Abstract
Little is known about the function of most non-coding RNAs (ncRNAs). The majority of long ncRNAs (lncRNAs) is expressed at very low levels and it is a matter of intense debate whether these can be of functional relevance. Here, we identified lncRNAs regulating the viability of lung cancer cells in a high-throughput RNA interference screen. Based on our previous expression profiling, we designed an siRNA library targeting 638 lncRNAs upregulated in human cancer. In a functional siRNA screen analyzing the viability of lung cancer cells, the most prominent hit was a novel lncRNA which we called Viability Enhancing LUng Cancer Transcript (VELUCT). In silico analyses confirmed the non-coding properties of the transcript. Surprisingly, VELUCT was below the detection limit in total RNA from NCI-H460 cells by RT-qPCR as well as RNA-Seq, but was robustly detected in the chromatin-associated RNA fraction. It is an extremely low abundant lncRNA with an RNA copy number of less than one copy per cell. Blocking transcription with actinomycin D revealed that VELUCT RNA was highly unstable which may partially explain its low steady-state concentration. Despite its extremely low abundance, loss-of-function of VELUCT with three independent experimental approaches in three different lung cancer cell lines led to a significant reduction of cell viability: Next to four individual siRNAs, also two complex siPOOLs as well as two antisense oligonucleotides confirmed the strong and specific phenotype. In summary, the extremely low abundant lncRNA VELUCT is essential for regulation of cell viability in several lung cancer cell lines. Hence, VELUCT is the first example for a lncRNA that is expressed at a very low level, but has a strong loss-of-function phenotype. Thus, our study proves that at least individual low-abundant lncRNAs can play an important functional role.
INTRODUCTION
Protein-coding genes were long assumed to be the main and probably only molecular drivers in a cell and RNA molecules were viewed as mediator molecules and servants for processes in protein synthesis. However, deep sequencing methods revealed that a large part of the human genome (∼75%) is transcribed, whereas only 1.5% encode for proteins (1). Long ncRNAs (lncRNAs) form a highly diverse non-coding RNA (ncRNA) class with >200 nucleotides in length that lack an open reading frame of significant length (2). The number of lncRNA genes in the human genome is still increasing, but recent analyses suggest at least numbers similar to protein-coding genes (3). The expression of lncRNAs is highly regulated and depends on the developmental stage (4), on the tissue (5,6) and on cell subtypes (7,8). So far, only a small fraction of lncRNAs is functionally characterized, but several examples are shown to play a critical role in physiological and pathological processes such as cancer. Transcripts such as HOTAIR (HOx Transcript Antisense Intergenic RNA) or MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) are upregulated in lung cancer (9,10) and associated with enhanced proliferation, metastasis and poor prognosis (9–12). However, knowledge is mostly limited to the most abundant lncRNAs. Contrarily, the majority of lncRNAs is of very low abundance, with many lncRNAs having a copy number of even lower than one per cell (13,14). Thus, an intense debate arose whether any of these many low abundant lncRNAs could play important physiological roles in a cell. Opponents of this theory argue that most of the low abundant lncRNAs are non-functional ‘junk’ (15,16), i.e. spurious RNAs that might derive from leaky transcription. Those junk transcripts might be rapidly removed because of quality control mechanisms resulting in their low abundance (17). Proponents of this theory argue in contrast, that the majority of lncRNAs—although expressed at a low level—could indeed play an important role in a cell (18,19). Small amounts of RNAs might be sufficient to trigger downstream effects, e.g. if acting directly on the genome at a unique allele.
Here, we identify lncRNAs that regulate the viability in lung cancer cell lines in a high-throughput RNA interference screen. We identified the novel lncRNA VELUCT (Viability Enhancing LUng Cancer Transcript) which was extremely low abundant and only reproducibly detectable in the chromatin-associated RNA fraction. Nonetheless, the functional importance of this transcript for cell viability and proliferation was validated using multiple independent silencing approaches. Thus, VELUCT is an example for a lncRNA that is expressed at a very low level, but has a strong phenotype upon knockdown.
MATERIALS AND METHODS
Cell lines and actinomycin D treatment
NCI-H460, NCI-H1944 and NCI-H1437 lung cancer cells were propagated in RPMI + 10% FCS. H1944 and H1437 were purchased from ATCC. H460 were authenticated using Multiplex Cell Authentication by Multiplexion (Heidelberg, Germany) as described recently (20). The SNP profiles matched known profiles or were unique. Cells were regularly tested for mycoplasma. For actinomycin D (actD) treatment, 2 × 106 H460 cells were seeded in a 10 cm dish and incubated for 24 h. The medium was aspirated and 7.5 ml complete medium was added containing 10 μg/ml actD (resuspended in DMSO) or the same volume of DMSO as a control.
siRNA Library
The library contained approx. 3100 single Silencer Select siRNAs (Life Technologies) targeting 638 lncRNAs that were upregulated in lung, liver and breast cancer samples according to previous studies. The library was arrayed in white 384-well plates using a Biomek FX200 liquid handling system (Beckman Coulter). Each well contained 5 μl of 300 nM siRNA. Column 23 and 24 of all screening plates contained three positive siRNA controls (siCOPB2, siKIF11, siPLK1; each in duplicates per plate) and non-targeting negative siRNA controls (NC_1, NC_2, NC_3; each in quadruplicates per plate).
Transfection with silencing reagents
Cells were reverse transfected with 10 or 30 nM RNAi reagents (siRNAs, siPOOLs or ASOs) in different plate formats. H460 cells were transfected with Dharmafect1 (Dharmacon); H1944 and H1437 with RNAiMAX (Life Technologies). For transfection in 384-well plates, 0.05 μl transfection reagent per well were diluted in 4.95 μl RPMI (Sigma) and incubated for 10 min (for siPOOLs (siTOOLs Biotech) and siRNAs) or 5 min (for ASOs (Exiqon)). 10 μl RPMI were added and incubated with 5 μl silencing reagent for 30 min (for siPOOLs and siRNAs) or 20 min (for ASOs). Cells (1000 cells/well) in 30 μl complete medium were added and incubated at 37°C, 5% CO2. Volumes were multiplied by two for transfection in 96-well plates. If subcellular fractionation of transfected cells was performed, 3 × 106 cells were transfected in a 10 cm dish (7.5 ml final volume) with 15 μl Dharmafect1 in 2.25 ml RPMI. See Supplementary Tables S3, S4 and S5 for sequences of siRNAs, siPOOLs and ASOs, respectively.
