Abstract
In eukaryotes, the synthesis of ribosomal subunits, which involves the maturation of the ribosomal (r)RNAs and assembly of ribosomal proteins, requires the co-ordinated action of a plethora of ribosome biogenesis factors. Many of these cofactors remain to be characterized in human cells. Here, we demonstrate that the human G-patch protein NF-κB-repressing factor (NKRF) forms a pre-ribosomal subcomplex with the DEAH-box RNA helicase DHX15 and the 5΄-3΄ exonuclease XRN2. Using UV crosslinking and analysis of cDNA (CRAC), we reveal that NKRF binds to the transcribed spacer regions of the pre-rRNA transcript. Consistent with this, we find that depletion of NKRF, XRN2 or DHX15 impairs an early pre-rRNA cleavage step (A’). The catalytic activity of DHX15, which we demonstrate is stimulated by NKRF functioning as a cofactor, is required for efficient A’ cleavage, suggesting that a structural remodelling event may facilitate processing at this site. In addition, we show that depletion of NKRF or XRN2 also leads to the accumulation of excised pre-rRNA spacer fragments and that NKRF is essential for recruitment of the exonuclease to nucleolar pre-ribosomal complexes. Our findings therefore reveal a novel pre-ribosomal subcomplex that plays distinct roles in the processing of pre-rRNAs and the turnover of excised spacer fragments.
INTRODUCTION
Ribosomes are essential ribonucleoprotein complexes (RNPs) that are responsible for all the cellular protein synthesis. The production of eukaryotic ribosomes is a highly energy consuming process that involves the synthesis and maturation of four ribosomal (r)RNAs, and the hierarchical recruitment and assembly of ∼80 ribosomal proteins (1–3). Ribosome biogenesis is initiated by the RNA polymerase I-mediated transcription of a single precursor rRNA transcript (47S in humans), which contains the sequences of the mature 18S, 5.8S and 28S rRNAs, as well as 5΄ and 3΄ external transcribed spacers (5΄ETS and 3΄ETS) and internal transcribed spacers 1 and 2 (ITS1 and ITS2; 4,5). During their transcription and maturation, the rRNAs are extensively modified by small nucleolar (sno)RNPs that introduce 2΄-O-methylations and pseudouridines, as well as stand-alone modification enzymes, which mediate various base modifications (6,7). In addition, the pre-rRNA spacer sequences are removed by a series of endonucleolytic cleavages and exonucleolytic processing events to produce the mature rRNAs (5). This also generates several excised spacer fragments that are subjected to exonucleolytic degradation, which is suggested to be important for the release and recycling of associated ribosome biogenesis factors (5,8). Cleavage in ITS1 leads to the separation of the precursors of small (SSU, 40S) and large (LSU, 60S) ribosomal subunits, which undergo independent maturation in the nucleolus and nucleoplasm before nuclear export followed by final maturation and quality control events in the cytoplasm (2,9,10). Along this pathway, many enzymatic proteins, such as RNA helicases, GTPases and AAA-ATPases, catalyse important structural remodelling steps that help establish the rRNA tertiary structure present in the mature ribosome and facilitate the recruitment or release of ribosome biogenesis factors and the assembly of ribosomal proteins (11,12; and, e.g. 13). Such irreversible steps drive the directionality of the pathway and require careful spatial and temporal regulation of the enzymes that catalyse them. In several cases, the activity of such remodelling factors is known to be regulated by dedicated protein cofactors; for example, in yeast, the activity of the RNA helicase Prp43, which functions in release of snoRNAs from pre-60S complexes and in late steps of pre-40S biogenesis, is regulated by the G-patch protein cofactors Sqs1/Pfa1 and Pxr1/Gno1 (14–17), while Rrp5 serves as an RNA helicase cofactor by modulating the activity of the SSU biogenesis factor Rok1 (18–21).
The pathway of ribosome biogenesis is best characterized in the yeast Saccharomyces cerevisiae, however, the importance of understanding this process in human cells has been highlighted by a growing body of evidence linking ribosome production to human disease. A number of genetic diseases, termed ribosomopathies, are caused by mutations in ribosomal proteins or ribosome biogenesis factors. These diseases, which often manifest in severe haematological disorders and craniofacial defects, include Diamond Blackfan anaemia, Treacher Collins syndrome and Bowen–Conradi syndrome (22,23). Furthermore, ribosome production is closely coupled to cellular proliferation and up-regulated in cancer via direct regulation by several tumour suppressors and oncogenes, such as c-MYC, RAS and p14ARF (24,25). Similarly, defects in ribosome assembly have been shown to trigger a nucleolar stress response pathway that leads to inhibition of HDM2 and activation of p53 (26,27). Understanding the molecular basis of these links between ribosome assembly and disease relies on the characterisation of the functions of the factors involved in human ribosome assembly. Although the roles of many of the human orthologues of yeast ribosome biogenesis factors appear to be conserved, several proteins have been shown to have additional or different functions, and variations in the pre-rRNA processing pathway, such as an additional cleavage step in the 5΄ETS are observed (4,5,28–30). Furthermore, the increased size and complexity of human ribosomes (31), which likely reflects the increased diversity of mRNA substrates they are required to translate as well as the need for greater levels of translational regulation in human cells, necessitates a more complex ribosome biogenesis pathway and, compared to yeast, many additional factors have been shown to participate in ribosome assembly in human cells (32 and see for example 33–36). Indeed, proteomic analyses of human nucleoli have yielded inventories of up to 700 putative human ribosome biogenesis factors (37–39) and large-scale RNAi-based screens have confirmed the requirement for many of these proteins for ribosome maturation (33,40–41). However, for numerous nucleolar proteins, it remains unknown whether they are involved in ribosome maturation and for many others, a detailed characterization of their functions in the pathway is still lacking.
Among the list of human nucleolar proteins is the NF-κB-repressing factor (NKRF), which has previously been implicated in transcriptional repression of NF-κB-regulated genes, such as IFN-β, IL-8, IP10 and hiNOS, through its ability to bind to a specific 11 nt sequence (NRE) in their promoter sequences (42–45). However, NKRF contains an RNA binding domain (R3H) and a glycine-rich G-patch domain, which in other proteins have been shown to mediate interactions with RNA or RNA helicases, suggesting that NKRF could have additional functions in the nucleolus. Here, we show that NKRF forms a nucleolar subcomplex with the RNA helicase DHX15 and the 5΄-3΄ exonuclease XRN2. Using crosslinking and analysis of cDNA (CRAC), NKRF was demonstrated to crosslink to pre-rRNAs in the internal and external transcribed spacer regions and within the 28S rRNA sequence. Consistent with this and similar to XRN2, we reveal that NKRF is required for the turnover of excised pre-rRNA fragments and demonstrate that NKRF is responsible for the recruitment of XRN2 to pre-ribosomal complexes in the nucleolus. Interestingly, we find that NKRF, DHX15 and XRN2 are required for efficient cleavage at the metazoan-specific A’ site in the 5΄ETS. Our data further show that the catalytic activity of DHX15 is required for this processing step and that NKRF functions as a cofactor of DHX15 and stimulates its ATPase and unwinding activity. Together, these results identify NKRF as a novel ribosome biogenesis factor in human cells that functions in both the initial processing of the pre-rRNA transcript and in the degradation of excised pre-rRNA spacer fragments through its interactions with DHX15 and XRN2.
MATERIALS AND METHODS
Generation of stable cell lines, cell culture and RNAi
The coding sequences for NKRF (NM_001173487.1), XRN2 (NM_001317960.1), DHX15 (NM_001358.2) and truncated versions of these sequences were cloned into a pcDNA5 vector with an N-terminal 2xFlag-PreScission protease site-His6 (Flag) tag. Stable HEK293 cell lines expressing these proteins were generated using the Flp-In T-Rex Expression System (Invitrogen) according to manufacturer's instructions. Site-directed mutagenesis was used to produce siRNA-resistant DHX15 constructs that contain five silent mutations in the siRNA target site, the DHX15E261Q catalytic mutant and an NKRF G-patch domain mutant (NKRFG1-6A) in which G570, G578, G581, G583, G585 and G590 were changed to A. HEK293 Flp-In T-Rex and HeLa CCL2 cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% foetal calf serum. For RNAi, cells were transfected with siRNAs (50 nM) listed in Supplementary Table S1 using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions and were harvested after 96 h.
Immunoprecipitation of complexes
Immunoprecipitation experiments with extracts from HeLa or HEK293 cell lines were performed essentially as previously described (34). For immunoprecipitation with an antibody against NKRF (Supplementary Table S2), HeLa cells were lysed by sonication in a buffer containing 20 mM HEPES pH 8.0, 150 mM KCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA) and centrifuged to remove insoluble material. The cleared lysate, supplemented with 10% glycerol, 1.5 mM MgCl2 and 0.2% Triton X-100, was incubated with Protein G Sepharose beads (GE Healthcare) coupled to anti-NKRF antibody for 2 h at 4°C. Bound proteins were eluted from the beads at 95°C with sodium dodecyl sulphate (SDS) loading dye, separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analysed by mass spectrometry or by western blotting (Supplementary Table S2). Alternatively, expression of Flag-tagged proteins in HEK293 cell lines was induced for 24 h with tetracycline and extracts were prepared as above. The lysate was incubated with anti-FLAG M2 magnetic beads (Sigma-Aldrich) for 2 h at 4°C. For RNase-treated samples, a mixture of RNase A (2.5 U) and RNase T1 (50 U) was added during the binding step. Bound complexes were eluted with 250 μg/ml FLAG Peptide (Sigma-Aldrich), precipitated with 20% trichloracetic acid (TCA) and analysed by western blotting with the indicated antibodies (Supplementary Table S2). RNAi-mediated knockdowns that preceded the immunoprecipitation experiments were done for 96 h with 50 nM siRNAs and expression of Flag-tagged proteins was induced for the last 24 h before harvesting.
