Abstract
The Mre11–Rad50–Nbs1(Xrs2) (MRN/X) complex is critical for the repair and signaling of DNA double strand breaks. The catalytic core of MRN/X comprised of the Mre11 nuclease and Rad50 adenosine triphosphatase (ATPase) active sites dimerizes through association between the Rad50 ATPase catalytic domains and undergoes extensive conformational changes upon ATP binding. This ATP-bound ‘closed’ state promotes binding to DNA, tethering DNA ends and ATM activation, but prevents nucleolytic processing of DNA ends, while ATP hydrolysis is essential for Mre11 endonuclease activity at blocked DNA ends. Here we investigate the regulation of ATP hydrolysis as well as the interdependence of the two functional active sites. We find that double-stranded DNA stimulates ATP hydrolysis by hMRN over ∼20-fold in an end-dependent manner. Using catalytic site mutants to create Rad50 dimers with only one functional ATPase site, we find that both ATPase sites are required for the stimulation by DNA. MRN-mediated endonucleolytic cleavage of DNA at sites of protein adducts requires ATP hydrolysis at both sites, as does the stimulation of ATM kinase activity. These observations suggest that symmetrical engagement of the Rad50 catalytic head domains with ATP bound at both sites is important for MRN functions in eukaryotic cells.
INTRODUCTION
The Mre11–Rad50–Nbs1 (Xrs2) (MRN/X) complex with its several enzymatic and scaffolding properties plays a critical role in DNA damage repair and response (1–4). Deletion of any of the three components results in embryonic lethality in mice and loss of proliferative activity in embryonic stem cells (5–7). Hypomorphic mutations in Mre11, Nbs1 and Rad50 also result in developmental and neurodegenerative disorders in humans, including Ataxia-Telangiectasia-like disorder, Nijmegen breakage syndrome (NBS) and NBS-like syndrome, respectively (8–11).
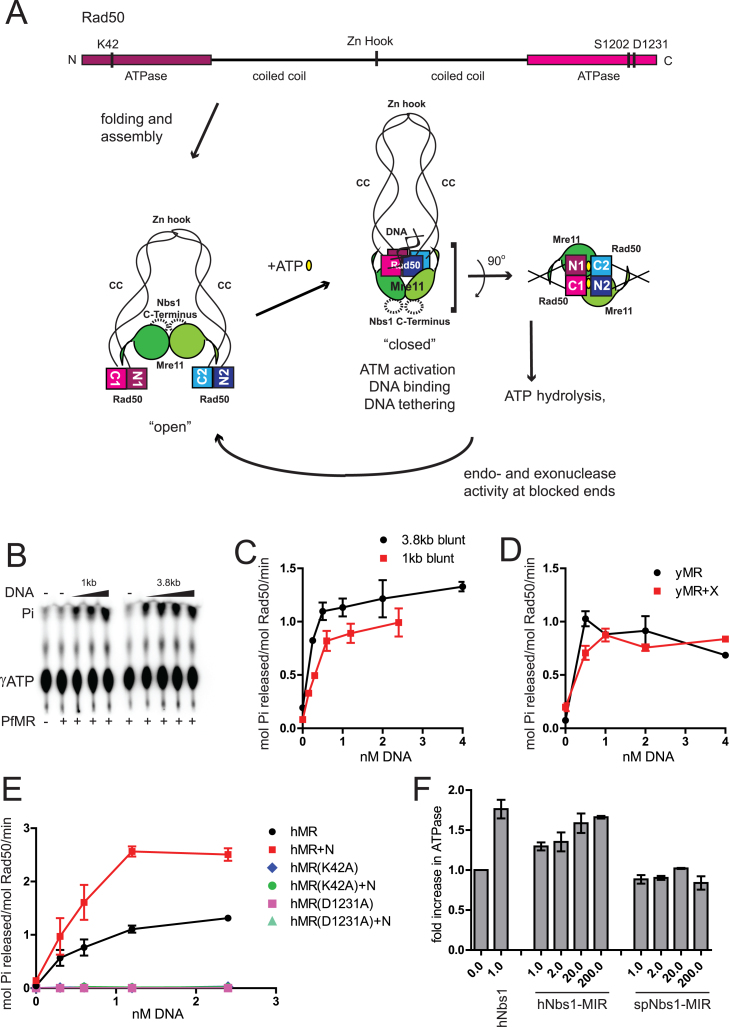
Rad50 belongs to the adenosine triphosphate (ATP) binding cassette (ABC) transporter family of ATPases and contains the conserved motifs found in all ABC nucleotide binding domains (NBD) (12). The domain arrangement and structural features of Rad50 are similar to the structural maintenance of chromosome (SMC) proteins including condensins and cohesins, which are also DNA-associated ABC transporter family proteins and not integral to membranes. Both Rad50 and SMC proteins have ATP binding domains located at N and C-termini separated by a long, intramolecular coiled coil, bringing the ATP binding domains together and forming two incomplete ATPase sites (13,14) (Figure 1A). A functional site is formed only when ATPase domains from two Rad50 molecules dimerize in trans through ATP, such that the N-terminal ATPase domain from one Rad50 and the C-terminal ATPase domain from the other molecule form one complete ATPase site. This kind of dimerization leads to formation of two complete and symmetric ATPase sites on a Rad50 dimer and brings the Walker A motif from one molecule face to face with the signature motif from the other molecule. The coiled coils from two Rad50 molecules also interact through the Zn-hook motif at their tips via coordination of a Zn atom (15) and adjacent interfaces (16).
Figure 1.
DNA stimulates adenosine triphosphate (ATP) hydrolysis by MR/MRN(X) complexes. (A) A schematic of the MRN complex and ATP-induced changes in MRN conformation. A linear map of human Rad50 shows the positions of important conserved residues that were mutated in this study. Intramolecular folding of coiled coil brings the terminally placed ATPase domains together. Coiled coils (CC) from two Rad50 molecules interact through their Zn-hook motifs (Zn Hook). Mre11 nuclease/dimerization domain and the capping domain connected to the linker binds to the coiled coils of Rad50. The binding of ATP induces dimerization of ATPase domains and extensive conformational changes in the MRN complex. This ‘closed’ state exhibits higher affinity for DNA ends and promotes DNA tethering. A DNA helix is bound to the Rad50 ATPase domains as shown (20,22). ATP hydrolysis leading back to an ‘open’ state is required for Mre11 endonuclease activity and promotion of 5΄ to 3΄ resection. The Rad50 ATPase domains, Mre11 nuclease domains and Nbs1 Mre11-interacting-region constitute the catalytic core of the complex. In the absence of ATP, the catalytic head is in an ‘open’ conformation and the Mre11 nuclease sites are accessible. Rad50 ATPase domains dimerize in trans in the presence of ATP (yellow ovals), forming two functional sites as shown. ATP is engaged by Walker A motif K42 from one Rad50 and signature motif S1202 from the other Rad50. (B) PfMR (50 nM) was incubated with [γ- 32P]ATP in reactions containing 1–4 nM dsDNA (either 1 or 3.8 kb long, as indicated), 50 μM total ATP and 5 mM MgCl2 at 65°C for 1 h. Released Pi was separated by thin layer chromatography and analyzed by phosphorimager. (C) Quantitation of ATP hydrolysis assays as shown in (B). (D) yMR (50 nM) was assayed for ATP hydrolysis with equimolar Xrs2 as indicated in (B) but at 30°C for 1 h. Reactions contained 0.5, 1.0, 2.0 or 4.0 nM 1 kb dsDNA. (E) Human MR (50 nM) was assayed for ATP hydrolysis as in (B) with equimolar Nbs1 as indicated using 0.3, 0.6, 1.2 or 2.4 nM 1 kb dsDNA at 37°C for 1 h. hMR mutant complexes with K42A or D1231A mutations were assayed similar to wild-type (WT) hMR as indicated. Error bars indicate standard deviation from three replicates. (F) ATP hydrolysis by hMR (50 nM) was measured in the presence of 2.4 nM 1 kb linear DNA and either full-length or truncated versions of the hNbs1 (a.a. 637–686) or spNbs1 polypeptides (a.a. 474–531). The molar ratio of the Nbs1 protein compared to hMR is as indicated. Results are from three independent experiments and error bars indicate standard deviation.
Based on data from X-ray crystal structures and protein–protein interaction studies, current models suggest that Mre11 binds Rad50 at the base of the coiled-coils where they connect to the ATPase domains (17,18). The ATPase domains engaging ATP are rearranged and positioned above the nuclease active site of Mre11 (17,19–22). Nbs1 interacts with Mre11 opposite to the position of Rad50 interaction but extends around the outside of the nuclease domain of Mre11 (23). In the absence of ATP, this MRN catalytic head is in an ‘open’ state, and the Mre11 nuclease active site is accessible (Figure 1A). In the presence of ATP, dimerization of Rad50 ATPase domains induces a ‘closed’ conformation (17,19). This ATP-bound ‘closed’ state promotes binding to DNA, tethering of DNA ends and Ataxia Telangiectasia Mutated (ATM) activation (24,25), but the Mre11 nucleolytic center is inaccessible to DNA in this ATP-bound closed conformation. Linear DNA molecules are bound to the groove on the top of the Rad50 globular domains, opposite from the Mre11 active sites (20,22). We and others have hypothesized that transient separation of ATPase domains following ATP hydrolysis must be essential for subsequent access of the Mre11 active sites to the ends of DNA (19–22,24).