Subcellular fractionation, RNA isolation and DNase treatment
Subcellular fractionation was performed according to Gagnon et al. (21). RNA was isolated using TRI reagent (Sigma) according to the manufacturer's protocol. Whole cell RNA that was used for RNA-seq experiments was isolated using RNeasy Mini columns (Qiagen). DNase treatment of RNA was performed with Turbo DNase (Thermo Scientific) with subsequent RNA purification using Phenol:Chloroform:Isoamyl Alcohol (25:24:1 [v/v/v]) (Roth).
rRNA depletion and RNA-seq
5 μg RNA were depleted of rRNA using the Ribo-Zero Gold rRNA Removal Kit for human, mouse and rat RNA (Illumina). RNA-seq libraries were generated using the Agilent Sure Select Strand Specific RNA Library Prep for Illumina Multiplex Sequencing Version C.0 (Illumina). The RNA input to generate the library was 20 ng. The single-stranded sequences were analyzed on a HiSeq 2000 V4 (Illumina) with 125 bp paired-end reads. The quality of the reads was assessed with the pipeline EvalRSeq that checks rRNA contamination and computes quality metrics. After adapter sequence removal, the reads were sorted for a length between 50 and 126 bp and uniquely aligned to the human genome v37 using Tophat2, allowing for up to two mismatches (22).
Statistical analysis
Luminescence data of the siRNA screens was statistically analyzed using the R package cellHTS2 (23,24). All other statistical analyses were performed using Excel. For statistical analysis of VELUCT expression in lung cancer samples according to the microarray profiling data, a paired t-test of the 26 paired normal and cancer tissue data was performed. Otherwise, significance was assessed using t-tests after determination of the variance equality using an f-test.
RESULTS
An siRNA screen to identify lncRNAs regulating cell viability
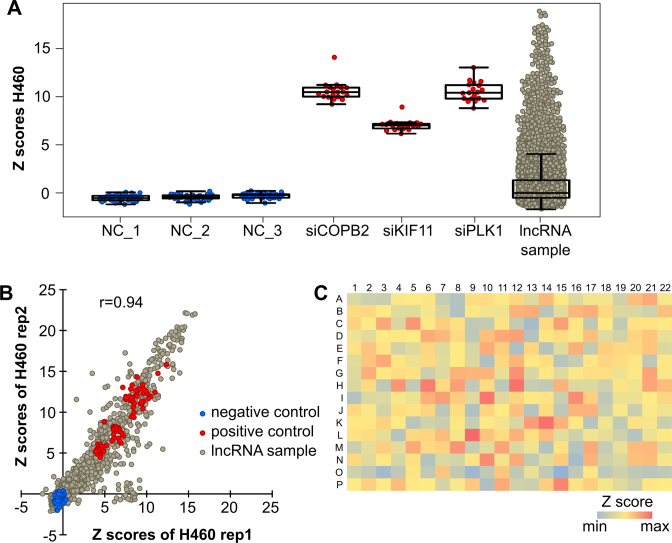
To identify lncRNAs that regulate the cell viability in lung cancer cells, an siRNA screen was performed using a custom-made siRNA library targeting cancer-associated lncRNAs. In order to identify tumor-related lncRNAs, a microarray-based expression profiling of 17 000 polyadenylated ncRNAs was carried out in lung, liver and breast cancer samples (Polycarpou et al., Roth et al., in preparation). Based on the profiling analysis, we identified 638 ncRNAs of interest, most of them lncRNAs that were significantly and at least two-fold upregulated in at least one tumor entity compared to normal tissue. These genes were subsequently used as targets of a custom-made siRNA library. Up to five single siRNAs per target were provided, resulting in a library that consisted of more than 3100 siRNAs. The siRNA screen was performed in the lung cancer cell line NCI-H460 (H460) and the cell viability was analyzed 72 h after transfection. Robust Z scores were calculated using the R package cellHTS2 and assigned to each single reaction. The Z score negatively correlated with the relative cell viability and indicated the strength and significance of the phenotype. Z scores of the replicates were averaged (Supplementary Table S1). All screening plates additionally contained three negative and three positive siRNA controls. As expected, Z scores of the negative controls (NCs) were close to zero indicating no change of cell viability (Figure 1A). Positive siRNA controls targeting COPB2, KIF11 and PLK1 resulted in high median Z scores of 10.8, 7.0 and 10.7, respectively, correlating with a strong reduction of cell viability. Thus, the controls indicated an excellent dynamic range and good performance of the screen. The lncRNA-targeting siRNAs triggered many high Z scores. In sum, there were many more and higher positive Z scores than negative ones. This was expected since the siRNA library mainly targeted lncRNAs that were overexpressed in human cancer and potentially of oncogenic function. The Pearson correlation coefficient between the two replicate screens was 0.94 indicating strong reproducibility (Figure 1B). To analyze edge effects of the screening plates, summarized Z scores were averaged over each well position of the nine screening plates (Figure 1C). The plates showed a rather random distribution of averaged Z scores over the entire plate indicating independence from their location and the absence of strong edge effects. To identify transcripts that potentially played a role in cell viability regulation, 44 hit candidates were selected with more than 60% of siRNAs per gene having a Z score of at least 2 (Supplementary Table S2). The number was further narrowed down to 17 genes with a stricter cutoff of at least 60% of siRNAs (e.g. 3 out of 5) per gene having a Z score of at least 4. Out of these, four genes were targeted by siRNAs for which even 80% had a Z score of minimum 4 (see Supplementary Table S2).
Figure 1.
Quality assessment of siRNA screens addressing viability of H460 cells. H460 cells were screened for cell viability 72 h after transfection (n = 2). Raw values were normalized to the plate median and Z scores were calculated for each reaction. Replicates were summarized by averaging the Z scores. (A) Boxplots with summarized Z scores of the negative controls (blue), the respective positive controls (red) and siRNAs targeting lncRNAs (gray). (B) Z scores of the two replicates were plotted against each other. R indicates the Pearson correlation coefficient. (C) Image plot of summarized Z scores that were averaged over each well position of the nine screening plates. Red indicates the maximum value, blue the minimum value. Positive and negative controls were excluded for this analysis and were not depicted on the plot.
VELUCT —Viability Enhancing in LUng Cancer Transript
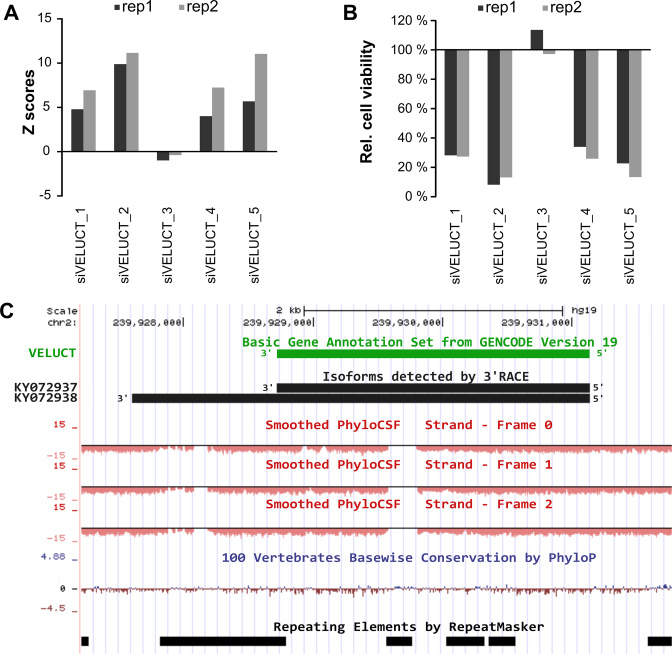
Out of these four hit candidates, ENST00000567151 was selected as the only intergenic long non-coding RNA (lincRNA). The other candidates were the snoRNA NR_003350 and the lncRNAs NR_036472 and U90903 which were not intergenic but overlapping with protein-coding genes. Thus, we focused on the lincRNA ENST00000567151 which was referred to as VELUCT. Its phenotype was extraordinarily strong with four siRNAs out of five having a mean Z score higher than 5 and one siRNA even higher than 10 (Figure 2A). The Z scores corresponded to a relative cell viability of 34–8% in those samples (Figure 2B).