Isolation of nucleoli and preparation of nucleolar lysates
Nucleoli were isolated from HEK293 stable cell lines as previously described (46) with a few modifications. Expression of Flag-DHX15 or the Flag-tag alone was induced for 24 h by addition of tetracycline and then cells were harvested and washed two times in ice-cold phosphate buffered saline (PBS). Cells were lysed for 10 min on ice in 5 ml of lysis buffer (20 mM Tris-HCl pH 7.4, 10 mM KCl, 3 mM MgCl2, 0.1% NP-40, 10% glycerol) and centrifuged at 1350 g for 10 min. The pellet containing intact nuclei was resuspended in 3 ml of solution S1 (0.25 M sucrose, 10 mM MgCl2), layered on top of 3 ml of solution S2 (0.35 M sucrose, 0.5 mM MgCl2) and centrifuged at 1430 g for 5 min. The nuclear pellet was then resuspended in 3 ml of solution S2 and sonicated to release nucleoli. The lysed nuclei were layered on top of 3 ml solution S3 (0.88 M sucrose, 0.5 mM MgCl2) and centrifuged at 3000 g for 10 min. The upper phase was retained as the nucleoplasmic fraction and the pellet containing nucleoli was resuspended in 500 μl of solution S2 and centrifuged again at 1430 g for 5 min. To prepare nucleolar lysate, the isolated nucleoli were incubated on ice for 30 min in 400 μl high-salt buffer (20 mM HEPES pH 8.0, 500 mM KCl, 0.5 mM EDTA, 0.8% Triton X-100, 0.4% CHAPS) supplemented with 16 U TURBO DNase (Ambion), and disrupted by sonication. Samples were then diluted four-fold with a buffer containing 20 mM HEPES pH 8.0, 2 mM MgCl2 and 13.3% glycerol, and immunoprecipitations with anti-FLAG M2 beads were performed as described above. Equal protein amounts (1.5 μg) of whole cells, nucleoplasmic and nucleolar fractions were analysed for purity by western blotting.
Sucrose density gradients
HeLa cells that were either untreated or transfected with siRNAs for 96 h were lysed by sonication in a buffer containing 50 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT. The cleared, whole cell extracts were separated on 10–45% sucrose gradients in an SW-40Ti rotor for 16 h at 23,500 rpm as previously described (34). Fractions comprising 530 μl were precipitated with 20% TCA, pellets resuspended in SDS-loading dye, proteins separated by SDS-PAGE and analysed by western blotting.
Immunofluorescence
Fluorescence microscopy was performed as previously described (47). In brief, HeLa cells were grown on coverslips and fixed using 4% paraformaldehyde in PBS for 20 min at room temperature before permeabilization using 0.1% Triton-X-100 in PBS for 15 min. Cells were blocked using 10% foetal calf serum in PBS with 0.1% Triton-X-100 for 1 h before incubation with antibodies against NKRF, DHX15 or XRN2, and NSUN5 (Supplementary Table S2) for 2 h at room temperature. Cells were washed and incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies for 2 h at room temperature. Coverslips were mounted onto slides using mounting medium containing DAPI and fluorescence was detected by confocal microscopy.
Crosslinking and analysis of cDNA (CRAC)
UV crosslinking and analysis of cDNA (CRAC) experiments were performed as previously described (34,48). For PAR-CRAC, cells were grown in media supplemented with 100 μM 4-thiouridine for 6 h prior to crosslinking at 365 nm in a Stratalinker (Agilent) using 2 cycles of 180 mJ/cm2. Samples were then processed as for UV-CRAC and the obtained sequencing data were analysed as previously described (49) with the exception that for PAR-CRAC samples, only sequence reads containing T-to-C mutations were mapped to the human genome.
Pre-rRNA analysis
Total RNA from control or siRNA-treated cells was extracted with TRI reagent (Sigma-Aldrich), separated on a 1.2% agarose-glyoxal gel, transferred to a nylon membrane and analysed by northern blotting (50). Detection of pre-rRNAs and spacer fragments was done using 32P-labelled oligonucleotide probes listed in Supplementary Table S3.
Purification of recombinant proteins and in vitro binding assays
Expression of ZZ-DHX15-His10, MBP-XRN2-His14, GST-NKRF-His7, MBP-NKRF-His14, MBP-DHX15-His14 or MBP-DHX15E261Q-His14 was induced in BL21 cells with 0.2 mM IPTG for 16 h at 18°C. The cell pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 7.4, 600 mM NaCl, 1% Triton X-100, 10% glycerol, 0.5 mM EDTA, 30 mM imidazole) and lysed with an EmulsiFlex-C3 (Avestin) by four passes at 10000 psi. After centrifugation at 50000 g for 30 min, the lysate was incubated with cOmplete His-tag purification resin (Roche) for 1 h at 4°C. Following several washes with low-salt and high-salt wash buffers (50 mM Tris-HCl pH 7.4, 150/1000 mM NaCl, 30 mM imidazole), proteins were eluted with 250 mM imidazole. Buffer exchange was done either by dialysis overnight at 4°C or with a Spin-X UF concentrator (Corning), and proteins were stored in a buffer with 50 mM Tris-HCl pH 7.4, 150 mM NaCl and 20% glycerol.
For in vitro binding assays, bait proteins (ZZ-DHX15-His10 or MBP-XRN2-His14) were incubated with 2.5-fold excess of the prey proteins for 1 h at 4°C in binding buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 0.01% NP-40, 2 mM DTT, 20 μg/ml RNase A). The mixture was then added to pre-equilibrated IgG Sepharose (GE Healthcare) or amylose (NEB) beads and binding was carried out for 45 min at 4°C. Bound proteins were eluted, separated by SDS-PAGE and analysed by Coomassie staining.
ATPase and unwinding assays
ATP hydrolysis was measured in an NADH-coupled enzymatic assay as reported previously (51). In brief, reactions containing 45 mM Tris-HCl pH 7.4, 25 mM NaCl, 2 mM MgCl2, 1 mM phosphoenolpyruvate, 300 μM NADH, 20 U/ml pyruvate kinase/lactic dehydrogenase (Sigma-Aldrich) and 4 mM ATP were started by addition of 250 nM helicase. Reactions were further supplemented with 2 μM RNA oligonucleotide (5΄-UAGAGGAACUAAAAGUCGAAACAAAACAAAAC-3΄) and 250 nM NKRF as stated. The ATPase activity was determined by measuring the decrease in absorbance at 340 nm using a BioTEK Synergy plate reader.
For unwinding assays, a 5’ 32P-labelled DNA oligonucleotide (5΄-GCTGATCATCTCTGTATTG-3΄) was annealed to a 118 nt in vitro transcribed RNA sequence (5΄-AGC UUGUUCGCGAAGUAACCCUUCGUGGA CAUUUGGUCAAUUUGAAACAAUACAGAGAU GAUCAGCAGUUCCCCUGCAUAAGGAUGAAC CGUUUUACAAAGAGAUUUAUUUCGUUUUG-3΄) obtained as previously described (49). The RNA-DNA duplex was prepared at a 1:1 molar ratio in a buffer with 30 mM HEPES pH 7.5 and 100 mM KAc by heating at 95°C for 3 min, incubating at 65°C for 5 min and then slowly cooling to room temperature. Unwinding reactions contained 50 mM HEPES pH 7.5, 50 mM KCl, 0.5 mM MgCl2, 100 μg/ml BSA, 5% glycerol, 2 mM DTT, 20 U RiboLock (Thermo Fisher), 1 nM duplex and 2.5 nM helicase with or without 2.5 nM NKRF. A 50-fold excess of unlabelled complementary DNA oligonucleotide was added to prevent re-annealing. Reactions were started with 2 mM ATP/MgCl2 and were carried out for 10 min and 20 min at 30°C. Samples were then mixed with 4x quenching buffer (50 mM Tris-HCl pH 8.0, 2.5% SDS, 50 mM EDTA, 25% glycerol) and separated on a 10% native PAGE followed by detection with a phosphorimager.
RESULTS
NKRF interacts with the RNA helicase DHX15 and the 5΄-3΄ exonuclease XRN2
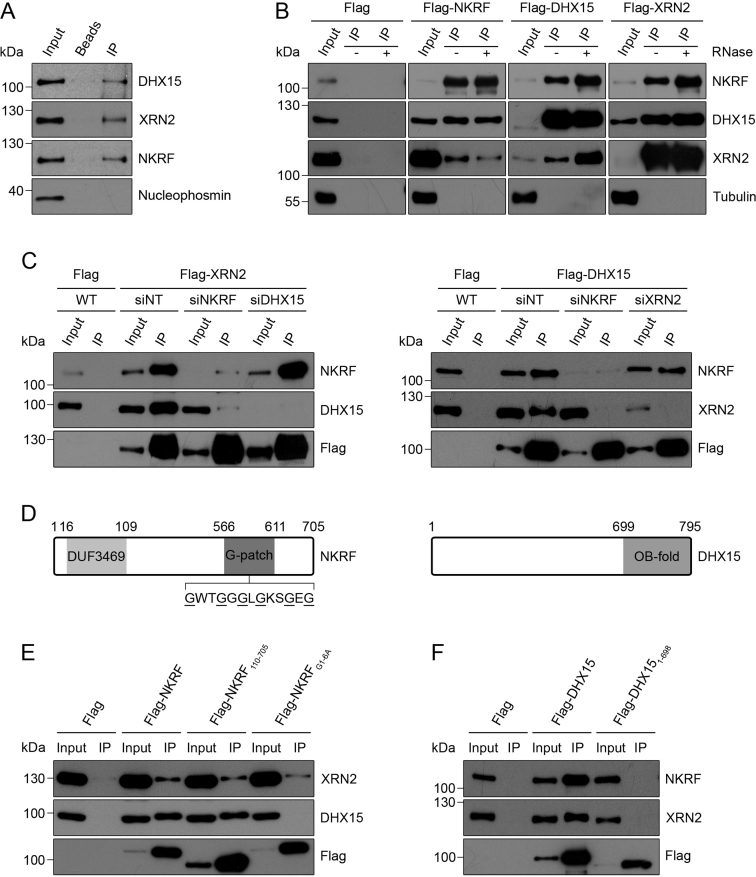
Inventories of the human nucleolar proteome have led to the identification of >700 nucleolar proteins (37,39). Among these proteins is NKRF that has previously been characterized as a transcriptional repressor of NF-κB-regulated genes, but which contains several domains that can bind RNA, suggesting that this protein may play additional roles in the nucleolus. To investigate the functions of NKRF, we first set out to identify protein interaction partners by performing immunoprecipitation (IP) experiments from HeLa cell extract using an antibody against the endogenous NKRF protein and analysing the eluate by mass spectrometry. Compared to a control sample from which the antibody was omitted, this revealed efficient recovery of NKRF and a significant enrichment of the RNA helicase DHX15 and the 5΄-3΄ exonuclease XRN2. Further analysis of the eluates by western blotting using antibodies against these proteins confirmed their association with NKRF and the specificity of these interactions is supported by the lack of the abundant nucleolar protein Nucleophosmin in the NKRF immunoprecipitation eluates (Figure 1A). To verify if these interactions also occur in other cell types and to determine if XRN2 and DHX15 also interact, HEK293 cell lines stably expressing 2xFlag-PreScission protease site-His6 (Flag)-tagged versions of these proteins from a tetracycline-inducible promoter were generated. These cells were then used for IP experiments via the Flag tag and the eluates were analysed by western blotting. Both XRN2 and DHX15 were co-precipitated from cells expressing Flag-tagged NKRF, but not the Flag-tag alone (Figure 1B). Similarly, NKRF and XRN2 were co-precipitated (even more efficiently) from cells expressing Flag-DHX15 and both NKRF and DHX15 were enriched in the eluates from Flag-XRN2 cells (Figure 1B), suggesting that DHX15 and XRN2 also interact and that these three proteins may form a complex. Furthermore, treatment of the cell lysates with RNase during the IP did not affect these interactions, implying that they represent protein–protein interactions.