Mre11 nuclease activity is essential for repair of protein bound meiotic DSBs but not for enzymatically generated clean DSBs or for ATM activation (25–30). Mre11 nuclease activity appears to be more important for mammalian cells than for yeast because Mre11 nuclease-deficient mammalian cells are inviable (31,32) while the same mutation in budding yeast shows only a hypomorphic DNA damage sensitivity phenotype (30). In vitro, purified recombinant Mre11 from many diverse organisms possesses both 3΄ to 5΄ exonuclease activity as well as endonuclease activity (1). In prokaryotes and archaea, Mre11 exonuclease activity requires Rad50 and the hydrolysis of ATP (33,34). In contrast, eukaryotic Mre11 exhibits exonuclease activity apart from Rad50 that is ATP-independent and also shows structure-specific endonuclease activity that is promoted by Rad50 and ATP (35–37). Lastly, MRN(X) complexes cleave DNA at the sites of 5΄ protein blocks on DNA ends, an Mre11-dependent endonuclease activity that is strictly dependent on the protein block, Rad50 and ATP hydrolysis (24,38–40).
The ATP-dependent activities of the Rad50 protein are essential for all of its roles in DNA repair and signaling, as mutations of residues involved in ATP binding and hydrolysis exhibit phenotypes equivalent to a rad50 deletion in vivo (41–43). However, the estimated rates of ATP hydrolysis for Mre11–Rad50 complexes are generally low compared to other ATPases, likely indicating a fairly stable ‘closed’ conformation (44–47). These observations suggest that there may be molecular events that trigger ATP hydrolysis. Here we investigate the regulation of Rad50 ATP hydrolysis as well as the importance of the two functional active sites for the human MRN (hMRN) complex. We find that DNA ends do in fact stimulate ATP hydrolysis by hMRN. The hMRN complex exhibits ATP-dependent endonuclease activity that removes 5’ blocks from DNA ends in vitro. Both of the ATPase sites formed upon ATP binding are required for efficient ATP hydrolysis, as well as for endonuclease activity and ATM kinase activation. With the human complex, Nbs1 appears to exert a regulatory role over ATP hydrolysis as well. These results show that DNA ends control the hydrolysis of ATP by MR/MRN(X) complexes and that the binding and/or hydrolysis of ATP must be concerted between the two active sites to promote the functions of the complex.
MATERIALS AND METHODS
Plasmids and strains
Wild-type (WT) hMR and hMRN complexes were expressed in Sf21 insect cells by co-expression with baculovirus prepared from the transfer vectors pTP2620 (WT Rad50), pTP813 (WT Mre11), pTP288(WT Nbs1), pTP2983 (Rad50-K42A), pTP2984 (Rad50-D1231A), pTP3191 (Rad50-S1202W) as described previously (35,36,44). Transfer vectors were converted into bacmids pTP2730 (WT Rad50), pTP814 (WT Mre11), pTP289 (WT Nbs1), pTP2987 (Rad50-K42A), pTP2988 (Rad50-D1231A), pTP3192 (Rad50-S1202W) and were used to make virus according to manufacturer instructions for the Bac-to-Bac system (Invitrogen). Transfer vectors pTP3090 (WT Rad50-His6), pTP3035 (WT Rad50-HA), pTP3091 (Rad50-K42A-His6) and pTP3092 (Rad50-K42A-HA) were used for making bacmids pTP3096, pTP3038, pTP3097 and pTP3098, respectively. Saccharomyces cerevisiae MR complex (yMR) and Xrs2 were expressed in Sf21 insect cells by co-expression with baculovirus prepared from pTP847 (WT Rad50) and the transfer vectors pTP391 (WT Mre11), pTP694 (WT Xrs2) as described previously (44). Transfer vectors were converted into bacmids pTP404 (WT yMre11), pTP701 (WT Xrs2) and were used to make virus according to manufacturer instructions for the Bac-to-Bac system (Invitrogen). ATM and the Escherichia coli expression constructs for GST-p53 were described previously (48).
Protein purification
WT and mutant hMR and yMR, were purified as described previously (25) with modifications. The complexes of His6-tagged Rad50 and FLAG-tagged Mre11 were purified using Ni-NTA, SP and anti-FLAG resin. Insect cells expressing complexes were lysed in Ni-A buffer (50 mM KH2PO4, 10% glycerol, 2.5 mM imidazole, 20 mM β-mercaptoethanol, 0.5 M KCl) using homogenization followed by sonication. Lysates were cleared by ultracentrifugation and applied to 3 ml Ni-NTA resin (Qiagen), washed and eluted with Ni-B buffer (50 mM KH2PO4, 10% glycerol, 250 mM imidazole, 20 mM β-mercaptoethanol, 50 mM KCl). The eluate from Ni-NTA column was then loaded onto a Hi-Trap SP column (GE), washed and eluted with 60% Buffer B (25 mM Tris pH 8.0, 1000 mM NaCl, 10% glycerol, 1 mM dithiothreitol (DTT)). The eluate was diluted with Buffer A (25 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT) to 150 mM NaCl, and applied to 1 ml anti-FLAG M2 antibody resin (Sigma), washed with Buffer A, with five volumes of 0.5 M LiCl and eluted with 0.1 mg/ml FLAG peptide (Sigma) in Buffer A. Aliquots were flash frozen and stored at −70°C. FLAG-Nbs1 and FLAG-Xrs2 were purified by lysing insect cells expressing either FLAG-Nbs1 or FLAG-Xrs2 in Buffer A using homogenization followed by sonication. Lysates were cleared by ultracentrifugation and applied to 5 ml SP-sepharose resin (GE), washed and eluted with 50% Buffer B. The eluate was then loaded onto 1 ml anti-FLAG M2 antibody resin (Sigma), washed with Buffer A, with five volumes of 0.5 M LiCl and eluted with 0.1 mg/ml FLAG peptide (Sigma) in Buffer A. Aliquots were flash frozen and stored at −70°C.
The heterodimeric hMR complexes (K42A–S1202W), (WT–WT) and (KA–KA) were prepared by co-expressing Rad50-His6, Rad50-HA and Mre11-FLAG baculovirus in insect cells. Complexes were purified by a sequential purification on Ni-NTA and anti-HA magnetic beads. Insect cells expressing complexes were lysed in Ni-A buffer (50 mM KH2PO4, 10% glycerol, 2.5 mM imidazole, 20 mM β-mercaptoethanol, 0.5 M KCl) using homogenization followed by sonication. Lysates were cleared by ultracentrifugation and applied to 3 ml Ni-NTA resin (Qiagen), washed and eluted with Ni-B buffer (50 mM KH2PO4, 10% glycerol, 250 mM imidazole, 20 mM β-mercaptoethanol, 50 mM KCl). The eluate from Ni-NTA was applied to 1 ml anti-HA magnetic beads (MBL), incubated for 2h at 4°C, washed and eluted with 0.1 mg/ml HA peptide (Sigma) in Buffer A (25 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT). Aliquots were flash frozen and stored at −70°C. hMR(S1202W) homodimer purification was carried out using anti-FLAG resin followed by gel filtration. The eluate from the anti-FLAG column was loaded onto a Superose 6 column (GE) equilibrated in Buffer A. Fractions containing S1202W hMR were flash frozen and stored at −70°C. The Pyrococcus furiosus MR complex was purified as described (34). The human Ku70/Ku80 dimer was expressed and purified as described (28) and purified human DNA–PKcs was a gift from Susan Lees-Miller. Recombinant ATM was made by transient transfection of Flag-tagged pcDNA3 expression construct into 293T cells using calcium phosphate and purified as described previously (48). GST-p53 protein was expressed and purified in E. coli as described previously (49). Protein concentrations were determined by quantification of protein preparations with standards on Coomassie-stained sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels using the Odyssey system (LiCor) and Imagequant software.
The Mre11 interacting region (MIR) of human Nbs1 (a.a. 637–686) and Schizosaccharomyces pombe Nbs1 (a.a. 474–531) were obtained from Biomatik and Peptide 2.0, Inc, respectively.
ATP hydrolysis
ATP hydrolysis was measured by release of Pi, essentially as described in (44). In a 10 μl reaction volume, 50 nM MR complex was incubated with 50 μM cold ATP in 25 mM 3-(N-morpholino) propanesulfonic acid (MOPS) pH 7.0, 20 mM Tris, pH 8.0, 80 mM NaCl, 8% glycerol, 1 mM DTT, 0.1 mg/ml bovine serum albumin (BSA) (human and yeast reactions only), 5 mM MgCl2 and 50 nM [γ-32P] ATP at 37°C for human, 30°C for yeast and 65°C for P. furiosus proteins for 1 h unless mentioned otherwise. In reactions with hMR and Nbs1, the proteins were pre-mixed on ice for 10 min prior to addition to the reaction. Reactions were stopped by the addition of 0.2% SDS and 10 mM ethylenediaminetetraacetic acid (EDTA). 0.5-1 μl of the reactions were spotted on PEI-cellulose TLC plates (SAI), developed in 0.75 M KH2PO4 followed by phosphorimager analysis. Released Pi was quantitated from the phosphorimager scans using ImageQuant software (GE) and data were analyzed using Graphpad Prism 5. Equimolar Nbs1 and Xrs2 were mixed with hMR and yMR complexes, respectively, prior to addition in assays where indicated. In ATPase assays containing the Ku70/Ku80 dimer and DNA–PKcs, these proteins (16 and 32 nM) with or without 200 μM NU7026 were allowed to bind 1.2 nM 1 kb DNA for 5 min at RT prior to addition of hMR and Nbs1.