Figure 2.
Identification of the novel oncogenic lncRNA VELUCT regulating the cell viability. (A) Z scores for siRNAs targeting VELUCT in both screening replicates of H460 cells. (B) Relative cell viability (normalized to plate median) for siRNAs targeting VELUCT in both screening replicates of H460 cells. (C) Genomic location of the annotated and the VELUCT isoforms that were detected by 3΄ RACE using chromatin-associated H460 RNA, PhyloCSF scores for all three frames on the respective strand, conservation by PhyloP and repeating elements by RepeatMasker are shown on the UCSC genome browser version hg19.
The transcript was 5.2-fold upregulated in lung cancer samples according to the microarray analysis (Supplementary Figure S1). The expression difference between normal and tumor samples was highly significant (P = 3.6 × 10−8).
3΄ RACE analyses confirmed the annotated 3΄-end and revealed another, less abundant isoform with a non-annotated 3΄-end (Figure 2C). The obtained sequences have been deposited into GenBank as VELUCT transcript isoform 1: KY072937 and VELUCT transcript isoform 2: KY072938. The gene for VELUCT was poorly conserved at the sequence level and transcribed into a 2418 nt long lincRNA according to Gencode version 19 (Figure 2C). The neighboring genes of VELUCT on chromosome 2 are histone deacetylase 4 (HDAC4) in upstream direction (∼40 kb distance) and TWIST2 (∼170 kb distance) in downstream direction.
To analyze the coding potential of VELUCT, the CPAT analysis revealed a coding probability of only 4.6% suggesting a non-coding nature of the transcript and PhyloCSF scores were negative (Figure 2C). Both bioinformatical analysis tools, CPAT and PhyloCSF, provide a robust prediction of the coding potential and also incorporate the conservation of the codon usage. Furthermore, there were no peptides found in PeptideAtlas.
VELUCT is a low abundant, chromatin-associated lncRNA
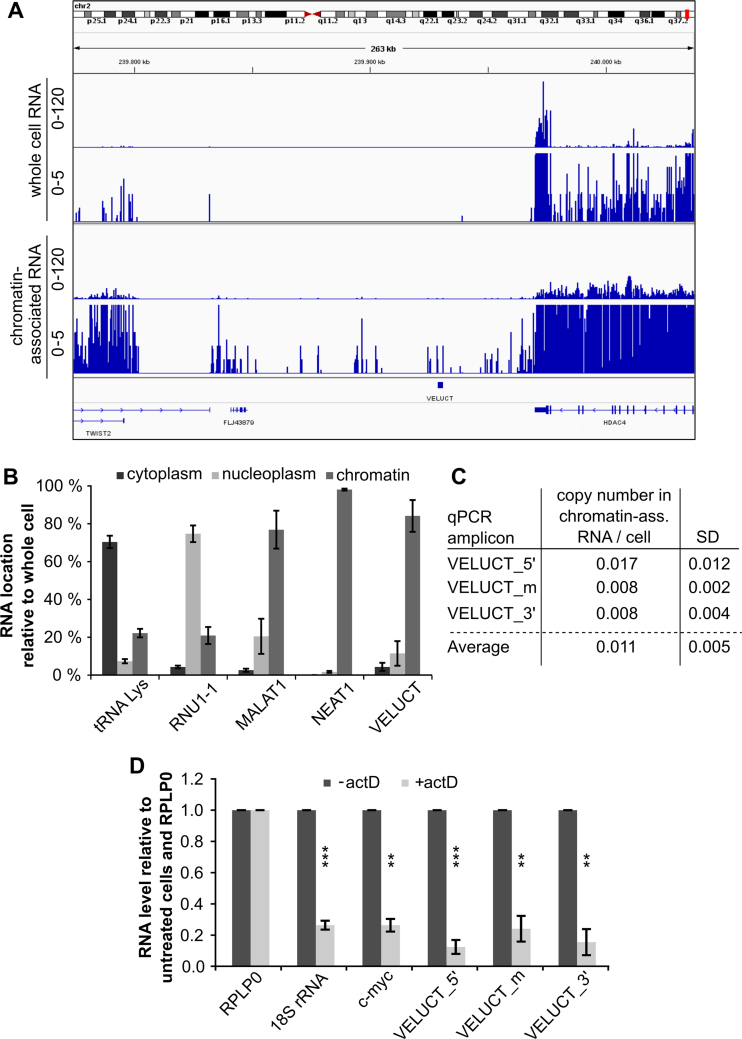
Since VELUCT was a novel RNA, no published expression data were found for this gene. Thus, we characterized the expression of VELUCT in the cell line H460 using multiple detection methods. Surprisingly, there were no reads detectable in the VELUCT region after deep sequencing of H460 whole cell RNA (Figure 3A). Using RT-qPCR, two out of three amplicons for VELUCT resulted in undetermined cycle threshold (CT) values or unspecific melt curves indicating unspecific products (Supplementary Figure S2). Only one out of three amplicons revealed measureable CT values: nevertheless, the CT values were very high (CT 33–34.3) and had a high standard deviation corresponding to a linear 2.5-fold expression difference in technical replicates (Supplementary Figure S2). It was not possible to reliably quantify VELUCT expression despite several reverse transcription and qPCR modifications and multiple qPCR amplicons. Also, northern blot analyses with 10 to 100 μg whole cell RNA did not reveal any signals (data not shown). In order to enrich RNAs of the single subcellular compartments, a subcellular fractionation of H460 cells into cytoplasmic, nucleoplasmic and chromatin-associated fractions was performed (Figure 3B). The cytoplasmic lysine-tRNA, nucleoplasmic RNU1-1 and chromatin-associated MALAT1 and NEAT1 validated the fractionation method. Remarkably, VELUCT was reproducibly detectable particularly in the chromatin fraction. Controls lacking reverse transcriptase (-RT) confirmed that qPCR signals of all amplicons of the monoexonic VELUCT were exclusively triggered by cDNA and not by gDNA. RNA deep sequencing of H460 cells validated the low abundance of VELUCT in chromatin by the presence of a few single reads (Figure 3A). To analyze the VELUCT level per cell, the VELUCT expression in the chromatin-associated fraction was analyzed via qPCR and compared to standard curve generated by serial dilutions of a plasmid that contained the annotated VELUCT sequence. Three amplicons were measured and an average copy number of ∼0.01 per cell was calculated as a conservative estimate (Figure 3C).