Figure 1.
The NF-κB repressing factor (NKRF) interacts with XRN2 and DHX15. (A) A HeLa cell extract was used for immunoprecipitation with an anti-NKRF antibody immobilized on Protein G sepharose. Inputs (10%) and eluates containing co-precipitated proteins were analysed by western blotting using antibodies against the indicated proteins. (B) Extracts from HEK293 cell lines expressing Flag-tagged NKRF, XRN2 or DHX15, or the Flag-tag alone were used in immunoprecipitation (IP) experiments in the presence (+) or absence (−) of RNase. Inputs (1%) and eluates containing co-precipitated proteins were analysed by western blotting using antibodies against NKRF, DHX15, XRN2 and Tubulin as indicated. Different western blot exposures are presented to enable the relative levels of the input and eluate to be compared in all cases. (C) HEK293 stable cell lines were transfected with non-target siRNAs (siNT) or siRNAs targeting NKRF (siNKRF), XRN2 (siXRN2) or DHX15 (siDHX15) as indicated and RNAi was performed for 96 h. During the final 24 h, expression of Flag-tagged DHX15 or XRN2 or the Flag-tag alone was induced using tetracycline. After harvesting, extracts were prepared and used in immunoprecipitation experiments as in (B). (D) Schematic view of the domains of NKRF and DHX15. Amino acid numbers corresponding to the boundaries of specific domains are indicated above. The amino acid sequence of a region of the G-patch domain containing six glycine residues (underlined) is shown. (E) Immunoprecipitation experiments were performed as in (B) using extracts from HEK293 cell lines expressing the Flag-tag alone or Flag-tagged versions of NKRF, truncated NKRF lacking the DUF3469 domain (NKRF110–705) or NKRF in which six glycine residues in the G-patch domain were substituted for alanine (NKRFG1-6A). (F) Extracts from HEK293 cell lines expressing Flag-tagged DHX15 or DHX151–698 that lacks the C-terminal OB-fold were used in immunoprecipitation experiments and analysed as in (B). All experiments presented in this figure were performed at least three times and representative data are shown.
To determine if the interactions between these proteins are direct, recombinant DHX15, NKRF and XRN2, purified with different affinity tags, were used for in vitro binding assays. Firstly, ZZ-tagged DHX15 was incubated with MBP-tagged NKRF and/or MBP-tagged XRN2 and complexes formed were retrieved on IgG sepharose. In addition to DHX15, both NKRF and XRN2 were recovered when added either individually or together (Supplementary Figure S1A). This demonstrates that DHX15 forms direct contacts with both these proteins and suggests that these interactions are not mutually exclusive. Similarly, after pre-incubation of MBP-tagged XRN2 with GST-tagged NKRF and/or ZZ-tagged DHX15, XRN2 could be isolated on the amylose resin together with either NKRF, DHX15 or both proteins (Supplementary Figure S1B), further supporting the model that these proteins form a complex. To gain insight into how this complex is assembled in vivo, we next established RNAi against each of these proteins to determine the effect of lack of one component on the interaction between the other two proteins. After confirming that siRNA treatment efficiently reduced the levels of each of the target proteins (Supplementary Figure S2), expression of Flag-tagged XRN2 was induced in cells that had been treated with non-target siRNAs or those targeting NKRF or DHX15 and the cell extracts were used for IPs. Similar amounts of the Flag-tagged bait protein were recovered in each case and depletion of DHX15 did not affect the interaction between Flag-XRN2 and NKRF. Interestingly, however, depletion of NKRF significantly reduced the amount of DHX15 co-precipitated with Flag-XRN2 (Figure 1C, left panel). Similarly, the interaction between Flag-DHX15 and XRN2 was strongly reduced when NKRF was knocked down while the interaction between Flag-DHX15 and NKRF was unaffected by depletion of XRN2 (Figure 1C, right panel). Taken together, these data suggest that NKRF, DHX15 and XRN2 can form direct protein–protein interactions with each other in vitro but that NKRF enhances the interaction between DHX15 and XRN2 in vivo.
NKRF contains two domains that can bind to RNA: an R3H domain and a glycine-rich G-patch domain. G-patch domains have been shown in other proteins to mediate interactions with RNA helicases such as Prp43, the yeast homologue of DHX15, often via the C-terminal OB-fold of the helicase (Figure 1D; 51–53). In addition, NKRF contains a domain of unknown function (DUF3469; Figure 1D), which in other proteins facilitates interactions with XRN2 (XTBD; 54–56). To assess the requirement for each of these domains for assembly of the NKRF-DHX15-XRN2 subcomplex, we generated tetracycline-inducible HEK293 cell lines for expression of Flag-tagged versions of a truncated form of DHX15 lacking the OB-fold (Flag-DHX151–698), a truncated form of NKRF missing the DUF3469 (Flag-NKRF110–705) and full length NKRF in which six glycine residues in the G-patch domain (G570, G578, G581, G583, G585 and G590) had been converted to alanine (Flag-NKRFG1-6A). Extracts from these cells, expressing wild-type and mutant proteins at similar levels (Figure 1E and F), were used in IP experiments, which demonstrated that NKRF lacking the DUF3469 was still able to interact with XRN2 (Figure 1E), suggesting that in NKRF, this domain does not mediate interactions with XRN2 or that such contacts are only formed in the context of other interactions between these two proteins. Furthermore, these experiments revealed that the G-patch domain of NKRF as well as the OB-fold of DHX15 are required for the NKRF-DHX15 interaction (Figure 1E and F), implying that the mode of interaction between these two proteins is similar to that of other RNA helicases with their corresponding G-patch proteins. Similarly, the truncated version of DHX15 lacking the OB-fold was no longer able to co-precipitate XRN2, further supporting the finding that NKRF is required for the interaction between DHX15 and XRN2 in vivo.
NKRF, DHX15 and XRN2 interact in the nucleolus and associate with pre-ribosomal complexes
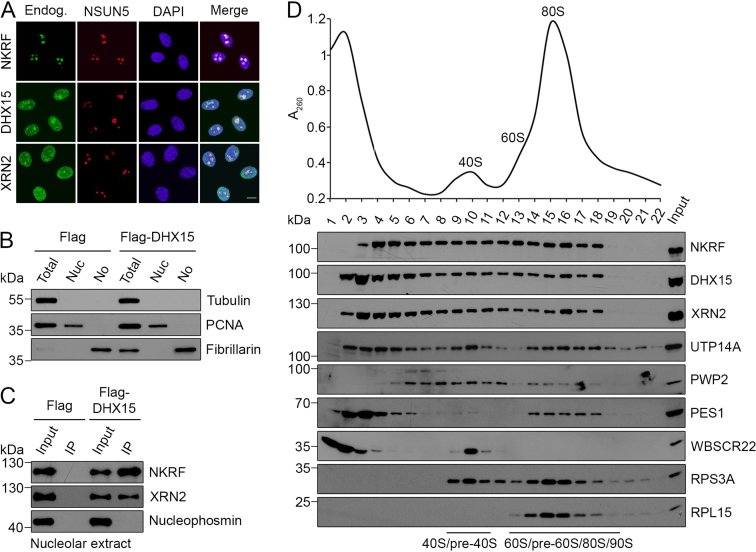
DHX15 is a multifunctional RNA helicase that is known to play several roles in pre-mRNA splicing as well as being implicated in the immune response to viral infection (57–59) and XRN2 has diverse functions in the degradation and processing of numerous RNA substrates (60). This raises the question of where in the cell the interactions between NKRF, DHX15 and XRN2 are formed and the localization of each of these proteins was therefore analysed by immunofluorescence (Figure 2A). This demonstrated that DHX15 and XRN2 are present in both the nucleoplasm and the nucleolus, whereas NKRF was predominantly localized to nucleoli (Figure 2A), which is consistent with the previously reported localization of NKRF and the identification of all three proteins in proteomic analyses of human nucleoli (39, 44). To analyse whether the three proteins are found in complex in nucleoli, we isolated nucleoli from HEK293 cells expressing Flag-tagged DHX15 or the Flag-tag alone and after verification of the purity of the isolated nucleoli by western blotting using antibodies against marker proteins for the cytoplasm (Tubulin), nucleoplasm (PCNA) and nucleoli (Fibrillarin; Figure 2B), the nucleolar extracts were used in IP experiments. Both NKRF and XRN2 were efficiently co-precipitated with Flag-DHX15 but not with the Flag-tag alone, demonstrating that these proteins interact in the nucleolus (Figure 2C). The abundant nucleolar protein Nucleophosmin was not detected in the IP eluates, further supporting the specificity of these interactions.
