, Inset). For example, when treated with 3 μM DNA, Δ1a yielded an absorbance hypochromism of 33.8%. However, just 19.5% was observed for Λ1a. This suggested that Δ1a could bind stronger to Tel22 than Λ1a. According to previous reported methods (34,35), the binding constant of Δ1a with Tel22 was estimated to be 2.8 × 107 M−1, whereas 4.5 × 105 M−1 for Λ1a (Table 2). This indicates that Δ1a can specifically bind to Tel22, however, the interaction of Λ1a with Tel22 may be just non-specific electrostatic interaction. It should be also noted that, upon addition of high enough concentration of Tel22, the two enantiomers displayed a comparable hypochromism. These results were understandable because both enantiomers were highly positive-charged and DNA was highly negative-charged. Thus, in the presence of high concentration of DNA, mutiple non-specific electrostatic interactions and stacking between DNA and Δ1a and Λ1a were occurred, which could cause hypochromism.
Figure 3.
Absorption spectra of Δ1a (A) and Λ1a (B) in the presence of Tel 22 DNA. Enantiomers was 5 μM, and Tel22 was varied from 0.5 to 10 μM. Arrow showed the absorption change of the enantiomer along with the addition of Tel22. Inset: plot of (εa-εf)/(εb-εf) versus concentration of Tel22. The assays were measured in 10 mM Tris buffer containing 10 mM KCl, pH = 7.2. (C) Schematic illustration of the individual 2-Ap position in Tel22 and the two types of Tel22 G-quadruplex conformations. (D) Plot of normalized fluorescence intensity at 370 nm of 2-Ap individually labeled Tel22 versus molar ratio of [Δ1a]/[DNA] in 10 mM KCl, 10 mM Tris buffer, pH = 7.2. 2-Ap labeled Tel22 was 0.5 μM in strand. (E) Job plot for complexation of Δ1a and Ap19-Tel22 in 10 mM KCl, 10 mM Tris buffer, pH = 7.2. [Δ1a]+[Ap19-Tel22] = 0.3 μM.
Table 2. Binding constants of Tel22 DNA with Δ1a and Λ1aa.
| Complex | 1 K a (M−1) | 2 K a (M−1) | 3 K a (M−1) |
|---|---|---|---|
| Δ1a | 2.8(±0.2) × 107 | 3.5(±0.5) × 107 | 0.92(±0.4) × 107 |
| Λ1a | 4.5(±0.8) × 105 | 6.9(±07) × 105 | 1.81(±0.3) × 105 |
aBinding constant 1Ka was measured by UV titration method. Binding constant 2Ka was measured by fluorescence titration method. Binding constant 3Ka was measured by ITC method. The values were the average of three independent measurements.
Modifying 2-aminopurine (2-Ap) in different loops of quadruplex DNA is a widely used strategy to estimate the binding mode of DNA with ligand (38). Herein, for exploring how the two enantiomers binding to hybrid Tel22 G-quadruplex, fluorescence experiments were carried out using Tel22 with different 2-Ap substitutions at adenine residue position 7, 13 and 19 (Figure 3C). First, CD studies showed that 2-Ap modified Tel22 had almost an identical CD feature with the original unmodified Tel22, indicating that 2-Ap modified Tel22 had the same conformation as the wild-type Tel22 (Supplementary Figure S9). In addition, after interacting with Δ1a, the CD signal of Tel22 was slightly changed, indicating that Δ1a did not disrupt the structure of Tel22 and Tel22 maintained hybrid conformation (Figure 2A). These offered the prerequisite for investigating the binding mode by using 2-Ap fluorescence assay. As shown in Figure 3D, addition of Δ1a decreased the 2-Ap fluorescence significantly and the order was Ap19 > Ap7 > Ap13. These indicated that, after binding with Tel22, Δ1a had a close contact with Ap19 and Ap7, not with Ap13. As previously reported, in K+ solution, Tel22 adopts two distinct (3 + 1) topologies: hybrid-1 and hybrid-2 form (13). The two forms are coexisted in a certain state of equilibrium, as shown in Figure 3C. Based on the results of 2-Ap fluorescence studies, we proposed that Δ1a preferably bound to hybrid-1 form rather than hybrid-2 form and Δ1a could bind to the end G-quartet near 5΄ end of hybrid-1 form by external stacking mode. This binding mode can well response to the order of Ap19 > Ap7 > Ap13. The binding mode can be benefited from the compatible size between Δ1a (diameter ∼1.2 nm) and G-quartet (length ∼1.4 nm, width ∼1.1 nm) (39).
