Abstract
CRISPR-Cas systems allow bacteria and archaea to acquire sequence-specific immunity against selfish genetic elements such as viruses and plasmids, by specific degradation of invader DNA or RNA. However, this involves the risk of autoimmunity if immune memory against host DNA is mistakenly acquired. Such autoimmunity has been shown to be highly toxic in several bacteria and is believed to be one of the major costs of maintaining these defense systems. Here we generated an experimental system in which a non-essential gene, required for pigment production and the reddish colony color, is targeted by the CRISPR-Cas I-B system of the halophilic archaeon Haloferax volcanii. We show that under native conditions, where both the self-targeting and native crRNAs are expressed, self-targeting by CRISPR-Cas causes no reduction in transformation efficiency of the plasmid encoding the self-targeting crRNA. Furthermore, under such conditions, no effect on organismal growth rate or loss of the reddish colony phenotype due to mutations in the targeted region could be observed. In contrast, in cells deleted for the pre-crRNA processing gene cas6, where only the self-targeting crRNA exists as mature crRNA, self-targeting leads to moderate toxicity and the emergence of deletion mutants. Sequencing of the deletions caused by CRISPR-Cas self targeting indicated DNA repair via microhomology-mediated end joining.
INTRODUCTION
CRISPR-Cas systems provide microorganisms with acquired and heritable immunity to selfish genetic elements such as viruses and plasmids (for reviews see (1–3)). This protective immunity is directed against foreign DNA that is identical or near-identical to previously acquired sequences known as spacers, which are characteristically flanked by repeats; this arrangement is denoted CRISPR (clustered regularly interspaced short palindromic repeats). The CRISPR-associated Cas enzymes mediate the degradation of the invasive DNA elements. However, as with every defense system, there exists a risk for auto-immunity—in this case, cleavage of host chromosomal DNA when self-targeting spacers (sequences identical to the host chromosome) are mistakenly acquired. This phenomenon has been shown to be common in microbial genomes and usually is resolved by mutational events that either eliminate the recognition of the self sequence (4) or disrupt the activity of the CRISPR-Cas system, resulting in ‘escape mutants’ (4,5). In Escherichia coli BW25113Δhns, where the CRISPR type I-E system is active, plasmids that contain spacers that match a prophage within its genome are only rarely maintained, indicating that self-targeting genome degradation is subject to strong negative selection (6). Similarly, self-targeting of the chromosome has been shown to be highly toxic and often causes large deletions in type I-F (7) and type II (8) CRISPR-Cas systems. In Sulfolobus islandicus, a member of the Crenarchaeota that contains a type I-A and two type III-B CRISPR-Cas systems, self-targeting of the chromosome by a plasmid resulted in lethality, and all cells that acquired the plasmid had mutations in the targeted region (9). Unlike type I and type II systems, the type III-A CRISPR-Cas system of Staphylococcus epidermidis has been shown to degrade DNA targets only in the presence of corresponding transcripts, ensuring that genomic regions that are not transcribed are effectively protected (10,11).
Some CRISPR-Cas systems in archaea are plasmid-encoded (12). Of the haloarchaea documented in CRISPRdb (13), a little over 50% (13/23) of those strains whose genomes encode CRISPR-Cas systems have either CRISPR arrays, cas genes or both located on mega-plasmids/mini-chromosomes. Because CRISPR arrays of many archaea contain spacers that target chromosomal genes of related species (14) or spacers that target integrated proviruses or plasmids (15), there is the danger that a plasmid that is horizontally acquired may target the host genome due to a spacer that had been acquired in one of its previous hosts. Similarly, a self-targeting spacer can be ‘acquired’, without any cas gene activity but rather via homologous recombination (HR) between CRISPR arrays, located either on chromosomes or plasmids, of two cells that engage in lateral gene transfer. Nevertheless, CRISPR-Cas systems are highly abundant in Euryarchaeota, including haloarchaea, where they are present in nearly 80% of genomes contained in CRISPRdb, despite the potential for acquisition of self-targeting spacers. This abundance implies that mechanisms exist to curtail the damage caused by plasmids containing spacers that target the chromosome, or in the very least ‘buy time’ so that mutational events can enable escape from auto-immunity.
To study how Euryarchaeota may tolerate auto-immunity, we focused on the type I-B system of Haloferax volcanii, a halophilic archaeon that contains a well-characterized plasmid-encoded CRISPR-Cas system (5,16,17). The plasmid encoding that system, pHV4, was previously shown to successfully invade during natural mating between H. volcanii and Haloferax mediterranei (18). pHV4 was found to replace the native H. mediterranei CRISPR-encoding plasmid pHM500 in multiple recombinant lineages, all of which had chromosomes that were over 85% H. mediterranei with the rest of their genomes originating from the H. volcanii parent (18). By generating a spacer that targets an actively-transcribed but non-essential gene, we studied the effects of self-targeting of the chromosome on cellular fitness.
MATERIALS AND METHODS
Strains and culture conditions
Strains and plasmids used in this work are detailed in Supplementary Table S1. Strains H119, Δcas6, HV30 (Δcas6 Δcas3) and HV31 (ΔpyrE2, ΔleuB, ΔtrpA, ΔHVO_2,385,045-2,386,660::trpA, ΔHVO_pHV4: 204,834-218,566) were grown aerobically at 45°C in Hv-YPC medium (19). Strains with plasmids were grown in Hv-Ca or Hv-Min medium with the necessary supplements. E. coli strains DH5α and GM121 were grown aerobically at 37°C in 2YT medium.