Figure 2.
NKRF, XRN2 and DHX15 interact in the nucleolus and associate with pre-ribosomal subunits. (A) HeLa cells were fixed and immunofluorescence was performed using antibodies against the endogenous NKRF, DHX15 or XRN2 (Endog.; green) and NSUN5 (nucleolar marker; red), and nuclei were visualized by DAPI staining (blue). Overlays of the different channels are shown on the right (Merge). The scale bar indicates 10 μm. (B) HEK293 cell lines expressing the Flag-tag alone (Flag) or Flag-DHX15 were lysed and fractionated to produce a whole cell lysate (Total), a nucleoplasmic extract (Nuc) and a nucleolar extract (No). Samples were analysed by western blotting using antibodies against Tubulin, PCNA and Fibrillarin as cytoplasmic, nucleoplasmic and nucleolar markers respectively. (C) Immunoprecipitation experiments were performed using nucleolar extracts (B) and co-precipitated proteins were detected using the indicated antibodies. (D) A HeLa cell extract was separated by sucrose density gradient centrifugation and the distributions of NKRF, XRN2 and DHX15, the ribosomal proteins RPS3A and RPL15, and several marker proteins for pre-ribosomal complexes were determined by western blotting using antibodies against the endogenous proteins. A260 measurements of fractions were used to generate a profile on which the positions of the 40S and 60S ribosomal subunits as well as 80S monosomes are indicated. All experiments presented in this figure were performed at least three times and representative data are shown.
The identification of the interactions in nucleoli, the major site for ribosome assembly, together with the known roles of XRN2 in pre-rRNA processing suggested that NKRF and DHX15 might also associate with pre-ribosomal subunits. We therefore used sucrose density gradients to separate large complexes, such as ribosomal and pre-ribosomal subunits from non-ribosome-associated proteins within whole cell extracts (61–62). The fractions containing ribosomal particles were identified by A260 measurements that indicate the positions of 80S monosomes and free 40S and 60S subunits (Figure 2D, upper panel) and by western blotting using antibodies against the ribosomal proteins RPL15 and RPS3A (Figure 2D, lower panel). The presence of pre-ribosomal complexes co-migrating in these fractions was confirmed by western blotting using antibodies against the SSU processome components PWP2 and UTP14A (90S/pre-40S; 34,63–64), PES1 (pre-60S; 65) and WBSCR22 (pre-40S; 50,66). Western blotting was then used to examine the distribution of NKRF, DHX15 and XRN2 in such gradients and although a portion of each protein was found in fractions containing non-ribosomal complexes, all three proteins also co-migrated in fractions containing pre-ribosomes (Figure 2D). Taken together with the nucleolar/nuclear localisation of these proteins, this implies that NKRF, DHX15 and XRN2 are indeed associated with pre-ribosomal complexes.
NKRF crosslinks to pre-rRNA in the spacer regions and within the 28S rRNA sequence
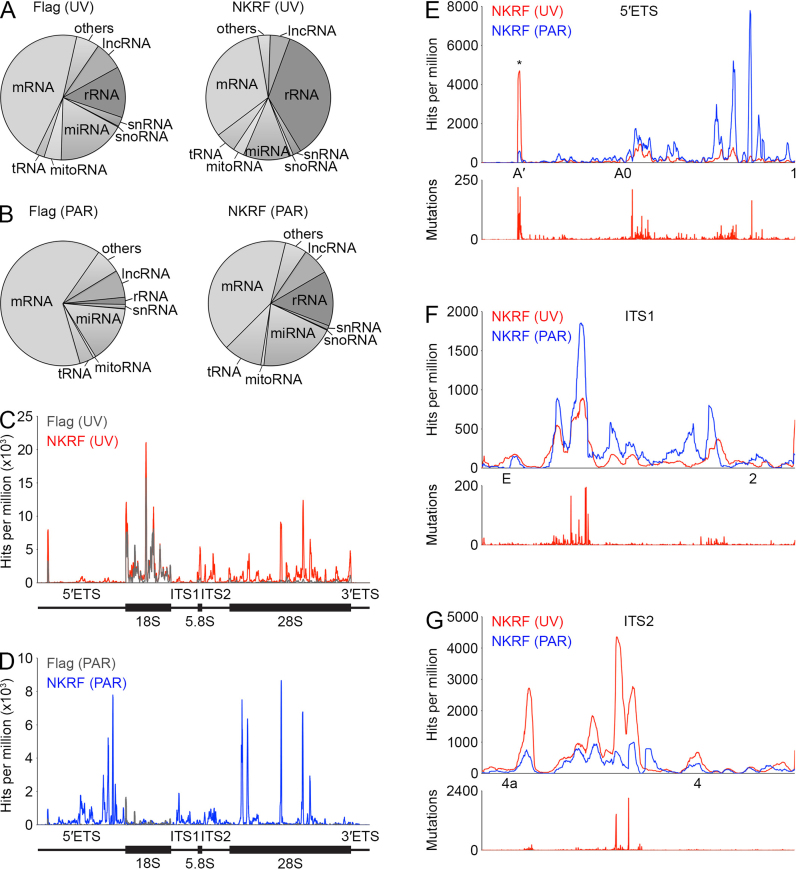
To gain insight into the function of the XRN2-NKRF-DHX15 complex in ribosome maturation, we set out to identify the pre-ribosomal binding site(s) of the core component of the complex, NKRF. For this, we performed the UV crosslinking and analysis of cDNA (CRAC; 34,48) approach using the HEK293 cell line expressing Flag-tagged NKRF, or the tag alone as a control. In addition, we used a PAR-CRAC approach in which, similar to photoactivatable ribonucleoside-enhanced (PAR)-CLIP (67, 68), the cells were first treated with 4-thiouridine, which is incorporated into nascent RNA and can then be specifically crosslinked to proteins using light at 365 nm. After crosslinking, RNA–protein complexes were isolated, an RNase digestion was performed to produce a protein footprint on target RNAs and these were ligated to adapters. Co-purified RNAs were then extracted, reverse transcribed and amplified by PCR to generate a cDNA library that was subjected to Illumina deep sequencing. The obtained sequence reads were mapped onto the human genome and in the case of data from the PAR-CRAC approach, which leads to the generation of T-to-C mutations at the crosslinking sites, only reads containing T-to-C mutations were mapped. Consistent with the nucleolar localization of (Flag-)NKRF (Figure 2A and Supplementary Figure S3) and its co-migration with pre-ribosomal complexes (Figure 2D), analysis of the distribution of reads between the major classes of RNA revealed an enrichment of (pre)-rRNA sequences in both Flag-NKRF samples compared to the corresponding controls (Figure 3A and B; UV – Flag 13% and Flag-NKRF 36%; PAR – Flag 2% and Flag-NKRF 14%). Investigation of the distribution of reads mapped to the rDNA revealed specific crosslinking of NKRF to various regions of the pre-rRNA transcript including within the 28S rRNA sequence (Figure 3C and D). Closer examination of these sites revealed that they lie in eukaryotic expansion segments that form flexible regions on the surface of the mature LSU, but it remains unknown what contacts or structure these sequences form in the pre-ribosomal complexes with which NKRF associates. Interestingly, NKRF also crosslinked to sequences in the 5΄ external transcribed spacer (5΄ETS) upstream of the 18S rRNA and the internal transcribed spacers 1 and 2 (ITS1 and ITS2) that lie between the 18S and 5.8S, and 5.8S and 28S rRNA sequences in the initial 47S pre-rRNA transcript (Figure 3C–G). During ribosome biogenesis, these spacers are removed by a series of endonucleolytic cleavages at specific sites that release fragments and can be followed by exonucleolytic processing to produce the mature ends of rRNAs. Closer inspection of the NKRF crosslinking sites in these spacer regions showed that the majority of reads maps upstream of site 1 in the 5΄ETS (Figure 3E), downstream of site E in ITS1 (Figure 3F) and between sites 4a and 4 in ITS2 (Figure 3G). That these crosslinking sites of NKRF represent bona fide protein binding sites is further supported by the detection of mutations/microdeletions (Figure 3E–G, lower panels), which are often introduced during cDNA preparation at nucleotides UV crosslinked to amino acids and therefore indicate direct RNA–protein contacts at these positions. Together these data indicate that NKRF specifically interacts with the 5΄ETS, ITS1 and ITS2 regions that are only present in early pre-rRNAs and that are removed during the synthesis of the ribosomal subunits.
Figure 3.
NKRF crosslinks to 28S rRNA sequences and the pre-rRNA spacer regions. (A–B) HEK293 cells expressing 2xFlag-PreScission protease cleavage site-His6 (Flag)-tagged NKRF or the tag alone (Flag) were UV crosslinked (UV; A) or treated with 4-thiouridine and crosslinked at 365 nm (PAR; B). RNA–protein complexes were isolated, bound RNAs were trimmed and adapters were ligated. RNAs were amplified by RT-PCR to produce a cDNA library, which was subjected to Illumina deep sequencing. The relative distributions of sequencing reads between different RNA classes after mapping onto the human genome are shown as pie charts. Abbreviations: mRNA – messenger RNA, rRNA – ribosomal RNA, miRNA – microRNA, lncRNA – long non-coding RNA, tRNA – transfer RNA, others – miscellaneous RNAs, snRNA – small nuclear RNA, mitoRNA – mitochondrial encoded RNA, snoRNA – small nucleolar RNA. (C–D) The number of reads per nucleotide mapped to the rDNA gene encoding the 47S pre-rRNA is shown for the Flag and Flag-NKRF samples prepared using (C) UV-CRAC and (D) PAR-CRAC. Schematic views of the 47S pre-rRNA are shown below. (E–G) Magnified views of the reads per nucleotide mapped to the 5΄ external transcribed spacer (5΄ETS; E), internal transcribed spacer 1 (ITS1; F) and 2 (ITS2; G) are shown. The approximate positions of pre-rRNA cleavage sites are indicated below and an asterisk indicates a peak that is present in both the Flag-NKRF and Flag tag control samples. Lower panels indicate the number and position of mutations that arise at sites of UV crosslinking during reverse transcription.