According to Job plot, a 1:2 binding ratio was found between Δ1a and Tel22 G-quadruplex, indicating that two Tel22 bind with one Δ1a (Figure 3E). The binding stoichiometry was also confirmed by ITC studies (Supplementary Figure S11 and Table 3). In our previous work, we reported that a di-nickel helicate, [Ni2L3]4+, exhibited 1:1 binding with Tel22. We propose that two aspects may cause different binding ratio for Δ1a and [Ni2L3]4+. First, Δ1a (length ∼2.2 nm, diameter ∼1.2 nm) has a larger size than [Ni2L3]4+ (length ∼1.8 nm, diameter ∼0.8 nm). Second, there are six phenyls in middle part of [Ni2L3]4+, where provide a tight stacking platform for Tel22 G-quadruplex by π-π interaction. Thus, Tel22 possibly locates in the middle part of [Ni2L3]4+. In contrast, there are only three phenyls in the middle part of Δ1a, which may not be suitable for providing tight π-stacking with Tel22. Thereby, Tel22 may bind to the two identical ends of Δ1a, thus resulting in 1:2 binding ratio.
Table 3. Binding stoichiometry and thermodynamic parameters for the interaction of Tel22 DNA with Δ1a and Λ1aa.
| Δ1a | Λ1a | |
|---|---|---|
| n | 0.5 ± 0.1 | 0.6 ± 0.2 |
| ΔG025 (kcal mol −1) | −9.4 ± 0.4 | −7.2 ± 0.5 |
| ΔH0 (kcal mol −1) | −15.4 ± 1.4 | −11.8 ± 0.7 |
| TΔS0 (kcal mol−1 K −1) | −6.0 ± 0.4 | −4.6 ± 0.5 |
aAll data were derived from ITC experiments. ΔH0 and n (stoichiometry) was directly obtained from ITC. ΔG025 was obtained from the relation ΔG0 = -RTlnKa (Ka was listed in Table 1). TΔS0 obtained from the relation TΔS0 = ΔH0-ΔG0.
Non-linear least-squares analysis of the fluorescence titration data of Ap19 by Δ1a yielded a binding constant of 3.5 × 107 M−1 (Table 2). In contrast, for Λ1a, the binding constant was 6.9 × 105 M−1 (Supplementary Figure S10). The results were in good agreement with ITC and UV titration data (Table 2). In addition, the thermodynamic parameters for Tel22 binding with Δ1a were estimated by ITC and the data were listed in Table 3. The results indicated that Δ1a binding to Tel22 G-quadruplex DNA was favorable enthalpy-driven.