Construction of plasmids expressing crRNAs targeting crtI
crRNAs against the open reading frame of crtI were generated by inverse polymerase chain reaction (PCR) with pMA-RQ-telecrRNA as template (primers for crRNA#1: crtI1/crtI2; for crRNA#2:crtI3/crtI4; for crRNA#3:crtI5/crtI6). Primers omit the original spacer and contain the new spacer sequence. Obtained plasmids contain a synthetic Haloferax promoter (A. Sabag-Daigle and C. J. Daniels, in preparation), the crRNA flanked by t-elements and a synthetic Haloferax terminator (A. Sabag-Daigle and C. J. Daniels, in preparation). The complete insert was excised from plasmids using KpnI and BamHI, purified and ligated into the Haloferax shuttle vector pTA232 (19). The resulting plasmids were termed pTA232–crRNA#1, pTA232–crRNA#2 and pTA232–crRNA#3. The insert containing crRNA#3 was additionally ligated into the Haloferax shuttle vector pTA352 (20), yielding plasmid pTA352–crRNA#3. Spacer sequences were as follows (5΄ to 3΄): crRNA#1:
GGCAAGCGGCCCGAGGACTACTACGAACTGACGCGG, crRNA#2: GAGTCCTACGAACCCGGCGCGGGCGACAGGCTCGAC, and
crRNA#3: CTCTGCGACCAGGTCGTCTCCGACGCCGACTACGCC. All spacers targeted sequences flanked by the TTC protospacer-adjacent motif (PAM) (5).
Northern analyzes for crRNA expression
Total RNA was isolated from exponentially growing H. volcanii cells as described in (26). After separation of 10 μg RNA (total RNA) on an 8% polyacrylamide gel electrophoresis (PAGE), RNA molecules were transferred to nylon membranes (Hybond-XL, GE Healthcare) and incubated with radioactively labeled DNA oligonucleotides specific for the crRNA or the 5S rRNA. Oligonucleotides used as hybridization probes were radioactively labeled at the 5΄ end with γ-32P-adenosine triphosphate.
Determination of transformation efficiencies and determination of white clones
Before transformation of Haloferax cells using the polyethylene glycol (PEG) method, all plasmids were passaged through E. coli dam− strain GM121 to avoid methylation. Transformations were carried out as described earlier (21). Haloferax strains H119 and Δcas6 were transformed with a plasmid expressing a self-targeting crRNA (pTA232–crRNA#1-#3, pTA352–crRNA#3). As a control, strains were transformed with the respective vector without insert (pTA232 or pTA352). After growth of colonies, the total number of colonies as well as the number of white and red colonies were determined. Each transformation reaction was repeated at least four times.
To determine transformation efficiency under expression of Cas3 variant D444A (22), Haloferax strain HV30 was transformed with a plasmid carrying the gene for the mutated Cas3 (22). This strain was subsequently transformed with pTA232–crRNA#3 and pTA232 as a control.
Determination of cell fitness during self-targeting
To determine cell fitness during self-targeting, strain Δcas6 was transformed with the plasmid carrying the self-targeting crRNA (pTA232–crRNA#3); as a control, cells were transformed with the plasmid without insert (pTA232). Plasmids were passaged through E. coli strain GM121 to avoid methylation and subsequently introduced into Haloferax cells using the PEG method. After transformation, 3 × 4 ml of Hv-Min + Trp + Ura medium were inoculated as follows: (i) with 100 μl of transformation culture Δcas6xpTA232, (ii) with 100 μl of transformation culture Δcas6xpTA232–crRNA#3 and (iii) with 50 μl each of both transformation cultures. Aliquots of cultures were taken at different optical density (ODs) (0.1–0.4; 0.4–0.6; 0.6–0.9) and plated in appropriate dilutions. Single colonies were counted and the number of white colonies was set into relation to the number of red colonies for culture number three (growth in head to head competition). To confirm that the vast majority of red colonies harbor the plasmid pTA232 without insert and are not a result of escape mutations, PCR was performed on 50 red colonies with primers that bind in the multiple cloning site of the vector. To analyze growth behavior during self-targeting, cultures (i)–(iii) were inoculated to an OD650nm of 0.01 and were grown to stationary phase while their OD650nm was being monitored every 2 h. Additionally, strain HV31 and five separate clones of Δcas6 expressing crRNA#3 were grown to exponential phase as precultures. These precultures were used to inoculate fresh cultures to an OD650nm of 0.01 that were grown to stationary phase while their OD650nm was being monitored every 3 h.
Determination of escape mutations
PCR was performed on red colonies using primers P1-Promi-up (forward-primer binding upstream of CRISPR-locus P1) and P2-do (reverse-primer binding downstream of CRISPR-locus P2). Since a product consisting of both loci and the cas gene cassette would have a size of about 12 kbp and is therefore too large to be amplified in a standard PCR, a PCR-product is only obtained if the cas gene cassette is deleted.
Determination of deletions in crtI
PCR was performed on white colonies with primers crtI#11 and crtI#12, amplifying the entire open reading frame of crtI and ca. a total of 100 bp upstream and downstream of the gene. Obtained PCR-products were purified and ligated into an EcoRV digested pBluescriptII (Stratagene). Plasmids containing PCR-products were sequenced at Seqlab Sequence Laboratories using the Sanger method.
Analysis of sequences surrounding the recombination site
For the 14 identified deletions, sequences surrounding the recombination site were scanned for homologous sequences. Microhomologies were defined as at least three homologous base pairs, with some substitutions, insertions or deletions tolerated.