NKRF, XRN2 and DHX15 are required for pre-rRNA processing at the A΄ site in the 5΄ETS
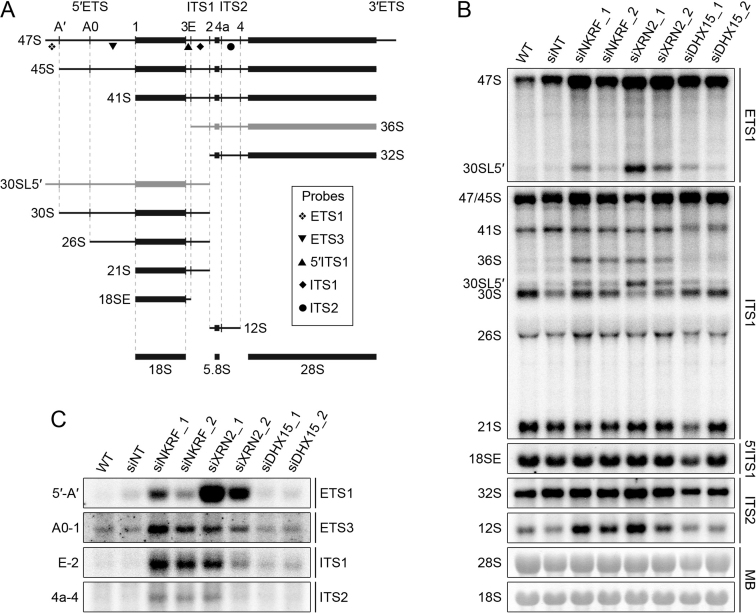
The detection of NKRF crosslinking sites in the pre-rRNA spacer regions as well as its interaction with the exonuclease XRN2 suggests that this complex, also containing DHX15, may be required for maturation of pre-rRNAs. We therefore used northern blotting to analyse the levels of different pre-rRNA species in HeLa cells after and without depletion of NKRF, XRN2 or DHX15 by RNAi (Figure 4A and B; Supplementary Figure S2). Compared to RNA from wild-type cells or those treated with non-target siRNAs, depletion of either NKRF, XRN2 or DHX15 caused accumulation of the initial 47S pre-rRNA transcript and an aberrant, 5΄-extended form of the 30S pre-rRNA, termed 30SL5΄, which together imply that A΄ cleavage is impeded in the knockdown cells and that in some cases, it can be preceded by cleavage at site 2 in ITS1. A΄ cleavage is specific to metazoans where it is the only pre-rRNA processing step that is proposed to take place co-transcriptionally (69). In the light of this, we investigated the effects of depletion of components of the subcomplex on the de novo synthesis of pre-rRNAs using pulse chase metabolic labelling. HeLa cells were transfected with non-target siRNAs or siRNAs targeting NKRF, DHX15 or XRN2 and grown in media containing 32P-labelled orthophosphate, which is incorporated into the backbone of newly synthesized RNAs, and then harvested at different time points after addition of unlabelled media. Total RNA was extracted, separated by agarose-glyoxal gel electrophoresis, transferred to a nylon membrane and labelled mature and specific pre-rRNA species were visualized using a phosphorimager (Supplementary Figure S4). Compared to cells transfected with non-target siRNAs, no significant alterations in the levels of the newly-synthesized mature 28S and 18S rRNAs were observed after depletion of NKRF, DHX15 or XRN2 (Supplementary Figure S4A and B). This is consistent with the observation that depletion of these factors does not fully abolish A΄ cleavage and is also in line with the previously proposed model that XRN2 regulates the order of pre-rRNA cleavage events, but that, through the use of alternative pre-rRNA processing pathways, this does not affect the production of the mature rRNAs (28). Furthermore, although depletion of DHX15 lead to a mild decrease in the levels of the newly synthesised 47/45S pre-rRNAs, neither depletion of XRN2 nor NKRF altered the rate of 47/45S synthesis (Supplementary Figure S4C and D), suggesting that this complex does not play an important role in pre-rRNA transcription by RNA polymerase I.
Figure 4.
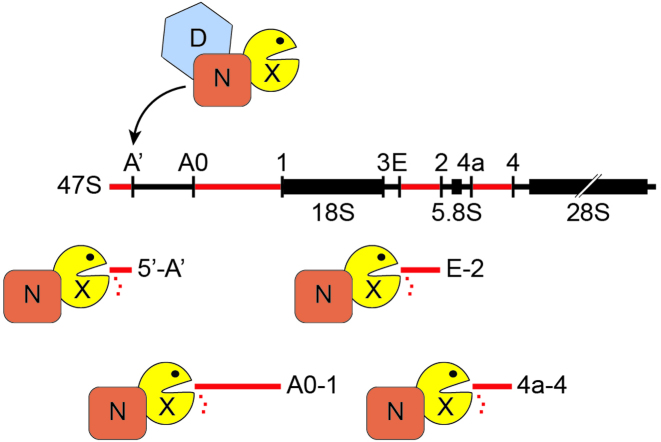
NKRF functions with XRN2 in turnover of excised pre-rRNA spacer fragments and with both XRN2 and DHX15 in A’ cleavage. (A) Schematic view of pre-rRNA processing intermediates in human cells. Mature rRNAs are shown as black rectangles and spacer regions are indicated by black lines (ETS – external transcribed spacer, ITS – internal transcribed spacer). Pre-rRNA cleavage sites are marked by vertical lines and are named above the initial 47S pre-rRNA transcript. Aberrant pre-rRNA species detected when the processing pathway is perturbed are shown in grey. The relative positions of probes used in northern blotting are indicated by the symbols depicted within the box. (B) HeLa cells were transfected with non-target siRNAs (siNT) or siRNAs targeting NKRF (siNKRF_1 or siNKRF_2), XRN2 (siXRN2_1 or siXRN2_2) or DHX15 (siDHX15_1 or siDHX15_2) and harvested after 96 h. Total RNA was extracted and separated by agarose–glyoxal gel electrophoresis and transferred to a nylon membrane. Pre-rRNA species were detected by northern blotting using probes (indicated to the right of the panel) hybridising to the different regions of the pre-rRNA transcript. Mature 28S and 18S rRNAs were visualized by methylene blue staining (MB). (C) Excised pre-rRNA spacer fragments (indicated to the left of each panel), generated by serial endonucleolytic cleavages were also detected on the membrane as in (B) by northern blotting using the probes indicated on the right of each panel. All experiments presented in this figure were performed at least three times and representative data are shown.
Northern blotting of RNA from siRNA-treated cells also revealed that reducing the levels of NKRF or XRN2, but not DHX15, lead to accumulation of the 36S pre-rRNA species that is detected when site 2 cleavage in ITS1 is inhibited, as well as an increase in the levels of the 12S precursor of the mature 5.8S rRNA (Figure 4B). In the case of XRN2, these defects are consistent with previous observations (28). Furthermore, similar to the depletion of XRN2, knockdown of NKRF lead to the accumulation of spacer fragments generated by cleavages in the 5΄ETS (5΄-A΄ and A0-1), ITS1 (E-2) and ITS2 (4a-4), suggesting that NKRF is required for the 5΄-3΄ degradation of these fragments by the exonuclease XRN2 (Figure 4C; 28,30,70,71). Together, these results show that all three factors are required for efficient A΄ cleavage and that NKRF and XRN2, but not DHX15, also function in the turnover of excised spacer fragments generated by serial endonucleolytic cleavages during pre-rRNA maturation.