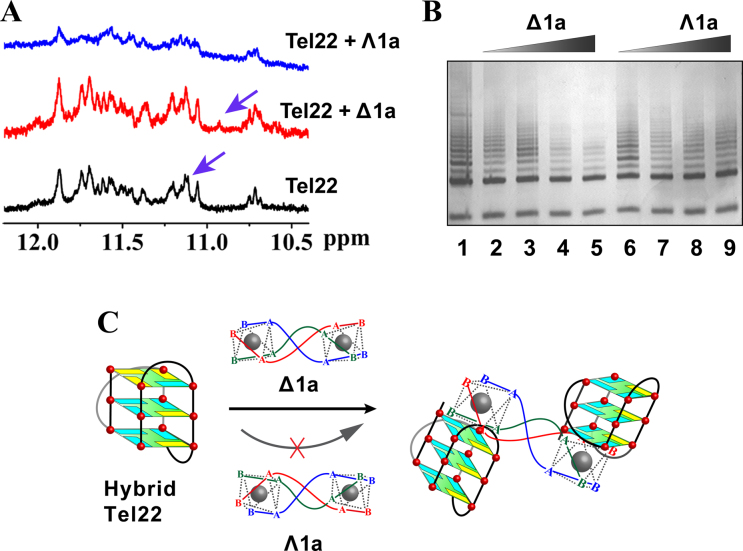
The interactions between the enantiomers and Tel22 were further studied by 1H-NMR experiments (Figure 4A and Supplementary Figure S12). In K+ buffer the guanine imino protons of the G-quadruplex were observed at characteristic chemical shifts (10–12 ppm) (14,16). With addition of Δ1a, the signal at 11.11 ppm was shifted to 10.9 ppm, indicating a strong interaction with the G-quartets (40–42). In contrast, addition of Λ1a just resulted in broadening of the signals, implying a labile interaction (39). This further supportsΔ1a specificically binds to Tel22 G-quadruplex. However, unfortunately, as we mentioned above (43), Tel22 adopts two hybrid conformations, hybrid-1 and hybrid-2, in K+ condition and the two forms are coexisted. This conformational diversity prevented us to clearly resolve the 1H-NMR of Tel22 upon binding with Δ1a/Λ1a. The above 2-Ap titration assays indicated that Δ1a preferred binding to hybrid-1 G-quadruplex of Tel22. As previously reported, another human telomeric DNA sequence, HT, can just form hybrid-1 G-quadruplex in K+ buffer (15). Therefore, we studied the interaction between HT and Δ1a by NMR. As shown in Supplementary Figures S13 and 14, after adding Δ1a, HT guanine imino protons peaks displayed obvious chemical shifts. These results confirmed the strong binding between Δ1a and hybrid-1 G-quadruplex. The imino peaks corresponding to G3 and G5 underwent the largest chemical-shift changes upon Δ1a binding, indicating Δ1a preferentially binding to the top G-quartet (Supplementary Figure S15), in good agreement with the results of 2-Ap titration assays (44). The NOESY data of HT binding with Δ1a was shown in Supplementary Figure S16. It can be seen that imino proton peaks in G3 and G5 are shifted remarkably, further supporting the strong interaction between HT and Δ1a. However, it should be pointed out that we can not provide a clear assignment of the NOESY data because of the complexity.
Figure 4.
(A) 1H NMR spectra of Tel22 in the absence and presence of Δ1a/Λ1a. The assays were measured in 10 mM Tris–KCl buffer containing 10% D2O. (B) Telomerase inhibition by Δ1a/Λ1a by using TRAP assay. Line 1: no enantiomer; Line 2–5: PCR products in the presence of 0.2, 0.5, 1, 2 μM Δ1a, respectively; Line 6–9: PCR products in the presence of 0.2, 0.5, 1, 2 μM Λ1a, respectively. (C) Schematic illustration of the chiral enantiomer selectively interacts with human telomeric hybrid G-quadruplex DNA.
On the basis of all the above results, the binding between Tel22 hybrid G-quadruplex and Δ1a shows the following characteristics. First, Δ1a can stack on the end G-quartet of the hybrid G-quadruplex with a preference for hybrid-1 form. Second, the terminal structure of Δ1a has strong influence on its binding to G-quadruplex. Third, one Δ1a binds with two Tel22 G-quadruplex units. A possible binding mode was shown in Figure 4C.
Discrimination between G-quadruplexes
To further explore the selectivity of the metallohelicates for human telomeric G-quadruplex, we studied the interactions of Δ1a/Λ1a with tetrahymena and oxytricha telomeric G-quadruplexes. CD spectra of the tetrahymena and oxytricha telomeric G-quadruplexes were not substantially affected by the metallohelicate enantiomers, indicating weak conformation changes of the G-quadruplexes (Supplementary Figure S17). Melting studies showed that no matter in Na+ or K+ condition, both Δ1a and Λ1a could not stabilize the two telomeric G-quadruplexes (Table 4 and Supplementary Table S1). These results confirmed the chiral selectivity of Δ1a for human telomeric hybrid G-quadruplex.