The pCDF-1b plasmid (Novagen) was used in ATPase assays either as a supercoiled closed circular DNA or linear fragments. pCDF-1b plasmid was digested with SspI and HincII to obtain 1–1.2 kb fragments with blunt ends. pCDF-1b plasmid was linearized with SspI, PstI and BamHI to create 3.8 kb DNA with blunt, 4 nt 3΄- overhangs or 4 nt 5΄- overhangs, respectively. Restriction enzyme digested DNA was added to ATPase reactions without further purification. TP124 (CATCTGGCCTGTCTTACACAGTGCTACAGACTGGAACAAAAACCCTGCAG) annealed with its complement TP125 was used as the 50 bp dsDNA in ATPase assays as indicated. The DNA values correspond to nM DNA molecules added to the reaction. The 197 bp DNA was made by polymerase chain reaction using TP4373 (TGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTT) and TP542 (GGTTTTCCCAGTCACGACGTTG) using pTP3718 (40) and purified on an 0.7% agarose gel. DNA with 5’ biotin on both ends was made by using same oligos with a 5’ dT- biotin. Biotinylated DNA was incubated with 20–40-fold molar excess of streptavidin for 10 min at room temperature prior to addition in assay reaction.
Nuclease assays
The endonuclease assay was carried out using a 50 bp dsDNA oligonucleotide substrate. TP4536 (Bio-TGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTT) with 5’ biotin triethylene glycol (TEG) was radiolabeled at the 3’ end using TdT and [α-32P]-cordycepin. Radiolabeled oligonucleotide was then annealed to the complement TP4559 (AACGTCATAGACGATTACATTGCTAGGACATCTTTGCCCACGTTG*A*C*C*C*A) containing five phosphorothioate bonds at the 3’ end as indicated. The substrate was incubated with a 20–40-fold molar excess of streptavidin at RT for 10 min prior to addition to the reaction. In reactions with hMR and Nbs1, the proteins were pre-mixed on ice for 10 min prior to addition to the reaction. In a 10 μl reaction volume, 50 nM hMR and 50 nM Nbs1 as indicated were incubated with 1 nM DNA in 25 mM MOPS pH 7.0, 20 mM Tris, pH 8.0, 80 mM NaCl, 8% glycerol, 1 mM DTT, 1 mM ATP, 20–40 nM streptavidin, 5 mM MgCl2, 1 mM MnCl2 and 0.2 mg/ml BSA at 37°C for 30 min. Reactions were stopped with 0.2% SDS and 10 mM EDTA. Samples were lyophilized, dissolved in formamide, boiled at 100°C for 4 min, loaded on denaturing polyacrylamide gels containing 16% acrylamide, 20% formamide and 6 M urea, separated at 40 W for 1.5 h, followed by phosphorimager analysis.
DNA binding assays
Gel shift assays were performed using 3’ radiolabeled TP4536 annealed to TP4559 in the absence of streptavidin. The reaction was identical to the endonuclease assay except for a shorter incubation at 37°C for 15 min. Samples were separated by electrophoresis on a 1.2% agarose gel in 0.5X TB (50 mM Tris-Borate) at 80V for 30–40 min, dried and analyzed by phosphorimager.
ATM kinase assays
ATM kinase assays were performed in kinase buffer: 50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM ATP and 1 mM DTT for 90 min at 30°C in 40 μl as described previously (48). A total of 10 ng DNA was used in kinase assays with 50 nM GST–p53, 0.2 nM ATM and 20 nM MR and Nbs1. Phosphorylated p53(ser15) was detected as described previously (48) using phospho-specific antibody from Calbiochem (PC461). Westerns were probed for ATM and Rad50 using Santa Cruz 1B10 and GeneTex 13B3 antibodies, respectively.
RESULTS
DNA stimulates ATP hydrolysis by Rad50
Although essential for hMRN activities in vivo, ATP hydrolysis by the hMRN complex and its regulation are not well understood. The reported rates of ATP hydrolysis by Rad50 are very slow, especially for the eukaryotic complexes, e.g. 0.1 ATP/min by yeast MR (45) and 0.026 ATP/min by human MR (44,46). In vitro examination of human Rad50 ATPase activity has proved especially difficult owing to its extremely slow rates of ATP hydrolysis, requiring high protein concentrations and long incubation periods for detection of activity (44). A stimulatory effect of DNA on Rad50 hydrolysis has been reported for T4 MR (47) and yMRX (45). Here we measured ATP hydrolysis by hMRN, archeal and yeast complexes in the presence of blunt-ended restriction enzyme generated DNA fragments 1 kb in length and observed significant increases in ATP hydrolysis upon addition of DNA as measured by the release of Pi (Figure 1). For instance, addition of 0.5 nM DNA to the purified P. furiosus MR complex (PfMR) in vitro generated a 12–14-fold increase in ATP hydrolysis by 50 nM PfMR while further addition of DNA did not have additional effect on ATPase activity (Figure 1B and C). A similar effect was seen with a longer 3.8 kb DNA (Figure 1B and C). The S. cerevisiae MR (yMR) complex showed up to a 10-fold increase in ATP hydrolysis with addition of linear DNA with no additional effect of Xrs2 (Figure 1D), generally agreeing with an earlier report (45) although we observe a larger stimulation.
With the human MR and MRN complexes, a 20-fold increase in ATPase activity was observed upon addition of 1 kb linear DNA (Figure 1E). The rate of ATP hydrolysis by human MR improved from 0.05 ATP per Rad50 per minute in the absence of DNA to 1 ATP per Rad50 per minute in the presence of 1.2 nM DNA. Addition of Nbs1 stimulated ∼2-fold increase in ATP hydrolysis by human MR complex under all assay conditions (Figure 1E), with the rate increasing to 2.6 ATP per Rad50 per minute in the presence of both Nbs1 and 1.2 nM DNA. With the human MR complex, maximum stimulation was achieved at a DNA concentration 40-fold lower than Rad50 concentration. To verify that the ATPase activity observed is in fact due to Rad50, we also tested ATPase mutants of human Rad50 containing K42A (Walker A) and D1231A (Walker B) mutations. MRN complexes containing these mutants failed to show ATPase activity or stimulation by linear DNA, confirming that Rad50 is responsible for the ATP hydrolysis (Figure 1E).
Nbs1 interacts with the Mre11-Rad50 head domain through a C-terminal MIR, as seen in a recent crystal structure of S. pombe Mre11 (23). We synthesized the MIR peptide from human and S. pombe Nbs1 and added these to the hMR ATPase reaction in 1–200-fold molar excess of hMR in the presence of linear DNA with blunt ends (Figure 1F). The human MIR stimulated hMR ATPase activity by ∼1.7-fold at a 200-fold molar excess of MIR suggesting that the MIR is sufficient to support ATPase stimulation, although much less efficient compared to full-length Nbs1. The S. pombe MIR failed to stimulate human MR at these concentrations, indicating a species-specific interaction between the human proteins.
These experiments show that eukaryotic MR complexes are DNA-dependent ATPases. In the reactions shown here, 8% glycerol was included in the buffer as well as at least 80 mM NaCl (see ‘Materials and Methods’ section). These levels of glycerol and salt are essential for the observed stimulation of both archaea and eukaryotic MR complexes (data not shown).