NKRF is required for the recruitment of XRN2 to nucleolar pre-ribosomal complexes
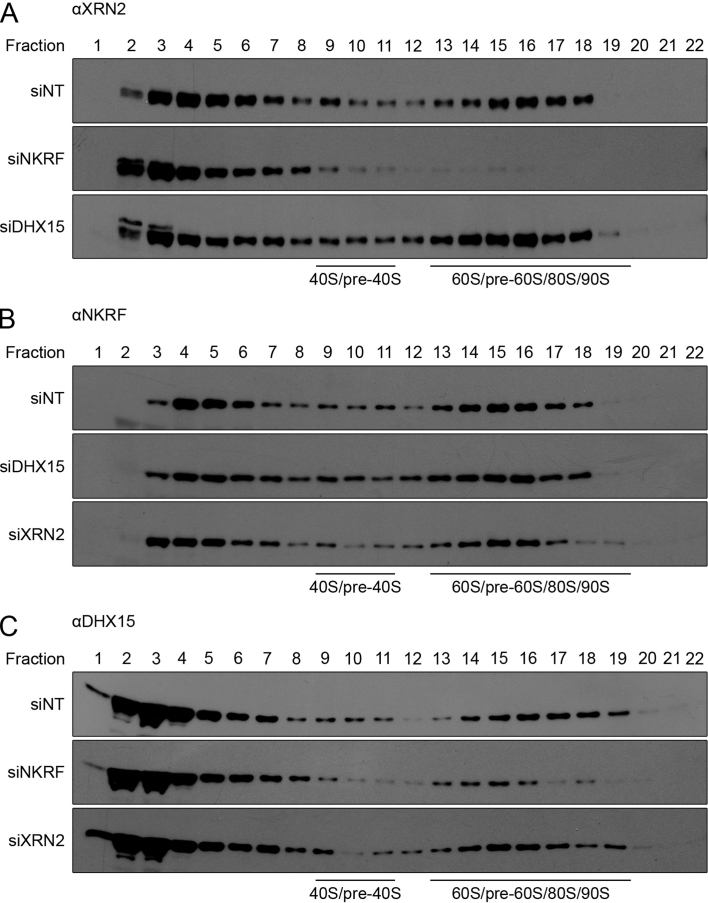
To understand the basis of the similar pre-rRNA processing defects observed upon depletion of NKRF, XRN2 and DHX15 (impaired A΄ cleavage) or NKRF and XRN2 (accumulation of excised spacer fragments), we next investigated how each member of the subcomplex influenced the stability and pre-ribosomal recruitment of the other components. Depletion of any individual protein did not affect the levels of the other two (Supplementary Figure S2), demonstrating that the pre-rRNA processing defects do not arise due to destabilization of the subcomplex as a whole. To investigate the role of NKRF, XRN2 and DHX15 in recruitment of their interaction partners to pre-ribosomal complexes, extracts from siRNA-treated cells were separated by sucrose density gradient centrifugation (Supplementary Figure S5) and the distribution of NKRF, XRN2 and DHX15 between fractions containing pre-ribosomal complexes and non-ribosome-associated proteins was determined by western blotting (Figure 5). As previously observed in wild-type cells (Figure 2D), a significant proportion of all three proteins also co-migrated with pre-ribosomal complexes in cells that had been transfected with non-target siRNAs (Figure 5A–C, upper panels). In contrast, depletion of NKRF, but not DHX15, lead to a lack of XRN2 in fractions containing pre-ribosomal complexes, demonstrating that NKRF is required for the association of XRN2 with pre-ribosomes (Figure 5A, middle and lower panel). Depletion of XRN2 did not, however, influence the amount of NKRF present in fractions containing pre-ribosomal complexes (Figure 5B, lower panel), implying that NKRF can be recruited to pre-ribosomes, independent of its interaction with XRN2. Similarly, depletion of DHX15 did not significantly affect the amount of NKRF co-migrating with pre-ribosomal complexes (Figure 5B, middle panel) and, while depletion of NKRF only mildly affected the recruitment of DHX15 to pre-ribosomal complexes (Figure 5C, middle panel), knockdown of XRN2 did not influence the pre-ribosomal association of DHX15 (Figure 5C, lower panel).
Figure 5.
XRN2 is recruited to pre-ribosomal complexes by NKRF. (A–C) HeLa cells were transfected with non-target siRNAs (siNT) or siRNAs targeting NKRF (siNKRF), DHX15 (siDHX15) or XRN2 (siXRN2) and harvested after 96 h. Extracts were prepared, separated by sucrose density gradient centrifugation and the distribution of XRN2 (A), NKRF (B) and DHX15 (C) were monitored by western blotting using antibodies against the endogenous proteins. Fraction numbers and the positions of ribosomal complexes are indicated above and below each panel respectively. All experiments presented in this figure were performed at least three times and representative data are shown.
It has been shown recently that XRN2 is retained in the nucleoplasm by an interaction with the collaborator of alternative reading frame protein (CARF; 55). Furthermore, overexpression of CARF leads to the accumulation of excised spacer fragments and defects in pre-rRNA processing (55) that are also observed upon depletion of either XRN2 or NKRF (Figure 4B and C). Together with the finding that NKRF is needed for recruitment of XRN2 to pre-ribosomal complexes, this suggests that the interaction between CARF and XRN2 maintains a nucleoplasmic pool of XRN2 and that the association of XRN2 with NKRF may enable localization of XRN2 to the nucleolus. To test this, we used RNAi to deplete NKRF (or DHX15) from a HEK293 cell line in which expression of GFP-XRN2 was induced during the last 24 h of the knockdown, and then examined the localization of XRN2 by fluorescence microscopy (Supplementary Figure S6). In cells transfected with non-target siRNAs, XRN2 was localized throughout the nucleoplasm and nucleolus. Upon depletion of NKRF but not DHX15, XRN2 was, however, excluded from nucleoli and was exclusively present in the nucleoplasm. Together, these data show that NKRF functions to recruit XRN2 to the nucleolus and to pre-ribosomal complexes, indicating that the accumulation of excised pre-rRNA spacer fragments upon depletion of NKRF likely arises due to a lack of XRN2 in the nucleolus.
The catalytic activity of DHX15 is stimulated by NKRF and is required for efficient A΄ cleavage
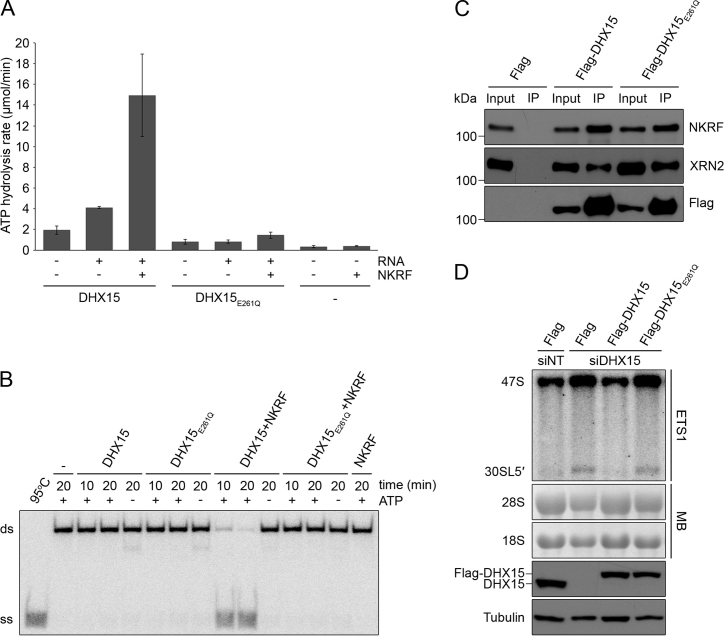
Our findings that both NKRF and DHX15 are required for efficient cleavage at the A΄ site (Figure 4B) and that the G-patch domain of NKRF and C-terminal region of DHX15 containing the OB-fold mediate the interaction between these proteins (Figure 1E and F) raises the possibility that, similar to other G-patch protein–RNA helicase partners, NKRF influences the activity of DHX15. We therefore purified recombinant MBP- and His14-tagged NKRF (Supplementary Figure S1) and DHX15, as well as DHX15 carrying a glutamate to glutamine substitution (DHX15E261Q; Supplementary Figure S7) in the conserved DEAH motif that is proposed to function in NTP binding and hydrolysis (52,72), and performed in vitro ATPase assays. DHX15 was weakly able to hydrolyse ATP in the absence of RNA and this activity was stimulated more than two-fold by the addition of RNA (Figure 6A). Addition of NKRF, which does not itself hydrolyse ATP, in the presence of RNA, further stimulated the ATPase activity of DHX15 by more than 3-fold. In contrast, the basal activity of DHX15E261Q in the absence of RNA was lower than that of the wild-type protein and the activity of this protein remained very low even in the presence of RNA or NKRF (Figure 6A). Next, we investigated the effect of NKRF on the helicase activity of DHX15 by performing unwinding assays. A radiolabelled RNA–DNA duplex served as a substrate for DHX15 and unwinding assays were carried out in the presence or absence of ATP, and with or without addition of NKRF. No unwinding was observed in the absence of the helicase or in the presence of DHX15 alone with or without ATP, which is consistent with previous reports for Prp43 (Figure 6B; see for example, 53). In contrast, addition of DHX15 together with NKRF caused significant unwinding of the duplex in the presence of ATP and the extent of duplex unwinding was increased by prolonged incubation with the helicase and G-patch protein (Figure 6B). In line with the lack of ATP hydrolysis by DHX15E261Q, no unwinding was observed by this protein upon addition of ATP or NKRF, confirming that the E261Q substitution in the DEAH motif renders DHX15 catalytically inactive. Together these results demonstrate that NKRF functions as a cofactor for DHX15 that stimulates its ATPase and helicase activities.
Figure 6.
The catalytic activity of DHX15 is stimulated by NKRF and is required for A’ cleavage. (A) The ATPase activity of recombinantly expressed, purified DHX15 and DHX15 carrying an E261Q mutation in the conserved DEAH-box (DHX15E261Q) was determined in the presence (+) and absence (−) of RNA and NKRF using an NADH-coupled assay. The ATPase activity of NKRF alone was also measured as a control. Data from three independent experiments are shown as mean ± SD. (B) Unwinding assays were performed using no helicase (−), DHX15 or DHX15E261Q and a radiolabelled RNA–DNA duplex in the presence (+) or absence (−) of ATP and NKRF. After incubation for the indicated times, samples were analysed by native PAGE and visualized using a phosphorimager. 95°C – denatured duplex, ds – double stranded RNA–DNA substrate, ss – single stranded DNA oligonucleotide. (C) Extracts prepared from HEK293 cell lines expressing Flag-tagged DHX15, DHX15E261Q or the Flag-tag alone (Flag) were used in immunoprecipitation experiments and input samples (1%) and eluates (IP) were analysed by western blotting using the indicated antibodies. (D) HEK293 cell lines capable of expressing Flag-tagged, siRNA-resistant DHX15, DHX15E261Q or the Flag-tag alone were transfected with non-target siRNAs (siNT) or siRNAs targeting the endogenous DHX15 mRNA and RNAi was performed for 96 h. Expression of Flag-tagged proteins was induced by addition of tetracycline 24 h before harvesting. The levels of Flag-tagged and endogenous DHX15 protein were determined by western blotting using an antibody against the endogenous protein and Tubulin served as a loading control. RNA was extracted and pre-rRNA levels were determined by northern blotting using a probe hybridizing upstream of the A’ cleavage site in the 5΄ETS (ETS1). Mature 28S and 18S rRNAs were visualized by methylene blue staining (MB). The experiments presented in panels B–D were performed at least three times and representative data are shown.