Determination of growth rates of H119 H. volcanii strains with pTA232 and pTA232–cRNA#3 low phosphate media
Low phosphate medium used was based on the minimal medium Hv-Min (19), by adding potassium phosphate buffer (pH −7.5), the only phosphate source, to a final concentration of either 0.1 or 1 mM phosphate. To compare growth rates, four transformed colonies from each H119 pTA232/crRNA#3 and control transformation were grown in Hv-YPC medium at 42°C in shaking incubator for three days to reach OD600 > 0.4. Each culture was then washed and re-suspended three times in 0.1 mM phosphate Hv-Min medium before being diluted to OD < 0.15, using the defined growth media with different phosphate concentration. The growth measurements were done using Biotek ELX808IU-PC in 96-well plates at 42°C with continuous shaking, measuring optical density at a wavelength of 595 nm every 30 min for 3 days.
Determination of fraction of white colonies following growth in 0.1 and 1 mM phosphate concentration
Five colonies of strain H119, that were successfully transformed with pTA232–crRNA#3, were plated on Hv-YPC plates after three days growth (42°C, shaking conditions) from each phosphate concentration 0.1 mM/1 mM phosphate Hv-Min medium. The ratio of white to red colonies was calculated by enumerating the relative number of colonies on Hv-YPC plates.
RESULTS
Haloferax volcanii survives self-targeting by its endogenous CRISPR-Cas system
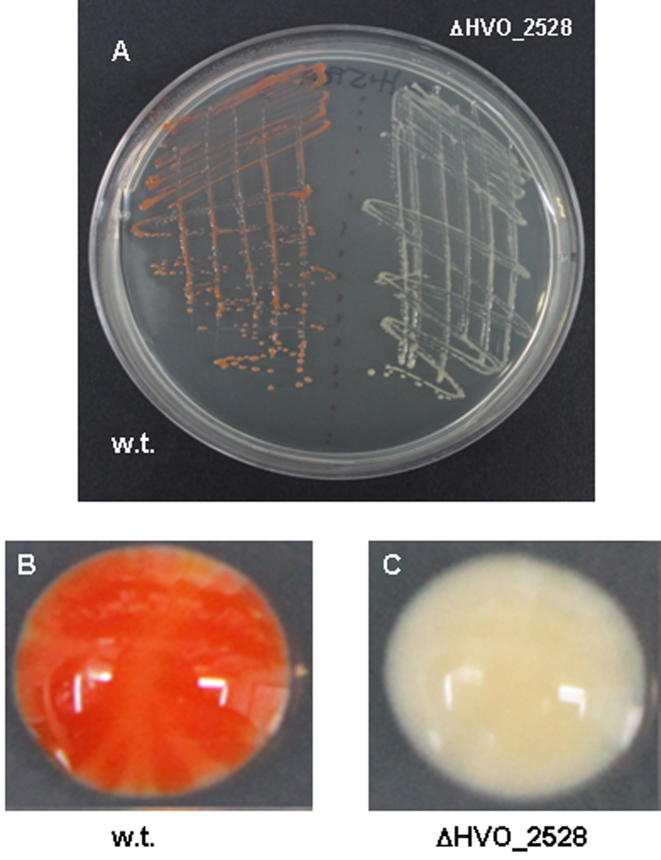
To investigate the effects of self targeting in H. volcanii, we chose the crtI gene (HVO_2528) as target. This gene is not critical for growth under lab conditions but the inactivation of its protein function can be easily monitored by changes in cell or colony color. The crtI gene is located on the main chromosome and encodes phytoene dehydrogenase, a protein involved in carotenoid biosynthesis (23). It was suggested in a previous work based on transposition mutagenesis that this enzymatic function could be provided by other proteins in H. volcanii (24). However, in our hands a complete deletion of this gene results in white colonies, in contrast to the orange-red wild-type (Figure 1). To target the crtI gene we used the single endogenous CRISPR-Cas system of H. volcanii (Type I-B). This system has been studied in detail and shown by plasmid transformation to be active in interference (17,25,26).
Figure 1.
Deletion of the crtI gene (HVO_2528), which is involved in the synthesis of the carotenoid pigment bacterioruberin, results in a white colony phenotype. (A) Wild-type Haloferax volcanii cells (DS2) and the ΔHVO_2528 strain on the same plate. (B) Wild-type Haloferax cells have a red-pink color, (C) ΔHVO_2528 cells are white.
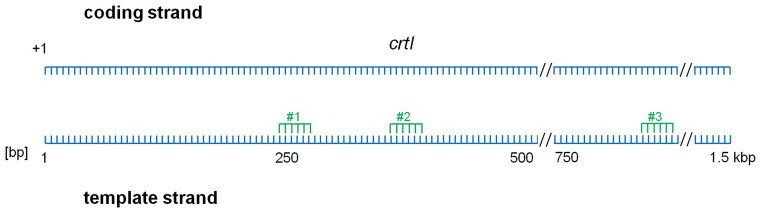
We designed three crRNAs that target the chromosomal crtI gene at different locations in the template strand (Figure 2). Previous experiments with the Haloferax CRISPR-Cas system showed that crRNAs are most effective in targeting a specific sequence when no competing crRNAs are present in the cell (22). Therefore, we first used a Haloferax Δcas6 strain to test the effectiveness of the three self-targeting crRNAs. Notably, because this strain does not express the Cas6b protein, it cannot generate endogenous crRNAs (27). Consequently, every interference complex (known as Cascade) in the cell can only be loaded with the crtI-targeting crRNA.