Double-stranded DNA ends are important for stimulation of Rad50 ATP hydrolysis
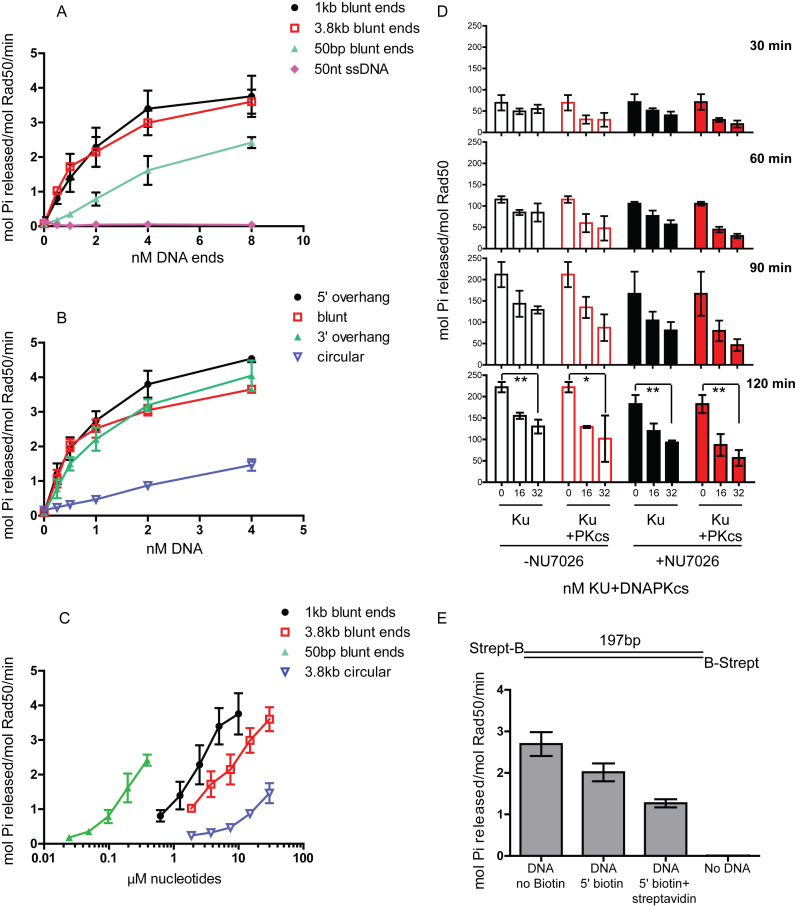
To determine the minimal requirements for DNA structures that stimulate hRad50 ATPase activity, we tested several DNA substrates with varying length and end structure. The stimulation of ATPase activity requires double-stranded DNA (dsDNA), as 50 nt ssDNA failed to stimulate ATP hydrolysis by human MRN (Figure 2A). In contrast, a 50 bp dsDNA increases ATP hydrolysis by 20-fold in the presence of Nbs1. Interestingly, using similar concentrations of DNA ends, the stimulation increases with the length of DNA, with a maximum 40-fold increase observed with DNA 1 kb in length. Further increase in DNA length to 3.8 kb had no additional effect (Figure 2A). DNA ends were important for this stimulation, as a closed circular plasmid DNA showed 10-fold stimulation compared to the 40-fold increase induced by a linear fragment (Figure 2B). We also tested ATP hydrolysis by hMR in the presence of Nbs1 and 3.8 kb DNA with either 5’ or 3’ overhangs and blunt ends, and found that all types of DNA end structures caused a similar increase in ATP hydrolysis (Figure 2B). Using the same data as shown in Figure 2A, the ATPase activity was plotted against the nucleotide concentration in the reaction (Figure 2C). From this analysis it is clear that the 50 bp dsDNA stimulates ATPase activity at 60–90-fold lower nucleotide concentrations compared to 1 kb linear DNA. Overall, we conclude that MR complexes from archaea and eukaryotes are stimulated to hydrolyze ATP in response to DNA ends, but that longer DNA is a more efficient activator. This preference for longer DNA has also been observed with ATM activation via the MRN complex (50), and is likely due to the tendency of MRN to load onto DNA at internal sites rather than directly at ends (51). In the presence of linear DNA, hMRN has an approximate Km for ATP of 31 μM (Supplementary Figure S1), similar to a previously measured Km for AMP–PNP binding by P. furiosus MR (43).
Figure 2.
DNA stimulation of ATPase activity is dependent on DNA ends. (A) Human MR complex (50 nM) was assayed at 37°C for 1 h as in Figure 1D with equimolar Nbs1 and various types of double-stranded DNA (1 kb with blunt ends, 3.8 kb with blunt ends, 50 bp with blunt ends) or 50 nt single-stranded DNA as indicated, with moles of DNA ends as shown (nM DNA ends). (B) The human MR complex (50 nM) was assayed at 37°C for 1 h as in (A) with equimolar Nbs1 using 3.8 kb circular and linearized plasmid DNA carrying 5΄ or 3΄ overhang ends or blunt ends as indicated. (C) Data from (A and B) were plotted with normalization to moles of nucleotides. (D) The human MR complex (50 nM) was assayed at 37°C for 30–120 min as indicated, with equimolar Nbs1 using 1 kb dsDNA (1.2 nM). A total of 16 and 32 nM Ku and DNA–PKcs were added to reactions with 200 μM NU7026 as indicated. The moles of Pi released per mole of Rad50 is shown for each time point. Error bars indicate standard deviation from three replicates. P-values were calculated using unpaired two-tailed t-tests and are shown as asterisks. Both * (P-value 0.02) and ** (P-value 0.0014–0.002) are <0.05 and suggest significant difference in activity. (E) hMR ATP hydrolysis was measured as in (A) in the absence of DNA or the presence of 2.5 nM 197 bp double-stranded DNA with no biotin, biotin at both 5΄ ends or both biotin and streptavidin on both 5΄ ends, as indicated. Error bars indicate standard deviation from three replicates.
In eukaryotes, the Ku70/Ku80 heterodimer competes with MRN(X) complexes for access to DNA ends (52–58). To determine how Ku affects ATP hydrolysis by Rad50, we blocked the DNA ends using 16 and 32 nM recombinant human Ku70/Ku80 (Ku), and observed a 30–40% reduction in ATP hydrolysis with MRN concentration constant at 50 nM and 1 kb DNA at 1.2 nM (Figure 2D). The Ku70/Ku80 complex is also suggested to slide on DNA (59,60). To prevent this sliding and to fix the complex to ends, we added its binding partner the DNA-dependent protein kinase catalytic subunit (DNA–PKcs), which has been shown to bind Ku loaded on DNA ends (59). With the addition of equimolar amounts of DNA–PKcs to Ku, we observed a further 10% decrease in stimulation compared to Ku alone (Figure 2D). With DNA–PKcs addition, the ATP hydrolysis by MRN was reduced by up to 60%, particularly at early time points (Figure 2D, 30 and 60 min).
After binding to Ku on DNA ends, DNA–PKcs autophosphorylates, resulting in dissociation of the kinase and increasing access of other enzymes to the ends (61–63). Here we used the DNA–PKcs inhibitor NU7026 to inhibit the ATPase activity of DNA–PKcs and prevent its dissociation from DNA ends. We observed that the addition of NU7026 results in further reduction of ATP hydrolysis by MRN with only 30% activity seen after 2 h in presence of 32 nM DNA–PK complex (Figure 2D, 120 min right panel, filled bars). These results indicate that access to DNA ends plays an important role in ATPase stimulation.
We have recently shown that blocking of DNA ends with streptavidin induces specific endonucleolytic cleavage of the DNA by human and yeast MRN(X) complexes (40). ATP hydrolysis is required for this cleavage as well as subsequent processing events by MRN. To address whether biotin-streptavidin blocked DNA ends affect ATP hydrolysis by Rad50, we generated 197 bp DNA with no modifications, or with biotin on both 5΄ ends (Figure 2E). The 197 bp DNA without biotin stimulated ATPase activity by hMRN similarly to the 1 kb DNA. The presence of biotin on both ends showed ∼20% reduction in stimulation, while the addition of streptavidin further reduced the stimulation by 50% (Figure 2E), confirming the importance of free DNA ends for efficient stimulation of ATPase by Rad50. It is important to note, however, that even with blocked ends Rad50 is still active in ATP hydrolysis albeit at a lower level.
An Mre11/Rad50 dimer with only one functional site is not capable of ATP hydrolysis
Rad50 is a member of the ABC ATPase family, where the ATPase domains are found at the N- terminus and C-terminus of the protein separated by a long coiled coil, which folds intramolecularly to bring the N- and C-terminal domains together (Figure 1A). In the presence of ATP, Rad50 dimerizes through its ATPase domains, forming two functional ATP binding and hydrolysis sites. This dimerization occurs in trans such that the N-terminal domain from one Rad50 molecule and the C-terminal domain from the other Rad50 molecule form one complete ATPase site. Many members of the ABC ATPase family are integral membrane proteins and function to transport ions or small molecules across the membrane (64). In some cases, structures and biochemical characterization of these transporters have suggested assymetric regulation of the ATPase activities of the active sites (65,66).
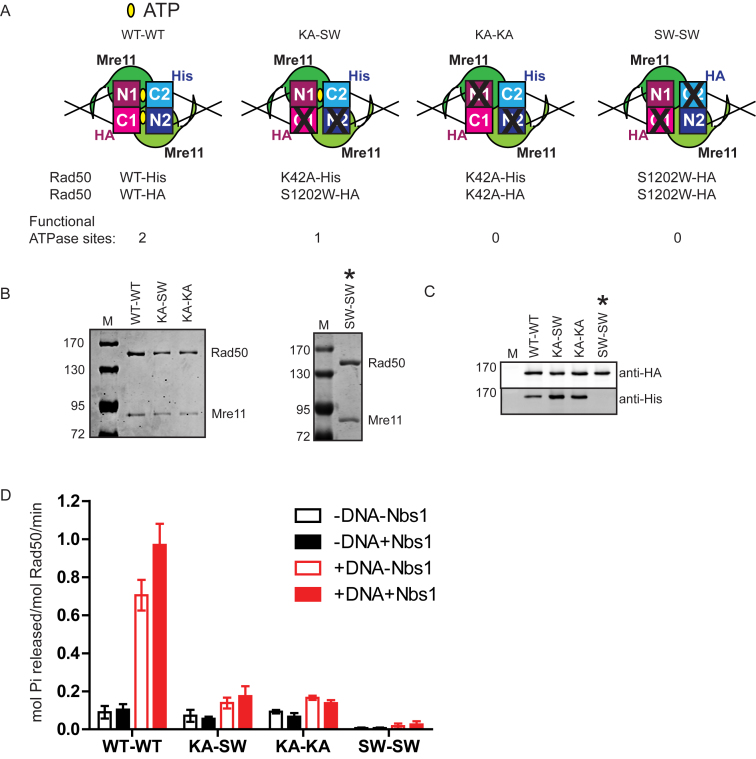
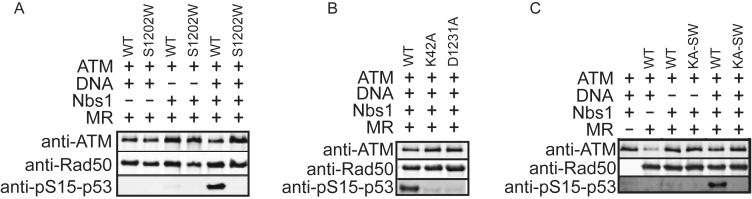
To determine if the stimulation of MR ATPase activity by DNA ends requires the coordinated function of both active sites in the Rad50 dimer, we sought to generate MR complexes containing only a single functional ATPase site. Since the active site forms in trans, the only way to do this is to generate a heterodimer of Rad50 within the MR complex using sequential affinity purification. To eliminate the possibility of active WT–WT homodimers re-assembling after purification, we used Rad50 C- or N-terminal ATPase mutants which are deficient for ATPase activity as homodimers (see Figure 3A). The N-terminal mutation K42A disables ATP binding at the Walker A motif whereas mutations at the S1202 residue disrupt the signature motif in the C-terminal domain (43,44,50). The resulting heterodimer (His-K42A/HA-S1202W) would then contain one WT active site and one site disabled by both K42A and S1202W mutations (Figure 3A). We also made His/HA heterodimer forms of the WT and K42A complexes purified in the same way as positive and negative controls, respectively, as well as a homodimeric HA-tagged S1202W complex for comparison. We found that complex formation with Mre11 was not affected in any of these ATPase mutants (Figure 3B and C).