To test if the catalytic activity of DHX15 is required for efficient A΄ cleavage in the 5΄ETS of the pre-rRNA transcript in vivo, we generated an RNAi-based rescue system in HEK293 cells in which endogenous DHX15 could be depleted by RNAi and expression of Flag-tagged, siRNA-insensitive DHX15 or DHX15E261Q could be induced to similar levels by addition of tetracycline. To verify that the inactive DHX15E261Q protein can associate with the other members of the subcomplex, we first performed IPs using lysate prepared from these cell lines, which demonstrated that the interactions of DHX15 with NKRF and XRN2 are not perturbed by this mutation (Figure 6C). The efficiency of the knockdown and expression of the tagged proteins to the endogenous level were verified by western blotting using an antibody against DHX15 that is also able to detect the Flag-tagged isoforms, which migrate slower in the gel (Figure 6D, lower panels). Examination of the levels of the 47S and 30SL5΄ pre-rRNAs revealed that, consistent with our earlier results (Figure 4B), depletion of DHX15 in cells expressing the Flag-tag alone, lead to accumulation of these precursors compared to cells treated with non-target siRNAs. These defects were rescued by expression of Flag-DHX15, confirming that they are specific for lack of this helicase (Figure 6D). In contrast, expression of the Flag-DHX15E261Q in the context of the DHX15 knockdown, did not rescue the defects, indicating that efficient processing at the A΄ site requires the catalytic activity of DHX15. Together these results demonstrate that NKRF functions as a cofactor for DHX15 by stimulating its ATPase and unwinding activities and that this catalytic activity of DHX15 is required for efficient cleavage at the A΄ site during pre-rRNA maturation.
DISCUSSION
A key aspect of the production of cytoplasmic ribosomes in eukaryotic cells is the processing of three of the mature rRNAs from the initial pre-rRNA transcript. This rRNA maturation is achieved by a series of endonucleolytic cleavages at specific sites within the transcribed spacer regions that flank and are inserted in between the mature rRNA sequences. In some cases these cleavages are co-ordinated with exonucleolytic processing to produce the mature 5΄ or 3΄ ends of the rRNAs (5). While the rRNA maturation pathway is broadly conserved in eukaryotes, an additional cleavage event, compared to yeast, occurs at the A΄ site in the 5΄ETS during processing of the human pre-rRNA transcript. In addition, there appears to be a greater degree of flexibility in the order of pre-rRNA cleavages leading to several alternative human pre-rRNA processing pathways. Here, we identify a nucleolar subcomplex composed of the G-patch protein NKRF, the DEAH-box helicase DHX15 and the 5΄-3΄ exonuclease XRN2, and show that interactions and functions of these proteins play important roles in various aspects of pre-rRNA processing in human cells (Figure 7).
Figure 7.
Interactions between NKRF, XRN2 and DHX15 are required for pre-rRNA processing and turnover of excised spacer fragments. NKRF (N), XRN2 (X) and DHX15 (D) form a complex and all three components are required for efficient cleavage at site A΄ in the 5΄ external transcribed spacer (5΄ETS) of the nascent 47S pre-rRNA transcript. Furthermore, NKRF recruits XRN2 to pre-ribosomal complexes for degradation of pre-rRNA fragments that are excised by serial endonucleolytic cleavages during rRNA maturation. Cleavage sites are indicated above the 47S pre-rRNA transcript and spacer fragments are named accordingly.
Although the yeast orthologue of DHX15, Prp43, is implicated in the release of a cluster of snoRNAs from pre-60S complexes (14) and in the cleavage of the 3΄ end of the 18S rRNA by Nob1 (73), to date, a function of DHX15 in ribosome biogenesis in human cells has not been described. Our data show that DHX15 is associated with pre-ribosomal complexes and furthermore, that its catalytic activity is required for efficient cleavage at site A΄. By analogy to the proposed role for Prp43 in facilitating Nob1-mediated cleavage in yeast, this could suggest a model in which structural remodelling of 5΄ETS regions of pre-rRNA transcripts by DHX15 is required for the currently elusive A΄ endonuclease to gain access to its cleavage site. Alternatively, it is possible that the helicase activity of DHX15 contributes to the recruitment or release of other proteins and that this is important for A΄ cleavage to take place efficiently. In yeast, the activity of Prp43 is regulated by a number of dedicated cofactors that contain a G-patch domain (17), including Pxr1 and Sqs1 that act together with Prp43 in ribosome biogenesis (15,16). Similarly, in human cells several G-patch proteins including TFIP11 and RBM5, have been shown to interact with and stimulate the activity of DHX15 for its functions in (alternative) pre-mRNA splicing (58,74). However, although the interaction of Pxr1-Prp43 is conserved in humans (PINX1-DHX15; 16), an RNAi-based screen for identification of human ribosome biogenesis factors did not reveal any significant pre-rRNA processing defects in cells depleted of PINX1 (40). We find that DHX15 also interacts with the human G-patch protein NKRF and that, as for other G-patch protein-helicase interactions (e.g. Spp382-Prp43), the G-patch domain of NKRF and the C-terminal OB-fold of DHX15 primarily mediate the association between these proteins. The functional relevance of this interaction is supported by the finding that NKRF stimulates the ATPase and unwinding activity of DHX15 and therefore serves as a bona fide cofactor of the RNA helicase. In vivo, similar to DHX15, NKRF is also required for A’ cleavage, implying that NKRF likely stimulates the catalytic activity of DHX15 to enable this pre-rRNA processing step to occur efficiently.
Interestingly, NKRF, but not DHX15, is also required for the turnover of pre-rRNA spacer fragments that are excised from the 5΄ETS (5΄-A΄ and A0-1), ITS1 and ITS2 during pre-rRNA processing. Here, NKRF functions together with XRN2, which has previously been identified as the major exonuclease responsible for the degradation of these fragments in various different organisms (28,30,70,71,75,76). Depletion of NKRF or XRN2 also leads to accumulation of the 30SL5΄ and 36S pre-rRNAs, indicating defects in the cleavages at the A΄ site and site 2 when either of these proteins is lacking. While these pre-rRNA processing defects may reflect a direct requirement for these factors in the cleavage steps, they may also arise due to impaired degradation of the spacer fragments leading to failure to recycle biogenesis factors that are important for these processing events.
Similar pre-rRNA processing defects have also been observed previously upon overexpression of another XRN2 interaction partner, CARF, and it was shown that increased levels of CARF lead to nucleoplasmic accumulation of XRN2, implying that this causes withdrawal of the exonuclease from its nucleolar functions in pre-rRNA processing and turnover (55). The finding that NKRF is responsible for the recruitment of XRN2 to nucleoli, suggests that NKRF and CARF work antagonistically to regulate the distribution of this multifunctional exonuclease between its nucleolar and nucleoplasmic substrates. In humans, CARF and in C. elegans, PAXT-1, have been shown to have an ‘XRN-Two-Binding-Domain’ (XTBD) that is similar to the DUF3469 that is present in NKRF (54–56). Interestingly, we observe, however, that the DUF3469 domain of NKRF is not essential for binding of XRN2. Consistent with this, an independent study recently reported that a region downstream of the DUF3469 is important for the interaction between NKRF and XRN2 (77). Our results therefore suggest that although the DUF3469 domain of NKRF may form contacts with XRN2, the nature of the XRN2-NKRF and XRN2-CARF interactions might differ, with other regions of NKRF likely contributing to its association with XRN2.
The identification of binding sites of NKRF in the spacer regions of the pre-rRNA transcript together with the detection of stable interactions between NKRF and both DHX15 and XRN2 highlight NKRF as the central hub of a novel pre-ribosomal subcomplex that functions in the early stages of ribosomal subunit assembly in human cells. Our data further suggest that, on the one hand, the interaction between the G-patch protein NKRF and the DEAH-box helicase DHX15 stimulates the catalytic activity of the helicase for a remodelling event that may facilitate the initial pre-rRNA cleavage event at the A΄ site. On the other hand, the association of NKRF and XRN2 is essential for the recruitment of the exonuclease to nucleolar pre-ribosomal complexes where in addition to A΄ cleavage, these two proteins also function together in the degradation of excised spacer fragments.
ACCESSION NUMBERS
GEO database (http://www.ncbi.nlm.nih.gov/geo/): identifier GSE92733.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Uwe Plessmann and Henning Urlaub for mass spectrometry, Annika Heininger for initial help with ATPase assays and Jens Kretschmer for help with bioinformatics analysis.