Figure 2.
Location of targeting sites of the crRNAs in the crtI gene. Three crRNAs were designed to target the crtI gene. crRNA#3 was most effective in targeting the gene, inducing gene deletions and mutations, while crRNAs #1 and #2 were less effective.
Transformation of Haloferax cells with the crRNA-expressing plasmids yielded many colonies on selective media. Nearly all experiments yielded some white colonies (Supplementary Figure S1), indicating mutations in the targeted gene, presumably due to the CRISPR-mediated targeting. Interestingly, compared to transformations with the control plasmids that did not contain a targeting spacer, the number of transformants obtained was reduced by a factor of two at most, for both high copy and low copy plasmids (Table 1). This is in stark contrast to findings from a similar experiment in Sulfolobus (9), where transformation was reduced by four orders of magnitude when the plasmid encoded a self-targeting crRNA matching a non-essential gene (encoding a β-galactosidase). The targeting crRNA#3 resulted in the strongest reduction in transformation rates and the highest fraction of white colonies. The high fraction of white colonies obtained in the absence of reduced transformation efficiency implies that CRISPR-Cas activity is not immediately lethal, but rather has a constant ratchet-like effect that eventually results in mutations in the targeted-region.
Table 1. Targeting of a chromosomal gene only slightly reduces transformation rates.
| Strain | crRNA | Transformation efficiency compared to control plasmid | Percentage of white clones | Percentage of red clones | Percentage of clones of intermediate phenotype |
|---|---|---|---|---|---|
| Δcas6 | #1 | 1.3 | 32% | 25.8% | 42.2% |
| n = 5 | |||||
| Δcas6 | #2 | 1.2 | 14.4% | 24.3% | 61.3% |
| n = 4 | |||||
| Δcas6 | #3 | 0.5 | 99% | 1% | 0% |
| n = 7 | |||||
| Δcas6 | #3 (pTA352, low copy) | 0.6 | 0.07% | 99.93% | 0% |
| n = 8 | |||||
| H119 | #3 | 1.6 | 0% | 100% | 0% |
| n = 4 |
Number of biological repeats per strain is provided (n). All transformations were with the high copy plasmid pTA232, unless otherwise noted.
Notably, the targeting crRNA had to be expressed from a high copy plasmid (see Table 1) for a high fraction of white colonies, while its expression from a low copy plasmid resulted in a very low number of such colonies (Table 1, see Figure 3 for differences in crRNA expression levels between the two different plasmid vectors). Thus, a high concentration of effector complexes containing the self-targeting crRNA is likely to be required for effective self-targeting.
Figure 3.
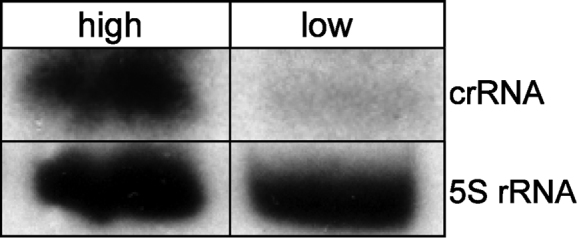
Expression from a high copy number plasmid results in higher amounts of crRNAs in the cell. RNA was isolated from cells expressing crRNA#3 from a high copy number plasmid (lane high) or a low copy number plasmid (lane low), separated by an 8% polyacrylamide gel electrophoresis (PAGE) and transferred to a membrane that was subsequently hybridized with a probe against the crRNA (upper panel crRNA) or a probe against the 5S rRNA (lower panel).
Self-targeting DNA degradation activity is Cas3-dependent
To confirm the CRISPR-Cas specificity of the experiments described above, and validate that the activity is via the known CRISPR-Cas mechanisms, we replaced the wild-type Cas3 protein with an inactive Cas3 variant. This Cas3 variant has a D444A mutation that results in failure of interference (22). Using this Cas3 variant in the assay did not generate white colonies, confirming that self-targeting DNA degradation in our system requires an active Cas3.
Different mechanisms may underlie escape from self-targeting
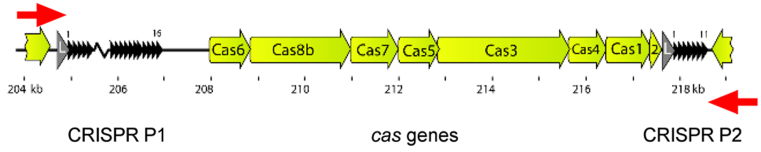
Mutational events that result in escape from self-targeting can occur either in the target locus or in the CRISPR-Cas system itself. The phenotype of the white colonies obtained indicates deletions or other mutations in the targeted crtI gene. In contrast, the red colonies are likely to still express an active CrtI. To investigate how these red colonies maintained an intact crtI we analyzed whether they had deleted or mutated components of the CRISPR-Cas system (28). Earlier studies of the H. volcanii CRISPR-Cas system showed that the most common way to escape CRISPR-Cas targeting is via recombination between repeats of the two CRISPR loci (5) flanking the cas genes (Figure 4). PCR with primers covering this region revealed that 13 out of 20 red clones indeed had a deletion in this region; as expected, none of the 20 white colonies showed such a deletion.
Figure 4.
Deletion of the cas genes mediated by a recombination event between the two CRISPR loci P1 and P2. To determine how many Haloferax cells escaped targeting by inactivation of the CRISPR-Cas system the presence and length of the CRISPR-Cas locus in each colony was tested by PCR using primers that bind to the leader region of locus P1 and directly downstream of locus P2. For simplification only the repeats of the CRISPR array are shown (black triangles).