Table 4. Stabilization effect of Δ1a and Λ1a on different telomeric G-quadruplex DNA in K+ containing buffera.
| Telomeric DNA | Structure | DNA Tm (°C) | +Δ1a ΔTm (°C) | +Λ1a ΔTm (°C) |
|---|---|---|---|---|
| Human (Tel22) | Hybrid | 54.6 ± 0.3 | 10.8 ± 0.3 | 0.2 ± 0.2 |
| Oxytrichab | Antiparallel | 61.6 ± 0.2 | −5.0 ± 0.5 | −4.7 ± 0.4 |
| Tetrahymenab | Hybrid | 58.2 ± 0.1 | 0.7 ± 0.3 | 0.8 ± 0.3 |
| Tel22-TTT | Hybrid | 64.8 ± 0.3 | 0.2 ± 0.3 | 0.4 ± 0.2 |
aMelting assays were carried out in 10 mM Tris buffer containing 10 mM KCl at pH 7.2. DNA is 3 μM in strand. The concentration of the Ligands is equivalent with corresponding DNA. The values were the average of three independent measurements.
bThe Tm of the was measured in 1 mM K+ containing buffer.
It should be emphasized that, although both Tel22 and tetrahymena telomeric G-quadruplex adopted hybrid conformation in K+ condition, Δ1a cannot stabilize tetrahymena telomeric G-quadruplex (Table 4). This indicated that the topology of Tel22 was not the only determinant for its selectivity of Δ1a. Previous studies have demonstrated that the loop sequence of G-quadruplex DNA plays a crucial role in DNA binding (45,46). Evidently, a significant difference between Tel22 and tetrahymena telomeric G-quadruplex is the adenine base in the loop region. Thus, to investigate how the loop sequences influence the binding behavior of Tel22, we substituted the TTA loops with TTT (abbreviated to Tel22-TTT), in which the adenine was replaced by a thymine. CD indicated that Tel22-TTT adopted the same hybrid conformation as the wild-type Tel22 (Supplementary Figure S9B). However, intriguingly, neither Δ1a nor Λ1a can stabilize Tel22-TTT (Table 4). This indicates that adenine in the loop of Tel22 plays important roles in chiral selection for hybrid Tel22 G-quadruplex. Thus, based on discussion mentioned above, both the hybrid topology of G-quadruplex and TTA loop sequence of Tel22 are crucial for determination the chiral recognition.
Inhibition of telomerase activity
Human telomeric G-quadruplex is associated with the activity regulation of telomerase, which is activated in 80–90% of human tumors and is considered to be a specific target for cancer therapy (47–51). Our results indicate that Δ1a can selectively and strongly bind to human telomeric hybrid G-quadruplex DNA. This intriguing finding promoted us to investigate telomerase inhibition effect of Δ1a by using TRAP assay. As shown in Figure 4B, Δ1a can inhibit telomerase efficiently with an IC50 of 600 nM. In contrast, Λ1a shows weak inhibition on telomerase. These results encouraged us to examine the behavior of tumor cell treated with Δ1a and Λ1a. Further studies are undergoing and will be reported in due course.