Figure 3.
The instable, low abundant VELUCT was only detectable in chromatin-associated lncRNA. (A) RNA deep sequencing was performed with whole cell RNA and chromatin-associated RNA of H460 cells. The read coverage of both runs is shown on a scale from 0 to 120 and from 0 to 5, respectively. The human genome version hg19 was used for alignment of reads. (B) Expression levels in cytoplasmic, nucleoplasmic and chromatin-associated fractions of H460 cells were analyzed by RT-qPCR and normalized to the expression in whole cell RNA. The VELUCT_m amplicon was used for detection of VELUCT. Bars show mean ± SD (n = 3). (C) Determination of VELUCT copy number in chromatin-associated H460 RNA with three qPCR amplicons (location at 5΄ end, middle or 3΄ end) (n = 3). (D) H460 cells were treated with actD or DMSO (-actD) for 1 h and subsequently subcellularly fractionated. Chromatin-associated RNA levels of indicated genes were normalized to RPLP0 and relative to DMSO-treated cells. Bars show mean ± SD (n = 3). Asterisks indicate significant expression difference between treated and untreated cells. *P < 0.05, **P < 0.01, ***P < 0.001.
VELUCT is an unstable transcript
The abundance of a transcript is determined by the balance between RNA synthesis and degradation. Thus, H460 cells were treated with actinomycin D (actD) in order to reveal whether the low abundance levels of VELUCT in the chromatin-associated fraction could be evoked by low RNA stability. Since actD blocked transcription of all three classes of polymerases, the total amount of chromatin-associated RNA was reduced to 52% upon actD treatment for 1 h (Supplementary Figure S3A). To analyze the transcript stability, its RNA levels should be normalized to a transcript which was stable upon inhibition of transcription by actD treatment (Supplementary Figure S3B). Hence, RNA levels of three housekeeping genes (cyclophilin A, GAPDH and RPLP0) were measured which are known stable transcripts (25–27) in order to determine a suitable normalization gene for this experiment. RNA levels were analyzed in relation to untreated cells (2ΔCT analysis) corrected for the loss of total RNA upon actD treatment. RPLP0 was used as normalization gene due to its high stability and little variance in detection. Although 18S rRNA is generally a very stable RNA (28), the rapid decrease of chromatin-associated 18S rRNA levels to 26% reflected the sensitivity of RNA polymerase I to actD (Figure 3D). The C-MYC mRNA has a half-life of only <30 min and is often used as a control for short-lived RNAs, also in the chromatin-associated fraction (29,30). The C-MYC levels in this experiment were consistent with these publications showing a strong reduction of RNA levels to 26% within one hour. RNA levels of three independent VELUCT amplicons were analyzed located close to the 5΄ end, the middle and the 3΄ end of the transcript. They were significantly reduced to 12–24% in the chromatin fraction upon actD treatment resulting in an RNA half-life of 20–30 min. Notably, the reduction was strongest and hence the half-life the shortest compared to all analyzed genes.
Chromatin-associated VELUCT knock-down efficiency
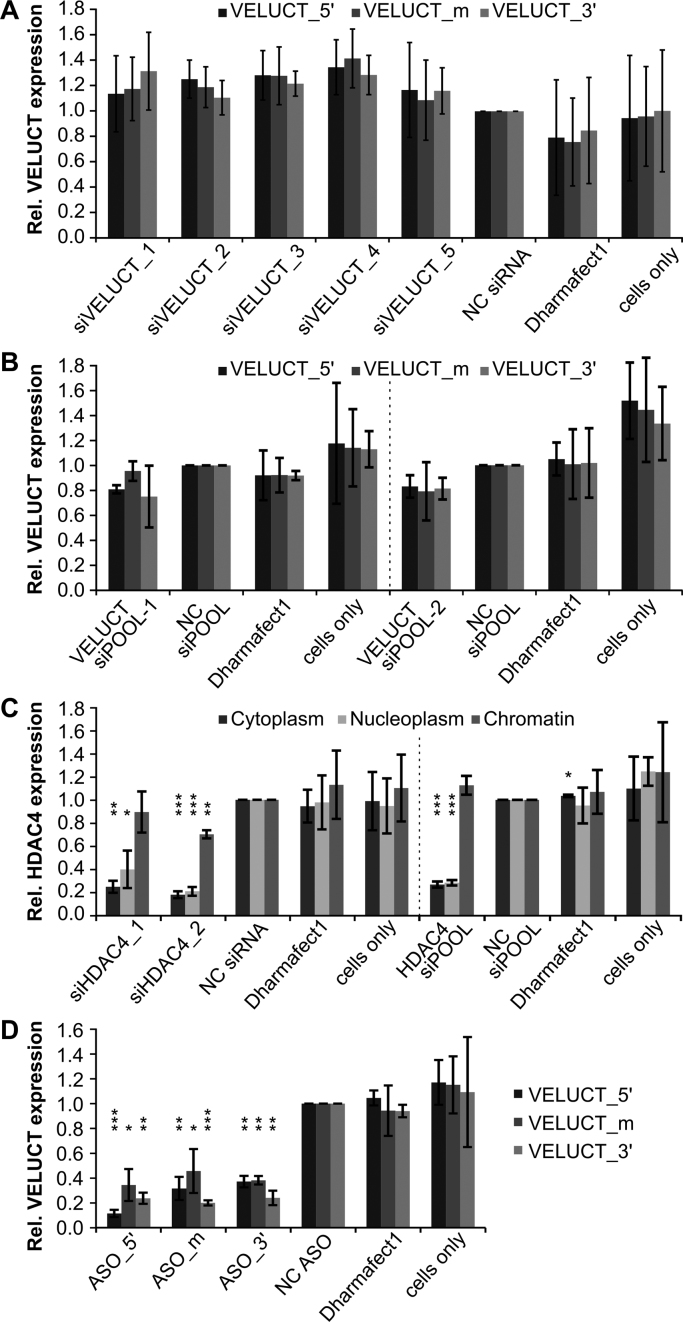
To analyze the downregulation of VELUCT upon transfection, H460 cells were transfected with five individual siRNAs targeting VELUCT, a subcellular fractionation was performed and gene expression was analyzed in the chromatin-associated RNA. A knockdown of VELUCT using three qPCR amplicons was not detectable after siRNA transfection of H460 cells in the chromatin fraction (Figure 4A). Additionally, H460 cells were transfected with two different VELUCT siPOOLs. SiPOOLs are a mixture of 30 independent siRNAs targeting one transcript and are a highly specific RNAi tool (31). Also these RNAi reagents did not evoke a significant detectable downregulation in the chromatin-associated fraction (Figure 4B). We hypothesized that this finding could either be due to a lack of knock-down efficiency or due to a lack of detectability of RNAi effects in the chromatin fraction. In order to analyze the general knockdown of chromatin-associated RNAs by RNAi reagents, the upstream neighboring gene HDAC4 was selected for comparison. It was targeted using two independent siRNAs and one siPOOL (Figure 4C). All reagents significantly knocked down HDAC4 expression in the cytoplasm and nucleoplasm to 20–40%. Only one siRNA caused a slight, significant knockdown of HDAC4 by 30% in the chromatin fraction, while the other two reagents also did not show an effect in the chromatin-associated fraction. Hence, the detectable knock-down in the cytoplasm and nucleoplasm was much stronger than in the chromatin fractions for all the reagents. This leads to the hypothesis that RNAi reagents post-transcriptionally regulate gene expression of cytoplasmic and nucleoplasmic, but not as efficiently of chromatin-bound RNA. This could explain the lack of a detectable RNAi-induced knockdown of VELUCT in the chromatin fraction, whereas the potential downregulation