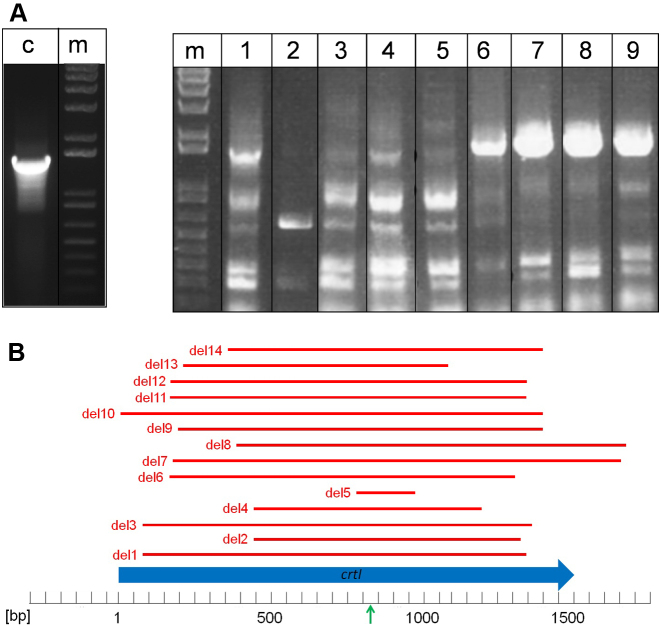
To investigate how H. volcanii repairs the chromosomal DNA damaged by CRISPR-Cas in the targeted locus, 10 white colonies were picked after transformation with the crRNA expressing plasmids (Supplementary Figure S1) and the crtI gene was amplified by PCR (Figure 5A). PCR fragments with different lengths were obtained from single colonies (for PCR amplicons of 9 of the 10 colonies see Figure 5), which is unsurprising given that a colony comprises many different cells that may be heterogeneous with respect to the crtI allele. A total of 19 different PCR fragments from these 10 colonies were cloned and analyzed by sequencing, revealing the exact size and location of 14 individual deletions (Figure 5B, Supplementary Table S2). The length of the deletions observed ranged in size from under 200 to over 1500 bp and some deletions were observed in multiple colonies.
Figure 5.
Deletions in the crtI gene, caused by self-targeting by CRISPR-Cas, have different lengths. (A). The deletions in the crtI gene in different cells in nine different colonies targeted by crRNA#3 are shown. An intact crtI gene would result in a 1.5 kb long PCR fragment, as present in colonies 6–9. Several fragments of shorter length are visible in all colonies analyzed. Lane m: DNA size marker, lane c: control PCR reaction using chromosomal DNA from a wild-type Haloferax H119 strain, which was not targeted, as a template. Lanes 1–9: PCR with DNA from nine different targeted Haloferax colonies (colonies 1–9). (B). Location and length of the different deletions are shown, based on DNA sequencing. Since primers used were located at the ends of the crtI gene, deletions spanning these regions could not be detected.
Deletion products of CRISPR-Cas targeting show a pattern of repair consistent with microhomology-mediated end joining (MMEJ)
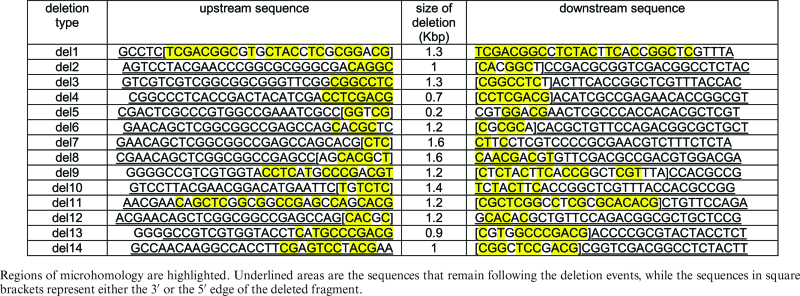
Analysis of the sequences surrounding the repair site revealed short homologous sequences that were from 3 to 19 bp long at the joined ends (Table 2). These patterns strongly suggest repair by microhomology-mediated end joining (MMEJ). In general, double strand breaks in H. volcanii tend to be repaired first by MMEJ, followed by HR (29) to restore the functional allele. However, in the context of CRISPR-mediated attack of a non-essential region, MMEJ in the absence of HR will result in immunity from future attack. Additionally, recent evidence from the E. coli type I-E system has suggested that Cas3-mediated DNA degradation may be primarily single-stranded (30) and how single-stranded gaps are repaired in H. volcanii is unknown.
Table 2. Sequences flanking the deletions in crtI.
|
The effect of self-targeting crRNA on host fitness in H. volcanii cells lacking Cas6
Next we investigated whether damage and subsequent repair of genomic DNA in cells expressing a self-targeting crRNA reduce cell fitness. We transformed the Δcas6 strain with either the crRNA-expressing plasmid or the plasmid without an insert. After transformation, a culture was inoculated with cells transformed with (i) the plasmid without the insert, (ii) the plasmid expressing the self-targeting crRNA and (iii) equal amounts of both transformations (i.e. pairwise competition). Cultures were plated at early exponential phase, mid exponential phase and stationary phase, and the fraction of white colonies was determined (Table 3). The culture inoculated with both transformations yielded only a very small fraction of white colonies. This indicates that cells forced to target and subsequently repair their genomic DNA face a fitness disadvantage and are very quickly outgrown by the control strain. PCR on red colonies confirmed that more than 90% indeed harbor the plasmid without an insert, rather than being ‘escape mutants’ where the CRISPR-Cas machinery or the self-targeting sequence have been inactivated by secondary mutations. In agreement with these results, growth curve analysis indicated that Δcas6 cells expressing the self-targeting spacer grew slower than those expressing the control plasmid, showing a much longer lag phase; however, both reached the same final cell density at stationary phase (Supplementary Figures S2 and S3).