CONCLUSION
In summary, our water-stable metallohelices, synthesized directly via diastereoselective self-assembly, are capable of discriminating human telomeric hybrid G-quadruplex with high chiral selectivity. One enantiomer, Δ1a, selectively stabilizes hybrid human telomeric G-quadruplex and shows no effect on the antiparallel conformer. In contrast, the mirror image compound Λ1a showed no stabilization on both hybrid and antiparallel G-quadruplex. To the best of our knowledge, this is the first example of one pair of enantiomers showing such distinct chiral selectivity for human telomeric hybrid G-quadruplex. In addition, only Δ1a was found to effectively inhibit telomerase activity. These findings provide new insights into rational design and synthesis of specific G-quadruplex targeting metal complexes.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge financial support from 973 Project and NSFC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
973 Project [2012CB720602]; National Natural Science Foundation of China (NSFC) [21210002, 21431007, 21533008, 21572216]. Funding for open access charge: 973 Project; NSFC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008; 37:1375–1384. [DOI] [PubMed] [Google Scholar]
- 2. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lam E.Y.N., Beraldi D., Tannahill D., Balasubramanian S.. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013; 4:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Y., Komiyama M.. Evidence for G-Quadruplex DNA in human cells. Chembiochem. 2013; 14:927–928. [DOI] [PubMed] [Google Scholar]
- 5. Yu H., Gu X., Nakano S.-I., Miyoshi D., Sugimoto N.. Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 2012; 134:20060–20069. [DOI] [PubMed] [Google Scholar]
- 6. Liu H.-Y., Zhao Q., Zhang T.-P., Wu Y., Xiong Y.-X., Wang S.-K., Ge Y.-L., He J.-H., Lv P., Ou T.-M. et al. . Conformation selective antibody enables genome profiling and leads to discovery of parallel G-quadruplex in human telomeres. Cell Chem. Biol. 2016; 23:1261–1270. [DOI] [PubMed] [Google Scholar]
- 7. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tarsounas M., Tijsterman M.. Genomes and G-Quadruplexes: for better or for worse. J. Mol. Biol. 2013; 425:4782–4789. [DOI] [PubMed] [Google Scholar]
- 9. Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burge S., Parkinson Gn Fau-Hazel P., Hazel P, Fau-Todd A.K., Todd Ak Fau-Neidle S., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyoshi D., Karimata H., Sugimoto N.. Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. 2005; 44:3740–3744. [DOI] [PubMed] [Google Scholar]
- 12. Koirala D., Ghimire C., Bohrer C., Sannohe Y., Sugiyama H., Mao H.. Long-Loop G-Quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013; 135:2235–2241. [DOI] [PubMed] [Google Scholar]
- 13. Hu M.H., Chen X., Chen S.B., Ou T.M., Yao M., Gu L.Q., Huang Z.S., Tan J.H.. A new application of click chemistry in situ: development of fluorescent probe for specific G-quadruplex topology. Sci. Rep. 2015; 5:17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D.. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006; 34:2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J.. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-Quadruplex scaffold. J. Am. Chem. Soc. 2006; 128:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dai J., Carver M., Punchihewa C., Jones R.A., Yang D.. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007; 35:4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K + solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray R.D., Chaires J.B.. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008; 36:4191–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamon F., Largy E., Guédin-Beaurepaire A., Rouchon-Dagois M., Sidibe A., Monchaud D., Mergny J.-L., Riou J.-F., Nguyen C.-H., Teulade-Fichou M.-P.. An acyclic oligoheteroaryle that discriminates strongly between diverse G-Quadruplex topologies. Angew. Chem. Int. Ed. 2011; 50:8745–8749. [DOI] [PubMed] [Google Scholar]