Figure 3.
Human MRN complex with only one functional ATPase site shows loss of ATP hydrolysis. (A) The catalytic globular domains of the MRN complex are shown, depicting the association of N- and C- terminal ATPase domains in the presence of ATP, forming two functional sites in the WT complex. A heterodimer of His-K42A and HA-S1202W mutants (KA–SW) disables one ATPase site. A heterodimer of His-K42A and HA-K42A (KA–KA) or a homodimer of HA-S1202W (SW–SW) will contain two non-functional sites. (B) WT and mutant human MR complexes were purified by sequential purification on Ni-NTA and anti-HA resins. SDS polyacrylamide gels stained with Coomassie blue show formation of MR complex was not affected in the heterodimer. (C) The purified complexes were transferred to PVDF membranes which were probed with anti-HA and anti-His antibodies to show the presence of both His and HA tagged Rad50 after the final purification. The S1202W mutation was made only as a HA tagged version, so the anti-His blot does not show a band in the S1202W lane (asterisk). (D) The ATPase activity of these complexes was assayed using 50 nM MR in the presence or absence of equimolar Nbs1 and 4.8 nM 1 kb dsDNA at 37°C for 2 h as in Figure 2A. Error bars indicate standard deviation from three replicates.
The WT heterodimer showed DNA-stimulated ATP hydrolysis, whereas the S1202W and KA–KA complexes with both sites mutated each showed very low levels of ATP hydrolysis, as expected (Figure 3D). The K42A–S1202W heterodimer was very similar in its activity to the KA–KA heterodimer. Both complexes were stimulated ∼2-fold by DNA but the rate of hydrolysis by the K42A–S1202W complex was at least 5-fold lower than the WT complex with two functional sites, much lower than the theoretical 50% of the WT activity that would be observed if the two ATPase sites function independently. We conclude from this experiment that one functional site is not sufficient for efficient Rad50 ATP hydrolysis.
Both ATPase sites are required to be functional for Mre11 endonuclease activity
The MRN(X) complex exhibits manganese-dependent 3΄ to 5΄ exonuclease activity on DNA ends but has also been implicated in the removal of protein–DNA adducts in eukaryotic cells through endonucleolytic activity (39,67–69). We recently characterized this activity with hMRN in vitro on streptavidin-blocked DNA ends, showing that an initial endonucleolytic cleavage is made by Mre11 on the strand with the adduct, followed by 3΄ to 5΄ exonuclease activity on the same strand and also endonucleolytic cleavage on the opposite strand to create a clean double-stranded break end (40). Cejka et al. also reported a similar activity in hMRN that is stimulated by human phosphorylated CtIP (38).
To determine if both catalytic sites in Rad50 are necessary for ATP-dependent endonucleolytic cleavage by hMRN, we tested the WT and mutant MR complexes on the biotinylated DNA substrate in the presence of Nbs1, ATP, magnesium, manganese and streptavidin (Figure 4). WT hMR with Nbs1 generates a single endonucleolytic cleavage product 18–20 nt from the streptavidin block, similar to previously reported results (40). Assays with the mutant complexes show that the ATPase mutants affecting both functional sites, the KA–KA and SW–SW complexes, failed to support 5΄ endonuclease activity, consistent with the important role of ATP in this assay (lanes 7–10). Similarly, the KA–SW heterodimer with a single functional site also failed to show this activity, indicating a requirement for both sites to be functional for endonucleolytic activity of human MRN (lanes 5 and 6). The 3΄ labeled nucleotide on the bottom strand is also removed by WT hMRN on this substrate, an activity which is ATP dependent (40). The mutant complexes also failed to show 3΄ nucleotide removal, consistent with our observations that the mutant complexes fail to support ATP hydrolysis by Rad50.
Figure 4.
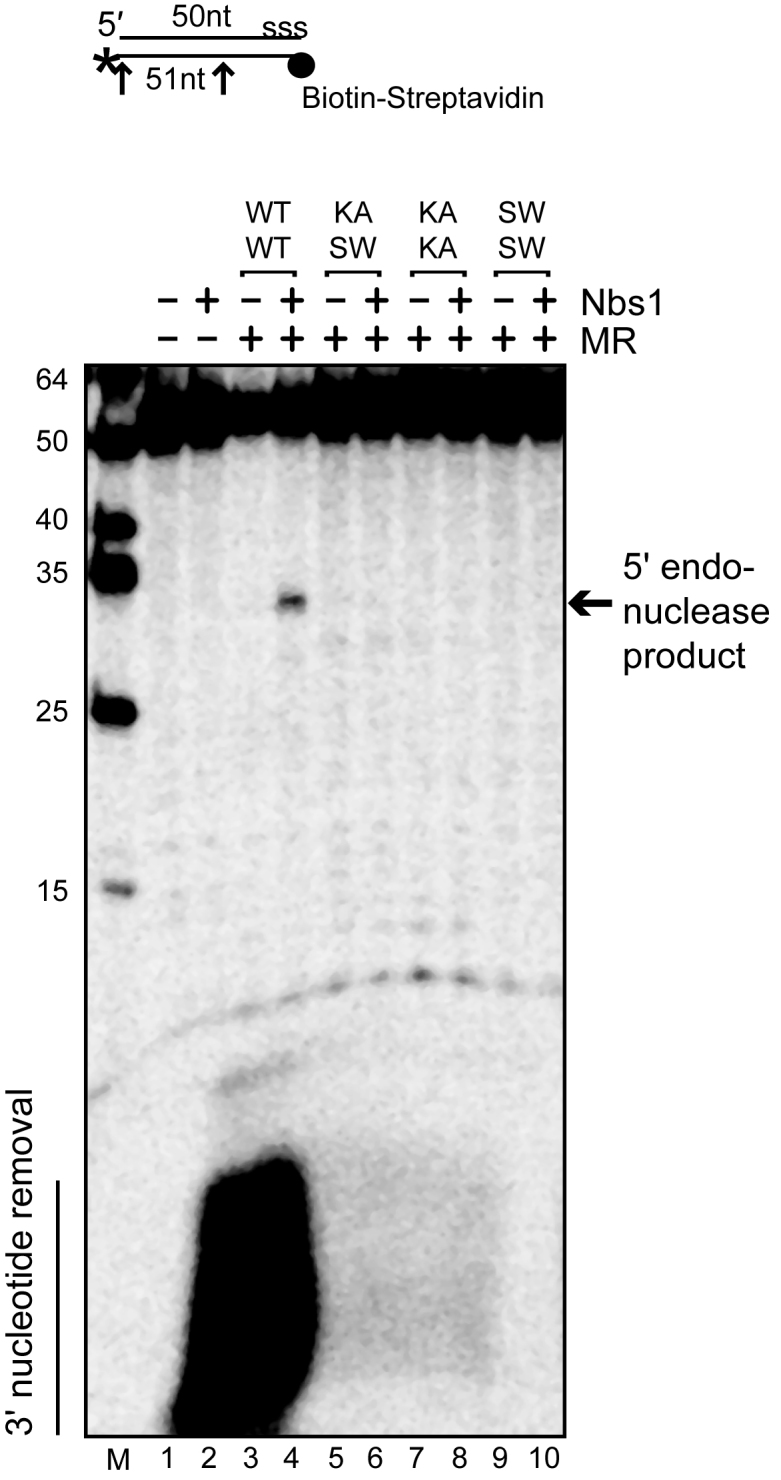
The human MRN complex shows endonuclease activity that requires coordination of the two ATPase sites. Human MR complexes (50 nM) containing either two (WT–WT), one (KA–SW) or zero (KA–KA, SW–SW) functional ATPase sites were incubated with a 50 bp dsDNA substrate containing a 5΄ biotin-streptavidin block and 3΄ [32P] label (*) as indicated. ‘s-s-s’ on the top strand denotes five phosphorothioate bonds at the 3΄ end to prevent exonucleolytic degradation. Reactions contained Nbs1 at equimolar ratio to hMR, 5 mM MgCl2, 1 mM MnCl2, streptavidin and 1.0 mM ATP as indicated and were incubated at 37°C for 30 min. Reaction products were separated on a 16% acrylamide, 20% formamide, 6 M urea gel and analyzed by phosphorimager. Arrows indicate sites of cleavage.