in the nucleoplasm or cytoplasm was not detectable due to its low abundance. Since nuclear transcripts are more effectively knocked down by antisense oligonucleotides (ASOs) (32), H460 cells were also transfected with three independent ASOs targeting VELUCT. They were designed to bind to sites close to the 5΄-end, in the middle and close to the 3΄-end of the transcript (ASO_5΄/m/3΄). Gene expression analyses in the chromatin-associated RNA fraction revealed a significant knockdown of VELUCT (Figure 4D). The VELUCT levels were significantly reduced by all three ASOs to 12–46% for all three tested amplicons. Interestingly, the single ASOs evoked deviating RNA levels for each amplicon, which were consistent for the single replicates. For example, ASO_5΄ also led to the strongest knock-down at the 5΄-end, while ASO_3΄ showed its strongest impact on the 3΄-end of VELUCT. To analyze a possible in cis effect of VELUCT on the expression regulation of the immediate neighboring genes HDAC4 and TWIST2, we analyzed their chromatin-associated RNA levels upon transfection of H460 cells with RNAi reagents targeting VELUCT. Notably, there was no significant deregulation of the neighboring genes HDAC4 and TWIST2 (Supplementary Figure S4). Also expression of p21 which is negatively regulated by HDAC4 (33) did not change significantly.
Figure 4.
VELUCT knockdown was not detectable with siRNAs and siPOOLs, but with ASOs. H460 cells were transfected with 30 nM VELUCT-specific siRNAs (A) or siPOOLs (B), 10 nM HDAC4-specific siRNAs or siPOOLs (C) or 30 nM VELUCT-specific ASOs (D) for 24 h. VELUCT RNA levels were analyzed in the chromatin fraction using three independent qPCR amplicons. HDAC4 levels were determined in all three subcellular fractions. All RNA levels were normalized to cyclophilin A and relative to the respective NC. Bars show mean ± SD (n = 3–4). *P < 0.05, **P < 0.01, ***P < 0.001.
Multiple independent VELUCT-specific silencing reagents strongly reduce viability
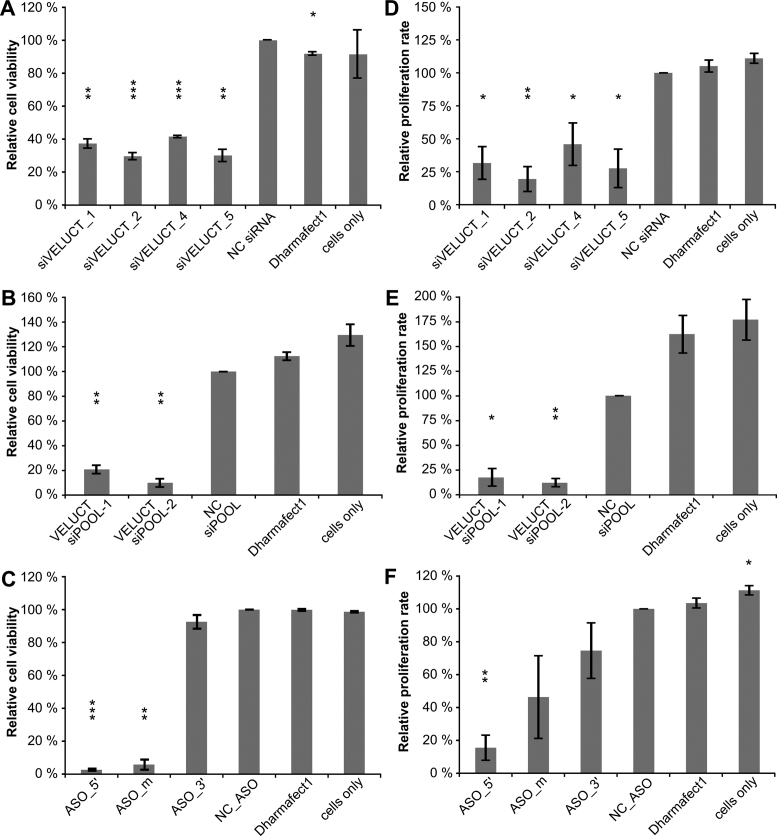
Since the siRNA screen already pointed to a pivotal role of VELUCT in cell viability, more detailed experiments were performed in order to validate the screen. To analyze the effect of VELUCT overexpression on H460 cell viability, a plasmid was generated that contained the annotated gene sequence of VELUCT. Despite high upregulation of exogenous VELUCT expression upon plasmid transfection (Supplementary Figure S5A), cell viability was not affected 24, 48 or 72 h after transfection (Supplementary Figure S5B). In order to validate the loss-of-function screening results, independent knockdown experiments were performed with the same settings as in the screen. Since siVELUCT_3 did not show any phenotype in the screen, it was omitted from further analyses. The screening results were validated as viability of H460 cells was significantly reduced to 30–40% upon siRNA-mediated loss-of-function of VELUCT (Figure 5A). To show that this effect was not only siRNA-dependent, the same experiment was performed with two VELUCT-specific siPOOLs. Both siPOOLs significantly decreased the cell viability even stronger to 10–20% (Figure 5B). H460 cells were also transfected with VELUCT-specific ASOs in order to have a silencing approach that was independent of the RNAi machinery. Compared to all silencing reagents, two out of three ASOs triggered the strongest phenotype with a reduction of cell viability to 3–6 % (Figure 5C). Only the ASO that was located close to the 3΄ end of VELUCT did not affect cell viability. The phenotype was time-dependent with a modest, but significant viability decrease after 24 h and reaching its maximum 48 or 72 h after transfection with all silencing reagents tested (Supplementary Figure S6A–C). Moreover, the effect on cell viability was significant, but slightly weaker using 10 nM silencing reagent concentration (Supplementary Figure S6D-E), except for ASOs for which only a concentration of 30 nM triggered a phenotype (Supplementary Figure S6F). To validate that the phenotype was not assay-specific, an independent readout was performed that measured the cell proliferation upon loss-of-function of VELUCT by BrdU incorporation. Most VELUCT-specific silencing reagents significantly decreased the cell proliferation to approx. 25% and the phenotypic pattern was similar between proliferation and viability (Figure 5D–F). Although the cell proliferation was reproducibly reduced upon transfection of ASO_m, the deregulation was not significant due to a high deviation between the biological replicates (Figure 5F). ASO_3΄ did not trigger a proliferation phenotype recapitulating the viability readout. A time-dependent analysis of the proliferation revealed that the phenotype was significantly reduced to ∼50% already 24 h after transfection, whereas the phenotype was more prominent after 48 and 72 h (Supplementary Figure S7). The analysis of the apoptosis rate revealed that cell death did not contribute to the reduction of viability upon siRNA transfection (Supplementary Figure S8).