- 20. Nicoludis J.M., Miller S.T., Jeffrey P.D., Barrett S.P., Rablen P.R., Lawton T.J., Yatsunyk L.A.. Optimized end-stacking provides specificity of N-Methyl mesoporphyrin IX for Human telomeric G-Quadruplex DNA. J. Am. Chem. Soc. 2012; 134:20446–20456. [DOI] [PubMed] [Google Scholar]
- 21. Nicoludis J.M., Barrett S.P., Mergny J.-L., Yatsunyk L.A.. Interaction of human telomeric DNA with N- methyl mesoporphyrin IX. Nucleic Acids Res. 2012; 40:5432–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howson S.E., Bolhuis A., Brabec V., Clarkson G.J., Malina J., Rodger A., Scott P.. Optically pure, water-stable metallo-helical ‘flexicate’ assemblies with antibiotic activity. Nat. Chem. 2012; 4:31–36. [DOI] [PubMed] [Google Scholar]
- 23. Kaner R.A., Scott P.. Metallohelices: potential mimetics of α-helical peptides in cancer treatment?. Future Med. Chem. 2015; 7:1–4. [DOI] [PubMed] [Google Scholar]
- 24. Faulkner A.D., Kaner R.A., AbdallahQasem M.A., Clarkson G., Fox D.J., Gurnani P., Howson S.E., Phillips R.M., Roper D.I., Simpson D.H. et al. . Asymmetric triplex metallohelices with high and selective activity against cancer cells. Nat. Chem. 2014; 6:797–803. [DOI] [PubMed] [Google Scholar]
- 25. Meistermann I., Moreno V., Prieto M.J., Moldrheim E., Sletten E., Khalid S., Rodger P.M., Peberdy J.C., Isaac C.J., Rodger A. et al. . Intramolecular DNA coiling mediated by metallo-supramolecular cylinders: Differential binding of P and M helical enantiomers. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5069–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oleksi A., Blanco A.G., Boer R., Usón I., Aymamí J., Rodger A., Hannon M.J., Coll M.. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. 2006; 45:1227–1231. [DOI] [PubMed] [Google Scholar]
- 27. Yu H., Wang X., Fu M., Ren J., Qu X.. Chiral metallo-supramolecular complexes selectively recognize human telomeric G-quadruplex DNA. Nucleic Acids Res. 2008; 36:5695–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao C., Wu L., Ren J., Xu Y., Qu X.. Targeting human telomeric higher-order DNA: dimeric G-Quadruplex units serve as preferred binding site. J. Am. Chem. Soc. 2013; 135:18786–18789. [DOI] [PubMed] [Google Scholar]
- 29. Xu B., Zhao C., Chen Y., Tateishi-Karimata H., Ren J., Sugimoto N., Qu X.. Methyl substitution regulates the enantioselectivity of supramolecular complex binding to human telomeric G-Quadruplex DNA. Chem. Eur. J. 2014; 20:16467–16472. [DOI] [PubMed] [Google Scholar]
- 30. Georgiades S.N., Abd Karim N.H., Suntharalingam K., Vilar R.. Interaction of metal complexes with G-Quadruplex DNA. Angew. Chem. Int. Ed. 2010; 49:4020–4034. [DOI] [PubMed] [Google Scholar]
- 31. Li M., Howson S.E., Dong K., Gao N., Ren J., Scott P., Qu X.. Chiral metallohelical complexes enantioselectively target amyloid β for treating Alzheimer's disease. J. Am. Chem. Soc. 2014; 136:11655–11663. [DOI] [PubMed] [Google Scholar]
- 32. Schlamadinger D.E., Kats D.I., Kim J.E.. Quenching of tryptophan fluorescence in unfolded cytochrome c: a biophysics experiment for physical chemistry students. J. Chem. Educ. 2010; 87:961–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Puchalski M.M., Morra M.J., von Wandruszka R.. Assessment of inner filter effect corrections in fluorimetry. Fresenius J. Anal. Chem. 1991; 340:341–344. [Google Scholar]