Coordinated nucleotide binding is essential for DNA binding by MRN
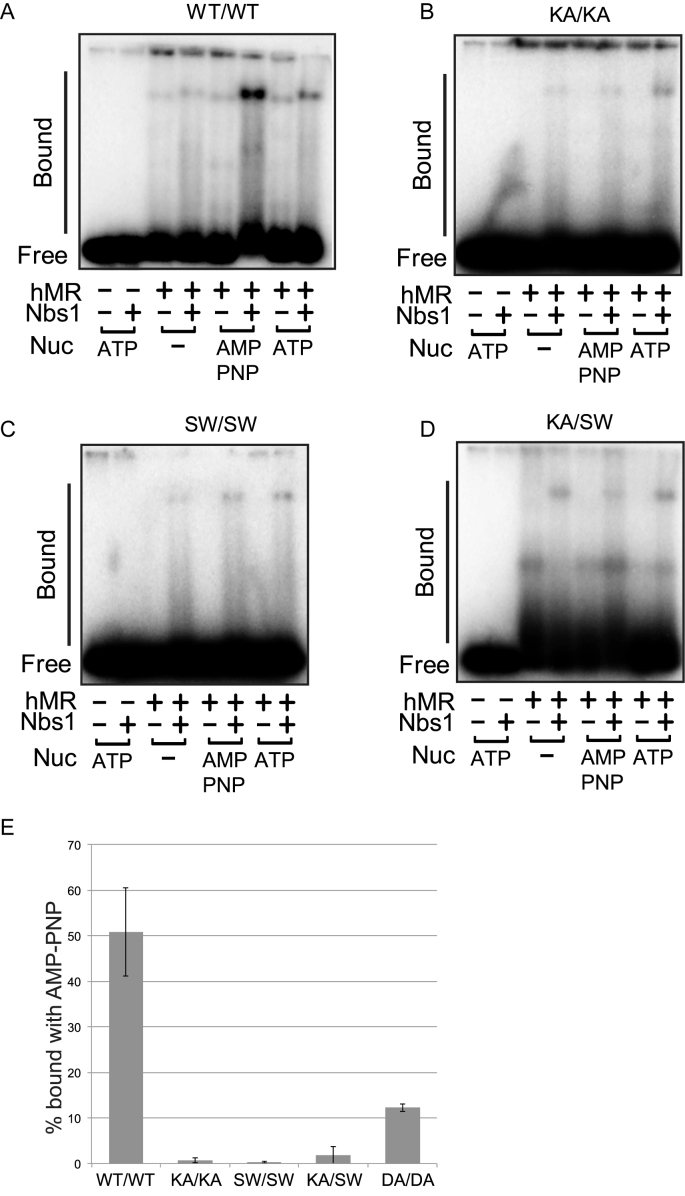
We have previously shown that binding to a non-hydrolyzable form of ATP such as AMP–PNP promotes stable DNA binding by human MRN (70). To determine the effect of the heterodimeric ATPase site on DNA binding, the WT and mutant complexes were also tested for DNA binding using a gel mobility shift assay (Figure 5). The WT complex showed an Nbs1-dependent shift in DNA migration, and in the presence of AMP–PNP generated a larger Nbs1-dependent complex (Figure 5A). The K42A and S1202W mutants also exhibited Nbs1-dependent binding, although this was weak compared to WT complex and the addition of AMP–PNP did not improve the binding (Figure 5B and C). The K42A–S1202W heterodimer likewise exhibited Nbs1-dependent binding to DNA, but at a low efficiency and the binding was similar in the absence and presence of ATP (Figure 5D). The heterodimer containing one normal active site as well as the other mutant complexes all form Nbs1-stimulated complexes with DNA but in no case are these dependent on nucleotide. Quantitation of the AMP–PNP-dependent complex from replicate experiments confirms that only the WT–WT complex exhibits efficient nucleotide-stimulated, Nbs1-dependent binding (Figure 5E).
Figure 5.
(A) DNA binding assays were performed with hMR complexes at 37°C for 15 min. Reactions contained 25 nM hMR, 25 nM Nbs1 and 1 mM ATP or AMP–PNP as indicated, as well as 5 mM MgCl2. The bound species were separated on a 1.2% agarose gel and analyzed by phosphorimager. (B) DNA binding assays were performed with KA–KA hMR complexes as in (A). (C) DNA binding assays were performed with SW–SW hMR complexes as in (A). (D) DNA binding assays were performed with KA–SW hMR complexes as in (A). (E) DNA binding assays were performed with WT and mutant hMR complexes as in (A). The level of protein–DNA bound species formed by recombinant MRN complexes in the presence of Nbs1 and AMP–PNP were quantitated. Results are shown from three independent experiments and error bars indicate standard deviation.
Both catalytic sites in Rad50 are required for stimulation of ATM kinase activity
The checkpoint signaling kinase ATM is activated by MRN upon DNA damage (49,71,72). Activation of ATM is dependent on Nbs1 association and also on the binding and local unwinding of DNA ends by hMR that is ATP-dependent (50,73). Recently we showed that the ATP-bound closed state of Rad50 is essential for ATM kinase stimulation, but ATP hydrolysis and Mre11 nuclease activity are not (25). To test whether ATP binding to both Rad50 active sites is important for ATM stimulation we performed an in vitro ATM kinase assay using a p53 fragment as a model substrate (Figure 6A). The WT complex showed activation of ATM that is dependent on Nbs1 and DNA whereas the ATPase deficient K42A, D1231A and S1202W complexes failed to activate ATM as expected (Figure 6A and B). The KA–SW heterodimeric mutant also completely failed to activate ATM (Figure 6C), indicating that one functional ATP-binding site in Rad50 is not sufficient for ATM activation.
Figure 6.
Coordination of Rad50 ATPase sites is required for ATM activation. (A) In vitro kinase assays with recombinant human ATM (0.2 nM) were performed with WT or S1202W hMR complexes (20 nM), Nbs1 (20 nM) and linear DNA using GST-p53 (50 nM) as a substrate. Reactions were separated by polyacrylamide gel electrophoresis, transferred to a PVDF membrane and probed with an antibody specific for phospho-Ser15 phosphorylation. Membranes were also probed for ATM and Rad50 for comparison as shown. (B) In vitro kinase assays were performed as in (A) but with homodimeric hMR (WT, K42A or D1231A) complexes. (C) In vitro kinase assays were performed as in (A) but with WT and heterodimeric (KA–SW) hMR.
DISCUSSION
Recent advances in structural studies and biochemical analyses of the domains of Rad50 depict an ATP-bound ‘closed’ conformation (Figure 1A) (17,19–22). We have previously shown that this conformation promotes ATP-dependent processes that include DNA end binding, DNA tethering and stimulation of ATM kinase activity (24,25). At the same time, this closed conformation is inhibitory to the nucleolytic processing of DNA by Mre11, and downstream processing complexes. We and others have hypothesized that occlusion of the nuclease center of Mre11 in the ATP-bound closed state of the complex necessitates ATP hydrolysis and subsequent separation of the Rad50 ATPase domains for Mre11 nuclease activity (17,19,20,22,24). However, the events that follow ATP hydrolysis leading to nuclease activity are still not understood. In addition, ATPase rates for Mre11/Rad50 complexes, particularly the eukaryotic complexes, are very low (44–46) and suggest that there may be factors that stimulate ATP hydrolysis. Rad50 complexes exist minimally as homodimeric units and thus contain two nucleotide binding sites per complex, but it is not known if hydrolysis of ATP is concerted between the two active sites, or whether one or both sites must bind or hydrolyze ATP to promote the functions of the complex.
Here we investigated the regulation of human Rad50 ATPase activity and how this is coordinated between the two ATP binding sites. We find that DNA ends are in fact stimulatory for ATP hydrolysis, which is also essential for the endonuclease activity of Mre11. The third component of the complex, Nbs1, which is essential for MRN-mediated 5’ endonucleolytic cleavage of dsDNA substrates with 5’ blocked ends, also stimulates ATP hydrolysis. In addition, our results show that concerted hydrolysis at the two ATPase sites in the Rad50 ATP-bound dimer is essential for endonuclease activity as well as for ATM kinase activation.
DNA ends stimulate Rad50 ATP hydrolysis
We observed DNA stimulation of ATP hydrolysis by MR/MRN(X) complexes from archaea (20-fold), yeast (10-fold) and humans (up to 40-fold) which is also dependent on DNA ends. A similar stimulation of ATP hydrolysis has been reported by other groups for the T4 MR complex (∼22-fold) (47) and yeast MR/MRX complexes (∼6-fold) (45) using linear DNA. We have previously examined ATP hydrolysis by MRN(X) complexes using oligonucleotide DNA substrates and not observed any stimulation (data not shown), although from this work it is clear that long DNA duplexes (>1 kb) are significantly more efficient in inducing ATP hydrolysis by all the complexes we tested. A preference for long DNA molecules is also observed with MRN stimulation of ATM (50). It appears that this feature is conserved in all MR/MRN(X) complexes although each complex varies in the extent of stimulation. In addition, stabilizing factors including glycerol and a high ionic strength buffer are also essential for the observation of DNA stimulation of Rad50 ATPase activity in vitro.