Table 3. Cells that carry a self-targeting plasmid are outgrown by non-targeted cells.
| % of white colonies in strains with plasmids | ||||||
|---|---|---|---|---|---|---|
| OD650nm range | pTA232 | pTA232–crRNA#3 | pTA232 + pTA232–crRNA#3 | |||
| Mean | SD | Mean | SD | Mean | SD | |
| 0.1–0.4 | 0 | 0 | 98.2 | 1.7 | 0.51 | 0.4 |
| 0.4–0.6 | 0 | 0 | 98.7 | 1.2 | 0.67 | 0.5 |
| 0.6–0.9 | 0 | 0 | 99.4 | 0 | 0.83 | 0.8 |
Tolerance to self-targeting crRNA by wild-type H. volcanii cells
The situation of over-expressing a single crRNA in a Δcas6 background is an extreme one, because not only is the level of expression high, but also all interference complexes in the cell are charged with only one type of crRNA, namely the one targeting crtI. In contrast, H. volcanii has 51 spacers in its three CRISPR arrays (5), and so the number of self-targeting complexes in a wild-type cell is expected to be about 50 times lower. To analyze self-targeting in a native context, we repeated the above experiments using a strain with wild-type CRISPR-Cas that expresses all endogenous crRNAs (H. volcanii strain H119) instead of the Δcas6 strain. Notably, transformation of H119 with the most efficient targeting crRNA#3 (plasmid pTA232–crRNA#3) showed no reduction in transformation efficiency and no white colonies were obtained after transformation, in contrast to the results obtained with the Δcas6 strain (Table 1). In addition, H119 cells transformed with pTA232–crRNA#3 grew as well as cells transformed with the empty vector pTA232 (Supplementary Figure S4). Taken together these results imply that H. volcanii is resistant or at least tolerant to self-targeting spacers that match non-essential genes, even when such spacers are expressed from a multi-copy plasmid [the origin of replication is derived from pHV2, which is present at six copies per chromosome (31)]. Thus, in a natural situation, acquisition of a plasmid containing a CRISPR-array with a spacer that targets the chromosome may be well-tolerated by Haloferax cells.
Self-targeting by CRISPR-Cas affects growth under severe phosphate limitation
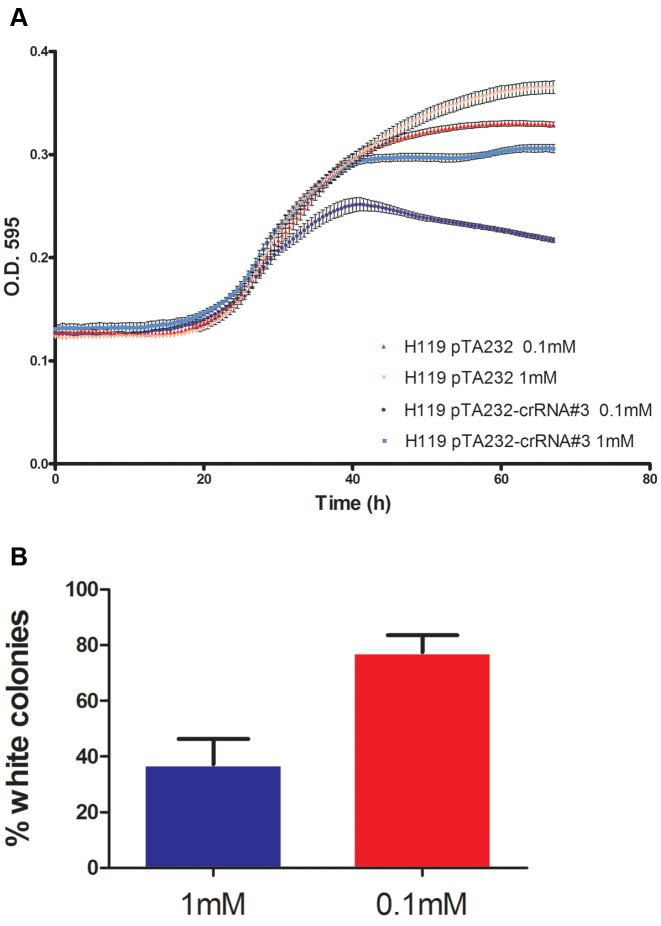
The lack of discernible effect of expressing pTA232–crRNA#3 on growth and lack of mutational events in the wild-type background indicate that whatever damage to chromosomal DNA is caused by CRISPR-Cas, this damage is repaired efficiently and with high fidelity, consistent with HR rather than the mutagenic MMEJ. Indeed, if the level of self-targeting complexes in the cell is low, only rare DNA breaks will occur and these can be repaired by HR with one of many intact copies of the genome, since H. volcanii can have over 20 copies of the chromosome per cell (32). If this is indeed the case, drastically lowering the phosphate concentration, which is known to reduce ploidy in H. volcanii (32), should make HR-dependent repair more difficult for the cells that experience self-targeting by CRISPR-Cas. Indeed, when we transferred H119 cells carrying pTA232–crRNA#3 from a minimal medium containing 1 mM phosphate to a limiting (0.1 mM) phosphate version of the same medium, we observed a substantial growth impairment. Notably, cells carrying pTA232–crRNA#3 entered stationary phase earlier and rather than leveling off at a constant optical density, experienced a steady decline during stationary phase (Figure 6A). Furthermore, when plating pTA232–crRNA#3-containing cultures from the stationary phase, the fraction of white colonies was over three-fold higher for cultures grown in 0.1 mM phosphate compared to 1 mM grown pTA232–crRNA#3-containing, indicating more MMEJ due to CRISPR-Cas targeting at the lower phosphate concentration (Figure 6B). These findings support a role for polyploidy for enabling high tolerance to self-targeting of the chromosome by CRISPR-Cas, although additional effects of low phosphate on DNA stability cannot be excluded.