Figure 5.
Multiple independent VELUCT-specific silencing reagents reduced viability and proliferation of H460 cells. H460 cells were transfected with 30 nM VELUCT-specific siRNAs (A, D), siPOOLs (B, E) or ASOs (C, F). Cell viability (A–C) and proliferation (D–F) was measured 72 h after transfection and normalized to the respective NC. Bars show mean ± SD (n = 3–4). *P < 0.05, **P < 0.01, ***P < 0.001.
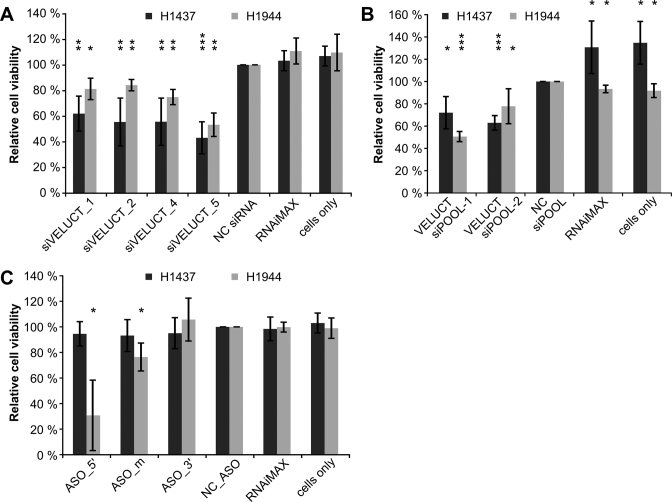
VELUCT knockdown impairs the viability of multiple lung cancer cell lines
In order to analyze whether these results were not only representative for H460 cells, but for several lung cancer cell lines, we tested multiple lung cancer cell lines for their viability phenotype upon loss-of-function of VELUCT. Not all cell lines tested revealed any phenotype, but all analyzed siRNAs targeting VELUCT significantly reduced the viability in H1437 and H1944 cells. This observation is concordant with the observation that lncRNAs are in general expressed and function in a tissue- and cell subtype-specific manner (5,8). H1437 cells showed a stronger phenotype than H1944 with a median relative cell viability of 56% versus 78%, respectively (Figure 6A). The VELUCT siPOOLs-1 and -2 significantly reduced the viability in both cell lines to 51 % to 78 %, respectively (Figure 6B). Strikingly, cell viability was not significantly altered upon transfection of H1437 with all three ASOs (Figure 6C). The viability of H1944 cells was significantly reduced by ASO_5΄ and ASO_m to 31% and 76%, respectively, but not by ASO_3΄ which was consistent with the cell line H460. Hence, the effect on cell viability due to transfection with VELUCT-specific siRNAs, siPOOLs and partially ASOs was validated in the lung cancer cell lines H1437 and H1944 and was thus not restricted to the cell line H460.
Figure 6.
Multiple independent VELUCT-specific silencing reagents reduced viability of other lung cancer cell lines. H1437 and H1944 cells were transfected with 30 nM siRNAs (A), siPOOLs (B) or ASOs (C) targeting VELUCT. Cell viability was measured 72 h after transfection and normalized to the respective NC. Bars show mean ± SD (n = 4–5). *P < 0.05, **P < 0.01, ***P < 0.001.
In summary, eight different knockdown reagents targeting VELUCT in three different cell lines reduced cell growth as determined in two different assays—despite the extremely low abundance of VELUCT.
DISCUSSION
Although thousands of lncRNAs exist, only a minor fraction has been functionally characterized. A common tool to analyze several hundreds of genes for their phenotype upon loss-of-function is an siRNA screen. However, so far, only a few studies described siRNA screens targeting lncRNAs (34–36). According to our knowledge, no siRNA screen has been performed that analyzed tumor-associated lncRNAs in cancer cell lines. Thus, we designed a custom-made siRNA library that was targeting 638 tumor-associated lncRNAs. In order to reduce the impact of off-target effects on hit identification, mostly five siRNAs were designed for each target. Since each siRNA has a distinct range of off-target effects, but the same on-target, a phenotype that is observed with multiple individual siRNAs increases the confidence that it is due to downregulation of the intended target (37,38). One of the most prominent hits was a novel gene that that we called VELUCT. Similar to most lncRNAs (13,14,39), VELUCT was a low abundant transcript. It was 5.2-fold upregulated in lung cancer adenocarcinoma according to microarray analyses. However, it was not reproducibly detectable in whole cell RNA by RT-qPCR. This signal difference between microarray and RT-qPCR might be due to a low correlation as determined before (40). Upon cellular fractionation of cells, VELUCT was quantifiable in the chromatin-associated RNA fraction of H460 cells. This implied that VELUCT might act directly on chromatin and might, e.g. regulate gene transcription as it was reported for many lncRNAs (41). On average and as a conservative estimate, only 0.01 VELUCT RNA copies were present in the chromatin fraction of each cell. While an underestimation for technical reasons is possible, it is highly likely that the copy number of VELUCT will remain even in a less conservative analysis lower than one copy per cell. The low abundance suggested that VELUCT likely acted only on single alleles. One reason for the low abundance of VELUCT was its low stability with a half-life of ∼20–30 min. For comparison, the median half-life of all mammalian RNAs is 5–9 h (27,42–44), while only a minor fraction of mammalian RNAs have a half-life of <1 h (e.g. MYC or the transcription factor FOXA2) (27,45). Short-lived transcripts are reported to be tightly regulated (42) and play a major role in processes such as transcription, cell cycle progression and apoptosis (27,45). Accordingly, the low stability of VELUCT might indicate a precise temporal regulation of this transcript.