DNA-stimulated activation of ATPase activity was also observed with the structurally similar SMC proteins constituting cohesin or condensins. When assayed in the presence of dsDNA oligonucleotides, Pyrococcus furiosus SMC catalytic domains exhibit a 13-fold increase in ATPase rates (74) and the Xenopus 13S condensin complex, which creates positive supercoils in a closed circular DNA in the presence of topoisomerases, exhibits a 5-fold increase in ATPase activity in the presence of relaxed circular dsDNA (75). The Deinococcus radiodurans recombination protein RecN, which is similar to SMC proteins except for a shorter coiled coil, exhibits a 3–4-fold increase in ATPase upon addition of linear plasmid DNA (76,77) and a DNA-dependent ATPase activity was shown for DrRecF which possesses conserved ABC ATPase domains but lacks the coiled coil domains of Rad50 or SMC proteins (78). The human mismatch repair protein complex MSH2–MSH6, which also lacks the coiled coil domain but has ATP-binding domains analogous to that of Rad50, is relatively inactive in the absence of DNA (0.9 ATP min−1), whereas the presence of a homoduplex causes a 7-fold increase in ATP hydrolysis (7.4 ATP min−1) and a mismatch in the duplex generates a further 3.5-fold increase (26 ATP min−1) (79).
The MRN complex is responsible for initiating DSB repair via homologous recombination pathways, the first step being the initiation of 5΄ end resection (68,80,81). MRN functions in homologous recombination are dependent on ATP binding and hydrolysis by Rad50 (41–43). We observed that Rad50 ATPase stimulation is at least partially dependent on direct contact with DNA ends, indicating that MR/MRN(X) complexes will be triggered into a DNA end processing mode only at DSB sites. In addition, we found that this stimulation is strongly inhibited by DNA–PK, which blocks DNA end-dependent ATP hydrolysis by Rad50 and forms an even stronger block in the absence of DNA–PKcs autophosphorylation. These results are consistent with reports showing an interplay between the DNA-PK and MRN complexes, where Ku70/80 dimers were observed to rapidly accumulate at DSBs (82,83). Based on these results and the in vitro data, it is clear that DNA–PK imposes a limitation on Rad50 ATP hydrolysis and MRN activities in general. Nevertheless, the fact that the endonucleolytic activity of MRN requires both blocked ends and ATP hydrolysis (40) also suggests that residual hydrolysis by Rad50 that takes place with protein blocks on the ends may be functionally distinct from hydrolysis stimulated by open ends. The regulation of MRN by end joining factors is an interesting area for future study, as well as the potential regulation of end joining factors by MRN.
Interdependence of the two ATPase sites
Rad50 belongs to the ABC transporter family of ATPases which have NBDs that dimerize to form two functional ATPase sites (84). The majority of the proteins in this family function as transporters for small molecules; however, a few non-transporter members are involved in DNA repair, e.g. Rad50, SMC proteins and MutS-related proteins (12). All proteins from these families show conserved features of NBDs, with the major differences found in other domains linked to the NBDs (85). The transporters possess transmembrane domains (TMDs) involved in import/export of molecules across the cell membrane, which are replaced in non-transporters by long coiled coils (Rad50 and SMC family) or DNA/mismatch binding domains (MutS family). Despite these differences, a common theme is seen in these proteins where ATP hydrolysis is utilized to bring about conformational changes in TMDs or the coiled coil domains (86). These conformational changes result in opening/closing of TMDs in ABC transporter ATPases for passage of small molecules across the cell membrane.
In the case of Rad50, we found that the two identical sites are interdependent and that hydrolysis by both sites is essential. Inactivating just one of the sites resulted in severe loss of ATP hydrolysis at the intact site and loss of ATP-dependent functions of MRN complex, including endonuclease activity and ATM activation. A similar interdependence of the two sites has also been seen for several other ABC ATPases. With the vitamin B12 transporter (BtuC2D2), mutation of the Walker B motif in the BtuD protein at only one site caused a 95% loss of ATP hydrolysis and deficiency in vitamin B12 transport (87). The maltose transporter MalK also forms symmetric ATPase sites and a heterodimer of WT with a Walker A mutant protein showed a severe loss of function (6% of the WT activities), indicating that the two sites do not act independently (88).
The closest relatives of Rad50 with respect to domain arrangement and protein architecture are the SMC subfamily of proteins involved in chromatin condensation and sister chromatid cohesion. Unlike Rad50, the cohesin and condensin complexes are heterodimers formed by SMC1/SMC3 and SMC2/SMC4 proteins respectively, although homodimers of yeast SMC1 and SMC3 catalytic domains were also observed in vitro (89). Coordination between the two ATP binding sites was also seen with human cohesin complexes containing one disabled site that were prepared by a tandem affinity purification protocol, resembling our purification strategy, with Smc1 and Smc3 bearing different tags (90). These complexes carrying single Smc3 ATPase mutations of Walker A, Walker B or signature motif affecting only one site in the Smc1-Smc3 dimer resulted in >90% loss of ATPase activity. With yeast cohesins, Walker A lysine mutations in either Smc1 or Smc3 similarly resulted in inactivation of ATP hydrolysis (89).
In previous studies we have shown that ATP hydrolysis stimulates a limited unwinding of DNA ends that extends ∼15 nt from the end of the DNA (36,73), consistent with recent structural and biochemical analysis of DNA unwinding by the Methanocaldococcus jannaschii MR complex (20). Our findings in this study suggest that ATP-driven unwinding of DNA ends is also dependent on both active sites, although this is not shown directly.
Nbs1 controls the MRN complex
Besides its vital role in checkpoint activation, Nbs1 plays an important role in MR function regulating the enzymatic activities of the complex. Nbs1 was shown to promote binding of the human MR complex to DNA (36,70) and the nucleolytic cleavage of a fully paired hairpin by the human MR complex, both activities increased in presence of ATP (36,70). Previously we reported that Nbs1 stabilizes the ATP-bound closed state of MR catalytic head that was responsible for ATM kinase activation (25). Nbs1 also regulates the endonucleolytic cutting of protein blocked ends at DSBs by human MR complex (38,40). In the present study we find that ATP hydrolysis by Rad50, which is essential for MR endonuclease activity as well, is also improved by addition of Nbs1 to human MR complex. Nbs1 is thus a key component of the MRN complex and regulates many if not all of its important biological activities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Susan Lees-Miller for DNA–PKcs protein and members of the Paull laboratory for helpful suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Prevention and Research Institute of Texas [RP110465-P4, in part]. Funding for open access charge: HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1. Paull T.T., Deshpande R.A.. The Mre11/Rad50/Nbs1 complex: recent insights into catalytic activities and ATP-driven conformational changes. Exp. Cell Res. 2014; 329:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamarche B.J., Orazio N.I., Weitzman M.D.. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010; 584:3682–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stracker T.H., Petrini J.H.. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011; 12:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams G.J., Lees-Miller S.P., Tainer J.A.. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair. 2010; 9:1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo G., Yao M.S., Bender C.F., Mills M., Bladl A.R., Bradley A., Petrini J.H.. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiao Y., Weaver D.T.. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997; 25:2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu J., Petersen S., Tessarollo L., Nussenzweig A.. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001; 11:105–109. [DOI] [PubMed] [Google Scholar]
- 8. Matsumoto Y., Miyamoto T., Sakamoto H., Izumi H., Nakazawa Y., Ogi T., Tahara H., Oku S., Hiramoto A., Shiiki T. et al. . Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair. 2011; 10:314–321. [DOI] [PubMed] [Google Scholar]
- 9. Stewart G.S., Maser R.S., Stankovic T., Bressan D.A., Kaplan M.I., Jaspers N.G., Raams A., Byrd P.J., Petrini J.H., Taylor A.M.. The DNA double-strand break repair gene hMre11 is mutated in individuals with an Ataxia-Telangiectasia-like disorder. Cell. 1999; 99:577–587. [DOI] [PubMed] [Google Scholar]
- 10. Varon R., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J. et al. . Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998; 93:467–476. [DOI] [PubMed] [Google Scholar]
- 11. Waltes R., Kalb R., Gatei M., Kijas A.W., Stumm M., Sobeck A., Wieland B., Varon R., Lerenthal Y., Lavin M.F. et al. . Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am. J. Hum. Genet. 2009; 84:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hopfner K.P., Tainer J.A.. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003; 13:249–255. [DOI] [PubMed] [Google Scholar]
- 13. Hopfner K.P., Karcher A., Shin D.S., Craig L., Arthur L.M., Carney J.P., Tainer J.A.. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000; 101:789–800. [DOI] [PubMed] [Google Scholar]
- 14. Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A.. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001; 105:473–485. [DOI] [PubMed] [Google Scholar]
- 15. Hopfner K.P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A., Karcher A., Henderson B., Bodmer J.L., McMurray C.T. et al. . The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002; 418:562–566. [DOI] [PubMed] [Google Scholar]
- 16. Park Y.B., Hohl M., Padjasek M., Jeong E., Jin K.S., Krezel A., Petrini J.H., Cho Y.. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 2017; 24:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lammens K., Bemeleit D.J., Mockel C., Clausing E., Schele A., Hartung S., Schiller C.B., Lucas M., Angermuller C., Soding J. et al. . The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011; 145:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams G.J., Williams R.S., Williams J.S., Moncalian G., Arvai A.S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P. et al. . ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011; 18:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lim H.S., Kim J.S., Park Y.B., Gwon G.H., Cho Y.. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011; 25:1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Sung S., Kim Y., Li F., Gwon G., Jo A., Kim A.K., Kim T., Song O.K., Lee S.E. et al. . ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016; 35:743–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mockel C., Lammens K., Schele A., Hopfner K.P.. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2011; 40:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seifert F.U., Lammens K., Stoehr G., Kessler B., Hopfner K.P.. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016; 35:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schiller C.B., Lammens K., Guerini I., Coordes B., Feldmann H., Schlauderer F., Mockel C., Schele A., Strasser K., Jackson S.P. et al. . Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012; 19:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deshpande R.A., Williams G.J., Limbo O., Williams R.S., Kuhnlein J., Lee J.H., Classen S., Guenther G., Russell P., Tainer J.A. et al. . ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014; 33:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J.H., Mand M.R., Deshpande R.A., Kinoshita E., Yang S.H., Wyman C., Paull T.T.. Ataxia Telangiectasia-Mutated (ATM) Kinase Activity Is Regulated by ATP-driven Conformational Changes in the Mre11/Rad50/Nbs1 (MRN) Complex. J. Biol. Chem. 2013; 288:12840–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lobachev K.S., Gordenin D.A., Resnick M.A.. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002; 108:183–193. [DOI] [PubMed] [Google Scholar]