Figure 6.
Low phosphate increases the effects of self-targeting by CRISPR-Cas in the wild-type cas6 background. (A) Growth curve comparing cells containing the self-targeting plasmid (or an empty vector) when grown in moderate (1 mM) versus low (0.1 mM) phosphate concentrations (see ‘Materials and Methods’ section). (B) Fraction of white colonies obtained from stationary cultures of H119 pTA232–crRNA#3- cells when grown in moderate (1 mM) versus low (0.1 mM) phosphate concentrations. Results shown represent the mean of five biological replicates.
DISCUSSION
Upon invasion by a new plasmid, the acquisition of a CRISPR array containing a self-targeting spacer is a highly likely event in halophilic archaea. Firstly, many archaea have CRISPR spacers that match different species from the same genus or family (14). Secondly, Haloferax has been shown to mate with relatively high efficiency with other species and acquire large plasmids in the process, including ones containing CRISPR loci (18). Thirdly, repeats of CRISPR subtype I-B of different halophilic archaea are highly conserved (33) across genetically distant haloarchaea (such as Haloarcula and Haloferax). Thus, even when a CRISPR array is acquired without any cas genes, the host's pre-existing CRISPR system may successfully process a newly acquired spacer and Cascade will become charged with the crRNA that the array encodes. However, despite the ease of acquiring such potentially suicidal spacers, the overwhelming majority of haloarchaea have CRISPR-Cas systems; this implies substantial tolerance to such events.
Here we show that when the self-targeting crRNA is but one of many in the cell (i.e. in a Cas6-proficient wild-type strain), there is no discernible cost to growth. H. volcanii is not especially resistant to DNA damage (29) and so tolerance to CRISPR-Cas-mediated DNA damage is not a byproduct of general resistance. While the lack of toxicity that we observe is in sharp contrast to what has been observed in bacteria (6–8) and Crenarchaeota (9) for CRISPR-mediated targeting, this is not completely unexpected. When a repair template was available, homology-directed repair was able to counteract CRISPR-Cas auto-immunity in E. coli expressing the Cas9 CRISPR system (34). H. volcanii and other halophilic archaea have over 20 copies of the chromosome per cell and therefore it is unlikely that a small number of self-targeting interference complexes would do much harm. When the chromosome copy number is high and Cascade levels are relatively modest, few DNA degradation events will occur and there will be multiple intact copies of the genome to mediate accurate repair via HR, without incurring a physiological cost. This is in contrast to infection by a selfish element such as a virus or plasmid, where a single copy of the invader DNA and no repair template will be present, and therefore the invading DNA will be much more sensitive to CRISPR-mediated degradation. Furthermore, during viral infection cas genes have been shown to be induced to much higher levels (35), likely resulting in higher levels of the Cascade effector complex.
Polyploidy is characteristic of diverse lineages of Euryarchaeota [for a recent review see (36)], all of which are known to carry CRISPR-Cas systems. Indeed, when we starved the cells for phosphate, a process known to drastically reduce ploidy (32), cell viability was reduced and increased mutagenic repair was observed. We speculate that polyploidy may buffer against autoimmunity by minimizing Cas3-mediated damage until subsequent mutational events can disarm the toxic self-targeting spacer, which helps explain the pervasiveness of these defense systems in the Euryarchaeal phylum. Nevertheless, there appears to be no direct association between maximal ploidy and the number of spacers or arrays in a given genome (Supplementary Table S3).
The most common event that resolves self-targeting in H. volcanii is recombination between two CRISPR arrays, resulting in the deletion of the cas operon that is nested between them ((5) and Figure 4). While such an event does not result in any mutational damage to the targeted gene it does entail complete loss of the anti-viral defense provided by CRISPR-Cas. The fact that the same genomic organization, where the cas operon is located between two CRISPR arrays, is seen in other halophilic archaea such as H. mediterranei (37), implies that such events are rare in the absence of self-targeting, and possibly also balanced by lateral acquisition of cas genes that are either compatible with existing CRISPR arrays or come with their own arrays. Thus, self-targeting following lateral acquisition of a new CRISPR-Cas system can stimulate the subsequent loss of an existing one, resulting in the pattern of extensive lateral gene transfer observed for these systems (38).