Cell viability was not altered upon overexpression of VELUCT. This might be explained by several reasons: First, VELUCT might act in cis, such as enhancer lncRNAs (46), XIST (47) or ANRIL (48). Since cis-acting genes regulate the expression of other genes exclusively on the same chromosome from which they are derived, a plasmid-based overexpression could not recapitulate its effect. Second, since only single VELUCT copies likely directly acted on the genome, only single alleles might be bound by lncRNAs in order to exert their function. In case the binding sites were already occupied, a further increase of VELUCT levels would not promote the phenotype.
To confirm that the observed phenotype in the siRNA screen was elicited by targeting of the VELUCT transcript, VELUCT expression was analyzed upon siRNA transfection of H460 cells. However, there was no RNAi-mediated knockdown of VELUCT detectable in the chromatin-associated fraction. Interestingly, RNAi reagents targeting the upstream neighboring gene HDAC4 as control also failed to induce a detectable knockdown in the chromatin fraction, while it significantly and efficiently knocked down HDAC4 expression in the cytoplasm and nucleoplasm. The fact that there is no detectable knockdown of chromatin-associated VELUCT might be explained by several reasons: On the one hand, technical limitations of RT-qPCR might contribute to the non-detectable knockdown. Although VELUCT is detectable in the chromatin-associated RNA fraction, its low abundance might still be hindering a precise measure of RNAi-mediated downregulation. On the other hand, we hypothesize that the RNAi reagents could bind to and thereby block, but do not downregulate the chromatin-bound transcript as also observed for HDAC4. Thus, the formation of an alternative secondary structure of VELUCT and / or the binding of other effector proteins to VELUCT could be prevented. In contrast, transfection of H460 cells with independent ASOs targeting VELUCT evoked a significant knockdown of VELUCT to 12–46%. ASOs - in contrast to RNAi reagents - might induce knockdown of chromatin-bound RNAs due to their gene silencing mechanism via RNase H (49,50). The localization of effector proteins differs between subcellular compartments: since RNase H is mainly localized in the nucleus (51), ASOs have a superior silencing efficiency in the nucleus than RNAi reagents (32). In contrast, since the RNA degrading RISC complex is primarily localized in the cytoplasm, RNA interference is predominantly active in the cytoplasm (52,53).
The VELUCT-specific phenotype that was observed in the siRNA screen was confirmed in multiple validation experiments. To further exclude the possibility of off-target effects, two siPOOLs comprised of 30 independent siRNAs confirmed the phenotype. Off-target effects are diluted out by the low concentration of each single siRNA making siPOOLs highly specific with strong on-target effects (31). Also, lower siRNA concentrations can reduce off-target effects (54), and the VELUCT-specific phenotype was also recapitulated at a 10 nM RNAi reagent concentration. RNAi reagents can nevertheless lead to a global perturbation of miRNA-mediated regulation due to saturation of the RNAi machinery (55). Again, the RNAi-independent silencing mechanism of ASOs confirmed that no saturation of the RNAi pathway caused the observed loss of cell viability. Lastly, the effect of VELUCT loss-of-function was not restricted to H460 cells, but also found in the lung cancer cell lines H1437 and H1944 and was also reproduced with a proliferation readout. This observation may hypothesize that VELUCT may be involved in the cell cycle. Unfortunately, the low expression of VELUCT precludes more detailed mechanistic studies or the identification of interaction partners for technical reasons.
Eight different knockdown reagents using two different assays and three different cell lines make it very unlikely that the observed phenotypes are not specific for VELUCT. The central question arising from the data presented is how to reconcile the extremely low abundance of VELUCT with its apparent functional importance. Multiple provoking hypotheses could be raised: first, the low abundance and chromatin association could indicate that VELUCT binds only to very few and specific sites in the chromatin requiring only very few copies to cover all sites. Second, VELUCT could only be needed in a very specific state of the cell, e.g. during cell cycle or during replicative or oxidative stress or another condition. The very short half-life of VELUCT then leads to the rapid clearance and low steady-state expression of VELUCT. Lastly, VELUCT could be involved in non-cell-autonomous processes. In this case, single copies in one cell could have an impact on multiple cells amplifying its effect. Although the low copy number of 0.01 could also be explained by a small subpopulation of H460 expressing VELUCT, e.g. the rare cancer stem cells, this is less likely since the VELUCT-specific phenotype is already observable after 24 h upon inhibition of the proliferative characteristics by loss-of-function of VELUCT.
Low abundant, unstable transcripts have already been identified in yeast, where they can play important regulatory roles, e.g. in modifications of histones (56) or chromatin remodeling (57). Although some lncRNAs have a low conservation level, they are nevertheless functional and essential, such as X-inactive specific transcript (XIST) and Antisense IGF2 receptor RNA (Air) (58). Furthermore, genes are not only conserved on the level of their sequence, but also in their secondary structure (59).
To our knowledge, VELUCT is the first example of a lncRNA that is expressed at a very low level, but nevertheless has a strong phenotype upon knockdown. Thus, our study corroborates that - at least individual - lncRNAs of low abundance can execute important functions in the cell. This sheds new light on the large majority of lncRNAs which are present at low copy numbers and have remained understudied so far. Well-controlled studies for each individual lncRNA are required to distinguish between the ‘transcriptional noise’ of aberrant transcripts and functionally important lncRNAs like VELUCT.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Matthias Groß and Jeanette Seiler for excellent technical assistance and the DKFZ Genomics and Proteomics Core Facility (GPCF) for deep sequencing. This study is part of the Ph.D. thesis of J.S.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in the Diederichs labs is supported by the German Research Foundation [DFG Di 1421/7-1, EXC81 CellNetworks EcTop5, SFB 850]; RNA@DKFZ Cross Program Topic and the National Center for Tumor Diseases Heidelberg (NCT 3.0 Integrative Projects in Basic Cancer Research). Funding for open access charge: Core funding.
Conflict of interest statement. S.D. is co-owner of siTOOLs Biotech GmbH, Martinsried, Germany.