Footnotes
Present address: Carmen Doebele, Hematology/Oncology, Department of Medicine, Johann Wolfgang Goethe University, 60590 Frankfurt, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [BO 3442/1-1 to M.T.B.]; Göttingen University Medical Department [to M.T.B.]; Alexander von Humboldt postdoctoral fellowship [to K.E.S.]. Funding for open access charge: Göttingen University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 2. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomson E., Ferreira-Cerca S., Hurt E.. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013; 126:4815–4821. [DOI] [PubMed] [Google Scholar]
- 4. Mullineux S.T., Lafontaine D.L.. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand?. Biochimie. 2012; 94:1521–1532. [DOI] [PubMed] [Google Scholar]
- 5. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 7. Sharma S., Lafontaine D.L.. 'View From A Bridge': a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 8. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R. et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 9. Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Böttcher B., Granneman S., Watkins N.J., Tollervey D.. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012; 19:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sloan K.E., Gleizes P.E., Bohnsack M.T.. Nucleocytoplasmic transport of RNAs and RNA-Protein complexes. J. Mol. Biol. 2016; 428:2040–2059. [DOI] [PubMed] [Google Scholar]
- 11. Strunk B.S., Karbstein K.. Powering through ribosome assembly. RNA. 2009; 15:2083–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kressler D., Hurt E., Bassler J.. Driving ribosome assembly. Biochim. Biophys. Acta. 2010; 1803:673–683. [DOI] [PubMed] [Google Scholar]
- 13. Matsuo Y., Granneman S., Thoms M., Manikas R.G., Tollervey D., Hurt E.. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014; 505:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bohnsack M.T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D.. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009; 36:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lebaron S., Papin C., Capeyrou R., Chen Y.L., Froment C., Monsarrat B., Caizergues-Ferrer M., Grigoriev M., Henry Y.. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009; 28:3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y.L., Capeyrou R., Humbert O., Mouffok S., Kadri Y.A., Lebaron S., Henras A.K., Henry Y.. The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res. 2014; 42:7330–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robert-Paganin J., Réty S., Leulliot N.. Regulation of DEAH/RHA helicases by G-patch proteins. Biomed. Res. Int. 2015; 931857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bohnsack M.T., Kos M., Tollervey D.. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008; 9:1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young C.L., Khoshnevis S., Karbstein K.. Cofactor-dependent specificity of a DEAD-box protein. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2668–E2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin R., Hackert P., Ruprecht M., Simm S., Brüning L., Mirus O., Sloan K.E., Kudla G., Schleiff E., Bohnsack M.T.. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA. 2014; 20:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khoshnevis S., Askenasy I., Johnson M.C., Dattolo M.D., Young-Erdos C.L., Stroupe M.E., Karbstein K.. The DEAD-box protein Rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from Pre-40S ribosomes to allow for 60S maturation. PLoS Biol. 2016; 14:e1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narla A., Ebert B.L.. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010; 115:3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sondalle S.B., Baserga S.J.. Human diseases of the SSU processome. Biochim. Biophys. Acta. 2014; 1842:758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stumpf C.R., Ruggero D.. The cancerous translation apparatus. Curr. Opin. Genet. Dev. 2011; 21:474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dai M.S., Challagundla K.B., Sun X.X., Palam L.R., Zeng S.X., Wek R.C., Lu H.. Physical and functional interaction between ribosomal protein L11 and the tumor suppressor ARF. J. Biol. Chem. 2012; 287:17120–17129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bursac S., Brdovcak M.C., Donati G., Volarevic S.. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta. 2014; 1842:817–830. [DOI] [PubMed] [Google Scholar]
- 27. Sloan K.E., Bohnsack M.T., Watkins N.J.. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013; 5:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J., Watkins N.J.. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013; 200:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Preti M., O’Donohue M.F., Montel-Lehry N., Bortolin-Cavaillé M.L., Choesmel V., Gleizes P.E.. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013; 41:4709–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sloan K.E., Bohnsack M.T., Schneider C., Watkins N.J.. The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA. 2014; 20:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P.. Structure of the human 80S ribosome. Nature. 2015; 520:640–645. [DOI] [PubMed] [Google Scholar]
- 32. Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E.. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014; 42:1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U.. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010; 8:e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sloan K.E., Leisegang M.S., Doebele C., Ramírez A.S., Simm S., Safferthal C., Kretschmer J., Schorge T., Markoutsa S., Haag S. et al. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res. 2015; 43:553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wandrey F., Montellese C., Koos K., Badertscher L., Bammert L., Cook A.G., Zemp I., Horvath P., Kutay U.. The NF45/NF90 heterodimer contributes to the biogenesis of 60S ribosomal subunits and influences nucleolar morphology. Mol. Cell. Biol. 2015; 35:3491–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larburu N., Montellese C., O’Donohue M.F., Kutay U., Gleizes P.E., Plisson-Chastang C.. Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res. 2016; 44:8465–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I.. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002; 12:1–11. [DOI] [PubMed] [Google Scholar]
- 38. Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J.. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002; 13:4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M.. Nucleolar proteome dynamics. Nature. 2005; 433:77–83. [DOI] [PubMed] [Google Scholar]
- 40. Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013; 51:539–551. [DOI] [PubMed] [Google Scholar]
- 41. Badertscher L., Wild T., Montellese C., Alexander L.T., Bammert L., Sarazova M., Stebler M., Csucs G., Mayer T.U., Zamboni N. et al. Genome-wide RNAi Screening Identifies Protein Modules Required for 40S Subunit Synthesis in Human Cells. Cell Rep. 2016; 13:2879–2891. [DOI] [PubMed] [Google Scholar]
- 42. Nourbakhsh M., Hauser H.. Constitutive silencing of IFN-beta promoter is mediated by NRF (NF-kappaB-repressing factor), a nuclear inhibitor of NF-kappaB. EMBO J. 1999; 18:6415–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng X., Guo Z., Nourbakhsh M., Hauser H., Ganster R., Shao L., Geller D.A.. Identification of a negative response element in the human inducible nitric-oxide synthase (hiNOS) promoter: The role of NF-kappa B-repressing factor (NRF) in basal repression of the hiNOS gene. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:14212–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Niedick I., Froese N., Oumard A., Mueller P.P., Nourbakhsh M., Hauser H., Köster M.. Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J. Cell Sci. 2004; 117:3447–3458. [DOI] [PubMed] [Google Scholar]
- 45. Huang K.H., Wang C.H., Lin C.H., Kuo HP.. NF-κB repressing factor downregulates basal expression and mycobacterium tuberculosis induced IP-10 and IL-8 synthesis via interference with NF-κB in monocytes. J. Biomed. Sci. 2014; 21:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chamousset D., Mamane S., Boisvert F.M., Trinkle-Mulcahy L.. Efficient extraction of nucleolar proteins for interactome analyses. Proteomics. 2010; 10:3045–3050. [DOI] [PubMed] [Google Scholar]
- 47. Warda A.S., Freytag B., Haag S., Sloan K.E., Görlich D., Bohnsack M.T.. Effects of the Bowen-Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum. Mol. Genet. 2016; doi:10.1093/hmg/ddw351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bohnsack M.T., Tollervey D., Granneman S.. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzymol. 2012; 511:275–288. [DOI] [PubMed] [Google Scholar]
- 49. Haag S., Warda A.S., Kretschmer J., Günnigmann M.A., Höbartner C., Bohnsack M.T.. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015; 21:1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haag S., Kretschmer J., Bohnsack M.T.. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA. 2015; 21:180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heininger A.U., Hackert P., Andreou A.Z., Boon K.L., Memet I., Prior M., Clancy A., Schmidt B., Urlaub H., Schleiff E. et al. Protein cofactor competition regulates the action of a multifunctional RNA helicase in different pathways. RNA Biol. 2016; 13:320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walbott H., Mouffok S., Capeyrou R., Lebaron S., Humbert O., van Tilbeurgh H., Henry Y., Leulliot N.. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010; 29:2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christian H., Hofele R.V., Urlaub H., Ficner R.. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014; 42:1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miki T.S., Richter H., Rüegger S., Großhans H.. PAXT-1 promotes XRN2 activity by stabilizing it through a conserved domain. Mol. Cell. 2014; 53:351–360. [DOI] [PubMed] [Google Scholar]
- 55. Sato S., Ishikawa H., Yoshikawa H., Izumikawa K., Simpson R.J., Takahashi N.. Collaborator of alternative reading frame protein (CARF) regulates early processing of pre-ribosomal RNA by retaining XRN2 (5΄-3΄ exoribonuclease) in the nucleoplasm. Nucleic Acids Res. 2015; 43:10397–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richter H., Katic I., Gut H., Großhans H.. Structural basis and function of XRN2 binding by XTB domains. Nat. Struct. Mol. Biol. 2016; 23:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fouraux M.A., Kolkman M.J., Van der Heijden A., De Jong A.S., Van Venrooij W.J., Pruijn G.J.. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002; 8:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wen X., Tannukit S., Paine M.L.. TFIP11 interacts with mDEAH9, an RNA helicase involved in spliceosome disassembly. Int. J. Mol. Sci. 2008; 9:2105–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mosallanejad K., Sekine Y., Ishikura-Kinoshita S., Kumagai K., Nagano T., Matsuzawa A., Takeda K., Naguro I., Ichijo H.. The DEAH-box RNA helicase DHX15 activates NF-κB and MAPK signaling downstream of MAVS during antiviral responses. Sci. Signal. 2014; 7:ra40. [DOI] [PubMed] [Google Scholar]
- 60. Miki T.S., Großhans H.. The multifunctional RNase XRN2. Biochem. Soc. Trans. 2013; 41:825–830. [DOI] [PubMed] [Google Scholar]
- 61. Kos M., Tollervey D.. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in saccharomyces cerevisiae. Mol. Cell. 2005; 20:53–64. [DOI] [PubMed] [Google Scholar]
- 62. Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dosil M., Bustelo X.R.. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004; 279:37385–37397. [DOI] [PubMed] [Google Scholar]
- 64. Hu L., Wang J., Liu Y., Zhang Y., Zhang L., Kong R., Zheng Z., Du X., Ke Y.. A small ribosomal subunit (SSU) processome component, the human U3 protein 14A (hUTP14A) binds p53 and promotes p53 degradation. J. Biol. Chem. 2010; 286:3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D.. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2007; 27:3682–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Õunap K., Käsper L., Kurg A., Kurg R.. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS One. 2013; 8:e75686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al. PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 2010; 41:e2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lazdins I.B., Delannoy M., Sollner-Webb B.. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997; 105:481–495. [DOI] [PubMed] [Google Scholar]
- 70. Schillewaert S., Wacheul L., Lhomme F., Lafontaine D.L.. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell. Biol. 2012; 32:430–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang M., Pestov D.G.. 5΄-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 2011; 39:1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 73. Pertschy B., Schneider C., Gnädig M., Schäfer T., Tollervey D., Hurt E.. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009; 284:35079–35091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Niu Z., Jin W., Zhang L., Li X.. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012; 586:977–983. [DOI] [PubMed] [Google Scholar]
- 75. Petfalski E., Dandekar T., Henry Y., Tollervey D.. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998; 18:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zakrzewska-Placzek M., Souret F.F., Sobczyk G.J., Green P.J., Kufel J.. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010; 38:4487–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rother S., Bartels M., Schweda A.T., Resch K., Pallua N., Nourbakhsh M.. NF-κB-repressing factor phosphorylation regulates transcription elongation via its interactions with 5΄→3΄ exoribonuclease 2 and negative elongation factor. FASEB J. 2016; 30:174–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.