- 34. Nair R.B., Teng E.S., Kirkland S.L., Murphy C.J.. Synthesis and DNA-Binding properties of [Ru(NH3)4dppz]2+. Inorg. Chem. 1998; 37:139–141. [DOI] [PubMed] [Google Scholar]
- 35. Carter M.T., Rodriguez M., Bard A.J.. Voltammetric studies of the interaction of metal chelates with DNA. 2. Tris-chelated complexes of cobalt(III) and iron(II) with 1,10-phenanthroline and 2,2΄-bipyridine. J. Am. Chem. Soc. 1989; 111:8901–8911. [Google Scholar]
- 36. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 37. Zhao C., Geng J., Feng L., Ren J., Qu X.. Chiral Metallo–Supramolecular complexes selectively induce human telomeric G-Quadruplex formation under salt-deficient conditions. Chem. Eur. J. 2011; 17:8209–8215. [DOI] [PubMed] [Google Scholar]
- 38. Kimura T., Kawai K., Fujitsuka M., Majima T.. Detection of the G-quadruplex-TMPyP4 complex by 2-aminopurine modified human telomeric DNA. Chem. Commun. 2006; 4:401–402. [DOI] [PubMed] [Google Scholar]
- 39. De Cian A., DeLemos E., Mergny J.-L., Teulade-Fichou M.-P., Monchaud D.. Highly efficient G-Quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007; 129:1856–1857. [DOI] [PubMed] [Google Scholar]
- 40. Gavathiotis E., Heald R.A., Stevens M.F.G., Searle M.S.. Recognition and stabilization of quadruplex DNA by a potent new telomerase inhibitor: NMR studies of the 2:1 complex of a pentacyclic methylacridinium cation with d(TTAGGGT)4. Angew. Chem. Int. Ed. 2001; 40:4749–4751. [DOI] [PubMed] [Google Scholar]
- 41. Haudecoeur R., Stefan L., Denat F., Monchaud D.. A model of smart G-Quadruplex ligand. J. Am. Chem. Soc. 2013; 135:550–553. [DOI] [PubMed] [Google Scholar]
- 42. Jaumot J., Gargallo R.. Experimental methods for studying the interactions between G-Quadruplex structures and ligands. Curr. Pharm. Des. 2012; 18:1900–1916. [DOI] [PubMed] [Google Scholar]
- 43. Singh V., Azarkh M., Exner T.E., Hartig J.S., Drescher M.. Human telomeric quadruplex conformations studied by pulsed EPR. Angew. Chem. Int. Ed. 2009; 48:9728–9730. [DOI] [PubMed] [Google Scholar]
- 44. Li Q., Xiang J., Li X., Chen L., Xu X., Tang Y., Zhou Q., Li L., Zhang H., Sun H. et al. . Stabilizing parallel G-quadruplex DNA by a new class of ligands: two non-planar alkaloids through interaction in lateral grooves. Biochimie. 2009; 91:811–819. [DOI] [PubMed] [Google Scholar]
- 45. Shi S., Xu J.-H., Gao X., Huang H.-L., Yao T.-M.. Binding behaviors for different types of DNA G-Quadruplexes: Enantiomers of [Ru(bpy)2(L)]2+ (L=dppz, dppz-idzo). Chem. Eur. J. 2015; 21:11435–11445. [DOI] [PubMed] [Google Scholar]
- 46. Yu H., Zhao C., Chen Y., Fu M., Ren J., Qu X.. DNA Loop Sequence as the determinant for chiral supramolecular compound G-Quadruplex selectivity. J. Med. Chem. 2009; 53:492–498. [DOI] [PubMed] [Google Scholar]
- 47. Riou J.F., Guittat L., Mailliet P., Laoui A., Renou E., Petitgenet O., Megnin-Chanet F., Helene C., Mergny J.L.. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q., Liu J.Q., Chen Z., Zheng K.W., Chen C.Y., Hao Y.H., Tan Z.. G-quadruplex formation at the 3΄ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011; 39:6229–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henderson E., Hardin C.C., Walk S.K., Tinoco I. Jr, Blackburn E.H.. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987; 51:899–908. [DOI] [PubMed] [Google Scholar]
- 50. Xu Y. Chemistry in human telomere biology: structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011; 40:2719–2740. [DOI] [PubMed] [Google Scholar]
- 51. Tan J.H., Ou T.M., Hou J.Q., Lu Y.J., Huang S.L., Luo H.B., Wu J.Y., Huang Z.S., Wong K.Y., Gu L.Q.. Isaindigotone derivatives: a new class of highly selective ligands for telomeric G-quadruplex DNA. J. Med. Chem. 2009; 52:2825–2835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.