REFERENCES
- 1. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. . Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L. et al. . RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007; 316:1484–1488. [DOI] [PubMed] [Google Scholar]
- 3. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al. . Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- 5. Tsoi L.C., Iyer M.K., Stuart P.E., Swindell W.R., Gudjonsson J.E., Tejasvi T., Sarkar M.K., Li B., Ding J., Voorhees J.J. et al. . Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015; 16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu S.J., Nowakowski T.J., Pollen A.A., Lui J.H., Horlbeck M.A., Attenello F.J., He D., Weissman J.S., Kriegstein A.R., Diaz A.A. et al. . Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016; 17:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D.H., Marinov G.K., Pepke S., Singer Z.S., He P., Williams B., Schroth G.P., Elowitz M.B., Wold B.J.. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell. 2015; 16:88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al. . MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 10. Liu X.-h., Liu Z.-l., Sun M., Liu J., Wang Z.-x., De W.. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013; 13:464–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang R., Shi Y., Chen L., Jiang Y., Mao C., Yan B., Liu S., Shan B., Tao Y., Wang X.. The ratio of FoxA1 to FoxA2 in lung adenocarcinoma is regulated by LncRNA HOTAIR and chromatin remodeling factor LSH. Scientific Rep. 2015; 5:17826–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gutschner T., Hämmerle M., Eißmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Groß M. et al. . The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. . The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J.a., Mattick J.S., Rinn J.L.. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2011; 30:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eddy S.R., Doolittle W.F., Sapienza C., Kidwell M.G., Lynch M., Ohno S., Orgel L.E., Crick F.H.C., Thomas C.A.. The C-value paradox, junk DNA and ENCODE. Curr. Biol. 2012; 22:R898–R899. [DOI] [PubMed] [Google Scholar]
- 16. van Bakel H., Nislow C., Blencowe B.J., Hughes T.R.. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010; 8:e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palazzo A.F., Lee E.S.. Non-coding RNA: what is functional and what is junk. Front. Genet. 2015; 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mercer T.R., Dinger M.E., Mattick J.S.. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009; 10:155–159. [DOI] [PubMed] [Google Scholar]
- 19. Clark M.B., Amaral P.P., Schlesinger F.J., Dinger M.E., Taft R.J., Rinn J.L., Ponting C.P., Stadler P.F., Morris K.V., Morillon A. et al. . The reality of pervasive transcription. PLoS Biol. 2011; 9:e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castro F., Dirks W.G., Fähnrich S., Hotz-Wagenblatt A., Pawlita M., Schmitt M.. High-throughput SNP-based authentication of human cell lines. Int. J. Cancer. 2013; 132:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gagnon K.T., Li L., Janowski B.A., Corey D.R.. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014; 9:2045–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boutros M., Brás L.P., Huber W.. Analysis of cell-based RNAi screens. Genome Biol. 2006; 7:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pelz O., Gilsdorf M., Boutros M.. web cellHTS2: a web-application for the analysis of high-throughput screening data. BMC Bioinformatics. 2010; 11:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuwano Y., Rabinovic A., Srikantan S., Gorospe M., Demple B., No H.. Analysis of nitric oxide-stabilized mRNAs in human fibroblasts reveals HuR-dependent heme oxygenase 1 upregulation. Mol. Cell. Biol. 2009; 29:2622–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersen J.B., Mazan-Mamczarz K., Zhan M., Gorospe M., Hassel B.A.. Ribosomal protein mRNAs are primary targets of regulation in RNase-L-induced senescence. RNA Biol. 2009; 6:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharova L.V., Sharov A.A., Nedorezov T., Piao Y., Shaik N., Ko M.S.H.. Database of mRNA half-life of 19977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells supplementary data. DNA Res. 2009; 16:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abelson H.T., Johnson L.F., Penman S., Green H.. Changes in RNA in relation to growth of the fibroblast: II. The lifetime of mRNA, rRNA, and tRNA in resting and growing cells. Cell. 1974; 1:161–165. [DOI] [PubMed] [Google Scholar]
- 29. Berteaux N., Aptel N., Cathala G., Genton C., Coll J., Daccache A., Spruyt N., Hondermarck H., Dugimont T., Curgy J.J. et al. . A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol. Cell Biol. 2008; 28:6731–6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mondal T., Rasmussen M., Pandey G.K., Isaksson A., Kanduri C.. Characterization of the RNA content of chromatin. Genome Res. 2010; 20:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hannus M., Beitzinger M., Engelmann J.C., Weickert M.T., Spang R., Hannus S., Meister G.. SiPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014; 42:8049–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lennox K.A., Behlke M.A.. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016; 44:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mottet D., Pirotte S., Lamour V., Hagedorn M., Javerzat S., Bikfalvi A., Bellahcene A., Verdin E., Castronovo V.. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009; 28:243–256. [DOI] [PubMed] [Google Scholar]
- 34. Chakraborty D., Kappei D., Theis M., Nitzsche A., Ding L., Paszkowski-Rogacz M., Surendranath V., Berger N., Schulz H., Saar K. et al. . Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat. Methods. 2012; 9:360–362. [DOI] [PubMed] [Google Scholar]
- 35. Theis M., Chakraborty D., Weisswange I., Paszkowski-Rogacz M., Buchholz F.. Targeting human long noncoding transcripts by endoribonuclease prepared siRNAs. J. Biomol. Screen. 2015; 20:1018–1026. [DOI] [PubMed] [Google Scholar]
- 36. Negishi M., Wongpalee S.P., Sarkar S., Park J., Lee K.Y., Shibata Y., Reon B.J., Abounader R., Suzuki Y., Sugano S. et al. . A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014; 9:e95216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cullen B.R. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods. 2006; 3:677–681. [DOI] [PubMed] [Google Scholar]
- 38. Echeverri C.J., Beachy P.a., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N. et al. . Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006; 3:777–779. [DOI] [PubMed] [Google Scholar]
- 39. Werner M.S., Ruthenburg A.J.. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 2015; 12:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camarillo C., Swerdel M., Hart R.P.. 2011; 698:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vance K.W., Ponting C.P.. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014; 30:348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tani H., Mizutani R., Salam K.A., Tano K., Ijiri K., Wakamatsu A., Isogai T., Suzuki Y., Akimitsu N.. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012; 22:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 44. Friedel C.C., Dolken L., Ruzsics Z., Koszinowski U.H., Zimmer R.. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009; 37:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raghavan A., Ogilvie R.L., Reilly C., Abelson M.L., Raghavan S., Vasdewani J., Krathwohl M., Bohjanen P.R.. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002; 30:5529–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. et al. . Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010; 143:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pontier D.B., Gribnau J.. Xist regulation and function eXplored. Hum. Genet. 2011; 130:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Congrains A., Kamide K., Ohishi M., Rakugi H.. ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013; 14:1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jepsen J.S., Wengel J.. LNA-antisense rivals siRNA for gene silencing. Curr. Opin. Drug Discov. Dev. 2004; 7:188–194. [PubMed] [Google Scholar]
- 50. Hutvágner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 51. Vickers T.A., Crooke S.T.. Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS One. 2014; 9:e108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zeng Y., Cullen B.R.. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002; 8:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F.. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003; 278:7108–7118. [DOI] [PubMed] [Google Scholar]
- 54. Caffrey D.R., Zhao J., Song Z., Schaffer M.E., Haney S.A., Subramanian R.R., Seymour A.B., Hughes J.D.. SiRNA off-target effects can be reduced at concentrations that match their individual potency. PLoS One. 2011; 6:e21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S.. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009; 27:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F.. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007; 131:706–717. [DOI] [PubMed] [Google Scholar]
- 57. Uhler J.P., Hertel C., Svejstrup J.Q.. A role for noncoding transcription in activation of the yeast PHO5 gene. PNAS. 2007; 104:8011–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pang K.C., Frith M.C., Mattick J.S.. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006; 22:1–5. [DOI] [PubMed] [Google Scholar]
- 59. Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014; 30:121–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.