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: A.M., U.G. and T.A. conceived the study, E.S.S. and T.A. designed, generated and characterized mutant strains and plasmids, A.M., A.E.S. and U.G. designed experiments, A.E.S. and I.T.G. performed experiments and analyzed data, U.G., A.M. and A.E.S. wrote the manuscript. All authors read and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [MA1538/16-2]; Israel Science Foundation [535/15]. Funding for open access charge: DFG internal funds; TAU internal funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Marraffini L.A.. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014; 54:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B.. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014; 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stern A., Keren L., Wurtzel O., Amitai G., Sorek R.. Self-targeting by CRISPR: gene regulation or autoimmunity?. Trends Genet. 2010; 26:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer S., Maier L.K., Stoll B., Brendel J., Fischer E., Pfeiffer F., Dyall-Smith M., Marchfelder A.. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012; 287:33351–33363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yosef I., Goren M.G., Kiro R., Edgar R., Qimron U.. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20136–20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vercoe R.B., Chang J.T., Dy R.L., Taylor C., Gristwood T., Clulow J.S., Richter C., Przybilski R., Pitman A.R., Fineran P.C.. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013; 9:e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Selle K., Klaenhammer T.R., Barrangou R.. CRISPR-based screening of genomic island excision events in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing type I and type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldberg G.W., Jiang W., Bikard D., Marraffini L.A.. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014; 514:633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samai P., Pyenson N., Jiang W., Goldberg G.W., Hatoum-Aslan A., Marraffini L.A.. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015; 161:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lillestol R.K., Shah S.A., Brugger K., Redder P., Phan H., Christiansen J., Garrett R.A.. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009; 72:259–272. [DOI] [PubMed] [Google Scholar]
- 13. Grissa I., Vergnaud G., Pourcel C.. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007; 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brodt A., Lurie-Weinberger M.N., Gophna U.. CRISPR loci reveal networks of gene exchange in archaea. Biol. Direct. 2011; 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu G., She Q., Garrett R.A.. Diverse CRISPR-Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res. 2016; 44:4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brendel J., Stoll B., Lange S.J., Sharma K., Lenz C., Stachler A.E., Maier L.K., Richter H., Nickel L., Schmitz R.A. et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J. Biol. Chem. 2014; 289:7164–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stoll B., Maier L.K., Lange S.J., Brendel J., Fischer S., Backofen R., Marchfelder A.. Requirements for a successful defence reaction by the CRISPR-Cas subtype I-B system. Biochem. Soc. Trans. 2013; 41:1444–1448. [DOI] [PubMed] [Google Scholar]
- 18. Naor A., Lapierre P., Mevarech M., Papke R.T., Gophna U.. Low species barriers in Halophilic Archaea and the formation of recombinant hybrids. Curr. Biol. 2012; 22:1444–1448. [DOI] [PubMed] [Google Scholar]
- 19. Allers T., Ngo H.P., Mevarech M., Lloyd R.G.. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004; 70:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norais C., Hawkins M., Hartman A.L., Eisen J.A., Myllykallio H., Allers T.. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007; 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allers T., Barak S., Liddell S., Wardell K., Mevarech M.. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 2010; 76:1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stachler A.E., Marchfelder A.. Gene repression in Haloarchaea using the CRISPR (clustered regularly interspaced short palindromic repeats) - Cas I-B system. J. Biol. Chem. 2016; 291:15226–15242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hartman A.L., Norais C., Badger J.H., Delmas S., Haldenby S., Madupu R., Robinson J., Khouri H., Ren Q., Lowe T.M. et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010; 5:e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kiljunen S., Pajunen M.I., Dilks K., Storf S., Pohlschroder M., Savilahti H.. Generation of comprehensive transposon insertion mutant library for the model archaeon, Haloferax volcanii, and its use for gene discovery. BMC Biol. 2014; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cass S.D., Haas K.A., Stoll B., Alkhnbashi O.S., Sharma K., Urlaub H., Backofen R., Marchfelder A., Bolt E.L.. The role of Cas8 in type I CRISPR interference. Biosci. Rep. 2015; 35:e00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maier L.K., Lange S.J., Stoll B., Haas K.A., Fischer S., Fischer E., Duchardt-Ferner E., Wohnert J., Backofen R., Marchfelder A.. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013; 10:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maier L.K., Stachler A.E., Saunders S.J., Backofen R., Marchfelder A.. An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein. J. Biol. Chem. 2015; 290:4192–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gudbergsdottir S., Deng L., Chen Z., Jensen J.V., Jensen L.R., She Q., Garrett R.A.. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011; 79:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delmas S., Shunburne L., Ngo H.P., Allers T.. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 2009; 5:e1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Redding S., Sternberg S.H., Marshall M., Gibb B., Bhat P., Guegler C.K., Wiedenheft B., Doudna J.A., Greene E.C.. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas System. Cell. 2015; 163:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Charlebois R.L., Lam W.L., Cline S.W., Doolittle W.F.. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:8530–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zerulla K., Chimileski S., Nather D., Gophna U., Papke R.T., Soppa J.. DNA as a phosphate storage polymer and the alternative advantages of polyploidy for growth or survival. PLoS One. 2014; 9:e94819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maier L.K., Fischer S., Stoll B., Brendel J., Pfeiffer F., Dyall-Smith M., Marchfelder A.. The immune system of halophilic archaea. Mob. Genet. Elem. 2012; 2:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui L., Bikard D.. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 2016; 44:4243–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quax T.E., Voet M., Sismeiro O., Dillies M.A., Jagla B., Coppee J.Y., Sezonov G., Forterre P., van der Oost J., Lavigne R. et al. Massive activation of archaeal defense genes during viral infection. J. Virol. 2013; 87:8419–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barilla D. Driving apart and segregating genomes in Archaea. Trends Microbiol. 2016; 24:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M., Liu H., Han J., Liu J., Wang R., Zhao D., Zhou J., Xiang H.. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J. Bacteriol. 2013; 195:867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vestergaard G., Garrett R.A., Shah S.A.. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014; 11:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.