- 27. Rattray A.J., McGill C.B., Shafer B.K., Strathern J.N.. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001; 158:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang S.H., Zhou R., Campbell J., Chen J., Ha T., Paull T.T.. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013; 32:126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y., Paull T.T.. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and Ataxia Telangiectasia-mutated (ATM). J. Biol. Chem. 2013; 288:37112–37125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moreau S., Ferguson J.R., Symington L.S.. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O.. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008; 135:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsusui K. et al. . Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell. 2016; 64:580–592. [DOI] [PubMed] [Google Scholar]
- 33. Connelly J.C., Kirkham L.A., Leach D.R.. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:7969–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hopkins B., Paull T.T.. The P. furiosus Mre11/Rad50 complex promotes 5΄ strand resection at a DNA double-strand break. Cell. 2008; 135:250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paull T.T., Gellert M.. The 3΄ to 5΄ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998; 1:969–979. [DOI] [PubMed] [Google Scholar]
- 36. Paull T.T., Gellert M.. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999; 13:1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trujillo K.M., Sung P.. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J. Biol. Chem. 2001; 276:35458–35464. [DOI] [PubMed] [Google Scholar]
- 38. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 39. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 40. Deshpande R.A., Lee J.H., Arora S., Paull T.T.. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell. 2016; 64:593–606. [DOI] [PubMed] [Google Scholar]
- 41. Alani E., Padmore R., Kleckner N.. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990; 61:419–436. [DOI] [PubMed] [Google Scholar]
- 42. Chen L., Trujillo K.M., Van Komen S., Roh D.H., Krejci L., Lewis L.K., Resnick M.A., Sung P., Tomkinson A.E.. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J. Biol. Chem. 2005; 280:2620–2627. [DOI] [PubMed] [Google Scholar]
- 43. Moncalian G., Lengsfeld B., Bhaskara V., Hopfner K.P., Karcher A., Alden E., Tainer J.A., Paull T.T.. The rad50 signature motif: essential to ATP binding and biological function. J. Mol. Biol. 2004; 335:937–951. [DOI] [PubMed] [Google Scholar]
- 44. Bhaskara V., Dupre A., Lengsfeld B., Hopkins B.B., Chan A., Lee J.H., Zhang X., Gautier J., Zakian V.A., Paull T.T.. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol. Cell. 2007; 25:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trujillo K.M., Roh D.H., Chen L., Van Komen S., Tomkinson A., Sung P.. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J. Biol. Chem. 2003; 278:48957–48964. [DOI] [PubMed] [Google Scholar]
- 46. de Jager M., Wyman C., van Gent D.C., Kanaar R.. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002; 30:4425–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herdendorf T.J., Albrecht D.W., Benkovic S.J., Nelson S.W.. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J. Biol. Chem. 2011; 286:2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee J.H., Paull T.T.. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006; 408:529–539. [DOI] [PubMed] [Google Scholar]
- 49. Lee J.H., Paull T.T.. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004; 304:93–96. [DOI] [PubMed] [Google Scholar]
- 50. Lee J.H., Paull T.T.. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005; 308:551–554. [DOI] [PubMed] [Google Scholar]
- 51. Myler L.R., Gallardo I.F., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. 2016; submitted [DOI] [PMC free article] [PubMed]
- 52. Wu D., Topper L.M., Wilson T.E.. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008; 178:1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., Hefferin M.L., Chen L., Shim E.Y., Tseng H.M., Kwon Y., Sung P., Lee S.E., Tomkinson A.E.. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007; 14:639–646. [DOI] [PubMed] [Google Scholar]
- 54. Allen C., Kurimasa A., Brenneman M.A., Chen D.J., Nickoloff J.A.. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shim E.Y., Chung W.H., Nicolette M.L., Zhang Y., Davis M., Zhu Z., Paull T.T., Ira G., Lee S.E.. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010; 29:3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fukushima T., Takata M., Morrison C., Araki R., Fujimori A., Abe M., Tatsumi K., Jasin M., Dhar P.K., Sonoda E. et al. . Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 2001; 276:44413–44418. [DOI] [PubMed] [Google Scholar]
- 57. Mimitou E.P., Symington L.S.. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010; 29:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foster S.S., Balestrini A., Petrini J.H.. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol. Cell. Biol. 2011; 31:4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yaneva M., Kowalewski T., Lieber M.R.. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997; 16:5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Vries E., van Driel W., Bergsma W.G., Arnberg A.C., van der Vliet P.C.. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 1989; 208:65–78. [DOI] [PubMed] [Google Scholar]
- 61. Chan D.W., Chen B.P., Prithivirajsingh S., Kurimasa A., Story M.D., Qin J., Chen D.J.. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002; 16:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ding Q., Reddy Y.V., Wang W., Woods T., Douglas P., Ramsden D.A., Lees-Miller S.P., Meek K.. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003; 23:5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chan D.W., Lees-Miller S.P.. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996; 271:8936–8941. [DOI] [PubMed] [Google Scholar]
- 64. Beis K. Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 2015; 43:889–893. [DOI] [PubMed] [Google Scholar]
- 65. Procko E., Ferrin-O’Connell I., Ng S.L., Gaudet R.. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell. 2006; 24:51–62. [DOI] [PubMed] [Google Scholar]
- 66. Zaitseva J., Oswald C., Jumpertz T., Jenewein S., Wiedenmann A., Holland I.B., Schmitt L.. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 2006; 25:3432–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aparicio T., Baer R., Gottesman M., Gautier J.. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016; 212:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 69. Garcia V., Phelps S.E., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee J.-H., Ghirlando R., Bhaskara V., Hoffmeyer M.R., Gu J., Paull T.T.. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ATLD mutant complexes. J. Biol. Chem. 2003; 278:45171–45181. [DOI] [PubMed] [Google Scholar]
- 71. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y.. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003; 22:5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Paull T.T. Mechanisms of ATM activation. Annu. Rev. Biochem. 2015; 84:711–738. [DOI] [PubMed] [Google Scholar]
- 73. Cannon B., Kuhnlein J., Yang S.H., Cheng A., Schindler D., Stark J.M., Russell R., Paull T.T.. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:18868–18873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lammens A., Schele A., Hopfner K.P.. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 2004; 14:1778–1782. [DOI] [PubMed] [Google Scholar]
- 75. Kimura K., Hirano T.. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997; 90:625–634. [DOI] [PubMed] [Google Scholar]
- 76. Pellegrino S., Radzimanowski J., de Sanctis D., Boeri Erba E., McSweeney S., Timmins J.. Structural and functional characterization of an SMC-like protein RecN: new insights into double-strand break repair. Structure. 2012; 20:2076–2089. [DOI] [PubMed] [Google Scholar]
- 77. Reyes E.D., Patidar P.L., Uranga L.A., Bortoletto A.S., Lusetti S.L.. RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. J. Biol. Chem. 2010; 285:16521–16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Koroleva O., Makharashvili N., Courcelle C.T., Courcelle J., Korolev S.. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007; 26:867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gradia S., Acharya S., Fishel R.. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997; 91:995–1005. [DOI] [PubMed] [Google Scholar]
- 80. Sun H., Treco D., Szostak J.W.. Extensive 3΄-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991; 64:1155–1161. [DOI] [PubMed] [Google Scholar]
- 81. Paques F., Haber J.E.. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999; 63:349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Britton S., Coates J., Jackson S.P.. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013; 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reid D.A., Keegan S., Leo-Macias A., Watanabe G., Strande N.T., Chang H.H., Oksuz B.A., Fenyo D., Lieber M.R., Ramsden D.A. et al. . Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hollenstein K., Dawson R.J., Locher K.P.. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007; 17:412–418. [DOI] [PubMed] [Google Scholar]
- 85. Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015; 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Higgins C.F., Linton K.J.. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004; 11:918–926. [DOI] [PubMed] [Google Scholar]
- 87. Tal N., Ovcharenko E., Lewinson O.. A single intact ATPase site of the ABC transporter BtuCD drives 5% transport activity yet supports full in vivo vitamin B12 utilization. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5434–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Davidson A.L., Sharma S.. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J. Bacteriol. 1997; 179:5458–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Arumugam P., Nishino T., Haering C.H., Gruber S., Nasmyth K.. Cohesin's ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr. Biol. 2006; 16:1998–2008. [DOI] [PubMed] [Google Scholar]
- 90. Ladurner R., Bhaskara V., Huis in 't Veld P.J., Davidson I.F., Kreidl E., Petzold G., Peters J.M.. Cohesin's ATPase activity couples cohesin loading onto DNA with Smc3 acetylation. Curr. Biol. 2014; 24:2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.